Abstract
Streptococcus suis serotype 2 is known to cause severe infections (meningitis, endocarditis, and septicemia) in pigs and is considered an emerging zoonotic agent. Antibiotics have long been used in the swine industry for disease treatment/prevention and growth promoters. This pattern of utilization resulted in the spread of antibiotic resistance in S. suis worldwide. Interestingly, pigs may harbor S. suis in their tonsils without developing diseases, while North American strains belonging to the sequence type 28 (ST28) are nonvirulent in animal models. Consequently, the aim of this study was to purify and characterize a bacteriocin produced by a nonvirulent strain of S. suis serotype 2, with a view to a potential therapeutic and preventive application. S. suis 90-1330 belonging to ST28 and previously shown to be nonvirulent in an animal model exhibited antibacterial activity toward all S. suis pathogenic isolates tested. The bacteriocin produced by this strain was purified to homogeneity by cationic exchange and reversed-phase fast protein liquid chromatography. Given its properties (molecular mass of <4 kDa, heat, pH and protease stability, and the presence of modified amino acids), the bacteriocin, named suicin 90-1330, belongs to the lantibiotic class. Using a DNA-binding fluorophore, the bacteriocin was found to possess a membrane permeabilization activity. When tested on other swine pathogens, the suicin showed activity against Staphylococcus hyicus and Staphylococcus aureus, whereas it was inactive against all Gram-negative bacteria tested. Amino acid sequencing of the purified bacteriocin showed homology (90.9% identity) with nisin U produced by Streptococcus uberis. The putative gene cluster involved in suicin production was amplified by PCR and sequence analysis revealed the presence of 11 open reading frames, including the structural gene and those required for the modification of amino acids, export, regulation, and immunity. Further studies will evaluate the ability of suicin 90-1330 or the producing strain to prevent experimental S. suis infections in pigs.
INTRODUCTION
Streptococcus suis has been associated with severe swine infections worldwide including, but not limited to, meningitis, arthritis, endocarditis, and septicemia (1). In addition, this Gram-positive bacterium is considered an emerging zoonotic agent that has caused severe outbreaks in Asia that affected hundreds of people (2). To date, 35 serotypes of S. suis have been described and serotype 2 is the most commonly isolated from diseased pigs and humans (1). Moreover, S. suis is classified into numerous sequence types (STs) by multilocus sequence typing (MLST) (3). A recent study showed that, depending on the geographical area, European isolates of S. suis serotype 2 are mainly highly virulent ST1, whereas North American isolates are moderately virulent ST25 or low-virulence ST28 (4). Over the last decade, a number of putative virulence factors produced by S. suis have been proposed, which allows the bacterium to colonize and invade the host tissues, to avoid destruction or neutralization by host defenses, and to promote inflammatory processes (5).
Antibiotics have long been used in the swine industry for disease treatment/prevention as well as growth promoters (6, 7). This pattern of utilization has likely contributed to the spread of antibiotic resistance and consequently resulted in increased regulation regarding the use of antibiotics in the swine industry. Although most strains of S. suis are still highly sensitive to penicillin and amoxicillin, resistances to macrolides, lincosamides, sulfonamides, and tetracyclines have been reported in up to 85% of strains (7, 8). In addition, Wang et al. (9) recently identified a plasmid-borne cfr (chloramphenicol-florfenicol resistance) gene in a S. suis isolate. This gene encodes a 23S rRNA methyltransferase causing resistance to five chemically unrelated classes of antibiotics (10). Resistance genes found in S. suis can be transmitted to other bacteria of the same species or across species. Moreover, through its zoonotic potential, S. suis might transfer resistance genes to human pathogens. Considering this threat, it has become a priority to identify alternative therapeutic and preventive strategies for infections caused by S. suis.
Bacteriocins, which are ribosomally synthesized antimicrobial peptides of bacterial origin, have been proposed as promising new agents for the treatment of diseases caused by pathogenic bacteria (11, 12). Although bacteriocins may be active against a number of different bacterial species, they usually have a narrow spectrum of activity and target specific bacteria by inducing the formation of membrane pores (12). Consequently, they offer the advantage of not perturbing the commensal microbial populations, a major side effect of classical antibiotics. Bacteriocins are also considered nontoxic for eukaryotic cells, and susceptible bacteria do not appear to be capable of developing effective mechanisms to resist these antimicrobial peptides (13). Lantibiotics (class I bacteriocins) are a large family of low-molecular-weight, heat-stable bacteriocins containing unusual posttranslationally modified amino acids, such as lanthionine, methyllanthionine, didehydroalanine, and didehydrobutyrine, with thioether linkages that contribute to their high stability (14). Lantibiotic biosynthesis is encoded by a gene cluster, which includes a structural gene for a prelantibiotic peptide, as well as genes required for the modification of amino acids, export, regulation, and immunity (14). In a recent study, LeBel et al. (15) showed that all strains of S. suis tested were susceptible to the lantibiotic nisin A, which is currently the most important bacteriocin used commercially as a food preservative specially in dairy products in more than 50 countries (16). Interestingly, synergistic effects of nisin A in combination with conventional antibiotics, including penicillin, amoxicillin, and ceftiofur, currently used in the swine industry were reported (15).
Based on the fact that pigs may harbor S. suis in their tonsils without developing diseases (17, 18) and that strains belonging to ST28 and tested thus far are poorly virulent in animal models (4; unpublished observations), the aim of the present study was to purify and characterize a bacteriocin produced by a nonvirulent ST28 strain of S. suis serotype 2 in view of a potential therapeutic and preventive application.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains of S. suis serotype 2 used in the present study, as well as their origins and STs, are listed in Table 1. Bacteria were routinely grown aerobically under static conditions at 37°C in Todd-Hewitt broth (THB; BBL Microbiology Systems, Cockeysville, MD).
TABLE 1.
Strains of S. suis serotype 2 used in this study
| Strain | Country | Origin | Sequence type |
|---|---|---|---|
| 24 | France | Septicemia | 1 |
| 31533 | France | Meningitis | 1 |
| DAT229 | Japan | Endocarditis | 1 |
| DAT264 | Japan | Meningitis | 1 |
| MGGUS2 | United States | Meningitis | 1 |
| MGGUS3 | United States | Meningitis | 1 |
| MNCM01 | Thailand | Endocarditis | 1 |
| MNCM06 | Thailand | Meningitis | 1 |
| P1/7 | United Kingdom | Meningitis | 1 |
| 1043248 | Canada | Meningitis | 25 |
| 1043629 | Canada | Pneumonia | 25 |
| 1053253 | Canada | Pneumonia | 25 |
| 1085543 | Canada | Meningitis | 25 |
| 1102864 | Canada | Septicemia | 25 |
| LPH4 | Thailand | Septicemia | 25 |
| MGGUS4 | United States | Septicemia | 25 |
| MNCM51 | Thailand | Septicemia | 25 |
| 1054471 | Canada | Meningitis | 28 |
| 1057906 | Canada | Meningitis | 28 |
| 1088563 | Canada | Meningitis | 28 |
| 90-1330 | Canada | Pneumonia | 28 |
| DAT245 | Japan | Meningitis | 28 |
| DAT292 | Japan | Unknown | 28 |
| MGGUS9 | United States | Endocarditis | 28 |
| MGGUS10 | United States | Pneumonia | 28 |
| MGGUS11 | United States | Pneumonia | 28 |
| MGGUS12 | United States | Pneumonia | 28 |
| MGGUS13 | United States | Meningitis | 28 |
| MNCM43 | Thailand | Endocarditis | 28 |
Plate diffusion assay for bacteriocin production.
