Abstract
Within wounds, microorganisms predominantly exist as biofilms. Biofilms are associated with chronic infections and represent a tremendous clinical challenge. As antibiotics are often ineffective against biofilms, use of dispersal agents as adjunctive, topical therapies for the treatment of wound infections involving biofilms has gained interest. We evaluated in vitro the dispersive activity of d-amino acids (d-AAs) on biofilms from clinical wound isolates of Staphylococcus aureus and Pseudomonas aeruginosa; moreover, we determined whether combinations of d-AAs and antibiotics (clindamycin, cefazolin, oxacillin, rifampin, and vancomycin for S. aureus and amikacin, colistin, ciprofloxacin, imipenem, and ceftazidime for P. aeruginosa) enhance activity against biofilms. d-Met, d-Phe, and d-Trp at concentrations of ≥5 mM effectively dispersed preformed biofilms of S. aureus and P. aeruginosa clinical isolates, an effect that was enhanced when they were combined as an equimolar mixture (d-Met/d-Phe/d-Trp). When combined with d-AAs, the activity of rifampin was significantly enhanced against biofilms of clinical isolates of S. aureus, as indicated by a reduction in the minimum biofilm inhibitory concentration (MBIC) (from 32 to 8 μg/ml) and a >2-log reduction of viable biofilm bacteria compared to treatment with antibiotic alone. The addition of d-AAs was also observed to enhance the activity of colistin and ciprofloxacin against biofilms of P. aeruginosa, reducing the observed MBIC and the number of viable bacteria by >2 logs and 1 log at 64 and 32 μg/ml in contrast to antibiotics alone. These findings indicate that the biofilm dispersal activity of d-AAs may represent an effective strategy, in combination with antimicrobials, to release bacteria from biofilms, subsequently enhancing antimicrobial activity.
INTRODUCTION
Chronic wounds are common in individuals with underlying medical conditions, such as diabetes mellitus, as well as in wounds resulting from traumatic injury, and significantly contribute to patient morbidity (1–3). A major factor contributing to the development of chronic wounds is colonization and subsequent infection by microorganisms. Recent studies evaluating the wound microbiota of chronic wounds of various etiologies have demonstrated that chronic wounds are often colonized by multiple bacterial species, of which Staphylococcus spp. and Pseudomonas spp. are two of the most commonly isolated organisms (4, 5). Within wounds, bacteria predominantly adopt a surface-attached mode of growth known as a biofilm. In brief, biofilms are an association of single or multiple microbial species surrounded by a self-produced, extracellular polymeric matrix, constituting a protected mode of growth (6–8). In contrast to their planktonic counterparts, biofilm-derived bacteria have a distinctive phenotype in regard to metabolic activity and gene expression, conferring an inherent resistance to antimicrobial agents as well as mechanisms of host clearance, making the treatment of biofilm-associated infections extremely difficult (9, 10).
The presence of bacterial biofilms within wounds is cited as a significant factor contributing to the chronicity and pathogenesis of wound infections (7, 11–13). For both Staphylococcus aureus and Pseudomonas aeruginosa, biofilm formation has been extensively documented in vitro and in vivo within chronic wounds (12, 14, 15). Importantly, the development and establishment of biofilms by both of these wound pathogens have been shown to directly impede wound healing and contribute to the development of chronic wounds (16–19). Given the importance of the biofilm phenotype in wound pathogenesis and the limitations of conventional antimicrobials against this phenotype, new strategies are needed for the treatment of chronic wounds.
Biofilm dispersal is a highly coordinated process, dependent on multiple factors, including cell density as well as responses to environmental cues, such as quorum-sensing signals and nutrient availability. To date, studies evaluating the late stages of biofilm growth and dispersal for a number of organisms, including S. aureus and P. aeruginosa, have identified multiple mechanisms that contribute this process (20–25). As a result, there have been tremendous research interest and focus of efforts in the identification of dispersive molecules that can be used to inhibit/disperse bacterial biofilms (20). Recently, for the soil bacterium Bacillus subtilis, the d-isoforms of various amino acids, including d-Leu, d-Met, d-Trp, and d-Tyr, were reported to have both inhibitory and dispersive activity against biofilms of B. subtilis (26, 27). In contrast to other biofilm dispersal agents that act to interfere with a single process essential for biofilm development, the dispersive activities of d-amino acids (d-AAs) have been attributed to multiple mechanisms, including (i) inhibition of growth and expression of genes involved in biofilm matrix production (28) as well as (ii) diminished surface expression of fibers involved in biofilm formation, resulting from incorporation of d-AAs into the bacterial cell wall (26). In addition to their activity against B. subtilis biofilms, d-AAs have also been shown to have dispersive activity against biofilms of S. aureus and P. aeruginosa in vitro (27, 29) and biofilms of S. aureus in vivo when incorporated into a modified bone graft (30).
Given these observations, we hypothesized that combining dispersal agents with antimicrobials may be an effective therapeutic strategy for biofilms, functionally restoring susceptibility of biofilms to antimicrobials through the release of bacteria from the biofilm. To explore this hypothesis, we evaluated the dispersal activity of d-AAs on biofilms of clinical wound isolates of S. aureus and P. aeruginosa and investigated whether combining d-AAs with various classes of antibiotics enhances the activity against biofilm-producing bacteria in vitro.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The clinical isolates utilized in this study were selected from a strain collection at the Brooke Army Medical Center/San Antonio Military Medical Center (BAMC/SAMMC) (Fort Sam Houston, TX, USA) and previously characterized for biofilm formation (Table 1) (31). Bacterial isolates from this strain collection were collected from patients in the course of routine clinical care not related to research. S. aureus strain UAMS-1 (ATCC 25943) is a methicillin-susceptible osteomyelitis isolate (32, 33). P. aeruginosa strain PAO1 is a well characterized wound isolate widely used as a laboratory strain (34, 35). For planktonic growth, clinical strains of S. aureus and P. aeruginosa were cultured in tryptic soy broth (TSB) and Luria-Bertani broth (LB), respectively, at 37°C. Bacteria were subcultured on blood agar plates (Remel, Lenexa, KS, USA) overnight at 37°C.
TABLE 1.
