Abstract
Increasing reports of cylindrospermopsins (CYNs) in freshwater ecosystems have promoted the demand for identifying all of the potential CYN-producing cyanobacterial species. The present study explored the phylogenetic distribution and evolution of cyr genes in cyanobacterial strains and water samples from China. Four Cylindrospermopsis strains and two Raphidiopsis strains were confirmed to produce CYNs. Mutant cyrI and cyrK genes were observed in these strains. Cloned cyr gene sequences from eight water bodies were clustered with cyr genes from Cylindrospermopsis and Raphidiopsis (C/R group) in the phylogenetic trees with high similarities (99%). Four cyrI sequence types and three cyrJ sequence types were observed to have different sequence insertions and repeats. Phylogenetic analysis of the rpoC1 sequences of the C/R group revealed four conserved clades, namely, clade I, clade II, clade III, and clade V. High sequence similarities (>97%) in each clade and a divergent clade IV were observed. Therefore, CYN producers were sporadically distributed in congeneric and paraphyletic C/R group species in Chinese freshwater ecosystems. In the evolution of cyr genes, intragenomic translocations and intergenomic transfer between local Cylindrospermopsis and Raphidiopsis were emphasized and probably mediated by transposases. This research confirms the existence of CYN-producing Cylindrospermopsis in China and reveals the distinctive variations of cyr genes.
INTRODUCTION
Harmful cyanobacterial blooms, along with eutrophication in freshwater ecosystems, global warming, and worldwide spread of invasive cyanobacterial species, have drawn great attention in recent years (1–4). Cyanotoxins, such as saxitoxins, anatoxins, microcystins, and cylindrospermopsins (CYNs), are toxic metabolites produced by cyanobacteria, and their syntheses are regulated by a series of genetic and environmental factors (5–7). The outbreak of hepatoenteritis in Palm Island (Queensland, Australia) in 1979 led to the discovery of CYN, which was first isolated from bloom-forming Cylindrospermopsis raciborskii and proved to be mainly hepatotoxic (8–10). CYN is a sulfate ester with high solubility in water and comprises a tricyclic guanidine group and a hydroxymethyluracil moiety (10). Two analogues of CYN have been described: 7-epi-CYN, an enantiomer of CYN (11), and 7-deoxy-CYN with no hydroxylation on C-7 (12).
CYN can damage the liver, thymus, kidney, and heart (13). The cytotoxicity of CYN may be mediated by inhibiting the syntheses of protein (14) and glutathione (15). CYN is also a potential carcinogen because of its genotoxic effects by inhibiting pyrimidine nucleotide synthesis (16) and inducing DNA strand breakage (17, 18). An assay performed in mice revealed that 7-epi-CYN has severe toxicity similar to CYN and that uracil moiety is required for their toxicity (19). However, 7-deoxy-CYN shows no toxicity to mouse, and thus hydroxylation at C-7 is also crucial for the toxicity of CYNs (12). The bioaccumulation of CYNs in the tissues of vertebrates and invertebrates has been reported (20, 21) as a great health risk for humans and animals.
To date, CYNs have been detected in Nostocales and Oscillatoriales species, including Cylindrospermopsis, Raphidiopsis (22, 23), Aphanizomenon (24–26), Anabaena (27), Umezakia (28, 29), Oscillatoria (30), and Lyngbya (31) spp. CYN-producing Cylindrospermopsis from Australia and Asia have been reported, whereas Cylindrospermopsis strains isolated from Europe and America are incapable of CYN production (32–36). However, no conclusion can be drawn about the geographic distribution of the CYN-producing genotype of Cylindrospermopsis before additional samples from each continent are investigated by molecular and chemical methods.
The cyr gene cluster that encodes amidinotransferase, peptide synthetase (PS), polyketide synthase (PKS), and tailoring enzymes involved in CYN production has been described in C. raciborskii (37), R. curvata (38), Aphanizomenon sp. (39), and Oscillatoria sp. (30). The amidinotransferase CyrA catalyzes a transfer of an amidino group from arginine to glycine, which results in the first-product guanidinoacetate (40). Five cyr genes (cyrB through cyrF) that encode multi-enzymatic PSs and PKSs are probably involved in the polyketide chain synthesis that incorporates five units of acetate (41). The uracil moiety results from de novo synthesis possibly catalyzed by CyrG and CyrH. The sulfate group is incorporated by a sulfotransferase CyrJ with a suggested adenylylsulfate kinase CyrN providing the phosphoadenylyl sulfate pool (37). CyrI has been proven to catalyze hydroxylation at the C-7 of 7-deoxy-CYN (42), and CyrK has been proposed to be a potential transporter. Although cyr genes are highly conserved, the rearrangements of the cyr gene cluster and the insertion mutation of the cyrI gene have been reported (38). The cyrN and cyrO genes are found only in the end of the cyr gene cluster of C. raciborskii and are suggested to be excluded from the core set of cyr genes (30, 38).
An AbrB-like protein has been reported to be involved in the transcription regulation of cyr genes in Aphanizomenon ovalisporum (43). However, the protein-DNA interaction has not been verified in other CYN-producing species. The effects of temperature, light, nitrogen, phosphate, and sulfate on CYN production are inconclusive because of uncertainties in strain dependence, the release of CYNs, heterocyst formation, and combined effects of multiple factors in different experimental conditions (43–51). Davis et al. (51) highlighted the effects of the genetic diversity of CYN producers on the concentration and composition of CYNs in aquatic ecosystems. Moreover, CYN-producing and non-CYN-producing genotypes often coexist in the same populations. Therefore, an overview of CYN-producing species in total phytoplankton is essential for the risk assessment of CYNs. Furthermore, a systematic investigation of the diversity of cyr genes has not been performed and is thus necessary.
Cyanobacterial blooms occur perennially in numerous freshwater ecosystems, and CYNs have been detected in some urban reservoirs of China (52). Therefore, a comprehensive understanding of the diversity and distribution of CYN producers is essential. We illustrate this issue here by investigating the presence of cyr genes in cyanobacterial strains and environmental samples from different parts of China. Specifically, phylogenetic analysis was performed to explore the diversity and evolution of CYN producers. The conservation and variation of cyr gene sequences were also characterized.
MATERIALS AND METHODS
Cyanobacterial strains and culture conditions.
Cyanobacterial strains isolated from Chinese freshwater bodies were used for molecular and chemical analysis of CYNs (see Table S1 in the supplemental material). Three strains—C. raciborskii AWT205, C. raciborskii cyDB-1, and Aphanizomenon ovalisporum ILC-164—were isolated from Australia, Brazil, and Israel, respectively. Pure cultures of the cyanobacterial strains were grown in liquid MA medium (53) at 25°C under a 12-h/12-h light/dark cycle with constant white light intensity of 30 μmol of photons m−2 s−1. Cyanobacterial cells were harvested at the exponential phase (optical density at 680 nm [OD680] = 0.8) by centrifugation (12,000 × g) and stored at −80°C before further processing.
Collection of environmental samples.
Water samples were collected in lakes and reservoirs of China from 2006 to 2013 (Table 1). These water bodies were located between 22°N and 47°N in subtropical and temperate regions (see Fig. S1 and Table S2 in the supplemental material). A volume of 300 to 500 ml of water was filtered using a membrane filter (MF-Millipore, 0.22-μm pore size) in quadruplicate for each water body at each collection period. The filters were also stored at −80°C before DNA extraction.
TABLE 1.
Gene detection of environmental DNA samples from Chinese freshwater bodies
| Geographic origin | Abbreviation | Date of sample | Gene regionsa |
|||
|---|---|---|---|---|---|---|
| rpoC1 | cyrA | cyrI | cyrJ | |||
| Longhu Lake, Daqing, Heilongjiang | LH | July, 2012 | ND | − | − | ND |
| Jinyang Lake, Taiyuan, Shanxi | JY | Aug., 2010 | 4 | − | − | ND |
| Fish pond, Qingdao, Shandong | FQ | Nov., 2013 | 7 | − | − | ND |
| Taihu Lake, Wuxi, Jiangsu | TH | Aug., 2011 | ND | − | − | ND |
| Nov., 2011 | ND | − | − | ND | ||
| Fish pond, Nanjing, Jiangsu | FN | Nov., 2007 | 3 | − | − | ND |
| Qiandao Lake, Hangzhou, Zhejiang | QA | Oct., 2012 | 3 | 3 | 10 | 10 |
| Xianghu Lake, Hangzhou, Zhejiang | XH | Oct., 2012 | 5 | 1 | 7 | 8 |
| Xihu Lake, Hangzhou, Zhejiang | XL | Oct., 2012 | 5 | − | − | ND |
| Dongqian Lake, Ningbo, Zhejiang | DQ | July, 2009 | 3 | − | − | ND |
| Donghu Lake, Wuhan, Hubei | DH | Nov., 2006 | 3 | − | − | ND |
| Tangxun Lake, Wuhan, Hubei | TX | Oct., 2012 | ND | − | − | ND |
| Liangzi Lake, Ezhou, Hubei | LZ | Sept., 2011 | 4 | 2 | 2 | D |
| Qiaodun Lake, Daye, Hubei | QD | Sept., 2011 | 9 | 1 | 5 | 8 |
| Chidong Lake, Qichun, Hubei | CD | Aug., 2006 | 6 | 1 | 11 | 5 |
| Lushui Reservoir, Chibi, Hubei | LS | May, 2006 | 5 | − | − | ND |
| Poyang Lake, Nanchang, Jiangxi | PO | Aug., 2012 | ND | − | − | ND |
| Oct., 2012 | ND | − | − | ND | ||
| Erhai Lake, Dali, Yunnan | EH | Aug., 2010 | ND | − | − | ND |
| Sept., 2010 | ND | − | − | ND | ||
| Oct., 2010 | ND | − | − | ND | ||
| Fish pond, Kunming, Yunnan | FK | Oct., 2006 | 3 | − | − | ND |
| Dongzhen Reservoir, Putian, Fujian | DZ | Sept., 2011 | 2 | 2 | 8 | 6 |
| Fish pond, Panyu, Guangdong | FP | May, 2012 | 5 | − | − | ND |
| Shiyan Reservoir, Shenzhen | SY | June, 2007 | 5 | D | 6 | 3 |
| Qiankeng Reservoir, Shenzhen | QK | June, 2007 | 5 | − | − | ND |
| Tiegang Reservoir, Shenzhen | TG | June, 2007 | 9 | 3 | 10 | 9 |
| Luotian Reservoir, Shenzhen | LT | June, 2007 | 2 | − | − | ND |
| Changliupi Reservoir, Shenzhen | CL | June, 2007 | ND | − | − | ND |
The number of unique sequences is indicated where applicable. D, detected; ND, not detected; −, not tested.
