Abstract
Vibrio (Aliivibrio) salmonicida is the etiological agent of cold water vibriosis, a disease in farmed Atlantic salmon (Salmo salar) that is kept under control due to an effective vaccine. A seawater temperature below 12°C is normally required for disease development. Quorum sensing (QS) is a cell density-regulated communication system that bacteria use to coordinate activities involved in colonization and pathogenesis, and we have previously shown that inactivation of the QS master regulator LitR attenuates the V. salmonicida strain LFI1238 in a fish model. We show here that strain LFI1238 and a panel of naturally occurring V. salmonicida strains are poor biofilm producers. Inactivation of litR in the LFI1238 strain enhances medium- and temperature-dependent adhesion, rugose colony morphology, and biofilm formation. Chemical treatment and electron microscopy of the biofilm identified an extracellular matrix consisting mainly of a fibrous network, proteins, and polysaccharides. Further, by microarray analysis of planktonic and biofilm cells, we identified a number of genes regulated by LitR and, among these, were homologues of the Vibrio fischeri symbiosis polysaccharide (syp) genes. The syp genes were regulated by LitR in both planktonic and biofilm lifestyle analyses. Disruption of syp genes in the V. salmonicida ΔlitR mutant alleviated adhesion, rugose colony morphology, and biofilm formation. Hence, LitR is a repressor of syp transcription that is necessary for expression of the phenotypes examined. The regulatory effect of LitR on colony morphology and biofilm formation is temperature sensitive and weak or absent at temperatures above the bacterium's upper threshold for pathogenicity.
INTRODUCTION
To protect themselves from environmental stress, bacteria can grow as surface-associated biofilms in their natural environment, as well as on artificial surfaces (reviewed by Karatan and Watnick [1]). The first step in biofilm formation is the attachment of single cells or bacterial aggregates to surfaces. Attachment requires that some adhesive properties to the surface are expressed. The resulting microcolony forms and grows in a self-produced extracellular matrix consisting mainly of secreted polysaccharides, proteins and sometimes also extracellular DNA (eDNA) (2–5). Other environmental factors such as temperature, pH, ion, nutrition, or water current may change the morphology and composition of the biofilm (1, 6).
Members of the vibrios are found in aquatic environments, as planktonic cells or attached to surfaces as bacterial aggregates or biofilms. Some vibrios have evolved to be pathogens, whereas others have developed symbiotic relationships with their hosts (7). Since many Vibrio species switch between growth within hosts and aquatic habitats, adaptation to changing environments is critical for their survival and colonization. A key factor for survival is the ability to produce a protective biofilm (8). An example of this is Vibrio cholerae, which forms biofilm in the aquatic environment, as well as in the intestines of the infected host (9–12). Similarly, biofilm formation plays a major role in the host colonization of Vibrio fischeri (13, 14), and it is also suggested to be an important feature of Vibrio anguillarum, Vibrio vulnificus, and Vibrio parahaemolyticus (15–18).
Exopolysaccharides (EPS) are major components of the vibrio biofilm matrix, and their expression is frequently correlated with changes in colony morphology (8). The EPS production in V. cholerae depends on two linked polysaccharide loci (vpsI and vpsII) and the positive regulators VpsT and VpsR (9, 19, 20). For V. fischeri, the formation of biofilm aggregates outside the squid light organ is crucial for proper initiation of host colonization. Both biofilm formation and the symbiotic relationship of V. fischeri with the squid host depend upon a cluster of 18 polysaccharide biosynthetic genes referred to as the symbiosis polysaccharide (syp) locus (14). Regulation of the syp locus involves a complex network of regulatory proteins, including the hybrid sensor kinase RscS and the downstream response regulator SypG (21, 22). V. fischeri strains overexpressing SypG show a dramatic increase in biofilm formation and have an advantage in colonizing the host (14, 21).
Quorum sensing (QS) is a cell-to-cell communication system that regulates a number of activities in vibrios, including biofilm formation, which is important for successful colonization and survival in environment. The majority of the signal molecules or autoinducers in QS are acylated homoserine lactones (AHLs) (23). A central regulator in QS is the transcription factor LitR, which in V. fischeri regulates bioluminescence and host colonization (24, 25). Homologs of V. fischeri LitR are found in many vibrios and their activities are often species specific (23). For example, the LitR homolog (HapR) in V. cholerae is a negative regulator of biofilm formation, colony rugosity, and vps expression (19, 26–28), whereas the corresponding homologs in V. parahaemolyticus (OpaR) and V. vulnificus (SmcR) are positive regulators of colony opacity and biofilm formation (17, 29, 30). Similarly, the V. anguillarum LitR homolog (VanT) positively regulates biofilm formation (15).
Vibrio (Aliivibrio) salmonicida causes cold-water vibriosis in farmed salmonid fish, and the disease occurs mainly in late autumn, winter, and early spring when the water temperature is below 12°C (31–35). The environmental lifestyle and transmission route of V. salmonicida is unclear but may be through the seawater either as bacterioplankton or on the surfaces of particles (32). Natural isolates of V. salmonicida have been characterized with regard to plasmid profiles and are grouped into at least four plasmid categories (36–38). Although differences in biochemical properties are found between strains, no correlation between biochemical reaction patterns, plasmid profiles, and virulence has been reported (37).
We have recently shown that LitR regulates virulence, as well as motility, cryptic bioluminescence, adhesion, and aggregation, in V. salmonicida. By light microscopy we found that LitR regulates negatively the formation of a loosely attached biofilm and speculated that a high content of polysaccharides prohibited quantification using traditional staining methods (39). In the present study, we have optimized the biofilm assay and show that V. salmonicida strains isolated at different geographic locations and with different plasmid profiles are poor biofilm producers at the chosen conditions. Inactivation of litR in the LFI1238 strain results in formation of a thick biofilm mainly composed of polysaccharides and proteins and to a much lesser extent also eDNA. Furthermore, a correlation between rugose colonies, adhesive properties, and biofilm formation was found, and these phenotypes were more strongly expressed at lower temperatures. Similar to V. fischeri, V. salmonicida LFI1238 harbors the syp locus with all 18 syp genes present (VSAL_II0295 to VSAL_II0312), and microarray analyses of cells grown in suspension or biofilm show that LitR is a strong repressor of syp transcription. Inactivation of syp genes alleviated biofilm, adhesion, and rugose colony morphology in the ΔlitR mutant.
MATERIALS AND METHODS
Strains and culture conditions.
