Abstract
Amixicile shows efficacy in the treatment of Clostridium difficile infections (CDI) in a mouse model, with no recurrence of CDI. Since amixicile selectively inhibits the action of a B vitamin (thiamine pyrophosphate) cofactor of pyruvate:ferredoxin oxidoreductase (PFOR), it may both escape mutation-based drug resistance and spare beneficial probiotic gut bacteria that do not express this enzyme. Amixicile is a water-soluble derivative of nitazoxanide (NTZ), an antiparasitic therapeutic that also shows efficacy against CDI in humans. In comparative studies, amixicile showed no toxicity to hepatocytes at 200 μM (NTZ was toxic above 10 μM); was not metabolized by human, dog, or rat liver microsomes; showed equivalence or superiority to NTZ in cytochrome P450 assays; and did not activate efflux pumps (breast cancer resistance protein, P glycoprotein). A maximum dose (300 mg/kg) of amixicile given by the oral or intraperitoneal route was well tolerated by mice and rats. Plasma exposure (rats) based on the area under the plasma concentration-time curve was 79.3 h · μg/ml (30 mg/kg dose) to 328 h · μg/ml (100 mg/kg dose), the maximum concentration of the drug in serum was 20 μg/ml, the time to the maximum concentration of the drug in serum was 0.5 to 1 h, and the half-life was 5.6 h. Amixicile did not concentrate in mouse feces or adversely affect gut populations of Bacteroides species, Firmicutes, segmented filamentous bacteria, or Lactobacillus species. Systemic bioavailability was demonstrated through eradication of Helicobacter pylori in a mouse infection model. In summary, the efficacy of amixicile in treating CDI and other infections, together with low toxicity, an absence of mutation-based drug resistance, and excellent drug metabolism and pharmacokinetic metrics, suggests a potential for broad application in the treatment of infections caused by PFOR-expressing microbial pathogens in addition to CDI.
INTRODUCTION
Clostridium difficile, a Gram-positive, spore-forming obligate anaerobe present in the intestinal microflora of most humans and animals, is an important cause of antibiotic-associated infectious diarrhea and pseudomembranous colitis. C. difficile infection (CDI) is the leading cause of health care-associated infectious diarrhea, which is attributed to the emergence of hypervirulent, binary-toxin-producing strains such as North American pulsed-field type 1 (NAP1/BI/027) (1–4). Antibiotic interventions with oral vancomycin and metronidazole (MTZ) are effective treatments for severe and mild forms of the disease, respectively (5, 6); but recurrence rates of 25% or higher are common and the risk of chronic CDI episodes increases to 60% (2). Whether recurrence is the result of eradication failure or reinfection, it is generally believed that susceptibility to CDI is a function of the species diversity and type of resident gut microflora, which serve as a protective barrier to colonization (7). Even standard CDI therapies with MTZ, vancomycin, and fidaxomicin impair intestinal flora and thereby contribute to continued susceptibility to and recurrence of CDI (6, 8, 9). The importance of fecal microflora is underscored by the success rates of 80 to 90% achieved with fecal transplants from healthy volunteers (10) and with the implementation of probiotic preventive measures that lower the incidence of recurrence by 66% (11). Among the attributes believed to be important in the development of newer therapeutics to treat CDI, selectivity for C. difficile and retention of the drug within the intestine are emphasized.
We initially developed amixicile, a bioavailable derivative of nitazoxanide (NTZ), to treat systemic infections caused by strictly anaerobic bacteria or anaerobic parasites and gastrointestinal infections caused by Helicobacter pylori and Campylobacter jejuni, all of which express pyruvate:ferredoxin oxidoreductase (PFOR) and related enzymes (12–15). NTZ shows good in vitro efficacy against these microorganisms but is limited clinically to the treatment of intestinal infections caused by Cryptosporidium parvum and Giardia lamblia (16). However, since NTZ was found to be noninferior to MTZ in the treatment of CDI in a randomized, double-blind, prospective patient trial (8, 17), we evaluated the efficacy of amixicile in a mouse CDI model (15). In this model, infected mice develop diarrhea, lose weight, and succumb on days 2 to 6 after oral inoculation with 104 to 105 CFU of C. difficile (6, 15). Amixicile proved superior to NTZ in this model. In an optimized CDI mouse model, amixicile showed equivalence to vancomycin and fidaxomicin at day 5 and superiority by day 12 (15). Recurrence was common in mice treated with vancomycin or fidaxomicin, whereas no recurrence was observed in mice receiving amixicile (6, 15). In fact, in all of our studies with mice treated with NTZ or tested analogues of NTZ, none of the surviving animals relapsed (6, 15). We concluded that gut repopulation with beneficial (non-PFOR) bacteria, considered essential for protection against CDI, rebounds much sooner with amixicile therapy than with vancomycin or fidaxomicin (15). McVay and Rolfe reported that NTZ was active against CDI in a hamster model and noted that unlike vancomycin and MTZ, pretreatment of hamsters with NTZ did not induce CDI, which suggested that NTZ did not suppress protective resident flora (18). This conclusion is supported by several MIC-based comparative studies showing that NTZ is not inhibitory to various species of Lactobacillus, Propionibacterium, and Bifidobacterium that lack the PFOR target (19, 20). In contrast, many of these bacteria are susceptible to fidaxomicin and vancomycin (21). Several studies found a greater abundance of members of the genus Bifidobacterium in the feces of individuals considered resistant to CDI infection than in the feces of those susceptible to recurrence (22, 23). Many of these human microbiome studies of CDI and controls reveal substantial changes in the complex gut microflora but have yet to correlate these changes with specific antibiotic therapy.
Here we report results of preclinical studies that showed that amixicile was not cytotoxic or metabolized by liver microsome fractions and that both dose range findings and pharmacokinetic (PK) studies with rats indicated that amixicile was safe and well tolerated and achieved levels in serum well in excess of the MIC for C. difficile. Therapeutic bioavailability was further demonstrated by successful eradication of H. pylori infection in a mouse model. Our studies challenge the conventional wisdom that a pathogen-specific therapeutic that concentrates in the gut is the most effective strategy for developing new CDI therapeutics. In contrast, we suggest that a systemic therapeutic like amixicile, by concentrating in areas of mucosal inflammation caused by C. difficile or H. pylori, would act locally and spare susceptible flora that are not associated with disease. Such therapeutics might prove beneficial when administered together with probiotic or fecal transplant treatments.
MATERIALS AND METHODS
Scale-up synthesis of amixicile.
Scale-up synthesis of amixicile-related derivatives has been previously described (13, 14). The chemical structures of amixicile and the other analogues used in this study are depicted in Fig. 1. NTZ was purchased commercially from Waterstone Technology (Carmel, IN). The purity of all of the compounds used in these studies was assessed by spectrophotometry, nuclear magnetic resonance, and mass spectrometry (MS) protocols (12, 14, 15).
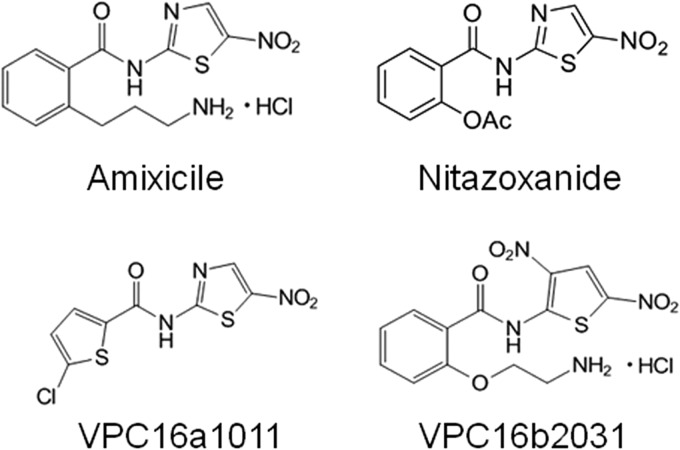
FIG 1.
Chemical structures of amixicile, NTZ, VPC16a1011, and VPC16b2031.
Bacterial strains.
H. pylori strains were grown on brucella-based medium supplemented with 7.5% newborn serum and grown in a microaerobic environment as previously described (12). The MICs for H. pylori strains SS1 and 26695 were determined by microdilution in brain heart infusion broth (14, 15).
Metabolic stability.
