Abstract
Accurate knowledge of fungemia epidemiology requires identification of strains to the molecular level. Various studies have shown that the rate of resistance to fluconazole ranges from 2.5% to 9% in Candida spp. isolated from blood samples. However, trends in antifungal resistance have received little attention and have been studied only using CLSI M27-A3 methodology. We assessed the fungemia epidemiology in a large tertiary care institution in Madrid, Spain, by identifying isolates to the molecular level and performing antifungal susceptibility testing according to the updated breakpoints of European Committee for Antimicrobial Susceptibility Testing (EUCAST) definitive document (EDef) 7.2. We studied 613 isolates causing 598 episodes of fungemia in 544 patients admitted to our hospital (January 2007 to December 2013). Strains were identified after amplification and sequencing of the ITS1-5.8S-ITS2 region and further tested for in vitro susceptibility to amphotericin B, fluconazole, posaconazole, voriconazole, micafungin, and anidulafungin. Resistance was defined using EUCAST species-specific breakpoints, and epidemiological cutoff values (ECOFFs) were applied as tentative breakpoints. Most episodes were caused by Candida albicans (46%), Candida parapsilosis (28.7%), Candida glabrata (9.8%), and Candida tropicalis (8%). Molecular identification enabled us to better detect cryptic species of Candida guilliermondii and C. parapsilosis complexes and episodes of polyfungal fungemia. The overall percentage of fluconazole-resistant isolates was 5%, although it was higher in C. glabrata (8.6%) and non-Candida yeast isolates (47.4%). The rate of resistance to echinocandins was 4.4% and was mainly due to the presence of intrinsically resistant non-Candida species. Resistance mainly affected non-Candida yeasts. The rate of resistance to fluconazole and echinocandins did not change considerably during the study period.
INTRODUCTION
Fungemia is a major cause of morbidity and mortality in both critically ill and non-critically ill patients (1, 2). Although Candida albicans is the main species causing fungemia, other nonalbicans Candida and non-Candida species showing diminished antifungal susceptibility are emerging (2, 3). Knowledge of the epidemiology of the species causing fungemia is clinically relevant, particularly when starting empirical antifungal treatment, since antifungal susceptibility patterns are species specific (4, 5).
The distribution of Candida spp. causing fungemia varies with the geographic region studied (6–9). However, identification of strains to the molecular level is necessary to obtain an accurate picture of the species causing fungemia, because cryptic species in Candida parapsilosis, Candida glabrata, and Candida guilliermondii complexes often go undetected by conventional identification procedures, such as the ID 32C system (10).
Previous reports have shown that the rate of resistance to fluconazole ranges from 2.5% to 9% in Candida spp. isolated from blood samples (3, 11). Echinocandins are fast becoming first-line antifungal agents for the treatment of fungemia (12, 13); however, increasing use of these agents can promote the emergence of resistance in Candida and non-Candida isolates (14). Reports on trends in the rate of antifungal resistance in isolates causing fungemia are scarce, and the few studies performed were based only on the CLSI M27-A3 method (4, 5). In contrast, the European Committee for Antimicrobial Susceptibility Testing (EUCAST) procedure has rarely been used to study trends in antifungal resistance in single institutions over long periods.
The aim of the present study was to assess the epidemiology of fungemia in a large tertiary care institution located in Madrid, Spain, over a 7-year period after identifying isolates to the molecular level and performing antifungal susceptibility testing according to the updated breakpoints of EUCAST definitive document (EDef) 7.2 (15).
(This study was presented in part at the 22nd European Congress on Clinical Microbiology and Infectious Diseases, London, United Kingdom, 31 March to 3 April 2012 [abstr. P-834] [16] and at the 24th European Congress on Clinical Microbiology and Infectious Diseases, Barcelona, Spain, 10 to 13 March 2014 [abstr. eP234] [17].)
MATERIALS AND METHODS
Hospital description, definition of fungemia episodes, and patients studied.
Hospital Gregorio Marañón serves a population of approximately 715,000 inhabitants in the city of Madrid, Spain, and cares for patients at high risk of fungemia, such as those admitted to medical and surgical intensive care units (ICUs), neonates, patients with hematological malignancies, solid-organ transplant recipients, and patients with central venous catheters.
We studied the episodes of fungemia detected in patients admitted to the hospital from January 2007 to December 2013. Multiple episodes in a single patient were defined as isolation of the same fungal species in further blood cultures taken ≥7 days after the last positive blood culture. Episodes in which 2 different Candida species were detected were considered polyfungal fungemia.
Blood cultures and identification of isolates.
Blood samples for culture were obtained by standard procedures and incubated in the automated Bactec 9240 (from 2007 to 2011) and Bactec-FX system (from 2011 onward) (Becton, Dickinson, Cockeysville, MD, USA). Blood cultures with presumptive visualization of yeasts in the Gram stain were subcultured on Chromagar (Chromagar, Paris, France) and incubated for 36 to 48 h at 35°C. If 2 different species were detected in the Chromagar, 1 colony representing each species was independently subcultured. Isolates were identified by means of the ID 32C system (bioMérieux, Marcy l'Etoile, France) and confirmed by amplification and sequencing of the internal transcribed spacer 1 (ITS1)-5.8S-ITS2 region (18).
Antifungal susceptibility testing.
We used the EUCAST EDef 7.2 microdilution procedure to test isolates for in vitro susceptibility to the following drugs: amphotericin B (Sigma-Aldrich, Madrid, Spain); fluconazole, voriconazole, and anidulafungin (Pfizer Pharmaceutical Group, New York, NY, USA); posaconazole and caspofungin (Merck & Co., Inc., Rahway, NJ, USA); and micafungin (Astellas Pharma, Inc., Tokyo, Japan) (15, 19). The antifungal agents were tested at concentrations ranging from 0.015 to 8 μg/ml (amphotericin B, voriconazole, posaconazole, caspofungin, anidulafungin, and micafungin) and 0.062 to 64 μg/ml (fluconazole). Inoculated plates were incubated for 24 h at 35°C. MIC values, which were determined spectrophotometrically at 530 nm (Multiskan FC microplate photometer; Thermo Scientific, Madrid, Spain), were defined as the lowest concentration of drug that resulted in inhibition of ≥50% of growth in comparison with a drug-free control growth well for fluconazole, voriconazole, posaconazole, and echinocandins or inhibition of ≥90% for amphotericin B. Plates with a drug-free control growth well showing an optical density threshold of <0.3 were reincubated for an additional 24 h or more until the drug-free control well reached the threshold. Candida krusei ATCC 6258 and C. parapsilosis ATCC 22019 isolates were used as quality control strains.