Overnight cultures of 12 strains of S. suis belonging to ST28 were spotted (2 μl) onto Todd-Hewitt agar (THA; BBL Microbiology Systems) plates, which were incubated at 37°C for 24 h. The plates were then overlaid with THB soft agar (0.75% [wt/vol]) that had been inoculated (750 μl in 7 ml) with a 24-h culture of indicator pathogenic strains of S. suis, and were further incubated at 37°C for 24 h. The zones of inhibition were measured from the edge of the growth of S. suis to the margin of the inhibitory zone.
Effect of carbon source and concentration on bacteriocin production by S. suis 90-1330.
The nonvirulent strain 90-1330 was chosen among the tested strains (see Results). The culture medium for bacteriocin production by S. suis 90-1330 was optimized using the above plate diffusion assay and both S. suis 24 and MGGUS2 as indicator virulent strains. A basal culture medium made of 2% proteose-peptone, 1% yeast extract, 0.25% NaCl, 0.3% K2HPO4, 0.2% KH2PO4, 0.01% MgSO4·7H2O, and 0.002% MnSO4·7H2O at pH 7.0 was supplemented with various carbohydrates (fructose, glucose, lactose, and sucrose) at 1% (wt/vol). The carbohydrate showing the largest inhibitory zones was further tested at concentrations ranging from 5 to 0.0625%.
Purification of bacteriocin produced by S. suis 90-1330.
S. suis 90-1330 was cultivated in 2 liters of the basal culture medium supplemented with 0.25% glucose and 0.01% Tween 80 (sorbitan polyoxyethylene monooleate; Sigma-Aldrich Canada Co., Oakville, Ontario, Canada). Tween 80 was added since it was shown to prevent bacteriocin adsorption to glassware and bacterial cells and thus to minimize bacteriocin loss (19). After incubation at 37°C under aerobic conditions for 24 h, bacterial cells were removed by centrifugation (10,000 × g for 15 min). Ammonium sulfate was slowly added to the culture supernatant to obtain 50% saturation, and the mixture was stirred at 4°C for 3 h. After centrifugation (14,000 × g for 10 min), the precipitate was suspended in 30 ml of 50 mM phosphate-buffered saline (pH 7.2; PBS) and dialyzed (1,000-Da cutoff) overnight against 20 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 5.5) containing 0.01% Tween 80. The sample was then subjected to cationic exchange fast protein liquid chromatography (FPLC; MonoS 5/50 GL column; GE Healthcare, Baie d'Urfé, Quebec, Canada) using an ÄKTA Purifier system (GE Healthcare). Elution was performed at a flow rate of 1 ml/min using a linear gradient of KCl from 0 to 0.5 M. The active fractions were detected by spotting 5 μl on the surfaces of THA plates inoculated with a lawn of S. suis virulent MGGUS2 strain (spot test plate assay). After growth at 37°C for 24 h, positive fractions showing an inhibitory zone were pooled and further dialyzed (1,000-Da cutoff) overnight against 0.01% trifluoroacetic acid (TFA) plus 10% acetonitrile. The resulting fraction was then subjected to reversed-phase FPLC (Source 15RPC column; GE Healthcare). Elution was performed at a flow rate of 1 ml/min using a linear gradient of acetonitrile from 10 to 60%. The acetonitrile and TFA were removed by a rotary evaporator prior to analyzing the fractions for bacteriocin activity using the spot test plate assay as described above. The active fractions were pooled, and 15% glycerol was added; the samples were then divided into aliquots (100 μl) and stored at −80°C. Total purified bacteriocin was quantified as arbitrary units, which correspond to the reciprocal of the highest 2-fold serial dilution giving a clear inhibitory zone following application of 5 μl of the bacteriocin solution on a lawn of S. suis MGGUS2. The total purified bacteriocin was also quantified by using a protein assay kit (Bio-Rad Laboratories, Mississauga, Ontario, Canada) with bovine serum albumin as a control.
SDS-PAGE analysis.
The purified bacteriocin (0.3 μg) was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 16.5% Tris-tricine gel (Bio-Rad Laboratories) and stained with silver nitrate. A gel was also fixed in 10% acetic acid–20% propanol (30 min) and washed thoroughly in sterile distilled water (three times for 30 min each time), and the bacteriocin activity was detected by using an overlay of soft agar medium inoculated with the indicator strain S. suis MGGUS2. Nisin A (Sigma-Aldrich Canada Co.) was used as a positive control.
Bacteriocin characterization.
The susceptibility of the purified bacteriocin to temperature, pH, and enzymatic treatments was investigated using the spot test plate assay and S. suis virulent MGGUS2 strain as the indicator bacteria. To evaluate temperature stability, the purified bacteriocin was incubated at 45, 70, 100, or 121°C for 15 min prior to determining the bacteriocin activity. The effect of storing the bacteriocin at 4°C or room temperature for 1 week was also tested. To investigate the susceptibility to extreme pHs, the bacteriocin solution was adjusted to pH 2 or 11 by using 0.125 N HCl or 0.125 N NaOH, respectively. After 15 min at room temperature, the bacteriocin solution was diluted 1:2 in PBS to neutralize pH, and the bacteriocin activity was determined. Lastly, the proteolytic enzymes trypsin, α-chymotrypsin, and proteinase K (Sigma-Aldrich Canada Co.), each at a final concentration of 500 μg/ml, were used to evaluate susceptibility of the bacteriocin to proteolytic cleavage. After incubation at 37°C for 1 h, the samples were treated for 5 min at 68°C to inactivate the enzymes, and the bacteriocin activity was determined. Lastly, the antibacterial activity of the purified bacteriocin was compared to that of commercial nisin A (Sigma-Aldrich Canada Co.) by determination of minimal inhibitory concentration (MIC; in μg/ml) for S. suis MGGUS2.
Membrane permeabilization assay.
The ability of the purified bacteriocin to permeabilize the cytoplasmic membrane of S. suis MGGUS2 was evaluated using the SYTOX Green dye (Life Technologies, Inc., Burlington, Ontario, Canada), which binds to the nucleic acid of bacterial cells once the cytoplasmic membrane is compromised. Briefly, 1.25 μM SYTOX Green dye was added to S. suis cells suspended in 10 mM HEPES (pH 7.0) to an optical density at 660 nm (OD660) of 0.4. Aliquots of 100 μl were added to wells of a 96-well black microplate prior to adding 10 μl of a 1:10 dilution of the purified bacteriocin fraction. The incubation was carried out in a microplate reader (Synergy 2; BioTek Instruments, Winooski, VT) at 37°C for 20 min, and the fluorescence resulting from the binding of the dye to bacterial DNA was recorded every 2 min following excitation at 485 nm and emission at 528 nm. Ethanol (70%) and boiled bacteria were used as positive controls. A reaction mixture containing HEPES instead of purified bacteriocin was used for the negative control.