Characteristics of strains used in this study
| Bacterial species and strains | Pulsed-field type | Phenotypea | Isolate source | Site of isolation |
|---|---|---|---|---|
| Pseudomonas aeruginosa | ||||
| SAMMC-604 | 1 | MDR | Wound culture | Tissue deep |
| SAMMC-015 | 2 | MDR | Blood | Blood |
| SAMMC-886 | 2 | MDR | Wound culture | Tissue deep |
| SAMMC-418 | 18 | MDR | Wound | Tissue deep |
| SAMMC-189 | 18 | MDR | Blood | Blood |
| PAO1 (ATCC 15692)b | Unknown | Unknown | Wound | Unknown |
| Staphylococcus aureus | ||||
| SAMMC-700 | USA 100 | MRSA | Wound culture | Tissue deep |
| SAMMC-641 | USA 200 | MRSA | Wound culture | Tissue deep |
| SAMMC-474 | USA 800 | MRSA | Wound culture | Tissue deep |
| SAMMC-446 | USA 300 | MRSA | Wound culture | Tissue deep |
| SAMMC-240 | USA 300 | MRSA | Wound culture | Tissue deep |
| UAMS-1 (ATCC 43290)c | USA 200 | MSSA | Wound culture | Bone |
A multidrug-resistant (MDR) organism was defined as an organism resistant to antimicrobials in ≥3 classes of antimicrobial agents (penicillins/cephalosporins, carbapenems, aminoglycosides, and quinolones), not including tetracyclines or colistin. MRSA, methicillin-resistant Staphylococcus aureus.
Antibiotics and d-amino acids.
For S. aureus clinical strains the following antibiotics and concentrations were used; clindamycin (CLI) (0.25 to 1,024 μg/ml), cefazolin (CFZ) (0.25 to 1,024 μg/ml), oxacillin (OXA) (0.125 to 1,024 μg/ml), vancomycin (VANC) (0.125 to 1,024 μg/ml), and rifampin (RIF) (0.125 to 1,024 μg/ml). For P. aeruginosa, amikacin (AMK) (0.5 to 1,024 μg/ml), colistin (CST) (0.25 to 1,024 μg/ml), ciprofloxacin (CIP) (0.125 to 1,024 μg/ml), imipenem (IPM) (0.25 to 1,024 μg/ml), and ceftazidime (CAZ) (0.25 to 1,024 μg/ml) were used. Antibiotics were purchased from Sigma-Aldrich (St. Louis, MO). Stock solutions were prepared as recommended and diluted to the appropriate concentrations in cation-adjusted Mueller-Hinton broth (MHB-II). Quality control strains included S. aureus ATCC 29213 and Pseudomonas aeruginosa ATCC 27853. d-amino acids were purchased from Sigma-Aldrich and prepared as concentrated stock solutions in water or 1.0 N HCl, followed by filter sterilization. From the prepared stock solutions, d-AAs were diluted into MHB-II to a final concentration of 50 mM and neutralized when necessary with NaOH (1 M) (pH 7 to 7.4). All subsequent working concentrations of d-AAs were prepared by diluting the neutralized 50 mM stock into MHB to yield final working concentrations.
Biofilm formation in 96-well plates and biofilm dispersal assays.
Biofilm formation was performed under static conditions for 24 h in polystyrene 96-well plates (Corning, Inc., Corning, NY) as described previously (31). Briefly, following overnight incubation, medium was removed from individual wells and washed with 1× phosphate-buffered saline (PBS), and 200 μl of media without or supplemented with d-AAs at the designated concentrations in MHB-II was added to each well for an additional 24 h. Following overnight exposure, cells were washed as above and biofilm biomass was determined by measuring the optical density at 570 nm (OD570) of crystal violet solubilized in ethanol. Experimental assays were performed in triplicate.
Confocal scanning laser microscopy.
S. aureus and P. aeruginosa biofilms were visualized using a FluoView confocal laser-scanning microscope (Olympus, Pittsburgh, PA). Biofilms were grown as described above in 8-well glass chamber slides (36). Biofilms were stained with dual combinations of biofilm ruby matrix stain and biofilm cell stain (Molecular Probes, Eugene, OR) to visualize the extracellular polymeric matrix and bacterial cells, respectively, according to the manufacturer's instructions. Confocal scanning laser microscopy (CLSM) images were acquired at ×20 magnification using a HeNe-G laser at 543 nm for the matrix stain and an argon laser (488 nm) for bacteria. Image analysis and z-stacks were acquired using the Olympus FluoView software. Images were taken from three distinct regions on the slide and representative images were selected for each treatment group.
Antibiotic susceptibility of planktonic bacteria.
Antibiotic susceptibilities of selected clinical strains were evaluated by determining the MIC as recommended by the Clinical and Laboratory Standards Institute (37). Test performance for antimicrobial agents was monitored using P. aeruginosa ATCC 27853 and S. aureus ATCC 29213 as control strains. For each antibiotic tested, the MIC of planktonic organisms was determined in the presence or absence of the d-AA mixture (1:1:1 d-Met/d-Phe/d-Trp). Antimicrobial susceptibility assays were performed in duplicate.
Antibiotic susceptibility of biofilm bacteria and determination of the minimal biofilm inhibitory concentration.
The method for determination of the MBIC for biofilm bacteria following antimicrobial treatment was adapted from previously described studies using modified minimum biofilm eradication concentration high-throughput (MBEC-HTP) assay plates (Innovotech, Canada) for biofilm antimicrobial susceptibility testing (38, 39). Briefly, bacteria were inoculated into wells containing either TSB or LB, for S. aureus and P. aeruginosa, respectively, covered with a lid containing pegs for the attachment of the bacteria, and incubated at 37°C for 48 h with agitation. Following incubation, plate lids containing the pegs with the attached biofilms were washed with 1× PBS and submerged in 2-fold serial dilutions of antibiotics diluted in MHB-II in 96-well plates (i.e., challenge plate) alone or in combination with the d-AA mixture (1:1:1 d-Met/d-Phe/d-Trp) overnight at 37°C. Plate lids were then removed, washed, and transferred to a new 96-well recovery plate with new culture media for determining the MBIC. The MBIC was defined as the lowest concentration of antibiotics at which no visible growth was observed after 6 h of recovery. Viability of bacteria on pegs immediately following antimicrobial exposure was determined by enumerating serial dilutions on blood agar plates (Remel, Lenexa, KS) following removal of bacteria by sonication. Antimicrobial susceptibility assays were performed in triplicate.
Statistical analysis.
Multigroup comparisons were performed using a one-way ANOVA followed by a Dunnett's post hoc test for comparisons between test and control groups using GraphPad Prism version 5. Differences were considered to be statistically significant at P values of <0.05.