The origins of the bacterial strains are listed in Table 1. Cells from glycerol stocks were streaked on blood agar with 2.5% NaCl (wt/vol) and incubated at 12°C for 4 days. The LFI1238 or the ΔlitR strains were used as parental strains for mutant construction. The derived V. salmonicida mutants were grown in lysogeny broth with 2.5% (wt/vol) NaCl (LB2.5) at 12°C unless otherwise indicated. The Escherichia coli strains S17-1 and JM109 were cultured in Luria-Bertani medium with 1% (wt/vol) NaCl at 37°C. The suicide plasmid pNQ705 was propagated in E. coli S17-1 cells. For selection of E. coli S17-1 transformants or V. salmonicida transconjugants, 25 or 2 μg of chloramphenicol/ml was added to the medium, respectively. For selection of E. coli JM109 transformants, 100 μg of ampicillin/ml was added.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description or sequence (5′–3′)a | Source or reference |
|---|---|---|
| Strains | ||
| V. salmonicida | ||
| LFI1238 | Wild type; isolated from Atlantic cod, sequenced strain | 40 |
| NCMB2262 | Wild type; isolated from Atlantic salmon, type strain | 33 |
| VS12 | Wild type; isolated from Atlantic salmon, Strønstad (1982) | 36 |
| VS26 | Wild type; isolated from Atlantic salmon, Seløy (1983) | 36 |
| VS103 | Wild type; isolated from Atlantic salmon, Valevåg (1987) | 36 |
| VS201 | Wild type; isolated from Atlantic salmon, Aukra (1987) | 36 |
| VS224 | Wild type; isolated from Atlantic salmon, Kristiansund (1987) | 36 |
| VS295 | Wild type; isolated from Atlantic salmon, Trangereid (1987) | 36 |
| VS370 | Wild type; isolated from Atlantic salmon, Krokelvdalen (1987) | 36 |
| VS446 | Wild type; isolated from Atlantic salmon, Sjøvegen (1988) | 36 |
| VS498 | Wild type; isolated from Atlantic cod, Hammerfest (1988) | 36 |
| ΔlitR mutant | LFI1238 containing an in-frame deletion in litR | 39 |
| ΔlitRc mutant | ΔlitR strain complemented with the wild-type litR gene; Cmr | 39 |
| ΔlitR sypC mutant | ΔlitR mutant with an insertional disruption in sypC; Cmr | This study |
| ΔlitR sypP mutant | ΔlitR mutant with an insertional disruption in sypP; Cmr | This study |
| ΔlitR sypQ mutant | ΔlitR mutant with an insertional disruption in sypQ; Cmr | This study |
| ΔlitR tadV mutant | ΔlitR mutant with an insertional disruption in tadV; Cmr | This study |
| ΔlitR rcpC mutant | ΔlitR mutant with an insertional disruption in rcpC; Cmr | This study |
| ΔlitR ptsIIB mutant | ΔlitR mutant with an insertional disruption in ptsIIB; Cmr | This study |
| ΔlitR ptsIIC mutant | ΔlitR mutantwith an insertional disruption in ptsIIC; Cmr | This study |
| ΔlitR ptsIIA mutant | ΔlitR mutantwith an insertional disruption in ptsIIA; Cmr | This study |
| E. coli | ||
| S17-1 | Donor strain for conjugation; λ pir | 41 |
| JM109 | Strain for subcloning pGEM-T Easy constructs | 42 |
| Plasmids | ||
| pNQ705 | Cmr; suicide vector with an R6K origin (pir requiring) | 43 |
| pGEM-T Easy | Ampr; TA cloning vector, white/blue screening | Promega |
| pNQ705-sypC | pNQ705 containing an internal fragment of VSAL_II0310 | This study |
| pNQ705-sypP | pNQ705 containing an internal fragment of VSAL_II0297 | This study |
| pNQ705-sypQ | pNQ705 containing an internal fragment of VSAL_II0296 | This study |
| pNQ705-tadV | pNQ705 containing an internal fragment of VSAL_II0367 | This study |
| pNQ705-rcpC | pNQ705 containing an internal fragment of VSAL_II0368 | This study |
| pNQ705-ptsIIB | pNQ705 containing an internal fragment of VSAL_II0823 | This study |
| pNQ705-ptsIIC | pNQ705 containing an internal fragment of VSAL_II0824 | This study |
| pNQ705-ptsIIA | pNQ705 containing an internal fragment of VSAL_II0825 | This study |
| Primers | ||
| II0823F | CTCGAGAGATTTTAGTGGTATGTGGCA | This study |
| II0823R | ACTAGTCAGCGTCTTCTAATTGAACG | This study |
| II0824F | CTCGAGCAGCAATCATGGTAGGTCTT | This study |
| II0824R | ACTAGTGGAGTGATACGAGCCAATA | This study |
| II0825F | CTCGAGTTGGTAACGACGGCATCG | This study |
| II0825R | ACTAGTTGACGCCATTCTTAAACACA | This study |
| II0296F | CTCGAGATTGCAGTAACCTCTGTCAGT | This study |
| II0296R | ACTAGTTTACGGTATCGTCGGTACA | This study |
| II0297F | CTCGAGTCATGTTGTTCAACATCTTTC | This study |
| II0297R | ACTAGTGATATGGTGTGAATGCACG | This study |
| II0310F | CTCGAGTCTACTGTCTCAATGGTGCA | This study |
| II0310R | ACTAGTGGAATCACCATCTGATAATGT | This study |
| II0367F | CTCGAGTTAGCCGCCATTTCTTATT | This study |
| II0367R | ACTAGTTTGGGTTAACTGCAATGG | This study |
| II0368F | CTCGAGTATGGCTTAGGGGAAAGC | This study |
| II0368R | ACTAGTATCGCTCATTGAAATATAAGTG | This study |
| LitR-G fwd | ACCAACGGCAGGACTTAGAC | 39 |
| LitR-H rev | TTGATAACAATCGAGCAGAGC | 39 |
Cmr, chloramphenicol resistance; Ampr, ampicillin resistance. For most of the wild-type strains, the source site is indicated, with the year of isolation specified in parentheses. Sequence underlining indicates the SpeI or XhoI restriction site added to the 5′ region of the primer.
Two different media were used for the biofilm assays: (i) Leibovitz L15 cell culture medium (Fisher Scientific) supplemented with NaCl to 380 mosM and further modified by the addition of MgCl2 to a final concentration of 10 mM, and the pH was adjusted to 8 (referred to as L15-M). (ii) A seawater-based (SWT) medium containing 5 g of Bacto peptone (BD), 3 g of yeast extract (Sigma), and 28 g of marine sea salt (Tetra) per liter. Solid medium was prepared by adding 1.5% (wt/vol) agar (Fluka).
Construction of V. salmonicida ΔlitR double mutants.
DNA extraction, general recombinant DNA techniques, and transformations were performed according to standard protocols (44). Restriction enzyme digestion, ligation, and PCR product and plasmid purifications were performed as recommended by the manufacturers (NEB Biolabs and OMEGA Bio-Tek). PCR (Phusion, Finnzyme) and BigDye sequencing (Applied Biosystems) were performed with custom-made primers (Sigma and Operon).
Construction of V. salmonicida LFI1238 containing a litR in-frame deletion (ΔlitR) and a complemented strain (ΔlitRc) have been described elsewhere (39). The ΔlitR double mutants used in the present study were made by chromosomal insertions mainly as described by others (43). Briefly, pNQ705-derived suicidal plasmids (Table 1) were used for chromosomal integration, and these were made by cloning a PCR-amplified part (∼250 bp) of the targeted genes into the suicide vector pNQ705. The PCR primers used for amplification are listed in Table 1, and their names correspond to the locus tag of the gene of interest, followed by the suffix “F” or “R” to indicate a forward or reverse primer direction, respectively. The restriction enzyme sites SpeI or XhoI were added to the 5′ end of the forward and reverse primers, respectively, to enable cloning of the PCR product into the corresponding sites in pNQ705. However, before being cloned into pNQ705, A′ overhangs were added to the PCR products and ligated into the pGEM-T Easy vector, followed by transformation into E. coli JM109 (Promega). The pGEM-T-derived plasmids were purified and digested with SpeI and XhoI before being electrophoresed in an agarose gel. Finally, the inserts were excised from the agarose gel, purified, and ligated into the corresponding restriction sites of pNQ705 before the transformation of E. coli S17-1. The different pNQ705 suicidal constructs were transferred from the S17-1 donor cells to V. salmonicida recipient cells by conjugation as described elsewhere (39). It should be noted that chromosomal plasmid insertions are likely to create polar effects of downstream genes in an operon (43).
Identification of the litR gene and prediction of σ54 binding sites in V. salmonicida.
The litR open reading frame in the V. salmonicida wild-type isolates (Table 1) were amplified by a standard PCR using the LitR-G forward and LitR-H reverse primers, followed by sequencing using BigDye chemistry (39).
Fuzznuc (http://emboss.sourceforge.net/apps/release/6.0/emboss/apps/fuzznuc.html) was used to predict the putative σ54 binding sites within the syp locus of V. salmonicida. To this end, a region spanning the genes VSAL_II0295 to VSAL_II0312 was extracted (GenBank accession number NC_011313.1) and used as input, together with the σ54 consensus sequence 5′-TGGCACGNNNNTTGCA-3′ (45). Three mismatches were allowed in the analysis.
Static biofilm assay.
Precultures of the different strains were made by adding one bacterial colony to 2 ml of medium and grown at 12°C with 200 rpm. After 2 days of incubation, the precultures were diluted 1:20 and incubated overnight. These overnight cultures were then diluted to an optical density at 600 nm (OD600) of 1.3 in LB2.5 before being further diluted 1:10 in L15-M or SWT medium. A total volume of 300 μl of each dilution was then added to each well in flat-bottom, non-tissue-culture-treated Falcon 24-well plates (BD Biosciences). The plates were incubated statically at different temperatures (4, 8, 12, 14, and 16°C). Biofilm formation was monitored by light phase-contrast microscopy, and pictures were taken by a photo camera installed with the microscope (Leica DM IRB or Zeiss Primo Vert). To quantify the biomass of the biofilm, the medium was removed, and 300 μl of 0.1% (wt/vol) crystal violet in H2O was added gently, followed by incubation for 30 min. The crystal violet stain was removed, and the wells were washed twice with 0.5 ml of H2O. The wells were air dried, and the biofilm was dissolved in 0.5 ml of 96% ethanol with strong agitation for 1 to 2 days. The dissolved biofilm was diluted 1:10 in 96% ethanol and transferred to a 96-well plate (100 μl/well). The absorbance was read at 590 nm (Vmax kinetic microplate reader; Molecular Devices).
Colony morphology assay.
The assay was performed mainly as described by others (46) and may allow differentiation of bacteria based on characteristics such shape, size, and color, as well as the texture of the colony. In brief, the different bacterial strains were grown overnight in LB2.5 broth before being diluted 1:10 in SWT medium. Then, 2 μl of each dilution was spotted onto blood agar plates supplemented with 2.5% (wt/vol) NaCl on LB2.5 and on SWT agar plates. The blood agar and LB2.5 plates were incubated at 4°C. The SWT plates were incubated at 4, 8, and 14°C. The colonies were viewed macroscopically and photographed with a Canon D400 digital camera. The colonies were also viewed microscopically (Zeiss Primo Vert) and photographed with AxioCam ERc5s.
Transmission electron microscopy.