Data analysis.
Candida isolates were classified as resistant (R), intermediate (I), or susceptible (S) according to species-specific clinical breakpoints or as wild type or non-wild type according to the epidemiological cutoff values (ECOFFs) proposed by EUCAST (20). The EUCAST document does not provide breakpoints or ECOFFs for caspofungin owing to the high interlaboratory variations reported (21, 22); therefore, the rate of resistance to caspofungin is not shown. The R breakpoint for amphotericin B was >1 μg/ml for Candida albicans, C. parapsilosis, Candida tropicalis, C. glabrata, and C. krusei. The non-species-related breakpoints for fluconazole (all Candida species, with the exception of C. glabrata and C. krusei) were as follows: S, ≤2 μg/ml; I, 4 μg/ml; and R, ≥8 μg/ml. The C. glabrata breakpoints were as follows: I, 0.062 to 32 μg/ml (when the lower limit of tested concentrations is taken into consideration), and R, >32 μg/ml. C. krusei was considered intrinsically fluconazole resistant. For voriconazole, the breakpoints were as follows: R, >0.125 μg/ml (C. albicans, C. parapsilosis, and C. tropicalis) and >1 μg/ml (C. glabrata and C. krusei; tentatively based on the ECOFFs) (23). The breakpoints for posaconazole were as follows: R, >0.06 μg/ml (C. albicans, C. parapsilosis, and C. tropicalis), >0.5 μg/ml (C. krusei; tentatively based on the ECOFFs), and >1 μg/ml (C. glabrata; tentatively based on the ECOFFs) (24). In the case of micafungin, the breakpoints were as follows: R, >0.015 μg/ml (C. albicans), >0.03 μg/ml (C. glabrata), >2 μg/ml (C. parapsilosis), >0.06 μg/ml (C. tropicalis), and >0.25 μg/ml (C. krusei; tentatively based on the ECOFFs) (25). The breakpoints for anidulafungin were as follows: R, >0.03 μg/ml (C. albicans), >0.06 μg/ml (C. glabrata, C. krusei, and C. tropicalis), and >4 μg/ml (C. parapsilosis).
In the absence of breakpoints for non-Candida species, we used tentative non-species-related fluconazole breakpoints, except for Cryptococcus neoformans, which was considered fluconazole resistant if its MIC was ≥16 μg/ml (26). Rhodotorula spp., Trichosporon spp., Arxula spp., and C. neoformans were considered intrinsically echinocandin resistant. Rates of resistance were not calculated for the remaining drug-species combinations. Isolates showing resistance to one or more antifungal agents were retested, and the MIC was confirmed.
The antifungal resistance rate was calculated based on isolates from both intrinsically resistant species and normally susceptible species that showed MIC values above the breakpoints used.
Identification of fks mutations.
We obtained the sequence of the HS1 and HS2 regions of the fks gene in Candida isolates with MICs for anidulafungin and/or micafungin greater than the breakpoints or ECOFFs, as previously described (27, 28).
RESULTS
Epidemiology of species causing fungemia.
During the study period (January 2007 to December 2013), we recorded 612 episodes of fungemia. The isolates from 14 episodes were not available (C. albicans [n = 7], C. parapsilosis [n = 5], C. glabrata [n = 1], and Blastoschyzomyces capitatus [n = 1]). We studied the isolates (n = 613) from the remaining 598 episodes diagnosed in 544 patients admitted to medical wards (25%), the ICU (20%), surgical wards (19%), neonatology (15%), oncology-hematology (14%), and other wards (7%) at the moment of blood sample collection.
The distribution of the species found is shown in Table 1. C. albicans was the main cause of fungemia (46%), followed by C. parapsilosis complex (27.8%), C. glabrata (9.8%), C. tropicalis (8%), C. krusei (1.6%), other Candida spp. (3.4%), and other, non-Candida yeasts (3.4%). The combination of Chromagar and the ID 32C system yielded an accurate identification of most isolates. However, in 7%, sequencing of the ITS region was necessary to ensure correct identification, because the species were not included in the ID 32C system or were from polyfungal fungemia not detected in Chromagar. Polyfungal fungemia was detected in 15 of the patients (3%) (Table 2), although Chromagar failed to detect both species in 3 out of the 15. Molecular identification performed on apparently pure-culture isolates showed a mixture of sequences representing a coinfection that was unraveled only after prolonged incubation of the Chromagar plates for up to 5 days.
TABLE 1.
Distribution of species causing fungemia in the 544 patients studied after amplification and further sequencing of the ITS1-5.8S-ITS2 region
| Species | n | % |
|---|---|---|
| Candida albicans | 282 | 46 |
| Candida parapsilosis complex | ||
| Candida parapsilosis sensu stricto | 165 | 26.9 |
| Candida orthopsilosis | 4 | 0.6 |
| Candida metapsilosis | 2 | 0.3 |
| Candida glabrata | 60 | 9.8 |
| Candida tropicalis | 49 | 8.0 |
| Candida krusei | 10 | 1.6 |
| Other Candida spp. | ||
| Candida guilliermondii sensu stricto | 7 | 1.1 |
| Pichia caribbica | 3 | 0.5 |
| Kodamaea ohmeri | 1 | 0.2 |
| Candida dubliniensis | 5 | 0.8 |
| Candida lusitaniae | 2 | 0.3 |
| Candida kefyr | 2 | 0.3 |
| Pichia anomala | 1 | 0.2 |
| Other yeasts | ||
| Rhodotorula mucilaginosa | 7 | 1.1 |
| C. neoformans var grubii | 4 | 0.7 |
| Trichosporon asahii | 2 | 0.3 |
| C. neoformans var neoformans | 2 | 0.3 |
| Saccharomyces cerevisiae | 1 | 0.2 |
| Trichosporon dermatis | 1 | 0.2 |
| Trichosporon japonicum | 1 | 0.2 |
| Trichosporon inkin | 1 | 0.2 |
| Arxula adeninivorans | 1 | 0.2 |
TABLE 2.