RESULTS
Dose-dependent effect of d-amino acids on S. aureus and P. aeruginosa biofilms.
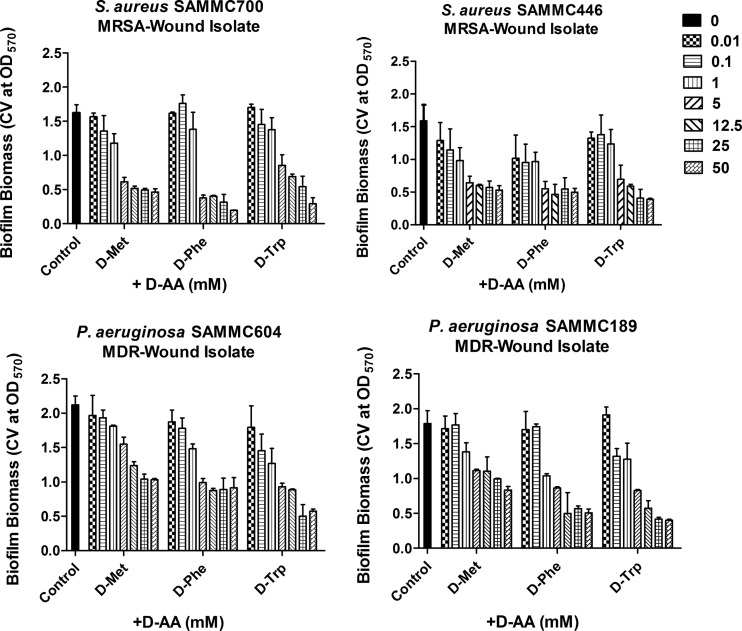
To evaluate the potential clinical application of d-AAs, we tested whether d-AAs effectively dispersed preformed biofilms of clinical isolates of S. aureus and P. aeruginosa. Prescreening of eight individual d-AAs identified three (d-Met, d-Phe, and d-Trp) that had potent activity at dispersing preformed biofilms of representative wound isolates of both S. aureus and P. aeruginosa (Fig. 1). In contrast, the other d-AAs tested (d-Ala, d-Ile, d-Leu, d-Tyr, and d-Val) had variable-to-minimal dispersive activity (data not shown). Dispersal activities of d-Met, d-Phe, and d-Trp were significant at concentrations of ≥5 mM. Of note, the pH of all tested concentrations of d-AAs diluted in MHB-II was neutral, indicating that the observed activity was independent of an effect from the pH of the solution. d-Met, d-Phe, and d-Trp were all observed to be effective against S. aureus biofilms (Fig. 1, top panels), whereas for P. aeruginosa, d-Trp and d-Phe had greater dispersive activity than d-Met (Fig. 1, bottom panels). Based on the initial screening of d-AA dispersal activity, 5 mM was chosen as the concentration for use in all subsequent in vitro assays.
FIG 1.
Dose-dependent effects of d-AAs against S. aureus and P. aeruginosa biofilms. Screening of dispersive activity of individual d-amino acids, d-methionine (d-Met), d-phenylalanine (d-Phe), and d-tryptophan (d-Trp) (at concentrations ranging from 0.01 to 50 mM) against biofilms of two clinical methicillin-resistant S. aureus (MRSA) isolates (top panels) and two multidrug-resistant (MDR) isolates of P. aeruginosa (bottom panels). Biofilm dispersal was assessed by measuring the absorbance of solubilized crystal violet from stained biofilms following overnight treatment with d-AAs at 570 nm. *, P < 0.05 versus untreated control. CV, crystal violet.
d-Amino acids disperse biofilms of clinical wound isolates of S. aureus and P. aeruginosa.
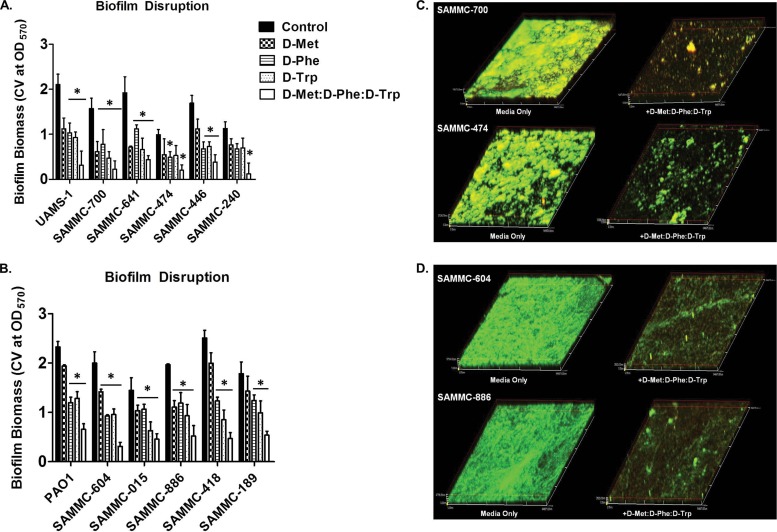
When tested against biofilms of genetically diverse clinical isolates of S. aureus (n = 5) and P. aeruginosa (n = 5), d-Met, d-Phe, and d-Trp at 5 mM were also observed to have significant dispersal activity, as indicated by the reductions in biofilm biomass as determined by crystal violet assay (Fig. 2A and B). As expected, the activity of d-AAs against biofilms was partly strain dependent, in particular for isolates of P. aeruginosa, although for each strain tested more than one of the d-AAs reduced biofilm to >50% of the untreated control. Notably, when combined as an equimolar mixture (1:1:1 d-Met/d-Phe/d-Trp), biofilm dispersal activity was enhanced compared to the exposure to individual d-AAs (Fig. 2A and B). Consistent with these results, confocal microscopy analysis of biofilms treated with the d-AA mixture demonstrated a significant reduction in biofilm biomass compared to that of untreated controls (Fig. 2C and D).
FIG 2.
Biofilm dispersive activity of d-AAs on clinical wound isolates. Activity of d-AAs d-Met, d-Phe, and d-Trp individually at 5 mM and as an equimolar mixture (1:1:1 d-Met/d-Phe/d-Trp) against biofilms of methicillin-resistant (SAMMC-700, SAMMC-641, SAMMC-474, SAMMC-446 and SAMMC-240) and methicillin-susceptible (UAMS-1) S. aureus isolates (A) and multidrug-resistant (SAMMC-604, SAMMC-015, SAMMC-886, SAMMC-418, and SAMMC-189) strains of P. aeruginosa (B). Biofilm dispersal was assessed by measuring the absorbance of solubilized crystal violet from stained biofilms following treatment with d-AA at 570 nm. *, P < 0.05 versus untreated control. Representative CLSM images of biofilms of (C) S. aureus and (D) P. aeruginosa clinical isolates treated with an equimolar mixture of d-AAs for 24 h. Biofilms were stained with dual combinations of biofilm ruby matrix stain (red) and biofilm cell stain (green) and images were taken at a magnification of ×20.
