Abstract
Streptomycin is a bactericidal antibiotic that induces translational errors. It binds to the 30S ribosomal subunit, interacting with ribosomal protein S12 and with 16S rRNA through contacts with the phosphodiester backbone. To explore the structural basis for streptomycin resistance, we determined the X-ray crystal structures of 30S ribosomal subunits from six streptomycin-resistant mutants of Thermus thermophilus both in the apo form and in complex with streptomycin. Base substitutions at highly conserved residues in the central pseudoknot of 16S rRNA produce novel hydrogen-bonding and base-stacking interactions. These rearrangements in secondary structure produce only minor adjustments in the three-dimensional fold of the pseudoknot. These results illustrate how antibiotic resistance can occur as a result of small changes in binding site conformation.
INTRODUCTION
The ribosome is the macromolecular machine responsible for the translation of genetic information and is the target of numerous antimicrobial agents (reviewed in references 1 and 2). Chemical footprinting experiments (3) suggested that many antibiotics bind primarily or exclusively to rRNA. This was shown to be the case for aminoglycoside antibiotics by nuclear magnetic resonance and crystallography experiments using oligonucleotide analogs of their 16S rRNA binding site (4, 5). X-ray crystallography of intact bacterial and archaeal ribosomes has shown that antibiotic-rRNA interactions are indeed the predominant mode of antibiotic binding (reviewed in reference 6); from such studies it is possible to infer the structural basis for the mechanisms of antibiotic action, which include the stabilization of nonproductive ribosome conformations, the impairment of essential conformational transitions, and the physical obstruction of substrate binding sites. Some antibiotics, such as the aminoglycosides, induce translational errors, apparently by stabilizing productive end-state conformations (7–9). Ribosome structural studies can also be used to infer mechanisms of antibiotic resistance that arise from mutations affecting components of the ribosome and have the potential to provide the basis for the rational design of novel antimicrobial agents effective against resistant pathogens.
Mutations conferring resistance to aminoglycoside antibiotics were first identified in ribosomal protein genes (10, 11) due to the ease of detecting protein alterations biochemically. The recognition of the primary involvement of rRNA in antibiotic binding was delayed partly because of the multicopy nature of rRNA genes in many experimental organisms that can mask resistance phenotypes and also because there was no way to rapidly identify the presence of rRNA mutations prior to the availability of rRNA gene sequences and facile DNA sequencing methods. Aminoglycoside resistance mutations were subsequently identified in the single-copy rRNA genes of chloroplasts of Euglena gracilis (12), Chlamydomonas reinhardtii (13, 14), and Nicotiana plumbaginifolia (15). These studies laid the foundation for later identification of streptomycin resistance (Strr) mutations in the single rRNA gene copies of the bacterium Mycobacterium tuberculosis (16–18). The use of multicopy plasmids expressing rRNA has allowed an analysis of Strr mutations in Escherichia coli 16S rRNA (19–23). Similarly, developments in the genetic manipulation of the rRNA genes of the extremophile Thermus thermophilus (24), used in many ribosome structural studies, have facilitated determination of the structure of resistant ribosomes. While strains producing pure populations of mutant ribosomes are a great advantage crystallographically, they also allow the identification of resistance mutations conferring a recessive resistance phenotype.
Some mutations conferring antibiotic resistance produce perturbations in ribosome structure beyond the antibiotic binding site, while others replace chemical groups that are directly involved in antibiotic-ribosome contacts and produce only minimal effects on ribosome structure (25, 26). Amino acid substitutions in ribosomal protein S12 conferring streptomycin dependence cause distortions of the decoding site of 16S rRNA (27). Thus, while some mutations confer antibiotic resistance by causing significant conformational deformities, others can act with only minimal effects on the structure.
The aminoglycoside antibiotic streptomycin induces misreading of mRNA (28) by binding near the site of codon recognition in the 30S subunit (8). The streptomycin binding site is rich in rRNA tertiary structure, including two pseudoknot interactions and multiple helix-packing interactions; the drug makes extensive backbone contacts with 16S rRNA helices 1, 18, 27, and 44 (Fig. 1A and B), and base substitutions in these structures can cause resistance. Binding of streptomycin also destabilizes a tertiary interaction between 16S rRNA helices 44 and 45 (29). Streptomycin is in position to contact several lysine ε-amino groups of ribosomal protein S12, including K42, K43, and K87 (8), and resistance is frequently conferred by amino acid substitutions at any one of these positions (30–32). Deficiency in N-7 methylation of G527 of 16S rRNA by the RsmG methyltransferase produces low-level streptomycin resistance (33, 34). How substitution of K42, K43, or K87 of S12 confers streptomycin resistance is somewhat self-evident, whereas predicting with any precision the mechanism of resistance conferred by base substitutions or a deficiency in base methylation is made difficult by the absence of any base-specific contacts between streptomycin and 16S rRNA.
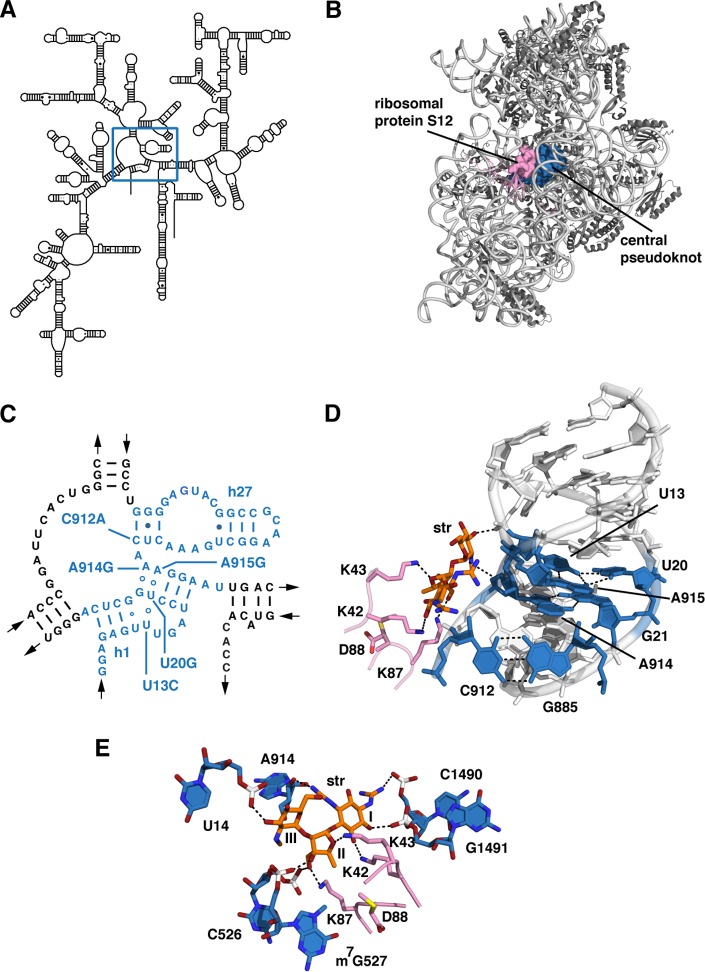
FIG 1.
