Abstract
Resistance to daptomycin in enterococcal clinical isolates remains rare but is being increasingly reported in the United States and worldwide. There are limited data on the genetic relatedness and microbiological and clinical characteristics of daptomycin-nonsusceptible enterococcal clinical isolates. In this study, we assessed the population genetics of daptomycin-nonsusceptible Enterococcus faecium (DNSE) clinical isolates by multilocus sequence typing (MLST) and whole-genome sequencing analysis. Forty-two nonduplicate DNSE isolates and 43 randomly selected daptomycin-susceptible E. faecium isolates were included in the analysis. All E. faecium isolates were recovered from patients at a tertiary care medical center in suburban New York City from May 2009 through December 2013. The daptomycin MICs of the DNSE isolates ranged from 6 to >256 μg/ml. Three major clones of E. faecium (ST18, ST412, and ST736) were identified among these clinical isolates by MLST and whole-genome sequence-based analysis. A newly recognized clone, ST736, was seen in 32 of 42 (76.2%) DNSE isolates and in only 14 of 43 (32.6%) daptomycin-susceptible E. faecium isolates (P < 0.0001). This report provides evidence of the association between E. faecium clone ST736 and daptomycin nonsusceptibility. The identification and potential spread of this novel E. faecium clone and its association with daptomycin nonsusceptibility constitute a challenge for patient management and infection control at our medical center.
INTRODUCTION
Daptomycin is a cyclic lipopeptide with in vitro bactericidal activity against Gram-positive bacteria. Daptomycin is approved by the U.S. Food and Drug Administration (FDA) for the treatment of complicated skin and soft-tissue infections, bacteremia, and right-sided infective endocarditis caused by certain Gram-positive microorganisms (1–3). Given its potent bactericidal activity against enterococci, low risk of serious side effects, and minimal drug-drug interactions, daptomycin is increasingly used in the United States and other countries to treat serious staphylococcal and enterococcal infections. Daptomycin is also used “off label” as the antibiotic of choice for serious infections caused by vancomycin-resistant enterococci (VRE) (1, 4–6), despite the high cost of the drug and some toxicity issues such as myopathy and eosinophilic pneumonia (7).
In vitro antimicrobial susceptibility testing of Gram-positive microorganisms worldwide suggests that resistance to daptomycin in Enterococcus spp. is rare (8–12). Daptomycin-nonsusceptible enterococci, however, are emerging as a cause of health care-associated infection (4, 6) and may be fairly common; they comprised 3.9% of 2,029 Enterococcus faecium isolates from U.S. hospitals over the period 2007 to 2010 (13) and 15.2% among VRE blood isolates at a New York cancer center (14). In addition to some earlier case reports (reviewed in reference 4), a notable increase in the number of patients infected or colonized with daptomycin-nonsusceptible enterococci has been reported from different geographic regions in the United States (14–19).
The majority of daptomycin-nonsusceptible enterococci documented in the literature are vancomycin-resistant E. faecium, although vancomycin-resistant E. faecalis and vancomycin-susceptible E. faecium have been reported (4, 6). Most patients develop resistance during daptomycin therapy (18, 20, 21). De novo resistance is also reported for some patients without prior use of daptomycin (16, 22). The results of a recent case-control study suggested that immunosuppression, the presence of comorbid conditions, and prior exposure to antimicrobials are independent predictors of infections caused by daptomycin-nonsusceptible enterococci (17). The mechanism of daptomycin resistance in enterococci, especially in E. faecium, remains to be fully elucidated. Based on whole-genome analysis of a very few clinical and laboratory-derived strains, mutations in several genes have been implicated in the development of daptomycin resistance in enterococci (15, 23–26). The emergence and spread of daptomycin-nonsusceptible enterococci in distinct patient populations or geographic regions are likely caused by a diverse population of enterococci (14, 18). Such studies are largely restricted due to the lack of sufficient daptomycin-nonsusceptible clinical isolates. In this study, we determined the genetic relatedness and clinical and microbiological characteristics of daptomycin-nonsusceptible E. faecium (DNSE) isolates from patients in a tertiary care medical center in suburban New York City. A novel E. faecium clone associated with daptomycin nonsusceptibility was identified and characterized.
(Part of this work was presented at the 113th General Meeting of the American Society for Microbiology at San Francisco, CA, May 2013.)
MATERIALS AND METHODS
Bacterial isolates.
