Abstract
The usefulness of atovaquone-proguanil (AP) as an antimalarial treatment is compromised by the emergence of atovaquone resistance during therapy. However, the origin of the parasite mitochondrial DNA (mtDNA) mutation conferring atovaquone resistance remains elusive. Here, we report a patient-based stochastic model that tracks the intrahost emergence of mutations in the multicopy mtDNA during the first erythrocytic parasite cycles leading to the malaria febrile episode. The effect of mtDNA copy number, mutation rate, mutation cost, and total parasite load on the mutant parasite load per patient was evaluated. Computer simulations showed that almost any infected patient carried, after four to seven erythrocytic cycles, de novo mutant parasites at low frequency, with varied frequencies of parasites carrying varied numbers of mutant mtDNA copies. A large interpatient variability in the size of this mutant reservoir was found; this variability was due to the different parameters tested but also to the relaxed replication and partitioning of mtDNA copies during mitosis. We also report seven clinical cases in which AP-resistant infections were treated by AP. These provided evidence that parasiticidal drug concentrations against AP-resistant parasites were transiently obtained within days after treatment initiation. Altogether, these results suggest that each patient carries new mtDNA mutant parasites that emerge before treatment but are killed by high starting drug concentrations. However, because the size of this mutant reservoir is highly variable from patient to patient, we propose that some patients fail to eliminate all of the mutant parasites, repeatedly producing de novo AP treatment failures.
INTRODUCTION
The recent emergence of artemisinin-resistant Plasmodium falciparum malaria in Southeast Asia may soon lead to major public health consequences. There is an urgent need to identify treatment regimens that maximize the therapeutically useful life span of current antimalarial drugs (1). Among available alternatives to artemisinin-based combination therapies, the role of atovaquone-proguanil (AP; initially licensed as Malarone) has been questioned (2–4). AP is a popular prophylactic drug and also shows high efficacy in the treatment of uncomplicated falciparum malaria in travelers. Recently, it has been used in the Greater Mekong Subregion to treat individuals detected as P. falciparum carriers as part of a plan to contain artemisinin resistance (4). In addition, generic forms of Malarone have been recently licensed, and the marketing and use of AP could increase in endemic settings (5).
Here, we focused on atovaquone resistance, which develops easily during the treatment of uncomplicated P. falciparum malaria when using atovaquone as a monotherapy (6). This high failure rate prompted the introduction of the AP combination therapy. Atovaquone is a potent inhibitor of the cytochrome bc1 complex or complex III in the mitochondrial electron transport chain (7). The addition of proguanil significantly increases the ability of atovaquone to collapse the mitochondrial membrane potential (8) and also generates in vivo the metabolite cycloguanil, which antagonizes the parasite dihydrofolate reductase in the folate pathway (9, 10). Resistance of P. falciparum to atovaquone is primarily determined by one point mutation in the mitochondrial and multicopy cytochrome b (cytb) gene (11, 12), which encodes a subunit of the cytochrome bc1 complex. In addition, P. falciparum parasites harboring dihydrofolate reductase gene (dhfr) mutations exhibit cross-resistance to the antifolates pyrimethamine and cycloguanil (9, 13, 14). Nowadays, a high rate of antifolate resistance is found in P. falciparum parasites across major areas of endemicity (15). Hence, infecting parasites often carry antifolate resistance and just a single evolutionary step—the acquisition of the cytb mutation—is sufficient to make parasites resistant to both cycloguanil and atovaquone. Although the cytb atovaquone resistance mutation is not detected in areas of endemicity (16–18), about 1 in 100 nonimmune travelers returning to the United Kingdom and who received AP to treat a P. falciparum malaria episode had a late treatment failure associated with atovaquone resistance (19). Genetic studies suggest that the atovaquone resistance mutation evolves de novo during the primary infection or its treatment and is selected for by AP treatment (20–22).
However, the mechanism underlying the rapid evolution of the cytb mutation during AP treatment remains elusive. Various hypotheses suggest a role for an increased rate of mitochondrial DNA (mtDNA) mutations induced by atovaquone (23) and for pharmacokinetic-pharmacodynamic considerations (5). Here, another explanation was considered for the intrahost acquisition of atovaquone resistance. We hypothesize that the multiple mtDNA copies per parasite, along with the complex replication of mtDNA (24, 25), could favor a large mutational input before treatment. While addressing experimentally the intrahost dynamics of mtDNA mutations is currently not possible in the P. falciparum model, mainly because of its intractability, modeling offers an opportunity to explore this question.
Therefore, we developed for the first time a patient-based stochastic model that tracks the intrahost emergence of malarial mtDNA mutations during the few rounds of asexual blood-stage reproduction leading to the malaria febrile episode and diagnosis. Briefly, this model describes the appearance of spontaneous mtDNA mutations and their propagation within replicating mitochondrial genomes and parasites according to intra- and intercellular random drift, in the absence of drug selection. We evaluated the effect of several parameters on the intrahost frequency of mtDNA-mutated parasites: the mtDNA mutation rate, the fitness cost associated with the mtDNA mutation, the number of mtDNA copies per parasite, and the total parasite load per patient. We also report clinical evidence showing for the first time how AP-resistant parasites responded to AP treatment. Finally, we integrated these new computational and clinical findings to propose a mechanistic framework describing how atovaquone resistance emerges during malaria blood stages and how it translates into AP treatment failure, and we discuss the implications of these results for drug usage in the field.
MATERIALS AND METHODS
We used a two-stage process: we first developed a model to calculate the probability that new mtDNA mutations will be present among the malaria parasites at the time of treatment initiation, and then we evaluated whether any such mtDNA-mutated parasites could survive AP treatment and recrudesce as a new resistant infection. This two-step strategy follows a logic similar to the one used for nuclear genes encoding resistance (26).
Model framework.
The mathematical description of the model is provided in the supplemental material. The model simulates the replication and partitioning of parasitic mitochondrial genomes within a replicating blood-stage parasite lineage. It tracks the production and propagation of parasite mtDNA mutations within a single infected human host, starting from the delivery of hepatic parasitic forms into the blood to the time at which the patient will seek medical care and treatment. We assumed that the infection takes place in a nonimmune host, that all parasites delivered from hepatocytes into the blood have the wild-type mitochondrial genotype, and that the mutation is a single nucleotide replacement that arises through random and recurrent mtDNA replication errors during the erythrocytic asexual parasite growth.
Parasite growth.
We assumed that all initial parasites originated from a single release from the liver and that each parasite infected a single erythrocyte. In each infected erythrocyte, the nuclear and mitochondrial genomes of the parasite each are replicated through five consecutive endomitosis steps (within one parasite cell) during one erythrocytic 48-h cycle, eventually leading to 32 daughter parasites. Upon egress from the erythrocyte, each daughter parasite released in the blood has a probability “p_surv” to survive and initiate a new erythrocytic cycle. We assumed parasite growth and cycles to be synchronous within a single host infection.
Replication and partitioning to daughter cells of mitochondrial genomes.
The detailed mechanisms by which the parasite mitochondrion and its genome replicate and are passed to daughter cells are largely unknown, although important features have been described (24, 27). Hence, we used a sequence of events with minimum mechanistic assumptions. Eukaryotic organelle genomes, such as mtDNA, undergo relaxed replication and partitioning, compared to the stringent nuclear genome (25, 28); this means that multiple mtDNA copies are replicated and partitioned randomly, with respect to genotype.
We assumed that each parasite contains one mitochondrion (29), that one mitochondrion contains n copies of its genome (24), that n is constant within an infection, and that at each mtDNA replication, the template copies to be duplicated will be selected by n random draws with replacement, allowing some mtDNA copies to be replicated several times, while other copies are not replicated. This leads to a parasite containing one mitochondrion with 2n mtDNA copies. The mitochondrion containing 2n mtDNA copies is then partitioned into two independent mitochondrial units, each containing n mtDNA copies selected at random among the 2n mtDNA copies. The duplication/partitioning cycle of the mitochondrial genome repeats five times during one erythrocytic 48-h cycle. Each of these 25 = 32 mitochondrial units constitutes a single mitochondrion of one of the 32 daughter parasites. Finally, the resistance mutation arises spontaneously in one mtDNA copy with a probability μ (per nucleotide per replication). Because the atovaquone resistance mutation is a single nucleotide substitution, we assumed that the probability of a resistance mutation to appear equals the per-nucleotide mitochondrial mutation rate. During the course of the erythrocytic phase, n different mutant parasite genotypes can be produced within a single host, with parasites having from 1 to n of their n mtDNA copies mutated. We did not consider back-mutation because its effect is unlikely to alter significantly the intrahost frequency of the resistance mutation, specifically with the short time scale that our study addresses (one to seven cycles of 48-h intra-erythrocytic growth).
Parasite definitions according to mitochondrial genotype.
Parasites having none of their mtDNA copies mutated were defined as wild type. Parasites having from 1 to n−1 of their n mtDNA copies mutated were defined as heteroplasmic parasites. Parasites having all of their mtDNA copies mutated were defined as homoplasmic-mutant parasites. The group including both heteroplasmic and homoplasmic-mutant parasites was defined as all-mutant parasites.
Fitness cost of the mutation.
In some simulations, we assumed that, in the absence of drug, carrying the resistance mutation results in decreased parasite fitness (12, 30, 31). We defined the fitness as the proportion of daughter parasites of a given mitochondrial genotype that will succeed in invading new erythrocytes. However, the mutation does not alter the probability of a mutant mtDNA copy to be replicated or partitioned (25). In addition to the neutral situation (no decrease in fitness associated with the mutation), two other situations were considered. First, the mutation is recessive relative to the decrease in fitness: only homoplasmic-mutant parasites have a decreased survival probability compared to any other genotype. Second, the mutation is dominant relative to the decrease in fitness: the survival probabilities of heteroplasmic and homoplasmic-mutant parasites are the same and are lower than for the wild-type parasites. The recessive and dominant effects explored here represent the two ends of a spectrum, in which parasite fitness would be a function of the number of mutated mitochondrial genes.
