Abstract
Four new primers and one published primer were used to PCR amplify hypervariable regions within the protozoal 18S rRNA gene to determine which primer pair provided the best identification and statistical analysis. PCR amplicons of 394 to 498 bases were generated from three primer sets, sequenced using Roche 454 pyrosequencing with Titanium, and analyzed using the BLAST database (NCBI) and MOTHUR version 1.29. The protozoal diversity of rumen contents from moose in Alaska was assessed. In the present study, primer set 1, P-SSU-316F and GIC758R (amplicon of 482 bases), gave the best representation of diversity using BLAST classification, and the set amplified Entodinium simplex and Ostracodinium spp., which were not amplified by the other two primer sets. Primer set 2, GIC1080F and GIC1578R (amplicon of 498 bases), had similar BLAST results and a slightly higher percentage of sequences that were identified with a higher sequence identity. Primer sets 1 and 2 are recommended for use in ruminants. However, primer set 1 may be inadequate to determine protozoal diversity in nonruminants. The amplicons created by primer set 1 were indistinguishable for certain species within the genera Bandia, Blepharocorys, Polycosta, and Tetratoxum and between Hemiprorodon gymnoprosthium and Prorodonopsis coli, none of which are normally found in the rumen.
INTRODUCTION
Rumen ciliate protozoa represent important functional members of the rumen environment, as most have some cellulolytic or amylolytic abilities (1–3). Most studies of rumen ciliate protozoa are performed using microscopy and traditional culturing techniques (2, 4–10), quantitative PCR (11, 12), denaturing gradient gel electrophoresis (13), and full-length 18S rRNA clone libraries (13, 14). A few studies of rumen ciliate protozoa use high-throughput sequencing, although primer selection remains a problem, as some studies use universal eukaryotic primers, primers which target only one ciliate protozoon signature region, or primers which produce long amplicons that are unsuitable for current high-throughput technology (15–20).
18S rRNA genes range from 1.5 kb to more than 4.5 kb (21), and in rumen ciliate protozoa, they are generally 1.5 kb to 1.8 kb in length. Like the 16S rRNA genes of prokaryotes, the 18S rRNA genes of eukaryotes have nine hypervariable regions (V1 to V9) which can be used for genus/species identification. Four gut ciliate signature regions exist within rumen protozoal 18S rRNA genes which represent areas of high variability that can improve identification down to the species level (22, 23). Signature region 1 occurs between bp 440 and 460 (within V3), signature region 2 occurs between bp 590 and 620 (between V3 and V4), signature region 3 occurs between bp 1220 and 1260 (within V6), and signature region 4 occurs between bp 1560 and 1580 (after V8) (Fig. 1). Additionally, rumen ciliate protozoa have a slightly different 18S rRNA secondary structure from nonrumen ciliates, in that rumen protozoa are missing helix E23-5 from the V4 region and other helices in the region are shorter (21–23). Previously, the V9 region (15) or the V5-to-V7 regions (13) have been amplified for phylogenetic analysis using high-throughput techniques. Ciliated protozoa found in the gastrointestinal tract of animals belong to phylum Ciliophora, class Litostomatea, subclass Trichostomatia, and orders Entodiniomorphida and Vestibuliferida. The vast majority of rumen ciliated protozoa belong to the Ophryoscolecidae, the largest family (both in numbers of species and genera) within the Entodiniomorphida.
FIG 1.
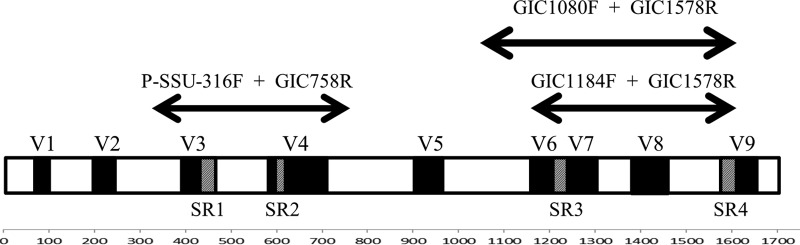
A map of the full-length protozoal 18S rRNA gene, including variable (V1 to V9) and rumen ciliate signature regions (SR1 to SR4) and showing the respective amplicons of the three primer sets used in the present study.