DNA extraction, PCR, and sequencing.
Genomic DNA of cyanobacterial cells were extracted by using sodium dodecyl sulfate lysis and a phenol-chloroform-isoamyl alcohol extraction method described previously (54). Environmental DNA was extracted from membrane filters using a water DNA extraction kit according to the manufacturer's protocol (Omega Bio-Tek, USA). The filters were cut into pieces first and then subjected to the extraction process of the kit. The purified DNA were dissolved in Tris-EDTA buffer (pH 8.0) and stored at −20°C. The purity and concentrations of DNA samples were determined by a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, USA).
The primer pair PCβF/PCαR (54) with specificity targeting the phycocyanin operon (cpc) of cyanobacteria was used to confirm the validity of DNA templates for PCRs. Primers specific for cyrA, cyrI, and cyrJ genes of current known CYN-producing species were designed (see Fig. S2A in the supplemental material). Another primer set, rpoC1F53/rpoC1R739, was designed to selectively amplify the rpoC1 genes of Cylindrospermopsis and Raphidiopsis (see Fig. S2B in the supplemental material). PCR mix were prepared in 50-μl volumes containing 5 μl of 10× PCR buffer (TaKaRa, Japan), 10 nmol of each deoxynucleotide triphosphate, 10 pmol of each primer, 1 U of LA Taq (TaKaRa), and 100 ng of DNA templates. The cycling conditions were as follows: 94°C for 3 min; 35 cycles of 94°C for 45 s, 50°C to 60°C for 1 min, and 72°C for 2 min; 72°C for 10 min; and a 4°C hold. The annealing temperatures depended on the Tm values of primers (Table 2).
TABLE 2.
Characteristics of primer pairs used for gene detection
| Gene | Primer | Sequence (5′-3′) | Tm (°C) |
|---|---|---|---|
| cpcBA-IGS | PCβF | GGCTGCTTGTTTACGCGACA | 50 |
| PCαR | CCAGTACCACCAGCAACTAA | 50 | |
| cyrA | cyrAF51a | GATGGTTGTCGGGATTGCAGAT | 57 |
| cyrAR1167 | GAAGCGAGAAACGCCATTGGT | 57 | |
| cyrI | cyrIFb | CAGGCTTATCTGCAACAACATTTCT | 56 |
| cyrIR813 | CGGTTTATCAGTTCCAGAGTATCCA | 56 | |
| cyrJ | cyrJF13 | CGAATCGCAATGTGGTCTGTGC | 59 |
| cyrJR720 | GACAAGATATAGCGGCAACGACTCA | 59 | |
| cyrK | RTcyrKF991 | GGAGCGTGTTGGCTATTTC | 55 |
| RTcyrKR1379 | TGAGTCAAGGCACGAGAAG | 55 | |
| rpoC1 | rpoC1F53 | CACCAGAACGTATCCGCGCT | 60 |
| rpoC1R739 | GGTGGAATGACTGGAATGGCTGA | 60 |
C. raciborskii CS-505 numbering.
Targeting flanking sequence upstream cyrI gene.
The positive PCR products were amplified in triplicate and purified using a gel extraction kit (Omega Bio-Tek, USA). Purified gene fragments from environmental DNA were cloned into pMD18-T vector (TaKaRa). Recombinant plasmids of 5 to 15 positive bacterial clones were extracted, and the gene fragments were sequenced using an ABI 3730 automated sequencer (Applied Biosystems) in both directions. The primer regions of obtained sequences were deserted, and duplicated sequences in each water body were removed. The gene fragments from cyanobacterial strains were sequenced directly by using PCR primers in double directions.
Two methods were utilized to obtain the whole cyr gene clusters of cyanobacterial strains. First, the cyr genes and flanking sequences were amplified and sequenced according to the PCR methods described earlier (38). Second, genome sequencing was performed using a Hiseq 2000 (Illumina, USA) according to the manufacturer's instructions. A sequence library of 300 bp was constructed, and paired-end sequencing was carried out. After removing the low-quality reads, genome sequences were assembled by two software programs, including SOAPdenovo (v1.05) and Velvet (v1.0.09). The conservation of gene and protein sequences was verified by homologous search using BLAST on the website of the National Center for Biotechnology Information (NCBI). Open reading frames (ORFs) were determined by the ORF Finder tool implemented on the NCBI website.
Transcription detection.
Cyanobacterial cells from 2 ml of culture at the exponential phase were harvested by centrifugation. RNA extraction, DNase digestion and cDNA synthesis were performed as described previously (38). The DNase-digested RNA extracts and cDNA were used as the templates for transcription detection. The cyrI and cyrK genes were amplified by using the primer sets cyrIF/cyrIR813 and RTcyrKF991/RTcyrKR1379 (Table 2), respectively. A negative control without cyanobacterial cells was also subjected to the extraction and detection procedures. The genomic DNA of C. raciborskii AWT205 was used for positive PCR templates.
Phylogenetic assignment.
Four data sets, namely, cyrA, cyrI, cyrJ, and rpoC1, were constructed, including environmental sequences and reference gene sequences from cyanobacterial strains. Multiple sequence alignments were created by using the CLUSTAL W (v1.4) option in Bioedit v7.0.9.0 software and manually corrected. The best substitution models for gene evolution were selected by Modeltest v3.7 (55) and used for the inference of phylogenetic trees. Maximum-likelihood (ML) algorithm was used to carry out phylogenetic analysis by PHYML v3.0 (56) and PAUP v4.0b10 with 1,000 bootstrap replicates. Bayesian phylogenetic inference was performed using MrBayes v3.1.2 (57), and the parameters were set as described earlier (38). Neighbor-joining (NJ) trees were constructed by MEGA v4 (58) using Kimura two-parameter model with 1,000 bootstrap replicates. The GenBank accession numbers of reference gene sequences were displayed in Table S3 in the supplemental material. Selection analysis of environmental cyrA, cyrI, and cyrJ sequences were also performed as described previously (38). The secondary structures of protein sequences were predicted by PSIPRED v3.3 available online (59).
Toxin extraction and analysis.
Intracellular CYNs were extracted from lyophilized cyanobacterial cells by a modification of a method reported previously (60). Briefly, 30 mg of dry cells were mixed with 1 ml of Millipore water, sonicated for 20 min in an ice bath and shaken for 1 h at room temperature, followed by centrifugation. A total of 2-ml supernatants were collected after the extraction step was repeated. The supernatants were further subjected to solid-phase extraction (SPE) as described previously (61). Carbograph SPE cartridges (6.0 ml, 250 mg) were pretreated with 10 ml of elution solvent (dichloromethane-methanol, 1:4 [vol/vol]) acidified with 5% formic acid (vol/vol) and washed with 10 ml of water. The extracts were acidified with formic acid (1% [vol/vol]), and the ionic strength was adjusted with 0.1% sodium chloride (wt/vol) before application to the cartridges. The cartridges were then washed with 10 ml of water, followed by air to remove excess liquid. The absorbed toxins were eluted by 10 ml of elution solvent, and the solvent was removed by rotary evaporation thereafter. The precipitate was redissolved in 2 ml of water, and the solution was filtered through a Millipore ultracentrifugal filter (100 kDa). Extracellular CYNs were also extracted from cell-free spent culture medium by the SPE method. A volume of 100 ml of acidified medium was applied with a flow rate of 5 ml min−1, and the toxins were eluted by 20 ml of elution solvent.
CYNs were analyzed using two methods. First, CYNs were detected by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using an ESI-Q-TOF 6530 coupled with Infinity UHPLC 1290 (Agilent, USA). For LC conditions, a C18 column (4.6 mm by 250 mm, 5 μm) was applied with a temperature of 35°C. Compounds were separated by two linear gradient stages, 5 to 15% methanol in water during 0 min to 10 min, and 15 to 50% methanol in water during 10 min to 20 min with a flow rate of 0.25 ml min−1. The injection volume was 20 μl. The parameters of the mass spectrometer were set as follows: gas temperature, 300°C; flow rate, 11 liters min−1; nebulizer pressure, 45 lb/in2; capillary voltage, 3,500 V; nozzle voltage, 1,000 V; and fragmentor voltage, 175 V. Positive ions of m/z 100 to m/z 2,500 were monitored, and toxin analogues were determined by parent ions (m/z 416.1 for CYN and m/z 400.1 for 7-deoxy-CYN) and corresponding fragments (m/z 336.1, 274.1, and 194.1 for CYN and m/z 320.1, 274.1, and 194.1 for 7-deoxy-CYN). CYN and 7-epi-CYN could not be discriminated in the present study and therefore CYN represented these two analogues.