Bacterial colonies grown at 4°C were removed from the agar plate by a scalpel and fixed in McDowell fixative (4% [wt/vol] formaldehyde and 1% [wt/vol] glutaraldehyde in phosphate buffer with an osmolarity of 320 mosM; pH 7.4) (47). The bacterial cells were postfixed with 1% (wt/vol) OsO4, dehydrated in graded series of ethanol, and embedded in an Epon substitute (AGAR [AGAR 100, MNA, DDSA, and DMP-30]) with propylene oxide as a transitional solvent. Ultrathin sections were cut using a Leica Ultracut S and a diamond knife (Diatome) and mounted on Formvar-coated copper grids, stained with 5% (wt/vol) uranyl acetate and Reynolds lead citrate (48), before being examined in a JEOL JEM 1010 transmission electron microscope with a Morada camera system (Olympus Soft Imaging System).
Scanning electron microscopy (SEM).
The static biofilm assay was performed as described above in SWT medium with Thermanox plastic coverslips with a 13-mm diameter (Nunc) soaked in the wells. The coverslips with the biofilm were removed from the medium at different time points and washed gently in phosphate-buffered saline (PBS) before being fixed and stained in 1.2% (wt/vol) glutaraldehyde, 0.45 M cacodylate buffer (pH 7.3), and 0.1% (wt/vol) ruthenium red. The biofilms were postfixed in 1% (wt/vol) OsO4, washed in PBS, and dehydrated in a graded series of ethanol. The specimens were dried using hexamethyldisilazane (Sigma-Aldrich) as a drying agent. The samples were mounted on specimen holders and coated with gold-palladium in a Polaron Sputter Coater. Pictures were taken on a JEOL JSM 6300 scanning electron microscope. At day 6, it was impossible to remove the slide without tearing the ΔlitR biofilm away from the slide. The ΔlitR biofilm was therefore harvested with a forceps before being fixed and processed as described above.
Biofilm inhibition and detachment assay.
In order to characterize the composition of the biofilm proteinase K, DNase I and sodium meta-periodate (NaIO4) were added to the wells, either before (biofilm inhibition assay) or after (detachment assay) biofilm formation. NaIO4 degrades β-1,6-linked polysaccharides. Two different endonucleases were used for degradation of eDNA. For the biofilm inhibition assay, a cold-tolerant and salt-active nuclease (SAN) was used. The SAN enzyme was kept in 20 mM Tris-HCl (pH 7.5), 2 mM MgCl2, 10 mM NaCl, 0.01% (vol/vol) Triton X-100, and 50% (wt/vol) glycerol (SAN buffer). The following reagents were added to the SWT medium to final concentrations of 40 μg of proteinase K/ml, 50 μg of SAN/ml, and 4 mM NaIO4. These concentrations did not inhibit the growth of the bacterium (data not shown). The biofilm was stained and quantified as described above.
The detachment assay was performed mainly as described elsewhere (49). Briefly, the biofilm was allowed to form in SWT for 72 h before being washed twice with PBS and incubated for 24 h at 37°C with 300 μl of (i) 10 mM NaIO4 dissolved in 0.1 M sodium acetate (pH 5.5), (ii) 0.1 mg/ml proteinase K (Sigma) in 20 mM Tris-HCl (pH 7.5)–100 mM NaCl, or (iii) 0.5 mg/ml bovine DNase I (DN25; Sigma) in 5 mM MgCl2. Detachment or dissolution of biofilm was monitored macroscopically by adding 5 μl of 0.1% (wt/vol) crystal violet to each well after incubation.
Microarray analysis.
Microarray analysis was performed on RNA isolated from planktonic bacteria grown in suspension and from bacteria grown in biofilm under static conditions as described above. In order to reveal potential targets for LitR regulation, we chose to grow the cells at low temperatures, when the differences in phenotypes between the wild type and ΔlitR mutant strains are most prominent. In addition, since the litR mutant is prone to aggregation in SWT medium (unpublished observation), as well as in other liquid media when grown at 4°C (39), it was necessary to grow planktonic cells at 8°C to monitor the cell density properly before harvest. Thus, for bacteria in suspension, the wild-type LFI1238 and the ΔlitR mutant were grown at 8°C in SWT medium with 200 rpm before being harvested at an OD600 of 0.8. The biofilms were grown in SWT medium in 72-cm2 cell culture flasks (Nunc) and harvested by a cell scraper after 72-h static incubation at 4°C. The RNA was extracted using RNA-isol (Fischer Scientific). DNA was removed by using a DNA-free kit (Ambion) and further cleaned up using the RNeasy MinElute Cleanup kit (Qiagen). cDNA was generated from 15 μg of RNA before being labeled by using an aminoallyl cDNA labeling kit (Ambion) and a CyDye postlabeling reactive dye pack (GE Healthcare). The samples were hybridized to “Vibrio salmonicida V1.0.1 AROS” slides, washed, and scanned as described elsewhere (50). Each microarray experiment was performed with four biological replicates, including one dye swap. The data were normalized within and between data sets using quantile normalization (51). To identify differentially expressed genes and calculate the fold change log2 values and P values, a linear model was implemented using R and the LIMMA package (52).
Accession numbers.
The V. salmonicida litR gene sequences have been deposited in the GenBank nucleotide sequence database (accession numbers KJ575525 to KJ575534). The microarray data are available at the NCBI Gene Expression Omnibus database (accession numbers GSE42994 and GSE42995).
RESULTS
Biofilm morphology changes with medium composition.
We previously showed that LitR in V. salmonicida regulates biofilm formation, and by using static conditions at a low temperature (4°C) the ΔlitR mutant formed a biofilm in the cell culture medium L15. The biofilm formed was viscous and loosely attached to the substratum, and attempts to quantify this biomass were unsuccessful (39). We therefore sought other medium combinations or culture media that could change the properties of the biofilm. Depending on culture media the biofilm showed different morphologies and thicknesses after 48- and 96-h incubations at 4°C. In L15-M the ΔlitR mutant produced a flat and regular biofilm, whereas in the SWT medium the ΔlitR mutant produced a much thicker, multilayered biofilm with irregular mushroom-like structures (Fig. 1A). The adhesion of the biofilm to the substratum improved in SWT and L15-M. Although care had to be taken, the biofilms were successfully stained with crystal violet and quantified, and the highest biomass content was found after growth in SWT medium (Fig. 1B).
FIG 1.
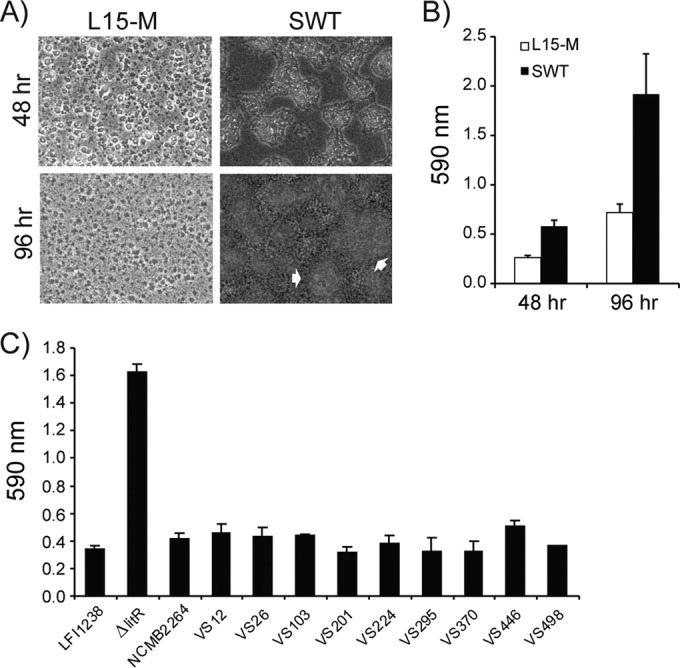
Biofilm formation of V. salmonicida wild-type strains and the LFI1238-derived ΔlitR mutant. (A and B) Morphology (A) and quantitation (B) of biofilms formed by the ΔlitR mutant in L15-M and SWT after 48 and 96 h of static incubation at 4°C. The morphology was monitored by phase-contrast microscopy (Leica), and the white arrows point to irregular mushroom-like structures in the biofilm. (C) Quantification of the biofilm formed by the different wild-type strains and the ΔlitR mutant in SWT medium after 72 h of incubation at 4°C. The biofilms were stained with crystal violet, and the absorbance was read at 590 nm. The error bars represent the standard deviations of three biological replicates.
Naturally occurring V. salmonicida strains are poor biofilm producers.
To find out whether our reference strain LFI1238, which is an Atlantic cod (Gadus morhua) isolate, is typical with regard to biofilm formation, we analyzed a collection of naturally occurring V. salmonicida strains isolated from Atlantic salmon (Salmo salar) (Table 1). This collection represents isolates from outbreaks at different geographical locations along the coast of Norway, as well as representing isolates with different plasmid profiles (see Table S1 in the supplemental material). Moreover, the isolates encode a complete litR open reading frame identical to the one in the LFI1238 strain (data not shown). Since bacterial growth and biofilm formation were more efficient in SWT medium, it was chosen as the standard medium. All wild-type strains produced significantly less biofilm than the ΔlitR mutant, irrespective of the geographical location or plasmid profile (Fig. 1C). This is in accordance with former assumptions that strains of V. salmonicida are very homogeneous with respect to biochemical reactions and virulence properties (37, 38).
Temperature-sensitive regulation of LitR on biofilm formation.