In the present study, four new primers were designed to specifically target conserved 18S rRNA gene regions for ciliate protozoa, which are normally found in the gastrointestinal tract of herbivores. Using our protocol, these primers did not amplify other eukaryotic, bacterial, or archaeal species or nonciliated protozoa, which are not normally found in a healthy gastrointestinal tract environment. The strategy for the pairing of the forward and reverse primers was to create amplicons that included at least one of the four signature regions of the rumen ciliate protozoal 18S rRNA genes. Current limitations of the Roche 454 and MiSeq version 3.0 (with 2 × 300 paired end reads) platforms, along with few reliable conserved regions exclusive to ciliate protozoa, prevent the inclusion of all four signature regions, which was possible when the full 18S rRNA genes were sequenced using Sanger sequencing technology.
The first objective of the present study was to test these primer pairs on rumen samples from the North American moose (Alces alces) to determine their suitability and validity for rumen ciliate identification. The second objective was to compare the primer sets using the CHAO, ACE, Shannon-Weaver, inverse Simpson, Good's coverage, and UniFrac methods in order to make a recommendation of the most suitable primer set for rumen protozoal 18S rRNA gene amplification.
MATERIALS AND METHODS
Sample collection and DNA extraction.
On 31 August 2012, whole rumen contents were collected via esophageal tubing from three captive, free-range wild moose at the Moose Research Center, Soldotna, AK (IACUC protocol 11-021, University of Vermont, and ACUC protocol 2011-026, Alaska Department of Fish and Game). All three moose were females between 10 and 11 years of age. Rumen samples were mixed with 70% ethanol and shipped to the University of Vermont (Burlington, VT, USA), where they were stored at 4°C. To confirm the sequencing results, all three moose samples were inspected visually using light microscopy to identify the genera of rumen ciliates.
To extract DNA, a 5-ml aliquot of whole rumen contents in ethanol was centrifuged for 5 min at 16,000 × g, and ethanol was removed by pouring off the liquid fraction. From the remaining whole contents of all the samples, 0.25-g aliquots of whole contents (liquid and associated particles) were used for extraction. DNA was extracted from the three rumen samples using the repeated bead-beating plus column (RBB+C) method (24) combined with the QIAamp DNA stool minikit (Qiagen, Maryland). The final elutions were made using 200 μl of TE buffer (1 M Tris-HCl, 0.5 M EDTA, pH 8.0), and eluted DNA was quantified using a NanoDrop 2000C spectrophotometer (Thermo Scientific).
Primer design.
The new forward and reverse primers were designed to target signature regions unique to gastrointestinal tract ciliate protozoa within the 18S rRNA gene. Protozoal 18S rRNA gene reference alignments were created and used to select areas which were highly conserved among the rumen ciliate protozoa. Four conserved regions were selected, and a potential primer sequence identified from each of those regions. The four new primers were given the prefix “GIC” for gastrointestinal ciliates and are listed in Table 1 with specifications. Along with a previously described rumen protozoal primer, P-SSU-316F (5′-GCTTTCGWTGGTAGTGTATT-3′) (12), primer sequences were compared to known sequences of gastrointestinal ciliates in GenBank (NCBI) (Table 2) to determine their specificity to amplify only gastrointestinal tract ciliate protozoa.
TABLE 1.
Gastrointestinal tract ciliate protozoal primer specifications
| Primer | Sequence (5′→3′)a | Tmb (°C) | Size (bp) of: |
End stability | |
|---|---|---|---|---|---|
| Dimer formation | Hairpin formation | ||||
| GIC758R | CAACTGTCTCTATKAAYCG | 47.4 | 2 | 2 | Medium |
| GIC1080F | GGGRAACTTACCAGGTCC | 53.6 | 3 | 3 | Medium |
| GIC1184F | TGTCTGGTTAATTCCGA | 47.2 | 4 | 3 | High |
| GIC1578R | GTGATRWGRTTTACTTRT | 42.1 | 2 | 2 | High |
Residues in boldface are as follows: K = G/T; Y = C/T; R = A/G; W = A/T.
Tm, melting temperature, determined at 50 mM NaCl.
TABLE 2.