Importantly, d-AAs alone or in combination as a mixture had no significant effect on the growth of S. aureus and P. aeruginosa cells, indicating that biofilm dispersal was not the result of growth inhibition (see Fig. S1 in the supplemental material).
d-Amino acids enhance the activity of antimicrobials against biofilm bacteria but not planktonic bacteria.
To determine if d-AAs could enhance the effect of conventional antimicrobials against bacteria within biofilms, antimicrobial susceptibility assays with and without the d-AA mixture (1:1:1 d-Met/d-Phe/d-Trp) were initially performed on both the planktonic (i.e., grown in liquid culture) and biofilm phenotype of two strains of S. aureus (UAMS-1 and SAMMC-700) and P. aeruginosa (SAMMC-418 and SAMMC-189). The MICs of antimicrobials for planktonic S. aureus strains (UAMS-1 and SAMMC-700, respectively) were determined for clindamycin (0.5 μg/ml), cefazolin (0.5 μg/ml and 16 μg/ml), oxacillin (1 μg/ml and 8 μg/ml), rifampin (0.5 μg/ml), and vancomycin (0.5 μg/ml and 1 μg/ml) (Table 2). Similarly, the MICs for the P. aeruginosa strains, SAMMC-418 and SAMMC-189, respectively, were determined for amikacin (16 μg/ml), colistin (1 μg/ml), ciprofloxacin (4 μg/ml; 8 μg/ml), imipenem (128 μg/ml and 64 μg/ml), and ceftazidime (16 μg/ml and 2 μg/ml) (Table 3). Notably, the combination of the d-AA mixture with antibiotics did not alter the susceptibilities of planktonic bacteria.
TABLE 2.
Minimal inhibitory concentration and minimal biofilm inhibition concentration of antibiotics alone or in combination with d-amino acids
| Antimicrobial agenta | Class | Planktonic MICb (μg/ml) for: |
Biofilm MBICb (μg/ml) for: |
||
|---|---|---|---|---|---|
| S. aureus UAMS-1 | S. aureus SAMMC-700 | S. aureus UAMS-1 | S. aureus SAMMC-700 | ||
| CLI | Lincosamide | 0.5 | 0.5 | 256 | 512 |
| CLI + d-AA | 0.25 | 0.5 | 32 | 128 | |
| CFZ | Cephem | 0.5 | 16 | 512 | 1,024 |
| CFZ + d-AA | 0.5 | 16 | 512 | 1,024 | |
| OXA | Penicillin | 1 | 8 | 256 | >1,024 |
| OXA + d-AA | 1 | 8 | 256 | >1,024 | |
| RIF | Ansamycin | 0.5 | 0.5 | 32 | 64 |
| RIF + d-AA | 0.5 | 0.5 | 8 | 8 | |
| VAN | Glycopeptide | 0.5 | 1 | 256 | 512 |
| VAN + d-AA | 0.5 | 1 | 128 | 128 | |
CLI, clindamycin; CFZ, cefazolin; OXA, oxacillin; RIF, rifampin; VAN, vancomycin.
MIC and minimum biofilm inhibition concentration (MBIC) of the antibiotic (μg/ml) alone or in combination with an equimolar mixture of d-Phe/d-Trp/d-Met (5 mM).
TABLE 3.
MIC and minimal biofilm inhibition concentration of antibiotics alone or in combination with d-amino acids
| Antimicrobial agenta | Class | Planktonic MICb (μg/ml) for: |
Biofilm MBICb (μg/ml) for: |
||
|---|---|---|---|---|---|
| SAMMC-189 | SAMMC-418 | SAMMC-189 | SAMMC-418 | ||
| AMK | Aminoglycoside | 16 | 16 | 512 | >1,024 |
| AMK + d-AA | 8 | 16 | 512 | >1,024 | |
| CS | Lipopeptide | 1 | 1 | 256 | 256 |
| CS + d-AA | 1 | 1 | 32 | 64 | |
| CIP | Fluoroquinolone | 4 | 8 | 128 | 256 |
| CIP + d-AA | 4 | 8 | 32 | 32 | |
| IMI | Carbapenem | 128 | 64 | 256 | 256 |
| IMI + d-AA | 128 | 64 | 256 | 128 | |
| TAZ | Cephem | 16 | 2 | 512 | >1,024 |
| TAZ + d-AA | 8 | 2 | 512 | 512 | |
AMK, amikacin; CS, colistin; CIP, ciprofloxacin; IMI, imipenem; TAZ, ceftazidime.
MIC and minimum biofilm inhibition concentration (MBIC) of the antibiotic (μg/ml) alone or in combination with an equimolar mixture of d-Phe/d-Trp/d-Met (5 mM).