An efficient high-pressure liquid chromatography (HPLC) method was established by optimization of the HPLC-PDA method reported by Welker et al. (60). In brief, the SSI 1500 series system (SSI, USA) and a Synergi Polar-RP column (4.6 mm by 250 mm, 4 μm) maintained at 30°C was used. The elution conditions were as follows: a linear gradient of 10 to 30% solution B (0.05% trifluoroacetic acid [vol/vol] in 50% aqueous methanol [vol/vol]) in solution A (0.05% aqueous trifluoroacetic acid [vol/vol]) for 0 to 10 min, an isocratic stage of 30% solution B for 5 min, ramping to 100% solution B in 5 min, and final equilibration for 15 min. An injection volume of 20 μl and a flow rate of 0.8 ml min−1 were applied. UV absorption was detected at 262 nm. Standard CYNs were prepared by manual collection from elution fractions and then confirmed by LC-MS/MS. The standards were used for the identification of potential analogues in samples. In addition, commercial standard CYN (Enzo Life Sciences, USA) was used for quantification analysis, and the concentration of 7-deoxy-CYN was calculated as CYN equivalents.
Detection of toxin production in growth cultures.
CYN-producing cyanobacterial strains were first cultured to obtain original biomass (OD680 = 0.2 to 0.4). The cyanobacterial cells were harvested onto a glass fiber filter (Whatman, GF/C) by gentle filtration (<5 lb/in2) at sterile conditions and washed three times using MA medium. Afterward, the cells were resuspended and diluted into six parallel cultures (100 ml of MA for each) in 500-ml Erlenmeyer flasks with a cell density of OD680 = 0.13. The cultures were shaken manually three times every day. After inoculation, random cultures of each strain were used for toxin detection in triplicate at day 3 and 7, respectively. The cells and spent medium were separated by gentle filtration (<5 lb/in2) using membrane filters (MF-Millipore, 0.22-μm pore size) and used for toxin extraction and detection as described above. Statistical analyses were performed by independent-sample t test with SPSS 21.0 for Windows, and the differences were taken as significant at a P of <0.05.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in the present study are available under GenBank accession numbers KJ139686 to KJ139955.
RESULTS
Phylogenetic and geographic distribution of CYN genes.
All DNA templates from cyanobacterial strains and the environmental samples were confirmed to be efficient for cpc gene amplification. A total of 362 cyanobacterial strains, belonging to 10 genera of three orders, namely, Chroococcales, Nostocales, and Oscillatoriales, were examined for the presence of cyrJ gene. Positive strains were then detected for cyrA and cyrI genes. These strains were collected from 38 freshwater bodies across China, except for several Lyngbya strains obtained from swards and hot springs. Four Cylindrospermopsis strains and two Raphidiopsis strains were confirmed to contain cyr genes. CYNs were detected in the cell extracts of these strains by LC-MS/MS (Table 3). C. raciborskii CHAB3438 and C. raciborskii CHAB3440 contained both CYN and 7-deoxy-CYN, but the other four strains produced only 7-deoxy-CYN. C. raciborskii CHAB357, C. raciborskii CHAB3440, and R. curvata CHAB114 were isolated from the same cyanobacterial populations as and shared highly similar cyr sequences and toxin production to C. raciborskii CHAB358, C. raciborskii CHAB3438, and R. curvata CHAB1150, respectively. In addition, cyr genes, CYN, and 7-deoxy-CYN were also detected in C. raciborskii cyDB-1.
TABLE 3.
CYN-producing cyanobacterial strains isolated from Chinese freshwater bodies
| Strain | Geographic origin | Resulta |
Source or reference | |||
|---|---|---|---|---|---|---|
| cyrI | cyrK | CYN | 7-Deoxy-CYN | |||
| C. raciborskii | ||||||
| CHAB357 | Wenshan Lake | M | M | ND | D | This study |
| CHAB358 | Wenshan Lake | M | M | ND | D | This study |
| CHAB3438 | Xianghu Lake | H | M | D | D | This study |
| CHAB3440 | Xianghu Lake | H | M | D | D | This study |
| R. curvata | ||||||
| CHAB114 | Chidong Lake | M | H | ND | D | This study |
| CHAB1150 | Chidong Lake | M | H | ND | D | Jiang et al. (38) |
| CHAB3416 | Qiaodun Lake | M | H | ND | D | This study |
| HB1 | Guanqiao Pond | M | H | D | D | Li et al. (22) |
M, mutant sequences compared to cyr genes of C. raciborskii AWT205; H, homologous to cyr genes of C. raciborskii AWT205; D, detected; ND, not detected.
The presence of cyr genes was also examined in environmental DNA samples from 25 freshwater bodies. Finally, 13 cyrA, 59 cyrI, and 49 cyrJ sequences were obtained from samples collected from eight lakes and reservoirs (Table 1). A BLAST search revealed high similarities between environmental cyr sequences and corresponding cyr genes from Cylindrospermopsis and Raphidiopsis (C/R group, 99%). The environmental cyrA and cyrJ sequences were also found to be highly similar to the cyr genes of Aphanizomenon sp. strain 10E6 (99%). In contrast, the cyrI sequences were found to have low similarities to the cyrI gene of Aphanizomenon sp. strain 10E6 (97%). The environmental cyr sequences and cyr genes from the C/R group and Aphanizomenon sp. strain 10E6 were clustered into an independent clade in phylogenetic trees (data not shown). This clade was separated from the cyr genes of other species by high bootstrap values in the trees of the cyrI and cyrJ genes (97 to 100%).
Sequence analysis.
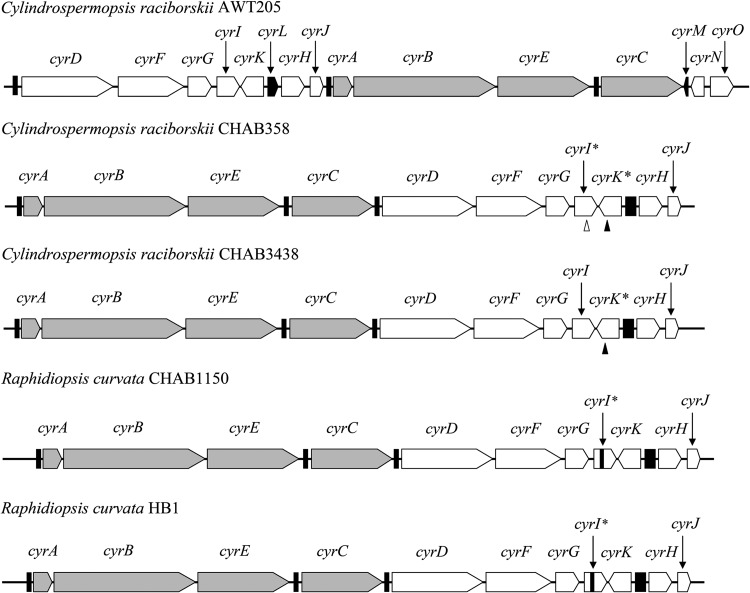
The cyr genes of C. raciborskii CHAB358 and R. curvata HB1 were sequenced and assembled into two complete gene clusters (Fig. 1). The genome of C. raciborskii CHAB3438 was assembled using high-quality data with an average coverage of 220, and the cyr gene cluster was found to be located in two contigs. The gap was closed by PCR amplification and Sanger sequencing. The final contig had a length of 50,355 bp and contained the whole cyr gene cluster (Fig. 1). The cyr genes in these three gene clusters showed high similarities to those of R. curvata CHAB1150 (>99%). The gene arrangement patterns of the cyr gene clusters of the C/R group strains from China were conserved and divergent from that of C. raciborskii AWT205 from Australia. The cyrN and cyrO genes were absent in the cyr gene clusters of Chinese strains (Fig. 1).
FIG 1.
Schematic structure of cyr gene clusters from CYN-producing cyanobacterial strains. Gray and white bars, cyr genes; black bar, transposase sequences or vestiges thereof; open triangles, base mutation in this position; solid triangles, nucleotide deletion in this position.