We next analyzed biofilm formation of the LFI1238, ΔlitR, and ΔlitRc strains in SWT medium at different temperatures. Increasing the temperature from 4 to 8°C accelerated biofilm formation, and the quantitative differences in biofilm formation between the wild-type LFI1238 strain and the ΔlitR strain were largest at 4 and 8°C, whereas at 16°C the wild-type and ΔlitR mutant strains produced similar amounts of biomass (Fig. 2A). Microscopic inspection of the ΔlitR biofilms revealed that there is a shift from the irregular mushroom-like structures formed at 4, 8, and 12°C to more flat and regular structures at 14°C and, at 16°C, biofilms with mushroom-like structures are no longer formed (Fig. 2B).
FIG 2.
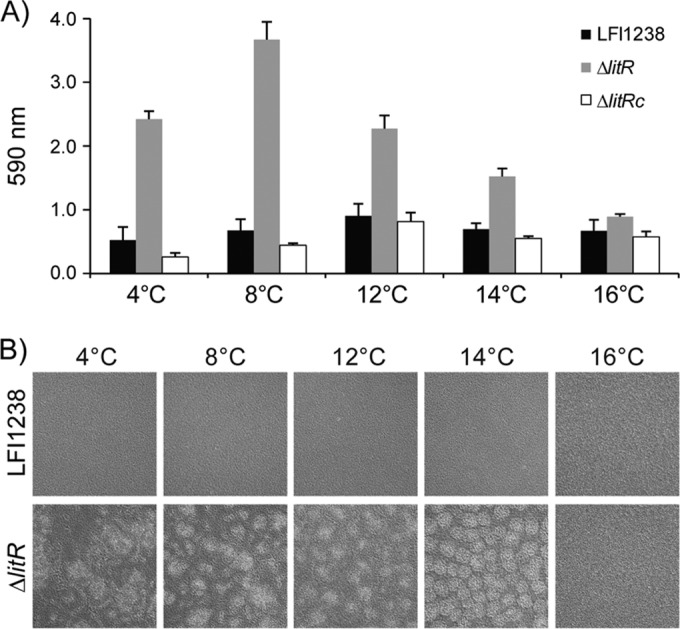
Biofilm formation in SWT medium at different temperatures. (A) Quantitation of crystal violet stained biofilms formed by wild-type LFI1238, ΔlitR, and ΔlitRc strains at the indicated temperatures. The biofilms were grown statically in the medium for 72 h before being quantified. The error bars represent the standard deviations of three biological replicates. (B) Morphologies of the biofilms formed by wild-type LFI1238 and the ΔlitR mutant at the indicated temperatures. The morphologies of biofilms were inspected in a Zeiss Primo Vert microscope (10× magnification) and photographed after 72 h of growth.
V. salmonicida LitR regulates medium- and temperature-dependent colony morphology.
The morphology of the colony often correlates with the ability to form biofilm, and biofilm producers usually show a wrinkled or rugose morphology (28, 46). Although the ΔlitR mutant grew as highly adhesive colonies at 4°C (compared to at 12°C) on blood agar, the colony phenotype seemed otherwise similar to the wild-type LFI1238 when grown from single cells (39). In order to more directly compare the colonies, we performed a spot colony analysis on different agar media. At 4°C, the LFI1238 strain produced smooth colonies on the three different media, whereas the ΔlitR mutant produced colonies that had a rough phenotype on blood and SWT agar but not on LB agar (Fig. 3A). The rugose colonies were highly adhesive and impossible to remove from the agar, whereas the smooth colonies had a creamy consistence. Temperature was important for the development of the rugose colony morphology; at 14°C the wrinkling of ΔlitR colonies is absent, and the morphology is similar to the one produced by the wild type (Fig. 3B). The complemented strain (ΔlitRc) behaves like the wild type in this assay (data not shown).
FIG 3.

Colony morphology of wild-type LFI1238 and ΔlitR strains. (A) The colonies were allowed to form on blood agar (BL), SWT, or LB agar plates for 10 days at 4°C before being photographed with a Canon camera. (B) The central and peripheral parts of colonies grown on SWT agar at 4, 8, and 14°C were viewed in a Zeiss Primo Vert microscope at 4× magnification. (C) Thin sections of wild-type and ΔlitR colonies grown on BL and analyzed by transmission electron microscopy. Scale bar, 2 μm.
Thin sections of colonies grown on blood agar plates at 4°C were prepared after the difference in morphology had manifested (8 to 10 days). As shown in Fig. 3C, the bacteria in the ΔlitR colonies contain an extracellular matrix (with thin filaments) not found in wild-type colonies.
SEM of ΔlitR biofilm reveals a high content of extracellular matrix.
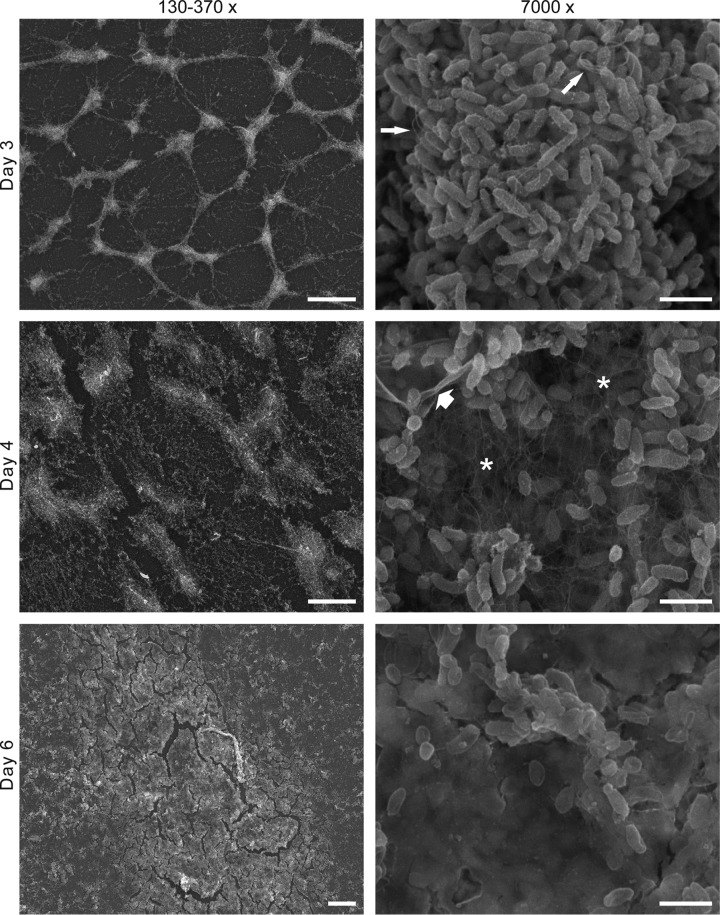
To investigate the formation and architecture of the biofilm produced by the ΔlitR mutant, we performed SEM analysis. The biofilm was allowed to form on the coverslips for 3 to 6 days in SWT medium before being analyzed. The biofilm formed slower on the slides than to the plastic in the cell culture wells. However, the time course of the biofilm development shows that the biofilm starts as a network of connected microcolonies that progress to a thick biofilm (Fig. 4). Using a higher magnification at day 4, we observe that the ΔlitR biofilm contained numerous long thin filaments that are likely to mediate the cell-to-cell interactions. Lastly, at day 6 the bacterial cells (in the collapsed biofilm) are completely embedded in a thick matrix. The wild-type strain showed significantly smaller amounts of attached bacteria (see Fig. S1 in the supplemental material).
FIG 4.
SEM analyses of ΔlitR biofilms after 3, 4, and 6 days. The biofilms were grown in SWT medium at 4°C before being fixed. The panel to the left shows the biofilms using a low magnification (scale bar, 100 μm for days 3 and 4 and 20 μm for day 6). The panel to the right shows the biofilm at a higher magnification (scale bar, 2 μm). At day 4 a network of fibers (*) is clearly visible, and at day 6 the bacteria are completely encapsulated in the extracellular matrix. The thin arrows point at some flagellum-like structures, and the thick arrow points to a sheet of collapsed extracellular matrix.
The major components of the extracellular matrix are polysaccharide and protein.
The composition of the extracellular matrix was determined by using a biofilm inhibition assay and a detachment assay. The biofilms were grown for 72 h in SWT media supplemented with SAN, proteinase K, or NaIO4 to analyze the contribution of eDNA, proteins, and polysaccharide in the extracellular matrix. Proteinase K and NaIO4 inhibited the biofilm formation almost completely, whereas biofilms grown in the presence of SAN produced a biofilm mass corresponding to ∼55% of the untreated control. The SAN buffer also showed some effect, suggesting that the reduction may not be entirely due to the action of SAN (Fig. 5A).
FIG 5.