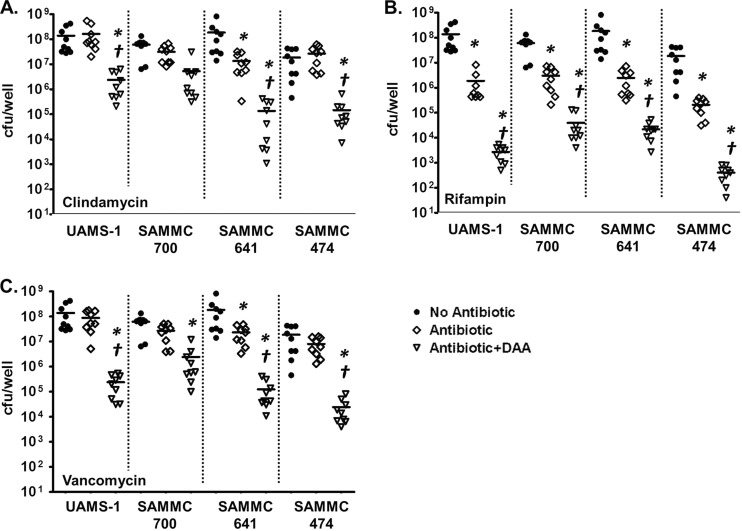
As anticipated, the majority of antimicrobial agents tested against biofilms of S. aureus, including clindamycin, cefazolin, oxacillin, and vancomycin, were ineffective at the tested concentrations against the biofilm phenotype, with observed MBICs exceeding the antimicrobial test range in most instances (Table 2). In contrast, rifampin was active against the biofilms of S. aureus at 32 μg/ml and 64 μg/ml for UAMS-1 and SAMMC-700, respectively. Interestingly, exposure to antimicrobials in combination with the d-AA mixture enhanced the activity of rifampin, clindamycin, and vancomycin against the biofilm phenotype (Table 2). Combined exposure resulted in reductions of the observed MBICs of 4- and 8-fold (2 and 3 2-fold dilutions) for rifampin, 6- and 4-fold (3 and 2 2-fold dilutions) for clindamycin, and 2- to 4-fold (2 2-fold dilutions) for vancomycin against S. aureus UAMS-1 and SAMMC-700, respectively. Notably, exposure of biofilms of other clinical isolates of S. aureus to combined treatments at the experimentally determined MBICs above for clindamycin, vancomycin, and rifampin (64 μg/ml for clindamycin and vancomycin and 8 μg/ml rifampin) also resulted in a greater reduction of viable bacteria from biofilms than treatment with antimicrobials alone (Fig. 3). Exposure of biofilms of genetically distinct staphylococcal isolates to combined treatments reduced viable bacterial counts between 1.5 and 2 logs for clindamycin and vancomycin, whereas combination treatment resulted in a >2-log reduction for rifampin, compared to antimicrobial treatment alone.
FIG 3.
Effects of combinations of d-AAs and antibiotics against biofilms of clinical isolates of S. aureus. Biofilms of clinical methicillin-resistant (SAMMC-700, SAMMC-641, SAMMC-474) and methicillin-susceptible (UAMS-1) S. aureus were developed on pegs of MBEC-HTP plates (Innovotech) for 24 h, followed by exposure to clindamycin (64 μg/ml) (A), rifampin (8 μg/ml) (B), and vancomycin (64 μg/ml) (C) in the absence (◇) or presence of the d-AA mixture (1:1:1 d-Met/d-Phe/d-Trp) (▽) for 24 h. Viable bacteria from biofilms were determined by plating serial dilutions, following removal of adherent bacteria by sonication. Values are expressed as log10 (CFU/well). *, P < 0.05 versus control; †, P < 0.05 versus antibiotic alone.
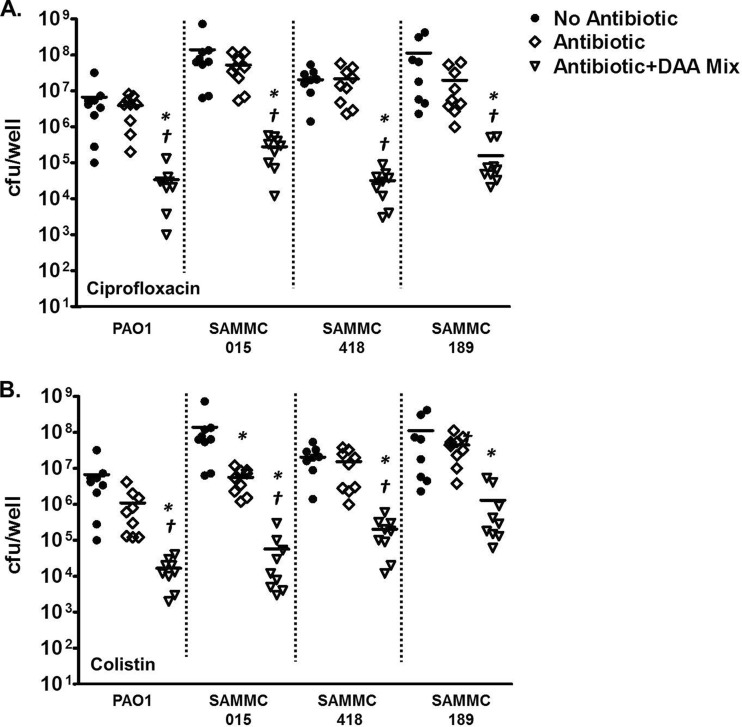
Similarly, for P. aeruginosa, treatment of the biofilms with the selected panel of antimicrobials alone at the test ranges was ineffective against the biofilms (Table 3). However, as observed with S. aureus, the addition of the d-AAs enhanced the activity of colistin and ciprofloxacin against multiple clinical strains of P. aeruginosa, reducing the observed MBICs (Table 3) and bacterial viability (Fig. 4A and B) compared to antimicrobials alone.
FIG 4.
Effect of combinations of d-AAs and antibiotics against biofilms of clinical isolates of P. aeruginosa. Biofilms of multidrug-resistant clinical strains (SAMMC-015, SAMMC-418, and SAMMC-189) and a laboratory strain (PAO1) of P. aeruginosa were developed on pegs of MBEC-HTP plates (Innovotech) for 24 h, followed by exposure to ciprofloxacin (32 μg/ml) (A) or colistin (64 μg/ml) (B) in the absence (◇) or presence of the d-AA mixture (1:1:1 d-Met/d-Phe/d-Trp) (▽) for 24 h. Viable bacteria from biofilms were determined by plating serial dilutions, following removal of adherent bacteria by sonication. Values are expressed as log10 (CFU/well). *, P < 0.05 versus control; †, P < 0.05 versus antibiotic alone.
DISCUSSION
Wound infections constitute a major burden of disease, with multiple factors complicating antimicrobial therapy, including antimicrobial resistance and biofilm formation. Biofilm formation is considered to be a significant pathogenic attribute for both invasive and opportunistic pathogens, such as S. aureus and P. aeruginosa. The importance of biofilms to clinical disease is illustrated by estimates indicating that more than 80% of all bacterial infections, in particular chronic infections, may involve a biofilm etiology (7, 8, 40). Although clinical evidence linking biofilms to recurrent or persistent clinical wound infections is scarce, we have recently observed biofilm formation in 61.4% of bacterial isolates of multiple bacterial species associated with clinical wound infections, demonstrating a higher degree of biofilm formation among tissue, bone, and respiratory isolates (31). Furthermore, we have also shown through a clinical case-control study that in vitro biofilm formation by infecting pathogens carries an odds ratio of 29.5 for persistent wound infection (41). Importantly, and as a result of growth within a biofilm, conventional antibiotics are often ineffective against biofilms, which require in most cases 10- to 1,000-fold-higher concentrations than their planktonic counterparts. Consequently, the biofilm phase of growth can further complicate therapy by conferring a high degree of innate resistance to antimicrobials, even those to which the causative bacteria are susceptible as planktonic organisms (9, 10, 42). Given the importance of the biofilms to disease and the limitation of conventional antimicrobials against this phenotype, herein we assessed whether the use of d-AAs, a biofilm dispersive agent, could enhance the activity of antimicrobials against biofilms.