Input parameters.
The input parameters included the number of parasites delivered into the blood after the hepatic phase (x = 5 × 104) and the mtDNA mutation rate (μ = 10−9, 10−10, and 10−11/nucleotide/replication). To our knowledge, there is no direct and unbiased estimate for the probability of an mtDNA change per nucleotide per mtDNA replication in P. falciparum. Therefore, we used very conservative values based on values estimated in other species; these values range from 0.7 × 10−8 to 17 × 10−8/nucleotide/mtDNA replication (with one DNA replication/cell division [32–35]). In P. falciparum, the nuclear mutation rate was estimated to be 1.7 × 10−9/nucleotide/48-h generation during blood-stage growth (36). The parasite survival probability associated with the distinct mitochondrial genotypes (12, 37) was as follows: wild type, p_surv = 12/32; homoplasmic-mutant, p_surv_m = 6/32; and heteroplasmic, p_surv_ht = 12/32 when the mutation cost is recessive and p_surv_ht = 6/32 when it is dominant. The numbers of mtDNA copies/parasite were n = 5, 10, 20, and 40. The copy number was estimated to be about 20 copies/cell for P. falciparum, based on an analysis of a single parasite strain (24). The number of erythrocytic 48-h cycles following delivery from the liver ranged from 1 to 7 (this corresponds to 2 to 14 days after the hepatic phase; the median delay between onset of symptoms and treatment is 4 days in imported P. falciparum malaria [38]).
Model simulations.
Varying parameters included the mtDNA copy number, the mutation rate, the fitness cost of the mutation, and the number of erythrocytic 48-h cycles following parasite delivery from the liver. All other parameters were fixed. For each combination of parameters tested, 20,000 simulations were run. The simulations were performed using R (v2.12.2; R Foundation for Statistical Computing, Vienna, Austria).
Clinical case reports.
The French Malaria Reference Center Database was searched for late AP treatment failures in which the recrudescent infections were treated again with AP. A total of 6,247 imported P. falciparum infections diagnosed and treated in France with AP were recorded by the French Malaria Reference Center from 2002 to 2012. Basic demographic and epidemiologic data, clinical and parasitological information, treatments, and history of travel and of malaria infections were collected systematically. Pre- and posttreatment blood isolates were sent to the French Malaria Reference Center by French hospitals participating in the sentinel network for drug resistance and plasma drug testing. Analyzed samples were obtained by blood collections required according to standard medical care for all patients presenting with fever upon hospital admission in France.
Ethics statement.
According to the French legislation, bio-banking and secondary use for scientific purposes of human clinical samples are possible as long as the corresponding patients are informed and have not indicated any objections. This requirement was fulfilled here since information is given to every patient through a hospital notice entitled “Information for Patients,” and no immediate or delayed patient opposition was reported by the hospital clinicians to the French Malaria Reference Center. Moreover, samples received at the French Malaria Reference Center were registered and declared for research purposes as a bio-bank for both the Assistance Publique des Hôpitaux de Paris and the French National Institute of Health Survey. No institutional review board approval is required according to French legislation (article L. 1111-7 du Code de la Santé Publique, article L. 1211-2 du Code de Santé Publique, articles 39 et suivants de la loi 78-17 du 6 janvier 1978 modifiée en 2004 relative à l'informatique, aux fichiers, et aux libertés). The samples used were not anonymized.
Molecular analysis.
Parasite genomic DNA was extracted from whole blood, thin or thick blood smears, and plasma. The cytb gene was sequenced directly from PCR products to genotype the codon 268 (cytb268) that associates with atovaquone resistance. Five nuclear microsatellite loci (TAA81, TAA87, PfPK2, Ara2, and TAA60) dispersed on five different P. falciparum chromosomes were genotyped by capillary electrophoresis to determine the nuclear genetic background associated with each of these parasite isolates. All methods were as previously described (22). Codons 51, 59, and 108 of the dhfr gene associated with cycloguanil resistance were genotyped either by PCR-restriction fragment length polymorphism as described previously (39) or by direct sequencing of PCR products.
Estimation of the parasite reduction ratio for atovaquone-proguanil/cycloguanil-resistant parasites.
To evaluate the intrinsic in vivo efficacy of AP treatment on drug resistant parasites, we measured the parasite reduction ratio (PRR), which is defined as the fractional reduction in parasite number per asexual life cycle (i.e., 48 h) or time unit (day) during the treatment (40). The PRR is similar to the killing rate induced by a specific treatment. A PRR of 50 means that 1 out of 50 parasites survives drug treatment per asexual life cycle. Based on the clinical cases in which infected patients carrying AP-resistant parasites were treated with AP and for which two non-negative and consecutive blood parasite counts were recorded, we estimated the PRR to be (P0/Pt)2/t, where P0 and Pt are the parasite loads at days 0 and t, respectively (41, 42).
Estimation of the minimum time required to clear all of the cryptic de novo mutant parasites from an infection.
Using the intrahost distribution of mtDNA-mutated parasites generated through 20,000 simulated infections, we estimated the minimum time (t) in days during which blood drug concentrations should equal or exceed the minimum AP-resistant parasiticidal concentration (MPCAPR) to clear all of the cryptic mtDNA-mutated parasites. We assumed that the PRR of AP-resistant parasites (PRRAPR) is constant and that AP-resistant parasites are killed at the same rate whether they represent the minority or the dominant population within the infection. An estimate for t can be obtained using the following formula: t = 2 × lnBm/ln(PRRAPR), where Bm is the mutant parasite load per infection (41). We estimated t for simulations obtained with a recessive fitness cost, a copy number of 20, and mutation rate of 10−10/nucleotide/replication.
RESULTS
Model of mtDNA evolution for the pretreatment phase.
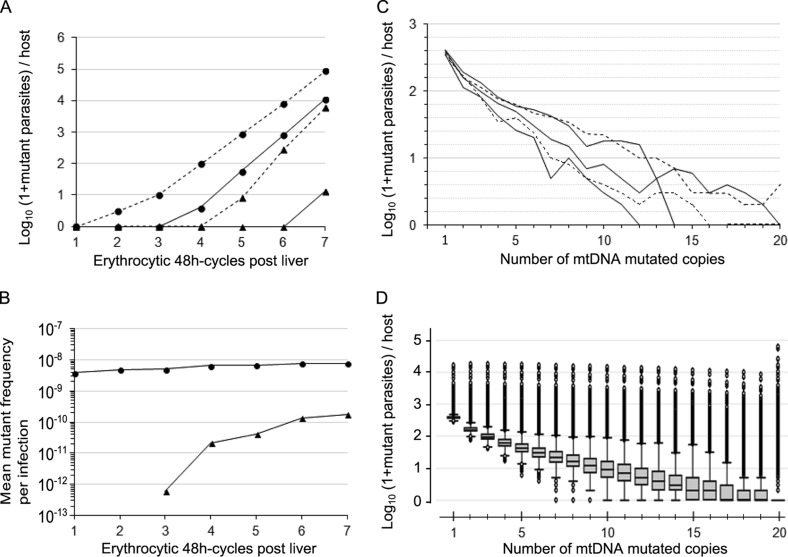
We modeled the evolution of de novo mtDNA mutations that arose during the intrahost expansion of replicating P. falciparum erythrocytic forms in the absence of drug selection. After the delivery of parasites from the liver into the bloodstream, the mean total parasite load per patient was multiplied by ∼12-fold/erythrocytic 48-h cycle and reached about 1.5 × 1011 parasites at the sixth cycle (corresponding to 12 days after the hepatic phase). Assuming a volume of 5 liters of blood per patient and an erythrocyte concentration of 5 × 1012/liter, this translates to 0.6% of the total host erythrocytes being infected. During this intense replicating phase, random recurrent mtDNA mutations occurred and then replicated and segregated according to random drift (combined or not with negative selection) within daughter mitochondrial genomes and parasites (Fig. 1). This produced heteroplasmic parasites and ultimately homoplasmic-mutant parasites, having all of their mtDNA copies mutated (Fig. 1A). For our reference model (neutral mutation, 20 mtDNA copies per parasite, and an mtDNA mutation rate of 10−10/nucleotide/replication), all-mutant parasites (having at least one mtDNA copy mutated) appeared on average at the first erythrocytic cycle with a mean frequency of ∼ 4 × 10−9 all-mutant parasites/infection, which then slightly increased with additional 48-h cycles to reach ∼75 times the mutation rate at the sixth cycle (Fig. 1B). Homoplasmic-mutant parasites appeared on average at the third cycle and then increased in frequency to a mean value close to the mutation rate at the sixth cycle (Fig. 1B). At the sixth cycle, the composition of the mtDNA mutant parasite population per patient was found to be complex, with each patient harboring at varied and low frequencies an array of genotypically distinct mutant parasites defined by the number of mtDNA copies mutated (Fig. 1C and D).
FIG 1.
