Abstract
We report here the emergence of VIM-2 and IMP-15 carbapenemases in a series of clinical isolates of carbapenem-resistant Pseudomonas aeruginosa in Lebanon. We also describe the disruption of the oprD gene by either mutations or insertion sequence (IS) elements ISPa1328 and ISPre2 isoform. Our study reemphasizes a rapid dissemination of the VIM-2 carbapenemase-encoding gene in clinical isolates of P. aeruginosa in the Mediterranean basin.
TEXT
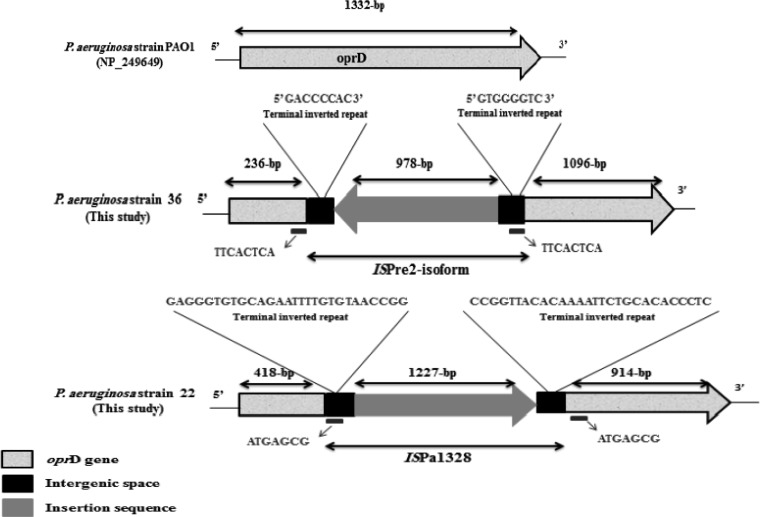
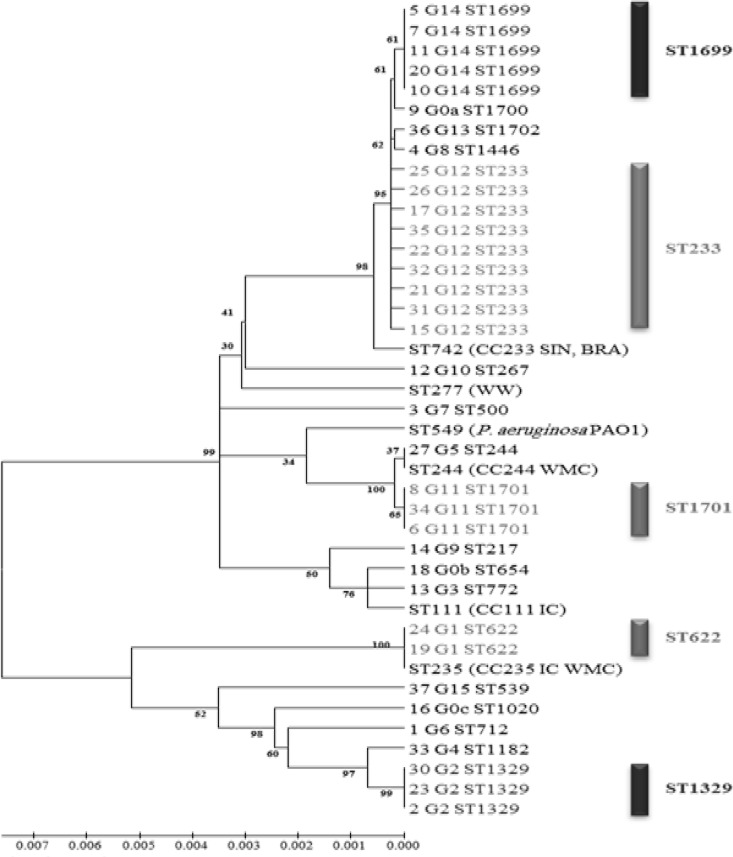
Pseudomonas aeruginosa is one of the most important nosocomial pathogens and is responsible for infections with a high mortality rate (1). Carbapenems remain the primary antimicrobial class for treating these infections; however, the emergence of resistance to these antibiotics may compromise their efficacy (1). The most common mechanism of resistance to carbapenems in P. aeruginosa is the loss or alteration of the outer membrane porin protein OprD, followed by the production of metallo-β-lactamases (MBLs), especially VIM and IMP (1, 2), and by the overexpression of efflux pumps (3). In Lebanon, there are no reports describing carbapenem resistance mechanisms in P. aeruginosa isolates. Here, we have characterized the molecular mechanism of resistance to carbapenems in 35 imipenem-resistant clinical isolates of P. aeruginosa collected between March 2006 and February 2013 from Nini Hospital in northern Lebanon. Standard laboratory procedures were used, including matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) for bacterial identification (4), antimicrobial susceptibility testing on Mueller-Hinton agar using a standard disk diffusion method, the determination of imipenem MICs using the Etest strip, and the molecular detection of carbapenemase-encoding genes, including blaVIM, blaIMP, blaGIM, blaSIM, and blaNDM, using specific primers, as described previously (5–7). PCR amplification and sequencing of the oprD gene and overproduction of the chromosomal cephalosporinase AmpC and efflux pumps (mexA, mexC, mexE, and mexX) were performed as described previously (8–10). Conjugal transfer using MBL-positive isolates as donors and azide-resistant Escherichia coli isolates as recipients was performed as described previously (11). The outer membrane profiles were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to check for the presence/absence