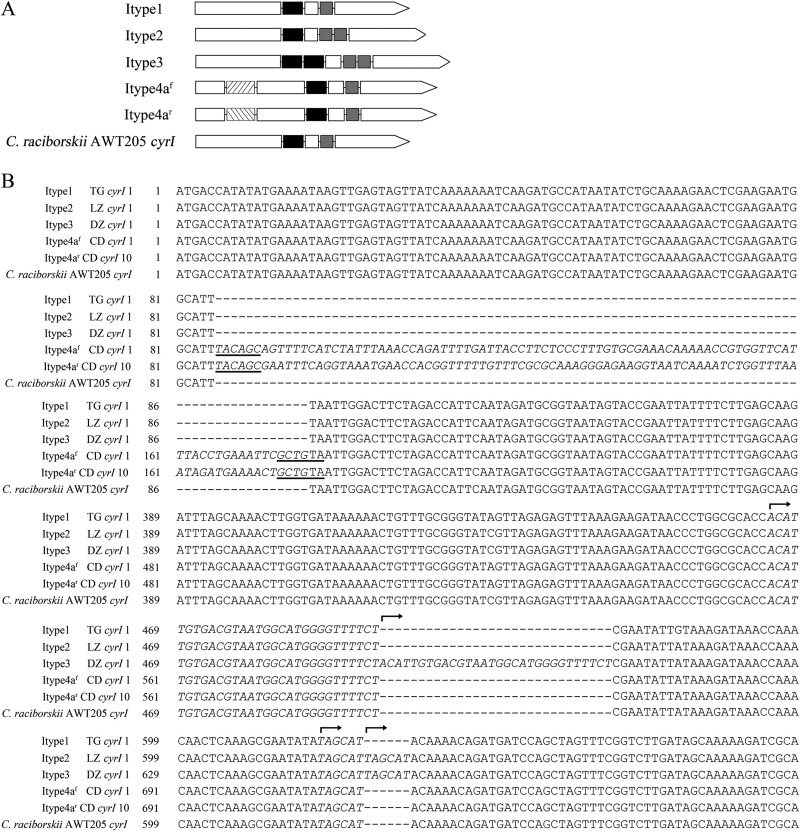
The CyrI of C. raciborskii CHAB358 was found to be truncated because of an intragenic stop codon caused by a base transition from cytosine to thymine at bp 529 (according to C. raciborskii AWT205 numbering, used here and below). Single-base mutations were also observed within cyrI sequences from TG and SY reservoirs (see Fig. S3 in the supplemental material). Six sequences had similar mutations to the cyrI gene of C. raciborskii CHAB358. Base transversions from guanine to thymine at bp 304 of two sequences were observed and also formed stop codons. In addition, four types of cyrI sequences were recognized according to intragenic sequence insertions compared to the cyrI gene of C. raciborskii AWT205, as depicted in Fig. 2. Itype1 contained no insertion sequence, but an insertion of a 6-nucleotide fragment, which is a repeat copy of its upstream sequence, was observed in Itype2 and Itype3 after bp 622. In addition, another insertion of a 30-nucleotide fragment, which is also a repeat copy of its upstream sequence, was observed in Itype3 after bp 494. Moreover, Itype4a included two kinds of sequences (i.e., Itype4af and Itype4ar) that contained reverse complementary insertion sequences of a 92-nucleotide fragment after bp 85, and the insertion sequences contained identical inverted terminal repeats (ITRs). The cyrI genes of R. curvata strains and those of other strains were classified into Itype4 and Itype1, respectively. In particular, both R. curvata CHAB1150 and R. curvata CHAB3416 contained the cyrI genes of Itype4af, and the cyrI gene of R. curvata HB1 was denominated as Itype4b with a long sequence insertion. Compared to Itype1, the deduced protein sequences of Itype2 and Itype3 were extended with repeated amino acids. The sequence insertions in Itype4 caused stop codons within the gene sequences and resulted in truncated protein sequences (see Fig. S4 in the supplemental material).
FIG 2.
Illustration of four sequence types of the cyrI gene. (A) Schematic structures of cyrI sequence types. White bar, cyrI sequences; black and gray bars, repeat sequences; slash and backslash bar, insertion sequences; C. raciborskii AWT205, reference strain. (B) Partial alignment of representative cyrI gene sequences. Repeat sequences and insertion sequences were italicized. Dashed line, gaps introduced into the alignment; bold line, ITRs; arrow, beginning of the repeat sequences.
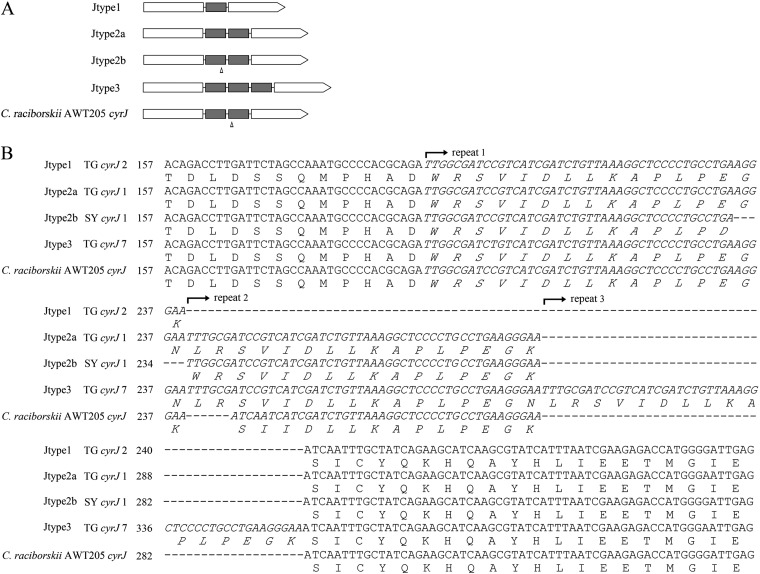
A 48-nucleotide fragment was found to be repeated within the cyrJ sequences. Three cyrJ sequence types—Jtype1, Jtype2, and Jtype3—were identified based on copy numbers 1, 2, and 3 of this sequence repeat, respectively (Fig. 3). Jtype2 contained two subtypes, namely, Jtype2a with two intact repeats and Jtype2b with a 6-nucleotide deletion in the first repeat. Most cyrJ genes from CYN-producing strains belong to Jtype2a, and those of three C. raciborskii strains (i.e., AWT205, CS-505, and cyDB-1) and R. mediterranea FSS-150 belong to a third subtype Jtype2c with a different 6-nucleotide deletion in the second repeat (Fig. 3). As displayed in Fig. S5 in the supplemental material, the sequence repeats within the cyrJ genes of the C/R group and Aphanizomenon sp. 10E6 were conserved. The second repeats in these species were divided into two groups based on nucleotide variations. One group contained the C/R group from Australia and Brazil, and the other contained the C/R group from China and Aphanizomenon sp. 10E6. Compared to Jtype1, the deduced protein sequences of Jtype2 and Jtype3 were extended and contained peptide repeats.
FIG 3.
Illustration of three sequence types of the cyrJ gene. (A) Schematic structures of cyrJ sequence types. White bar, cyrJ sequence; gray bar, repeat sequences; triangle, nucleotide deletions; C. raciborskii AWT205, reference strain. (B) Partial alignment of representative cyrJ gene sequences and deduced protein sequences. Repeat sequences were italicized. Dashed line, gaps introduced into the alignment; arrow, beginning of the repeat sequences.
The cyrK genes of C. raciborskii CHAB358 and C. raciborskii CHAB3438 lacked a thymine nucleotide at bp 1347 unlike those of C. raciborskii AWT205 (1,398-bp length). This lack of thymine nucleotide led to the truncation of the C-terminal sequence of CyrK (Fig. 4). Thus, the CyrK mutant (451 amino acids) was shorter than the original CyrK (465 amino acids).
FIG 4.
Partial alignment of mutant cyrK gene sequences and deduced protein sequences. Rectangle, nucleotide deletion; asterisk, stop codon.
Transcription analysis.
The transcriptions of cyrI and cyrK genes were examined for C. raciborskii CHAB358 and C. raciborskii CHAB3438. C. raciborskii AWT205 was used as a positive strain. Pure RNA extracts were not contaminated by genomic DNA, and cyr gene fragments were obtained from all cDNA samples. In addition, the amplicons covered the gene regions with nucleotide mutation and deletions.
Assessment of toxin release.
As depicted in Table S4 in the supplemental material, the cultures of four CYN-producing cyanobacterial strains maintained exponential growth from a low (OD680 = 0.13) to a high (OD680 = 0.34 to 0.61) cell density. The concentrations and extracellular percentages of CYNs were analyzed (Table 4; see also Table S4 in the supplemental material). Only 7-deoxy-CYN was detected in C. raciborskii CHAB358 and R. curvata CHAB1150, and a high percentage of CYN was observed in both extracellular (92 to 96%) and intracellular (95 to 98%) CYNs of C. raciborskii AWT205 and C. raciborskii CHAB3438. The extracellular percentages of CYN (30 to 39%), 7-deoxy-CYN (24 to 51%), and total CYNs (24 to 40%) on day 7 were significantly higher than the corresponding percentages on day 3 for all strains except C. raciborskii CHAB3438. The extracellular percentages of CYN and 7-deoxy-CYN between C. raciborskii AWT205 and C. raciborskii CHAB3438 were not significantly different except those of 7-deoxy-CYN on day 3. The extracellular percentages of 7-deoxy-CYN in C. raciborskii CHAB358 and R. curvata CHAB1150 were similar and significantly lower than those of other two strains, except between C. raciborskii AWT205 and C. raciborskii CHAB358 on day 3. For the extracellular percentages of the total CYNs, C. raciborskii CHAB358 and R. curvata CHAB1150 had lower values, with the significantly lowest percentage for R. curvata CHAB1150 on day 3 (15% ± 1.0%) and the significantly highest percentage for C. raciborskii AWT205 on day 7 (40% ± 3.0%).
TABLE 4.
Percent extracellular CYNs in cultures of four CYN-producing cyanobacterial strains
| CYN | Mean extracellular CYN content (%) ± SDa |
|||
|---|---|---|---|---|
| AWT205 | CHAB358 | CHAB3438 | CHAB1150 | |
| CYN | ||||
| Day 3 | 27 ± 6.0 | – | 24 ± 3.0 | – |
| Day 7 | 39 ± 3.0 | – | 30 ± 6.0 | – |
| 7-Deoxy-CYN | ||||
| Day 3 | 24 ± 4.0 | 21 ± 1.0 | 47 ± 4.0 | 15 ± 1.0 |
| Day 7 | 45 ± 2.0 | 24 ± 1.0 | 51 ± 9.0 | 26 ± 2.0 |
| Total CYNs | ||||
| Day 3 | 27 ± 6.0 | 21 ± 1.0 | 25 ± 3.0 | 15 ± 1.0 |
| Day 7 | 40 ± 3.0 | 24 ± 1.0 | 31 ± 6.0 | 26 ± 2.0 |
The four strains are described in the text. Data obtained on days 3 and 7 after inoculation are shown. −, not detected.