Metabolic stability was determined at 1 and 10 μM final concentrations of compounds incubated with pooled human and rat and dog liver microsomes (0.5 mg protein/ml) containing appropriate cofactors (2.5 mM NADPH and 3.3 mM MgCl2) in 0.1 M phosphate buffer, pH 7.4, in a 37°C water bath. The incubation mixture contained a final organic solvent concentration of 0.1% dimethyl sulfoxide (DMSO). Reactions were started with the addition of microsomes and stopped by removing 100-μl aliquots at selected times (0, 15, 30, and 60 min) and mixing them with 200 μl of acetonitrile containing an internal standard. Samples were transferred to a 96-well plate for further dilution, followed by liquid chromatography-tandem MS (LC-MS/MS) analysis. Controls for metabolism included 10 μM midazolam, a known substrate of cytochrome P450 3A4 (CYP3A4), and incubation of test compounds and midazolam with heat-inactivated microsomes for 0 and 60 min, as a negative control. All samples were assayed in triplicate.
In vitro CYP inhibition.
Pooled human liver microsomes (0.5 mg/ml) and cofactors (2.5 mM NADPH and 3.3 mM MgCl2) were incubated with test compounds (1 and 10 μM) and a cocktail of seven different CYP probe substrates in 0.1 M phosphate buffer, pH 7.4 (final volume, 200 μl). The substrates used included 25 μM phenacetin (CYP1A2), 25 μM bupropion (CYP2B6), 10 μM diclofenac (CYP2C9), 20 μM mephenytoin (CYP2C19), 10 μM bufuralol (CYP2D6), 50 μM testosterone (CYP3A4), and 4 μM midazolam (CYP3A4). Specific inhibitor control samples were incubated and analyzed in the same manner as test compounds but contained the following inhibitors in place of the test compound: 10 μM furafylline (CYP1A2), 10 μM TEPA (CYP2B6), 3 μM sulfaphenazole (CYP2C9), 10 μM nootkatone (CYP2C19), 2 μM quinidine (CYP2D6), and 5 μM ketoconazole (CYP3A4). Reactions were started with the addition of microsomes and terminated after 20 min of incubation at 37°C by the addition of 200 µl of ice-cold acetonitrile containing 2 μM dextrorphan (internal standard). Samples were centrifuged at 1,500 rpm for 20 min at 10°C and supernatants were collected and analyzed by LC-MS/MS with positive-ion electrospray ionization. The percentage of CYP activity in test compounds or specific inhibitor samples relative to the control samples not containing the test compound or controls was calculated as follows: [substrate metabolite response (peak area ratio, PAR) in the presence of inhibitor or test compound/substrate metabolite mean PAR in control] × 100. CYP enzyme activity in the presence of the test compound that was less than 70% of the control activity was considered significant inhibition in this assay.
In vitro bidirectional permeability of Caco-2 cells.
CacoReady HTS Transwell-24 plates consisting of differentiated Caco-2 cells plated on microporous polycarbonate filters (6.5-mm diameter, 0.33-cm2 growth area, 0.4-mm pore size) were obtained from ADMEcell, Inc. (Emeryville, CA). The cells were prepared for assay according to the manufacturer's instructions, the transport medium was replaced with Dulbecco's modified Eagle medium, and the cells were incubated for 72 h prior to incubation with test compounds. The integrity of the monolayer was assessed by measuring the transepithelial electrical resistance (TEER) with an epithelial volt-ohm meter (EVOM Instrument; World Precision Instruments; Sarasota, FL). A TEER value of >1,000 Ω/cm2 indicated that the barrier system was acceptable. For apical-to-basal (A-B) permeability determination, test and control compound solutions were prepared in HBSS (Hanks' balanced salt solution) at pH 6.0 or 7.4 and added to the apical side of the cell monolayer. For basal-to-apical (B-A) permeability determination, test and control compound solutions were prepared in HBSS at pH 7.4 and added to the basal side of the cell monolayer. Permeability was measured by testing aliquots from the receiving compartment. Samples in acetonitrile were stored at −80°C and analyzed by LC-MS/MS in multiple-reaction-monitoring mode with positive- or negative-ion electrospray ionization. All assays were done in triplicate, and results are presented as means and standard deviations (SDs).
Plasma protein binding studies.
Plasma and phosphate-buffered saline (PBS) calibration standards were prepared with blank human, rat, or dog plasma and PBS. Spiking solutions were prepared by diluting the test compound amixicile (10 mg/ml) or VPC16a1011 (5 mg/ml) DMSO stock to give spiking standards of 3, 5, 10, 30, 100, 300, 500, 800, 1,000, 1,200, and 1,500 μg/ml of DMSO. Five microliters of each spiking standard was used to spike 495 μl of plasma (or PBS) to give calibration standards of 0.03, 0.05, 0.1, 0.3, 1, 3, 5, 8, 10, 12, and 15 μg/ml. The lower limit of quantification for amixicile analysis was 0.05 μg/ml. Amixicile plasma samples for the dialysis experiments were prepared by using the spiking solutions, and 10 μl of each spiking solution (0.1, 1.0, or 10 mg/ml) was added to 990 μl of plasma (human, dog, or rat) to give samples of 1, 10, and 100 μg/ml. The samples were dialyzed against PBS at pH 7.4 with a Thermo Scientific rapid equilibrium dialysis plate system with a cutoff of 8,000 Da. The chambers were incubated at 37°C for 4 h. Upon completion, 50 μl of dialyzed plasma was diluted with an equal volume of PBS prior to protein precipitation. As a control for nonspecific binding to the filter, amixicile was tested in PBS in the absence of serum. Samples were prepared for quantification of amixicile in dialysis samples (100 μl) to which 300 μl of acetonitrile containing 10 μg/ml methyl nicotinate was added to precipitate plasma proteins. The supernatants following centrifugation were diluted in formic acid and analyzed by LC-MS/MS. A calibration standard curve was used to determine the concentrations of amixicile in the chambers, and they were compared as buffer chamber/sample chamber. The percent amixicile bound to plasma protein was determined as the % bound = 100 − (100 × [test compound]buffer chamber/mean value fequilibrium/[test compound]sample chamber), and the mean and SD were calculated by using Microsoft Excel software.
Induced mutation.
Escherichia coli tester strain CC103 [araΔ(lac proB)/F′ lacI lacZ proAB+)] (24) was grown for 6 h in the presence of test compounds, and then decimal dilutions were prepared in PBS and spread plated onto LB medium supplemented with 50 μg/ml of rifampin or onto LB medium without antibiotics (24). Two independent experiments, performed in triplicate, were averaged. The mutation frequency was determined as the number of rifampin-resistant CFU/total CFU and is presented as the number of mutations per 108 CFU.
Dose range studies with Sprague-Dawley rats.
Male Sprague-Dawley rats (350 to 376 g) were obtained from Charles River (Hollister, CA). Animals were randomly distributed into groups of three based on weight and on day 1 were given the indicated dose of amixicile (20, 100, 200, or 300 mg/kg in 1% methylcellulose) by oral gavage. Animals were monitored immediately and at 24 and 48 h, and all surviving animals were euthanized after 48 h. General procedures for animal care and housing were in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals.
Plasma PK studies.
Jugular-vein-catheterized Sprague-Dawley rats (300 to 350 g) were purchased from Charles River. Plasma amixicile levels in the rats (three per group) following a single oral dose (30 or 100 mg/kg in 1% methylcellulose in sterile water) were determined. Predose and postdose blood samples (50 μl) were collected at 5, 15, and 30 min and 1, 2, 4, 8, 12, and 24 h (50 ml) by jugular vein catheter, and plasma was prepared within 30 min of collection. All plasma samples were stored at −70°C until analyzed. Calibration standards of amixicile were prepared in rat plasma from a 1-mg/ml stock solution in DMSO over a range of 0 to 2,500 ng/ml. Rat plasma protein (sample volume, 50 μl) was precipitated with 200 μl of acetonitrile containing 1,000 ng of VPC16b2031. VPC16b2031 is a dinitrothiophene derivative used as an internal standard (Fig. 1). The suspensions were clarified by centrifugation, and 150 μl of each was used for LC-MS/MS analysis. The calibration standard curve for amixicile was prepared by performing weighted 1/x2 linear regression of the peak area (PA) of amixicile as the dependent variable (y axis) and concentration as the independent variable (x axis) as follows: PA = m × [amixicile] + b, where m is the slope and b is the y intercept. The goodness of fit of this standard curve is indicated by the coefficient of determination (R2) obtained by quadratic regression, with a perfect fit yielding an R2 value of 1.000.