of the oprD gene (12). Typing of the isolates was done by multilocus sequence typing (MLST), as described previously (13). The imipenem MICs ranged from 16 to >32 μg/ml for all isolates (Table 1) that also had a high prevalence of resistance to other classes of antibiotics (see Table S1 in the supplemental material). Eighteen out of 35 isolates were positive by the modified Hodge and EDTA tests and harbored the blaVIM-2 gene (16 isolates) or the blaIMP-15 gene (two isolates) (Table 1). All the isolates had modifications to their oprD gene sequence, including 16 with a stop codon, 10 with an insertion sequence (IS) element, four with mutations, and five in which the gene was not detected (Table 1). Nine strains harbored the IS element ISPa1328 inserted in OprD nucleotide position 419. This IS element was also described in P. aeruginosa in the United States (14), China (3), and Japan (15). This IS can be present in a different OprD location, as described previously (3). For isolate 36, an IS element was observed at nucleotide position 237 of the oprD gene that was predicted to be ISPre2 isoform by IS Finder (www-is.biotoul.fr), which was 1,191 bp in length, flanking a novel transposase of 978 bp. The detected ISPre2-like element showed 99% identity with the known ISPre2-like element of P. aeruginosa strain 208/2009 (GenBank accession no. KF682464). The IS was flanked by 8-bp terminal inverted repeats (IRs). We also detected an 8-bp short directly repeated sequence of target DNA flanking the IS (Fig. 1). The TnpA1-like transposase in this isolate was one amino acid shorter than the known transposase and showed 99% identity with those of Pseudomonas resinovorans, Pseudomonas putida, Pseudomonas stutzeri, Pseudomonas fluorescens, and Pseudomonas mendocina. AmpC hyperproduction was observed in eight isolates, and all isolates except isolate 3 showed a high level of expression for at least one efflux pump (Table 1). SDS-PAGE showed the absence of the OprD protein in the five isolates whose oprD gene was not detected by PCR, as well as in isolate 37, which had a 15-nucleotide deletion (Fig. 2). All the conjugal transfer attempts failed to yield E. coli transconjugants, suggesting that these MBL genes are chromosomally encoded. Finally, a total of 18 different sequence types (STs) were assigned (www.pubmlst.org) to the 35 carbapenem-resistant isolates (Table 1 and Fig. 3).
TABLE 1.