Location of Strr mutations in the central pseudoknot of the 30S subunit. (A) Secondary structure model of T. thermophilus 16S rRNA (GenBank accession number M26923) (48), with the central pseudoknot indicated by a square. (B) X-ray crystal structure of the wild-type T. thermophilus 30S subunit-streptomycin complex (PDB accession number 4DR3) (29). The central pseudoknot is shown as a blue surface, and ribosomal protein S12 is shown as a pink surface. (C) Secondary structure of the central pseudoknot, with helices 1 and 27 (h1 and h27, respectively) colored blue. The base substitutions examined in this study are labeled. (D) Partial structure of the central pseudoknot, indicating base pair and base triple interactions (blue) affected by mutations. Also shown are streptomycin (orange sticks) and residues of S12 involved in streptomycin binding (pink). (E) The streptomycin binding site, showing ribosome-streptomycin contacts.
We previously described the isolation and characterization of spontaneous Strr mutants of the thermophilic bacterium T. thermophilus with amino acid substitutions in ribosomal protein S12 (35) or base substitutions in 16S rRNA (24, 36). Mutant ribosomes with base substitutions in the central pseudoknot exhibit altered reactivity to base-specific chemical probes, suggesting an effect of the mutations on the hydrogen-bonding arrangement of the pseudoknot (24). In five of the mutants studied here, resistance results from base substitutions in and around the central pseudoknot formed by helices 1 and 27, at the junction of the three major domains of the 16S rRNA molecule (37–39) (Fig. 1C and D). We also previously identified mutants deficient in the modification at the 7-methyl-guanosine at position 527 (m7G527) due to a transposon insertion into rsmG (40) or deletion of rsmG (41). Here we describe the X-ray crystal structures of 30S ribosomal subunits bearing each of the five base substitutions or lacking the m7G527 modification. The results from these experiments indicate that alternative RNA secondary structure arrangements can adopt nearly native backbone conformations and that a series of small changes combines to abrogate streptomycin binding. They also suggest that conformational dynamics play a role in modulating antibiotic resistance.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All experiments were conducted with mutants derived from T. thermophilus HB8 (ATCC 27634). All strains used in this study have been described previously (24, 41). HG917 (ΔrsmG::htk1) is a derivative of HB8 in which the rsmG locus has been deleted and replaced with the htk gene encoding a thermostable kanamycin-adenyltransferase. HG286 (ΔrrsA::htk1) is a derivative of HB8 having a single gene encoding 16S rRNA (rrsB), with rrsA having been deleted and replaced with the htk gene. All other strains were derived from HG286 and included HG422 (ΔrrsA::htk1 rrsB-U13C), HG451 (ΔrrsA::htk1 rrsB-U20G), HG419 (ΔrrsA::htk1 rrsB-A915G), HG420 (ΔrrsA::htk1 rrsB-A914G), and HG426 (ΔrrsA::htk1 rrsB-C912A). Cultures were grown in Thermus enhanced medium (TEM; ATCC medium 1598) in liquid or on plates solidified with 2.8% Bacto agar (Difco). Liquid cultures were grown aerobically in an Innova 4200 shaker incubator (New Brunswick) at 72°C with shaking at 180 rpm. Strains were maintained as 25% glycerol stocks, stored at −80°C.
Purification and crystallization of ribosomes.
30S ribosomal subunits were purified and crystallized essentially as described previously (42), with the exception that crystallization growth times were extended to 4 to 8 months. Ribosome-streptomycin complexes were prepared by mixing 30S subunits with 400 μg/ml streptomycin sulfate, filtering with a 0.22-μm-pore-size filter, and incubating at 55°C for 10 min before cooling to 4°C and were then crystallized. All 30S crystals were sequentially transferred to a final buffer with 26% (vol/vol) 2-methyl-2,4-pentanediol (including 400 μg/ml streptomycin for cocrystals) for cryoprotection. Crystals were kept in the final cryoprotection buffer for at least 1 week. Crystals were frozen in liquid nitrogen before data collection.
Data collection and refinement.
Diffraction data for each mutant or mutant-streptomycin complex were collected from single crystals (U13C, U20G, C912A, A914G, and ΔrsmG mutants) or two crystals (A915G mutant) with an ADSC 315 detector at the Advanced Photon Source in Argonne National Laboratory. The data sets for the U13C, U20G, A914G, A915G, and ΔrsmG mutants (apo and streptomycin-bound forms) were collected at beam line ID-24-C, and data sets for the C912A mutant (apo and streptomycin-bound forms) were collected at beam line ID-24E. Diffraction data were processed with the HKL2000 package (43). Either the wild-type apo 30S structure (PDB accession number 4DR1) or the wild-type streptomycin-bound 30S structure (PDB accession number 4DR3) was used as the starting model for refinement with the PHENIX software package (44). After simulated-annealing refinement, individual coordinates and TLS parameters were refined. Potential positions of magnesium or potassium ions were compared with those in a high-resolution (2.5-Å) 30S subunit structure (PDB accession number 2VQE) [45] in the Coot program (46), and positions with strong difference densities were retained. All magnesium atoms were replaced with magnesium hexahydrate. Water molecules located outside significant electron density were manually removed.
Structural alignments.
Alignments were performed using the alignment algorithm of the PyMOL molecular graphics system with the default 2σ rejection criterion and 50 iterative alignment cycles. Alignments used phosphate atoms from residues 50 to 500, avoiding residues participating in streptomycin binding and regions known to be especially mobile. All figures were generated with PyMOL (47).
Protein structure accession numbers.
Atomic coordinates for all structures have been deposited in the RCSB Protein Data Bank (PDB; www.pdb.org) under accession numbers 4DUY (U13C, apo form), 4DUZ (U13C, streptomycin-bound form), 4DV0 (U20G, apo form), 4DV1 (U20G, streptomycin-bound form), 4DV2 (C912A, apo form), 4DV3 (C912A, streptomycin-bound form), 4DV4 (A914G, apo form), 4DV5 (A914G, streptomycin-bound form), 4DV6 (A915G, apo form), 4DV7 (A915G, streptomycin-bound form), 4NXM (ΔrsmG, apo form), and 4NXN (ΔrsmG, streptomycin-bound form).
RESULTS AND DISCUSSION
Structural effects of base substitutions on the central pseudoknot.
We examined 30S ribosomal subunits with one of five base substitutions in and around the central pseudoknot of T. thermophilus 16S rRNA (U13C, U20G, C912A, A914G, and A915G) or a 30S ribosomal subunit lacking the m7G527 modification due to a deletion of the rsmG locus (Fig. 1). Ribosomes from T. thermophilus have proven to be effective targets for structure determination by X-ray crystallography, and we were able to crystallize mutant 30S subunits using methods originally developed for wild-type T. thermophilus (38, 42). In total, we collected 12 X-ray diffraction data sets with resolutions ranging from 3.3 Å to 3.85 Å and with coordinate errors ranging from 0.28 Å to 0.37 Å (see Table S1 in the supplemental material). These included mutant 30S subunits in the apo form or cocrystallized with streptomycin.
Three of the five bases substituted in these mutants, U13, U20, and A915, form a base triple in the wild-type ribosome, as shown in Fig. 1D. The fourth mutated base, A914, pairs with G21 to form a sheared G·A base pair engaged in a stacking interaction with the U13-U20-A915 base triple. The fifth mutated base, C912, establishes a Watson-Crick pair with G885 to close helix 27. Across the three phylogenetic domains, U13, U20, A914, and A915 are found in 99.4%, 98.9%, 99.7%, and 99.8% of small-subunit rRNAs, respectively (48). Position 912 is conserved at the domain level; while a G885-C912 Watson-Crick base pair predominates in bacteria (96.8%), a G885-U912 wobble pair predominates in eukarya (99.6%) and archaea (99.1%), indicating that these base identities were fixed prior to the divergence of the three domains from their last common ancestor. The methylated base m7G527 is located in 16S rRNA helix 18 and is in close proximity to G530, which directly participates in codon recognition (9). This residue is also highly conserved (99.2% in bacteria, 100% in archaea, and 99.9% in eukarya), and its N-7 methylation has been found in the few bacterial, archaeal, and eukaryal species whose rRNA modifications have been systematically examined (49).