A noncompartmental model based on extravascular administration (oral gavage) was performed with WinNonlin (ver. 5.2) by using uniform weighting. The dosages were entered as mg/kg so that no adjustment for body weight was needed. The data collected included the maximum drug concentration in plasma (Cmax), the time at Cmax (Tmax), the mean area under the plasma concentration-time curve (AUC) up to the last measurable time point (AUClast) or to infinity (AUCinf), the terminal elimination half-life (t1/2), the apparent volume of distribution after oral administration (V/F), and the total clearance after oral administration (CL/F). Bioavailability was not determined since there was no intravenous dose used in this study. Terminal-phase parameters (t1/2, AUCinf, and CL/F) were reported only when the goodness-of-fit (R2) value of the best-fit line in the terminal elimination phase was ≥0.85.
PK analyses of serum and feces from C57BL/6 male mice receiving a single oral dose of 200 mg/kg of amixicile were also performed. A spectrophotometric assay was developed on the basis of the absorbance at 413 nm of amixicile (ε = 20 mM−1 cm−1) (12, 15). Serum samples prepared from collected blood (15 min to 4 h and 24 h) were mixed with equal volumes of methanol to precipitate proteins. Following centrifugation, supernatants were diluted to 50% in PBS (pH 7.4) and the concentration of amixicile was determined spectrophotometrically. Standard-curve determination and limit-of-detection assays were done by spiking mouse serum samples with a range of amixicile concentrations. The limit of detection in this assay was 0.1 μg/ml. Fecal samples were suspended in PBS and similarly treated with equal volumes of methanol. Supernatants were assayed for amixicile. The mean and SD of three samples, each obtained from two mice per time point, are reported.
Animal studies.
C57BL/6J mice were challenged with 3 doses of 5 × 107 CFU of the H. pylori SS1 wild-type strain in brucella broth on days 1, 3, and 5 as previously described (25, 26). At 2 weeks postinfection, groups of five mice each were treated by gavage with amixicile or MTZ at 20 mg/kg as previously described (15, 27). For amixicile-treated mice, a second group received two doses of 20 mg/kg each day. At 1 week posttreatment, mice were sacrificed, their stomachs were removed and homogenized, and CFU counts were determined by plate counting (25). Briefly, weighted gastric specimens were homogenized in PBS and plated in triplicate onto Columbia agar plates with the selective antibiotics vancomycin (10 μg/ml), trimethoprim (1 μg/ml), amphotericin B (5 μg/ml), and polymyxin B (5 μg/ml). Plates were incubated for 4 days at 37°C under microaerobic conditions (25, 27). Bacterial numbers are reported as the mean and SD of the number of CFU/g of stomach tissue. Statistically significant differences were determined by using the Student t test, with P values of <0.05 considered significant.
Microbiota qPCR.
C57BL/6 mice (5- to 8-week-old males and females) received 30 mg/kg (∼600 μg/100 μl in PBS) of amixicile (eight mice) or PBS (seven mice) once daily for 3 days by oral gavage. The mice were sacrificed on day 4, and the intestines were collected for quantitative real-time reverse transcription (RT)-PCR (qPCR) analysis for selected microbial flora (28). Gene expression in the terminal ileum was measured by real-time PCR with Sybr green and phylum and species level-specific primers as previously described (28, 29). Data were normalized to a conserved eubacterial 16S rRNA gene (EUB). The sequences of the EUB primers used are as follows: EUB forward, 5′-ACTCCTACGGGAGGCAGCAGT-3′; EUB reverse, 5′-ATTACCGCGGCTGCTGGC-3′. The sequences of the primers based on the segmented filamentous bacterial 16S rRNA gene are as follows: SFB forward, 5′-GACGCTGAGGCATGAGAGCAT-3′; SFB reverse, 5′-GACGGCACGGATTGTTATTCA-3′. The sequences of the primers based on the Lactobacillus 16S rRNA gene are as follows: Lactobacillus sp. F, 5′-AGCAGTAGGGAATCTTCCA-3′; Lactobacillus sp. R, 5′-CACCGCTACACATGGAG-3′. The sequences of the primers based on the Bacteroides 16S rRNA gene are as follows: Bacteroides F, 5′-GGTTCTGAGAGGAGGTCCC-3′; Bacteroides R, 5′-GCTGCCTCCCGTAGGAGT-3′. The sequences of the primers based on the Firmicutes 16S rRNA gene are as follows: Firmicutes F, 5′-GGAGYATGTGGTTTAATTCGAAGCA-3′; Firmicutes R, 5′-AGCTGACGACAACCATGCAC-3′. The 16S rRNA results were normalized to the total bacteria.
RESULTS
Drug metabolism.
The results of tests of the metabolic stability of amixicile incubated with human, rat, and dog liver microsome fractions are presented in Table 1. After 1 h, the remaining amixicile (10 μM) was 99% ± 5.2% for human liver microsomes, 96.5% ± 1.8% for rat liver microsomes, and 97.7% ± 2.4% for dog liver microsomes. These results were comparable to those of heat-inactivated microsome controls, indicating that amixicile is not appreciably metabolized by liver microsome fractions. In this assay, NTZ was deacetylated to the phenol (tizoxanide) as previously reported (16, 30, 31). In CYP inhibition assays (data not presented), amixicile showed inhibitory activities equivalent to or lower than those of NTZ against CYP1A2, CYP2B6, CYP2C9, CYP2C19, CYP2D6, and CYP3A4. The potential for induction of CYPs was determined by qPCR over a concentration range of 0.6 to 200 μM for amixicile but was limited to 10 μM for NTZ because of toxicity. At 10 μM, neither NTZ nor amixicile showed any potential to activate CYPs. Amixicile was tested at higher concentrations, and at the highest (200 μM), a wide variation was observed among the three sources of hepatocytes for CYP1A2 and CYP3A4, as one source was consistently high (up to 102-fold versus 10-fold for the other two sources). It is noteworthy that the absence of appreciable amixicile metabolism by liver microsome fractions is consistent with previous studies showing that the 5-nitro group of amixicile and NTZ is not susceptible to nitroreduction (15, 32). These findings support previous conclusions indicating that NTZ and amixicile were not substrates of the NsfB nitroreductase of E. coli (15).
TABLE 1.
Liver microsome fraction drug metabolisma
| Time (min) | Mean % of amixicile remaining ± SD |
|||||
|---|---|---|---|---|---|---|
| Human |
Rat |
Dog |
||||
| 1 μM | 10 μM | 1 μM | 10 μM | 1 μM | 10 μM | |
| 15 | 96.3 ± 4.3 | 96.5 ± 3.1 | 94.2 ± 1.8 | 98.6 ± 1.7 | 97.7 ± 1.1 | 102.6 ± 1.8 |
| 30 | 94.5 ± 1.7 | 101 ± 5.5 | 92.4 ± 3.5 | 98.1 ± 1.8 | 96.1 ± 1.1 | 101.2 ± 0.8 |
| 60 | 83.7 ± 8.8 | 99.9 ± 5.2 | 88.0 ± 0.4 | 96.5 ± 1.8 | 91.2 ± 3.6 | 97.7 ± 2.4 |
| 60 (HI)b | 96.1 ± 3.0 | 99.2 ± 3.5 | 100.5 ± 1.2 | 99.1 ± 1.6 | 96.3 ± 4.4 | 100.6 ± 3.4 |
Human, rat, and dog liver microsome fractions were prepared as detailed in the text. Amixicile was added at the concentrations indicated, and the percentages of the compound remaining and any metabolic products were determined by MS.
HI, heat-inactivated control.
Hepatocyte toxicity.
Hepatocyte viability was measured by the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) method, and the cytotoxicity results are depicted in Table 2. NTZ was the most toxic, with 99% of the monolayer destroyed at 25 μM (∼8 μg/ml). NTZ was not toxic at 10 μM. Amixicile exhibited little cytotoxicity over a concentration range of 10 to 200 μM, which was not considered significant since there was no dose-dependent increase in toxicity. A second analogue tested for cytotoxicity was VPC16a1011 in which the benzene propylamine was replaced with a chlorothiophene group (Fig. 1). VPC16a1011 was toxic to hepatocytes at 5 μM, though in previous studies, both VPC16a1011 and NTZ were shown to be less toxic to human foreskin cells (15).
TABLE 2.
Toxicity to human hepatocytes
| Treatment | Concn (μM) | Mean % viability ± SDa |
|---|---|---|
| DMSO | 0 | 100 ± 6 |
| NTZ | 10 | 98 ± 8 |
| NTZ | 25 | 1 ± 0 |
| Amixicile | 10 | 82 ± 14 |
| Amixicile | 25 | 81 ± 8 |
| Amixicile | 50 | 85 ± 10 |
| Amixicile | 100 | 85 ± 7 |
| Amixicile | 200 | 91 ± 7 |
| VPC16a1011 | 5 | 55 ± 2 |
| VPC16a1011 | 10 | 4 ± 1 |
| VPC16a1011 | 20 | 0 |
The percent viability of the monolayer was determined by the MTT assay at 24 h. The structures of the compounds used are presented in Fig. 1.