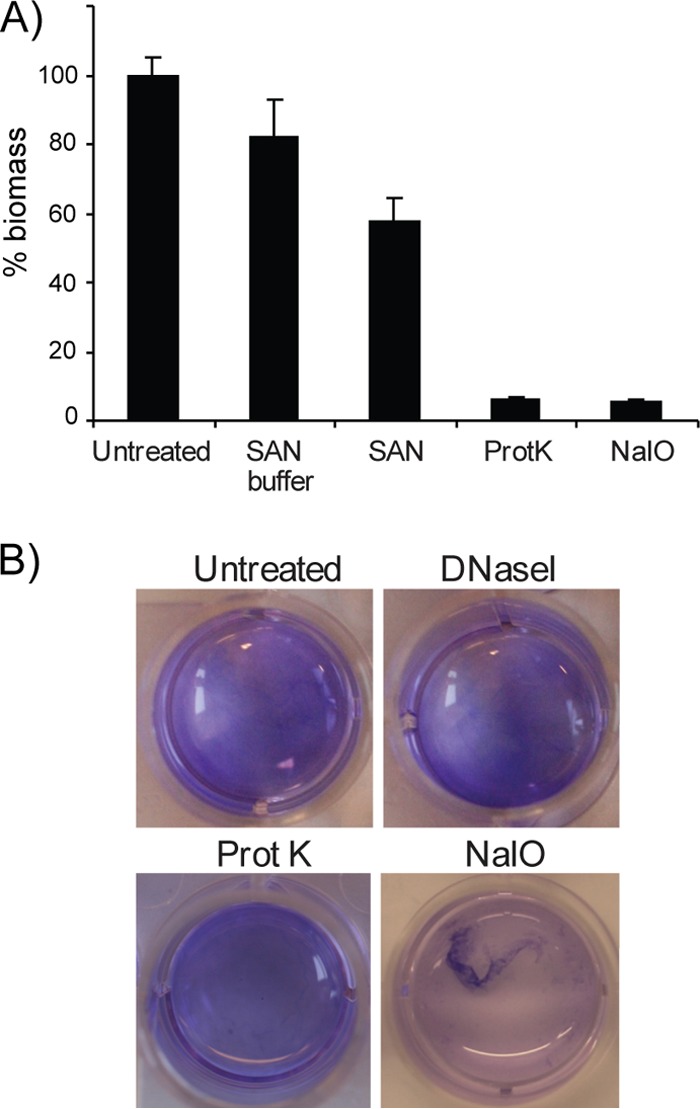
Characterization of extracellular matrix using a biofilm inhibition assay and a biofilm detachment assay. (A) For the inhibition assay, the different treatments (SAN, proteinase K, or NaIO4) were added directly to the SWT medium before the biofilms were allowed to form. After 3 days of static incubation at 4°C, the biofilms were stained with crystal violet, and the absorbance was read at 590 nm. The inhibition is shown as the percent biomass relative to the untreated control. (B) For the detachment assay, the biofilm was allowed to form for 3 days at 4°C before the medium (SWT) was removed, and the different treatments (bovine DNase I, proteinase K, or NaIO4) were added. The wells were incubated for 24 h at 37°C before dissolution of the biofilm was visualized by adding a small amount crystal violet into the wells. The error bars represent the standard deviations of three biological replicates.
Similarly, the composition was analyzed by adding proteinase K, bovine DNase I, and NaIO4 after the biofilms had formed. The incubation required in the posttreatment assay lead to detachment of the untreated biofilm, as well as for the treated biofilms. This prohibited standard quantification. However, the effects of the different treatments were visualized macroscopically by adding crystal violet directly to the wells. Dissolution of the biofilm was only detected after addition of NaIO4 and not when treated with bovine DNase I or proteinase K (Fig. 5B). The high quantity of polysaccharides in the matrix may have protected from the action of proteinase K.
LitR regulates syp expression in V. salmonicida.
To identify potential targets of LitR regulation, we performed microarray analyses with RNA isolated from the wild-type LFI1238 and the isogenic ΔlitR mutant grown in SWT medium either as static biofilm (4°C for 72 h) or in suspension (8°C, 200 rpm, OD600 = 0.8). The whole-genome expression data were analyzed, and genes showing a >1.0-fold log2 differential expression and a P value below 0.05 were extracted. With these cutoff values, a total of 123 genes were differently expressed in the biofilm experiment, while the number was 87 in the suspension experiment (see Tables S2 to S5 in the supplemental material). Interestingly, ca. 20% (biofilm, 18.7%; suspension, 19.5%) of the differently expressed genes were located in close proximity on chromosome II (Table 2). Among these were genes within the syp locus (VSAL_II0295 to VSAL_II0312) that were significantly upregulated in the ΔlitR strain compared to LFI1238 in both analyses. Apart from sypE (VSAL_II0308), the syp genes were more differentially expressed in cells grown as biofilms than in those grown as suspension. Immediately upstream the syp locus are two genes found to be highly upregulated in the ΔlitR mutant, one encoding a putative exported protein and the other a lipoprotein (VSAL_II0313 and VSAL_II0314), followed by a set of genes (VSAL_II0319 to VSAL_II0329) that are downregulated, including the recently described rpoQ (53) and a putative anti-sigma factor (Table 2). Moreover, among the upregulated genes in the ΔlitR mutant after growth in suspension were the phosphoenolpyruvate phosphotransferase system (pts) genes VSAL_II0825 and VSAL_II0823 (see Table S2 in the supplemental material). The homologous pts genes in V. cholerae have been shown to be involved in biofilm formation (54). We detected a fibrous network in the biofilm by SEM analysis, and although not abundant, some genes encoding pili, flagella, and curli were upregulated in the ΔlitR mutant (see Tables S2 and S4 in the supplemental material).
TABLE 2.
Subset of genes differently expressed between V. salmonicida wild-type LFI1238 and the ΔlitR mutanta
| CDS | Gene | Gene product | Biofilm |
Suspension |
||
|---|---|---|---|---|---|---|
| Log2 ratio | P (adjusted) | Log2 ratio | P (adjusted) | |||
| VSAL_II0296 | sypQ | Putative transmembrane glycosyltransferase | –2.45 | 9.51E–03 | –1.82 | 7.55E–03 |
| VSAL_II0297 | sypP | Putative glycosyltransferase | –5.17 | 7.86E–04 | –2.54 | 5.73E–04 |
| VSAL_II0298 | sypO | Putative membrane protein | –2.88 | 4.90E–03 | –1.62 | 3.16E–02 |
| VSAL_II0299 | sypN | Putative glycosyltransferases | –3.37 | 1.87E–03 | ||
| VSAL_II0300 | sypM | Hypothetical protein | –3.73 | 7.80E–03 | ||
| VSAL_II0301 | sypL | O-antigen polymerase | –1.01 | 5.30E–02 | ||
| VSAL_II0302 | sypK | Putative polysaccharide biosynthesis protein | –2.62 | 6.10E–04 | –1.70 | 4.27E–03 |
| VSAL_II0303 | sypJ | Putative glycosyltransferase | –2.46 | 3.57E–03 | ||
| VSAL_II0304 | sypI | Putative glycosyltransferase | –3.69 | 1.75E–05 | –2.13 | 2.98E–02 |
| VSAL_II0308 | sypE | Putative response regulator | –1.57 | 4.42E–02 | –1.80 | 5.44E–03 |
| VSAL_II0309 | sypD | Putative capsular polysaccharide synthesis protein | –1.71 | 2.78E–04 | ||
| VSAL_II0310 | sypC | Polysaccharide biosynthesis/export protein | –3.64 | 3.34E–05 | –1.42 | 7.54E–02 |
| VSAL_II0311 | sypB | Outer membrane protein, OmpA family | –4.15 | 1.88E–06 | –2.05 | 1.87E–03 |
| VSAL_II0312 | sypA | Hypothetical protein, putative anti-sigma factor antagonist | –4.34 | 5.76E–04 | –2.66 | 2.91E–04 |
| VSAL_II0313 | Putative exported protein | –4.38 | 5.87E–03 | |||
| VSAL_II0314 | Putative lipoprotein | –3.85 | 2.17E–04 | |||
| VSAL_II0316 | Response regulator, histidine kinase | 1.24 | 7.67E–02 | |||
| VSAL_II0319 | rpoQ | rpoS RNA polymerase sigma factor | 1.34 | 1.34E–02 | 2.27 | 5.34E–03 |
| VSAL_II0320 | Putative membrane-associated signaling protein | 1.36 | 2.07E–02 | |||
| VSAL_II0321 | Putative glycosyltransferase | 2.42 | 7.28E–03 | |||
| VSAL_II0322 | Putative membrane protein | 1.19 | 7.05E–02 | 3.48 | 2.21E–02 | |
| VSAL_II0323 | Putative lipoprotein | 2.05 | 3.81E–03 | 2.80 | 2.74E–02 | |
| VSAL_II0324 | Putative lipoprotein | 2.77 | 7.67E–03 | |||
| VSAL_II0325 | Putative exported protein | 1.13 | 5.86E–02 | |||
| VSAL_II0326 | Hypothetical protein | 1.17 | 6.41E–02 | 2.50 | 1.08E–02 | |
| VSAL_II0327 | Putative nucleotidyltransferase | 1.48 | 1.13E–02 | 2.13 | 5.39E–02 | |
| VSAL_II0328 | spoIIAA putative anti-sigma F factor antagonist | 1.10 | 2.74E–02 | 2.97 | 5.32E–03 | |
| VSAL_II0329 | Putative response regulator | 0.99 | 3.06E–02 | |||
| VSAL_II0332 | Putative hemolysin-type calcium-binding protein (fragment) | 1.55 | 3.41E–03 | 1.88 | 3.11E–02 | |
Positive fold log2 values represent genes downregulated in the ΔlitR mutant, and negative values represent genes upregulated in the ΔlitR mutant. The log2 ratios indicated in boldface were originally excluded from the analyses due to P values of >0.05.