Only recently have compounds and strategies been reported which specifically target biofilms, including d-AAs, promoting disassembly that may be used in combination with antimicrobials to enhance activity against biofilms (43, 44). The d-isoforms of amino acids represent one such example, and they have been previously shown to have dispersal activity against biofilms of S. aureus and P. aeruginosa (27, 45). Consistent with these studies, we observed that d-AAs, including d-Met, d-Phe, and d-Trp, when used individually, and to a greater extent as an equimolar combination, had significant dispersive activity on biofilms of multiple, genetically distinct clinical isolates at concentrations of ≥5 mM. This observed activity of individual d-AAs is consistent with previous reports indicating a range of activities for d-Met, d-Phe, and d-Trp of between 2 and 5 mM and 10 mM against biofilms of S. aureus and P. aeruginosa, respectively (27, 29, 45). Importantly, and in contrast to previous studies with B. subtilis, significant effects on either cell viability or bacterial growth were not observed at the effective concentrations (28). Collectively, these initial screening studies demonstrate that the dispersive activity of d-AAs functions in a strain-independent manner against common wound pathogens, such as S. aureus and P. aeruginosa, and are suggestive of the potential use in applications for infectious complications due to these agents.
Although the addition of d-AAs did not enhance the activity of antimicrobials against planktonic cells, the addition of d-AAs was observed to enhance the activity of several antibiotic classes against biofilms of genetically distinct isolates of methicillin-resistant S. aureus (MRSA) and multidrug-resistant (MDR) P. aeruginosa. The greatest enhancement of antimicrobial activity by d-AAs was observed with rifampin (against S. aureus) and ciprofloxacin (against P. aeruginosa).
For rifampin and ciprofloxacin, combinations with d-AA demonstrated near-bactericidal activity, with 2- to 3-log CFU decreases compared to each agent alone. Additionally, we also observed 1- to 2-log CFU reductions with vancomycin, clindamycin, and colistin when combined with d-AAs compared to each agent alone. Notably, we did not observe potentiation of activity of oxacillin or cefazolin against methicillin-susceptible strains of S. aureus (strain UAMS-1), nor of amikacin, imipenem, or ceftazidime against P. aeruginosa when combined with d-AAs. In line with our findings, previous studies have demonstrated that the activity of various antimicrobial agents can be significantly enhanced by the addition of the dispersal agents, such as quorum-sensing inhibitors. In one study, the use of a quorum-sensing analog significantly enhanced the activity of several antibiotics, including tobramycin, against P. aeruginosa biofilms, as well as enhanced clearance in a foreign-body infection model (46). Similar results were also observed in studies examining the use of quorum-sensing inhibitors (QSI) against biofilms of P. aeruginosa, Burkholderia cepacia, and S. aureus, whereby the combined use of an antibiotic and a QSI resulted in increased killing compared to antibiotic alone (47). For d-AAs, as well as the quorum-sensing inhibitors, the mechanisms through which the combinations with these agents enhanced the activity of antimicrobials have not been fully addressed. In contrast to QSI inhibitors, which may modulate metabolic activity of bacteria as signaling occurs through two-component signaling mechanisms, d-AAs may have augmented the activity of antimicrobials against biofilms in part following dispersal of bacteria from biofilms. However, we cannot exclude the possibility of other effects of d-AAs, such as changes in protein expression, cell wall integrity, and metabolic status, which may also contribute to observed enhanced susceptibility in vitro.
The explanation for potentiation by d-AAs of some agents, but not others, against established biofilms is not readily apparent and requires further examination. Biofilm penetration on the basis of hydrophobicity does not appear to be the explanation for the inactivity of some agents, as log P values describing the degree of hydrophobic/hydrophilic partitioning ranged from −7.9 (amikacin) to 2.4 (oxacillin) versus −3.3 (colistin) to 4 (rifampin). Furthermore, the remaining agents found to be inactive in our study target cell wall penicillin-binding proteins and may be less active due to decreased bacterial replication. However, vancomycin and colistin, which were potentiated in combination with d-AA, also target bacterial cell wall constituents. Future studies are warranted to fully characterize these phenomena.
Our observation that these primary antimicrobial therapies can be enhanced to a bactericidal level of activity against biofilms by addition of d-AAs introduces the possibility that local delivery of antibiotics combined with d-AAs might be used to effectively reduce infections with little systemic toxicity. The concentrations of antimicrobials tested (selected as the MBIC for each agent) in combination with d-AAs, although higher than what are safely achievable by systemic administration, can potentially be reached by local antibiotic delivery within an infected wound. However, the local elution of antibiotics at high concentrations raises concerns for tissue toxicity and impaired wound healing (48). However, since antimicrobial concentrations required for biofilm eradication may be reduced by codelivery with a dispersal agent, toxicity can potentially be mitigated with their inclusion. In support of this concept, recently we have demonstrated the potential benefits of local delivery of antibiofilm agents in vivo, utilizing d-AA elution from an impregnated polyurethane scaffold implanted in a contaminated rat segmental bone defect model, which prevented S. aureus adherence and reduced microbial burden within the bone and on the graft (30). Thus, a safe and effective local delivery vehicle for the antibiofilm treatment of infected wounds might be possible by including both dispersal agents and antimicrobials which elute at subtoxic concentrations.
Importantly, we observed antibiofilm activity for these agents in the setting of very high inoculums (∼108 CFU), which approximates the bacterial burden in an abscess or undebrided wounds at which the risk for development of resistance is increased (49). The recent observation of a “critical colonization” contamination threshold (∼105 CFU/g tissue) associated with local and systemic elevations in inflammatory cytokines (50), which have been linked to wound dehiscence (51, 52), suggests that postdebridement adjunctive use of dispersal agents such as d-AAs might shorten the required duration of antimicrobial therapy for these difficult infections, translating into substantial cost savings and limiting complications of therapy. Whether the use of dispersal agents could eliminate the need for a second antimicrobial agent to protect against resistance requires further investigation.