Cases of fungemia caused by 2 different species (polyfungal fungemia)
| Species 1 | Species 2 | No. of cases | Species identified by ID 32C system plus Chromagar |
|---|---|---|---|
| C. albicans | C. glabrata | 4 | Both species |
| C. glabrata | C. metapsilosis | 1a | Only C. parapsilosis |
| C. glabrata | C. parapsilosis | 1a | Only C. glabrata |
| C. albicans | C. parapsilosis | 5a | Both species |
| C. tropicalis | C. parapsilosis | 1 | Both species |
| C. parapsilosis | C. metapsilosis | 1 | Only C. parapsilosis |
| C. albicans | C. krusei | 1 | Both species |
| C. parapsilosis | C. guilliermondii | 1 | Both species |
In 3 out of the 15 patients, conventional identification was able to detect only 1 of the 2 species causing the infection. A total of 6 isolates (C. parapsilosis [n = 2], C. glabrata [n = 2], C. metapsilosis [n = 1], and C. albicans [n = 1]) from 3 patients with polyfungal fungemia were excluded from antifungal susceptibility testing because molecular identification revealed a mixture of different species after several attempts to obtain pure-culture isolates.
Overall, ID 32C performed well in the identification of isolates, with the exception of C. guilliermondii and C. parapsilosis complexes, polyfungal fungemia, and other yeasts. The proportion of episodes caused by cryptic species was low. We did not detect Candida nivariensis or Candida bracarensis among isolates of the C. glabrata complex. In the C. parapsilosis complex, 96.4% of isolates were C. parapsilosis sensu stricto, 2.4% Candida orthopsilosis, and 1.2% Candida metapsilosis. In the C. guilliermondii complex, 63.6% of the strains were confirmed as C. guilliermondii sensu stricto, 27.3% as Pichia caribbica, and 9.1% as Kodamaea ohmeri. Excluding C. parapsilosis sensu stricto and C. guilliermondii sensu stricto, cryptic species of these 2 complexes frequently infected patients with gastrointestinal involvement (78%) (e.g., solid cancer, abdominal surgery, and mucositis).
Antifungal susceptibility testing.
The antifungal activities of the 7 antifungal agents studied are shown in Table 3. The rate of antifungal resistance for each species-drug combination is shown in Table 4. All isolates were susceptible to amphotericin B. Overall resistance was 5% for fluconazole, 4.4% for micafungin, and 3.8% for anidulafungin. The percentage of fluconazole-resistant Candida isolates was low and varied from 0.7% (C. albicans) to 8.6% (C. glabrata). The C. parapsilosis and C. tropicalis isolates were susceptible to fluconazole and the remaining azoles. In contrast, although C. neoformans isolates were uniformly susceptible to fluconazole, Rhodotorula spp. (100%), Trichosporon spp. (25%), and Arxula adeninivorans (100%) showed a high percentage of resistance to the agent.
TABLE 3.
Antifungal activities of antifungal agents against the 606 isolates studied (EUCAST EDef 7.2 procedure)
| Speciesa | Parameterb | Valuec (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|---|
| AmB | FLU | VRC | POS | MYC | AND | CAS | ||
| C. albicans | Mode | 0.5 | 0.125 | ≤0.015 | ≤0.015 | ≤0.015 | ≤0.015 | 0.062 |
| MIC50 | 0.5 | 0.125 | ≤0.015 | ≤0.015 | ≤0.015 | ≤0.015 | 0.062 | |
| MIC90 | 1 | 0.25 | ≤0.015 | 0.031 | ≤0.015 | ≤0.015 | 0.125 | |
| Range | (0.062 to 1) | (≤0.125 to ≥64) | (≤0.015 to ≥16) | (≤0.015 to 8) | (≤0.015 to 1) | (≤0.015 to 2) | (≤0.015 to 0.5) | |
| C. parapsilosis | Mode | 0.25 | 0.25 | ≤0.015 | 0.031 | 1 | 2 | 0.5 |
| MIC50 | 0.25 | 0.25 | ≤0.015 | 0.031 | 1 | 2 | 0.5 | |
| MIC90 | 1 | 0.5 | ≤0.015 | 0.062 | 2 | 2 | 1 | |
| Range | (0.062 to 1) | (≤0.125 to 2) | (≤0.015 to 0.062) | (≤0.015 to 0.125) | (≤0.015 to 2) | (≤0.015 to 4) | (0.125 to 2) | |
| C. glabrata | Mode | 0.25 | 8 | 0.25 | 0.5 | ≤0.015 | 0.031 | 0.125 |
| MIC50 | 0.25 | 8 | 0.25 | 0.5 | ≤0.015 | 0.031 | 0.125 | |
| MIC90 | 1 | ≥64 | 1 | 1 | ≤0.015 | 0.031 | 0.125 | |
| Range | (0.125 to 1) | (2 to ≥64) | (0.062 to 4) | (0.125 to 2) | (≤0.015 to 0.031) | (≤0.015 to 0.031) | (0.062 to 0.125) | |
| C. tropicalis | Mode | 0.5 | 0.25 | ≤0.015 | ≤0.015 | 0.031 | ≤0.015 | 0.125 |
| MIC50 | 0.5 | 0.25 | ≤0.015 | ≤0.015 | 0.031 | ≤0.015 | 0.125 | |
| MIC90 | 1 | 0.5 | 0.031 | 0.031 | 0.031 | 0.031 | 0.125 | |
| Range | (0.25 to 1) | (≤0.125 to 1) | (≤0.015 to 0.062) | (≤0.015 to 0.062) | (≤0.015 to 0.25) | (≤0.015 to 0.125) | (0.031 to 0.125) | |
| C. krusei | Mode | 0.5 | ≥64 | 0.5 | 0.25 | 0.062 | 0.062 | 0.125 |
| MIC50 | 0.5 | ≥64 | 0.5 | 0.25 | 0.062 | 0.062 | 0.125 | |
| MIC90 | 1 | ≥64 | 0.5 | 0.25 | 1 | 2 | 0.5 | |
| Range | (0.5 to 1) | (16 to ≥64) | (0.125 to 0.5) | (0.062 to 0.25) | (≤0.015 to 1) | (≤0.015 to 2) | (0.062 to 0.5) | |
| Candida spp. | Mode | 1 | 0.25 | ≤0.015 | 0.031 | 0.5 | 1 | 0.25 |
| MIC50 | 0.5 | 2 | 0.062 | 0.062 | 0.125 | 0.5 | 0.25 | |
| MIC90 | 2 | 16 | 0.5 | 0.5 | 1 | 2 | 2 | |
| Range | (0.125 to 2) | (≤0.125 to 16) | (≤0.015 to 2) | (≤0.015 to 1) | (≤0.015 to 4) | (≤0.015 to 2) | (0.031 to 8) | |
| Other yeasts | Mode | 1 | ≥64 | 0.031 | 1 | 8 | 8 | 8 |
| MIC50 | 1 | 8 | 0.25 | 0.5 | 8 | 8 | 8 | |
| MIC90 | 8 | ≥64 | 8 | 2 | ≥16 | ≥16 | ≥16 | |
| Range | (0.5 to 8) | (0.5 to ≥64) | (0.031 to 8) | (0.031 to 4) | (0.125 to ≥16) | (0.5 to ≥16) | (0.25 to ≥16) | |
Six isolates were excluded from antifungal susceptibility testing because molecular identification showed a mixture of different species after several attempts to obtain pure-culture isolates, and the Saccharomyces cerevisiae isolate did not grow in RPMI. Isolates of C. orthopsilosis (n = 3) and C. metapsilosis (n = 1) were considered C. parapsilosis for analysis of the rate of resistance.