Activity spectrum of the purified bacteriocin.
Several swine pathogens were tested in the spot test plate assay to evaluate the activity spectrum of the purified bacteriocin. Actinobacillus pleuropneumoniae serotype 5b strain 81-750 was cultivated on THA plate supplemented with NAD (NAD; 20 μg/ml), while Actinobacillus suis JG-2 was grown on blood-supplemented THA plate. Haemophilus parasuis 99-9048-B and Bordetella bronchiseptica ATCC 19395 were cultivated on THA plates supplemented with NAD and hemin (10 μg/ml) or NAD and cysteine (400 μg/ml), respectively. Escherichia coli P82-862, Pasteurella multocida ATCC 12948, Staphylococcus aureus ATCC 25923, and Staphylococcus hyicus ATCC 11249 were grown on THA plates. All cultures were incubated for 24 h at 37°C in aerobiosis.
Amino acid sequencing.
The purified bacteriocin was subjected to SDS-PAGE as described above and then electroblotted onto a polyvinylidene difluoride (PVDF) membrane. The bacteriocin band, localized based on the migration of molecular mass markers, was excised and transferred into a microtube. Ethanethiol derivatization of posttranslationally modified amino acids of the PVDF-blotted bacteriocin was carried out as previously described by Meyer et al. (20). In an anaerobic chamber, 200 μl of a reaction mixture containing 280 μl of methanol, 200 μl of H2O, 65 μl of 5 M NaOH, and 60 μl of ethanethiol was added. After incubation at 50°C for 1 h, the solution was acidified by adding 66 μl of 70% (vol/vol) formic acid, and the bacteriocin-blotted PVDF membrane was vacuum dried. The treated bacteriocin was then sent to the SPARC BioCentre (The Hospital for Sick Children, Toronto, Ontario, Canada) and subjected to Edman degradation using an Applied Biosystems ABI 492 Procise cLC sequencer (Life Technologies, Inc.).
Identification of the putative gene cluster encoding the putative bacteriocin 90-1330.
The putative gene cluster encoding the bacteriocin 90-1330 was identified using the BActeriocin GEnome mining tooL (BAGEL3; http://bagel.molgenrug.nl) (21). Using this approach, public sequenced genomes of S. suis serotype 2 strains reported in National Center for Biotechnology (NCBI) databases (www.ncbi.nlm.nih.gov) were analyzed for the presence of genes related to previously published bacteriocins. The putative bacteriocin locus that was identified was used to design primers for gene amplification using genomic DNA extracted from S. suis 90-1330. Sequencing of the putative bacteriocin 90-1330 locus, as well as BLAST homology, was then performed.
Distribution of sslA gene in S. suis strains.
Using a PCR approach, selected S. suis strains were tested for the presence of the sslA gene identified in the bacteriocin-producing strain 90-1330 and which encodes for the prepeptide bacteriocin. PCRs consisted of 40.8 μl of PCR-grade water, 5 μl of 10× buffer, 1 μl of nucleotide mix, 0.6 μl each of the appropriate forward (G48, 5′-AAACAACTCAGGAGCTTCAC-3′) and reverse (G130R, 5′-CACAGGTCATCAAAATACCC-3′) primers, 1 μl of Taq polymerase (5U/μl), and 1 μl of genomic DNA as the template. The PCR was performed with a DNA Thermal Cycler 480 (Perkin-Elmer, Waltham, MA) according to the EconoTaq reaction protocol of Lucigen Corp. (Middleton, WI). The reaction was carried out for 30 cycles with the following temperature-time profile: 95°C for 1 min, 50°C for 1 min, and 72°C for 1 min. At the end of the amplification process, the samples were incubated at 72°C for 3 min. A 1% agarose gel was used to analyze the PCR products.
In vitro safety assessment of S. suis 90-1330.
First, the presence of virulence factor genes coding for suilysin (sly) and extracellular protein factor (epf) in S. suis 90-1330 was evaluated by PCR as described above. The pathogenic strain S. suis P1/7 (serotype 2) and the glutamate dehydrogenase (gdh) gene were used as positive controls. The sequences of the forward and reverse primers used for the PCR were as follows: SLY358 (5′-TTGAATATTGACATGAAGATTGCGA-3′) and SLY455R (5′-AAGCTGGAGAAGAAGTTTGGGAACC-3′), respectively, for sly; EF1274 (5′-CTAAACGTAACTTGGAATTTGTAAG-3′) and EF1435R (5′-AGCCATAAGTAAGATTATTTGATCC-3′), respectively, for epf; and GDH645 (5′-TTTGGTTTACTTCACTGATAACATG-3′) and GDH794R (5′-GAGTCTGAAACAGAAATAACTTTTG-3′), respectively, for gdh. The reaction was carried out for 30 cycles with the following temperature-time profile: 95°C for 1 min, 50°C for 1 min, and 72°C for 30 s. At the end of the amplification process, the samples were incubated at 72°C for 3 min. Second, the susceptibility of S. suis 90-1330 to common antibiotics (penicillin G and amoxicillin) used in the swine industry was determined as follows. Briefly, a 24-h bacterial culture in THB was diluted in fresh broth medium to obtain an OD660 of 0.2. Equal volumes (100 μl) of bacteria and serial dilutions of antibiotics in THB were mixed into the wells of 96-well plates. Control wells with no bacteria or no antibiotics were also prepared. After an incubation of 24 h at 37°C, bacterial growth was recorded visually. MIC values (μg/ml) were determined as the lowest concentration at which no growth occurred. To determine minimal bactericidal concentration (MBC) values (μg/ml), aliquots (5 μl) of each well showing no visible growth were spread on THA plates, which were incubated 24 h at 37°C. MBCs were determined as the lowest concentration at which no colony formation occurred. The MIC and MBC values were determined in three independent experiments.