Dynamics and loads of mtDNA mutant parasites per human host. The data are from 20,000 simulations, with one simulation representing one human infection. (A) Triangles represent homoplasmic-mutant parasites, and circles represent all-mutants (either heteroplasmic or homoplasmic-mutant); plain and dashed lines represent the median and the 99.5th upper percentile, respectively. (B) Triangles and circles represent homoplasmic-mutant and all-mutant parasites, respectively. (C and D) Data are from simulations stopped at the sixth erythrocytic 48-h cycle following parasite delivery from the liver (corresponding to a mean total parasite load per host of 1.5 × 1011). The load of mutant parasites per host is shown, according to the number of mtDNA mutant copies per parasite. In panel C, each line (either plain or dashed) represents one simulated infected host randomly chosen among 20,000 simulations. Five simulations are shown. In panel D, box plots show the medians and interquartile ranges (IQR), and the bottom and top whiskers show the lowest data still within the lower quartile minus 1.5 times the IQR (1.5×IQR) and the highest data still within the upper quartile plus 1.5×IQR, respectively. Outlier values are shown as dots. There is no mutation cost, the mtDNA copy number is 20, and the mutation rate is 10−10/nucleotide/replication for all of the simulations presented here.
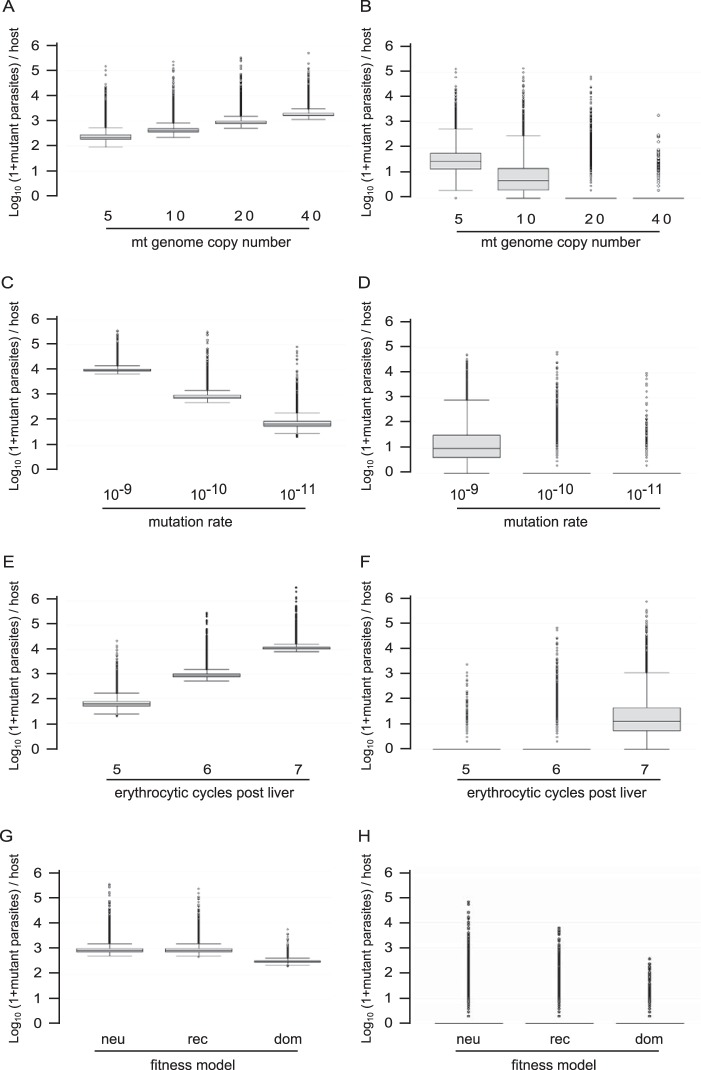
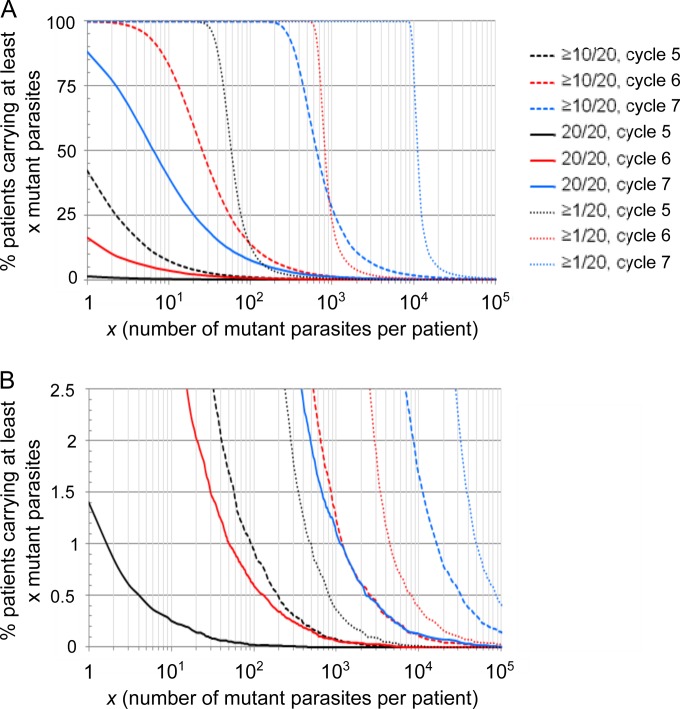
Next, we focused on the intrahost distribution of mutant parasites at the sixth cycle when the mean total parasite load per patient was ∼1.5 × 1011. The number of mtDNA copies per parasite had opposite effects on the intrahost distribution of homoplasmic-mutant and all-mutant parasite loads (Fig. 2A and B): the larger the copy number, the larger the number of all-mutant parasites but the lower the number of homoplasmic-mutant parasites per infection. The size of both the homoplasmic-mutant and all-mutant populations increased with the mutation rate (Fig. 2C and D) and with the total parasite load per patient (Fig. 2E and F). Finally, the fitness cost of the resistance mutation altered the number of mutant parasites per infection in a very expected way compared to a neutral model (Fig. 2G and H): a recessive mutation cost did not alter the all-mutant population but was associated with a slight decrease in the number and the variability of the homoplasmic-mutant population, whereas a dominant mutation cost markedly decreased both all-mutant and homoplasmic-mutant population sizes. Simulations also revealed as a general trend a right-skewed distribution of mutant parasites per infection (Fig. 1D and Fig. 2). One observation that was highly relevant to the outcome of malaria treatment is that infections containing mutant parasites before treatment were extremely frequent (Fig. 3). Even when considering a conservatively low mtDNA mutation rate of 10−10/nucleotide/replication and a parasite burden of 1.2 × 1010/body (∼0.05% of total host erythrocytes being infected), 100% of the simulated infections contained at least one parasite with at least one mtDNA copy mutated (corresponding to the fifth cycle in Fig. 3A; this proportion decreased to 93% for 109 parasites per host). Infections containing at least one homoplasmic-mutant parasite were also frequent: 1.4, 16.3, and 88.3% at the fifth, sixth and seventh cycles, respectively. The proportions of patients carrying high loads of mutant parasites were largely altered by the total parasite load per patient (Fig. 3).
FIG 2.
Load of mtDNA mutant parasites per human host according to different parameters. The data are from 20,000 simulations, with one simulation representing one infected host. (A, C, E, and G) All-mutant parasites, i.e., parasites having at least one of their n mtDNA copies mutated; (B, D, F, and H) homoplasmic-mutant parasites, i.e., parasites having all of their n mtDNA copies mutated. (E and F) At the fifth, sixth, and seventh erythrocytic 48-h cycles post liver, there were mean numbers of 1.2 × 1010, 1.5 × 1011, and 1.8 × 1012 parasites per host, respectively, in our simulations. Unless stated otherwise, the mutation rate was 10−10/nucleotide/replication, there was no resistance mutation cost (neutral), the mtDNA copy number was 20, and the parasite load per host was 1.5 × 1011 (corresponding to the sixth erythrocytic 48-h cycle following parasite delivery from the liver). Abbreviations: neu, neutral; rec, recessive; dom, dominant. Box plots show the medians and IQR, and the bottom and top whiskers show the lowest data still within the lower quartile minus 1.5×IQR and the highest data still within the upper quartile plus 1.5×IQR, respectively. Outlier values are shown as dots.
FIG 3.
Proportions of infections carrying mutant parasites. The data are from 20,000 simulations, with one simulation representing one human infected host. (A) The y axis scale ranges from 0 to 100; (B) same data as in panel A, but the y axis scale ranges from 0 to 2.5. Black, red, and blue lines represent infections after five, six, and seven erythrocytic 48-h cycles, respectively, following parasite release from the liver, corresponding to mean numbers of 1.2 × 1010, 1.5 × 1011, and 1.8 × 1012 parasites per host, respectively. The mtDNA copy number was 20, the mutation rate was 10−10/nucleotide/replication, and the mutation cost was recessive in the simulations presented here. A value of ≥1/20 refers to parasites with at least one of their 20 mtDNA copies mutated (i.e., all-mutant parasites), ≥10/20 refers to parasites with at least 10 of their 20 mtDNA copies mutated, and 20/20 refers to parasites with all of their 20 mtDNA copies mutated (i.e., homoplasmic-mutant parasites).
In summary, whatever the parameter values tested here, all simulations shared four essential results. (i) The mtDNA resistance mutation could occur de novo and propagate in most infected host during the erythrocytic parasite expansion phase before a treatment has been initiated. (ii) Each patient harbored a complex reservoir of cryptic mutant parasite populations, with varied frequencies of parasites having varied numbers of their mtDNA copies mutated. (iii) A marked interpatient variation in the size of this cryptic mutant reservoir was found, even for a unique set of parameters. (iv) Finally, a small fraction of the patients carried a high load of mutant parasites. The very high rate of patients found to carry mtDNA-mutated parasites suggests a very high rate of AP treatment failure, unless the current AP regimen retains therapeutic efficacy toward mtDNA-mutated parasites, thereby limiting the extent of treatment failure. Therefore, we sought evidence showing how AP-resistant parasites respond to AP therapy.