Mutation frequency.
We had previously shown that the parent drug NTZ did not induce mutations in E. coli (32). To ensure that amixicile was not mutagenic, E. coli strain CC103 was exposed to 32 μg/ml of amixicile (∼100 μM) for 6 h and then plated onto medium containing rifampin (Table 3). We also included NTZ and MTZ in this study. Nitrofurazone was included as a positive control, as we had previously shown the drug to induce mutations in E. coli (24). As a further control, we included VPC16b2031, which is a dinitrothiophene that showed weak activity in a nitroreductase assay. As shown in Table 3, the frequency of rifampin mutants was not significantly greater than that of controls upon exposure to amixicile or NTZ. In this assay, both VPC16b2031 and MTZ produced mutation frequency elevations (ca. 3- and 7-fold, respectively) and the mutation frequency was increased ∼20-fold by nitrofurazone. Neither amixicile nor NTZ is a substrate of nitroreductases, and we conclude that these compounds are not mutagenic.
TABLE 3.
Induced mutation to rifampin resistance in E. coli strain CC103
| Compound | Concn (μg/ml) | Avg mutation frequency/108 CFU ± SDa |
|---|---|---|
| Amixicile | 32 | 4.4 ± 2.7 |
| VPC162031 | 15 | 10 ± 3.5 |
| NTZ | 15 | 3.8 ± 3.1 |
| MTZ | 15 | 21 ± 1.4 |
| Nitrofurazone | 2.5 | 64 ± 32 |
| Control | 0 | 2.9 ± 1.3 |
Forward mutation to rifampin resistance was determined. Each value is the average of two independent experiments performed in triplicate.
Plasma binding studies.
Equilibrium dialysis was used to assess the extent of plasma binding by amixicile and VPC16a1011. As shown in Table 4, amixicile binding to plasma proteins was highest with human plasma (92% at 100 μg/ml) and somewhat less for dog and rat plasma. Relative aqueous solubility did not appear to be a factor, as the partition coefficient (cLogP) of amixicile is 1.1, while VPC16a1011, which binds plasma at >99%, is insoluble in water (cLogP, 3.0). The latter result is similar to that reported for NTZ (cLogP, 2.2), which is 99% bound to plasma proteins (30, 31), and it is known that high protein concentrations affect the potency of NTZ (MIC tests) against pathogens (33). We had previously reported that bovine serum albumin does not affect the MIC of amixicile (15).
TABLE 4.
Plasma binding studiesa
| Test compound and concn (μg/ml) | Mean % bound to plasma ± SD |
||
|---|---|---|---|
| Human | Dog | Rat | |
| Amixicile | |||
| 1 | 95.9 ± 0.437 | 84.3 ± 0.473 | 89.4 ± 0.945 |
| 10 | 94.5 ± 0.440 | 86.8 ± 0.208 | 87.5 ± 0.529 |
| 100 | 92.1 ± 0.640 | 79.0 ± 1.30 | 80.0 ± 0.954 |
| VPC16a1011 | |||
| 1 | 100 | 101 | 100 |
| 10 | 99.4 | 98.4 ± 0.577 | 99.2 |
| 50 | 99.8 | 98.8 ± 0.577 | 99.8 ± 0.058 |
Studies employed equilibrium dialysis. VPC16a1011 is a hydrophobic analogue of amixicile.
Absorption studies.
Bidirectional permeability was determined with established Caco-2 monolayers in Transwell chambers. The ratio of apparent A-B permeability (10−6 cm s−1) to B-A permeability was 3.6 at 1 μM and 3.3 at 10 μM, consistent with the likelihood that amixicile is subject to efflux. This possibility was supported by the inclusion of the efflux inhibitor ketoconazole, which increased A-B uptake. However, we noticed that the pH on the apical side was acidic at 6.0 and that on the basal side was 7.4. Since the pK for amixicile is ca. 6.2 (12), we considered the possibility that diffusion of the drug might be influenced by the anionic status. To test this possibility, the study was repeated with the apical pH set to 7.4 in HBSS. Under these conditions, the efflux ratio was 0.979. These studies also evaluated efflux via breast cancer resistance protein and P glycoprotein (P-gp) and found efflux ratios below 2, indicating that amixicile is not a substrate for efflux in the Caco-2 model. Further studies showed that amixicile is not a substrate of OAT1, OAT3, OCT2, OATP1B1, or OATP1B3 (data not presented). Taken together, these studies suggest that amixicile shows bidirectional permeability and is likely to be absorbed. These studies do raise the possibility that uptake of amixicile may be sensitive to changes in the local pH and to possible differences in absorption between the anion and base.
Dose range studies.
Previous dose range studies with mice had shown that amixicile was well tolerated when administered by the oral and intraperitoneal (i.p.) routes at 200 mg/kg (15). We noted in previous studies that many of the NTZ analogues, including NTZ, were lethal when injected by the i.p. route (34). To further evaluate the safety of amixicile, dose range studies were performed with rats. Amixicile was well tolerated at the highest concentrations (300 mg/kg) administered by oral gavage. The animals were observed for 48 h, and there were no changes in animal behavior or activity compared to that of controls.
PKs of amixicile in rats and mice.
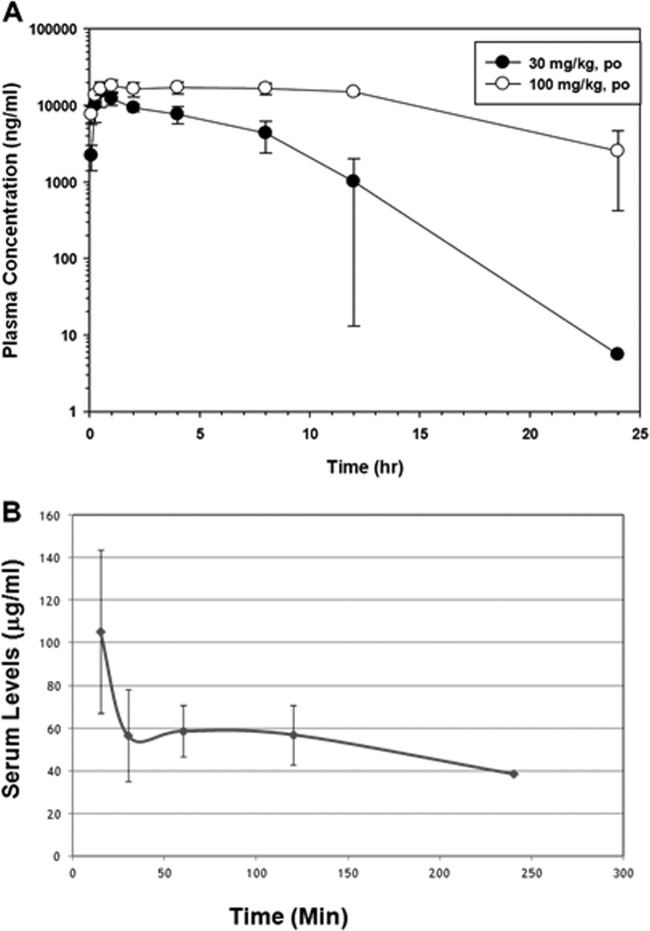
Plasma PKs were determined in rats following a single dose of amixicile (30 or 100 mg/kg) by oral gavage (Fig. 2A). The Tmax was 30 min to 1 h, and amixicile was quantifiable at 24 h postadministration. Plasma drug levels after the 100-mg/kg dose remained high throughout the study (Fig. 2A, open circles). The mean Cmax values for the two doses were 15.5 and 19.7 μg/ml, respectively, and were not statistically significantly different (Table 5). However, the two drug concentrations showed 3-fold differences in AUC values (79.2 versus 328 h · μg/ml) and t1/2 values (1.5 versus 5.6 h). These results may suggest that amixicile absorption decreases with the drug concentration or elimination might be slowed by saturation of clearance mechanisms. When similar plasma PKs were determined in mice receiving a single dose of 200 mg/kg in PBS, we noted that amixicile was more efficiently absorbed, with a Cmax of 179 μg/ml within 15 min, followed by a steady level between 40 and 60 μg/ml for 4 h (Fig. 2B). Since amixicile was suspended in 1% methylcellulose for oral gavage in the rat studies, we believe that the 100-mg/kg drug dose in the rat likely limited absorption by creating a time-release profile that was not seen in the mouse study in the absence of a carrier. In both studies, the high plasma drug levels are consistent with good absorption of amixicile and when the Cmax/MIC ratios are computed for the various susceptible pathogens, ratios above 150 would be predictive of clinical success. Consistent with good amixicile absorption metrics, the level of amixicile in the feces of mice receiving 200 mg/kg was below the limit of detection (∼0.1 μg/ml) compared to a standard curve produced with amixicile-spiked fecal samples. Routes of excretion were not investigated in this study.