Forty-two nonduplicate, daptomycin-nonsusceptible E. faecium (DNSE) clinical isolates were included in this study. These DNSE isolates were recovered from patients of a tertiary medical center in suburban New York City from May 2009 through December 2013. The Institutional Review Board of New York Medical College approved this study. For patients with multiple DNSE isolates, only the first confirmed isolate was selected. In this report, DNSE was defined as an E. faecium isolate with a daptomycin MIC of >4 μg/ml determined by a MicroScan WalkAway system (Siemens, Tarrytown, NY) and confirmed by Etest (bioMérieux, Durham, NC) and/or the reference broth microdilution method per the Clinical Laboratory and Standards Institute (CLSI) guidelines (27). Forty-three nonduplicate daptomycin-susceptible E. faecium (DSE) isolates from the same study period were randomly selected and analyzed for comparison. All DNSE and DSE clinical isolates were identified on the basis of colony morphology and conventional biochemical tests and were confirmed by sequence analysis of 16S rRNA genes. Antimicrobial susceptibility of enterococcal isolates to daptomycin and other antimicrobial agents was determined routinely using MicroScan dried Gram-positive panels by the Prompt inoculation method with a MicroScan WalkAway system. Confirmatory Etest was performed for DNSE isolates reported by MicroScan (i) if the isolates were recovered from blood or other sterile body sites, (ii) if the isolates were recovered from patients in the intensive care units and oncology wards, or (iii) if the test was requested by clinicians. Eleven DNSE isolates confirmed by Etest in 2009 and 2010 were also sent to Laboratory Specialists (Westlake, OH) and validated by the CLSI reference broth microdilution method (27).
DNA preparation.
Bacterial isolates were subcultured on blood agar or Trypticase soy broth from cryovials stored at −80°C or from refrigerated nutrient slants. DNA from pure bacterial culture was extracted by using a protocol for isolation of genomic DNA from Gram-positive bacteria with a QIAamp Mini DNA kit (Qiagen, Germantown, MD) per instructions of the manufacturer. DNA concentration was measured by using a Qubit fluorometric quantitation system (Life Technologies, Grand Island, NY).
Whole-genome sequencing (WGS).
DNA libraries from clinical isolates were prepared by using a Nextera XT sample preparation kit according to instructions of the manufacturer (Illumina, San Diego, CA). WGS was performed on an Illumina MiSeq system by using paired-end methods (2 × 250 bp) with either 24 or 96 indices incorporated. Taken as a whole, the data set had a mean fold coverage of 59× and a median fold coverage of 53× for the chromosomal DNA.
MLST analysis.
Multilocus sequence typing (MLST) data were obtained using both classic Sanger DNA sequencing and WGS methods. For classic MLST analysis, 7 loci (atpA, ddl, gdh, purK, gyd, pstS, and adk) of each E. faecium isolate were amplified using PCR primers and conditions as described by Homan et al. (28). PCR amplicons were sequenced using a BigDye Terminator v1.1 cycle sequencing kit (Life Technology, Foster City, CA) on an ABI 3500xl Genetic Analyzer.
For WGS-based MLST analysis, WGS sequence data were aligned to E. faecium sequence type 17 (ST17) reference strain Aus0004 (NC_017022.1) (29) using the BWA alignment software (30, 31). Consensus base calls for the aligned nucleotides were derived using SAMtools (32). MLST profiles for E. faecium were downloaded from an online MLST database (http://efaecium.mlst.net). The consensus genome sequences generated from the alignment were then analyzed using a locally configured version of the BIGSdb framework to call MLST types (33).
Phylogenetic analysis.
Single nucleotide variations (SNVs) were called from the BWA alignment to E. faecium Aus0004 chromosome 1 using varscan. Phylogenetic trees were obtained using a maximum-likelihood method (34). Trees were visualized and annotated using the Interactive Tree Of Life online tool (35). The genetic relatedness of different enterococcal isolates and STs was explored using the goeBURST program (36).
Statistical analysis.
The Fisher's exact test from the Prism software (version 5.0) was used to determine the statistical significance of the results of comparisons of different groups of enterococci with distinct clinical and microbiological characteristics.
RESULTS
Microbiological and clinical characteristics of DNSE isolates.
From May 2009 through December 2013, 42 nonduplicate E. faecium clinical isolates were confirmed as DNSE by Etest and/or reference broth microdilution. The 42 DNSE isolates were recovered from urine (n = 17, 40.5%), blood (n = 13, 30.9%), surgical wounds (n = 7, 16.7%), and peritoneal fluid (n = 5, 11.9%). Thirty-five of 42 (83.3%) isolates were from patients with distinct signs and symptoms suggestive of infection associated with these enterococcal isolates, including 13 patients with bacteremia. The average age of the patients was 56.5 (range, 23 to 84) years. Twenty-two (52.4%) of these patients were immunocompromised due to underlying malignancy and/or ongoing immunosuppressive therapies. Twenty-four of 42 (57.1%) patients had had exposure to treatment with daptomycin within the past 12 months prior to the DNSE isolation. It is noteworthy that 17 of 21 (81.0%) patients with DNSE colonization or infection from 2009 to 2012 had a prior exposure to daptomycin, in contrast to only 7 of 21 (33.3%) patients in 2013 (P = 0.0044). The microbiological characteristics of 42 DNSE isolates and clinical features of patients are summarized in Table 1.
TABLE 1.