18S rRNA protozoal reference sequences used to determine genetic distance cutoff

Primer set 1, P-SSU-316F (12) and GIC758R, created an amplicon of 482 bases (primers not included) that encompassed variable regions V3 and V4 and rumen ciliate signature regions 1 and 2 (Fig. 1). Primer set 2, GIC1080F and GIC1578R, created an amplicon of 498 bases that encompassed V6 to V8 and signature regions 3 and 4 (Fig. 1). Primer set 3, GIC1184F and GIC1578R, created an amplicon of 394 bases that also encompassed V6 to V8, along with rumen ciliate signature regions 3 and 4 (Fig. 1).
PCR amplification.
The Phusion high-fidelity DNA polymerase kit (Thermo Scientific) was used for PCR. The reaction mixtures contained 10 μl of 5× high-fidelity buffer (including MgCl2), 1.0 μl of 10 mM deoxynucleoside triphosphate mix, 2.5 μl each of forward and reverse primer at 10 mM concentration, 0.5 μl of Phusion DNA polymerase (2U/μl), and 31.5 μl of double-distilled water. DNA templates (2 μl of 10 to 50 ng/μl concentration) were added once the master mix had been aliquoted, to a total reaction volume of 50 μl. The PCR protocol was adapted from a previously published protocol (12) and was 94°C for a 4-min hot start, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, with a final extension of 72°C for 6 min on the last cycle. All PCR products were run on a 1% agarose gel at 100 V for 60 min and imaged on a ChemiDoc XRS+ gel imager (Bio-Rad, CA).
DNA bands of the correct amplicon size were excised out of the agarose gel for DNA purification, using the QIAquick gel extraction kit (Qiagen, MD) according to the manufacturer's instructions. For each primer set, all gel bands from each of the three moose samples were filtered through the same column. The gel-extracted DNA from each primer set was quantified using a NanoDrop 2000C spectrophotometer (Thermo Scientific) at a minimum final concentration of 20 ng/μl in a volume of 20 μl. The DNA amplicons from the three test primer sets were frozen and shipped overnight to Molecular Research LP (MR DNA), Shallowater, TX, USA, for Roche 454 pyrosequencing with Titanium chemistry.
Sequence analysis.
Sequences were deposited online in the NCBI Sequence Read Archive (SRA) under study accession number SRP034591. To analyze the DNA sequencing data, the open-source computer software program MOTHUR version 1.29 (25) was used. Sequences from all three primer sets were processed independently using the original standard flowgram format (sff) output file from the sequencer. Noise was removed from flow grams using the MOTHUR-integrated version of the PyroNoise algorithm (26), which also creates phylotypes, which are unique sequences representing multiple identical sequences. Unique sequences are not equivalent to singletons, which are operational taxonomic units (OTUs) containing a single sequence. The barcode and primer sequences were removed, and sequences that contained any of the following conditions were discarded: <400 bases for primer set 1 and 2 or <375 bases for primer set 3, >500 bases, homopolymer runs of >8 bases, or any mismatches in the barcode.
To determine the genetic distance cutoffs for species-level comparisons, 51 valid full-length 18S rRNA protozoal reference sequences were obtained from NCBI (Table 2; see also Fig. S4 in the supplemental material). These reference sequences were then trimmed using the three test primer sets as trim points. BLAST sequences were manually aligned, and pairwise distances were calculated with PHYLIP (version 3.69), using a Kimura 2-parameter model (Table 3). A total of 51 pairwise species (within a genus) and 2,926 pairwise distances between genera were compared using validly recognized species to determine genetic distances.
TABLE 3.
Comparison of statistical parameters and results for the three primer sets
| Parameter | P-SSU-316F and GIC758R | GIC1080F and GIC1578R | GIC1184F and GIC1578R |
|---|---|---|---|
| Total no. of sequences before quality assurance steps | 19,886 | 10,143 | 8,611 |
| Total no. of sequences after quality assurance steps | 12,326 | 6,070 | 8,265 |
| No. of unique sequences used for BLAST | 769 | 697 | 424 |
| Trimmed sequence length (bases) | 450 | 450 | 375 |
| No. of subsampled unique sequences used for statistical analysis | 424 | 424 | 424 |
| Species-level distance within generaa | 0.036 | 0.039 | 0.042 |
| Recommended % cutoff for species-level OTUs | 4 | 4 | 4 |
| Observed species-level OTUs | 48 | 25 | 20 |
| CHAO | 112 | 58 | 42 |
| ACE | 226 | 92 | 145 |
| Shannon-Weaver | 2.36 | 1.02 | 1.37 |
| Good's coverage | 0.94 | 0. 96 | 0.97 |
| Inverse Simpson | 4.69 | 1.71 | 2.59 |
| Genus-level distance across generab | 0.087 | 0.079 | 0.096 |
| Recommended % cutoff for genus-level OTUs | 9 | 8 | 10 |
| Observed genus-level OTUs | 15 | 14 | 12 |
Using nearly full-length 18S rRNA gene sequences from valid species, the species-level cutoff was calculated to be 0.031.