In the mutant crystal structures, each of the base substitutions eliminates the hydrogen-bonding and base-stacking interactions that are formed in the native structure. Similar observations for mutations in the peptidyltransferase active site have been made (26). Despite some substantial rearrangements in secondary structure, all mutants maintained a nearly native backbone conformation of the pseudoknot and the surrounding tertiary structure (Fig. 2 to 4). Ribosome-streptomycin complexes showed a clear difference in electron density attributable to the antibiotic (see Fig. S1 in the supplemental material), as well as the disruption of the contact between helix 45 and helix 44 that we recently reported for the wild-type 30S-streptomycin complex (29). This binding occurs at a concentration (400 μg/ml) that is below that needed to inhibit the growth of the mutants. From our previous studies (24, 41), 500 μg/ml completely inhibited the growth of the C912A mutant, partially inhibited the growth of the U20G, A914G, and A915G mutants, and had no effect on the growth of the U13C mutant. All mutants were completely inhibited by 1,000 μg/ml. The ΔrsmG mutant was only weakly resistant, being partially inhibited by 50 μg/ml and almost completely inhibited by 100 μg/ml. Binding of streptomycin largely reversed the structural changes induced by base substitutions. This suggests that effects on streptomycin binding are at least in part due to changes in local conformational dynamics. Each mutant exhibited individual structural characteristics, which are described below.
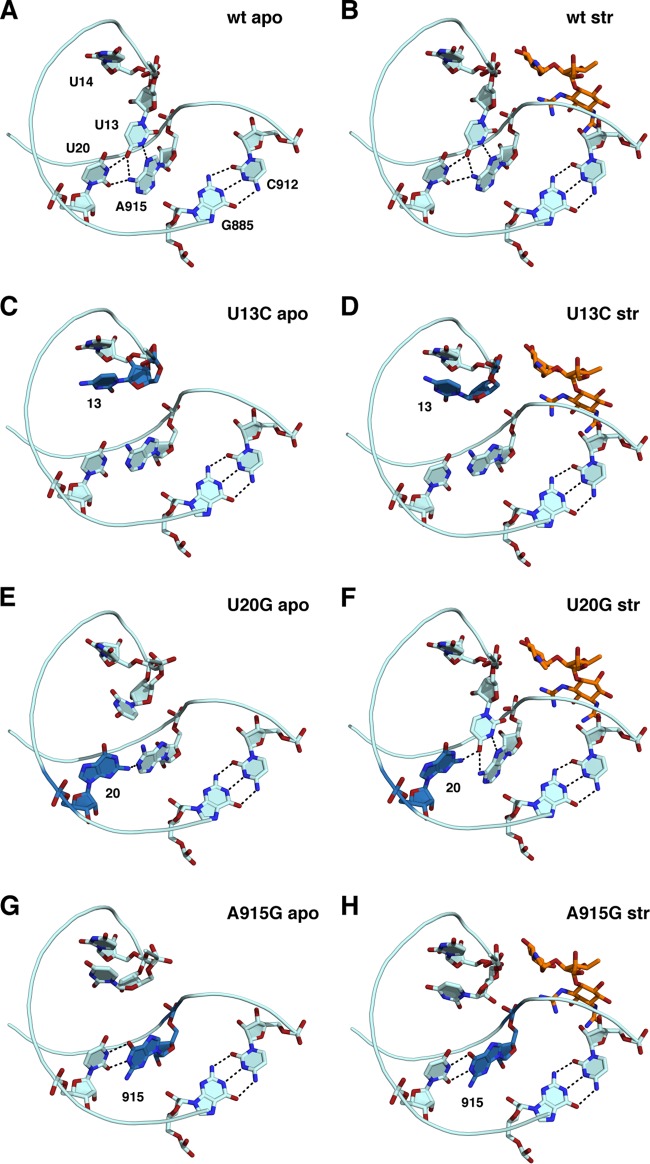
FIG 2.
Structural rearrangements in the U13C, U20G, and 915G mutants. (A and B) Wild type (wt); (C and D) U13C; (E and F) U20G; (G and H) A915G. (Left) apo structures; (right) structures cocrystallized with streptomycin (orange sticks). The mutated residue in each structure is shown in blue.
FIG 4.
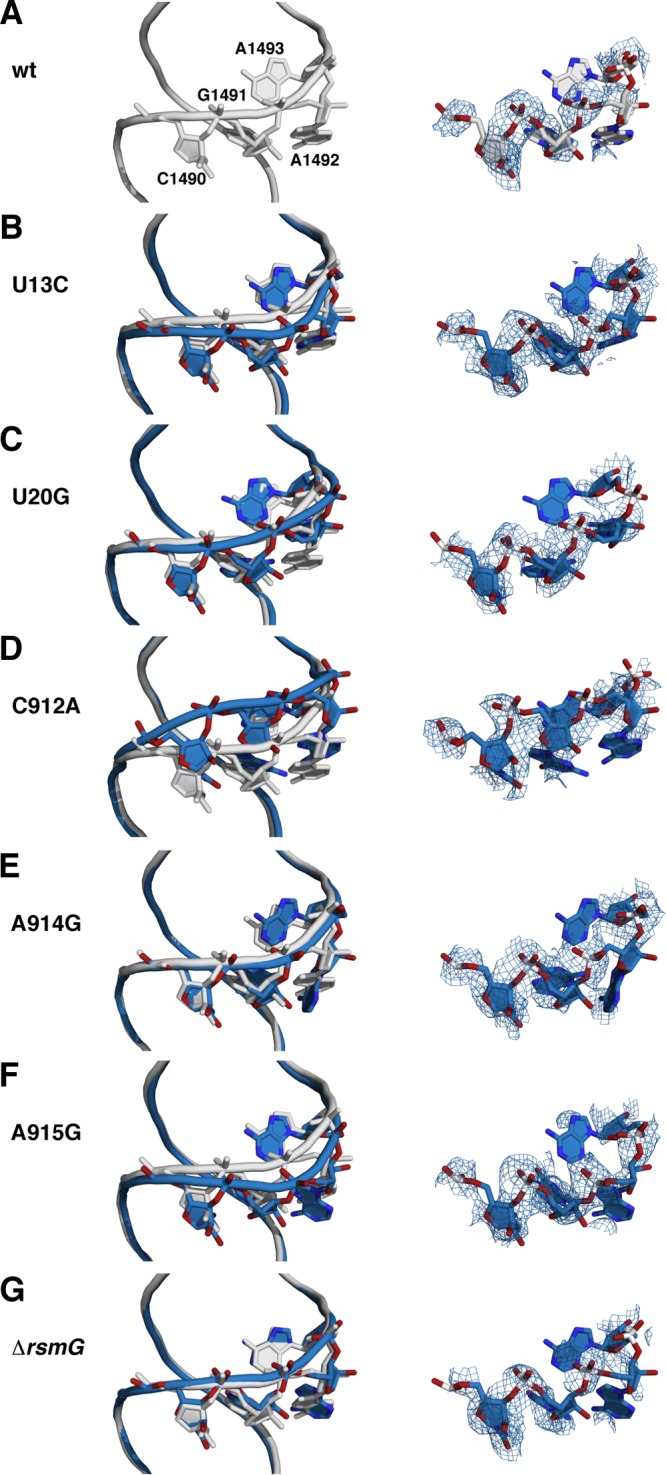
Backbone shifts in the decoding site. (Left) Wild-type and mutant structures were aligned using the phosphate atoms of 16S rRNA residues 50 to 500. Gray, wild type; pale cyan, mutants. Side chains for residues C1490, G1491, A1492, and A1493 are also shown. (Right) Composite omit 2mFo − DFc electron density maps for the same four residues. (A) Wild type; (B) wild type and U13C; (C) wild type and U20G; (D) wild type and C912A; (E) wild type and A914G; (F) wild type and A915G; (G) wild type and ΔrsmG.