Phenotypic and genotypic features of the 35 imipenem-resistant P. aeruginosa clinical isolates
| Isolate no. | IPM MIC (μg/ml)a | MEM resistanceb | Hodge test result | EDTA test result | VIM-2 | IMP-15 | AmpC hyperproduction | oprD mutational groupsc | mRNA expression ford: |
Sequence type | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mexA | mexC | mexE | mexX | ||||||||||
| 19 | >32 | R | − | − | − | − | − | G1 | 2.32* | 0.003 | 1.23 | 11.08* | ST622 |
| 24 | >32 | R | − | − | − | − | − | G1 | 2.14* | 0.004 | 1.04 | 10.48* | ST622 |
| 31 | >32 | R | + | + | + | − | − | G12 | 0.83 | 0.20 | 1.27 | 11.00* | ST233 |
| 26 | >32 | R | + | + | + | − | − | G12 | 1.13 | 0.58 | 1.09 | 29.65* | ST233 |
| 17 | >32 | R | + | + | + | − | − | G12 | 0.44 | 0.28 | 1.14 | 16.56* | ST233 |
| 35 | >32 | R | + | + | + | − | − | G12 | 1.87 | 0.34 | 1.23 | 28.44* | ST233 |
| 21 | >32 | R | + | + | + | − | − | G12 | 1.69 | 1.23 | 1.07 | 14.92* | ST233 |
| 15 | >32 | R | + | + | + | − | − | G12 | 0.43 | 0.43 | 0.64 | 14.82* | ST233 |
| 32 | >32 | R | + | + | + | − | − | G12 | 1.90 | 0.36 | 1.33 | 48.50* | ST233 |
| 25 | >32 | R | + | + | + | − | − | G12 | 1.07 | 0.27 | 1.05 | 14.52* | ST233 |
| 22 | >32 | R | + | + | + | − | − | G12 | 0.5 | 0.17 | 1.27 | 17.02* | ST233 |
| 27 | 16 | R | − | − | − | − | − | G5 | 0.46 | 11.15* | 0.53 | 65.34* | ST244 |
| 12 | >32 | R | − | − | − | − | − | G10 | 0.08 | 3.27* | 0.24 | 6.58 | ST267 |
| 2 | 32 | R | − | − | − | − | + | G2 | 0.57 | 3.13* | 1.04 | 1.23 | ST1329 |
| 30 | 16 | R | − | − | − | − | + | G2 | 1.94 | 61.81* | 1.23 | 0.30 | ST1329 |
| 23 | 16 | R | − | − | − | − | + | G2 | 1.97 | 2.63* | 0.98 | 0.06 | ST1329 |
| 4 | >32 | R | + | + | − | + | − | G8 | 1.09 | 0.51 | 0.11 | 12.29* | ST1446 |
| 11 | >32 | R | + | + | + | − | − | G14 | 9.78* | 0.67 | 0.047 | 24.08* | ST1699 |
| 10 | >32 | R | + | + | + | − | − | G14 | 4.19* | 0.54 | 0.05 | 51.26* | ST1699 |
| 7 | >32 | R | + | + | + | − | − | G14 | 4.53* | 0.30 | 0.05 | 17.02* | ST1699 |
| 20 | >32 | R | + | + | + | − | − | G14 | 5.97* | 1.90 | 0.06 | 19.05* | ST1699 |
| 5 | >32 | R | + | + | + | − | − | G14 | 4.43* | 1.32 | 0.05 | 11.15* | ST1699 |
| 9 | >32 | R | + | + | + | − | − | G0a | 0.97 | 0.66 | 0.30 | 27.47* | ST1700 |
| 14 | >32 | R | − | − | − | − | − | G9 | 0.76 | 4.89* | 0.56 | 0.54 | ST217 |
| 1 | >32 | R | − | − | − | − | + | G6 | 3.2* | 0.57 | 0.99 | 0.15 | ST712 |
| 37 | >32 | R | − | − | − | − | − | G15 | 0.33 | 0.15 | 0.26 | 14.42* | ST539 |
| 36 | >32 | R | − | − | − | − | − | G13 | 0.019 | 0.11 | 0.21 | 10.48* | ST1702 |
| 18 | >32 | R | + | + | − | + | − | G0b | 11.87* | 17.26* | 0.54 | 35.50* | ST654 |
| 6 | >32 | R | − | − | − | − | + | G11 | 0.3 | 5.09* | 0.06 | 0.84 | ST1701 |
| 8 | >32 | R | − | − | − | − | + | G11 | 0.33 | 4.69* | 0.06 | 0.93 | ST1701 |
| 34 | >32 | R | − | − | − | − | + | G11 | 0.68 | 8.33* | 0.10 | 1.07 | ST1701 |
| 33 | >32 | R | − | − | − | − | − | G4 | 0.75 | 9.12* | 1.02 | 91.77* | ST1182 |
| 16 | >32 | R | + | + | + | − | − | G0c | 3.11* | 0.85 | 0.15 | 0.26 | ST1020 |
| 13 | >32 | R | − | − | − | − | − | G3 | 4.14* | 28.64* | 0.99 | 37.53* | ST772 |
| 3 | 16 | S | − | − | − | − | + | G7 | 0.73 | 1.11 | 1.10 | 0.57 | ST500 |
IPM, imipenem.