Disruption of syp genes in the ΔlitR mutant alleviates rugosity and biofilm formation.
The organization of the syp locus in V. salmonicida is similar to the one in V. fischeri (14, 40) and contains four operons with putative σ54 binding sites (Fig. 6). To analyze whether expression of the syp locus was important for colony morphology and biofilm formation, we disrupted the sypC, sypP, and sypQ genes in the ΔlitR mutant by insertional inactivation (Fig. 6). We also disrupted the pts genes VSAL_II0823 to VSAL_II0825 and the tight adherence (tad) genes VSAL_II0367 and VSAL_II0368 since homologues of these have been shown to have an impact on biofilm formation in other species (54, 55). It should be noted that the tad genes were not differently expressed in the microarray analyses. Mutations of tad or pts genes did not show any effect on the colony morphology, and the colonies remained wrinkled and adhesive, similar to its parental ΔlitR strain, nor were there any significant differences in biofilm formation (Fig. 7A and B). On the other hand, for the ΔlitR syp double mutants the colony morphology reverted to smooth and nonadhesive, similar to the wild-type LFI1238 morphology (Fig. 7A). Moreover, mutation of the syp genes diminished biofilm formation in SWT medium, whereas in L15-M the ΔlitR syp mutants produced biofilm masses corresponding to wild-type LFI1238 levels (Fig. 7B). The difference in morphology between the ΔlitR and the ΔlitR syp mutants suggests that the initial attachment of the biofilm remains intact for the ΔlitR syp mutants in SWT medium, whereas building up the viscous mushroom-like structure is inhibited (Fig. 7C). Hence, disruption of syp expression in the ΔlitR mutant alleviates rugosity, adhesion and reduces biofilm formation.
FIG 6.
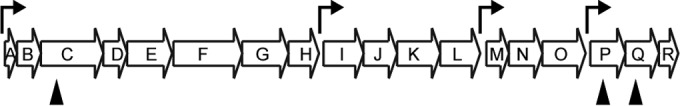
Genetic organization of the syp locus in V. salmonicida LFI1238. The locus consists of 18 genes (sypA to sypR) and four operons. Putative σ54 binding sites were predicted using Fuzznuc and are indicated by bent arrows. The syp genes that were interrupted by plasmid insertions in the present study are indicated by black arrowheads.
FIG 7.
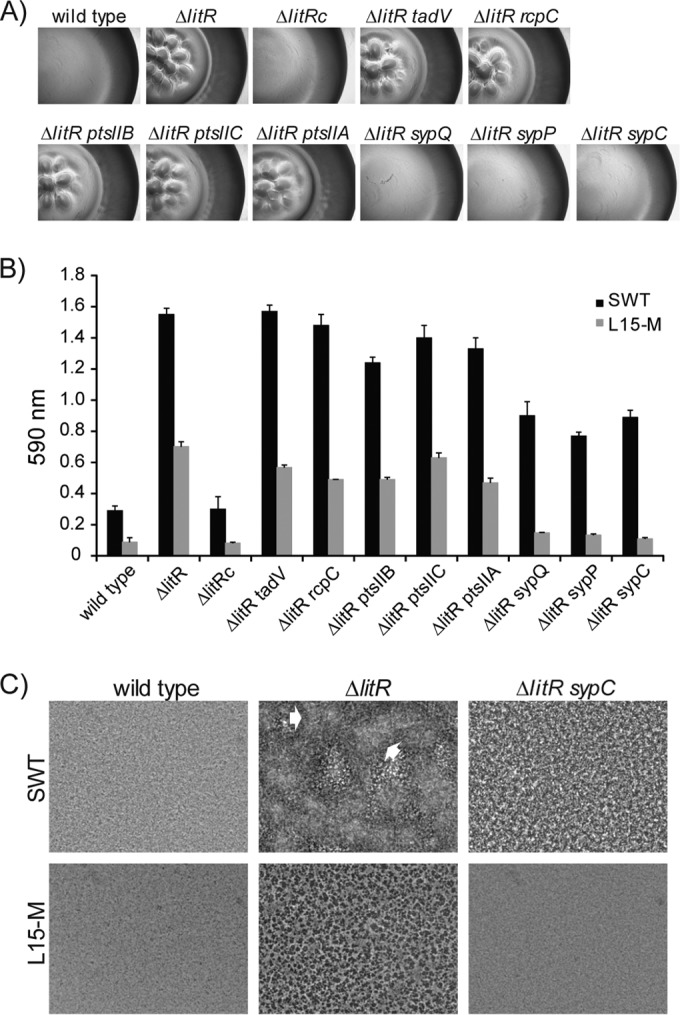
Colony morphology and biofilm formation of V. salmonicida wild-type LFI1238 and the different mutants. (A) The colony morphology of the different strains shows that the disruption of sypC, sypP, and sypQ in the ΔlitR mutant alleviates the rugose appearance. (B) Quantitation of the biofilms formed in L15-M and SWT medium after 3 days of static incubation. The error bars show the standard deviations of three biological replicates. (C) Phase-contrast microscopy (Leica) shows that disruption of syp results in a flat and regular biofilm when formed in SWT medium. The white arrows point to irregular mushroom-like structures in the biofilm. All experiments were performed at 4°C.
DISCUSSION
We have previously shown that inactivation of litR reduces the pathogenicity of V. salmonicida and enhances motility, adhesion, and biofilm formation in vitro (39). It is well established that environmental factors are important for biofilm formation, virulence, and the expression of other phenotypes (1). One of many environmental signals a microbe may encounter is a temperature change which may alter its growth, development, and pathogenesis (56).
V. salmonicida causes disease in the Atlantic salmon when the seawater temperature is below 10 to 12°C (35). In the laboratory, V. salmonicida grows at temperatures between 1 and 22°C, with a growth optimum at 12°C in liquid cultures and at 15°C on agar plates (33, 35). Furthermore, iron sequestration has been proposed to be temperature sensitive in V. salmonicida, being more efficient at temperatures of ≤10°C compared to at 15°C (57). Here we show that both temperature and medium composition are of importance for colony morphology, as well as biofilm formation in V. salmonicida. In L15-M medium the morphology of the biofilm was flat and regular biofilm, whereas in artificial seawater-based medium (SWT) thick biofilms with mushroom structures were formed. Thus, the ability of the ΔlitR mutant to produce biofilm and wrinkled colonies is both temperature and medium dependent. At low temperatures (4 to 8°C) the differences in phenotypes between the wild-type and ΔlitR strains are pronounced, in which expression of the litR inhibits biofilm formation (or activates biofilm dispersal), relieves adhesion, and inhibits the wrinkling of colonies. Clearly, biofilm formation in an artificial growth media may be very different from the natural ocean environment or fish tissues, and caution should be taken when extrapolating in vitro result to what may occur in vivo. However, the litR mutant mimics a low cell density (LCD) phenotype, and we speculate that LCD activities such as adhesion, motility and biofilm formation are important for colonization and survival in the environment. Expression of litR is regulated in a cell density-dependent manner (39). Thus, at LCD when litR is not expressed, the bacteria show increased adhesion to surfaces, produce an extracellular matrix, and acquire the ability to grow into a biofilm. A cold temperature is favorable for expressing LCD phenotypes and probably increases the ability of the bacteria to colonize a surface. The expression of litR is most likely expressed in response to accumulation of QS signals such as acylated homoserine lactones (AHLs), and several AHLs have been identified in the stationary growth phase of V. salmonicida (58, 59). Thus, as the cell density increases, the accumulation of AHLs and the expression of litR downregulates the biofilm, yielding planktonic cells that are able to escape and colonize new sites. At temperatures clearly above the limit for disease development, the V. salmonicida wild type and the ΔlitR mutant behaved more similarly to each other with regard to the expression of the different phenotypes investigated, and at 16°C neither strain produced biofilms with mushroom-like structures. Hence, this temperature may represent a cue that triggers exit of cells from the biofilm or show that the environment is not favorable for building up a three-dimensional biofilm. These two explanations are not mutually exclusive. If the results obtained at 16°C are due to QS, the lack of LitR regulation or other mechanisms remains to be resolved, and we are currently mapping the production of AHLs at different temperatures. It may be that the production of AHLs itself is subject to temperature regulation.