MICs of ≤0.015 or ≤0.125 were transformed to 0.015 and 0.125, respectively for purposes of analysis.
AmB, amphotericin B; FLU, fluconazole; VRC, voriconazole; POS, posaconazole; MYC, micafungin; AND, anidulafungin; CAS, caspofungin.
TABLE 4.
Resistance to the antifungal agents studied for Candida and non-Candida species
| Species | % intermediate/resistant or N-WT isolatese |
|||||
|---|---|---|---|---|---|---|
| AmB (R) | FLU (I/R) | VRC (R) | POS (R) | MYC (I/R) | AND (I/R) | |
| C. albicans | 0 | 0/0.7 | 0.7 | 0.7 | 0/1.4 | 0/0.4 |
| C. parapsilosis complexa | 0 | 0/0 | 0 | 0 | 100/0 | 100/0 |
| C. glabrata | 0 | 91.4/8.6 | 5.2 | 7 | 0/0 | 0/0 |
| C. tropicalis | 0 | 0/0 | 0 | 0 | 0/4.1 | 0/2 |
| C. kruseib | 0 | 0/100 | 0 | 0 | 0/10 | 0/10 |
| Candida spp. | NA | 0/19 | NA | NA | NA | NA |
| Other yeasts | NA | 0/47.4 | NA | NA | 0/100 | 0/100 |
| Overall (all isolates)d | NA | 8.7/5 | NA | NA | 28.7/4.4 | 28.7/3.8 |
| Overall (only Candida species)d | 0c | 9/3.6 | 0.9 | 1 | 29.6/1.2 | 29.6/0.5 |
Isolates of C. metapsilosis (n = 3) and C. orthopsilosis (n = 1) were considered C. parapsilosis for analysis of the rate of resistance.
C. krusei was considered fluconazole resistant. For the tentative breakpoints used for non-Candida isolates, see Materials and Methods.
The rate of resistance to amphotericin B was calculated only for C. albicans, C. parapsilosis, C. glabrata, C. krusei, and C. tropicalis.
Six isolates were excluded from antifungal susceptibility testing because molecular identification showed a mixture of different species after several attempts to obtain pure-culture isolates, and the S. cerevisiae isolate did not grow in RPMI.
NA, not applicable; I, intermediate (for fluconazole, MIC, 4 mg/liter, except for C. glabrata, where the MIC was >0.062 to 32 mg/liter); R, resistant; N-WT, non-wild type; AmB, amphotericin B; FLU, fluconazole; VRC, voriconazole; POS, posaconazole; MYC, micafungin; AND, anidulafungin.
We analyzed the trend in the rate of resistance to fluconazole and echinocandins throughout the study period (Table 5). The rate of fluconazole resistance for Candida ranged from 0% (2009) to 6.4% (2010), although it has remained stable at around 4% since 2011. The echinocandin resistance rate was low, and only 7 Candida strains showed phenotypic resistance to anidulafungin and/or micafungin (C. albicans [n = 4], C. tropicalis [n = 2], and C. krusei [n = 1]). One C. tropicalis strain had an MIC for micafungin of 0.25 μg/ml and harbored a point mutation (R647G) in the HS1 region of the fks1 gene; the remaining 6 isolates were wild type for fks genes, and most of them (n = 5/6) showed a slightly higher MIC (1- or 2-fold dilution) than the breakpoint or ECOFF for anidulafungin and micafungin (see Table 7). The rate of resistance to fluconazole and echinocandin antifungals was dependent on the number of episodes caused by species with diminished susceptibility to fluconazole or by intrinsically resistant species, such as C. neoformans, Trichosporon spp., or Rhodotorula spp.
TABLE 5.
Trends in resistance to fluconazole and echinocandins during the study period (2007 to 2013)
| Drug and species | Antifungal resistance (%)c |
||||||
|---|---|---|---|---|---|---|---|
| 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | |
| Fluconazole | |||||||
| C. albicans (n = 281) | 0 | 0 | 0 | 1.8 | 2.6 | 0 | 0 |
| C. parapsilosis (n = 168) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. glabrata (n = 58) | 10 | 12.5 | 0 | 28.5 | 8.3 | 0 | 0 |
| C. tropicalis (n = 49) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. krusei (n = 10) | 100 | 0 | 0 | 100 | 100 | 100 | 100 |
| Other Candida spp. (n = 21) | 50 | 20 | 0 | 16.6 | 0 | 0 | 25 |
| Other yeasts (n = 19) | 33.3 | 100 | 25 | 100 | 50 | 60 | 0 |
| Overall (only Candida spp.) (n = 587) | 3.3 | 2.3 | 0 | 6.4 | 4.7 | 4.2 | 3.4 |
| Overall (n = 606)a | 4 | 3.3 | 1.4 | 8.4 | 5.8 | 7.8 | 3.3 |
| Echinocandins | |||||||
| C. albicans (n = 281) | 1.7 | 0 | 0 | 1.8 | 0 | 6 | 0 |
| C. parapsilosis (n = 168) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. glabrata (n = 58) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. tropicalis (n = 49) | 0 | 14.2 | 0 | 0 | 16.6 | 0 | 0 |
| C. krusei (n = 10) | 50 | 0 | 0 | 0 | 0 | 0 | 0 |
| Other Candida spp. (n = 21) | NA | NA | NA | NA | NA | NA | NA |
| Other yeasts (n = 19) | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Overall (only Candida spp.) (n = 566)b | 1.7 | 1.2 | 0 | 1.1 | 1.2 | 2.8 | 0 |
| Overall (n = 585)a,b | 4.1 | 2.3 | 5.5 | 3.4 | 3.6 | 9.2 | 3.5 |
Six isolates were excluded from antifungal susceptibility testing because molecular identification showed a mixture of different species after several attempts to obtain pure-culture isolates, and the S. cerevisiae isolate did not grow in RPMI. No C. krusei strain was isolated in 2008 and 2009.