Clinical and microbiological characteristics of DNSE isolatesa
| Isolate no. | Year | Isolate designation | ST | VAN MICb (μg/ml) | DAP MICc (μg/ml) | Source | Underlying disease(s) or condition(s) | Age (yrs) | Sex | No. of days prior to DNSE isolationd | Prior DAP exposure |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2009 | E39 | ST736 | >16 | 96 | Blood | ALL | 28 | F | 79 | Y |
| 2 | 2009 | E49 | ST750 | ≤2 | 16 | Blood | Cholangiocarcinoma | 72 | F | 29 | Y |
| 3 | 2009 | E7 | ST18 | 1 | 32 | Urine | Liver transplant, abdominal abscess | 63 | F | 83 | Y |
| 4 | 2009 | E8 | ST282 | >16 | 16 | Urine | AML | 45 | M | 52 | Y |
| 5 | 2010 | E13 | ST736 | 1 | 16 | Blood | ALL | 72 | M | 17 | Y |
| 6 | 2010 | E14 | ST736 | >16 | 8 | Blood | ALL | 46 | F | 24 | Y |
| 7 | 2010 | E17 | ST736 | >16 | 16 | Wound | Hepatitis C, DM | 58 | M | 42 | Y |
| 8 | 2010 | E20 | ST893 | 2 | 6 | Urine | Hepatitis B, lymphoma | 58 | M | 43 | Y |
| 9 | 2010 | E34 | ST736 | >16 | 48 | Urine | Multiple myeloma, DM | 54 | M | 9 | Y |
| 10 | 2010 | E50 | ST412 | >16 | 64 | Blood | Heart transplant | 65 | F | 53 | Y |
| 11 | 2010 | E53 | ST736 | >16 | 24 | Peritoneum | Trauma, bowel perforation | 48 | M | 38 | Y |
| 12 | 2010 | E78 | ST18 | >16 | 6 | Peritoneum | Fulminant hepatic failure, liver transplant | 66 | M | 15 | N |
| 13 | 2011 | E91 | ST736 | >16 | 64 | Blood | ALL, HSCT, GVHD | 24 | M | 41 | N |
| 14 | 2011 | E146 | ST736 | >16 | 32 | Urine | Cholangitis/peritonitis | 69 | F | 88 | Y |
| 15 | 2011 | E165 | ST736 | >16 | 6 | Urine | Hepatitis C, DM, ovarian carcinoma | 58 | F | 22 | N |
| 16 | 2011 | E176 | ST736 | >16 | 12 | Blood | Chronic osteomyelitis, decubitus ulcer, paraplegia | 54 | M | 238 | Y |
| 17 | 2012 | E166 | ST736 | 2 | 8 | Urine | Cirrhosis, enterocutaneous fistula | 56 | M | 26 | Y |
| 18 | 2012 | E168 | ST736 | >16 | 32 | Urine | Scrotal/testicular cancer, spina bifida | 39 | M | 75 | Y |
| 19 | 2012 | E169 | ST736 | >16 | 8 | Blood | Endocarditis, necrotizing fasciitis | 61 | F | 262 | Y |
| 20 | 2012 | E173 | ST736 | >16 | 32 | Wound | Burn, osteomyelitis | 23 | M | 162 | Y |
| 21 | 2012 | E203 | ST736 | >16 | 8 | Blood | ALL | 41 | F | 11 | N |
| 22 | 2013 | E207 | ST736 | >16 | 12 | Urine | Cirrhosis, autoimmune hepatitis | 62 | F | 13 | N |
| 23 | 2013 | E208 | ST736 | >16 | 6 | Peritoneum | Cirrhosis, ESRD | 66 | F | 5 | N |
| 24 | 2013 | E209 | ST736 | >16 | 8 | Urine | AML, DM | 61 | M | 0 | N |
| 25 | 2013 | E211 | ST412 | >16 | 12 | Urine | AML, colon cancer | 38 | M | 86 | Y |
| 26 | 2013 | E214 | ST736 | >16 | 8 | Blood | Hepatitis C, liver/kidney transplant | 56 | M | 5 | N |
| 27 | 2013 | E218 | ST736 | >16 | 8 | Urine | CABG, MVR, ureteral stent | 84 | F | 19 | N |
| 28 | 2013 | E222 | ST736 | >16 | 8 | Urine | Lymphoma, HSCT | 48 | M | 1 | N |
| 29 | 2013 | E225 | ST736 | >16 | 6 | Blood | Cirrhosis, ESRD, liver transplant | 69 | F | 139 | Y |
| 30 | 2013 | E226 | ST736 | >16 | 6 | Peritoneum | Liver transplant | 71 | M | 5 | N |
| 31 | 2013 | E231 | ST736 | >16 | 6 | Wound | Necrotizing fasciitis | 41 | M | 40 | Y |
| 32 | 2013 | E236 | ST736 | >16 | 6 | Wound | s/p LVAD, osteomyelitis | 69 | M | 16 | N |
| 33 | 2013 | E237 | ST736 | >16 | 8 | Blood | Lymphoma | 66 | M | 12 | N |
| 34 | 2013 | E241 | ST18 | >16 | 12 | Urine | Multiple myeloma | 56 | F | 36 | N |
| 35 | 2013 | E242 | ST736 | >16 | 8 | Wound | Necrotizing fasciitis | 52 | M | 32 | Y |
| 36 | 2013 | E243 | ST736 | >16 | >256 | Peritoneum | Endometrial carcinoma | 56 | F | 21 | Y |
| 37 | 2013 | E245 | ST18 | >16 | 12 | Urine | Ovarian cancer, AML | 66 | F | 18 | N |
| 38 | 2013 | E249 | ST736 | >16 | 8 | Urine | Vulvar cancer, wound infection | 61 | F | 34 | N |
| 49 | 2013 | E251 | ST736 | >16 | 6 | Urine | Trauma, bowel perforation | 75 | F | 29 | N |
| 40 | 2013 | E252 | ST412 | >16 | 8 | Blood | AML | 56 | F | 30 | Y |
| 41 | 2013 | E253 | ST736 | >16 | >256 | Wound | ESRD, prosthetic knee infection | 74 | F | 83 | Y |
| 42 | 2013 | E261 | ST736 | >16 | 8 | Urine | SLE, kidney transplant, DM | 49 | F | 11 | N |
n = 42. ALL, acute lymphocytic leukemia; AML, acute myeloid leukemia; HSCT, hematopoietic stem cell transplant; DM, diabetes mellitus; ESRD, end-stage renal disease; GVHD, graft versus host disease; CNS, central nervous system; SLE, systemic lupus erythematosus; CABG, coronary artery bypass graft; MVR, mitral valve repair; s/p LVAD, status post-left ventricular assist device; CHF, congestive heart failure; F, female; M, male; Y, yes; N, no.