The ability to produce species-specific EPS and biofilm is probably important in niche selection (8). The ΔlitR mutant of V. salmonicida LFI1238 produced a slimy biofilm that contained an extracellular matrix consisting mainly of proteins and polysaccharides, and the microarray analyses demonstrated clearly that LitR regulates expression of the syp locus. Of the 18 syp genes, 14 were highly expressed in the biofilms produced by the ΔlitR mutant compared to similar samples of the wild type, whereas 9 syp genes were upregulated after the bacteria were grown in suspension. The difference may be due to time and temperature of growth, in addition to static incubation versus incubation with agitation. However, mutation of three syp genes (sypC, sypP, and sypQ) alleviated the wrinkled-colony phenotype and, depending on the medium, reduced or completely alleviated the biofilm formation of the ΔlitR mutant. Thus, the QS regulator LitR is a negative regulator of EPS in V. salmonicida, but the regulation cascade from LitR to the syp locus remains to be elucidated. Regulation of EPS expression and biofilm formation in vibrios involves numerous transcriptional regulators, two-component signal transduction, and QS regulators (8). HapR, the LitR homolog in V. cholerae, is also known to downregulate biofilm formation, and it does this by repressing the expression of VpsR and VpsT, which are positive regulators of the vps genes (19, 28, 60–62). In V. fischeri the syp locus is the major contributor to biofilm formation and colony wrinkling (14, 21). LitR has, to our knowledge, not been shown to regulate either biofilm or syp expression in V. fischeri. Instead, SypG is the direct activator of syp transcription, which itself is regulated by the sensor kinase RscS (22). V. salmonicida encodes homologues of RscS and the regulators SypG, VpsR, and VpsT (40), but none of these genes was found to be differentially expressed in our microarray analyses.
Interestingly, downstream the syp locus in V. salmonicida follows several genes that are downregulated in the ΔlitR mutant (VSAL_II0319 to VSAL_II0329). These are annotated as putative lipoproteins, response regulators, a sigma factor, and an anti-sigma factor antagonist (40). The sigma factor RpoQ (VSAL_II0319) was recently described in V. fischeri and found to be regulated by QS (53). Of the Vibrionaceae, only V. salmonicida and V. fischeri encode this sigma factor, and in both species LitR positively regulates rpoQ expression. Between the rpoQ and syp locus are two genes (VSAL_II0313 and II0314) located that are annotated as a putative export protein and a putative lipoprotein. Both are highly upregulated in the litR mutant, and further studies are needed to address the functions or biological activities of these gene products. Interestingly, the genes VSAL_II0317 and VSAL_II0318, as well as VSAL_II0321 to VSAL_II0329, have no apparent orthologues in V. fischeri but show similarity to genes of three Shewanella species (S. piezotolerans, S. pealeana, and S. halifaxensis) (data not shown). These three species form a distinct clade among the genus Shewanella (63). S. pealeana is a psychrotolerant species that has been isolated from the nidamental gland of the squid Loligo peali (64). Thus, this island of LitR-regulated genes located on chromosome II in V. salmonicida (Table 2) shares sequence similarities with V. fischeri and S. pealeana, both having squids as hosts. This may shed some light on the evolutionary history of V. salmonicida.
The ability to adhere to surfaces and form biofilms also depends upon structural components such as flagella and pili (55, 65, 66). We observed numerous long and thin filaments between the cells in the extracellular matrix of the biofilm and, in addition to flagella, the V. salmonicida genome encodes genes for two type IV pili, a Flp-type pilus (Tad pilus) and curli fibers (40). The tad locus is present in the genomes of a wide range of bacteria, including V. cholerae and V. vulnificus, and environmental conditions and QS are known to drastically modulate Flp-pilus expression (55, 67, 68). Since V. salmonicida encodes a complete tad locus, we inactivated two of the tad genes (VSAL_II0367 and VSAL_II0368) in the ΔlitR mutant. However, interruption of tad did not restore the smooth colony morphology, and amounts of biofilm similar to those produced by the parental strain were produced by these two double mutants. Nor did we detect any differential expression of the tad locus, suggesting that adhesion and biofilm formation in V. salmonicida is dispensable for the tad locus under the conditions used here. Lastly, the microarray showed upregulation of some flagella and the curli genes CsgF and CsgE (VSAL_I2856 and VSAL_I2857). Interestingly, the assembly of the E. coli curli pili is temperature regulated, and lower temperatures increase the formation of curli (69). However, the contribution of curli fibers in V. salmonicida biofilms remains to be verified.
We have shown here that LitR regulates biofilm formation, colony morphology, and syp expression. The differences in the phenotypes between wild-type and ΔlitR strains were more prominent at low temperatures. However, it remains to be demonstrated that litR expression per se is indeed regulated by temperature and in response to AHL accumulation. It also remains uncertain whether LitR regulates syp transcription directly or whether other factors are involved. The signaling cascade is likely complex and probably important to fine-tune activities under changing environmental conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Randi Olsen and Tom-Ivar Eilertsen at the Electron Microscopic Department, UiT–The Arctic University of Norway, for excellent technical assistance. We also thank Ruth H. Paulssen and The Microarray Resource Centre in Tromsø for offering facilities and equipment. We thank Henning Sørum at the Norwegian University of Life Sciences for V. salmonicida isolates, Olav Lanes at ArcticZymes for the SAN, and Debra Milton at Umeå University for the pNQ705 plasmid.
This study was supported by the Norwegian Research Council and UiT–The Arctic University of Norway.
Footnotes
Published ahead of print 27 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01239-14.
REFERENCES
- 1.Karatan E, Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73:310–347. 10.1128/MMBR.00041-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutherland IW. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9:222–227. 10.1016/S0966-842X(01)02012-1 [DOI] [PubMed] [Google Scholar]
- 3.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. 10.1126/science.295.5559.1487 [DOI] [PubMed] [Google Scholar]
- 4.Flemming HC, Neu TR, Wozniak DJ. 2007. The EPS matrix: the “house of biofilm cells.” J. Bacteriol. 189:7945–7947. 10.1128/JB.00858-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harmsen M, Lappann M, Knochel S, Molin S. 2010. Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Appl. Environ. Microbiol. 76:2271–2279. 10.1128/AEM.02361-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goller CC, Romeo T. 2008. Environmental influences on biofilm development. Curr. Top. Microbiol. Immunol. 322:37–66 [DOI] [PubMed] [Google Scholar]
- 7.Thompson FL, Iida T, Swings J. 2004. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68:403–431. 10.1128/MMBR.68.3.403-431.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yildiz FH, Visick KL. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17:109–118. 10.1016/j.tim.2008.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yildiz FH, Schoolnik GK. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 96:4028–4033. 10.1073/pnas.96.7.4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watnick PI, Kolter R. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586–595. 10.1046/j.1365-2958.1999.01624.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colwell RR, Huq A, Islam MS, Aziz KM, Yunus M, Khan NH, Mahmud A, Sack RB, Nair GB, Chakraborty J, Sack DA, Russek-Cohen E. 2003. Reduction of cholera in Bangladeshi villages by simple filtration. Proc. Natl. Acad. Sci. U. S. A. 100:1051–1055. 10.1073/pnas.0237386100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faruque SM, Biswas K, Udden SM, Ahmad QS, Sack DA, Nair GB, Mekalanos JJ. 2006. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. U. S. A. 103:6350–6355. 10.1073/pnas.0601277103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nyholm SV, Stabb EV, Ruby EG, Fall-Ngai MJ. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. U. S. A. 97:10231–10235. 10.1073/pnas.97.18.10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yip ES, Grublesky BT, Hussa EA, Visick KL. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and sigma-dependent biofilm formation by Vibrio fischeri. Mol. Microbiol. 57:1485–1498. 10.1111/j.1365-2958.2005.04784.x [DOI] [PubMed] [Google Scholar]
- 15.Croxatto A, Chalker VJ, Lauritz J, Jass J, Hardman A, Williams P, Camara M, Milton DL. 2002. VanT, a homologue of Vibrio harveyi LuxR, regulates serine, metalloprotease, pigment, and biofilm production in Vibrio anguillarum. J. Bacteriol. 184:1617–1629. 10.1128/JB.184.6.1617-1629.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gulig PA, Bourdage KL, Starks AM. 2005. Molecular pathogenesis of Vibrio vulnificus. J. Microbiol. 43:118–131. [PubMed] [Google Scholar]
- 17.Lee JH, Rhee JE, Park U, Ju HM, Lee BC, Kim TS, Jeong HS, Choi SH. 2007. Identification and functional analysis of Vibrio vulnificus SmcR, a novel global regulator. J. Microbiol. Biotechnol. 17:325–334 [PubMed] [Google Scholar]