Other Candida species isolates were excluded from the echinocandin-resistant rate analysis because of the lack of clinical breakpoints or ECOFF.
NA, not applicable. The rate of resistance was calculated overall and for Candida spp.
TABLE 7.
Characteristics of 7 patients infected with echinocandin-resistant Candida isolates
| Date of candidemia (day/mo/yr) | Candida species | Underlying condition(s) | Previous antifungals | Resistance | MIC (μg/ml) |
Antifungal treatment | Outcome | |
|---|---|---|---|---|---|---|---|---|
| AND | MYC | |||||||
| 01/01/2007 | C. albicans | Cardiac surgery | No | MYC, AND | 2 | 1 | FLU | Death at 30 days |
| 02/01/2007 | C. krusei | Diabetes, lithiasis | No | MYC, AND, FLU | 0.25 | 0.5 | AmB | Favorable |
| 02/05/2008 | C. tropicalis | Hypopharyngeal cancer | Azoles (FLU) | MYC, AND | 0.125 | 0.125 | CAS, FLU | Death at 30 days |
| 23/08/2010 | C. albicans | Diabetes, lithiasis | Azoles (VRC) | FLU, VRC, POS, MYC | 0.015 | 0.031 | MYC | Favorable |
| 31/10/2011a | C. tropicalis | Renal transplantation | Candins (MYC) | MYC | 0.062 | 0.25 | AmB | Death at 7 days |
| 19/08/2012 | C. albicans | Lymphoepithelioma | No | MYC | 0.015 | 0.031 | AND | Death at 7 days |
| 25/11/2012 | C. albicans | Hemangioblastoma | AmB and azoles (FLU) | MYC | 0.015 | 0.031 | AmB, FLU | Favorable |
Patient infected by a C. tropicalis strain harboring a point mutation (R647G) in the HS1 region of the fks1 gene.
The characteristics of patients infected with fluconazole-resistant Candida strains (n = 19; 3.5%) or echinocandin-resistant Candida strains (n = 7; 1.3%) are shown in Tables 6 and 7. Patients had severe underlying conditions, and many of them had cancer. Mortality was high (74%). Half of the patients infected by fluconazole-resistant isolates had previously received azoles (58%) and/or had cancer; in contrast, only 14% of patients infected by echinocandin-resistant strains had previously received echinocandins.
TABLE 6.
Characteristics of 19 patients infected with fluconazole-resistant Candida isolates
| Date of candidemia (day/mo/yr) | Candida species | Underlying condition(s) | Previous antifungal(s) | Resistance | Antifungal treatment | Outcome |
|---|---|---|---|---|---|---|
| 02/01/2007 | C. krusei | Diabetes, lithiasis | No | MYC, AND, FLU | AmB | Favorable |
| 20/05/2007 | C. guilliermondii | Colon adenocarcinoma | No | FLU | Unknown | Favorable |
| 27/11/2007 | C. krusei | Oropharyngeal cancer | Azoles (FLU) | FLU | AmB | Death at 7 days |
| 21/10/2007 | C. glabrata | Chronic lymphoid leukemia | Azoles (VRC) | FLU, VRC, POS | CAS | Favorable |
| 10/10/2008 | C. guilliermondii | Colon adenocarcinoma | No | FLU | VRC | Favorable |
| 22/12/2008 | C. glabrata | Diabetes | Azoles (FLU) | FLU | CAS | Favorable |
| 26/01/2010a | C. glabrata | Abdominal surgery | No | FLU, VRC, POS | AND, AmB | Favorable |
| 30/05/2010 | C. lusitaniae | Acute lymphoid leukemia | Azoles (FLU, POS, ITRAc) | FLU | AmB, VRC | Death at 30 days |
| 23/08/2010 | C. albicans | Diabetes, lithiasis | Azoles (VRC) | FLU, VRC, POS, MYC | MYC | Favorable |
| 28/09/2010 | C. krusei | Hepatitis | No | FLU | AmB, VRC | Death at 30 days |
| 27/12/2010b | C. krusei | Pelvic adenocarcinoma | Azoles (FLU) | FLU | MYC | Death at 7 days |
| 08/03/2011 | C. krusei | Retroperitoneal sarcoma | Azoles (FLU) and candins (CAS) | FLU | MYC/AND | Death at 7 days |
| 27/07/2011 | C. albicans | Abdominal surgery, newborn | No | FLU, VRC, POS | AmB | Favorable |
| 08/11/2011 | C. glabrata | Gastric adenocarcinoma | Azoles (FLU) | FLU | MYC | Favorable |
| 02/01/2012 | C. krusei | Esophageal cancer | Azoles (FLU) | FLU | MYC | Death at 30 days |
| 05/03/2012 | C. krusei | Liver cirrhosis | No | FLU | CAS | Favorable |
| 29/05/2012 | C. krusei | Renal transplantation | Azoles (FLU) | FLU | MYC | Favorable |
| 05/04/2013 | C. krusei | Acute lymphoid leukemia | Azoles (POS) | FLU | CAS, AmB | Favorable |
| 23/04/2013 | C. guilliermondii | Diabetes, pancreatitis | No | FLU | FLU, AmB/CAS, FLU | Favorable |
The patient had an additional episode of candidemia caused by C. glabrata diagnosed 1 month later.
The patient had an additional episode of candidemia caused by C. krusei diagnosed 10 days later.
ITRA, itraconazole.