FIG 2.
Time course of plasma amixicile concentrations following a single oral dose. (A) Male rats received amixicile at 30 or 100 mg/kg in methylcellulose by gavage. Each datum point represents the mean plasma amixicile concentration of up to three rats ± the SD. (B) Male mice received amixicile at 200 mg/kg in PBS by oral gavage, and at each time point, five mice were sacrificed for blood collection. The mean and SD are presented. The data collected were used to generate the PK information presented in Table 5.
TABLE 5.
PK data from rat single-dose experimentsa
| Amixicile dose (mg/kg) and rat no. or parameter | Cmax (μg/ml) | Tmax (h) | AUClast (h · μg/ml) | AUCinf (h · μg/ml) | t1/2 (h) | V/F (liters/kg) | CL/F (ml/h/kg) |
|---|---|---|---|---|---|---|---|
| 30 | |||||||
| 15 | 20.4 | 0.50 | 107 | 107 | 1.54 | 0.621 | 279 |
| 16 | 14.8 | 0.50 | 70.6 | 70.6 | 1.65 | 1.01 | 425 |
| 17 | 11.2 | 1.00 | 59.6 | 59.8 | 1.39 | 1.00 | 501 |
| Mean ± SD | 15.5 ± 4.6 | 0.67 ± 0.29 | 79.2 ± 25.0 | 79.3 ± 24.9 | 1.53 ± 0.13 | 0.879 ± 0.224 | 402 ± 113 |
| 100 | |||||||
| 18 | 18.7 | 8.0 | 298 | 331 | 6.50 | 2.84 | 303 |
| 19 | 21.8 | 1.00 | 323 | 323 | 2.04 | 0.91 | 309 |
| 20 | 18.6 | 0.50 | 283 | 330 | 8.25 | 3.62 | 303 |
| Mean ± SD | 19.7 ± 1.8 | 3.17 ± 4.19 | 301 ± 20 | 328 ± 4 | 5.60 ± 3.22 | 2.45 ± 1.39 | 305 ± 4 |
Single doses of amixicile (30 and 100 mg/kg) were administered to male rats. The results for individual rats are depicted with the means and SDs.
Therapeutic bioavailability.
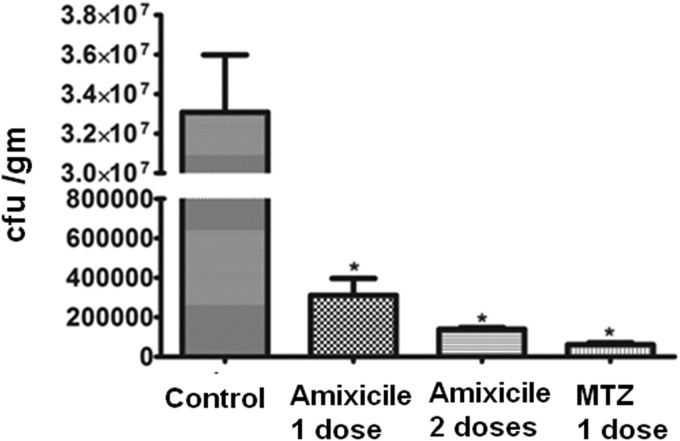
To test whether plasma protein binding might affect the bioavailability and therefore the therapeutic efficacy of amixicile, we used a mouse model of H. pylori infection where plasma drug levels above the MIC are required for eradication. Both human and animal studies have established that NTZ is not an effective therapy for H. pylori infections, despite excellent in vitro efficacy (15, 35). In contrast, MTZ is an effective therapeutic for eradication of H. pylori infection in mice but not as a monotherapy in humans or when strains are resistant to MTZ (27, 36). As shown in Fig. 3, a single 20-mg/kg dose of amixicile was sufficient to produce a >2-log decrease in bacterial numbers (CFU/g of stomach tissue), in comparison with a similar dose of MTZ, which produced a nearly 3-log decrease in bacterial numbers. Our studies showed that two 20-mg/kg doses of amixicile was nearly equivalent to the single dose of MTZ. These results show that plasma protein binding noted for amixicile does not appear to affect bioavailability or potency against H. pylori in the mouse model. On the basis of the Cmax determined in mice receiving amixicile at 200 mg/kg (179 μg/ml), the estimated levels in serum after a 20-mg/kg dose of amixicile (>17 μg/ml) would be in excess of the MIC for the SS1 strain of H. pylori (<1.0 μg/ml) and for C. difficile determined previously (15). We conclude that plasma protein binding does not affect the bioavailability or therapeutic efficacy of amixicile.
FIG 3.
Therapeutic efficacy of amixicile in mice. Mice were infected with the SS1 strain of H. pylori, and following 2 weeks to enable the infection to manifest itself, mice were divided into groups with one serving as an untreated control and the other receiving one or two doses of amixicile or MTZ of 20 mg/kg/day. One week later, animals were sacrificed and stomach material was collected for bacterial enumeration. The data are reported as CFU/g of stomach material. The mean and SD from five animals are presented. Asterisks indicate statistical significance (P < 0.001).
Effect of amixicile on resident mouse flora.
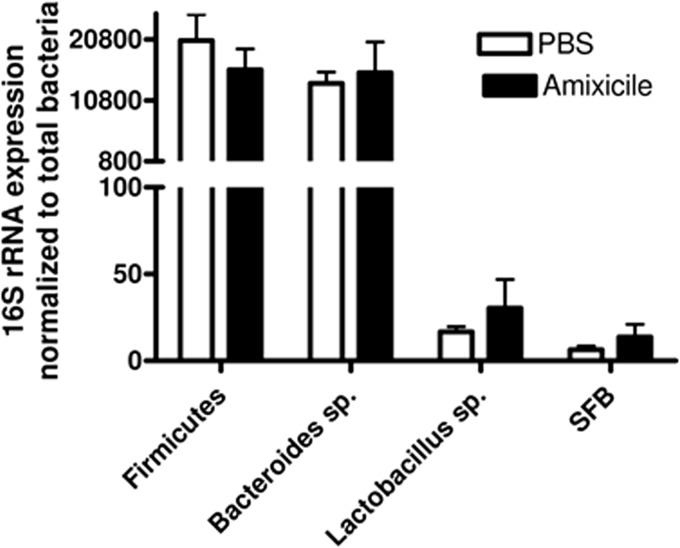
In vitro studies have shown that NTZ exhibits broad-spectrum inhibitory activity against strictly anaerobic bacteria, and our previous studies have shown that amixicile was particularly potent against Bacteroides fragilis in vitro (15). To evaluate the potential that amixicile might disrupt resident anaerobic flora, we randomized 15 mice of various ages into two groups, one group receiving amixicile at 30 mg/kg/day and one receiving PBS for 3 days. On day 4, the Bacteroides, segmented filamentous bacterium, firmicutes, and lactobacillus populations were analyzed by qPCR. As shown in Fig. 4, there were no significant changes in these gut microflora populations over controls. These findings are consistent with our findings that amixicile does not accumulate in detectable concentrations in the feces of mice (<0.1 μg/ml).
FIG 4.
Effect of amixicile on mouse gut microflora. Eight mice per group received either PBS or amixicile at 30 mg/kg/day by oral gavage. Primers for Firmicutes, Bacteroides species, Lactobacillus species, and segmented filamentous bacteria (SFB) were quantified by 16S rRNA expression that had been normalized to the total bacteria. The mean and SD are presented.
DISCUSSION
Amixicile, a water-soluble derivative of NTZ, selectively interferes with the biological function of thiamine pyrophosphate (TPP), the vitamin B1 cofactor of PFOR and related α-ketoacid:ferredoxin oxidoreductases (12). These essential enzymes are ubiquitous in obligate anaerobic bacteria, human intestinal parasites, archaea, and members of the epsilonproteobacteria (12, 13). Other TPP-containing enzymes, such as pyruvate dehydrogenase, are not inhibited by amixicile. Conceptually, drugs that target the function of vitamins, themselves small molecules, are unlikely to cause mutation to drug resistance without losing biological function. Such therapeutics might revolutionize treatment strategies for chronic infections that often require extended periods of antimicrobial intervention.