In conclusion, we have demonstrated in vitro enhancement of antibiofilm activity to near-bactericidal levels, resulting from biofilm dispersal by d-AAs. Biofilm dispersal by individual amino acids was dose dependent, and amino acids in combination potentiated the antibiofilm activity of multiple antimicrobial classes against genotypically diverse isolates of S. aureus and P. aeruginosa, including several agents recommended as first-line treatments for osteomyelitis and prosthetic joint infections. Additional studies should confirm these findings in vivo and explore potential delivery devices which could be used for the clinical treatment of osteoarticular infections mediated by bacterial biofilms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by intramural funding from the Combat Casualty Care Research Program, U.S. Army Medical Research and Materiel Command (to J.C.W.). C.J.S. is supported through a research fellowship through the National Research Council.
The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Department of the Army or the Department of Defense.
Footnotes
Published ahead of print 19 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02468-14.
REFERENCES
- 1.Fonder MA, Lazarus GS, Cowan DA, Aronson-Cook B, Kohli AR, Mamelak AJ. 2008. Treating the chronic wound: a practical approach to the care of nonhealing wounds and wound care dressings. J. Am. Acad. Dermatol. 58:185–206. 10.1016/j.jaad.2007.08.048 [DOI] [PubMed] [Google Scholar]
- 2.Gordois A, Scuffham P, Shearer A, Oglesby A, Tobian JA. 2003. The health care costs of diabetic peripheral neuropathy in the US. Diabetes Care 26:1790-1795. 10.2337/diacare.26.6.1790 [DOI] [PubMed] [Google Scholar]
- 3.Ramsey SD, Newton K, Blough D, McCulloch DK, Sandhu N, Reiber GE, Wagner EH. 1999. Incidence, outcomes, and cost of foot ulcers in patients with diabetes. Diabetes Care 22:382–387. 10.2337/diacare.22.3.382 [DOI] [PubMed] [Google Scholar]
- 4.Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD. 2008. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8:43. 10.1186/1471-2180-8-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. 2008. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One 3:e3326. 10.1371/journal.pone.0003326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton JW: 1999. Introduction to biofilm. Int. J. Antimicrob. Agents 11:217–221, 237–219 [DOI] [PubMed] [Google Scholar]
- 7.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- 8.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95–108. 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- 9.Anderson GG, O'Toole GA. 2008. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol. 322:85–105 [DOI] [PubMed] [Google Scholar]
- 10.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39. 10.1016/S0966-842X(00)01913-2 [DOI] [PubMed] [Google Scholar]
- 11.Bjarnsholt T, Kirketerp-Moller K, Jensen PO, Madsen KG, Phipps R, Krogfelt K, Hoiby N, Givskov M. 2008. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 16:2–10. 10.1111/j.1524-475X.2007.00283.x [DOI] [PubMed] [Google Scholar]
- 12.James GA, Swogger E, Wolcott R, Pulcini E, Secor P, Sestrich J, Costerton JW, Stewart PS. 2008. Biofilms in chronic wounds. Wound Repair Regen. 16:37–44. 10.1111/j.1524-475X.2007.00321.x [DOI] [PubMed] [Google Scholar]
- 13.Percival SL, Bowler PG. 2004. Biofilms and their potential role in wound healing. Wounds 16:234–240 [Google Scholar]
- 14.Davis SC, Ricotti C, Cazzaniga A, Welsh E, Eaglstein WH, Mertz PM. 2008. Microscopic and physiologic evidence for biofilm-associated wound colonization in vivo. Wound Repair Regen. 16:23–29. 10.1111/j.1524-475X.2007.00303.x [DOI] [PubMed] [Google Scholar]
- 15.Mulcahy LR, Isabella VM, Lewis K. 6 October 2013. Pseudomonas aeruginosa biofilms in disease. Microb. Ecol. 10.1007/s00248-013-0297-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roche ED, Renick PJ, Tetens SP, Ramsay SJ, Daniels EQ, Carson DL. 2012. Increasing the presence of biofilm and healing delay in a porcine model of MRSA-infected wounds. Wound Repair Regen. 20:537–543 [DOI] [PubMed] [Google Scholar]
- 17.Schierle CF, De la Garza M, Mustoe TA, Galiano RD. 2009. Staphylococcal biofilms impair wound healing by delaying reepithelialization in a murine cutaneous wound model. Wound Repair Regen. 17:354–359. 10.1111/j.1524-475X.2009.00489.x [DOI] [PubMed] [Google Scholar]
- 18.Zhao G, Hochwalt PC, Usui ML, Underwood RA, Singh PK, James GA, Stewart PS, Fleckman P, Olerud JE. 2010. Delayed wound healing in diabetic (db/db) mice with Pseudomonas aeruginosa biofilm challenge: a model for the study of chronic wounds. Wound Repair Regen. 18:467–477. 10.1111/j.1524-475X.2010.00608.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao G, Usui ML, Underwood RA, Singh PK, James GA, Stewart PS, Fleckman P, Olerud JE. 2012. Time course study of delayed wound healing in a biofilm-challenged diabetic mouse model. Wound Repair Regen. 20:342–352. 10.1111/j.1524-475X.2012.00793.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan JB. 2010. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 89:205–218. 10.1177/0022034509359403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kostakioti M, Hadjifrangiskou M, Hultgren SJ. 2013. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 3:a010306. 10.1101/cshperspect.a010306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boles BR, Horswill AR. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. 10.1371/journal.ppat.1000052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crossman L, Dow JM. 2004. Biofilm formation and dispersal in Xanthomonas campestris. Microbes Infect. 6:623–629. 10.1016/j.micinf.2004.01.013 [DOI] [PubMed] [Google Scholar]
- 24.Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T. 2010. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 59:253–268. 10.1111/j.1574-695X.2010.00690.x [DOI] [PubMed] [Google Scholar]