Atovaquone-proguanil/cycloguanil-resistant parasite response to AP treatment: clinical observations.
Among the ∼6,200 P. falciparum imported malaria cases that were treated with AP and reported to the French Malaria Reference Center between 2002 and 2012, we identified seven cases in which recrudescing AP-resistant infections were treated again with AP (Table 1). Full case reports are included in the supplemental material. These clinical observations were backed up with the genotyping of atovaquone and cycloguanil resistance markers and of microsatellites to assess the identity of the pretreatment and posttreatment recrudescing parasites. For each patient, the genetic data showed the selection after the first AP treatment of mutant cytb268 parasites having a nuclear genetic background identical to the one of parasites from the primary infection. This is consistent with our earlier findings (11, 12). For five of these patients (patients P1 to P5) the treatment of the AP-resistant infections with another standard cure of AP was associated with a late treatment failure with parasites recrudescing 8 to 30 days after therapy initiation (mean, 20.6 days), whereas for the other two patients the treatment was altered at days 3 and 5, although their initial parasitemia had significantly decreased. For five cases (P3 to P7), evidence for AP-resistant parasite clearance within days after treatment was obtained from microscopic examination of blood smear, and for three cases (patients P1, P4, and P7) no standard antimalarial drug in addition to atovaquone, proguanil, and cycloguanil was detected in the blood during treatment (this information was not available for the other cases).
TABLE 1.
Patients reported in the study
| Patienta | Sexb | Age (yr) | Wt (kg) | Isolate (day)c | Drugd | Parasitemia (%)e | Plasma drug concn (μM)e,f |
Parasite geneticse,g |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cytb | dhfr | Microsatellites |
|||||||||||||||
| Ato | Pro | Cyc | Others | TAA81 | TAA87 | PfPk2 | Ara2 | TAA 60 | |||||||||
| P1 | M | 24 | 55 | d0 | AP | <0.005 | – | – | – | – | Y | – | 179 | 93 | – | – | 94 |
| d25/d0 | AP | 0.1 | – | – | – | – | S | IRN | 179 | 93 | 169 | 103 | 94 | ||||
| d26/d1 | — | 1.20 | 0.48 | 0.18 | Neg | – | – | – | – | – | – | – | |||||
| d45/d20 | QN | 0.4 | – | – | – | – | S | IRN | 179 | 93 | 169 | 103 | 94 | ||||
| P2 | M | 12 | 40 | d0 | AP | 2.9 | Neg | Neg | Neg | Neg | Y | IRN | 173 | 84 | 172 | 106 | 82 |
| d8 | Neg | – | – | – | – | – | – | – | – | – | – | – | |||||
| d35/d0 | AP | 0.5 | Neg | Neg | Neg | Neg | S | IRN | 173 | 84 | 172 | 106 | 82 | ||||
| d65/d30 | MFQ | <0.1 | – | – | – | – | S | IRN | 173 | 84 | 172 | 106 | 82 | ||||
| P3 | M | 9 | 53 | d0 | AP | 6.6 | Neg | Neg | Neg | Neg | Y | IRN | 182 | – | 172 | 103 | 95 |
| d7 | Neg | 4.12 | 0.02 | Neg | Neg | Y | IRN | 182 | – | 172 | 103 | – | |||||
| d29/d0 | AP | 12.5 | – | – | – | – | C | IRN | 182 | – | 172 | 103 | 95 | ||||
| d32/d3 | 0.01 | – | – | – | – | C | – | – | – | 172 | 103 | 95 | |||||
| d49/d20 | QN | 3.5 | 0.39 | Neg | Neg | Neg | C | IRN | 182 | – | 172 | 103 | 95 | ||||
| P4 | F | 59 | 64 | d0 | AP | 1 | – | – | – | – | – | – | – | – | – | – | – |
| d12 | Neg | 2.39 | Neg | Neg | Neg | Y | – | – | – | – | – | – | |||||
| d24/d0 | AP | 1 | – | – | – | – | C | IRN | 179 | 81 | 163 | 109 | 95 | ||||
| d32/d8 | Neg | 7.98 | 0.04 | Neg | Neg | C | – | – | 81 | 163 | – | 95 | |||||
| d46/d22 | QN | 0.2 | 2.54 | Neg | Neg | Neg | C | IRN | 179 | 81 | 163 | 109 | 94 | ||||
| P5 | F | 14 | 52 | d0 | AP | 0.3 | – | – | – | – | Y | IRN | 179 | 84 | 169, 172 | 103 | 79 |
| d10 | Neg | – | – | – | – | – | – | – | – | – | – | – | |||||
| d23/d0 | AP | 0.1 | – | – | – | – | S | IRN | – | – | – | – | – | ||||
| d31/d8 | <0.1h | 4.10 | Neg | Neg | – | S | IRN | – | – | – | – | – | |||||
| d39/d16 | MFQ | 0.25 | – | – | – | – | S | IRN | 179 | 84 | 172 | 103 | 79 | ||||
| P6 | M | 0.5 | 8 | d0 | AP | 4 | – | – | – | – | – | – | – | – | – | – | – |
| d3 | <0.1 | – | – | – | – | Y | IRN | 179 | 96 | 190 | 112 | 91 | |||||
| d19/d0 | AP | 6 | – | – | – | – | Y+C | IRN | 179 | 96 | 190 | 112 | 91 | ||||
| d22/d3 | MFQ | 0.2 | – | – | – | – | – | – | – | – | – | – | – | ||||
| P7 | M | 58 | 70 | d0 | AP | 2 | Neg | Neg | Neg | Neg | Y | IRN | 164 | 93 | 166 | 127 | 91 |
| d22 | Neg | – | – | – | – | – | – | – | – | – | – | – | |||||
| d27/d0 | AP | 1.5 | 0.10 | Neg | Neg | Neg | C+Sj | IRN | 164 | 93 | 166 | 127 | 91 | ||||
| d32/d5 | QN | 0.0003i | 3.62 | 0.09 | 0.04 | Neg | C+S | IRN | 164 | 93 | 166 | 127 | 91 | ||||
Patients P1 to P7 were infected in The Union of the Comoros, Mali, Senegal, Gabon, Ivory Coast, The Union of the Comoros, and Togo, respectively. All were of African ethnicity except P7, who is Caucasian; patient P5 was already reported (11); additional information and a full case report are provided here.
M, male; F, female.
d0 corresponds to the day of diagnosis and treatment initiation.
That is, the drug used for treatment. AP, atovaquone-proguanil; MFQ, mefloquine; QN, quinine.
Neg, not detected; –, not done.
Ato, atovaquone; Pro, proguanil; Cyc, cycloguanil; Others, monodesethyl-chloroquine, chloroquine, doxycycline, sulfadoxine, pyrimethamine, mefloquine, carboxy-mefloquine, amodiaquine, monodesethyl-amodiaquine, desbutyl-halofantrine, lumefantrine, and dihydro-artemisinin. The lower limit of quantification was 5 μg/liter, which corresponds to 0.014, 0.020, and 0.020 μM for atovaquone, cycloguanil, and proguanil, respectively.
cytb, codon 268 was analyzed; the wild type is tyrosine (Y), and the mutants are serine (S) and cysteine (C). The mutations cytb Y268S and Y268C confer resistance to atovaquone. dhfr, the triple dhfr mutation N51I+C59R+S108N (IRN) confers resistance to cycloguanil. For microsatellites, the allele size (in base pairs) is reported.
A few trophozoites were detected on the thin and thick smears.
Calculated peripheral parasitemia, based on 14 parasites/μl detected on the thick smear and a measured erythrocyte concentration of 3.7 T/liter.
The presence of the two mutations Y268S and Y268C within the same isolate was confirmed by cloning the PCR product and subsequent sequencing of multiple clones (see Fig. S1 in the supplemental material).
Based on patients P3, P6, and P7, for whom two non-negative and consecutive blood parasite counts were available (at day 0 and day 3 or day 5; Table 1), we estimated the PRRs for AP-resistant infections (PRRAPR) during the first days of AP therapy to be 116, 10, and 30 (mean, 52), respectively. Altogether, these clinical findings showed that some AP-resistant parasites can be eliminated by AP as long as the drug blood concentrations are maintained above an as-yet-unknown threshold value, which we defined as the minimum parasiticidal concentration (MPCAPR).
Estimation of the minimum time required to clear the cryptic mtDNA-mutated parasites.
We estimated the time (t) during which drug concentrations in the blood should exceed or equal the MPCAPR to clear from any host all of the cryptic mutant parasites that emerged during the primary infection. Assuming a constant PRRAPR of 50, t = 0.51 lnBm (where Bm is the mutant parasite load per patient). In hyperparasitemic patients carrying about 1.8 × 1012 parasites (∼7% of total host erythrocytes being infected), the maximum load of homoplasmic-mutant parasites found before treatment among 20,000 simulations was 176,395 parasites/host (simulation parameters: recessive mutation, mutation rate of 10−10/nucleotide/replication, and mtDNA copy number of 20). The MPCAPR should then be maintained for at least 6.2 days to ensure the complete elimination of this homoplasmic-mutant intrahost population (Table 2). When considering mutant parasites with at least 1/20 or 10/20 of their mtDNA copies mutated, drug blood concentrations equal to or larger than the MPCAPR should be maintained for a longer period to produce a similar treatment efficacy. In nonhyperparasitemic patients (for example, those carrying about 1.5 × 1011 or 1.2 × 1010 parasites, which corresponds to about 0.6 or 0.05% of total host erythrocytes being infected, respectively), the MPCAPR should be maintained for less time to clear all of the mutant parasites (Table 2).