U13C.
The U13-U20-A915 base triple seen in the wild-type 30S subunit (Fig. 2A and B) is disrupted by the U13C base substitution, with a large-scale reorientation (a 7.2-Å movement of the pyrimidine N-3) of the mutated base that is stabilized by a stacking interaction with U14 (Fig. 2C). The hydrogen-bonding arrangement between U20 and A915 also changed to a wobble-like juxtaposition, with loss of the lone hydrogen bond between A915 N-6 and U20 O-2. This loss is apparently compensated for by a shift in the position of A915 toward the major groove that results in improved stacking of A915 over the N-6 of A914. Thus, the displacement of the C at position 13 is stabilized by alternative or improved stacking interactions of both the mutated residue and A915. These conformational changes induce only small changes in the backbone trajectory in the vicinity of these residues in the streptomycin binding site that are sufficient to confer resistance. Binding of streptomycin to the mutant 30S subunit stabilizes the rRNA backbone in the vicinity of positions 13 and 915 (Fig. 2D).
U20G.
In the U20G mutant structure, the 13-20-915 base triple is largely obliterated, with U13 being in a flipped-out configuration but not stacked on U14, as in the U13C mutant (Fig. 2E). It should be noted that the electron density for U13 is weakly defined in both the apo and streptomycin-bound U20G structures in omit difference electron density maps, indicating that this base is largely disordered as a consequence of the U20G substitution (see Fig. S2 in the supplemental material). The A915 base is shifted toward the native U13 position, as observed in the U13C mutant, and also displaced from the bulkier G now present at position 20. The N-2 of this G forms a single hydrogen bond with the N-1 of A915. The lateral displacement of A915 is reversed by binding of streptomycin, causing it to assume a position similar to its native orientation (Fig. 2F). U13 rotates back toward a native-like orientation in the presence of streptomycin, restoring a nearly isosteric 13-20-915 base triple.
A915G.
In contrast to the U20G data, electron density maps were well defined for the A915G mutant structures (see Fig. S2 in the supplemental material). The base triple conformation in the A915G apo form closely resembles that of the U13C mutant, with the structural rearrangement of the U13 base stacking on U14 (Fig. 2G). In this mutant, the G at 915 and U20 adopt a G·U wobble configuration, similar to the anomalous A·U wobble configuration observed for A915 and U20 in the U13C mutant. This reinforces the notion that the movement of U13 into its alternative stacking arrangement on U14 is sufficiently stable to compensate for the unfavorable configuration of A915 and U20 in the U13C mutant. Binding of streptomycin induces small changes in the backbone at positions 13 and 915 toward the native conformation (Fig. 2H).
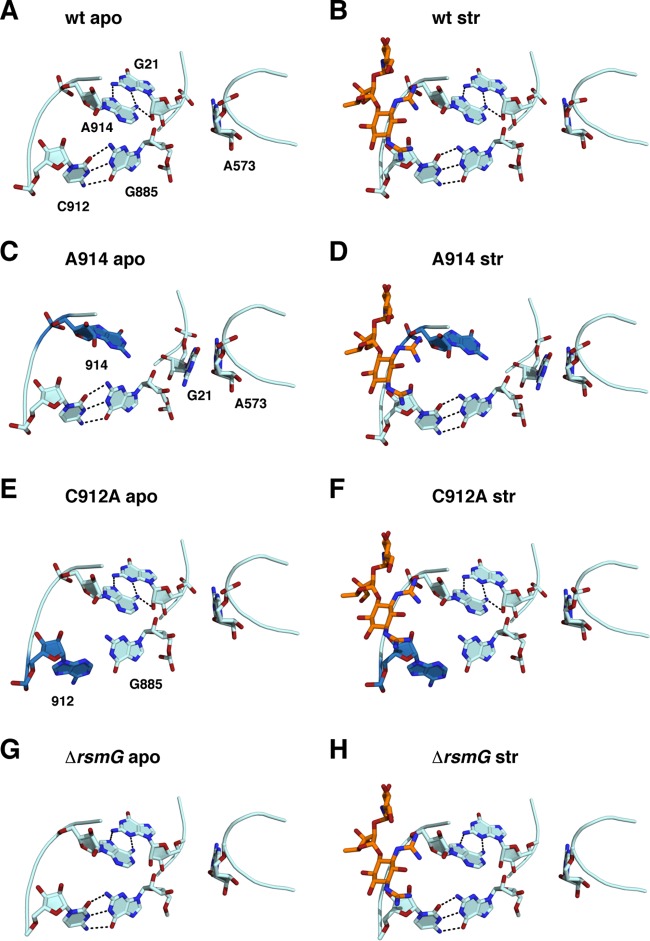
A914G.
Compared to the wild-type structure (Fig. 3A and B), the A914G mutant structure shows a dramatic rearrangement. The sheared pair between A914 and G21 is lost, with G21 flipping out of its native orientation to stack onto A573 (Fig. 3C). Similar to the reorientation of U13, this formation of a new stacking orientation compensates for the loss of native hydrogen-bonding interactions. The A914G mutant base is shifted toward the native position of G21, analogous to the compensatory shift of A915 in the base triple mutants. The U13-U20-A915 base triple interaction remains intact in this mutant structure, with a small rotational adjustment in the U20 base due to the loss of the stacking interaction with G21. The coordination of streptomycin to this mutant induces a further displacement of the A914G mutant base, which may reflect a higher degree of flexibility in the absence of any stabilizing hydrogen-bonding interactions (Fig. 3D).
FIG 3.
Structural rearrangement in the A914G, C912A, and ΔrsmG mutants. (A and B) Wild type; (C and D) A914G; (E and F) C912A; (G and H) ΔrsmG. (Left) apo structures; (right) structures cocrystallized with streptomycin (orange sticks). The mutated residue in each structure is shown in blue.
C912A.
Of the five bases altered in Strr mutants, the one that does not directly participate in the pseudoknot is C912. The C912A substitution replaces a Watson-Crick G-C pair (Fig. 3E and F; see Fig. S4C and D in the supplemental material) with a G·A mismatch having a geometry similar to that of the pseudo-Watson-Crick G·A base pair seen in tRNAPhe (50), but with the adenosine base displaced toward the minor groove of helix 27. There are no apparent hydrogen-bonding interactions between these two bases, consistent with the reactivity of the N-1 position to dimethyl sulfate (24). Only a minor shift in the backbone position of the adenosine is sufficient to accommodate it into the helix 27 structure. Shifts in the position of the mutant A at position 912 alter the streptomycin binding site and could also alter the position and orientation of the lysine residue in ribosomal protein S12 that interacts with the antibiotic. As with the other mutant structures, the binding of streptomycin reverses the small backbone shifts and induces small adjustments of the U911 and C912A base positions (see Fig. S4D in the supplemental material).
ΔrsmG.