Phylogenetics of potential CYN producers based on rpoC1 sequences.
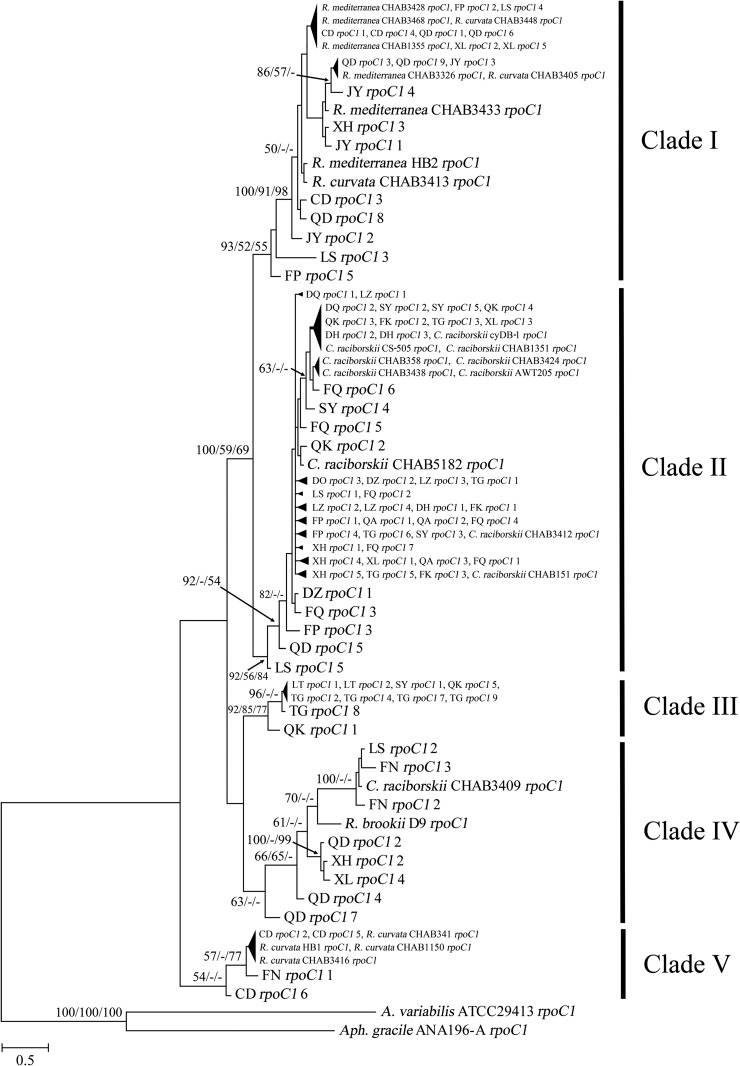
As displayed in Table 1, rpoC1 genes were detected in 19 lakes and reservoirs by C/R group specific primers, and 88 rpoC1 sequences were obtained. All of these sequences were confirmed to be derived from the C/R group by best BLASTn hits. Five independent clades were observed in the phylogenetic tree of rpoC1 sequences, and high support values were obtained for the divergence of clade I and clade II (Fig. 5). High sequence similarities were displayed within four clades: clade I (>97%), clade II (>99%), clade III (>98%), and clade V (>98%). However, clade IV comprised sequences with low to high similarities (95% to 100%), which is consistent with the long branches of this clade in the tree. Sequence similarities among clades were also calculated. The values between clade I and clade II (96 to 98%) were higher than those between these two and other clades (93 to 97%). In addition, median values were found between clade III and clade IV (95 to 96%). Also, clade V was the most divergent of all clades with lowest similarities (93 to 96%). Both clade I and clade V contained reference sequences from Raphidiopsis. However, the former was a Raphidiopsis-mix clade related to both R. mediterranea and R. curvata, whereas the latter was related to R. curvata only and thus was a R. curvata-like clade. Clade II contained reference sequences from Cylindrospermopsis and was denominated as a Cylindrospermopsis-like clade. For the closely related clade III and clade IV, no reference sequence was obtained for the former, but the latter included two reference sequences from C. raciborskii CHAB3409 and R. brookii D9. In addition, CYN-producing strains, along with non-CYN-producing strains, clustered together in clade II and clade V.
FIG 5.
Phylogenetic tree of rpoC1 gene sequences from environmental samples and cyanobacterial strains (topology based on a Bayesian tree). Bootstrap values above 50% are indicated at the nodes of the tree (Bayesian/ML/NJ). Aphanizomenon gracile ANA196-A and Anabaena variabilis ATCC 29413 were used as outgroups.
DISCUSSION
As displayed in Table 3, four C. raciborskii strains and four R. curvata strains from Chinese freshwater bodies were confirmed to contain both cyr genes and CYNs. However, CYN-producing strains constituted only a small percentage of the total cyanobacterial strains in the present study (1.7%). The cyr genes were also detected in eight freshwater bodies from which five CYN-producing strains were isolated. All of these aquatic ecosystems were located in the subtropical region.
Homologous and phylogenetic analyses revealed that the cloned cyr sequences from environmental samples were most likely to be derived from the C/R group. The mixed clade of cyr genes from the C/R group and Aphanizomenon sp. 10E6 was due to highly conserved sequences and few information sites (38, 39, 62).
The deduced protein sequences of Itype1 to Itype3 were conserved. The 6-nucleotide insertion in Itype2 and Itype3 formed two additional amino acids that belong to α-helix in the predicted secondary structures of CyrI proteins (see Fig. S6 in the supplemental material), and the 30-nucleotide insertion in Itype3 formed a duplicate peptide, including two residues involved in Fe2+ binding (42). The reverse complementary insertion sequences in Itype4af and Itype4ar provided more evidence for the transposon origin of these insertions. Similarly, the insertions of transposable elements within microcystin genes have also been reported (63, 64). The cyrI genes of two C. raciborskii strains contained base mutations, and those of four R. curvata strains denominated as Itype4 contained insertion mutations (Table 3). All of these mutations caused truncated protein sequences of CyrI, and therefore five strains synthesized only 7-deoxy-CYN due to the lack of CyrI function, as discussed earlier (38). Likewise, the cyrI gene variations may explain the high concentrations of 7-deoxy-CYN rather than CYN in L. wollei (31), C. raciborskii ISG9 (65), and the field populations of C. raciborskii (49).
The 48-nucleotide repeats in Jtype2 and Jtype3 caused duplicate peptides that belong to α-helix in the predicted secondary structures of CyrJ proteins (see Fig. S7 in the supplemental material). CYNs have been detected in cyanobacterial strains with the cyrJ genes of Jtype2a and Jtype2c. Therefore, nucleotide deletion in one repeat of Jtype2 does not lead to the deficiency of CyrJ function. The conservation of sequence repeats within cyrJ genes among the C/R group and Aphanizomenon sp. emphasized the horizontal gene transfer (HGT) among these species as described by Jiang et al. (38). According to the second repeat, the C/R group strains differed between China and Australia/Brazil, a finding which coincided with different arrangement patterns of the cyr gene clusters. Therefore, HGT events were hypothesized to have occurred between local Cylindrospermopsis and Raphidiopsis species.
Neutral evolution has been demonstrated for most cyr genes with low frequency of negatively selected codons (38), but purifying selection has also been found for the adenylation domain of Aphanizomenon ovalisporum-like cyrB sequences (66). Selection analysis of a large data set of environmental cyr sequences revealed evidence for neither recombination nor positive selection. Further, both cyrI and cyrJ sequences were not under neutral evolution (Tajima's test, P < 0.01) with 1 to 11 negatively selected codons. Thus, these two cyr genes from the C/R group may be under weak purifying selection. The sequence variations may be anciently created during the formation of these genes.
The CyrK sequences of four C. raciborskii strains were truncated at the C-terminal ends due to single-nucleotide deletions within the cyrK genes. However, the transcription of mutant cyrK and cyrI genes could still be detected. The transcription of cyrI genes may be ascribed to the cotranscription of polycistron (38), but cyrK gene was transcribed in the direction opposite to that of other cyr genes (Fig. 2). The release of CYNs in four strains with the CyrK of different lengths was investigated during a short culture period. A minor proportion of the total CYNs were extracellular for each strain (15 to 40%), but the accumulation of extracellular CYNs during the exponential growth phase must result from active release as proposed by Preussel et al. (47). The release was probably mediated by the transporter protein CyrK (37). The extracellular percentages of CYNs were strain dependent and did not correlate with CyrK lengths. Therefore, the mutant CyrK may function as the original CyrK.
Stucken et al. (67) found that the cyr gene cluster of Australian Cylindrospermopsis strains is inserted into a hydrogenase gene cluster (hyp). The genome sequencing of C. raciborskii CHAB3438 also revealed a hyp gene cluster (see Fig. S8 in the supplemental material). Four ORFs were observed between hypF and hupC genes, including two transposases (T1 and T2), as well as cyrN and cyrO genes. Intergenic sequences between hypF and hupC genes in other strains of the C/R group were also characterized (see Fig. S8 in the supplemental material). As a result, the cyrN gene was only found in CYN-producing strains, and the cyrO gene was observed in both CYN-producing and non-CYN-producing strains. Therefore, the cyrN gene, rather than the cyrO gene, likely belongs to the cyr gene cluster. The whole cyr gene cluster was probably originally inserted into the hyp gene cluster and then translocated to other genomic loci, with cyrN being a remnant. On the other hand, the cyr genes may have experienced acquisition, loss, and reacquisition in Chinese CYN-producing strains. The transfer of the cyr gene cluster was probably mediated by transposases observed to surround the gene cluster and between hyp genes.