TABLE 2.
Time required to clear the de novo mtDNA mutant parasites emerging from primary infections
| Total parasite load/patient | Fraction of mutated mtDNA copiesa | Mutant parasite load/patientb |
Mutant clearance time, t (days)c | ||
|---|---|---|---|---|---|
| Median | Minimum | Maximum | |||
| 1.8 × 1012 | 20/20 | 6 | 0 | 176,395 | 6.2 |
| ≥10/20 | 651 | 99 | 1,480,258 | 7.2 | |
| ≥1/20 | 11,132 | 8,418 | 3,049,318 | 7.6 | |
| 1.5 × 1011 | 20/20 | 0 | 0 | 6,157 | 4.5 |
| ≥10/20 | 25 | 0 | 79,843 | 5.8 | |
| ≥1/20 | 825 | 443 | 210,890 | 6.2 | |
| 1.2 × 1010 | 20/20 | 0 | 0 | 448 | 3.1 |
| ≥10/20 | 0 | 0 | 6,960 | 4.5 | |
| ≥1/20 | 58 | 17 | 18,352 | 5.0 | |
≥1/20 refers to parasites having at least one of their 20 mtDNA copies mutated (i.e., all-mutant), ≥10/20 refers to parasites having at least 10 of their 20 mtDNA copies mutated, and 20/20 refers to parasites having all of their 20 mtDNA copies mutated (i.e., homoplasmic-mutant).
Data are from 20,000 simulations performed with the following parameters: recessive mutation cost, mtDNA copy number of 20, and a mutation rate of 10−10/nucleotidic sites/replication.
Values were determined assuming a constant parasite reduction ratio for AP-resistant parasites of 50. Maximum mutant parasite loads were used for the estimation.
DISCUSSION
We report here new findings that help to explain the easy and repeated evolution of P. falciparum mtDNA mutations conferring atovaquone resistance and the high rates of atovaquone and AP treatment failures in nonimmune or partially immune patients (6, 19, 21, 43). Our work explores for the first time the intrahost dynamics of malarial mtDNA mutations through stochastic modeling at the microevolutionary time scale, i.e., during a few erythrocytic parasite cycles following the hepatic phase. Computational data show that almost any primary malarial infection in a nonimmune human host carries a cryptic population of de novo mtDNA-mutated parasites at the time of malaria diagnosis, before treatment has been initiated. Given the large parasite load per human body at malaria diagnosis (the median peripheral parasitemia of 0.4% in travelers roughly corresponds to 1.2 × 1011 parasites per patient) and that each parasite contains ∼20 copies of mtDNA (24), the intrahost population size of mtDNA molecules grows to extremely large values. This provides multiple opportunities for random mtDNA mutations to occur within each infected host. The extremely low intrahost frequency of these de novo mtDNA mutations (approximately 1 to 10 times the mutation rate; see Table S1 in the supplemental material) precludes their detection in patient blood samples with standard molecular techniques. Indeed, in hyperparasitemic patients carrying ∼1.8 × 1012 parasites and assuming 5 liters of blood per patient, the model indicated that a mutant allele is present at an intrahost median frequency of ∼10−9, which translates to ∼3 mutant allele copies/10 ml of blood (this increased to 36 allele copies/10 ml of blood in the top 1% of patients with a high mutant parasite burden). This is consistent with genetic tests that failed to detect the cytb mutation conferring atovaquone resistance in the pretreatment blood samples (19–21, 44, 45), even using a mutant-enrichment method (22). Highly sensitive methodologies based on next-generation sequencing that can detect one mutant copy among about 106 to 107 wild-type copies may be helpful in the future to test our computational results (46, 47). In summary, the model points to spontaneous mtDNA mutations occurring before treatment as a primary source of atovaquone resistance mutations, without the need to invoke a mutagenic effect of atovaquone (for further discussion, see the supplemental material). This proposal is in line with a previous study of Mycobacterium tuberculosis that reported the de novo acquisition of multidrug resistance before treatment as a risk factor for treatment failure (48) and with mathematical modeling of drug resistance in several pathogens (48–51). In addition to our modeling data, we report novel clinical cases showing that the drug blood concentrations attained during the first days of AP therapy are high enough to clear some AP-resistant parasites. We provide an estimate for the PRRAPR (mean, 52; see the supplemental material for further discussion associated with this estimate) that is not markedly different from the values obtained for AP-sensitive infections, which ranged from 46 to 261 in nonimmune adults (52). The reasons for this finding remain elusive, but we propose several hypotheses. At high concentrations, atovaquone can inhibit both wild-type and mutant cytochrome b (12) and was found also to be a more general ubiquinone antagonist, inhibiting the substrate-ubiquinone reductases of the respiratory chain distinct from the cytochrome bc1 complex (53). Furthermore, the net effect on parasite killing of high concentrations of atovaquone, proguanil, and cycloguanil in combination should be considered, an issue raised recently also for chloroquine (54). An estimate for the MPCAPR has yet to be determined, and the MPCAPR will likely be a complex interplay between atovaquone, proguanil, and cycloguanil blood concentrations and the threshold number of mutated mtDNA copies per parasite that confers resistance to atovaquone.
Together, our computational and clinical results support the following scenario regarding de novo AP treatment failures. AP treatment success relies in almost any patient on the clearance of cryptic mtDNA-mutated parasites that evolved during the erythrocytic growth phase of the primary infection. Clearance of mutants must occur within the first days of AP treatment, when drug blood concentrations are the highest and exceed the MPCAPR. We then refer to the time during which drug blood concentrations exceed or equal the MPCAPR as the window of mutant prevention. We suggest that the therapeutic margin of the current AP regimen, especially in the context of nonsupervised treatment, is not wide enough to clear all of the cryptic mutant parasites in any patient, hence repeatedly producing de novo AP treatment failures. The main reasons for this, based on our data and the literature, could be the interpatient variability of the following two parameters: the width of the mutant prevention window and the number of de novo mtDNA-mutated parasites that the treatment has to destroy. With regard to the width of the mutant prevention window, a major source of variability is surely the large interpatient variability in atovaquone blood concentration, which is driven by the slow and limited absorption of the drug (55, 56). In addition, this variability is likely made worse in the context of nonsupervised treatment because of potential vomiting and/or poor compliance with the 3-day course of treatment. Another source of variability could be the level of atovaquone resistance conferred by the cytb268 mutation, which will likely alter the MPCAPR and the width of the mutant prevention window. With regard to the de novo mutant parasite load, the model revealed a very large interpatient variability. This variability was contributed largely by some type of random drift specifically associated to mtDNA replication and partitioning during mitosis and also by the interpatient variability in total parasite load at diagnosis. As shown also by modeling studies for nuclear mutations (51, 57), the larger the total parasite load, the larger the de novo mutant load, and the longer it will take to eliminate all de novo resistant parasites from an infection. This is consistent with our clinical data and those already published (19–21, 45, 58–60), which together showed that 17 of 20 patients experiencing de novo AP treatment failure presented with parasitemia at diagnosis close to or greater than 0.4% (corresponding to the median peripheral parasitemia observed at the time of P. falciparum malaria diagnosis in travelers returning to France and the United Kingdom [38, 61]; mean parasitemia, 2.4%, median, 1.4%, minimum, <0.005%; maximum, 13%; see Table S2 in the supplemental material). Controlled studies will be needed to confirm this observation. Additional sources of mutant load variability may include mtDNA mutation rate (48, 62) and mtDNA copy number, but whether these parameters vary substantially between P. falciparum clinical isolates remains to be established.
We did not consider mutations occurring earlier during the primary infection, i.e., during the liver phase (63), which would further increase the probability of a patient carrying mutant parasites. Regarding the physical mechanisms controlling how Plasmodium organelles and their genomes replicate and are passed to daughter cells, this remains a largely unexplored area. During one intraerythrocytic 48-h cycle, the malarial mitochondrion elongates from a single organelle and then branches and ultimately divides to create multiple daughter organelles (27). Our model assumes five consecutive rounds of random mtDNA replication/partitioning during one intraerythrocytic cycle, in which each daughter pool of mtDNA copies behave as an independent unit of replication. We argue that this model mimics the effects of both the likely separation of mtDNA molecules within the elongating and branching mitochondrion and the random drift associated with the mtDNA replication (24, 25, 27). Variations of this standard model can be envisaged, such as a more panmictic behavior of mtDNA copies (24), but we do not expect they will produce qualitatively very different results. In its current version, our model is not designed to evaluate the long-term evolution of mitochondrial genetic diversity, either within a single-host infection that would last for several weeks or months or within an entire malaria parasite population across multiple rounds of host/vector transmission. This would require us to consider back-mutation and host-specific immunity and to take into account the large heterogeneity and complexity of parasite transmission (26, 64). Altogether, our model provides, to our knowledge, the first framework for studying the intrahost early evolution of malarial mtDNA mutations. It may provide a valuable addition to the current modeling methodologies that focus on nuclear mutations. Specifically, it could be helpful for studying the evolution of mitochondrial genes whose products are targeted by antimalarials under drug development (65–68).
It is currently not clear how many mutant cytb copies per parasite are necessary to confer high-grade atovaquone resistance. Our model did not simulate the fate of parasites with distinct mtDNA genotypes (wild type, heteroplasmic, and homoplasmic-mutant) after the initiation of treatment. Studies in yeast show that the effect of drug selection will largely overshadow the one of random genetic drift (69). We speculate that once AP treatment has started, intercellular selection within the host will likely favor the growth of parasites with the largest proportions of their mtDNA copies mutated. Should some heteroplasmic genotypes could survive AP treatment, the combination of random drift and strong drug selection would result in a progressive shift in the mutant parasite population, with the progressive accumulation of homoplasmic-mutant parasites (as shown experimentally in yeast [69]; see also the supplemental material).