Amixicile demonstrated improved selectivity for the PFOR drug target and had lost many of the off-target activities attributed to NTZ, including inhibition of chaperone-usher pilin biogenesis by E. coli and biofilm production by Staphylococcus epidermidis (15, 37, 38). Perhaps most importantly, amixicile had lost the intrinsic cytotoxicity to immortalized cell lines noted for NTZ (14, 39). Studies presented herein indicate that amixicile was well tolerated by mice and rats receiving maximum doses (200 or 300 mg/kg, respectively) and that in vitro assays revealed no cytotoxicity to hepatocytes or measurable metabolism by human, dog, or rat liver microsome fractions. Amixicile did not increase the mutation frequency in E. coli, as determined by forward mutation to rifampin resistance. Bidirectional permeability studies with Caco-2 cell monolayers showed amixicile to be readily translocated in both directions, though a low pH (6.0 versus 7.4) appeared to affect apical uptake. Our studies showed that amixicile was not a substrate or an inhibitor of OAT1, OCT2, and OATP1B3 or breast cancer resistance protein and P-gp transporters. CYP induction studies suggested that amixicile might induce CYP1A2 and CYP3A4 at the highest concentrations tested and showed little inhibitory activity against these enzymes. Our concerns that plasma protein binding might also affect the biological activity of amixicile (a noted problem with NTZ) seemed to be mitigated by MIC-based studies where additions of bovine serum albumin to MIC assays had no effect (15) and by demonstrating therapeutic efficacy against H. pylori in a mouse infection model. Consistent with efficient uptake of amixicile, fecal drug levels were below the level of detectability (<0.1 μg/ml). Finally, the administration of amixicile to healthy mice did not alter the levels of selected gut bacterial species, including Bacteroides species, which MIC tests had shown to be highly susceptible to this group of therapeutics (15, 19, 20). Taken together, the preclinical studies found no mitigating safety or toxicological concerns about amixicile that might preclude further development.
With the exception of MTZ, most of the therapeutics used to treat CDI are minimally absorbed, including vancomycin, fidaxomicin, NTZ, and experimental drugs like LFF571 (6, 40). This has led to the notion that therapeutics that concentrate in the gut and retain potency should be much more efficacious than systemic therapeutics. Presumably, systemic therapeutics require higher doses to achieve similar luminal concentrations. However, in a mouse acute CDI model, systemic amixicile showed equivalence to vancomycin and fidaxomicin and superiority to NTZ at 5 days and superiority to all by day 14 postinfection (15). Importantly, with amixicile and NTZ, there was no relapse of CDI in any of the surviving treated animals, whereas recurrence was observed in 70 to 80% of the animals receiving similar doses of fidaxomicin or vancomycin, respectively (6, 15). The apparent equivalence of a systemic therapeutic to minimally absorbed therapeutics raises more fundamental questions regarding the nature of an infection, its location, and the biological action of the therapeutic at that site (i.e., luminal or mucosal). Our studies suggest that C. difficile colonizes the intestinal mucosa and promotes local inflammation and that elimination of these organisms by treatment with amixicile leads to resolution of disease. This view is compatible with the notion that repopulation of the site by resident flora, naturally or through probiotics or fecal transplants, protects against reinfection (10, 11, 23).
To explore the nature of antibiotic action further, we tested amixicile in a mouse model of H. pylori infection. We had previously determined that amixicile and NTZ were inhibitory to H. pylori and C. jejuni in the 0.5- to 1-μg/ml range by MIC tests (14, 15). Moreover, we had tested NTZ for efficacy against H. pylori in a C57BL/6 mouse model and as in human studies, NTZ proved ineffective at eradicating the infection (35). In the mouse model, H. pylori resides in the gastric mucosa but does not appreciably invade gastric epithelial cells, as demonstrated by hematoxylin-and-eosin staining of stomach tissue (27). All therapeutics active against H. pylori (e.g., MTZ, amoxicillin, tetracycline, and clarithromycin) are systemic and must diffuse through the gastric epithelium in order to reach the bacteria. In general, minimally absorbed drugs are ineffective against H. pylori (36). Thus, drugs that concentrate in areas of inflammation (serum leakage) are likely to be the most effective against H. pylori, such as MTZ, which also tends to concentrate in gastric acid (41). Our group and others have shown that MTZ is the most effective monotherapy for the treatment of mice colonized by strains of H. pylori that are susceptible to MTZ (42, 43). The remarkable efficacy of amixicile as a monotherapy was unexpected, since neither amoxicillin nor clarithromycin, a mainline therapeutic used for the treatment of H. pylori in humans, shows any efficacy as a monotherapy in this model (42, 43). On the basis of these results, we suggest that amixicile most likely concentrates in areas of inflammation associated with active infections. By not accumulating in the colon, it avoids the collateral damage to the resident gut microflora that is problematic with all minimally absorbed antimicrobials. While luminal C. difficile might also be spared by systemic therapeutics like amixicile, both washout and competition with repopulated gut microflora as suggested previously (15) might mitigate a relapse of CDI.
While amixicile shows good efficacy against H. pylori in the mouse model, we have previously reported that some MTZ-resistant (MTZr) strains exhibit cross-resistance to NTZ and consequently to amixicile in vitro (32). This includes H. pylori strains 1061rdxAfrxA and G27rdxA (MIC of NTZ, 16 μg/ml), and resistance is not due to mutations of pforGDAB, since PFOR enzyme activity in cell extracts was essentially wild type and could be inhibited by amixicile (unpublished data). These strains lack a functional RdxA NAD(P)H MTZ-reducing nitroreductase that also exhibits potent NADPH oxidase activity (44). There is accumulating evidence now to suggest that nitroreductases are potent scavengers of cytoplasmic oxygen and are part of a cellular redox system that maintains an anoxic cytoplasm (44). Accordingly, loss of RdxA function contributes to oxidative stress, leading to activation of compensatory metabolic pathways that appear to be controlled by HsrA, a homeostatic oxidative stress regulator (45). While we do not completely understand the underlying mechanisms that render the PFOR drug target less essential, the phenomenon might be uniquely limited to H. pylori, as MTZr strains of C. jejuni retain susceptibility to amixicile (14, 15). In general, MTZ resistance is rare in strictly anaerobic bacteria and parasites. However, a few reports of resistance in C. difficile have appeared (46, 47), but since the clostridia express multiple α-ketoacid:ferredoxin oxidoreductases, resistance would likely require additional metabolic changes. Similarly, in Bacteroides species, MTZr strains often harbor nim genes whose products nitroreduce MTZ to nontoxic ammonia (48, 49), so cross-resistance with amixicile would be unlikely since the nitro groups of amixicile and NTZ are not susceptible to nitroreduction (15, 32). It is important to emphasize that MTZ is both mutagenic and selective for resistance, not only to MTZ but to other antimicrobials used therapeutically (24). While amixicile and NTZ have the same clinical spectrum as MTZ (8, 12, 14, 15, 19, 20, 31), both their modes of action and their drug targets are different.
PK studies show that amixicile is readily absorbed and Cmaxs of 15 and 20 μg/ml were obtained in rats receiving 30 and 100 mg/kg, respectively, and a Cmax of 179 μg/ml was obtained in mice receiving a single dose of 200 mg/kg. As shown in Fig. 2A, rats receiving 100 mg/kg did not show the expected concentration-dependent 3-fold increase in Cmax (assuming rapid absorption) but did show this increase when AUCs were compared. We interpret these results to suggest that the methylcellulose carrier caused a time release of amixicile at the higher concentration, which also extended the t1/2. This was not observed in the mouse study, where the drug was administered in PBS in the absence of methylcellulose. This knowledge might be useful in the future when formulations are optimized for humans. Taken together, the PK studies show that amixicile is readily absorbed and disseminated. On the basis of plasma drug levels in both the rat and mouse studies, amixicile readily achieved levels well in excess of the MIC for C. difficile and H. pylori. It is also likely that plasma amixicile levels of >20 μg/ml might show efficacy against MTZr strains of H. pylori in the mouse model (MIC, 16 μg/ml). Studies are in progress to test this possibility.