The wards of admission of the patients infected with Candida sp. strains that were resistant to fluconazole or echinocandins are shown in Fig. 1. Most patients infected with fluconazole-resistant isolates were admitted to medical wards, whereas patients infected by echinocandin-resistant isolates were mainly admitted to the ICU. The rate of fluconazole resistance was 5.4% in adult patients admitted to ICUs.
FIG 1.
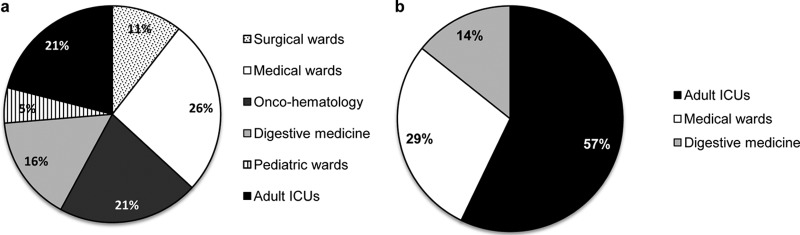
Wards of admission of patients infected with fluconazole-resistant (a) or echinocandin-resistant (b) Candida species isolates. Medical wards included geriatrics, urology, internal medicine, nephrology, infectious diseases, and otorhinolaryngology.
DISCUSSION
The study of the epidemiology of fungemia is clinically relevant, since it enables us to select accurate empirical antifungal treatment. Previous epidemiology studies of fungemia were based on isolates collected over a relatively short period, thus making it difficult to analyze trends in antifungal resistance (2, 7, 29). To our knowledge, this is the first study in Spain to analyze trends in resistance rates to fluconazole and echinocandins.
The distribution of species causing fungemia shows marked geographical differences. In northern Europe and North America, C. glabrata is the second most common species after C. albicans. Studies conducted in Spain, Italy, Greece, and Latin America show that the percentage of C. parapsilosis is higher than that of C. glabrata (6). Our results confirm that C. parapsilosis was the second most frequent species causing fungemia in patients admitted to hospitals in southern Europe. Identification to the molecular level is necessary to obtain a precise understanding of the epidemiology of fungemia, since several species go undetected with conventional identification combining Chromagar medium and the ID 32C system (10, 30–32). We found that the proportion of cryptic species in the C. parapsilosis complex was low (3.5%). In contrast, we found higher variability of species in the C. guilliermondii complex. Most patients infected by cryptic species of these complexes had digestive disorders, suggesting that the gastrointestinal tract was the source of infection. Conventional identification performed well, and only 7% of strains required ITS sequencing to confirm identification, particularly in the episodes caused by the C. parapsilosis complex and C. guilliermondii complex and in polyfungal fungemia.
Resistance to fluconazole in different studies conducted in Spain varied from 7% to 8% (1, 7, 29). The rate of resistance in Candida spp. reported here is lower than 4% and did not vary considerably during the study period, showing that development of secondary fluconazole resistance is not a problem in the clinical setting. When all the isolates were considered together, the rate of resistance was approximately 5%, which is in line with the findings of the recently published CANDIPOP multicenter study conducted in Spain (7). The rate of fluconazole resistance was very low in the most common species causing fungemia (C. albicans, C. parapsilosis, and C. tropicalis). Resistance to echinocandins in Candida remains low (<2%) and is similar to that reported previously in Spain (7). However, resistance to echinocandins is emerging in other regions, such as the United States (33). We found that the rate of resistance to echinocandins was mostly related to the presence of species that are intrinsically resistant to candins, different from the Candida spp. shown in Table 5. The rates of fluconazole and echinocandin resistance were similar to those reported after analyzing isolates collected from all over the world (34). Of the 7 Candida strains showing phenotypic resistance to echinocandins, only one isolate presented mutations in the fks genes, whereas most of the remaining isolates had MICs slightly greater than the breakpoints or ECOFFs, particularly for micafungin (Table 7). If the statistical ECOFFs for micafungin (0.031 μg/ml) (20) had been used, 3 out of 4 C. albicans isolates would have been classified as susceptible to both echinocandins.
We excluded 6 isolates from 3 patients with polyfungal fungemia from the antifungal susceptibility testing analysis. Sequence analyses enabled us to correctly identify the isolates present in a higher proportion, although a mixture of sequences was found, suggesting the presence of another species in a much lower proportion. Sequential cultures on agar plates failed to separate both species. Furthermore, isolates showed altered susceptibility (resistance to fluconazole [C. parapsilosis complex, n = 3] or to echinocandins [C. albicans, n = 1; C. glabrata, n = 2]). Isolates showing fluconazole resistance were from patients infected simultaneously with C. glabrata, and isolates showing echinocandin resistance were from patients also infected with C. parapsilosis complex strains. The inclusion of these isolates would have artificially altered the rate of resistance, thus proving the importance of molecular identification, not only for epidemiological purposes, but also when attempting to obtain an accurate rate of antifungal resistance.
Our study is subject to a series of limitations. First, we detected a high rate of echinocandin resistance in C. krusei (10%). However, the number of isolates was low, and the clinical impact of phenotypic resistance in isolates showing the wild-type fks1 gene sequence is unknown. Second, in the absence of species-specific breakpoints for all the species studied, we had to use ECOFFs as tentative breakpoints. For non-Candida species, we used the unrelated species breakpoints for fluconazole; therefore, we show the overall rate of resistance, as well as the rate of resistance exclusively for Candida spp. Finally, since we included only isolates from a single institution, we may not be able to extrapolate them to other hospitals. However, our data are comparable to those of the CANDIPOP study, in which 29 Spanish hospitals participated (7).
In conclusion, we showed that although the number of episodes of fungemia caused by cryptic species was low, molecular identification provided an accurate picture of the epidemiology of fungemia. The rate of resistance to fluconazole and echinocandins in yeast isolates causing fungemia was also low and does not show signs of increasing.
ACKNOWLEDGMENTS
We thank Thomas O'Boyle for editing and proofreading the article.
This work was supported by grants from Fondo de Investigación Sanitaria (FIS) (grant number PI11/00167). L. J. Marcos-Zambrano is supported by a predoctoral grant from FIS (FI12/00265). P. Escribano is supported by a Sara Borrell contract (CD09/00230) from FIS. J. Guinea is supported by a Miguel Servet contract from FIS (MS09/00055).
We have no conflicts of interest to report.