Vancomycin (VAN) MICs were determined using MicroScan Gram-positive MIC panels.
Daptomycin (DAP) MICs were determined by Etest. DNSE for isolates 1 to 11 was also confirmed by CLSI reference broth microdilution.
Duration of hospital stay prior to DNSE isolation.
Sequence types of DNSE and DSE isolates.
MLST data were derived from WGS of 85 E. faecium (42 DNSE and 43 DSE) isolates. Of these, the MLST sequence types of 25 E. faecium isolates were also determined using Sanger DNA sequencing. Identical alleles and STs were confirmed for all but one isolate, for which the Sanger DNA sequencing and the WGS method called different ddl alleles, and the ST determined by the Sanger DNA sequencing was used in the final data analysis.
For the 42 DNSE isolates, six different STs were observed (Table 2). Surprisingly, 32 of 42 (76.2%) DNSE isolates possessed a novel sequence type, ST736. Of the remaining DNSE isolates, ST18 (n = 4) and ST412 (n = 3) were observed in 7 of 10 non-ST736 DNSE isolates, while three other isolates each represented a distinct ST (ST282, ST750, and ST893).
TABLE 2.
Distribution of sequence types among DNSE and daptomycin-susceptible E. faecium clinical isolates determined by multilocus sequence typing analysis
| ST | No. (%) of isolates |
P value | |
|---|---|---|---|
| DNSE | DSE | ||
| ST736 | 32 (76.2) | 14 (32.6) | <0.0001 |
| ST18 | 4 (9.5) | 14 (32.6) | 0.0155 |
| ST412 | 3 (7.1) | 9 (20.9) | 0.1171 |
| ST282 | 1 (2.4) | 1 (2.3) | |
| ST750 | 1 (2.4) | 0 | |
| ST117 | 0 | 1 (2.3) | |
| ST584 | 0 | 1 (2.3) | |
| Newa | 1 (2.4) | 3 (7.0) | |
| Total | 42 (100) | 43 (100) | |
The four new STs first described in this study have been assigned the designations ST893 (isolate E20), ST894 (isolate E179), ST895 (isolate E238), and ST896 (isolate E293).
To determine if E. faecium ST736 is the predominant clone that is endemic among patients at our study site, we also sequenced the whole genomes of 43 additional DSE clinical isolates randomly selected from the same study period. The MLST data from these DSE isolates are summarized in Table 2. Three major E. faecium clones (ST18, ST412, and ST736) were identified among these DSE isolates. While ST736 was recognized as the predominant clone in DNSE isolates, it was seen in only 14 of 43 (32.6%) of the daptomycin-susceptible E. faecium clinical isolates. Twenty-nine of 43 (67.4%) DSE isolates were ST18 (n = 14), ST412 (n = 9), and other non-ST736 (n = 6) (DNSE versus DSE, P < 0.0001). This demonstrates that the ST736 is variably distributed among DNSE and DSE isolates and is not the sole dominant enterococcal clone at our study site.
Antimicrobial susceptibility of DNSE isolates.
As shown in Table 1, the daptomycin MIC of 42 DNSE isolates ranged from 6 to >256 μg/ml, with a geometric mean MIC of 14 μg/ml. Notably, 10 of 42 (23.8%) DNSE isolates had a high-level daptomycin MIC (> = 32 μg/ml). Two of these isolates (E243 and E253, both belonging to ST736) were resistant to daptomycin, with an in vitro MIC of >256 μg/ml. All 42 DNSE isolates examined were resistant to ampicillin as typically seen in North American E. faecium isolates (9). Thirty-seven (88.1%) DNSE isolates were resistant and 5 (11.9%) DNSE isolates were susceptible to vancomycin. Forty of 42 (95.2%) DNSE isolates were susceptible to linezolid and quinupristin-dalfopristin. Similar susceptibility profiles were seen among DNSE and DSE isolates for almost all antibiotics. The antimicrobial susceptibility profiles of DNSE and DSE isolates are summarized in Table S1 in the supplemental material.