MEM, meropenem; R, resistant; S, susceptible.
G0a or G0b, full-length oprD gene (mutations on the two groups are not identical); G0c, full-length oprD gene (silent mutation, C-to-T substitution in nucleotide position 309); G1, G-to-T substitution in nucleotide position 688 leading to a premature stop codon TAA in oprD, resulting in a truncated polypeptide of 229 amino acid residues; G2, G-to-A substitution in nucleotide position 830 leading to the premature stop codon TAG in oprD, resulting in a truncated polypeptide of 276 amino acid residues; G3, G-to-A substitution in nucleotide position 831 leading to the premature stop codon TGA in oprD, resulting in a truncated polypeptide of 276 amino acid residues; G4, A-to-T substitution in nucleotide position 613 leading to the premature stop codon TAG in oprD, resulting in a truncated polypeptide of 204 amino acid residues; G5, G-to-A substitution in nucleotide position 195 leading to the premature stop codon TGA in oprD, resulting in a truncated polypeptide of 64 amino acid residues; G6, several mutation types leading to the premature stop codon TGA in oprD, resulting in a truncated polypeptide of 264 amino acid residues; G7, C-to-T and A-to-G substitutions in nucleotide positions 490 and 491, respectively, leading to the premature stop codon TAG in oprD, resulting in a truncated polypeptide of 163 amino acid residues; G8, several mutation types leading to the premature stop codon TGA in oprD, resulting in a truncated polypeptide of 228 amino acid residues; G9, several mutation types leading to the premature stop codon TAA in oprD, resulting in a truncated polypeptide of 363 amino acid residues; G10, several mutation types leading to the premature stop codon TGA in oprD, resulting in a truncated polypeptide of 372 amino acid residues; G11, several mutation types leading to the premature stop codon TGA in oprD, resulting in a truncated polypeptide of 345 amino acid residues; G12, insertion sequence ISPa1328 in nucleotide position 419 of the oprD gene; G13, insertion sequence ISPre2 isoform in nucleotide position 237 of the oprD gene; G14, oprD gene not detected; G15, large deletion of 15 nucleotides in the oprD gene from the positions 1293 to 1307; IPM, imipenem.
Values are relative to the expression of P. aeruginosa PAO1, which is assigned a value of 1. Asterisks indicate overexpression of efflux pump compared to the PAO1 reference strain: ≥2-fold for mexA, mexC, and mexE and ≥10-fold for mexX (10).
FIG 1.
Schematic representation of the oprD gene sequence disrupted by the ISPre2 isoform and ISPa1328 compared to the oprD gene sequence of the P. aeruginosa PAO1 reference strain. P. aeruginosa strain 22 is one of the nine isolates that harbored this ISPa1328. The sequences shown immediately below the diagrams (and on the right and left) for strains 36 and 22 are direct repeats.
FIG 2.
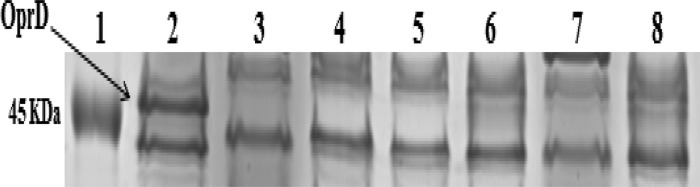
SDS-PAGE of outer membrane proteins of selected carbapenem-resistant isolates. Lane 1, molecular marker; lane 2, reference strain PAO1; lane 3, isolate 37; lane 4, isolate 5; lane 5, isolate 7; lane 6, isolate 10; lane 7, isolate 11; lane 8, isolate 20.
FIG 3.
Phylogenetic tree of the 35 imipenem-resistant isolates based on the MLST concatenated gene sequence of each isolate. Numbers at the nodes represent bootstrap values, and scale bar represents the degree of divergence between sequences. CC, clonal complex; WW, worldwide; WMC, west Mediterranean countries; IC, international clone; SIN, Singapore; BRA, Brazil.