In contrast to the Strr mutations in the central pseudoknot, the loss of the m7G527 modification due to the ΔrsmG mutation showed no detectable structural distortion (Fig. 3G and H). All base-base hydrogen-bonding interactions observed in the wild-type ribosome remain intact in this mutant. These results are consistent with the lower resistance levels in vivo.
Effects of base substitutions on dimensions of streptomycin binding site.
In several of the mutant structures, the most significant structural changes were located in helix 44, on the side of the streptomycin binding site opposite the central pseudoknot (Fig. 4; Table 1). In the wild-type 30S subunit, streptomycin contacts helix 44 via backbone interactions at C1490 and G1491 (Fig. 1E), and the shifts in phosphate positions at these residues could account for the decreased streptomycin binding affinity. This is particularly true for the C912A mutant, which shows a 3.4-Å shift of the G1491 phosphate. The U13C, U20G, and A915G mutants also show some distortion in this vicinity, while the A914G and ΔrsmG mutants show little or no detectable distortion of this region (Fig. 4).
TABLE 1.
Shifts in phosphate atom positions induced by Strr mutationsa
| Residue | Distance (Å) |
|||||
|---|---|---|---|---|---|---|
| U13C (4DUY) | U20G (4DV0) | C912A (4DV2) | A914G (4DV4) | A915G (4DV6) | ΔrsmG (4NXM) | |
| U13 | 0.5 | 0.8 | 0.2 | 0.5 | 0.6 | 0.2 |
| U14 | 0.9 | 0.6 | 0.3 | 0.2 | 1.0 | 0.0 |
| G15 | 0.5 | 0.6 | 0.3 | 0.1 | 0.5 | 0.1 |
| C526 | 0.3 | 0.6 | 0.4 | 0.2 | 0.2 | 0.1 |
| m7G527 | 1.1 | 0.3 | 0.2 | 0.7 | 0.8 | 0.2 |
| C912 | 0.6 | 0.5 | 1.4 | 0.3 | 0.8 | 0.2 |
| A913 | 1.1 | 1.1 | 0.3 | 0.4 | 1.2 | 0.1 |
| A914 | 0.7 | 0.4 | 0.5 | 1.0 | 0.5 | 0.4 |
| A915 | 0.8 | 1.0 | 0.2 | 0.5 | 0.8 | 0.2 |
| C1490 | 1.2 | 0.4 | 1.4 | 0.4 | 1.1 | 0.2 |
| G1491 | 1.3 | 0.6 | 3.4 | 0.6 | 1.4 | 0.4 |
| A1492 | 1.9 | 0.7 | 3.6 | 0.7 | 2.3 | 0.7 |
| A1493 | 0.8 | 1.8 | 2.9 | 0.9 | 1.5 | 0.4 |
Coordinate errors are provided in Table S1 in the supplemental material. Root mean square deviation values for the overall alignments were 0.219 (U13C), 0.262 (U20G), 0.247 (C912A), 0.204 (A914G), 0.250 (A915G), and 0.188 (ΔrsmG). Residues involved in streptomycin binding are U14, C526, m7G527, A914, C1490, and G1491. PDB accession numbers are given in parentheses.
Effects on the dimensions of the streptomycin binding site were also detectable (Table 2). Small changes in the distance between phosphates directly involved in streptomycin binding may combine to distort the binding site and decrease binding affinity. These small differences noticeable in the apo subunit structures were eliminated upon binding streptomycin (Table 3). These data suggest that streptomycin binding requires a precise three-dimensional structure of the binding site and that several subtle changes in binding site geometry (each on the order of 1 or 2 Å) are sufficient to confer resistance.
TABLE 2.
Streptomycin binding site dimensions in apo 30S structuresa
| Residue pair | Distance (Å) |
||||||
|---|---|---|---|---|---|---|---|
| Wild type (4DR)1 | U13C (4DUY) | U20G (4DV0) | C912A (4DV2) | A914G (4DV4) | A915G (4DV6) | ΔrsmG (4NXM) | |
| U14-C526 | 12.5 | 12.7 | 12.8 | 12.4 | 12.5 | 12.7 | 12.6 |
| U14-m7G527 | 8.7 | 9.3 | 9.1 | 8.7 | 9.0 | 9.2 | 8.9 |
| U14-A914 | 6.7 | 7.0 | 7.3 | 6.7 | 6.6 | 7.4 | 7.0 |
| U14-C1490 | 18.7 | 19.1 | 19.3 | 17.4 | 18.8 | 19.2 | 18.7 |
| U14-G1491 | 16.8 | 17.4 | 17.3 | 14.2 | 17.2 | 17.9 | 16.6 |
| C526-A914 | 6.6 | 6.4 | 6.5 | 6.5 | 6.3 | 6.3 | 6.6 |
| C526-C1490 | 17.9 | 17.1 | 17.7 | 17.4 | 17.9 | 17.1 | 17.9 |
| C526-G1491 | 15.0 | 14.3 | 14.7 | 15.2 | 15.3 | 14.5 | 15.2 |
| m7G527-A914 | 6.2 | 6.2 | 6.4 | 6.0 | 5.8 | 6.5 | 6.6 |
| m7G527-C1490 | 19.6 | 18.8 | 19.4 | 18.7 | 19.5 | 18.6 | 19.6 |
| m7G527-G1491 | 15.9 | 15.0 | 15.6 | 14.9 | 15.9 | 15.2 | 15.8 |
| A914-C1490 | 15.9 | 15.6 | 15.7 | 14.7 | 16.0 | 15.2 | 15.7 |
| A914-G1491 | 14.1 | 14.0 | 13.7 | 12.5 | 14.3 | 13.9 | 14.0 |
Distances between phosphate atoms of each pair of residues indicated. Coordinate errors are provided in Table S1 in the supplemental material. PDB accession numbers are given in parentheses.
TABLE 3.
Streptomycin binding site dimensions in ribosome-streptomycin complexesa
| Residue pair | Distance (Å) |
||||||
|---|---|---|---|---|---|---|---|
| Wild type (4DR3) | U13C (4DUZ) | U20G (4DV1) | C912A (4DV3) | A914G (4DV5) | A915G (4DV7) | ΔrsmG (4NXN) | |
| U14-C526 | 12.1 | 12.4 | 12.3 | 12.2 | 12.4 | 12.2 | 12.2 |
| U14-m7G527 | 8.8 | 9.0 | 9.0 | 8.9 | 9.0 | 8.8 | 8.6 |
| U14-A914 | 6.9 | 7.4 | 7.2 | 6.9 | 7.0 | 7.3 | 7.0 |
| U14-C1490 | 18.5 | 19.0 | 18.7 | 18.3 | 18.4 | 18.4 | 18.3 |
| U14-G1491 | 16.9 | 17.5 | 16.9 | 16.8 | 16.5 | 16.9 | 16.8 |
| C526-A914 | 6.1 | 5.9 | 6.1 | 6.1 | 6.1 | 5.9 | 6.2 |
| C526-C1490 | 17.0 | 16.7 | 17.0 | 17.2 | 17.1 | 17.0 | 16.9 |
| C526-G1491 | 14.0 | 14.0 | 13.9 | 14.0 | 14.0 | 14.2 | 13.8 |
| m7G527-A914 | 6.7 | 6.8 | 6.9 | 6.7 | 6.7 | 6.7 | 6.9 |
| m7G527-C1490 | 18.8 | 18.6 | 19.2 | 19.0 | 19.0 | 18.8 | 18.8 |
| m7G527-G1491 | 15.1 | 15.0 | 15.3 | 15.1 | 15.1 | 15.1 | 14.9 |
| A914-C1490 | 15.4 | 15.4 | 15.5 | 15.6 | 15.6 | 15.5 | 15.4 |
| A914-G1491 | 13.8 | 14.0 | 13.8 | 13.8 | 13.6 | 14.0 | 13.9 |
Distances between phosphate atoms are indicated for each pair of residues in the streptomycin binding site in 30S subunits complexed with streptomycin. Coordinate errors are provided in Table S1 in the supplemental material. PDB accession numbers are given in parentheses.