Our model for the emergence of atovaquone resistance suggests that the partner drug—here, proguanil together with its active metabolite cycloguanil—plays a critical role in clearing de novo mtDNA mutant parasites. This is consistent with the observation that most atovaquone resistance mutations reported to date were found to emerge in parasites carrying a mutant dhfr gene (see Table S2 in the supplemental material). Controlled studies with a large patient cohort will be necessary to confirm these observations. Speculating on the computational model findings and the high rate of de novo AP treatment failure reported in travelers, we warn against the risk of the rapid emergence of atovaquone resistance if AP is used massively with the current regimen in transmission areas where people are partially or not immune (2, 3). This could be particularly rapid in the many areas where the mutant dhfr gene is found at a high rate in parasite populations (15), as a consequence of a long history of sustained use of the antifolate pyrimethamine.
The framework proposed here suggests several ways to delay the clinical emergence of AP resistance. A first strategy could be to extend the window of mutant prevention by maintaining atovaquone and proguanil/cycloguanil blood concentrations at a higher level and for a longer period of time than those achieved with the current AP dosage regimen. The benefit of a more aggressive chemotherapy in reducing the chance of de novo resistance selection has been recently questioned (1, 70). However, the clinical findings that AP at high drug blood concentrations remains potent on AP-resistant parasites suggest such a strategy merits further attention. A second (and possibly complementary) strategy that we favor would be to minimize the number of de novo mutant parasites that AP has to clear. This could be achieved through combination therapy, employing an additional drug equally potent against AP-sensitive and AP-resistant parasites. For example, the triple combination of artesunate, atovaquone, and proguanil proved to be very efficient for the treatment of malaria in pregnancy and of uncomplicated malaria (71–73). Third, the use of AP could be restricted to treat infections originating in areas of endemicity where the frequency of mutant dhfr genes is low. Finally, an important prediction of our model is that patients with higher parasite burden at diagnosis face an increased risk for carrying larger de novo mutant parasites. The use of AP as a treatment could then be restricted to patients with lower parasitemia at diagnosis.
In summary, we report new computational and clinical findings describing the intrahost evolution of mtDNA mutations conferring atovaquone resistance. The mechanistic framework proposed here sheds light on the repeated evolution of de novo AP resistance and could help policy makers in designing AP-based treatment regimens that delay the clinical emergence of atovaquone resistance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Vivien Le Bras, Selva Pillai, Fanny Bernardon, Rich Eastman, Marcus Lee, and Sophie Adjalley for critical reading of the manuscript and Olivier Tenaillon for helpful discussions. The other members of the atovaquone-proguanil treatment failure study group involved in this study included Sandrine Houzé (Centre National de Référence du Paludisme, Laboratoire de Parasitologie-Mycologie, AP-HP, Hôpital Bichat-Claude Bernard, Paris, France), Céline Maréchal (Centre National de Référence du Paludisme, Laboratoire de Parasitologie-Mycologie, AP-HP, Hôpital Bichat-Claude Bernard, Paris, France), Pascal Houzé (Laboratoire de Biochimie, AP-HP, Hôpital Saint Louis, Paris, France), Marc Thellier (Centre National de Référence du Paludisme, Laboratoire de Parasitologie-Mycologie, AP-HP, Hôpital La Pitié-Salpêtrière, Paris, France), Eric Kendjo (Centre National de Référence du Paludisme, AP-HP, Hôpital La Pitié-Salpêtrière, Paris, France), Liliane Ciceron (Centre National de Référence du Paludisme, AP-HP, Hôpital La Pitié-Salpêtrière, Paris, France), Sophie Matheron (Maladies infectieuses et tropicales, AP-HP, Hôpital Bichat-Claude Bernard, Paris, France), Anne Delaval (Centre Hospitalier Intercommunal Robert Ballanger, Aulnay-sous-Bois, France), François Martin-Barbaz (Centre Hospitalier de Niort, Niort, France), Bernadette Buret (Laboratoire de Biologie, Centre Hospitalier de Niort, Niort, France), Gianandrea Borgherini (Maladies Infectieuses, Groupe Hospitalier Sud Réunion, Réunion, France), Sandrine Picot (Laboratoire de Bactériologie-Parasitologie, Groupe Hospitalier Sud Réunion, Réunion, France), Dieudonné Bemba (Laboratoire de Microbiologie, AP-HP, Hôpital Jean Verdier, Bondy, France), Isabelle Poilane (Laboratoire de Microbiologie, AP-HP, Hôpital Jean Verdier, Bondy, France), Thanh-Van Trieu (Urgences Pédiatriques, AP-HP, Hôpital Jean Verdier, Bondy, France), Marie Belloy (Centre Hospitalier Intercommunal Robert Ballanger, Aulnay-sous-Bois, France), and Martin Danis (Centre National de Référence du Paludisme, Laboratoire de Parasitologie-Mycologie, AP-HP, Hôpital La Pitié-Salpêtrière, Paris, France).
This study was supported by the Université Paris Descartes (G.C., J.C., and J.L.B.), the Institut de Recherche pour le Développement (G.C., J.C., and J.L.B.), the Assistance Publique des Hôpitaux de Paris (J.L.B.), and the French Malaria Reference Center (J.L.B., L.M., V.H., and J.C.). Since 2002, the French Malaria Reference Center has been supported by the Institut de Veille Sanitaire, Assistance Publique des Hôpitaux de Paris, and GlaxoSmithKline (grant to J.L.B. to survey atovaquone-proguanil side effects and treatment failures).
Footnotes
Published ahead of print 27 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02550-13.
REFERENCES
- 1.Read AF, Day T, Huijben S. 2011. The evolution of drug resistance and the curious orthodoxy of aggressive chemotherapy. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 2): S10871–S10877. 10.1073/pnas.1100299108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloland PB, Kazembe PN, Watkins WM, Doumbo OK, Nwanyanwu OC, Ruebush TK., II 1997. Malarone-donation programme in Africa. Lancet 350:1624–1625. 10.1016/S0140-6736(97)06082-0 [DOI] [PubMed] [Google Scholar]
- 3.Nosten F. 2000. Prophylactic effect of Malarone against malaria: all good news? Lancet 356:1864–1865. 10.1016/S0140-6736(00)03250-5 [DOI] [PubMed] [Google Scholar]
- 4.Hoyer S, Nguon S, Kim S, Habib N, Khim N, Sum S, Christophel EM, Bjorge S, Thomson A, Kheng S, Chea N, Yok S, Top-Ros SS, Sophal U, Thompson MM, Mellor S, Ariey F, Witkowski B, Yeang C, Yeung S, Duong S, Newman RD, Menard D. 2012. Focused screening and treatment (FSAT): a PCR-based strategy to detect malaria parasite carriers and contain drug resistant P. falciparum, Pailin, Cambodia. PLoS One 7:e45797. 10.1371/journal.pone.0045797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nixon GL, Moss DM, Shone AE, Lalloo DG, Fisher N, O'Neill PM, Ward SA, Biagini GA. 2013. Antimalarial pharmacology and therapeutics of atovaquone. J. Antimicrob. Chemother. 68:977–985. 10.1093/jac/dks504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Looareesuwan S, Viravan C, Webster HK, Kyle DE, Hutchinson DB, Canfield CJ. 1996. Clinical studies of atovaquone, alone or in combination with other antimalarial drugs, for treatment of acute uncomplicated malaria in Thailand. Am. J. Trop. Med. Hyg. 54:62–66 [DOI] [PubMed] [Google Scholar]
- 7.Srivastava IK, Rottenberg H, Vaidya AB. 1997. Atovaquone, a broad spectrum antiparasitic drug, collapses mitochondrial membrane potential in a malarial parasite. J. Biol. Chem. 272:3961–3966. 10.1074/jbc.272.7.3961 [DOI] [PubMed] [Google Scholar]
- 8.Srivastava IK, Vaidya AB. 1999. A mechanism for the synergistic antimalarial action of atovaquone and proguanil. Antimicrob. Agents Chemother. 43:1334–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson DS, Milhous WK, Wellems TE. 1990. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc. Natl. Acad. Sci. U. S. A. 87:3018–3022. 10.1073/pnas.87.8.3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foote SJ, Galatis D, Cowman AF. 1990. Amino acids in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum involved in cycloguanil resistance differ from those involved in pyrimethamine resistance. Proc. Natl. Acad. Sci. U. S. A. 87:3014–3017. 10.1073/pnas.87.8.3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korsinczky M, Chen N, Kotecka B, Saul A, Rieckmann K, Cheng Q. 2000. Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob. Agents Chemother. 44:2100–2108. 10.1128/AAC.44.8.2100-2108.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kessl JJ, Ha KH, Merritt AK, Lange BB, Hill P, Meunier B, Meshnick SR, Trumpower BL. 2005. Cytochrome b mutations that modify the ubiquinol-binding pocket of the cytochrome bc1 complex and confer antimalarial drug resistance in Saccharomyces cerevisiae. J. Biol. Chem. 280:17142–17148. 10.1074/jbc.M500388200 [DOI] [PubMed] [Google Scholar]