RESULTS
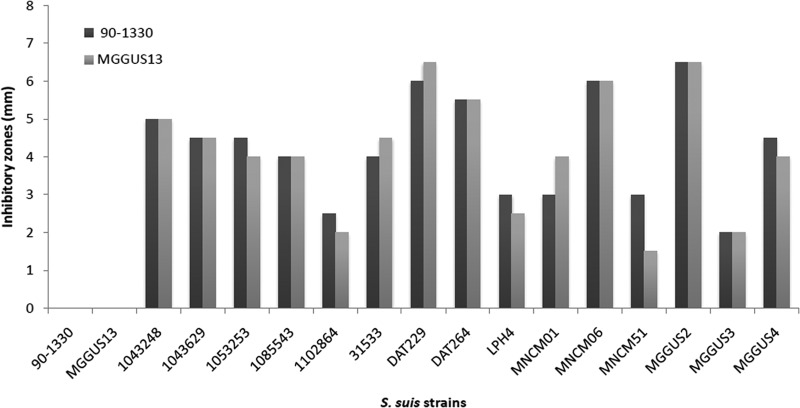
Since evidence has been brought that S. suis isolates belonging to ST28 present lower virulence potential (4), these strains have been chosen in the present study to be screened for the production of bacteriocin activity toward S. suis 24, a virulent ST1 isolate (22) used as indicator strain, using a plate diffusion assay. Two strains of S. suis (90-1330 and MGGUS13) exhibited a strong antibacterial activity (inhibitory zones of 5 mm). The strains 90-1330 and MGGUS13 were then further tested in a similar assay for their capacity to inhibit a large array of virulent S. suis strains isolated from diseased pigs. These latter strains belong to either sequence type 1 (ST1) or sequence type 25 (ST25) (Table 1), which are known to be highly or moderately virulent in animal models, respectively (4; unpublished data). As depicted in Fig. 1, both S. suis 90-1330 and MGGUS13 produced inhibitory zones of various degrees against all strains tested (n = 15). The inhibitory zones produced by S. suis 90-1330 and MGGUS13 were in most cases comparable. Interestingly, both strains showed cross-immunity to each other, suggesting that the bacteriocins produced may be similar or closely related. Given that a previous study by Quessy et al. (23) showed that S. suis 90-1330 is avirulent in both mouse and pig models of infection, this strain was selected for bacteriocin purification and characterization.
FIG 1.
Inhibitory zones produced by S. suis 90-1330 and MGGUS13, two ST28 strains, toward isolates of S. suis from diseased pigs and belonging to either ST1 or ST25.
Bacteriocin production by S. suis 90-1330 in a basal culture medium supplemented with various carbohydrates was evaluated. The data reported in Table 2 indicate that when carbohydrates were used at 1%, the largest inhibitory zones toward S. suis 24 and MGGUS2 were obtained with glucose. When glucose was used at various concentrations, few variations in the inhibitory zones were observed. More specifically, the basal medium supplemented with 0.25% glucose produced inhibitory zones of 9 mm for both indicator strains (Table 2). Therefore, this medium markedly increased bacteriocin production by S. suis 90-1330, considering that THA plates gave inhibitory zones of 5 and 7 mm for S. suis 24 and MGGUS2 (Table 2, Fig. 1), respectively.
TABLE 2.
Effect of carbohydrate source and concentration on inhibitory zones produced by the nonvirulent S. suis 90-1330 toward the indicator virulent S. suis strains 24 and MGGUS2
| Carbohydrate | Concn (%) | Inhibitory zones (mm) |
|
|---|---|---|---|
| 24 | MGGUS2 | ||
| Fructose | 1 | 8 | 6 |
| Glucose | 5 | 8 | 6 |
| 2 | 8 | 8 | |
| 1 | 9 | 8 | |
| 0.25 | 9 | 9 | |
| 0.125 | 8 | 6 | |
| 0.0625 | 8 | 6 | |
| Lactose | 1 | 8 | 7 |
| Sucrose | 1 | 8 | 8 |
S. suis 90-1330 was cultured in the above optimized medium (2 liters) and proteins present in the supernatant were precipitated with ammonium sulfate at 50% saturation. This fraction was subjected to cationic exchange and reversed-phase FPLC. Bacteriocin activity recovered from the reversed-phase FPLC eluted in a single peak and Tris-tricine–SDS-PAGE analysis of the purified fraction yielded a single band stained by silver nitrate and migrating similarly to the commercial lantibiotic nisin A, which is known to have a molecular mass of 3,354 Da (Fig. 2A). No evidential protein contaminants appear to be present in the purified fraction. An overlay with the indicator strain (MGGUS2) correlated bacteriocin activity with the protein band (Fig. 2B). From the 2-liter culture of S. suis 90-1330, the purification protocol allowed the recovery of 17,280 arbitrary units of bacteriocin, as defined in Materials and Methods, corresponding to 156 μg of protein.
FIG 2.
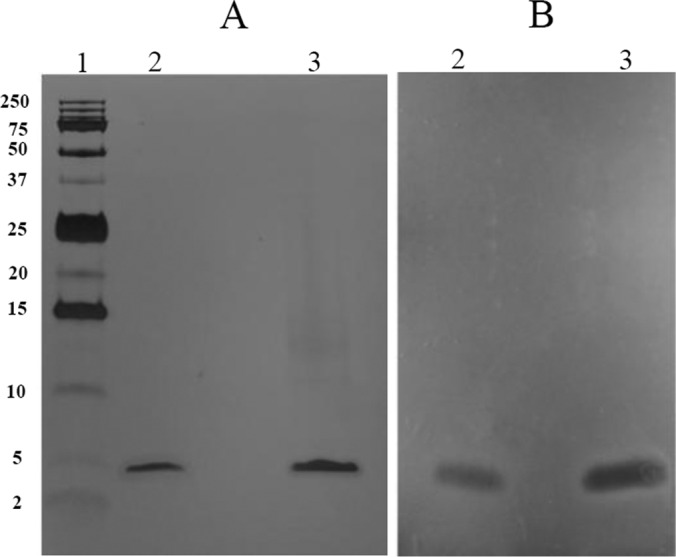
Tris-Tricine SDS-PAGE analysis of the purified bacteriocin produced by S. suis 90-1330. (A) Silver nitrate staining. (B) Antibacterial activity detected by an overlay with S. suis MGGUS2 as the indicator strain. Lane 1, molecular mass markers indicated to the left in kilodaltons; lane 2, nisin A; lane 3, purified bacteriocin 90-1330.
The purified bacteriocin was subjected to various treatments to determine its stability (Table 3). The results of the heat stability assay showed that the bacteriocin was highly heat stable as the antibacterial activity was still detected even after treatment at 121°C for 15 min. In addition, storage at room temperature for 1 week did not reduce antibacterial activity. The bacteriocin was also found to be stable under a wide range of pH since there was no reduction in antibacterial activity observed after exposure at pH 2 and 11. Lastly, treatment of the purified bacteriocin with proteolytic enzymes (trypsin, chymotrypsin, and proteinase K) did not show detectable reduction in its antibacterial activity. In order to assess the relative antibacterial activity of the purified bacteriocin, the MIC (S. suis MGGUS2) was determined and compared to that of commercial nisin A. The purified bacteriocin showed an MIC of 0.47 μg/ml, while that of nisin A was 2.5 μg/ml.
TABLE 3.