In summary, preclinical studies indicate that amixicile, which is in development for the treatment of CDI, is well tolerated by both rats and mice, is not appreciably metabolized by liver microsome fractions, and is not cytotoxic for hepatocytes or human foreskin cells (15). PK studies indicate that the drug is efficiently absorbed and is below detectable levels in feces samples. While amixicile, like NTZ, shows broad-spectrum action against strictly anaerobic bacteria, including Bacteroides fragilis in vitro, amixicile did not affect gut bacterial populations of susceptible anaerobes, including species of Bacteroides. These studies also confirmed that bacteria lacking the PFOR drug target (lactobacilli) are also unaffected by amixicile. In this regard, the key component microbes of probiotics (lactobacilli and bifidobacteria) do not contain the PFOR drug target and would not be affected by amixicile. While cross-resistance to MTZ might limit the use of amixicile for the treatment of H. pylori in humans, further efficacy studies with resistant strains that include combination therapies and a proton pump inhibitor might overcome resistance. Finally, our studies strongly suggest that a focus on minimally absorbed therapeutics to treat CDI should be reconsidered. Clearly, a systemic therapeutic that concentrates in areas of active infection, spares resident flora, and resists mutation-based drug resistance has potential application in treatment not only of CDI but for other infections where anaerobic microorganisms are involved, such as periodontal disease, Crohn's disease, inflammatory bowel disease, and a range of parasitic infections caused by C. parvum, G. lamblia, and Trichomonas vaginalis.
ACKNOWLEDGMENTS
This project was funded by National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services grant 1R21 AI111604-01 to P.S.H. and contract HHSN2722011000221.
Footnotes
Published ahead of print 2 June 2014
REFERENCES
- 1.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect. Control Hosp. Epidemiol. 31:431–455. 10.1086/651706 [DOI] [PubMed] [Google Scholar]
- 2.Surawicz CM, Alexander J. 2011. Treatment of refractory and recurrent Clostridium difficile infection. Nat. Rev. Gastroenterol. Hepatol. 8:330–339. 10.1038/nrgastro.2011.59 [DOI] [PubMed] [Google Scholar]
- 3.Hensgens MP, Goorhuis A, Dekkers OM, van Benthem BH, Kuijper EJ. 2013. All-cause and disease-specific mortality in hospitalized patients with Clostridium difficile infection: a multicenter cohort study. Clin. Infect. Dis. 56:1108–1116. 10.1093/cid/cis1209 [DOI] [PubMed] [Google Scholar]
- 4.See I, Mu Y, Cohen J, Beldavs ZG, Winston LG, Dumyati G, Holzbauer S, Dunn J, Farley MM, Lyons C, Johnston H, Phipps E, Perlmutter R, Anderson L, Gerding DN, Lessa FC. 2014. NAP1 strain type predicts outcomes from Clostridium difficile infection. Clin. Infect. Dis. 58:1394–1400. 10.1093/cid/ciu125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ritter AS, Petri WA., Jr 2013. New developments in chemotherapeutic options for Clostridium difficile colitis. Curr. Opin. Infect. Dis. 26:461–470. 10.1097/QCO.0b013e328363456e [DOI] [PubMed] [Google Scholar]
- 6.Warren CA, van Opstal EJ, Riggins MS, Li Y, Moore JH, Kolling GL, Guerrant RL, Hoffman PS. 2013. Vancomycin treatment's association with delayed intestinal tissue injury, clostridial overgrowth, and recurrence of Clostridium difficile infection in mice. Antimicrob. Agents Chemother. 57:689–696. 10.1128/AAC.00877-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buffie CG, Pamer EG. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13:790–801. 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musher DM, Logan N, Mehendiratta V, Melgarejo NA, Garud S, Hamill RJ. 2007. Clostridium difficile colitis that fails conventional metronidazole therapy: response to nitazoxanide. J. Antimicrob. Chemother. 59:705–710. 10.1093/jac/dkl553 [DOI] [PubMed] [Google Scholar]
- 9.Goldstein EJ, Citron DM, Sears P, Babakhani F, Sambol SP, Gerding DN. 2011. Comparative susceptibilities to fidaxomicin (OPT-80) of isolates collected at baseline, recurrence, and failure from patients in two phase III trials of fidaxomicin against Clostridium difficile infection. Antimicrob. Agents Chemother. 55:5194–5199. 10.1128/AAC.00625-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore T, Rodriguez A, Bakken JS. 2014. Fecal microbiota transplantation: a practical update for the infectious disease specialist. Clin. Infect. Dis. 58:541–545. 10.1093/cid/cit950 [DOI] [PubMed] [Google Scholar]
- 11.Johnston BC, Ma SS, Goldenberg JZ, Thorlund K, Vandvik PO, Loeb M, Guyatt GH. 2012. Probiotics for the prevention of Clostridium difficile-associated diarrhea: a systematic review and meta-analysis. Ann. Intern. Med. 157:878–888. 10.7326/0003-4819-157-12-201212180-00563 [DOI] [PubMed] [Google Scholar]
- 12.Hoffman PS, Sission G, Croxen MA, Welch K, Harman WD, Cremades N, Morash MG. 2007. Antiparasitic drug nitazoxanide inhibits the pyruvate oxidoreductases of Helicobacter pylori and selected anaerobic bacteria and parasites, and Campylobacter jejuni. Antimicrob. Agents Chemother. 51:868–876. 10.1128/AAC.01159-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.St Maurice M, Cremades N, Croxen MA, Sisson G, Sancho J, Hoffman PS. 2007. Flavodoxin:quinone reductase (FqrB): a redox partner of pyruvate:ferredoxin oxidoreductase that reversibly couples pyruvate oxidation to NADPH production in Helicobacter pylori and Campylobacter jejuni. J. Bacteriol. 189:4764–4773. 10.1128/JB.00287-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ballard TE, Wang X, Olekhnovich I, Koerner T, Seymour C, Salamoun J, Warthan M, Hoffman PS, Macdonald TL. 2011. Synthesis and antimicrobial evaluation of nitazoxanide-based analogues: identification of selective and broad spectrum activity. ChemMedChem. 6:362–377. 10.1002/cmdc.201000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warren CA, van Opstal E, Ballard TE, Kennedy A, Wang X, Riggins M, Olekhnovich I, Warthan M, Kolling GL, Guerrant RL, Macdonald TL, Hoffman PS. 2012. Amixicile, a novel inhibitor of pyruvate:ferredoxin oxidoreductase, shows efficacy against Clostridium difficile in a mouse infection model. Antimicrob. Agents Chemother. 56:4103–4111. 10.1128/AAC.00360-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilles HM, Hoffman PS. 2002. Treatment of intestinal parasitic infections: a review of nitazoxanide. Trends Parasitol. 18:95–97. 10.1016/S1471-4922(01)02205-X [DOI] [PubMed] [Google Scholar]
- 17.Musher DM, Logan N, Bressler AM, Johnson DP, Rossignol JF. 2009. Nitazoxanide versus vancomycin in Clostridium difficile infection: a randomized, double-blind study. Clin. Infect. Dis. 48(4):e41–46. 10.1086/596552 [DOI] [PubMed] [Google Scholar]
- 18.McVay CS, Rolfe RD. 2000. In vitro and in vivo activities of nitazoxanide against Clostridium difficile. Antimicrob. Agents Chemother. 44:2254–2258. 10.1128/AAC.44.9.2254-2258.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pankuch GA, Appelbaum PC. 2006. Activities of tizoxanide and nitazoxanide compared to those of five other thiazolides and three other agents against anaerobic species. Antimicrob. Agents Chemother. 50:1112–1117. 10.1128/AAC.50.3.1112-1117.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finegold SM, Molitoris D, Väisänen ML. 2009. Study of the in vitro activities of rifaximin and comparator agents against 536 anaerobic intestinal bacteria from the perspective of potential utility in pathology involving bowel flora. Antimicrob. Agents Chemother. 53:281–286. 10.1128/AAC.00441-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein EJ, Citron DM, Tyrrell KL, Merriam CV. 2013. Comparative in vitro activities of SMT19969, a new antimicrobial agent, against Clostridium difficile and 350 gram-positive and Gram-negative aerobic and anaerobic intestinal flora isolates. Antimicrob. Agents Chemother. 57:4872–4876. 10.1128/AAC.01136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skraban J, Dzeroski S, Zenko B, Mongus D, Gangl S, Rupnik M. 2013. Gut microbiota patterns associated with colonization of different Clostridium difficile ribotypes. PLoS One 8(2):e58005. 