Population genetics of DNSE and DSE isolates.
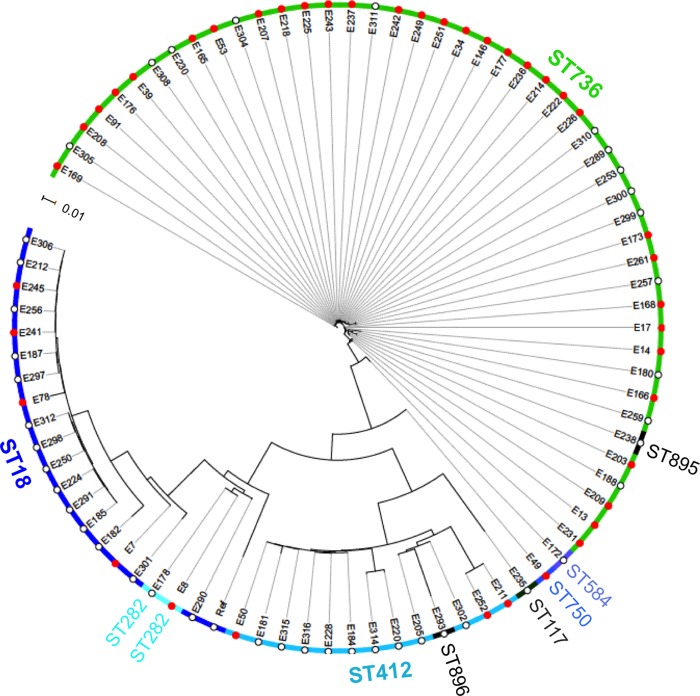
Based on MLST data, 76 of 85 (89.4%) E. faecium isolates analyzed belonged to three major STs (ST18, ST412, and ST736). Of the remaining 9 E. faecium isolates, 5 isolates each possessed a distinct ST, while 4 isolates had new STs (assigned as ST893 to ST896). To further determine the population genetics of these E. faecium clinical isolates, a phylogenetic tree based on the SNVs of WGS data, compared to the reference genome from isolate Aus0004, was generated. Collectively, the 85 isolates analyzed had a mean SNV call rate of 1.96 SNVs/kb on the reference genome. As shown in Fig. 1, three major clusters, corresponding to ST18, ST412, and ST736, were observed for the 83 E. faecium clinical isolates included in the analysis. DNSE isolate E20 and DSE isolate E179 (not shown in Fig. 1) showed a larger genetic distance from the reference isolate as well as from other E. faecium clinical isolates and may represent two unique E. faecium clones. Although bearing a new ST, DSE isolates E238 and E293 showed high genetic similarity and clustered in the ST736 and ST412 groups, respectively.
FIG 1.
Whole-genome-based phylogenetic tree of daptomycin-nonsusceptible (red solid circles; n = 42) and daptomycin-susceptible (white empty circles; n = 43) E. faecium clinical isolates. The phylogenetic tree was generated using a maximum-likelihood method based on single nucleotide variations of genomes compared to a reference strain, Aus0004 (29). Two outlier isolates (E20 and E179) were excluded from the phylogenetic tree for improved visualization of the genetic distance between different subgroups of E. faecium.
Genetic relatedness of DNSE with VRE isolates.
To explore the genetic relatedness of E. faecium isolates from this study to other VRE clones reported worldwide, comparative clustering analysis was performed using the goeBURST algorithm. Data from ∼500 different STs of E. faecium in clonal complex 17 (CC17) that are currently available in the MLST database were included in the analysis. Interestingly, the two major STs (ST18 and ST412) seen in DSE isolates have been widely reported in other geographic areas of North America and South America (37–39). E. faecium clone ST736 that was seen in 76.2% of DNSE and 32.6% of DSE isolates appears to represent a novel emerging group within CC17. E. faecium ST736 is most closely related to ST280, showing a single allele variation in 1 of the 7 alleles (purK). Two allelic differences (ddl and purK) between ST736 and ST17, the presumed founder of CC17 enterococci, were noticed (see Fig. S1 in the supplemental material).
DISCUSSION
Vancomycin-resistant enterococci belonging to CC17 have been emerging globally since the 1990s and are now in the predominant group of enterococci causing nosocomial infections (5, 40). Our findings in 42 DNSE clinical isolates from patients of a tertiary medical center in suburban New York City, together with those of several recent studies from other U.S. institutions (14–19), demonstrate the emergence and spread of DNSE in the U.S. health care setting. However, there are only limited data available on the population genetics of DNSE isolates. It is also unknown if an association exists between daptomycin nonsusceptibility and a particular genetic group or clone of enterococci.