In our wild-type structure, ribosomal protein S12 residues K42, K43, and K87 were all in position to contact streptomycin. This is consistent with the identification of Strr mutants with amino acid substitutions at each of these positions. While in composite omit maps we observed clear electron density for ribosomal protein S12 backbone atoms, side chain electron density was not consistently observed (see Fig. S5 and S6 in the supplemental material). Thus, it remains unclear how 16S rRNA mutations influence S12 contacts and how such defects might contribute to streptomycin resistance.
Structural basis for streptomycin resistance.
The nature of the streptomycin binding interaction indicated that the structural basis for the Strr phenotype would be distinct from that of other antibiotic resistance mutations (8). While most antibiotics make sequence-specific interactions with rRNA bases, streptomycin interacts exclusively with rRNA backbone atoms and lysine amino groups of ribosomal protein S12. The structural rearrangements caused by the pseudoknot mutations produce a series of small changes in the backbone conformation of the streptomycin binding site. Because the pseudoknot is engaged in extensive helix-packing interactions with the rest of the 30S subunit, the small backbone changes needed to confer resistance may require substantial disruption of base-base hydrogen-bonding interactions. These characteristics distinguish the Strr phenotype from resistance to antibiotics binding to the peptidyltransferase center, which make base-specific contacts and whose binding interaction can be disrupted by the loss of a single functional group without distortion of the rRNA fold (25). As observed in the crystal structure of the Haloarcula marismortui 50S subunit, the base substitutions C2452U and U2500A both eliminate antibiotic-rRNA contacts without greatly distorting the binding site conformation (26). The A2058G mutation introduces an N-2 amino group, blocking macrolide interaction (25). This is also true of mutations conferring resistance to aminoglycoside antibiotics that bind to the decoding site via both backbone and base-specific interactions; base substitutions can both remove functional group contacts and perturb the backbone trajectory (51).
Ribosomal protein S12 residues K42, K43, and K87 appear to make or are in a position to make direct contact with streptomycin. These positions are all also sights of Strr mutations identified in various organisms, including T. thermophilus (35). Our ability to draw definitive conclusions regarding the effects of base substitutions in the central pseudoknot is limited by the current resolution of 30S structures. Nevertheless, small changes in the positions of rRNA residues in the streptomycin binding site, together with the loss of one or more contacts with S12, could combine to reduce streptomycin binding. The effect of base substitutions in the pseudoknot could perturb S12-streptomycin contacts, as S12 also makes direct contact with the pseudoknot. Given that single amino acid substitutions at K42, K43, or K87 can confer high-level streptomycin resistance, disordering of these side chains indirectly through distortion of S12-rRNA contacts could provide a straightforward explanation for streptomycin resistance.
Thus, rather than being attributable to the loss of a specific single antibiotic-ribosome contact, resistance due to these 16S rRNA mutations results from a distortion of the streptomycin binding site brought about by a series of small local changes in backbone position. These changes are expected to increase the energetic cost of drug binding. Previous studies using E. coli ribosomes (21, 22) showed that similar or identical mutations (U13A, U13C, C912U, A913G, A914U, A914G, and A915G) effectively abolish streptomycin binding. This phenotype is therefore mechanistically distinct from streptomycin dependence (Strd), in which decoding requires binding of streptomycin to alleviate a structural distortion of the decoding site (27).
Nature of the restrictive phenotype of Strr mutants.
Since the 1960s it has been known that streptomycin causes misreading (28) and that many Strr mutations causing amino acid substitutions in ribosomal protein S12 produce hyperaccuracy or error-restrictive phenotypes (52). Such restrictive Strr mutations confer a fitness cost, at least in part due to the decreased catalytic efficiency of protein synthesis (53). Strr mutations in the pseudoknot, including some of the same base substitutions examined here, have been shown previously in E. coli to produce an error-restrictive phenotype (54), which is best explained by the data presented here, in particular, the perturbation of the critical decoding site residues A1492 and A1493. These two bases undergo a conformational change to directly monitor codon-anticodon base pairing (9). It seems quite probable that shifts in backbone position would influence the kinetics of this conformational change.
These data, together with our own, indicate that the central pseudoknot and the decoding site are conformationally coupled. C912 is in close proximity to positions A908 and A909 in helix 27 that engage in a packing interaction with A1413 and G1487 in helix 44, near the decoding site. This packing interaction has been shown by chemical probing of E. coli ribosomes to be influenced by restrictive S12 mutations and ram S4 mutations (55). What is not clear is if changes in the dynamics of A1492 and A1493 actually contribute to the resistance phenotype. We previously found that the A1408G base substitution confers streptomycin resistance in T. thermophilus (36). A1408 is located on the far side of helix 44 away from the central pseudoknot and the contacts with streptomycin, suggesting that changes in conformational dynamics could extend some distance.
Possible effects of conformational dynamics on streptomycin binding.
It is not yet known whether streptomycin binding occurs via an induced fit mechanism or by a stochastic gating mechanism, as indicated by molecular dynamics simulation of gentamicin binding (56). Comparison of the interphosphate distance between residues m7G527 and G1491 of the apo 30S structure with the streptomycin-bound structure indicates a closure upon drug binding. In a stochastic gating model, the dynamics of this transition could be slower in the mutant ribosomes, thus reducing the lifetime of the conformation optimum for binding. Such changes are likely to be missed using static structural approaches. Nevertheless, the ability of mutant ribosomes to adopt a native-like conformation upon streptomycin binding is consistent with increased conformational dynamics, rather than a rigid distortion of the binding site.
The decoding site, especially in the vicinity of helix 44, is known from structural studies to be conformationally dynamic; bases A1492 and A1493, in particular, can adopt two distinct conformations, a tucked-in conformation in the apo state of the ribosome or a flipped-out conformation in the actively decoding ribosome (9). The latter conformation is stabilized by aminoglycoside antibiotics, such as paromomycin (7, 8). That the decoding site is intrinsically dynamic is supported by molecular dynamics simulations (57).