The majority of the MBL-positive isolates produced blaVIM-2 (16 out of 18 isolates), and only 2 produced the blaIMP-15 gene; these data confirm previous studies done in the Mediterranean basin, where the main MBL produced by P. aeruginosa is VIM-2 (16). Although the VIM-2 carbapenemase in P. aeruginosa has been detected worldwide (17), to our knowledge, IMP-15 has never been detected in P. aeruginosa isolates from the west Mediterranean basin but has been detected in Europe (18), Mexico (19), and Thailand (GenBank accession no. AY553333). Moreover, we found that mutational inactivation of the oprD gene was a major determinant of resistance to imipenem, as previously described (2), and SDS-PAGE confirmed the absence of the protein in isolates whose oprD gene was not detected by PCR, as well as in isolate 37, which had a 15-nucleotide deletion, as previously described (15). We report here the first case of cooccurring blaVIM-2 and oprD porin loss in identical isolates of P. aeruginosa. To date, the presence of different IS elements disrupting the oprD gene leading to carbapenem resistance in P. aeruginosa has been reported in South Africa, the United States, Spain, China, and France (8). Our study describes the emergence of a new IS element in the oprD gene, the ISPre2 isoform. Carbapenem resistance was accompanied or not by the overexpression of AmpC β-lactamase and/or efflux pumps, as previously described in isolates from China (3). Finally, MLST analysis revealed the presence of multiple clones in our study, with clones belonging to ST233 and ST1699 (highly similar to ST233) being the most frequent; this ST233 was reported in Norway in a case imported from Ghana (11). MLST analysis showed that ST233 and ST235 described in western Mediterranean countries (20) are distinct in the phylogenetic tree, suggesting that there are two different successful epidemic clones present in the two sides of the Mediterranean basin. We also confirm our previous findings that oprD mutational group sequences can be used as a surrogate to MLST for typing isolates (21).
In conclusion, this study described the emergence of blaVIM-2 and blaIMP-15 carbapenemase-encoding genes in P. aeruginosa clinical isolates in Lebanon and also reported the multiple possibilities for becoming resistant to carbapenems, including the disruption of the oprD gene by either mutations, IS elements, gene loss, and/or overexpression of efflux pumps.
Supplementary Material
ACKNOWLEDGMENTS
We thank Linda Hadjadj, Saiid Azza, Taha Abdo, and Maryam Yehya for their technical assistance. We also thank Leanne McIlreavey for English corrections.
We declare no conflicts of interest.
This work was supported by the AZM Research Center for Biotechnology, Lebanese University, Lebanon. The research project was supported by the National Council for Scientific Research, Lebanon, and by IHU Méditerranée Infection and the French CNRS.
Footnotes
Published ahead of print 9 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02523-13.
REFERENCES
- 1.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front. Microbiol. 2:65. 10.3389/fmicb.2011.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JY, Ko KS. 2012. OprD mutations and inactivation, expression of efflux pumps and AmpC, and metallo-β-lactamases in carbapenem-resistant Pseudomonas aeruginosa isolates from South Korea. Int. J. Antimicrob. Agents 40:168–172. 10.1016/j.ijantimicag.2012.04.004 [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Zhou JY, Qu TT, Shen P, Wei ZQ, Yu YS, Li LJ. 2010. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Chinese hospitals. Int. J. Antimicrob. Agents 35:486–491. 10.1016/j.ijantimicag.2009.12.014 [DOI] [PubMed] [Google Scholar]
- 4.Seng P, Rolain JM, Fournier PE, La Scola B, Drancourt M, Raoult D. 2010. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol. 5:1733–1754. 10.2217/fmb.10.127 [DOI] [PubMed] [Google Scholar]