In this report, we describe the distribution of various STs among E. faecium clinical isolates and their distinct associations with daptomycin susceptibility. A total of six STs have been identified from the 42 DNSE isolates. Strikingly, 32 of 42 (76.2%) DNSE isolates exhibited high genetic similarity by MLST and WGS analysis and were defined as a single novel E. faecium clone, ST736. In contrast, among daptomycin-susceptible E. faecium isolates, three major clones (ST18, ST412, and ST736) were recognized. E. faecium ST18 and ST412 isolates have been well documented in North America, including the United States, Canada, and Mexico (37–39). They are among the most common STs found in VRE isolates from Canada since 2006 (37). Similarly, E. faecium ST18 and ST412, together with other non-ST736 isolates, account for approximately two-thirds of the DSE isolates analyzed in this study, demonstrating that E. faecium clone ST736 is not the overall dominant endemic clone at our study site. Conversely, ST736 is the predominant clone among DNSE isolates and is therefore strongly associated with daptomycin nonsusceptibility.
Ten of 42 (23.8%) DNSE isolates in this study showed a high level (MIC ≥ 32 μg/ml) of daptomycin resistance. Two of these DNSE isolates exhibited a daptomycin MIC of >256 μg/ml. Both de novo resistance and development of resistance during daptomycin treatment have been reported. A possible community source of de novo DNSE has also been proposed (41, 42). It is unclear if this predominance of the ST736 clone, especially among DNSE clinical isolates in our institution, is attributable to nosocomial clonal spread or whether the ST736 clone carries unique genetic and microbiological characteristics that predispose it to daptomycin nonsusceptibility. Twelve of 17 (70.6%) patients infected with DNSE ST736 isolates in 2013 had no prior exposure to daptomycin. It is likely that nosocomial transmission and clonal spread of E. faecium ST736 isolates occurs among some patients, if not all, at this study site. Our finding is similar to data from a study at a New York City hospital (14) but is distinct from data from a recent study in Ohio in which a diverse population of DNSE was reported (i.e., 16 pulsed-field gel electrophoresis [PFGE] types and 24 subtypes among the 29 patient isolates available for molecular typing) (18). Due to the lack of sufficient strain typing data for comparison, we cannot conclude if the observed differences between the two studies are due to the dissimilarity in the subtypes of the infecting enterococci, their transmission modes among discrete patient populations, or mechanisms of development of resistance to daptomycin.
Daptomycin nonsusceptibility has been shown in other non-ST736 E. faecium clones as well, including ST18, ST412 in this study, and ST203 (referred from the published whole-genome sequence of confirmed DNSE isolate R446 [GenBank accession numbers AMAJ01000001 to AMAJ01000323] from Texas). The limitations of our study are that all of the DNSE isolates were from a single institution and that most isolates were selected from patients in the intensive care and oncology units. It will be meaningful and worthwhile to assess the clonality of DNSE isolates from other institutions and geographic regions and compare them to a larger collection of DSE isolates to better define the population genetics of daptomycin-nonsusceptible enterococcal infections.
In conclusion, this report provides evidence of the association between E. faecium clone ST736 and daptomycin nonsusceptibility. Our findings offer new insights on the population genetics of DNSE clinical isolates and a possible direction in which to explore the mechanisms of daptomycin resistance in enterococci. The identification of novel E. faecium clone ST736 and its predominance in DNSE isolates may imply a potential clonal outbreak of this organism in our institution. Recently, E. faecium ST736 isolates have been recovered from the cerebrospinal fluids of two patients in a U.S. hospital in Bethesda, MD (http://efaecium.mlst.net/; accessed 15 May 2014). This also raises concern on its further spread in U.S. hospital settings as well as the associated patient management and infection control challenges. Further studies will be undertaken to determine the genomic similarities and differences between different STs of E. faecium clones and their relative likelihood in developing resistance to daptomycin.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by research grants from Cubist Pharmaceuticals (G.W.), the Department of Pathology of New York Medical College, and Philips Research North America.
We thank Ira Schwartz and Gary P. Wormser for providing constructive comments on the manuscript. We also thank the medical technologists in the Clinical Microbiology Laboratory at Westchester Medical Center for technical assistance and Laboratory Specialists, Inc. (Westlake, OH), for susceptibility testing by the reference method for selected isolates.
G.W. has received grant support from Cubist Pharmaceuticals. A.D. is on the speaker's bureau of and has received honoraria from Cubist Pharmaceuticals. All of the other authors report no conflicts of interest.
Footnotes
Published ahead of print 9 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02683-14.