Comparison of the T. thermophilus pseudoknot structure (PDB accession number 4DR1) (29) with that of the mesophile E. coli (PDB accession number 2AVY) (58) reveals no significant structural difference that would suggest differences in structural stability (see Fig. S7 in the supplemental material). The nucleotide sequences of the pseudoknots are identical, with the exception an A19-U916 base pair in E. coli versus a C19-G916 base pair in T. thermophilus. Nevertheless, we would expect the streptomycin binding site to be more conformationally dynamic at the physiological temperature for T. thermophilus (72°C) than at the physiological temperature for E. coli (37°C) or at the low temperatures at which crystals are prepared. This could in part account for the lower intrinsic streptomycin sensitivity of T. thermophilus compared to that of E. coli (although differences in streptomycin uptake could also contribute). Changes in conformational dynamics could therefore constitute an important component of the resistance mechanism, especially given the small magnitude of structural changes that we observed. This also provides a plausible explanation for the resistance phenotype of the rsmG mutant, for which no structural changes were detected. Notably, the hydrophobic N-7 methyl group introduced by the RsmG methyltransferase is in close proximity to the β-methylthio modification of ribosomal protein S12 residue D88 (59), forming a hydrophobic pocket. While mutants deficient in β-methylthio modification are viable (60, 61) and do not exhibit substantial streptomycin resistance, the hydrophobic interaction of these two modifications could nevertheless have the effect of stabilizing the conformation of the streptomycin binding site. Unfortunately, it is not possible to obtain high-resolution structural information at physiological temperature using currently available crystallographic approaches. However, recent developments in the application of X-ray-free electron lasers to obtain crystallographic data at room temperature (62) present one potential solution to addressing the role of conformational dynamics in antibiotic resistance.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants GM019756 (to A.E.D.) and GM094157 (to G.J. and S.T.G.) from the U.S. National Institutes of Health and RR-15301 (to F.V.M.) from the National Center for Research Resources at the National Institutes of Health. Part of this work was conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beam lines, which are supported by grants from the National Center for Research Resources (5P41RR015301-10) and the National Institute of General Medical Sciences (8 P41 GM103403-10) of the U.S. National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the DOE under contract no. DE-AC02-06CH11357.
Footnotes
Published ahead of print 12 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02857-14.
REFERENCES
- 1.Gale EF, Cundliffe E, Reynolds PE, Richmond MH, Waring MJ. 1981. The molecular basis of antibiotic action. John Wiley & Sons, London, United Kingdom [Google Scholar]
- 2.Poehlsgaard J, Douthwaite S. 2005. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 3:870–881. 10.1038/nrmicro1265 [DOI] [PubMed] [Google Scholar]
- 3.Moazed D, Noller HF. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389–394. 10.1038/327389a0 [DOI] [PubMed] [Google Scholar]
- 4.Fourmy D, Recht MI, Blanchard SC, Puglisi JD. 1996. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science 274:1367–1371. 10.1126/science.274.5291.1367 [DOI] [PubMed] [Google Scholar]
- 5.Vicens Q, Westhof E. 2001. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure 9:647–658. 10.1016/S0969-2126(01)00629-3 [DOI] [PubMed] [Google Scholar]
- 6.Blaha GM, Polikanov YS, Steitz TA. 2012. Elements of ribosomal drug resistance and specificity. Curr. Opin. Struct. Biol. 22:750–758. 10.1016/j.sbi.2012.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fourmy D, Yoshizawa S, Puglisi JD. 1998. Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA. J. Mol. Biol. 277:333–345. 10.1006/jmbi.1997.1551 [DOI] [PubMed] [Google Scholar]
- 8.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340–348. 10.1038/35030019 [DOI] [PubMed] [Google Scholar]
- 9.Ogle JM, Brodersen DE, Clemons WM, Tarry MJ, Carter AP, Ramakrishnan V. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897–902. 10.1126/science.1060612 [DOI] [PubMed] [Google Scholar]
- 10.Ozaki M, Mizushima S, Nomura M. 1969. Identification and functional characterization of the protein controlled by the streptomycin-resistant locus in E. coli. Nature 222:333–339. 10.1038/222333a0 [DOI] [PubMed] [Google Scholar]
- 11.Funatsu G, Wittmann HG. 1972. Ribosomal proteins. XXXIII. Location of amino-acid replacements in protein S12 isolated from Escherichia coli mutants resistant to streptomycin. J. Mol. Biol. 68:547–550 [DOI] [PubMed] [Google Scholar]
- 12.Montandon PE, Nicolas P, Schürmann P, Stutz E. 1985. Streptomycin-resistance of Euglena gracilis chloroplasts: identification of a point mutation in the 16S rRNA gene in an invariant position. Nucleic Acids Res. 13:4299–4310. 10.1093/nar/13.12.4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gauthier A, Turmel M, Lemieux C. 1988. Mapping of chloroplast mutations conferring resistance to antibiotics in Chlamydomonas: evidence for a novel site of streptomycin resistance in the small subunit rRNA. Mol. Gen. Genet. 214:192–197. 10.1007/BF00337710 [DOI] [PubMed] [Google Scholar]
- 14.Harris EH, Burkhart BD, Gillham NW, Boynton JE. 1989. Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh KC, To KY, Sun SW, Wu MC, Lin TY, Chen CC. 1994. Point mutations in the chloroplast 16S rRNA gene confer streptomycin resistance in Nicotiana plumbaginifolia. Curr. Genet. 26:132–135. 10.1007/BF00313800 [DOI] [PubMed] [Google Scholar]
- 16.Honoré N, Cole ST. 1994. Streptomycin resistance in mycobacteria. Antimicrob. Agents Chemother. 38:238–242. 10.1128/AAC.38.2.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meier A, Kirschner P, Bange FC, Vogel U, Böttger EC. 1994. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis: mapping of mutations conferring resistance. Antimicrob. Agents Chemother. 38:228–233. 10.1128/AAC.38.2.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Springer B, Kidan YG, Prammananan T, Ellrott K, Böttger EC, Sander P. 2001. Mechanisms of streptomycin resistance: selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob. Agents Chemother. 45:2877–2884. 10.1128/AAC.45.10.2877-2884.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melançon P, Lemieux C, Brakier-Gingras L. 1988. A mutation in the 530 loop of Escherichia coli 16S ribosomal RNA causes resistance to streptomycin. Nucleic Acids Res. 16:9631–9639. 10.1093/nar/16.20.9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frattali AL, Flynn MK, De Stasio EA, Dahlberg AE. 1990. Effects of mutagenesis of C912 in the streptomycin binding region of Escherichia coli 16S ribosomal RNA. Biochim. Biophys. Acta 1050:27–33. 10.1016/0167-4781(90)90136-P [DOI] [PubMed] [Google Scholar]
- 21.Leclerc D, Melançon P, Brakier-Gingras L. 1991. Mutations in the 915 region of Escherichia coli 16S ribosomal RNA reduce the binding of streptomycin to the ribosome. Nucleic Acids Res. 19:3973–3977. 10.1093/nar/19.14.3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinard R, Payant C, Melançon P, Brakier-Gingras L. 1993. The 5′ proximal helix of 16S rRNA is involved in the binding of streptomycin to the ribosome. FASEB J. 7:173–176 [DOI] [PubMed] [Google Scholar]
- 23.Powers T, Noller HF. 1991. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 10:2203–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gregory ST, Dahlberg AE. 2009. Genetic and structural analysis of base substitutions in the central pseudoknot of Thermus thermophilus 16S ribosomal RNA. RNA 15:215–223. 10.1261/rna.1374809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu D, Blaha G, Moore PB, Steitz TA. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121:257–270. 10.1016/j.cell.2005.02.005 [DOI] [PubMed] [Google Scholar]