Stability and activity spectrum of purified bacteriocin from S. suis 90-1330
| Treatment | Indicator strain | Inhibitory activity |
|---|---|---|
| 45°C/15 min | S. suis MGGUS2 | + |
| 70°C/15 min | S. suis MGGUS2 | + |
| 100°C/15 min | S. suis MGGUS2 | + |
| 121°C/15 min | S. suis MGGUS2 | + |
| 4°C/7 days | S. suis MGGUS2 | + |
| 25°C/7 days | S. suis MGGUS2 | + |
| pH 2/15 min | S. suis MGGUS2 | + |
| pH 11/15 min | S. suis MGGUS2 | + |
| Trypsin/60 min | S. suis MGGUS2 | + |
| Chymotrypsin/60 min | S. suis MGGUS2 | + |
| Proteinase K/60 min | S. suis MGGUS2 | + |
| None | S. aureus ATCC 25923 | + |
| S. hyicus ATCC 11249 | + | |
| A. pleuropneumoniae 81-750 | – | |
| A. suis JG-2 | – | |
| B. bronchiseptica ATCC 19395 | – | |
| E. coli P82-862 | – | |
| H. parasuis 99-9048-B | – | |
| P. multocida ATCC 12948 | – |
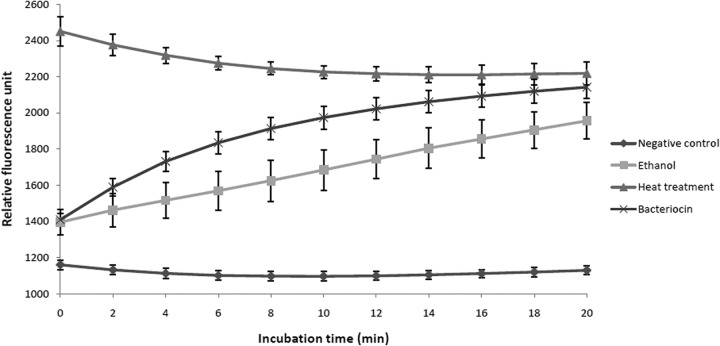
The ability of the purified bacteriocin to permeabilize the membrane of the indicator strain S. suis MGGUS2 was investigated using the SYTOX Green dye, which can penetrate damaged cytoplasmic membranes and has high affinity for DNA. As shown in Fig. 3, following the addition of either purified bacteriocin or ethanol (positive control), an increase in fluorescence was observed time dependently, indicating permeabilization of the membrane. No increase in fluorescence occurred in the negative control, while the entry of SYTOX Green dye occurred instantaneously for boiled cells.
FIG 3.
Effect of purified bacteriocin 90-1330 on membrane permeabilization of S. suis MGGUS2, as determined using SYTOX Green dye.
In order to evaluate its activity spectrum, the antibacterial effect of the purified bacteriocin was tested against different Gram-positive and Gram-negative bacterial species known to cause infections in swine. As reported in Table 3, the bacteriocin was active against both Gram-positive bacteria (S. aureus and S. hyicus), while it did not show any detectable activity against all Gram-negative bacteria tested.
Peptide sequencing of the purified bacteriocin was carried out by Edman degradation. When chemical treatment of the sample was not performed, the Edman degradation was blocked after the first residue, suggesting the presence of modified amino acids commonly found in bacteriocin belonging to the class of lantibiotics. Upon chemical derivatization of putative dehydrated amino acids and methyllanthionine/lanthionine bridges by alkaline ethanethiol, Edman degradation of the first 18 amino acids of the purified bacteriocin yielded the following sequence: Val1-Dhb or MeLan(Thr)2-X(Ser or Cys or Thr)3-Lys4-X(Ser or Cys or Thr)5-Leu6-X(Cys or Ser or Thr)7-Dhb or MeLan(Thr)8-Pro9-Gly10-X(Cys or Ser or Thr)11-Lys12-X(Thr)13-Gly14-Ile15-Leu16-Met17-Dhb(Thr)18. None of the modified amino acids (residues 2, 3, 5, 7, 8, 11, 13, and 18) blocked the Edman degradation. Residues 2, 8, and 18 yielded peaks close to the Phe peak, accompanied by relatively strong Leu signal, which is characteristic of Thr residues modified to either Dhb or a methyllanthionine (MeLan) moiety. The other modified amino acids that gave no signals in Edman degradation are either Ser, Cys, or Thr, which are involved in lanthionine or β-methyllanthionine bridges.
Using the UniPROT web-based platform (http://www.uniprot.org) (24), a strong similarity of the above 18-amino-acid sequence with the lantibiotic nisin U produced by Streptococcus uberis (25) was highlighted. Published genomes of S. suis were then analyzed with BAGEL3 for the presence of bacteriocin-related genes. One strain (JS14) showed the presence of a gene with a high homology (91%) with nsuA encoding nisin U in S. uberis. Further analysis of the JS14 genome revealed the presence of a complete lantibiotic biosynthesis locus. Based on this gene locus identified in strain JS14, primers were designed for PCR amplification in S. suis 90-1330. After sequencing, the complete sslA (for Streptococcus suis lantibiotic A) gene cluster of S. suis 90-1330 was identified and found to contain 11 open reading frames (ORFs) involved in bacteriocin production. As reported in Fig. 4, the locus encodes the suicin 90-1330 precursor (sslA), a dehydratase involved in lantibiotic synthesis (sslB), an ABC transporter (sslT), a cyclase involved in lantibiotic synthesis (sslC), a protease involved in proteolytic cleavage of the leader peptide (sslP), a response regulator (sslR), a sensor histidine kinase (sslK), and four immunity proteins (sslF, sslE, sslG, and sslI). A small ORF, likely unrelated to bacteriocin production, was located before the sslA gene. This gene showed 98% identity with a gene encoding a hypothetical protein identified in Streptococcus agalactiae ATCC 13813 (data not shown). Table 4 reports the percent identities of these S. suis genes with the corresponding genes found in S. uberis. For all 11 genes, the percent identities ranged from 79.2 to 94.2%.
FIG 4.

Genetic organization of the putative suicin 90-1330 gene cluster. X, gene (182 bp) unrelated to the bacteriocin cluster.
TABLE 4.
Percent identity in deducted amino acid sequences between the S. suis suicin 90-1330 gene cluster and the S. uberis nisin U gene cluster
From the structural gene of suicin 90-1330 (sslA), the inferred amino acid sequence of the leader peptide contains 24 amino acids, while that of mature unmodified peptide consists of 31 amino acids (Fig. 5). Comparison of the SslA amino acid sequence to previously characterized lantibiotics highlighted high levels of homology with nisin U and, to a lesser extent, with nisin A and nisin Q (Fig. 5). A much lower homology was observed with salivaricin D and N, more particularly for the mature unmodified peptide.
FIG 5.
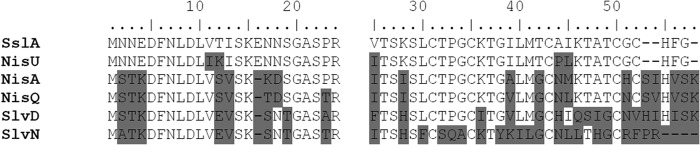
Comparison of the amino acid sequence of suicin 90-1330 to other lantibiotics produced by S. uberis (nisU), Lactococcus lactis (nisA and nisQ), and Streptococcus salivarius (slvD and slvN). The left and right blocks of the sequence refer to the leader and mature peptides, respectively.
Based on the inferred amino acid sequence, the amino acid sequencing of the purified bacteriocin, as well as its high homology with nisin U, the predicted structure of the suicin 90-1330 can be deducted. As shown in Fig. 6, the suicin 90-1330 contains 13 modified amino acids, as well as 1 lanthionine and 4 methyllanthionine bridges. Among the 18 nonmodified amino acids identified, 10 were hydrophobic amino acid residues (Ala, Ile, Leu, Met, Phe, Pro, and Val), and 4 were cationic amino acid residues (His and Lys).