REFERENCES
- 1.Carpenter CF, Chambers HF. 2004. Daptomycin: another novel agent for treating infections due to drug-resistant gram-positive pathogens. Clin. Infect. Dis. 38:994–1000. 10.1086/383472 [DOI] [PubMed] [Google Scholar]
- 2.Steenbergen JN, Alder J, Thorne GM, Tally FP. 2005. Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections. J. Antimicrob. Chemother. 55:283–288. 10.1093/jac/dkh546 [DOI] [PubMed] [Google Scholar]
- 3.Kullar R, Casapao AM, Davis SL, Levine DP, Zhao JJ, Crank CW, Segreti J, Sakoulas G, Cosgrove SE, Rybak MJ. 2013. A multicentre evaluation of the effectiveness and safety of high-dose daptomycin for the treatment of infective endocarditis. J. Antimicrob. Chemother. 68:2921–2926. 10.1093/jac/dkt294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelesidis T, Humphries R, Uslan DZ, Pegues DA. 2011. Daptomycin nonsusceptible enterococci: an emerging challenge for clinicians. Clin. Infect. Dis. 52:228–234. 10.1093/cid/ciq113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10:266–278. 10.1038/nrmicro2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humphries RM, Pollett S, Sakoulas G. 2013. A current perspective on daptomycin for the clinical microbiologist. Clin. Microbiol. Rev. 26:759–780. 10.1128/CMR.00030-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fowler VG, Jr, Boucher HW, Corey GR, Abrutyn E, Karchmer AW, Rupp ME, Levine DP, Chambers HF, Tally FP, Vigliani GA, Cabell CH, Link AS, Demeyer I, Filler SG, Zervos M, Cook P, Parsonnet J, Bernstein JM, Price CS, Forrest GN, Fatkenheuer G, Gareca M, Rehm SJ, Brodt HR, Tice A, Cosgrove SE. 2006. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N. Engl. J. Med. 355:653–665. 10.1056/NEJMoa053783 [DOI] [PubMed] [Google Scholar]
- 8.Sader HS, Jones RN. 2009. Antimicrobial susceptibility of Gram-positive bacteria isolated from US medical centers: results of the Daptomycin Surveillance Program (2007–2008). Diagn. Microbiol. Infect. Dis. 65:158–162. 10.1016/j.diagmicrobio.2009.06.016 [DOI] [PubMed] [Google Scholar]
- 9.Castanheira M, Jones RN, Sader HS. 2008. Update of the in vitro activity of daptomycin tested against 6710 Gram-positive cocci isolated in North America (2006). Diagn. Microbiol. Infect. Dis. 61:235–239. 10.1016/j.diagmicrobio.2008.01.016 [DOI] [PubMed] [Google Scholar]
- 10.Sader HS, Moet G, Jones RN. 2009. Update on the in vitro activity of daptomycin tested against 17,193 Gram-positive bacteria isolated from European medical centers (2005–2007). J. Chemother. 21:500–506. 10.1179/joc.2009.21.5.500 [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Ruiz A, Beiras-Fernandez A, Lehmkuhl H, Seaton RA, Loeffler J, Chaves RL. 2011. Clinical experience with daptomycin in Europe: the first 2.5 years. J. Antimicrob. Chemother. 66:912–919. 10.1093/jac/dkq528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sader HS, Flamm RK, Jones RN. 2013. Antimicrobial activity of daptomycin tested against Gram-positive pathogens collected in Europe, Latin America, and selected countries in the Asia-Pacific Region (2011). Diagn. Microbiol. Infect. Dis. 75:417–422. 10.1016/j.diagmicrobio.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 13.Edelsberg J, Weycker D, Barron R, Li X, Wu H, Oster G, Badre S, Langeberg WJ, Weber DJ. 2014. Prevalence of antibiotic resistance in US hospitals. Diagn. Microbiol. Infect. Dis. 78:255–262 [DOI] [PubMed] [Google Scholar]
- 14.Kamboj M, Cohen N, Gilhuley K, Babady NE, Seo SK, Sepkowitz KA. 2011. Emergence of daptomycin-resistant VRE: experience of a single institution. Infect. Control. Hosp. Epidemiol. 32:391–394. 10.1086/659152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphries RM, Kelesidis T, Tewhey R, Rose WE, Schork N, Nizet V, Sakoulas G. 2012. Genotypic and phenotypic evaluation of the evolution of high-level daptomycin nonsusceptibility in vancomycin-resistant Enterococcus faecium. Antimicrob. Agents Chemother. 56:6051–6053. 10.1128/AAC.01318-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelesidis T, Humphries R, Uslan DZ, Pegues D. 2012. De novo daptomycin-nonsusceptible enterococcal infections. Emerg. Infect. Dis. 18:674–676. 10.3201/eid1804.110932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Judge T, Pogue JM, Marchaim D, Ho K, Kamatam S, Parveen S, Tiwari N, Nanjireddy P, Bheemreddy S, Biedron C, Reddy SM, Khammam V, Chalana IK, Tumma RS, Collins V, Yousuf A, Lephart PR, Martin ET, Rybak MJ, Kaye KS, Hayakawa K. 2012. Epidemiology of vancomycin-resistant enterococci with reduced susceptibility to daptomycin. Infect. Control Hosp. Epidemiol. 33:1250–1254. 10.1086/668438 [DOI] [PubMed] [Google Scholar]
- 18.Storm JC, Diekema DJ, Kroeger JS, Johnson SJ, Johannsson B. 2012. Daptomycin exposure precedes infection and/or colonization with daptomycin non-susceptible enterococcus. Antimicrob. Resist. Infect. Control 1:19. 10.1186/2047-2994-1-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryant KA, Roberts AL, Rupp ME, Anderson JR, Lyden ER, Fey PD, Van Schooneveld TC. 2013. Susceptibility of enterococci to daptomycin is dependent upon testing methodology. Diagn. Microbiol. Infect. Dis. 76:497–501. 10.1016/j.diagmicrobio.2013.04.019 [DOI] [PubMed] [Google Scholar]