- 26.Blaha G, Gurel G, Schroeder SJ, Moore PB, Steitz TA. 2008. Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J. Mol. Biol. 379:505–519. 10.1016/j.jmb.2008.03.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demirci H, Wang L, Murphy FV, Murphy EL, Carr JF, Blanchard SC, Jogl G, Dahlberg AE, Gregory ST. 2013. The central role of protein S12 in organizing the structure of the decoding site of the ribosome. RNA 19:1791–1801. 10.1261/rna.040030.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies J, Gilbert W, Gorini L. 1964. Streptomycin, suppression, and the code. Proc. Natl. Acad. Sci. U. S. A. 51:883–890. 10.1073/pnas.51.5.883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demirci H, Murphy F, IV, Murphy E, Gregory ST, Dahlberg AE, Jogl G. 2013. A structural basis for streptomycin-induced misreading of the genetic code. Nat. Commun. 4:1355. 10.1038/ncomms2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timms AR, Steingrimsdottir H, Lehmann AR, Bridges BA. 1992. Mutant sequences in the rpsL gene of Escherichia coli B/r: mechanistic implications for spontaneous and ultraviolet light mutagenesis. Mol. Gen. Genet. 232:89–96. 10.1007/BF00299141 [DOI] [PubMed] [Google Scholar]
- 31.Timms AR, Bridges BA. 1993. Double, independent mutational events in the rpsL gene of Escherichia coli: an example of hypermutability? Mol. Microbiol. 9:335–342. 10.1111/j.1365-2958.1993.tb01694.x [DOI] [PubMed] [Google Scholar]
- 32.Björkman J, Samuelsson P, Andersson DI, Hughes D. 1999. Novel ribosomal mutations affecting translational accuracy, antibiotic resistance and virulence of Salmonella typhimurium. Mol. Microbiol. 31:53–58. 10.1046/j.1365-2958.1999.01142.x [DOI] [PubMed] [Google Scholar]
- 33.Okamoto S, Tamaru A, Nakajima C, Nishimura K, Tanaka Y, Tokuyama S, Suzuki Y, Ochi K. 2007. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 63:1096–1106. 10.1111/j.1365-2958.2006.05585.x [DOI] [PubMed] [Google Scholar]
- 34.Nishimura K, Johansen SK, Inaoka T, Hosaka T, Tokuyama S, Tahara Y, Okamoto S, Kawamura F, Douthwaite S, Ochi K. 2007. Identification of the RsmG methyltransferase target as 16S rRNA nucleotide G527 and characterization of Bacillus subtilis rsmG mutants. J. Bacteriol. 189:6068–6073. 10.1128/JB.00558-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gregory ST, Cate JHD, Dahlberg AE. 2001. Streptomycin-resistant and streptomycin-dependent mutants of the extreme thermophile Thermus thermophilus. J. Mol. Biol. 309:333–338. 10.1006/jmbi.2001.4676 [DOI] [PubMed] [Google Scholar]
- 36.Gregory ST, Carr JF, Dahlberg AE. 2005. A mutation in the decoding center of Thermus thermophilus 16S rRNA suggests a novel mechanism of streptomycin resistance. J. Bacteriol. 187:2200–2202. 10.1128/JB.187.6.2200-2202.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woese CR, Gutell R, Gupta R, Noller HF. 1983. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol. Rev. 47:621–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. 2000. Structure of the 30S ribosomal subunit. Nature 407:327–339. 10.1038/35030006 [DOI] [PubMed] [Google Scholar]
- 39.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102:615–623. 10.1016/S0092-8674(00)00084-2 [DOI] [PubMed] [Google Scholar]
- 40.Gregory ST, Dahlberg AE. 2008. Transposition of an insertion sequence, ISTth7, in the genome of the extreme thermophile Thermus thermophilus HB8. FEMS Microbiol. Lett. 289:187–192. 10.1111/j.1574-6968.2008.01389.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory ST, Demirci H, Belardinelli R, Monshupanee T, Gualerzi C, Dahlberg AE, Jogl G. 2009. Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG. RNA 15:1693–1704. 10.1261/rna.1652709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clemons WM, Jr, Brodersen DE, McCutcheon JP, May JLC, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2001. Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: purification, crystallization and structure determination. J. Mol. Biol. 310:827–843. 10.1006/jmbi.2001.4778 [DOI] [PubMed] [Google Scholar]
- 43.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326. 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurata S, Weixlbaumer A, Ohtsuki T, Shimazaki T, Wada T, Kirino Y, Takai K, Watanabe K, Ramakrishnan V, Suzuki T. 2008. Modified uridines with C5-methylene substituents at the first position of the tRNA stabilize U-G wobble pairing during decoding. J. Biol. Chem. 283:18801–18811. 10.1074/jbc.M800233200 [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schrödinger LLC. 2012. The PyMOL molecular graphics system, version 1.5. Schrödinger LLC, Mannheim, Germany [Google Scholar]
- 48.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Müller KM, Pande N, Shang Z, Yu N, Gutell RR. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. 10.1186/1471-2105-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guymon R, Pomerantz SC, Crain PF, McCloskey JA. 2006. Influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: the complete modification map of 16S rRNA of Thermus thermophilus. Biochemistry 45:4888–4899. 10.1021/bi052579p [DOI] [PubMed] [Google Scholar]
- 50.Saenger W. 1984. Principles of nucleic acid structure. Springer, New York, NY [Google Scholar]
- 51.Lynch SR, Puglisi JD. 2001. Structural origins of aminoglycoside specificity for prokaryotic ribosomes. J. Mol. Biol. 306:1037–1058. 10.1006/jmbi.2000.4420 [DOI] [PubMed] [Google Scholar]
- 52.Ruusala T, Andersson D, Ehrenberg M, Kurland CG. 1984. Hyper-accurate ribosomes inhibit growth. EMBO J. 3:2575–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurland CG. 1992. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 26:29–50. 10.1146/annurev.ge.26.120192.000333 [DOI] [PubMed] [Google Scholar]
- 54.Pinard R, Côté M, Payant C, Brakier-Gingras L. 1994. Positions 13 and 914 in Escherichia coli 16S ribosomal RNA are involved in the control of translational accuracy. Nucleic Acids Res. 22:619–624. 10.1093/nar/22.4.619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allen PN, Noller HF. 1989. Mutations in ribosomal proteins S4 and S12 influence the higher order structure of 16 S ribosomal RNA. J. Mol. Biol. 208:457–468. 10.1016/0022-2836(89)90509-3 [DOI] [PubMed] [Google Scholar]
- 56.Vaiana AC, Sanbonmatsu KY. 2009. Stochastic gating and drug–ribosome interactions. J. Mol. Biol. 386:648–661. 10.1016/j.jmb.2008.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Munro JB, Sanbonmatsu KY, Spahn CMT, Blanchard SC. 2009. Navigating the ribosome's metastable energy landscape. Trends Biochem. Sci. 34:390–400. 10.1016/j.tibs.2009.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310:827–834. 10.1126/science.1117230 [DOI] [PubMed] [Google Scholar]
- 59.Kowalak JA, Walsh KA. 1996. β-Methylthio-aspartic acid: identification of a novel posttranslational modification in ribosomal protein S12 from Escherichia coli. Protein Sci. 5:1625–1632. 10.1002/pro.5560050816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carr JF, Hamburg D-M, Gregory ST, Limbach PA, Dahlberg AE. 2006. Effects of streptomycin resistance mutations on posttranslational modification of ribosomal protein S12. J. Bacteriol. 188:2020–2023. 10.1128/JB.188.5.2020-2023.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anton BP, Saleh L, Benner JS, Raleigh EA, Kasif S, Roberts RJ. 2008. RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105:1826–1831. 10.1073/pnas.0708608105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Demirci H, Sierra RG, Laksmono H, Shoeman RL, Botha S, Barends TRM, Nass K, Schlichting I, Doak RB, Gati C, Williams GJ, Boutet S, Messerschmidt M, Jogl G, Dahlberg AE, Gregory ST, Bogan MJ. 2013. Serial femtosecond X-ray diffraction of 30S ribosomal subunit microcrystals in liquid suspension at ambient temperature using an X-ray free-electron laser. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69:1066–1069. 10.1107/S174430911302099X [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.