- 13.Sirawaraporn W, Sathitkul T, Sirawaraporn R, Yuthavong Y, Santi DV. 1997. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc. Natl. Acad. Sci. U. S. A. 94:1124–1129. 10.1073/pnas.94.4.1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basco LK, Ringwald P. 2000. Molecular epidemiology of malaria in Yaounde, Cameroon. VI. Sequence variations in the Plasmodium falciparum dihydrofolate reductase-thymidylate synthase gene and in vitro resistance to pyrimethamine and cycloguanil. Am. J. Trop. Med. Hyg. 62:271–276 [DOI] [PubMed] [Google Scholar]
- 15.Naidoo I, Roper C. 2013. Mapping ‘partially resistant,' ‘fully resistant,' and ‘super resistant' malaria. Trends Parasitol. 29:505–515. 10.1016/j.pt.2013.08.002 [DOI] [PubMed] [Google Scholar]
- 16.Musset L, Pradines B, Parzy D, Durand R, Bigot P, Le Bras J. 2006. Apparent absence of atovaquone/proguanil resistance in 477 Plasmodium falciparum isolates from untreated French travelers. J. Antimicrob. Chemother. 57:110–115. 10.1093/jac/dki420 [DOI] [PubMed] [Google Scholar]
- 17.Ekala MT, Khim N, Legrand E, Randrianarivelojosia M, Jambou R, Fandeur T, Menard D, Assi SB, Henry MC, Rogier C, Bouchier C, Mercereau-Puijalon O. 2007. Sequence analysis of Plasmodium falciparum cytochrome b in multiple geographic sites. Malar. J. 6:164. 10.1186/1475-2875-6-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanabe K, Jombart T, Horibe S, Palacpac NM, Honma H, Tachibana S, Nakamura M, Horii T, Kishino H, Mita T. 2013. Plasmodium falciparum mitochondrial genetic diversity exhibits isolation-by-distance patterns supporting a sub-Saharan African origin. Mitochondrion 13:630–636. 10.1016/j.mito.2013.08.008 [DOI] [PubMed] [Google Scholar]
- 19.Sutherland CJ, Laundy M, Price N, Burke M, Fivelman QL, Pasvol G, Klein JL, Chiodini PL. 2008. Mutations in the Plasmodium falciparum cytochrome b gene are associated with delayed parasite recrudescence in malaria patients treated with atovaquone-proguanil. Malar. J. 7:240. 10.1186/1475-2875-7-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz E, Bujanover S, Kain KC. 2003. Genetic confirmation of atovaquone-proguanil-resistant Plasmodium falciparum malaria acquired by a nonimmune traveler to East Africa. Clin. Infect. Dis. 37:450–451. 10.1086/375599 [DOI] [PubMed] [Google Scholar]
- 21.Musset L, Bouchaud O, Matheron S, Massias L, Le Bras J. 2006. Clinical atovaquone-proguanil resistance of Plasmodium falciparum associated with cytochrome b codon 268 mutations. Microbes Infect. 8:2599–2604. 10.1016/j.micinf.2006.07.011 [DOI] [PubMed] [Google Scholar]
- 22.Musset L, Le Bras J, Clain J. 2007. Parallel evolution of adaptive mutations in Plasmodium falciparum mitochondrial DNA during atovaquone-proguanil treatment. Mol. Biol. Evol. 24:1582–1585. 10.1093/molbev/msm087 [DOI] [PubMed] [Google Scholar]
- 23.Srivastava IK, Morrisey JM, Darrouzet E, Daldal F, Vaidya AB. 1999. Resistance mutations reveal the atovaquone-binding domain of cytochrome b in malaria parasites. Mol. Microbiol. 33:704–711. 10.1046/j.1365-2958.1999.01515.x [DOI] [PubMed] [Google Scholar]
- 24.Preiser PR, Wilson RJ, Moore PW, McCready S, Hajibagheri MA, Blight KJ, Strath M, Williamson DH. 1996. Recombination associated with replication of malarial mitochondrial DNA. EMBO J. 15:684–693 [PMC free article] [PubMed] [Google Scholar]
- 25.Birky CW., Jr 1994. Relaxed and stringent genomes: why cytoplasmic genes don't obey Mendel's laws. J. Hered. 85:355–365 [Google Scholar]
- 26.Hastings IM. 2004. The origins of antimalarial drug resistance. Trends Parasitol. 20:512–518. 10.1016/j.pt.2004.08.006 [DOI] [PubMed] [Google Scholar]
- 27.van Dooren GG, Marti M, Tonkin CJ, Stimmler LM, Cowman AF, McFadden GI. 2005. Development of the endoplasmic reticulum, mitochondrion and apicoplast during the asexual life cycle of Plasmodium falciparum. Mol. Microbiol. 57:405–419. 10.1111/j.1365-2958.2005.04699.x [DOI] [PubMed] [Google Scholar]
- 28.Coller HA, Khrapko K, Bodyak ND, Nekhaeva E, Herrero-Jimenez P, Thilly WG. 2001. High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nat. Genet. 28:147–150. 10.1038/88859 [DOI] [PubMed] [Google Scholar]
- 29.Slomianny C, Prensier G. 1986. Application of the serial sectioning and tridimensional reconstruction techniques to the morphological study of the Plasmodium falciparum mitochondrion. J. Parasitol. 72:595–598. 10.2307/3281516 [DOI] [PubMed] [Google Scholar]
- 30.Peters JM, Chen N, Gatton M, Korsinczky M, Fowler EV, Manzetti S, Saul A, Cheng Q. 2002. Mutations in cytochrome b resulting in atovaquone resistance are associated with loss of fitness in Plasmodium falciparum. Antimicrob. Agents Chemother. 46:2435–2441. 10.1128/AAC.46.8.2435-2441.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher N, Abd Majid R, Antoine T, Al-Helal M, Warman AJ, Johnson DJ, Lawrenson AS, Ranson H, O'Neill PM, Ward SA, Biagini GA. 2012. Cytochrome b mutation Y268S conferring atovaquone resistance phenotype in malaria parasite results in reduced parasite bc1 catalytic turnover and protein expression. J. Biol. Chem. 287:9731–9741. 10.1074/jbc.M111.324319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch M, Sung W, Morris K, Coffey N, Landry CR, Dopman EB, Dickinson WJ, Okamoto K, Kulkarni S, Hartl DL, Thomas WK. 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. U. S. A. 105:9272–9277. 10.1073/pnas.0803466105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haag-Liautard C, Coffey N, Houle D, Lynch M, Charlesworth B, Keightley PD. 2008. Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol. 6:e204. 10.1371/journal.pbio.0060204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saxer G, Havlak P, Fox SA, Quance MA, Gupta S, Fofanov Y, Strassmann JE, Queller DC. 2012. Whole genome sequencing of mutation accumulation lines reveals a low mutation rate in the social amoeba Dictyostelium discoideum. PLoS One 7:e46759. 10.1371/journal.pone.0046759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sung W, Tucker AE, Doak TG, Choi E, Thomas WK, Lynch M. 2012. Extraordinary genome stability in the ciliate Paramecium tetraurelia. Proc. Natl. Acad. Sci. U. S. A. 109:19339–19344. 10.1073/pnas.1210663109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bopp SE, Manary MJ, Bright AT, Johnston GL, Dharia NV, Luna FL, McCormack S, Plouffe D, McNamara CW, Walker JR, Fidock DA, Denchi EL, Winzeler EA. 2013. Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet. 9:e1003293. 10.1371/journal.pgen.1003293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roestenberg M, O'Hara GA, Duncan CJ, Epstein JE, Edwards NJ, Scholzen A, van der Ven AJ, Hermsen CC, Hill AV, Sauerwein RW. 2012. Comparison of clinical and parasitological data from controlled human malaria infection trials. PLoS One 7:e38434. 10.1371/journal.pone.0038434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Briand V, Bouchaud O, Tourret J, Behr C, Abgrall S, Ralaimazava P, Le Bras J, Fontanet A. 2007. Hospitalization criteria in imported falciparum malaria. J. Travel Med. 14:306–311. 10.1111/j.1708-8305.2007.00143.x [DOI] [PubMed] [Google Scholar]
- 39.Maiga-Ascofare O, Le Bras J, Mazmouz R, Renard E, Falcao S, Broussier E, Bustos D, Randrianarivelojosia M, Omar SA, Aubouy A, Lepere JF, Jean-Francois V, Djimde AA, Clain J. 2010. Adaptive differentiation of Plasmodium falciparum populations inferred from single-nucleotide polymorphisms (SNPs) conferring drug resistance and from neutral SNPs. J. Infect. Dis. 202:1095–1103. 10.1086/656142 [DOI] [PubMed] [Google Scholar]
- 40.White NJ. 2002. The assessment of antimalarial drug efficacy. Trends Parasitol. 18:458–464. 10.1016/S1471-4922(02)02373-5 [DOI] [PubMed] [Google Scholar]
- 41.White NJ. 1997. Assessment of the pharmacodynamic properties of antimalarial drugs in vivo. Antimicrob. Agents Chemother. 41:1413–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White NJ. 2013. Pharmacokinetic and pharmacodynamic considerations in antimalarial dose optimization. Antimicrob. Agents Chemother. 57:5792–5807. 10.1128/AAC.00287-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiodini PL, Conlon CP, Hutchinson DB, Farquhar JA, Hall AP, Peto TE, Birley H, Warrell DA. 1995. Evaluation of atovaquone in the treatment of patients with uncomplicated Plasmodium falciparum malaria. J. Antimicrob. Chemother. 36:1073–1078. 10.1093/jac/36.6.1073 [DOI] [PubMed] [Google Scholar]