- 18.Nair GB, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, Sack DA. 2007. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20:39–48. 10.1128/CMR.00025-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yildiz FH, Dolganov NA, Schoolnik GK. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPSETr-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716–1726. 10.1128/JB.183.5.1716-1726.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casper-Lindley C, Yildiz FH. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186:1574–1578. 10.1128/JB.186.5.1574-1578.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yip ES, Geszvain K, Loney-Marino CR, Visick KL. 2006. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol. 62:1586–1600. 10.1111/j.1365-2958.2006.05475.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussa EA, Darnell CL, Visick KL. 2008. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J. Bacteriol. 190:4576–4583. 10.1128/JB.00130-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milton DL. 2006. Quorum sensing in vibrios: complexity for diversification. Int. J. Med. Microbiol. 296:61–71. 10.1016/j.ijmm.2006.01.044 [DOI] [PubMed] [Google Scholar]
- 24.Fidopiastis PM, Miyamoto CM, Jobling MG, Meighen EA, Ruby EG. 2002. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 45:131–143. 10.1046/j.1365-2958.2002.02996.x [DOI] [PubMed] [Google Scholar]
- 25.Lupp C, Ruby EG. 2005. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 187:3620–3629. 10.1128/JB.187.11.3620-3629.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammer BK, Bassler BL. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101–104. 10.1046/j.1365-2958.2003.03688.x [DOI] [PubMed] [Google Scholar]
- 27.Zhu J, Mekalanos JJ. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647–656. 10.1016/S1534-5807(03)00295-8 [DOI] [PubMed] [Google Scholar]
- 28.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497–515. 10.1111/j.1365-2958.2004.04154.x [DOI] [PubMed] [Google Scholar]
- 29.McCarter LL. 1998. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J. Bacteriol. 180:3166–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enos-Berlage JL, McCarter LL. 2000. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J. Bacteriol. 182:5513–5520. 10.1128/JB.182.19.5513-5520.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egidius E, Andersen K, Clausen E, Raa J. 1981. Cold-water vibriosis or “Hitra disease” in Norwegian salmonid farming. J. Fish Dis. 4:353–354. 10.1111/j.1365-2761.1981.tb01143.x [DOI] [Google Scholar]
- 32.Holm KO, Strøm E, Stensvaag K, Raa J, Jørgensen T. 1985. Characteristics of a Vibrio sp. associated with the “Hitra disease” of Atlantic salmon in Norwegian fish farms. Fish Pathol. 20:125–129. 10.3147/jsfp.20.125 [DOI] [Google Scholar]
- 33.Egidius E, Wiik R, Andersen K, Hoff KA, Hjeltnes B. 1986. Vibrio salmonicida sp. nov., a new fish pathogen. Int. J. Syst. Bacteriol. 36:518–520. 10.1099/00207713-36-4-518 [DOI] [Google Scholar]
- 34.Jørgensen T. 1989. Microbiological and immunological aspects of “Hitra disease” of coldwater vibriosis (a summary), p 113–119 In Stenmark A, Malmberg G. (ed), Parasites and diseases in natural waters and aquaculture in Nordic countries. Naturhistoriska Riksmuseet, Stockholm, Sweden [Google Scholar]
- 35.Colquhoun DJ, Alvheim K, Dommarsnes K, Syvertsen C, Sørum H. 2002. Relevance of incubation temperature for Vibrio salmonicida vaccine production. J. Appl. Microbiol. 92:1087–1096. 10.1046/j.1365-2672.2002.01643.x [DOI] [PubMed] [Google Scholar]
- 36.Sørum H, Poppe TT, Olsvik O. 1988. Plasmids in Vibrio salmonicida isolated from salmonids with hemorrhagic syndrome (Hitra disease). J. Clin. Microbiol. 26:1679–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiik R, Andersen K, Daae FL, Hoff KA. 1989. Virulence studies based on plasmid profiles of the fish pathogen Vibrio salmonicida. Appl. Environ. Microbiol. 55:819–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sørum H, Hvaal AB, Heum M, Daae FL, Wiik R. 1990. Plasmid profiling of Vibrio salmonicida for epidemiological studies of cold-water vibriosis in Atlantic salmon (Salmo salar) and cod (Gadus morhua). Appl. Environ. Microbiol. 56:1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bjelland AM, Sørum H, Tegegne DA, Winther-Larsen HC, Willassen NP, Hansen H. 2012. LitR of Vibrio salmonicida is a salinity-sensitive quorum-sensing regulator of phenotypes involved in host interactions and virulence. Infect. Immun. 80:1681–1689. 10.1128/IAI.06038-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hjerde E, Lorentzen MS, Holden MT, Seeger K, Paulsen S, Bason N, Churcher C, Harris D, Norbertczak H, Quail MA, Sanders S, Thurston S, Parkhill J, Willassen NP, Thomson NR. 2008. The genome sequence of the fish pathogen Aliivibrio salmonicida strain LFI1238 shows extensive evidence of gene decay. BMC Genomics 9:616. 10.1186/1471-2164-9-616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat. Biotechnol. 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 42.Messing J, Crea R, Seeburg PH. 1981. A system for shotgun DNA sequencing. Nucleic Acids Res. 9:309–321. 10.1093/nar/9.2.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook JE, Fritsch F, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45.Barrios H, Valderrama B, Morett E. 1999. Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acids Res. 27:4305–4313. 10.1093/nar/27.22.4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim B, Beyhan S, Meir J, Yildiz FH. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60:331–348. 10.1111/j.1365-2958.2006.05106.x [DOI] [PubMed] [Google Scholar]
- 47.McDowell EM, Trump BF. 1976. Histologic fixatives suitable for diagnostic light and electron microscopy. Arch. Pathol. Lab. Med. 100:405–414 [PubMed] [Google Scholar]
- 48.Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212. 10.1083/jcb.17.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fredheim EG, Klingenberg C, Rohde H, Frankenberger S, Gaustad P, Flaegstad T, Sollid JE. 2009. Biofilm formation by Staphylococcus haemolyticus. J. Clin. Microbiol. 47:1172–1180. 10.1128/JCM.01891-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pedersen HL, Hjerde E, Paulsen SM, Hansen H, Olsen L, Thode SK, Dos Santos MT, Paulssen RH, Willassen NP, Haugen P. 2010. Global responses of Aliivibrio salmonicida to hydrogen peroxide as revealed by microarray analysis. Mar. Genomics 3:193–200. 10.1016/j.margen.2010.10.002 [DOI] [PubMed] [Google Scholar]
- 51.Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193. 10.1093/bioinformatics/19.2.185 [DOI] [PubMed] [Google Scholar]
- 52.Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- 53.Cao X, Studer SV, Wassarman K, Zhang Y, Ruby EG, Miyashiro T. 2012. The novel sigma factor-like regulator RpoQ controls luminescence, chitinase activity, and motility in Vibrio fischeri. mBio 3:e00285-11. 10.1128/mBio.00285-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Houot L, Chang S, Pickering BS, Absalon C, Watnick PI. 2010. The phosphoenolpyruvate phosphotransferase system regulates Vibrio cholerae biofilm formation through multiple independent pathways. J. Bacteriol. 192:3055–3067. 10.1128/JB.00213-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomich M, Planet PJ, Figurski DH. 2007. The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5:363–375. 10.1038/nrmicro1636 [DOI] [PubMed] [Google Scholar]
- 56.Shapiro RS, Cowen LE. 2012. Thermal control of microbial development and virulence: molecular mechanisms of microbial temperature sensing. mBio 3:e00238-12. 10.1128/mBio.00238-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colquhoun DJ, Sørum H. 2001. Temperature dependent siderophore production in Vibrio salmonicida. Microb. Pathog. 31:213–219. 10.1006/mpat.2001.0464 [DOI] [PubMed] [Google Scholar]
- 58.Bruhn JB, Dalsgaard I, Nielsen KF, Buchholtz C, Larsen JL, Gram L. 2005. Quorum sensing signal molecules (acylated homoserine lactones) in gram-negative fish pathogenic bacteria. Dis. Aquat. Organ. 65:43–52. 10.3354/dao065043 [DOI] [PubMed] [Google Scholar]
- 59.Purohit AA, Johansen JA, Hansen H, Leiros HK, Kashulin A, Karlsen C, Smalås A, Haugen P, Willassen NP. 2013. Presence of acyl-homoserine lactones in 57 members of the Vibrionaceae family. J. Appl. Microbiol. 115:835–847. 10.1111/jam.12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waters CM, Lu W, Rabinowitz JD, Bassler BL. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190:2527–2536. 10.1128/JB.01756-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shikuma NJ, Fong JC, Odell LS, Perchuk BS, Laub MT, Yildiz FH. 2009. Overexpression of VpsS, a hybrid sensor kinase, enhances biofilm formation in Vibrio cholerae. J. Bacteriol. 191:5147–5158. 10.1128/JB.00401-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868. 10.1126/science.1181185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dikow RB. 2011. Genome-level homology and phylogeny of Shewanella (Gammaproteobacteria: lteromonadales: Shewanellaceae). BMC Genomics 12:237. 10.1186/1471-2164-12-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leonardo MR, Moser DP, Barbieri E, Brantner CA, MacGregor BJ, Paster BJ, Stackebrandt E, Nealson KH. 1999. Shewanella pealeana sp. nov., a member of the microbial community associated with the accessory nidamental gland of the squid Loligo pealei. Int. J. Syst. Bacteriol. 49(Pt 4):1341–1351 [DOI] [PubMed] [Google Scholar]
- 65.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131–147. 10.1146/annurev.micro.60.080805.142106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Proft T, Baker EN. 2009. Pili in Gram-negative and Gram-positive bacteria: structure, assembly, and their role in disease. Cell. Mol. Life Sci. 66:613–635. 10.1007/s00018-008-8477-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066–2079. 10.1128/JB.185.7.2066-2079.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080–2095. 10.1128/JB.185.7.2080-2095.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olsen A, Jonsson A, Normark S. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652–655. 10.1038/338652a0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.