10.1371/journal.pone.0058005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abujamel T, Cadnum JL, Jury LA, Sunkesula VC, Kundrapu S, Jump RL, Stintzi AC, Donskey CJ. 2013. Defining the vulnerable period for re-establishment of Clostridium difficile colonization after treatment of C. difficile infection with oral vancomycin or metronidazole. PLoS One 8(10):e76269. 10.1371/journal.pone.0076269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sisson G, Jeong JY, Goodwin A, Bryden L, Rossler N, Lim-Morrison S, Raudonikiene A, Berg DE, Hoffman PS. 2000. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori RdxA(+) (nitroreductase) gene. J. Bacteriol. 182:5091–5096. 10.1128/JB.182.18.5091-5096.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carbo A, Bassaganya-Riera J, Pedragosa M, Viladomiu M, Marathe M, Eubank S, Wendelsdorf K, Bisset K, Hoops S, Deng X, Alam M, Kronsteiner B, Mei Y, Hontecillas R. 2013. Predictive computational modeling of the mucosal immune responses during Helicobacter pylori infection. PLoS One 8(9):e73365. 10.1371/journal.pone.0073365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bassaganya-Riera J, Dominguez-Bello MG, Kronsteiner B, Carbo A, Lu P, Viladomiu M, Pedragosa M, Zhang X, Sobral BW, Mane SP, Mohapatra SK, Horne WT, Guri AJ, Groeschl M, Lopez-Velasco G, Hontecillas R. 2012. Helicobacter pylori colonization ameliorates glucose homeostasis in mice through a PPAR γ-dependent mechanism. PLoS One 7(11):e50069. 10.1371/journal.pone.0050069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman PS, Vats N, Hutchison D, Butler J, Chisholm K, Sisson G, Raudonikiene A, Marshall JS, Veldhuyzen van Zanten SJ. 2003. Development of an interleukin-12-deficient mouse model that is permissive for colonization by a motile KE26695 strain of Helicobacter pylori. Infect. Immun. 71:2534–2541. 10.1128/IAI.71.5.2534-2541.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harley IT, Giles DA, Pfluger PT, Burgess SL, Walters S, Hembree J, Raver C, Rewerts CL, Downey J, Flick LM, Stankiewicz TE, McAlees JW, Wills-Karp M, Balfour Sartor R, Divanovic S, Tschöp MH, Karp CL. 2013. Differential colonization with segmented filamentous bacteria and Lactobacillus murinus do not drive divergent development of diet-induced obesity in C57BL/6 mice. Mol. Metab. 2:171–183. 10.1016/j.molmet.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. 2008. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect. Immun. 76:907–915. 10.1128/IAI.01432-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stockis A, Deroubaix X, Lins R, Jeanbaptiste B, Calderon P, Rossignol JF. 1996. Pharmacokinetics of nitazoxanide after single oral dose administration in 6 healthy volunteers. Int. J. Clin. Pharmacol. Ther. 34:349–351 [PubMed] [Google Scholar]
- 31.Hemphill A, Mueller J, Esposito M. 2006. Nitazoxanide, a broad-spectrum thiazolide anti-infective agent for the treatment of gastrointestinal infections. Expert Opin. Pharmacother. 7:953–964. 10.1517/14656566.7.7.953 [DOI] [PubMed] [Google Scholar]
- 32.Sisson G, Goodwin A, Raudonikiene A, Hughes NJ, Mukhopadhyay AK, Berg DE, Hoffman PS. 2002. Enzymes associated with reductive activation and action of nitazoxanide, nitrofurans, and metronidazole in Helicobacter pylori. Antimicrob. Agents Chemother. 46:2116–2123. 10.1128/AAC.46.7.2116-2123.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shigyo K, Ocheretina O, Merveille YM, Johnson WD, Pape JW, Nathan CF, Fitzgerald DW. 2013. Efficacy of nitazoxanide against clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57:2834–2837. 10.1128/AAC.02542-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Debache K, Guionaud C, Kropf C, Boykin D, Stephens CE, Hemphill A. 2011. Experimental treatment of Neospora caninum-infected mice with the arylimidamide DB750 and the thiazolide nitazoxanide. Exp. Parasitol. 129:95–100. 10.1016/j.exppara.2011.07.010 [DOI] [PubMed] [Google Scholar]
- 35.Guttner Y, Windsor HM, Viiala CH, Dusci L, Marshall BJ. 2003. Nitazoxanide in treatment of Helicobacter pylori: a clinical and in vitro study. Antimicrob. Agents Chemother. 47:3780–3783. 10.1128/AAC.47.12.3780-3783.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu JY, Liou JM, Graham DY. 2014. Evidence-based recommendations for successful Helicobacter pylori treatment. Expert Rev. Gastroenterol. Hepatol. 8:21–28. 10.1586/17474124.2014.859522 [DOI] [PubMed] [Google Scholar]
- 37.Shamir ER, Warthan M, Brown SP, Nataro JP, Guerrant RL, Hoffman PS. 2010. Nitazoxanide inhibits biofilm production and hemagglutination by enteroaggregative Escherichia coli strains by blocking assembly of AafA fimbriae. Antimicrob. Agents Chemother. 54:1526–1533. 10.1128/AAC.01279-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tchouaffi-Nana F, Ballard TE, Cary CH, Macdonald TL, Sifri CD, Hoffman PS. 2010. Nitazoxanide inhibits biofilm formation by Staphylococcus epidermidis by blocking accumulation on surfaces. Antimicrob. Agents Chemother. 54:2767–2774. 10.1128/AAC.00901-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller J, Sidler D, Nachbur U, Wastling J, Brunner T, Hemphill A. 2008. Thiazolides inhibit growth and induce glutathione-S-transferase Pi (GSTP1)-dependent cell death in human colon cancer cells. Int. J. Cancer 123:1797–1806. 10.1002/ijc.23755 [DOI] [PubMed] [Google Scholar]
- 40.Citron DM, Tyrrell KL, Merriam CV, Goldstein EJ. 2012. Comparative in vitro activities of LFF571 against Clostridium difficile and 630 other intestinal strains of aerobic and anaerobic bacteria. Antimicrob. Agents Chemother. 56:2493–2503. 10.1128/AAC.06305-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goddard AF, Jessa MJ, Barrett DA, Shaw PN, Idström JP, Cederberg C, Spiller RC. 1996. Effect of omeprazole on the distribution of metronidazole, amoxicillin, and clarithromycin in human gastric juice. Gastroenterology 111:358–367. 10.1053/gast.1996.v111.pm8690200 [DOI] [PubMed] [Google Scholar]
- 42.Sizemore CF, Quispe JD, Amsler KM, Modzelewski TC, Merrill JJ, Stevenson DA, Foster LA, Slee AM. 2002. Effects of metronidazole, tetracycline, and bismuth-metronidazole-tetracycline triple therapy in the Helicobacter pylori SS1 mouse model after 1 day of dosing: development of an H. pylori lead selection model. Antimicrob. Agents Chemother. 46:1435–1440. 10.1128/AAC.46.5.1435-1440.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Zanten SJ, Kolesnikow T, Leung V, O'Rourke JL, Lee A. 2003. Gastric transitional zones, areas where Helicobacter treatment fails: results of a treatment trial using the Sydney strain mouse model. Antimicrob. Agents Chemother. 47:2249–2255. 10.1128/AAC.47.7.2249-2255.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olekhnovich IN, Goodwin A, Hoffman PS. 2009. Characterization of the NAD(P)H oxidase and metronidazole reductase activities of the RdxA nitroreductase of Helicobacter pylori. FEBS J. 276:3354–3364. 10.1111/j.1742-4658.2009.07060.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olekhnovich IN, Vitko S, Valliere M, Hoffman PS. 2014. Response to metronidazole and oxidative stress is mediated through homeostatic regulator HsrA (HP1043) in Helicobacter pylori. J. Bacteriol. 196:729–739. 10.1128/JB.01047-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lynch T, Chong P, Zhang J, Hizon R, Du T, Graham MR, Beniac DR, Booth TF, Kibsey P, Miller M, Gravel D, Mulvey MR, Canadian Nosocomial Infection Surveillance Program (CNISP) 2013. Characterization of a stable, metronidazole-resistant Clostridium difficile clinical isolate. PLoS One 8(1):e53757. 10.1371/journal.pone.0053757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chong PM, Lynch T, McCorrister S, Kibsey P, Miller M, Gravel D, Westmacott GR, Mulvey MR, Canadian Nosocomial Infection Surveillance Program (CNISP) 2014. Proteomic analysis of a NAP1 Clostridium difficile clinical isolate resistant to metronidazole. PLoS One 9(1):e82622. 10.1371/journal.pone.0082622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Löfmark S, Fang H, Hedberg M, Edlund C. 2005. Inducible metronidazole resistance and nim genes in clinical Bacteroides fragilis group isolates. Antimicrob. Agents Chemother. 49:1253–1256. 10.1128/AAC.49.3.1253-1256.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leitsch D, Sóki J, Kolarich D, Urbán E, Nagy E. 2014. A study on Nim expression in Bacteroides fragilis. Microbiology 160:616–622. 10.1099/mic.0.074807-0 [DOI] [PMC free article] [PubMed] [Google Scholar]