The screening detection of potential CYN producers in the present study was performed with cyrJ gene as a molecular probe for its higher specificity to CYN-producing species than PS/PKS genes (37). However, Cylindrospermopsis-like cyr fragments except cyrJ were detected in Cylindrospermopsis strains from Brazil and water samples from Florida (34, 36). The Brazilian C. raciborskii cyDB-1 showed the presence of both cyr genes and CYNs and thus provides strong evidence for the distribution of CYN-producing Cylindrospermopsis in the American continent.
The aquatic ecosystems that contained rpoC1 genes of C/R group included those with cyr genes and are located in both subtropical and temperate regions. Thus, non-CYN-producing species were more widely distributed than were the CYN-producing species. The phylogenetics of potential CYN producers (C/R group) were analyzed based on the rpoC1 gene that displays higher discriminatory power at the genus and species levels than does the 16S rrn gene (68). The sequences in each clade were homogeneous but a little divergent in clade IV. Low support values were obtained for most of the five clades (Fig. 5), but sequence similarities among clades were lower than those within each clade. Raphidiopsis-mix clade I, Cylindrospermopsis-like clade II, and R. curvata-like clade V were also observed in a phylogenetic tree based on multigene sequences (69). Clade III and clade IV indicated cryptic and intricate evolutionary clades in C/R group. Clade III, clade IV, and clade V contained sequences from only subtropical regions, indicating the existence of warm-adaptive species in the C/R group. The distribution of CYN producers in these clades was sporadic, as reported previously (70). Cylindrospermopsis and Raphidiopsis might be congeneric as previously described (67). Meanwhile, both genera are suggested to be paraphyletic and taxonomic reconsideration of the C/R group is necessary.
Previous phylogeographic studies have suggested that Cylindrospermopsis strains were separated into three distinct groups, namely, strains from Australia, Europe, and America, with African strains and the former two groups being closely related (71–74). However, inconsistent phylogenetics have been observed for Tunisian and Spanish strains clustered into the America group (75, 76), and for clade II with strains from China, Australia, and Brazil without geographical separation. The present hypotheses suggested that the worldwide dispersion of Cylindrospermopsis originated from the tropical zones of Africa and Australia (77) or the warm refuge areas of each continent (73). The invasion success of Cylindrospermopsis has been attributed to phenotypic plasticity and different ecotypes (2, 74). On the contrary, the adaptability of Cylindrospermopsis and closely related Raphidiopsis in different environmental conditions may imply that the two species have similar cosmopolitan distribution to Microcystis (78), instead of invasive colonization. Furthermore, the coexistence of local and invasive species is a probable reason for the inconsistent results of phylogeographic analyses. For instance, R. curvata CHAB3413 and R. curvata CHAB3416 were isolated from the same water body with highly similar morphology and clustered into clade I and clade V, respectively. Worldwide cooperation is suggested for further phylogeographic study of Cylindrospermopsis and Raphidiopsis with strains from all climate conditions of each continent and through more effective methods, such as comparative genomics. Particularly, evidence for the distribution and growth conditions of Raphidiopsis should be provided in the future.
In conclusion, CYN biosynthesis genes were found to be sporadically distributed in cyanobacterial strains and freshwater ecosystems of China. All of the CYN-producing strains and environmental cyr sequences described here belong to congeneric and paraphyletic Cylindrospermopsis and Raphidiopsis species. Distinctive sequence variations, including base mutations, repeat sequences, and transposon insertions in the conserved cyr genes, are likely to be created during the formation of these genes. The C-terminal sequence of CyrK is probably not crucial for its function as a transporter. The cyrN gene is likely to be a member of the cyr gene cluster and distant from other cyr genes in Chinese CYN-producing strains. The intragenomic translocations and HGT of the cyr gene cluster are related to flanking transposases. The worldwide dispersion of Cylindrospermopsis may result from the simultaneous spread of local and invasive species.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Assaf Sukenik for the provision of the strain Aphanizomenon ovalisporum ILC-164.
This research was supported by the National Natural Science Foundation of China (31170189) and the National Water Science and Technology Projects (2012ZX07101-02-001-01 and 2012ZX07105-004).
Footnotes
Published ahead of print 13 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00551-14.
REFERENCES
- 1.Paerl HW, Huisman J. 2009. Climate change: a catalyst for global expansion of harmful cyanobacterial blooms. Environ. Microbiol. Rep. 1:27–37. 10.1111/j.1758-2229.2008.00004.x [DOI] [PubMed] [Google Scholar]
- 2.Bonilla S, Aubriot L, Soares MCS, González-Piana M, Fabre A, Huszar VLM, Lürling M, Antoniades D, Padisák J, Kruk C. 2012. What drives the distribution of the bloom-forming cyanobacteria Planktothrix agardhii and Cylindrospermopsis raciborskii? FEMS Microbiol. Ecol. 79:594–607. 10.1111/j.1574-6941.2011.01242.x [DOI] [PubMed] [Google Scholar]
- 3.Sinha R, Pearson LA, Davis TW, Burford MA, Orr PT, Neilan BA. 2012. Increased incidence of Cylindrospermopsis raciborskii in temperate zones: is climate change responsible? Water Res. 46:1408–1419. 10.1016/j.watres.2011.12.019 [DOI] [PubMed] [Google Scholar]
- 4.Paerl HW, Otten TG. 2013. Harmful cyanobacterial blooms: causes, consequences, and controls. Microb. Ecol. 65:995–1010. 10.1007/s00248-012-0159-y [DOI] [PubMed] [Google Scholar]
- 5.Pearson L, Mihali T, Moffitt M, Kellmann R, Neilan B. 2010. On the chemistry, toxicology and genetics of the cyanobacterial toxins, microcystin, nodularin, saxitoxin, and cylindrospermopsin. Mar. Drugs 8:1650–1680. 10.3390/md8051650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dittmann E, Fewer DP, Neilan BA. 2013. Cyanobacterial toxins: biosynthetic routes and evolutionary roots. FEMS Microbiol. Rev. 37:23–43. 10.1111/j.1574-6976.2012.12000.x [DOI] [PubMed] [Google Scholar]
- 7.Neilan BA, Pearson LA, Muenchhoff J, Moffitt MC, Dittmann E. 2013. Environmental conditions that influence toxin biosynthesis in cyanobacteria. Environ. Microbiol. 15:1239–1253. 10.1111/j.1462-2920.2012.02729.x [DOI] [PubMed] [Google Scholar]
- 8.Byth S. 1980. Palm Island mystery disease. Med. J. Aust. 2:40–42 [DOI] [PubMed] [Google Scholar]
- 9.Hawkins PR, Runnegar MTC, Jackson ARB, Falconer IR. 1985. Severe hepatotoxicity caused by the tropical cyanobacterium (blue-green alga) Cylindrospermopsis raciborskii (Woloszynska) Seenaya and Subba Raju isolated from a domestic water supply reservoir. Appl. Environ. Microbiol. 50:1292–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohtani I, Moore RE, Runnegar MTC. 1992. Cylindrospermopsin: a potent hepatotoxin from the blue-green alga Cylindrospermopsis raciborskii. J. Am. Chem. Soc. 114:7942–7944. 10.1021/ja00046a068 [DOI] [Google Scholar]
- 11.Banker R, Teltsch B, Sukenik A, Carmeli S. 2000. 7-Epicylindrospermopsin, a toxic minor metabolite of the cyanobacterium Aphanizomenon ovalisporum from Lake Kinneret, Israel. J. Nat. Prod. 63:387–389. 10.1021/np990498m [DOI] [PubMed] [Google Scholar]
- 12.Norris RL, Eaglesham GK, Pierens G, Shaw GR, Smith MJ, Chiswell RK, Seawright AA, Moore MR. 1999. Deoxycylindrospermopsin, an analog of cylindrospermopsin from Cylindrospermopsis raciborskii. Environ. Toxicol. 14:163–165 [DOI] [PubMed] [Google Scholar]
- 13.Terao K, Ohmori S, Igarashi K, Ohtani I, Watanabe MF, Harada KI, Ito E, Watanabe M. 1994. Electron microscopic studies on experimental poisoning in mice induced by cylindrospermopsin isolated from blue-green alga Umezakia natans. Toxicon 32:833–843. 10.1016/0041-0101(94)90008-6 [DOI] [PubMed] [Google Scholar]