FIG 6.
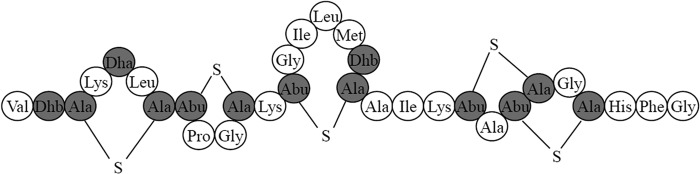
Predicted structure of suicin 90-1330. Mature peptide where Ser and Thr residues are posttranslationally dehydrated to Dha and Dhb, or involved in the formation of Lan and MeLan, respectively, with cysteine residues, are shaded in gray.
The presence of the structural bacteriocin gene sslA in various strains of S. suis was investigated by PCR. As shown in Fig. 7, in addition to 90-1330, the second ST28, bacteriocin-producing strain MGGUS13 also possessed the sslA gene, thus confirming that both strains are secreting the same bacteriocin. None of the other isolates of S. suis tested had the gene. All of these strains were also unable to produce bacteriocin activity (data not shown).
FIG 7.
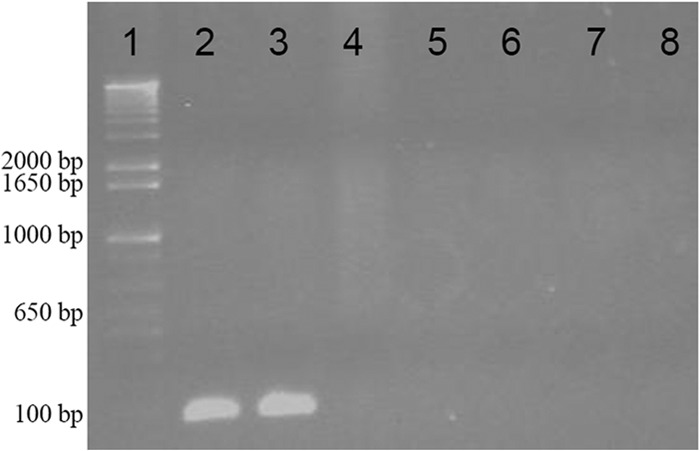
Detection of the structural suicin 90-1330 gene sslA in various isolates of S. suis. Lane 1, DNA molecular weight markers; lane 2, 90-1330; lane 3, MGGUS13; lane 4, P1/7; lane 5, DAT229; lane 6, DAT245; lane 7, MGGUS2; lane 8, MGGUS3.
Given that the bacteriocin-producing S. suis strain 90-1330 may have potential probiotic and protective applications, it was of interest to evaluate some safety parameters. First, the presence of two important virulence factor genes, namely, sly (suilysin) and epf (extracellular protein factor), was investigated. As shown in Fig. 8, PCR amplification using specific primers showed that strain 90-1330 did not possess sly and epf genes, while the virulence factor genes were present in the pathogenic isolate S. suis P1/7. The gdh gene used as control was detected in both strains. Second, the susceptibility of S. suis 90-1330 to antibiotics commonly used in the swine industry was evaluated. This strain was highly sensitive to both penicillin G and amoxicillin, with MIC and MBC values in the range of 0.0195 to 0.078 μg/ml.
FIG 8.
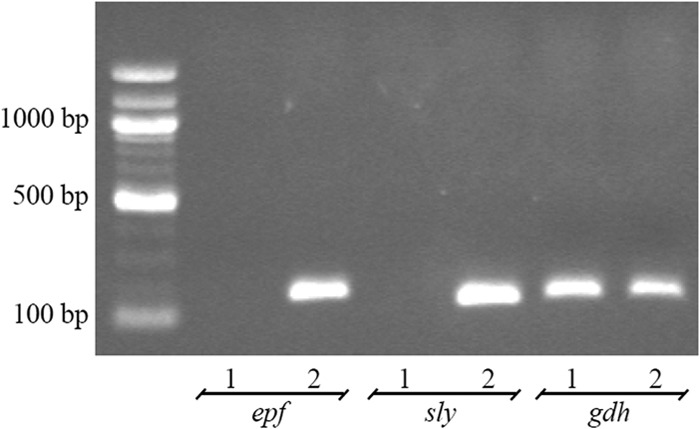
Detection of sly (suilysin), epf (extracellular protein factor), and gdh (glutamate dehydrogenase) genes in S. suis 90-1330 (lanes 1) and P1/7 (lanes 2).
DISCUSSION
S. suis is becoming increasingly resistant to several antibiotics, more specifically to macrolides, tetracyclines, and lincosamides (5, 6). One strategy to overcome this problem is to look for new antimicrobial agents. In this regard, bacteriocins, which are ribosomally synthesized antimicrobial peptides of bacterial origin, have been proposed to represent a promising alternative for the treatment of infectious diseases (9, 10). In a preliminary report, Mélançon and Grenier (26) were the first to report the ability of some isolates of S. suis to produce inhibitory substances not related to organic acids or hydrogen peroxide and showing characteristics related to classical bacteriocins. In the present study, we first sought to identify a nonpathogenic strain of S. suis with the capacity to produce a bacteriocin and then to purify and characterize the bacteriocin as well as the genes involved in its biosynthesis.
Two of twelve strains of S. suis belonging to ST28 were found to exert bacteriocin-mediated antibacterial activity toward pathogenic strains of S. suis serotype 2. Given that a previous study reported that S. suis 90-1330 was avirulent in an animal model (23), this strain was selected for further analysis. Bacteriocin production by S. suis 90-1330 was found to be modulated by the composition of the culture medium, and among the carbohydrates tested, glucose allowed the highest production. Although the production of many bacteriocins is controlled by quorum sensing and produced in larger amounts when bacteria were grown on solid media, the bacteriocin produced by S. suis 90-1330 was secreted in sufficient amounts when cultured in liquid media to allow a good yield of recovery. This bacteriocin produced by S. suis 90-1330 was purified to homogeneity by cationic exchange and reversed-phase chromatography from a supernatant of bacteria grown in the above optimal medium. The bacteriocin showed a molecular mass of ∼3.4 kDa and was highly stable to heat, pH, and proteolytic treatments. This high resistance to low pH and proteolytic enzymes is of great interest if the bacteriocin or the producing strain is used for therapy. Amino acid sequencing of the purified bacteriocin by Edman degradation revealed the presence of modified amino acid residues, indicating that it belongs to the lantibiotic class. These modified amino acids are involved in the high resistance of the peptide to extreme pHs and proteases.
We then assessed depolarization of the cytoplasmic membrane of susceptible S. suis induced by suicin 90-1330 using the DNA-binding SYTOX Green fluorescent dye. Treatment of bacteria with the bacteriocin caused permeabilization of the cell membrane, resulting in the uptake of fluorescent dye. This is in agreement with the mode of action previously reported for lantibiotics. Indeed, nisin A has a high affinity for lipid II, which is a major constituent of the cell membrane of Gram-positive bacteria that is involved in the biosynthesis of the peptidoglycan (27, 28). This binding induces pore formation, leading to the rapid depolarization of membrane potential, as well as efflux of the cytoplasmic components (29).