- 5.Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunel JM, Drissi M, Mesli E, Touati A, Rolain JM. 2012. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One 7:e31676. 10.1371/journal.pone.0031676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yong D, Lee K, Yum JH, Shin HB, Rossolini GM, Chong Y. 2002. Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 40:3798–3801. 10.1128/JCM.40.10.3798-3801.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Touati M, Diene SM, Dekhil M, Djahoudi A, Racherache A, Rolain JM. 2013. Dissemination of a class I integron carrying VIM-2 carbapenemase in Pseudomonas aeruginosa clinical isolates from a hospital intensive care unit in Annaba, Algeria. Antimicrob. Agents Chemother. 57:2426–2427. 10.1128/AAC.00032-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diene SM, L'homme T, Bellulo S, Stremler N, Dubus JC, Mely L, Leroy S, Degand N, Rolain JM. 2013. ISPa46, a novel insertion sequence in the oprD porin gene of an imipenem-resistant Pseudomonas aeruginosa isolate from a cystic fibrosis patient in Marseille, France. Int. J. Antimicrob. Agents 10.1016/j.ijantimicag.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 9.De Champs C, Poirel L, Bonnet R, Sirot D, Chanal C, Sirot J, Nordmann P. 2002. Prospective survey of beta-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolated in a French hospital in 2000. Antimicrob. Agents Chemother. 46:3031–3034. 10.1128/AAC.46.9.3031-3034.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quale J, Bratu S, Gupta J, Landman D. 2006. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 50:1633–1641. 10.1128/AAC.50.5.1633-1641.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samuelsen O, Toleman MA, Sundsfjord A, Rydberg J, Leegaard TM, Walder M, Lia A, Ranheim TE, Rajendra Y, Hermansen NO, Walsh TR, Giske CG. 2010. Molecular epidemiology of metallo-beta-lactamase-producing Pseudomonas aeruginosa isolates from Norway and Sweden shows import of international clones and local clonal expansion. Antimicrob. Agents Chemother. 54:346–352. 10.1128/AAC.00824-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tam VH, Schilling AN, Melnick DA, Coyle EA. 2005. Comparison of beta-lactams in counter-selecting resistance of Pseudomonas aeruginosa. Diagn. Microbiol. Infect. Dis. 52:145–151. 10.1016/j.diagmicrobio.2005.02.010 [DOI] [PubMed] [Google Scholar]
- 13.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42:5644–5649. 10.1128/JCM.42.12.5644-5649.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolter DJ, Acquazzino D, Goering RV, Sammut P, Khalaf N, Hanson ND. 2008. Emergence of carbapenem resistance in Pseudomonas aeruginosa isolates from a patient with cystic fibrosis in the absence of carbapenem therapy. Clin. Infect. Dis. 46:e137–e141. 10.1086/588484 [DOI] [PubMed] [Google Scholar]
- 15.Sanbongi Y, Shimizu A, Suzuki T, Nagaso H, Ida T, Maebashi K, Gotoh N. 2009. Classification of OprD sequence and correlation with antimicrobial activity of carbapenem agents in Pseudomonas aeruginosa clinical isolates collected in Japan. Microbiol. Immunol. 53:361–367. 10.1111/j.1348-0421.2009.00137.x [DOI] [PubMed] [Google Scholar]
- 16.Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect. Dis. 11:381–393. 10.1016/S1473-3099(11)70056-1 [DOI] [PubMed] [Google Scholar]
- 17.Patel G, Bonomo RA. 2013. “Stormy waters ahead”: global emergence of carbapenemases. Front. Microbiol. 4:48. 10.3389/fmicb.2013.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilarranz R, Juan C, Castillo-Vera J, Chamizo FJ, Artiles F, Álamo I, Oliver A. 2013. First detection in Europe of the metallo-β-lactamase IMP-15 in clinical strains of Pseudomonas putida and Pseudomonas aeruginosa. Clin. Microbiol. Infect. 10.1111/1469-0691.12248 [DOI] [PubMed] [Google Scholar]
- 19.Quinones-Falconi F, Galicia-Velasco M, Marchiaro P, Mussi MA, Ballerini V, Vila AJ, Viale AM, Bermejo-Morales K, Limansky AS. 2010. Emergence of Pseudomonas aeruginosa strains producing metallo-beta-lactamases of the IMP-15 and VIM-2 types in Mexico. Clin. Microbiol. Infect. 16:126–131. 10.1111/j.1469-0691.2009.02780.x [DOI] [PubMed] [Google Scholar]
- 20.Maatallah M, Cheriaa J, Backhrouf A, Iversen A, Grundmann H, Do T, Lanotte P, Mastouri M, Elghmati MS, Rojo F, Mejdi S, Giske CG. 2011. Population structure of Pseudomonas aeruginosa from five Mediterranean countries: evidence for frequent recombination and epidemic occurrence of CC235. PLoS One 6:e25617. 10.1371/journal.pone.0025617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sefraoui I, Berrazeg M, Drissi M, Rolain JM. 2013. Molecular epidemiology of carbapenem-resistant Pseudomonas aeruginosa clinical strains isolated from western Algeria between 2009 and 2012. Microb. Drug Resist. 10.1089/mdr.2013.0161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.