- 14.Froscio SM, Humpage AR, Wickramasinghe W, Shaw G, Falconer IR. 2008. Interaction of the cyanobacterial toxin cylindrospermopsin with the eukaryotic protein synthesis system. Toxicon 51:191–198. 10.1016/j.toxicon.2007.09.001 [DOI] [PubMed] [Google Scholar]
- 15.Runnegar MT, Kong SM, Zhong YZ, Lu SC. 1995. Inhibition of reduced glutathione synthesis by cyanobacterial alkaloid cylindrospermopsin in cultured rat hepatocytes. Biochem. Pharmacol. 49:219–225. 10.1016/S0006-2952(94)00466-8 [DOI] [PubMed] [Google Scholar]
- 16.Reisner M, Carmeli S, Werman M, Sukenik A. 2004. The cyanobacterial toxin cylindrospermopsin inhibits pyrimidine nucleotide synthesis and alters cholesterol distribution in mice. Toxicol. Sci. 82:620–627. 10.1093/toxsci/kfh267 [DOI] [PubMed] [Google Scholar]
- 17.Bazin E, Huet S, Jarry G, Hégarat LL, Munday JS, Humpage AR, Fessard V. 2012. Cytotoxic and genotoxic effects of cylindrospermopsin in mice treated by gavage or intraperitoneal injection. Environ. Toxicol. 27:277–284. 10.1002/tox.20640 [DOI] [PubMed] [Google Scholar]
- 18.Alja Š, Filipi č M, Novak M, Žegura B. 2013. Double strand breaks and cell-cycle arrest induced by the cyanobacterial toxin cylindrospermopsin in HepG2 cells. Mar. Drugs 11:3077–3090. 10.3390/md11083077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banker R, Carmeli S, Werman M, Teltsch B, Porat R, Sukenik A. 2001. Uracil moiety is required for toxicity of the cyanobacterial hepatotoxin cylindrospermopsin. J. Toxicol. Environ. Health A 62:281–288. 10.1080/009841001459432 [DOI] [PubMed] [Google Scholar]
- 20.Saker ML, Eaglesham GK. 1999. The accumulation of cylindrospermopsin from the cyanobacterium Cylindrospermopsis raciborskii in tissues of the redclaw crayfish Cherax quadricarinatus. Toxicon 37:1065–1077. 10.1016/S0041-0101(98)00240-2 [DOI] [PubMed] [Google Scholar]
- 21.Kinnear S. 2010. Cylindrospermopsin: a decade of progress on bioaccumulation research. Mar. drugs 8:542–564. 10.3390/md8030542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li R, Carmichael WW, Brittain S, Eaglesham GK, Shaw GR, Liu Y, Watanabe MM. 2001. First report of the cyanotoxins cylindrospermopsin and deoxycylindrospermopsin from Raphidiopsis curvata (Cyanobacteria). J. Phycol. 37:1121–1126. 10.1046/j.1529-8817.2001.01075.x [DOI] [Google Scholar]
- 23.McGregor GB, Sendall BC, Hunt LT, Eaglesham GK. 2011. Report of the cyanotoxins cylindrospermopsin and deoxy-cylindrospermopsin from Raphidiopsis mediterranea Skuja (Cyanobacteria/Nostocales). Harmful Algae 10:402–410. 10.1016/j.hal.2011.02.002 [DOI] [Google Scholar]
- 24.Banker R, Carmeli S, Hadas O, Teltsch B, Porat R, Sukenik A. 1997. Identification of cylindrospermopsin in Aphanizomenon ovalisporum (Cyanophyceae) isolated from Lake Kinneret, Israel. J. Phycol. 33:613–616. 10.1111/j.0022-3646.1997.00613.x [DOI] [Google Scholar]
- 25.Preussel K, Stüken A, Wiedner C, Chorus I, Fastner J. 2006. First report on cylindrospermopsin producing Aphanizomenon flos-aquae (Cyanobacteria) isolated from two German lakes. Toxicon 47:156–162. 10.1016/j.toxicon.2005.10.013 [DOI] [PubMed] [Google Scholar]
- 26.Kokociński M, Mankiewicz-Boczek J, Jurczak T, Spoof L, Meriluoto J, Rejmonczyk E, Hautala H, Vehniäinen M, Pawełczyk J, Soininen J. 2013. Aphanizomenon gracile (Nostocales), a cylindrospermopsin-producing cyanobacterium in Polish lakes. Environ. Sci. Pollut. Res. 20:5243–5264. 10.1007/s11356-012-1426-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spoof L, Berg KA, Rapala J, Lahti K, Lepistö L, Metcalf JS, Meriluoto J. 2006. First observation of cylindrospermopsin in Anabaena lapponica isolated from the boreal environment (Finland). Environ. Toxicol. 21:552–560. 10.1002/tox.20216 [DOI] [PubMed] [Google Scholar]
- 28.Harada KI, Ohtani I, Iwamoto K, Suzuki M, Watanabe MF, Watanabe M, Terao K. 1994. Isolation of cylindrospermopsin from a cyanobacterium Umezakia natans and its screening method. Toxicon 32:73–84. 10.1016/0041-0101(94)90023-X [DOI] [PubMed] [Google Scholar]
- 29.Niiyama Y, Tuji A, Tsujimura S. 2011. Umezakia natans M. Watan. does not belong to Stigonemataceae but to Nostocaceae. Fottea 11:163–169 [Google Scholar]
- 30.Mazmouz R, Chapuis-Hugon F, Mann S, Pichon V, Méjean A, Ploux O. 2010. Biosynthesis of cylindrospermopsin and 7-epicylindrospermopsin in Oscillatoria sp. strain PCC 6506: identification of the cyr gene cluster and toxin analysis. Appl. Environ. Microbiol. 76:4943–4949. 10.1128/AEM.00717-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seifert M, McGregor G, Eaglesham G, Wickramasinghe W, Shaw G. 2007. First evidence for the production of cylindrospermopsin and deoxy-cylindrospermopsin by the freshwater benthic cyanobacterium, Lyngbya wollei (Farlow ex Gomont) Speziale and Dyck. Harmful Algae 6:73–80. 10.1016/j.hal.2006.07.001 [DOI] [Google Scholar]
- 32.Fastner J, Heinze R, Humpage AR, Mischke U, Eaglesham GK, Chorus I. 2003. Cylindrospermopsin occurrence in two German lakes and preliminary assessment of toxicity and toxin production of Cylindrospermopsis raciborskii (Cyanobacteria) isolates. Toxicon. 42:313–321. 10.1016/S0041-0101(03)00150-8 [DOI] [PubMed] [Google Scholar]
- 33.Valério E, Pereira P, Saker ML, Franca S, Tenreiro R. 2005. Molecular characterization of Cylindrospermopsis raciborskii strains isolated from Portuguese freshwaters. Harmful Algae 4:1044–1052. 10.1016/j.hal.2005.03.002 [DOI] [Google Scholar]
- 34.Yilmaz M, Phlips EJ, Szabo NJ, Badylak S. 2008. A comparative study of Florida strains of Cylindrospermopsis and Aphanizomenon for cylindrospermopsin production. Toxicon 51:130–139. 10.1016/j.toxicon.2007.08.013 [DOI] [PubMed] [Google Scholar]
- 35.Mankiewicz-Boczek J, Kokociński M, Gagała I, Pawełczyk J, Jurczak T, Dziadek J. 2012. Preliminary molecular identification of cylindrospermopsin-producing cyanobacteria in two Polish lakes (Central Europe). FEMS Microbiol. Lett. 326:173–179. 10.1111/j.1574-6968.2011.02451.x [DOI] [PubMed] [Google Scholar]
- 36.Hoff-Risseti C, Dörr FA, Schaker PDC, Pinto E, Werner VR, Fiore MF. 2013. Cylindrospermopsin and saxitoxin synthetase genes in Cylindrospermopsis raciborskii strains from Brazilian freshwater. PLoS One 8:e74238. 10.1371/journal.pone.0074238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mihali TK, Kellmann R, Muenchhoff J, Barrow KD, Neilan BA. 2008. Characterization of the gene cluster responsible for cylindrospermopsin biosynthesis. Appl. Environ. Microbiol. 74:716–722. 10.1128/AEM.01988-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y, Xiao P, Yu G, Sano T, Pan Q, Li R. 2012. Molecular basis and phylogenetic implications of deoxycylindrospermopsin biosynthesis in the cyanobacterium Raphidiopsis curvata. Appl. Environ. Microbiol. 78:2256–2263. 10.1128/AEM.07321-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stüken A, Jakobsen KS. 2010. The cylindrospermopsin gene cluster of Aphanizomenon sp. strain 10E6: organization and recombination. Microbiology 156:2438–2451. 10.1099/mic.0.036988-0 [DOI] [PubMed] [Google Scholar]
- 40.Muenchhoff J, Siddiqui KS, Poljak A, Raftery MJ, Barrow KD, Neilan BA. 2010. A novel prokaryotic l-arginine: glycine amidinotransferase is involved in cylindrospermopsin biosynthesis. FEBS J. 277:3844–3860. 10.1111/j.1742-4658.2010.07788.x [DOI] [PubMed] [Google Scholar]
- 41.Burgoyne DL, Hemscheidt TK, Moore RE, Runnegar MTC. 2000. Biosynthesis of cylindrospermopsin. J. Org. Chem. 65:152–156. 10.1021/jo991257m [DOI] [PubMed] [Google Scholar]
- 42.Mazmouz R, Chapuis-Hugon F, Pichon V, Méjean A, Ploux O. 2011. The Last step of the biosynthesis of the cyanotoxins cylindrospermopsin and 7-epi-cylindrospermopsin is catalyzed by CyrI, a 2-oxoglutarate-dependent iron oxygenase. Chembiochem 12:858–862. 10.1002/cbic.201000726 [DOI] [PubMed] [Google Scholar]
- 43.Shalev-Malul G, Lieman-Hurwitz J, Viner-Mozzini Y, Sukenik A, Gaathon A, Lebendiker M, Kaplan A. 2008. An AbrB-like protein might be involved in the regulation of cylindrospermopsin production by Aphanizomenon ovalisporum. Environ. Microbiol. 10:988–999. 10.1111/j.1462-2920.2007.01519.x [DOI] [PubMed] [Google Scholar]
- 44.Saker ML, Griffiths DJ. 2000. The effect of temperature on growth and cylindrospermopsin content of seven isolates of Cylindrospermopsis raciborskii (Nostocales, Cyanophyceae) from water bodies in northern Australia. Phycologia 39:349–354. 10.2216/i0031-8884-39-4-349.1 [DOI] [Google Scholar]