The antibacterial spectrum of the suicin 90-1330 is not restricted to S. suis since the other Gram-positive swine pathogens tested (S. aureus and S. hyicus) were found to be susceptible. However, the bacteriocin did not show any detectable activity against the Gram-negative pathogens tested. This is in agreement with the fact that lantibiotics are well known to act mainly against Gram-positive bacteria (14). However, a larger array of bacterial species should be tested to confirm that the suicin 90-1330 has a narrow activity spectrum.
A comparative analysis of the amino acid sequence of suicin 90-1330 with previously characterized lantibiotics revealed a high homology with the nisin U produced by S. uberis (25). More specifically, it differs in only two amino acids in the leader peptide (24 residues) and in 3 amino acids in the unmodified mature peptide (31 residues). Given that the suicin 90-1330 shares a high homology with nisin U and to a lesser extent with nisin A and Q, it may be regarded as a variant of this family of lantibiotics. Natural variants usually have only a few amino acids substitutions and the same ring pattern, as observed for the bacteriocin 90-1330. Interestingly, using the plate diffusion assay, the nisin A-producing Lactococcus lactis (ATCC 11454) was found to be immunized against the suicin produced by S. suis 90-1330. On the other hand, S. suis 90-1330 did not show complete immunity against the nisin A produced by L. lactis, although it appears more resistant than strains negative for suicin production (data not shown).
Using a PCR approach, the complete gene locus of suicin 90-1330 was identified. More specifically, the cluster consists in a structural gene named sslA, together with genes sslC and sslB, involved in posttranslational modifications of the suicin 90-1330 prepeptide, sslP responsible for proteolytic cleavage of the prepeptide, and sslT coding for a transporter. The immunity genes sslE, sslF, sslG, and sslI, as well as the regulatory genes sslR, and sslK, were also present. The structural gene sslA encodes a mature peptide of 31 amino acids containing serine/threonine/cysteine residues capable of generating modified amino acids as well as lanthionine and methyllanthionine structures. The suicin 90-1330 locus contains all of the genes present in the nisin U locus in a similar arrangement.
Recently, Wang et al. (30) identified a putative lantibiotic locus containing nine genes in highly virulent serotype strains. However, no bacteriocin activity could be detected because the putative lantibiotic modification gene (suiM) was interrupted through insertion of a 7.9-kb integron, while other biosynthesis-related genes contained various frameshift mutations. The structural gene of this bacteriocin encoded a 57 amino acids precursor peptide consisting of a 24-amino-acid leader peptide and a 33-amino-acid mature peptide. The product of this structural gene showed a low level of homology (22%) with that of the sslA gene of S. suis 90-1330.
In the present study, a second strain of S. suis (MGGUS13) belonging to ST28 was also found to produce a bacteriocin. This bacteriocin is likely similar to the suicin 90-1330 since (i) the sslA gene was identified in strain MGGUS13 and (ii) cross-immunity was observed when each of the producing strain were tested for sensitivity with both strains, suggesting that the complete ssl operon, including the immunity proteins was present in the MGGUS13 strain also.
Since bacteriocin production is considered a probiotic feature, the strain S. suis 90-1330 can be regarded as a candidate for probiotic therapy. However, in order to be used, the safety of the strain should be demonstrated. Our study showed that this strain was devoid of two important virulence factors (suilysin and extracellular protein factor) that have been associated with the virulence of S. suis (5). To further support the safety of S. suis 90-1330, a previous study by Quessy et al. (23) showed that it is avirulent in both mouse and pig models of infection. Moreover, protection against S. suis serotype 2 infection was demonstrated in pigs after vaccination with the live avirulent strain 90-1330 (31). We also showed that this strain was highly susceptible to antibiotics (penicillin G and amoxicillin) currently used in the swine industry. Taken together, these characteristics suggest that S. suis 90-1330 may be regarded as a safe strain, although additional animal studies deserve to be carried out.
Although the use of bacteriocins for controlling swine infections has not been investigated, there are some studies regarding the potential of bacteriocins to reduce Campylobacter spp. from colonized chicken in order to reduce the risk of human exposure to this pathogen (32). More specifically, Svetoch et al. (33) reported that a bacteriocin produced by a strain of Enterococcus faecium (isolated from broiler chicken cecum) significantly reduced Campylobacter jejuni colonization in chicken intestine when added in drinking water. Moreover, Cao et al. (34) brought evidence regarding the effectiveness of nisin for treating bovine mastitis caused by S. aureus. Future research should evaluate whether the suicin 90-1330 or the producing strain could be used as a therapeutic agent targeting S. suis or other Gram-positive pathogens such as S. aureus and S. hyicus.
In conclusion, a nonvirulent ST28 strain (90-1330) of S. suis serotype 2 was found to produce a bacteriocin belonging to the lantibiotic family. Characterization of the purified bacteriocin (named suicin 90-1330) at both protein and genetic levels revealed a high homology with nisin U. Further studies will evaluate the ability of suicin 90-1330 or the producing strain to prevent experimental S. suis infections in pigs.
ACKNOWLEDGMENTS
This study was supported by a grant from the Ministère de l'Agriculture, des Pêcheries, et de l'Alimentation du Québec.
We thank Daisuke Takamatsu (National Institute of Animal Health, Tsukuba, Ibaraki, Japan) and Prasit Tharavichitkul (Chiang Mai University, Chiang Mai, Thailand) for providing some of the strains used in this study.