- 44.Kuhn S, Gill MJ, Kain KC. 2005. Emergence of atovaquone-proguanil resistance during treatment of Plasmodium falciparum malaria acquired by a non-immune north American traveler to west Africa. Am. J. Trop. Med. Hyg. 72:407–409 [PubMed] [Google Scholar]
- 45.Savini H, Bogreau H, Bertaux L, Bouchiba H, Kraemer P, Parzy D, Garnotel E, Rogier C, Simon F, Pradines B. 2008. First case of emergence of atovaquone-proguanil resistance in Plasmodium falciparum during treatment in a traveler in Comoros. Antimicrob. Agents Chemother. 52:2283–2284. 10.1128/AAC.00282-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. 2012. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 109:14508–14513. 10.1073/pnas.1208715109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. 2013. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 9:e1003794. 10.1371/journal.pgen.1003794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, Johnston JC, Gardy J, Lipsitch M, Fortune SM. 2013. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat. Genet. 45:784–790. 10.1038/ng.2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colijn C, Cohen T, Ganesh A, Murray M. 2011. Spontaneous emergence of multiple drug resistance in tuberculosis before and during therapy. PLoS One 6:e18327. 10.1371/journal.pone.0018327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pennings PS. 2012. Standing genetic variation and the evolution of drug resistance in HIV. PLoS Comput. Biol. 8:e1002527. 10.1371/journal.pcbi.1002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White NJ, Pongtavornpinyo W. 2003. The de novo selection of drug-resistant malaria parasites. Proc. Biol. Sci. 270:545–554. 10.1098/rspb.2002.2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCarthy JS, Sekuloski S, Griffin PM, Elliott S, Douglas N, Peatey C, Rockett R, O'Rourke P, Marquart L, Hermsen C, Duparc S, Mohrle J, Trenholme KR, Humberstone AJ. 2011. A pilot randomised trial of induced blood-stage Plasmodium falciparum infections in healthy volunteers for testing efficacy of new antimalarial drugs. PLoS One 6:e21914. 10.1371/journal.pone.0021914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fry M, Pudney M. 1992. Site of action of the antimalarial hydroxynaphthoquinone, 2-[trans-4-(4′-chlorophenyl) cyclohexyl]-3-hydroxy-1,4-naphthoquinone (566C80). Biochem. Pharmacol. 43:1545–1553. 10.1016/0006-2952(92)90213-3 [DOI] [PubMed] [Google Scholar]
- 54.Roepe PD. 2014. To kill or not to kill, that is the question: cytocidal antimalarial drug resistance. Trends Parasitol. 30:130–135. 10.1016/j.pt.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hussein Z, Eaves CJ, Hutchinson DB, Canfield CJ. 1996. Population pharmacokinetics of proguanil in patients with acute Plasmodium falciparum malaria after combined therapy with atovaquone. Br. J. Clin. Pharmacol. 42:589–597. 10.1111/j.1365-2125.1996.tb00053.x [DOI] [PubMed] [Google Scholar]
- 56.Hussein Z, Eaves J, Hutchinson DB, Canfield CJ. 1997. Population pharmacokinetics of atovaquone in patients with acute malaria caused by Plasmodium falciparum. Clin. Pharmacol. Ther. 61:518–530. 10.1016/S0009-9236(97)90132-6 [DOI] [PubMed] [Google Scholar]
- 57.White NJ, Pongtavornpinyo W, Maude RJ, Saralamba S, Aguas R, Stepniewska K, Lee SJ, Dondorp AM, White LJ, Day NP. 2009. Hyperparasitaemia and low dosing are an important source of anti-malarial drug resistance. Malar. J. 8:253. 10.1186/1475-2875-8-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fivelman QL, Butcher GA, Adagu IS, Warhurst DC, Pasvol G. 2002. Malarone treatment failure and in vitro confirmation of resistance of Plasmodium falciparum isolate from Lagos, Nigeria. Malar. J. 1:1. 10.1186/1475-2875-1-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perry TL, Pandey P, Grant JM, Kain KC. 2009. Severe atovaquone-resistant Plasmodium falciparum malaria in a Canadian traveler returned from the Indian subcontinent. Open Med. 3:e10–e16 [PMC free article] [PubMed] [Google Scholar]
- 60.Rose GW, Suh KN, Kain KC, Le Saux N, McCarthy AE. 2008. Atovaquone-proguanil resistance in imported falciparum malaria in a young child. Pediatr. Infect. Dis. J. 27:567–569. 10.1097/INF.0b013e318167918d [DOI] [PubMed] [Google Scholar]
- 61.Roberts CH, Armstrong M, Zatyka E, Boadi S, Warren S, Chiodini PL, Sutherland CJ, Doherty T. 2013. Gametocyte carriage in Plasmodium falciparum-infected travellers. Malar. J. 12:31. 10.1186/1475-2875-12-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rathod PK, McErlean T, Lee PC. 1997. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 94:9389–9393. 10.1073/pnas.94.17.9389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pongtavornpinyo W, Hastings IM, Dondorp A, White LJ, Maude RJ, Saralamba S, Day NP, White NJ, Boni MF. 2009. Probability of emergence of antimalarial resistance in different stages of the parasite life cycle. Evol. Appl. 2:52–61. 10.1111/j.1752-4571.2008.00067.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnston GL, Smith DL, Fidock DA. 2013. Malaria's missing number: calculating the human component of R0 by a within-host mechanistic model of Plasmodium falciparum infection and transmission. PLoS Comput. Biol. 9:e1003025. 10.1371/journal.pcbi.1003025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong CK, Urgaonkar S, Cortese JF, Gamo FJ, Garcia-Bustos JF, Lafuente MJ, Patel V, Ross L, Coleman BI, Derbyshire ER, Clish CB, Serrano AE, Cromwell M, Barker RH, Jr, Dvorin JD, Duraisingh MT, Wirth DF, Clardy J, Mazitschek R. 2011. Identification and validation of tetracyclic benzothiazepines as Plasmodium falciparum cytochrome bc1 inhibitors. Chem. Biol. 18:1602–1610. 10.1016/j.chembiol.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nam TG, McNamara CW, Bopp S, Dharia NV, Meister S, Bonamy GM, Plouffe DM, Kato N, McCormack S, Bursulaya B, Ke H, Vaidya AB, Schultz PG, Winzeler EA. 2011. A chemical genomic analysis of decoquinate, a Plasmodium falciparum cytochrome b inhibitor. ACS Chem. Biol. 6:1214–1222. 10.1021/cb200105d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Biagini GA, Fisher N, Shone AE, Mubaraki MA, Srivastava A, Hill A, Antoine T, Warman AJ, Davies J, Pidathala C, Amewu RK, Leung SC, Sharma R, Gibbons P, Hong DW, Pacorel B, Lawrenson AS, Charoensutthivarakul S, Taylor L, Berger O, Mbekeani A, Stocks PA, Nixon GL, Chadwick J, Hemingway J, Delves MJ, Sinden RE, Zeeman AM, Kocken CH, Berry NG, O'Neill PM, Ward SA. 2012. Generation of quinolone antimalarials targeting the Plasmodium falciparum mitochondrial respiratory chain for the treatment and prophylaxis of malaria. Proc. Natl. Acad. Sci. U. S. A. 109:8298–8303. 10.1073/pnas.1205651109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nilsen A, LaCrue AN, White KL, Forquer IP, Cross RM, Marfurt J, Mather MW, Delves MJ, Shackleford DM, Saenz FE, Morrisey JM, Steuten J, Mutka T, Li Y, Wirjanata G, Ryan E, Duffy S, Kelly JX, Sebayang BF, Zeeman AM, Noviyanti R, Sinden RE, Kocken CH, Price RN, Avery VM, Angulo-Barturen I, Jimenez-Diaz MB, Ferrer S, Herreros E, Sanz LM, Gamo FJ, Bathurst I, Burrows JN, Siegl P, Guy RK, Winter RW, Vaidya AB, Charman SA, Kyle DE, Manetsch R, Riscoe MK. 2013. Quinolone-3-diarylethers: a new class of antimalarial drug. Sci. Transl. Med. 5:177ra37. 10.1126/scitranslmed.3005029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Backer JS, Birky CW., Jr 1985. The origin of mutant cells: mechanisms by which Saccharomyces cerevisiae produces cells homoplasmic for new mitochondrial mutations. Curr. Genet. 9:627–640. 10.1007/BF00449815 [DOI] [PubMed] [Google Scholar]
- 70.Huijben S, Bell AS, Sim DG, Tomasello D, Mideo N, Day T, Read AF. 2013. Aggressive chemotherapy and the selection of drug-resistant pathogens. PLoS Pathog. 9:e1003578. 10.1371/journal.ppat.1003578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Vugt M, Leonardi E, Phaipun L, Slight T, Thway KL, McGready R, Brockman A, Villegas L, Looareesuwan S, White NJ, Nosten F. 2002. Treatment of uncomplicated multidrug-resistant falciparum malaria with artesunate-atovaquone-proguanil. Clin. Infect. Dis. 35:1498–1504. 10.1086/344901 [DOI] [PubMed] [Google Scholar]
- 72.McGready R, Keo NK, Villegas L, White NJ, Looareesuwan S, Nosten F. 2003. Artesunate-atovaquone-proguanil rescue treatment of multidrug-resistant Plasmodium falciparum malaria in pregnancy: a preliminary report. Trans. R. Soc. Trop. Med. Hyg. 97:592–594. 10.1016/S0035-9203(03)80040-8 [DOI] [PubMed] [Google Scholar]
- 73.McGready R, Ashley EA, Moo E, Cho T, Barends M, Hutagalung R, Looareesuwan S, White NJ, Nosten F. 2005. A randomized comparison of artesunate-atovaquone-proguanil versus quinine in treatment for uncomplicated falciparum malaria during pregnancy. J. Infect. Dis. 192:846–853. 10.1086/432551 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.