- 45.Saker ML, Neilan BA. 2001. Varied diazotrophies, morphologies, and toxicities of genetically similar isolates of Cylindrospermopsis raciborskii from northern Australia. Appl. Environ. Microbiol. 67:1839–1845. 10.1128/AEM.67.4.1839-1845.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bácsi I, Vasas G, Surányi G, Máthé C, Tóth E, Grigorszky I, Gáspár A, Tóth S, Borbely G. 2006. Alteration of cylindrospermopsin production in sulfate- or phosphate-starved cyanobacterium Aphanizomenon ovalisporum. FEMS Microbiol. Lett. 259:303–310. 10.1111/j.1574-6968.2006.00282.x [DOI] [PubMed] [Google Scholar]
- 47.Preussel K, Wessel G, Fastner J, Chorus I. 2009. Response of cylindrospermopsin production and release in Aphanizomenon flos-aquae (Cyanobacteria) to varying light and temperature conditions. Harmful Algae 8:645–650. 10.1016/j.hal.2008.10.009 [DOI] [Google Scholar]
- 48.Bar-Yosef Y, Sukenik A, Hadas O, Viner-Mozzini Y, Kaplan A. 2010. Enslavement in the water body by toxic Aphanizomenon ovalisporum, inducing alkaline phosphatase in phytoplanktons. Curr. Biol. 20:1557–1561. 10.1016/j.cub.2010.07.032 [DOI] [PubMed] [Google Scholar]
- 49.Orr PT, Rasmussen JP, Burford MA, Eaglesham GK, Lennox SM. 2010. Evaluation of quantitative real-time PCR to characterise spatial and temporal variations in cyanobacteria, Cylindrospermopsis raciborskii (Woloszynska) See-naya et Subba Raju and cylindrospermopsin concentrations in three subtropical Australian reservoirs. Harmful Algae 9:243–254. 10.1016/j.hal.2009.11.001 [DOI] [Google Scholar]
- 50.Cirés S, Wörmer L, Timón J, Wiedner C, Quesada A. 2011. Cylindrospermopsin production and release by the potentially invasive cyanobacterium Aphanizomenon ovalisporum under temperature and light gradients. Harmful Algae 10:668–675. 10.1016/j.hal.2011.05.002 [DOI] [Google Scholar]
- 51.Davis TW, Orr PT, Boyer GL, Burford MA. 2014. Investigating the production and release of cylindrospermopsin and deoxy-cylindrospermopsin by Cylindrospermopsis raciborskii over a natural growth cycle. Harmful Algae 31:18–25. 10.1016/j.hal.2013.09.007 [DOI] [PubMed] [Google Scholar]
- 52.Lei L, Peng L, Huang X, Han B. 2014. Occurrence and dominance of Cylindrospermopsis raciborskii and dissolved cylindrospermopsin in urban reservoirs used for drinking water supply, South China. Environ. Monit. Assess. 186:3079–3090. 10.1007/s10661-013-3602-8 [DOI] [PubMed] [Google Scholar]
- 53.Ichimura T. 1979. Media for the cultivation of algae, p 295–296 In Nishizawa K, Chihara M. (ed), Methods in phycological studies. Kyouritu Press, Tokyo, Japan: (In Japanese) [Google Scholar]
- 54.Neilan BA, Jacobs D, Goodman AE. 1995. Genetic diversity and phylogeny of toxic cyanobacteria determined by DNA polymorphisms within the phycocyanin locus. Appl. Environ. Microbiol. 61:3875–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- 56.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 57.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 58.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 59.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195–202. 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]
- 60.Welker M, Bickel H, Fastner J. 2002. HPLC-PDA detection of cylindrospermopsin: opportunities and limits. Water Res. 36:4659–4663. 10.1016/S0043-1354(02)00194-X [DOI] [PubMed] [Google Scholar]
- 61.Wormer L, Carrasco D, Cirés S, Quesada A. 2009. Advances in solid-phase extraction of the cyanobacterial toxin cylindrospermopsin. Limnol. Oceanogr. Methods 7:568–575. 10.4319/lom.2009.7.568 [DOI] [Google Scholar]
- 62.Kellmann R, Mills T, Neilan BA. 2006. Functional modeling and phylogenetic distribution of putative cylindrospermopsin biosynthesis enzymes. J. Mol. Evol. 62:267–280. 10.1007/s00239-005-0030-6 [DOI] [PubMed] [Google Scholar]
- 63.Christiansen G, Kurmayer R, Liu Q, Börner T. 2006. Transposons inactivate biosynthesis of the nonribosomal peptide microcystin in naturally occurring Planktothrix spp. Appl. Environ. Microbiol. 72:117–123. 10.1128/AEM.72.1.117-123.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fewer DP, Halinen K, Sipari H, Bernardová K, Mänttäri M, Eronen E, Sivonen K. 2011. Non-autonomous transposable elements associated with inactivation of microcystin gene clusters in strains of the genus Anabaena isolated from the Baltic Sea. Environ. Microbiol. Rep. 3:189–194. 10.1111/j.1758-2229.2010.00207.x [DOI] [PubMed] [Google Scholar]
- 65.Zarenezhad S, Sano T, Watanabe MM, Kawachi M. 2012. Evidence of the existence of a toxic form of Cylindrospermopsis raciborskii (Nostocales, Cyanobacteria) in Japan. Phycological Res. 60:98–104. 10.1111/j.1440-1835.2012.00639.x [DOI] [Google Scholar]
- 66.Yilmaz M, Phlips EJ. 2011. Diversity of and selection acting on cylindrospermopsin cyrB gene adenylation domain sequences in Florida. Appl. Environ. Microbiol. 77:2502–2507. 10.1128/AEM.02252-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stucken K, John U, Cembella A, Murillo AA, Soto-Liebe K, Fuentes-Valdéz JJ, Friedel M, Plominsky AM, Vásquez M, Glöckner G. 2010. The smallest known genomes of multicellular and toxic cyanobacteria: comparison, minimal gene sets for linked traits and the evolutionary implications. PLoS One 5:e9235. 10.1371/journal.pone.0009235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toledo G, Palenik B. 1997. Synechococcus diversity in the California current as seen by RNA polymerase (rpoC1) gene sequences of isolated strains. Appl. Environ. Microbiol. 63:4298–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu Z, Shi J, Xiao P, Liu Y, Li R. 2011. Phylogenetic analysis of two cyanobacterial genera Cylindrospermopsis and Raphidiopsis based on multi-gene sequences. Harmful Algae 10:419–425. 10.1016/j.hal.2010.05.001 [DOI] [Google Scholar]
- 70.Stucken K, Murillo AA, Soto-Liebe K, Fuentes-Valdés JJ, Méndez MA, Vásquez M. 2009. Toxicity phenotype does not correlate with phylogeny of Cylindrospermopsis raciborskii strains. Syst. Appl. Microbiol. 32:37–48. 10.1016/j.syapm.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 71.Dyble J, Paerl HW, Neilan B. 2002. Genetic characterization of Cylindrospermopsis raciborskii (Cyanobacteria) isolates from diverse geographical origins based on nifH and cpcBA-IGS nucleotide sequence analysis. Appl. Environ. Microbiol. 68:2567–2571. 10.1128/AEM.68.5.2567-2571.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neilan BA, Saker ML, Fastner J, Törökné A, Burns BP. 2003. Phylogeography of the invasive cyanobacterium Cylindrospermopsis raciborskii. Mol. Ecol. 12:133–140. 10.1046/j.1365-294X.2003.01709.x [DOI] [PubMed] [Google Scholar]
- 73.Gugger M, Molica R, Le Berre B, Dufour P, Bernard C, Humbert JF. 2005. Genetic diversity of Cylindrospermopsis strains (Cyanobacteria) isolated from four continents. Appl. Environ. Microbiol. 71:1097–1100. 10.1128/AEM.71.2.1097-1100.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Piccini C, Aubriot L, Fabre A, Amaral V, González-Piana M, Giani A, Figueredo CC, Vidal L, Kruk C, Bonilla S. 2011. Genetic and eco-physiological differences of South American Cylindrospermopsis raciborskii isolates support the hypothesis of multiple ecotypes. Harmful Algae 10:644–653. 10.1016/j.hal.2011.04.016 [DOI] [Google Scholar]
- 75.Fathalli A, Ben Rejeb Jenhani A, Moreira C, Welker M, Romdhane M, Antunes A, Vasconcelos V. 2011. Molecular and phylogenetic characterization of potentially toxic cyanobacteria in Tunisian freshwaters. Syst. Appl. Microbiol. 34:303–310. 10.1016/j.syapm.2010.12.003 [DOI] [PubMed] [Google Scholar]
- 76.Cirés S, Wörmer L, Ballot A, Agha R, Wiedner C, Velázquez D, Casero MC, Quesada A. 2014. Phylogeography of cylindrospermopsin and paralytic shellfish toxin-producing Nostocales cyanobacteria from Mediterranean Europe (Spain). Appl. Environ. Microbiol. 80:1359–1370. 10.1128/AEM.03002-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Padisák J. 1997. Cylindrospermopsis raciborskii (Woloszynska) Seenayya et Subba Raju, an expanding, highly adaptive cyanobacterium: worldwide distribution and review of its ecology. Arch. Hydrobiol. Suppl. 107:563–593 [Google Scholar]
- 78.van Gremberghe I, Leliaert F, Mergeay J, Vanormelingen P, Van der Gucht K, Debeer AE, Lacerot G, De Meester L, Vyverman W. 2011. Lack of phylogeographic structure in the freshwater cyanobacterium Microcystis aeruginosa suggests global dispersal. PLoS One 6:e19561. 10.1371/journal.pone.0019561 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.