Footnotes
Published ahead of print 27 June 2014
REFERENCES
- 1.Gottschalk M. 2012. Streptococcosis, p 841–855 In Karriker L, Ramirez A, Schwartz KJ, Stevenson G, Zimmerman J. (ed), Diseases of swine. Wiley Publishers, New York, NY [Google Scholar]
- 2.Gottschalk M, Xu J, Calzas C, Segura M. 2010. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol. 5:371–391. 10.2217/fmb.10.2 [DOI] [PubMed] [Google Scholar]
- 3.King SJ, Leigh JA, Heath PJ, Luque I, Tarradas C, Dowson CG, Whatmore AM. 2002. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 40:3671–3680. 10.1128/JCM.40.10.3671-3680.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fittipaldi N, Xu JG, Lacouture S, Tharavichitkul P, Osaki M, Sekizaki T, Takamatsu D, Gottschalk M. 2011. Lineage and virulence of Streptococcus suis serotype 2 isolates from North America. Emerg. Infect. Dis. 17:2239–2244. 10.3201/eid1712.110609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fittipaldi N, Segura M, Grenier D, Gottschalk M. 2012. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 7:259–279. 10.2217/fmb.11.149 [DOI] [PubMed] [Google Scholar]
- 6.Cromwell GL. 2002. Why and how antibiotics are used in the swine production. Anim. Biotechnol. 13:7–27. 10.1081/ABIO-120005767 [DOI] [PubMed] [Google Scholar]
- 7.Varela NP, Gadbois P, Thibault C, Gottschalk M, Dick P, Wilson J. 2013. Antimicrobial resistance and prudent use for Streptococcus suis. Anim. Health Res. Rev. 20:1–10. 10.1017/S1466252313000029 [DOI] [PubMed] [Google Scholar]
- 8.Callens BF, Haesebrouck F, Maes D, Butaye P, Dewulf J, Boyen F. 2013. Clinical resistance and decreased susceptibility in Streptococcus suis isolates from clinically healthy fattening pigs. Microb. Drug Resist. 19:146–151. 10.1089/mdr.2012.0131 [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Li D, Song L, Liu Y, He T, Liu H, Wu C, Schwarz S, Shen J. 2013. First report on the multiresistance gene cfr in Streptococcus suis. Antimicrob. Agents Chemother. 57:4061–4063. 10.1128/AAC.00713-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500–2505. 10.1128/AAC.00131-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammami R, Fernandez B, Lacroix C, Fliss I. 2013. Anti-infective properties of bacteriocins: an update. Cell. Mol. Life Sci. 70:2947–2967. 10.1007/s00018-012-1202-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins: a viable alternative to antibiotics? Nat. Rev. Microbiol. 11:95–105. 10.1038/nrmicro2937 [DOI] [PubMed] [Google Scholar]
- 13.Peschel A, Sahl HG. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4:529–536. 10.1038/nrmicro1441 [DOI] [PubMed] [Google Scholar]
- 14.Bierbaum G, Sahl HG. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 10:2–18. 10.2174/138920109787048616 [DOI] [PubMed] [Google Scholar]
- 15.LeBel G, Piché F, Frenette M, Gottschalk M, Grenier D. 2013. Antimicrobial activity of nisin against the swine pathogen Streptococcus suis and synergistic interaction with antibiotics. Peptides 50:19–23. 10.1016/j.peptides.2013.09.014 [DOI] [PubMed] [Google Scholar]
- 16.De Arauz LJ, Jozala AF, Mazzola PG, Penna TCV. 2009. Nisin biotechnological production and application: a review. Trends Food Sci. Technol. 20:146–154. 10.1016/j.tifs.2009.01.056 [DOI] [Google Scholar]
- 17.Luque I, Blume V, Borge C, Vela AI, Perea JA, Marquez JM, Fernandez-Garayzabal JF, Tarradas C. 2010. Genetic analysis of Streptococcus suis isolates recovered from diseased and healthy carrier pigs at different stages of production on a pig farm. Vet. J. 186:396–398. 10.1016/j.tvjl.2009.09.005 [DOI] [PubMed] [Google Scholar]
- 18.Zhang CP, Ning YB, Zhang ZQ, Song L, Qiu HS, Gao HY, Fan XZ. 2009. Prevalence of Streptococcus suis isolated from clinically healthy sows in China. Agr. Sci. China 8:638–642. 10.1016/S1671-2927(08)60257-6 [DOI] [PubMed] [Google Scholar]
- 19.Joosten HMLJ, Nunez M. 1995. Adsorption of nisin and enterocin 4 to polypropylene and glass surfaces and its prevention by Tween 80. Lett. Appl. Microbiol. 21:389–392. 10.1111/j.1472-765X.1995.tb01089.x [DOI] [Google Scholar]
- 20.Meyer HE, Heber M, Eisermann B, Korte H, Metzger JW, Jung G. 1994. Sequence analysis of lantibiotics: chemical derivatization procedures allow a fast access to complete Edman degradation. Anal. Biochem. 223:185–190. 10.1006/abio.1994.1571 [DOI] [PubMed] [Google Scholar]
- 21.Van Heel AJ, de Jong A, Montalban-Lopez M, Kok J, Kuipers OP. 2013. BAGEL 3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 41:448–453. 10.1093/nar/gkt391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berthelot-Hérault F, Cariolet R, Labbé A, Gottschalk M, Cardinal JY, Kobish M. 2001. Experimental infection of specific pathogen free piglets with French strains of Streptococcus suis capsular type 2. Can. J. Vet. Res. 65:196–200 [PMC free article] [PubMed] [Google Scholar]
- 23.Quessy S, Dubreuil JD, Caya M, Higgins R. 1995. Discrimination of virulent and avirulent Streptococcus suis capsular type 2 isolates from different geographical origins. Infect. Immun. 63:1975–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uniprot Consortium. 2008. The universal protein resource (UniProt). Nucleic Acids Res. 36:190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wirawan RE, Klesse NA, Jack RW, Tagg JR. 2006. Molecular and genetic characterization of a novel nisin variant produced by Streptococcus uberis. Appl. Environ. Microbiol. 72:1148–1156. 10.1128/AEM.72.2.1148-1156.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mélançon D, Grenier D. 2003. Production and properties of bacteriocin-like inhibitory substances from the swine pathogen Streptococcus suis serotype 2. Appl. Environ. Microbiol. 69:4482–4488. 10.1128/AEM.69.8.4482-4488.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lubelski J, Rink R, Khusainov R, Moll GN, Kuipers OP. 2008. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci. 65:455–476. 10.1007/s00018-007-7171-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauer R, Dicks LM. 2005. Mode of action of lipid II-targeting lantibiotics. Int. J. Food Microbiol. 101:201–216. 10.1016/j.ijfoodmicro.2004.11.007 [DOI] [PubMed] [Google Scholar]
- 29.Bonev BB, Breukink E, Swiezewska E, De Kruijff B, Watts A. 2004. Targeting extracellular pyrophosphates underpins the high selectivity of nisin. FASEB J. 18:1862–1869. 10.1096/fj.04-2358com [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Gao Y, Teng K, Zhang J, Sun S, Zhong J. 2014. Restoration of a bioactive lantibiotic suicin from a remnant lan locus of pathogenic Streptococcus suis serotype 2. Appl. Environ. Microbiol. 80:1062–1071. 10.1128/AEM.03213-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Busque P, Higgins R, Caya F, Quessy S. 1997. Immunization of pigs against Streptococcus suis serotype 2 infection using a live avirulent strain. Can. J. Vet. Res. 61:275–279 [PMC free article] [PubMed] [Google Scholar]
- 32.Lin J. 2009. Novel approaches for Campylobacter control in poultry. Foodborne Pathog. Dis. 6:755–765. 10.1089/fpd.2008.0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svetoch EA, Eruslanov BV, Perelygin VV, Mitsevich EV, Mitsevich IP, Borzenkov VN, Levchuk VP, Svetoch OE, Kovalev YN, Stepanshin YG, Siragusa GR, Seal BS, Stern NJ. 2008. Diverse antimicrobial killing by Enterococcus faecium E 50-52 bacteriocin. J. Agric. Food Chem. 56:1942–1948. 10.1021/jf073284g [DOI] [PubMed] [Google Scholar]
- 34.Cao LT, Wu JQ, Xie F, Hu SH, Mo Y. 2007. Efficacy of nisin in treatment of clinical mastitis in lactating dairy cows. J. Dairy Sci. 90:3980–3985. 10.3168/jds.2007-0153 [DOI] [PubMed] [Google Scholar]