- 25.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186:1838–1850. 10.1128/JB.186.6.1838-1850.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cava F, Lam H, de Pedro MA, Waldor MK. 2011. Emerging knowledge of regulatory roles of d-amino acids in bacteria. Cell. Mol. Life Sci. 68:817–831. 10.1007/s00018-010-0571-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. 2010. d-Amino acids trigger biofilm disassembly. Science 328:627–629. 10.1126/science.1188628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leiman SA, May JM, Lebar MD, Kahne D, Kolter R, Losick R. 2013. d-Amino acids indirectly inhibit biofilm formation in Bacillus subtilis by interfering with protein synthesis. J. Bacteriol. 195:5391–5395. 10.1128/JB.00975-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochbaum AI, Kolodkin-Gal I, Foulston L, Kolter R, Aizenberg J, Losick R. 2011. Inhibitory effects of d-amino acids on Staphylococcus aureus biofilm development. J. Bacteriol. 193:5616–5622. 10.1128/JB.05534-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez CJ, Jr, Prieto EM, Krueger CA, Zienkiewicz KJ, Romano DR, Ward CL, Akers KS, Guelcher SA, Wenke JC. 2013. Effects of local delivery of d-amino acids from biofilm-dispersive scaffolds on infection in contaminated rat segmental defects. Biomaterials 34:7533–7543. 10.1016/j.biomaterials.2013.06.026 [DOI] [PubMed] [Google Scholar]
- 31.Sanchez CJ, Jr, Mende K, Beckius ML, Akers KS, Romano DR, Wenke JC, Murray CK. 2013. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect. Dis. 13:47. 10.1186/1471-2334-13-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smeltzer MS, Thomas JR, Hickmon SG, Skinner RA, Nelson CL, Griffith D, Parr TR, Jr, Evans RP. 1997. Characterization of a rabbit model of staphylococcal osteomyelitis. J. Orthop. Res. 15:414–421. 10.1002/jor.1100150314 [DOI] [PubMed] [Google Scholar]
- 33.Weiss EC, Zielinska A, Beenken KE, Spencer HJ, Daily SJ, Smeltzer MS. 2009. Impact of sarA on daptomycin susceptibility of Staphylococcus aureus biofilms in vivo. Antimicrob. Agents Chemother. 53:4096–4102. 10.1128/AAC.00484-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holloway BW. 1955. Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13:572–581. 10.1099/00221287-13-3-572 [DOI] [PubMed] [Google Scholar]
- 35.Schmidt KD, Tummler B, Romling U. 1996. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J. Bacteriol. 178:85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker JN, Horswill AR. 2012. A coverslip-based technique for evaluating Staphylococcus aureus biofilm formation on human plasma. Front. Cell. Infect. Microbiol. 2:39. 10.3389/fcimb.2012.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing. M100-S22. CLSI, Wayne, PA [Google Scholar]
- 38.Coraca-Huber DC, Fille M, Hausdorfer J, Pfaller K, Nogler M. 2012. Evaluation of MBEC-HTP biofilm model for studies of implant associated infections. J. Orthop. Res. 30:1176–1180. 10.1002/jor.22065 [DOI] [PubMed] [Google Scholar]
- 39.Coraca-Huber DC, Fille M, Hausdorfer J, Pfaller K, Nogler M. 2012. Staphylococcus aureus biofilm formation and antibiotic susceptibility tests on polystyrene and metal surfaces. J. Appl. Microbiol. 112:1235–1243. 10.1111/j.1365-2672.2012.05288.x [DOI] [PubMed] [Google Scholar]
- 40.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167–193. 10.1128/CMR.15.2.167-193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akers KS, Mende K, Cheatle KA, Zera WC, Yu X, Beckius ML, Aggarwal D, Li P, Sanchez CJ, Wenke JC, Weintrob AC, Tribble DR, Murray CK, Infectious Disease Clinical Research Program Trauma Infectious Disease Outcomes Study Group 2014. Persistent wound infections in United States military trauma patients: a case-control analysis. BMC Infect. Dis. 14:190. 10.1186/1471-2334-14-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138. 10.1016/S0140-6736(01)05321-1 [DOI] [PubMed] [Google Scholar]
- 43.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. 10.1038/nature10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim JS, Heo P, Yang TJ, Lee KS, Cho DH, Kim BT, Suh JH, Lim HJ, Shin D, Kim SK, Kweon DH. 2011. Selective killing of bacterial persisters by a single chemical compound without affecting normal antibiotic-sensitive cells. Antimicrob. Agents Chemother. 55:5380–5383. 10.1128/AAC.00708-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brandenburg KS, Rodriguez KJ, McAnulty JF, Murphy CJ, Abbott NL, Schurr MJ, Czuprynski CJ. 2013. Tryptophan inhibits biofilm formation by Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57:1921–1925. 10.1128/AAC.00007-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christensen LD, van Gennip M, Jakobsen TH, Alhede M, Hougen HP, Hoiby N, Bjarnsholt T, Givskov M. 2012. Synergistic antibacterial efficacy of early combination treatment with tobramycin and quorum-sensing inhibitors against Pseudomonas aeruginosa in an intraperitoneal foreign-body infection mouse model. J. Antimicrob. Chemother. 67:1198–1206. 10.1093/jac/dks002 [DOI] [PubMed] [Google Scholar]
- 47.Brackman G, Cos P, Maes L, Nelis HJ, Coenye T. 2011. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 55:2655–2661. 10.1128/AAC.00045-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rathbone CR, Cross JD, Brown KV, Murray CK, Wenke JC. 2011. Effect of various concentrations of antibiotics on osteogenic cell viability and activity. J. Orthop. Res. 29:1070–1074. 10.1002/jor.21343 [DOI] [PubMed] [Google Scholar]
- 49.Achermann Y, Eigenmann K, Ledergerber B, Derksen L, Rafeiner P, Clauss M, Nuesch R, Zellweger C, Vogt M, Zimmerli W. 2013. Factors associated with rifampin resistance in staphylococcal periprosthetic joint infections (PJI): a matched case-control study. Infection 41:431–437. 10.1007/s15010-012-0325-7 [DOI] [PubMed] [Google Scholar]
- 50.Brown TS, Hawksworth JS, Sheppard FR, Tadaki DK, Elster E. 2011. Inflammatory response is associated with critical colonization in combat wounds. Surg. Infect. 12:351–357. 10.1089/sur.2010.110 [DOI] [PubMed] [Google Scholar]
- 51.Hawksworth JS, Stojadinovic A, Gage FA, Tadaki DK, Perdue PW, Forsberg J, Davis TA, Dunne JR, Denobile JW, Brown TS, Elster EA. 2009. Inflammatory biomarkers in combat wound healing. Ann. Surg. 250:1002–1007. 10.1097/SLA.0b013e3181b248d9 [DOI] [PubMed] [Google Scholar]
- 52.Utz ER, Elster EA, Tadaki DK, Gage F, Perdue PW, Forsberg JA, Stojadinovic A, Hawksworth JS, Brown TS. 2010. Metalloproteinase expression is associated with traumatic wound failure. J. Surg. Res. 159:633–639. 10.1016/j.jss.2009.08.021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.