- 20.Munoz-Price LS, Lolans K, Quinn JP. 2005. Emergence of resistance to daptomycin during treatment of vancomycin-resistant Enterococcus faecalis infection. Clin. Infect. Dis. 41:565–566. 10.1086/432121 [DOI] [PubMed] [Google Scholar]
- 21.Lewis JS, II, Owens A, Cadena J, Sabol K, Patterson JE, Jorgensen JH. 2005. Emergence of daptomycin resistance in Enterococcus faecium during daptomycin therapy. Antimicrob. Agents Chemother. 49:1664–1665. 10.1128/AAC.49.4.1664-1665.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lesho EP, Wortmann GW, Craft D, Moran KA. 2006. De novo daptomycin nonsusceptibility in a clinical isolate. J. Clin. Microbiol. 44:673. 10.1128/JCM.44.2.673.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arias CA, Panesso D, Mcgrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, Reyes J, Roh JH, Lobos E, Sodergren E, Pasqualini R, Arap W, Quinn JP, Shamoo Y, Murray BE, Weinstock GM. 2011. Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 365:892–900. 10.1056/NEJMoa1011138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer KL, Daniel A, Hardy C, Silverman J, Gilmore MS. 2011. Genetic basis for daptomycin resistance in enterococci. Antimicrob. Agents Chemother. 55:3345–3356. 10.1128/AAC.00207-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tran TT, Panesso D, Gao H, Roh JH, Munita JM, Reyes J, Diaz L, Lobos EA, Shamoo Y, Mishra NN, Bayer AS, Murray BE, Weinstock GM, Arias CA. 2013. Whole-genome analysis of a daptomycin-susceptible Enterococcus faecium strain and its daptomycin-resistant variant arising during therapy. Antimicrob. Agents Chemother. 57:261–268. 10.1128/AAC.01454-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munita JM, Panesso D, Diaz L, Tran TT, Reyes J, Wanger A, Murray BE, Arias CA. 2012. Correlation between mutations in liaFSR of Enterococcus faecium and MIC of daptomycin: revisiting daptomycin breakpoints. Antimicrob. Agents Chemother. 56:4354–4359. 10.1128/AAC.00509-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS-approved standard, vol M7-A7, Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 28.Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, Van Embden JD, Willems RJ. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963–1971. 10.1128/JCM.40.6.1963-1971.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam MM, Seemann T, Bulach DM, Gladman SL, Chen H, Haring V, Moore RJ, Ballard S, Grayson ML, Johnson PD, Howden BP, Stinear TP. 2012. Comparative analysis of the first complete Enterococcus faecium genome. J. Bacteriol. 194:2334–2341. 10.1128/JB.00259-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Letunic I, Bork P. 2011. Interactive Tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39:W475–W478. 10.1093/nar/gkr201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Francisco AP, Bugalho M, Ramirez M, Carrico JA. 2009. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10:152. 10.1186/1471-2105-10-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCracken M, Wong A, Mitchell R, Gravel D, Conly J, Embil J, Johnston L, Matlow A, Ormiston D, Simor AE, Smith S, Du T, Hizon R, Mulvey MR. 2013. Molecular epidemiology of vancomycin-resistant enterococcal bacteraemia: results from the Canadian Nosocomial Infection Surveillance Program, 1999–2009. J. Antimicrob. Chemother. 68:1505–1509. 10.1093/jac/dkt054 [DOI] [PubMed] [Google Scholar]
- 38.Panesso D, Reyes J, Rincon S, Diaz L, Galloway-Pena J, Zurita J, Carrillo C, Merentes A, Guzman M, Adachi JA, Murray BE, Arias CA. 2010. Molecular epidemiology of vancomycin-resistant Enterococcus faecium: a prospective, multicenter study in South American hospitals. J. Clin. Microbiol. 48:1562–1569. 10.1128/JCM.02526-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galloway-Peña JR, Nallapareddy SR, Arias CA, Eliopoulos GM, Murray BE. 2009. Analysis of clonality and antibiotic resistance among early clinical isolates of Enterococcus faecium in the United States. J. Infect. Dis. 200:1566–1573. 10.1086/644790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leavis HL, Bonten MJ, Willems RJ. 2006. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr. Opin. Microbiol. 9:454–460. 10.1016/j.mib.2006.07.001 [DOI] [PubMed] [Google Scholar]
- 41.Kelesidis T, Chow AL. 2014. Proximity to animal or crop operations may be associated with de novo daptomycin-non-susceptible Enterococcus infection. Epidemiol. Infect. 142:221–224. 10.1017/S0950268813000885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Wall SK, Xu L, Ebner PD. 2010. Contamination rates and antimicrobial resistance in bacteria isolated from “grass-fed” labeled beef products. Foodborne Pathog. Dis. 7:1331–1336. 10.1089/fpd.2010.0562 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.