Footnotes
Published ahead of print 27 May 2014
REFERENCES
- 1.Almirante B, Rodríguez D, Park BJ, Cuenca-Estrella M, Planes AM, Almela M, Mensa J, Sánchez F, Ayats J, Giménez M, Saballs P, Fridkin SK, Morgan J, Rodríguez-Tudela JL, Warnock DW, Pahissa A. 2005. Epidemiology and predictors of mortality in cases of Candida bloodstream infection: results from population-based surveillance, Barcelona, Spain, from 2002 to 2003. J. Clin. Microbiol. 43:1829–1835. 10.1128/JCM.43.4.1829-1835.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puig-Asensio M, Padilla B, Garnacho-Montero J, Zaragoza O, Aguado JM, Zaragoza R, Montejo M, Muñoz P, Ruiz-Camps I, Cuenca-Estrella M, Almirante B. 2014. Epidemiology and predictive factors for early and late mortality in Candida bloodstream infections: a population-based surveillance in Spain. Clin. Microbiol. Infect. 20:O245–O254. 10.1111/1469-0691.12380 [DOI] [PubMed] [Google Scholar]
- 3.Pemán J, Cantón E, Minana JJ, Flórez JA, Echeverría J, Ortega DN, Alarcón JM, Fontanals D, Sard BG, Moreno BB, Torroba L, Ayats J, Pérez MA, Fernández MA, Reus FS, Natal IF, García-Rodríguez J, Ezpeleta G, Martín-Mazuelos E, Iglesias I, Rezusta A, de Ocáriz IR, Nieto AG. 2011. Changes in the epidemiology of fungaemia and fluconazole susceptibility of blood isolates during the last 10 years in Spain: results from the FUNGEMYCA study. Rev. Iberoam Micol. 28:91–99. 10.1016/j.riam.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 4.Pfaller M, Boyken L, Hollis R, Kroeger J, Messer S, Tendolkar S, Diekema D. 2011. Use of epidemiological cutoff values to examine 9-year trends in susceptibility of Candida species to anidulafungin, caspofungin, and micafungin. J. Clin. Microbiol. 49:624–629. 10.1128/JCM.02120-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfaller MA, Andes D, Diekema DJ, Espinel-Ingroff A, Sheehan D. 2010. Wild-type MIC distributions, epidemiological cutoff values and species-specific clinical breakpoints for fluconazole and Candida: time for harmonization of CLSI and EUCAST broth microdilution methods. Drug Resist. Updat. 13:180–195. 10.1016/j.drup.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 6.Guinea J. 2014. Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect. 20(Suppl 6):5–10. 10.1111/1469-0691.12539 [DOI] [PubMed] [Google Scholar]
- 7.Guinea J, Zaragoza O, Escribano P, Martín-Mazuelos E, Pemán J, Sánchez-Reus F, Cuenca-Estrella M. 2014. Molecular identification and antifungal susceptibility of yeast isolates causing fungemia collected in a population-based study in Spain from 2010 to 2011. Antimicrob. Agents Chemother. 58:1529–1537. 10.1128/AAC.02155-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arendrup MC, Bruun B, Christensen JJ, Fuursted K, Johansen HK, Kjaeldgaard P, Knudsen JD, Kristensen L, Moller J, Nielsen L, Rosenvinge FS, Roder B, Schonheyder HC, Thomsen MK, Truberg K. 2011. National surveillance of fungemia in Denmark (2004 to 2009). J. Clin. Microbiol. 49:325–334. 10.1128/JCM.01811-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brito LR, Guimaraes T, Nucci M, Rosas RC, Paula Almeida L, Da Matta DA, Colombo AL. 2006. Clinical and microbiological aspects of candidemia due to Candida parapsilosis in Brazilian tertiary care hospitals. Med. Mycol. 44:261–266. 10.1080/13693780500421476 [DOI] [PubMed] [Google Scholar]
- 10.Tavanti A, Davidson AD, Gow NA, Maiden MC, Odds FC. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43:284–292. 10.1128/JCM.43.1.284-292.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfaller MA, Moet GJ, Messer SA, Jones RN, Castanheira M. 2011. Geographic variations in species distribution and echinocandin and azole antifungal resistance rates among Candida bloodstream infection isolates: report from the SENTRY Antimicrobial Surveillance Program (2008 to 2009). J. Clin. Microbiol. 49:396–399. 10.1128/JCM.01398-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pappas PG, Kauffman CA, Andes D, Benjamin DK, Jr, Calandra TF, Edwards JE, Jr, Filler SG, Fisher JF, Kullberg BJ, Ostrosky-Zeichner L, Reboli AC, Rex JH, Walsh TJ, Sobel JD. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update. Clin. Infect. Dis. 48:503–535. 10.1086/596757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornely OA, Bassetti M, Calandra T, Garbino J, Kullberg BJ, Lortholary O, Meersseman W, Akova M, Arendrup MC, Arikan-Akdagli S, Bille J, Castagnola E, Cuenca-Estrella M, Donnelly JP, Groll AH, Herbrecht R, Hope WW, Jensen HE, Lass-Florl C, Petrikkos G, Richardson MD, Roilides E, Verweij PE, Viscoli C, Ullmann AJ. 2012. ESCMID guideline for the diagnosis and management of Candida diseases 2012: non-neutropenic adult patients. Clin. Microbiol. Infect. 18(Suppl 7):S19–S37. 10.1111/1469-0691.12039 [DOI] [PubMed] [Google Scholar]
- 14.Lortholary O, Desnos-Ollivier M, Sitbon K, Fontanet A, Bretagne S, Dromer F. 2011. Recent exposure to caspofungin or fluconazole influences the epidemiology of candidemia: a prospective multicenter study involving 2,441 patients. Antimicrob. Agents Chemother. 55:532–538. 10.1128/AAC.01128-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST). 2008. EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin. Microbiol. Infect. 14:398–405. 10.1111/j.1469-0691.2007.01935.x [DOI] [PubMed] [Google Scholar]
- 16.Escribano P, Recio S, Peláez T, Sánchez-Carrillo C, Rodríguez-Créixems M, Muñoz P, Bouza E, Guinea J. 2012. Molecular identification of yeasts causing fungaemia: are cryptic species frequent? Abstr. 22nd Eur. Congr. Clin. Microbiol. Infect. Dis. (ECCMID), abstr. p 834 [Google Scholar]
- 17.Marcos-Zambrano LJ, Escribano P, Muñoz P, Bouza E, Guinea J. 2014. Low rate of cryptic species and antifungal resistance in Candida isolates causing candidaemia in Madrid Abstr. 24th Eur. Congr. Clin. Microbiol. Infect. Dis. (ECCMID), abstr. eP-234 . [Google Scholar]
- 18.White T, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 322 In PCR protocols: guide to methods and applications.Academic Press, San Diego, CA [Google Scholar]
- 19.Arendrup MC, Cuenca-Estrella M, Lass-Florl C, Hope W. 2012. EUCAST technical note on the EUCAST definitive document EDef 7.2: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts, EDef 7.2 (EUCAST-AFST). Clin. Microbiol. Infect. 18:E246–E247. 10.1111/j.1469-0691.2012.03880.x [DOI] [PubMed] [Google Scholar]
- 20.Arendrup MC, Cuenca-Estrella M, Lass-Florl C, Hope WW. 2013. Breakpoints for antifungal agents: an update from EUCAST focussing on echinocandins against Candida spp. and triazoles against Aspergillus spp. Drug Resist. Updat. 16:81–95. 10.1016/j.drup.2014.01.001 [DOI] [PubMed] [Google Scholar]
- 21.Arendrup MC, Rodriguez-Tudela JL, Park S, Garcia-Effron G, Delmas G, Cuenca-Estrella M, Gomez-Lopez A, Perlin DS. 2011. Echinocandin susceptibility testing of Candida spp. Using EUCAST EDef 7.1 and CLSI M27-A3 standard procedures: analysis of the influence of bovine serum albumin supplementation, storage time, and drug lots. Antimicrob. Agents Chemother. 55:1580–1587. 10.1128/AAC.01364-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espinel-Ingroff A, Arendrup MC, Pfaller MA, Bonfietti LX, Bustamante B, Canton E, Chryssanthou E, Cuenca-Estrella M, Dannaoui E, Fothergill A, Fuller J, Gaustad P, Gonzalez GM, Guarro J, Lass-Florl C, Lockhart SR, Meis JF, Moore CB, Ostrosky-Zeichner L, Pelaez T, Pukinskas SR, St-Germain G, Szeszs MW, Turnidge J. 2013. Interlaboratory variability of caspofungin MICs for Candida spp. using CLSI and EUCAST methods: should the clinical laboratory be testing this agent? Antimicrob. Agents Chemother. 57:5836–5842. 10.1128/AAC.01519-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.European Committee on Antimicrobial Susceptibility Testing. 2010. Voriconazole: rationale for the clinical breakpoints, version 2.0. http://www.eucast.org
- 24.European Committee on Antimicrobial Susceptibility Testing. 2010. Posaconazole: rationale for the clinical breakpoints, version 1.0. http://www.eucast.org
- 25.European Committee on Antimicrobial Susceptibility Testing. 2013. Micafungin and Candida spp.: rationale for the clinical breakpoints, version 1.0. http://www.eucast.org
- 26.Aller AI, Martin-Mazuelos E, Lozano F, Gomez-Mateos J, Steele-Moore L, Holloway WJ, Gutierrez MJ, Recio FJ, Espinel-Ingroff A. 2000. Correlation of fluconazole MICs with clinical outcome in cryptococcal infection. Antimicrob. Agents Chemother. 44:1544–1548. 10.1128/AAC.44.6.1544-1548.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desnos-Ollivier M, Bretagne S, Raoux D, Hoinard D, Dromer F, Dannaoui E. 2008. Mutations in the fks1 gene in Candida albicans, C. tropicalis, and C. krusei correlate with elevated caspofungin MICs uncovered in AM3 medium using the method of the European Committee on Antibiotic Susceptibility Testing. Antimicrob. Agents Chemother. 52:3092–3098. 10.1128/AAC.00088-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Effron G, Park S, Perlin DS. 2009. Correlating echinocandin MIC and kinetic inhibition of fks1 mutant glucan synthases for Candida albicans: implications for interpretive breakpoints. Antimicrob. Agents Chemother. 53:112–122. 10.1128/AAC.01162-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pemán J, Cantón E, Quindós G, Eraso E, Alcoba J, Guinea J, Merino P, Ruiz-Pérez-de-Pipaón MT, Pérez-del-Molino L, Linares-Sicilia MJ, Marco F, García J, Roselló EM, Gómez de la Pedrosa G, Borrell N, Porras A, Yague G. 2012. Epidemiology, species distribution and in vitro antifungal susceptibility of fungaemia in a Spanish multicentre prospective survey. J. Antimicrob. Chemother. 67:1181–1187. 10.1093/jac/dks019 [DOI] [PubMed] [Google Scholar]
- 30.Cornet M, Sendid B, Fradin C, Gaillardin C, Poulain D, Nguyen HV. 2011. Molecular identification of closely related Candida species using two ribosomal intergenic spacer fingerprinting methods. J. Mol. Diagn. 13:12–22. 10.1016/j.jmoldx.2010.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, Millar BC, Moore JE, McClurg R, Walker MJ, Evans J, Hedderwick S, McMullan R. 2002. Comparison of API20C with molecular identification of Candida spp. isolated from bloodstream infections. J. Clin. Pathol. 55:774–777. 10.1136/jcp.55.10.774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yong PV, Chong PP, Lau LY, Yeoh RS, Jamal F. 2008. Molecular identification of Candida orthopsilosis isolated from blood culture. Mycopathologia 165:81–87. 10.1007/s11046-007-9086-8 [DOI] [PubMed] [Google Scholar]
- 33.Alexander BD, Johnson MD, Pfeiffer CD, Jimenez-Ortigosa C, Catania J, Booker R, Castanheira M, Messer SA, Perlin DS, Pfaller MA. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 56:1724–1732. 10.1093/cid/cit136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfaller MA, Messer SA, Woosley LN, Jones RN, Castanheira M. 2013. Echinocandin and triazole antifungal susceptibility profiles of opportunistic yeast and mould clinical isolates (2010–2011): Application of new CLSI clinical breakpoints and epidemiological cutoff values to characterize geographic and temporal trends of antifungal resistance. J. Clin. Microbiol. 51:2571–2581. 10.1128/JCM.00308-13 [DOI] [PMC free article] [PubMed] [Google Scholar]