Using nearly full-length 18S rRNA gene sequences, the genus-level cutoff was calculated to be 0.071.
Sequences were aligned using the Needleman-Wunsch global alignment algorithm (27), 8-base kmer searching, match reward of +1, mismatch penalty of −1, and gap open/extend penalty of −2. An 18S rRNA gene reference alignment, featuring rumen and nonrumen ciliate protozoal sequences downloaded from NCBI, was created in the laboratory to provide a better alignment of candidate sequences. The reference alignment contained 219 full-length 18S rRNA sequences for all available gastrointestinal tract (rumen, forestomach, cecum, and colon) ciliate sequences, as well as nonruminant ciliates and nonciliate protozoal sequences from BLAST. The reference alignment contained all 51 sequences previously used to determine genetic distance cutoffs. The candidate alignment was then filtered to remove gap-only columns and any sequences which would not align (<10 sequences per data set). The candidate alignment was checked for chimeras with the MOTHUR-integrated version of the program UCHIME (28), using the ciliate 18S rRNA gene sequence reference alignment which was created in our laboratory. To determine the specificity of the primers, unique sequences were classified using BLAST.
Sequences from the three primer sets were trimmed to a uniform length per library (Table 3) and clustered using the nearest-neighbor method to determine the number of OTUs observed per library. Libraries were then subsampled equal to the smallest library (n = 424 sequences per library), and Shannon-Weaver diversity index (29), Good's coverage (30), inverse Simpson (31), CHAO (32), and ACE (33) values were calculated for each library based on the recommended genetic distance. Like Simpson's diversity, inverse Simpson measures the number and abundance of species. However, it weights rare species lower than Simpson's diversity does, to prevent a dramatic increase in diversity with the addition of rare species. Additionally, using MOTHUR, relaxed neighbor-joining trees were created from trimmed sequences (375 bases) using CLEARCUT, and trees were clustered using weighted and unweighted UniFrac as a measure of similarity of abundance and structure between libraries.
RESULTS
Primer set 1, P-SSU-316F and GIC758R.
A total of 12,326 sequences passed quality assurance measures and were used for sequence analysis with primer set 1, P-SSU-316F and GIC758R. Of these, 769 sequences were unique (Table 3). Using aligned sequences of valid protozoa to generate pairwise genetic distances (see Fig. S1 in the supplemental material), the species- and genus-level cutoffs were determined to be 0.036 and 0.087, respectively. So, a 4% species-level cutoff and a 9% genus-level cutoff were comparable to cutoffs for near full-length gene sequences. Sequences were trimmed to 450 bases, and various diversity indices calculated for the data set. At a 4% species-level cutoff, 48 species-level OTUs were observed, and at a 9% genus-level cutoff, 15 genus-level OTUs were observed (Table 3). ACE, CHAO, Shannon-Weaver, and inverse Simpson values can be found in Table 3.
Using BLAST, the most prevalent taxon was Polyplastron multivesiculatum, which represented nearly 60% of the unique sequences, followed by the genus Entodinium, which represented just over 20% of the unique sequences (Fig. 2). The most prevalent species of Entodinium were E. furca dilobum (7% of the unique sequences) and E. nanellum (5% of the unique sequences). Epidinium caudatum represented 5% of the unique sequences (Fig. 2). Primer set 1 amplified Entodinium simplex, Ostracodinium gracile, and other Ostracodinium spp., which were not amplified by the other primer sets. The percent identity to known sequences ranged from 95 to 100% for primer set 1, with 66% of sequences having a 99% identity to a known sequence in BLAST (Fig. 3). However, there were six sequences that had a 99 to 100% identity on only 93 to 95% of the query sequence. The average percent identity to Ostracodinium gracile was 98.2% (range 97 to 99%). There were 21 unique (63 total) sequences which had <96% identity to a known sequence.
FIG 2.
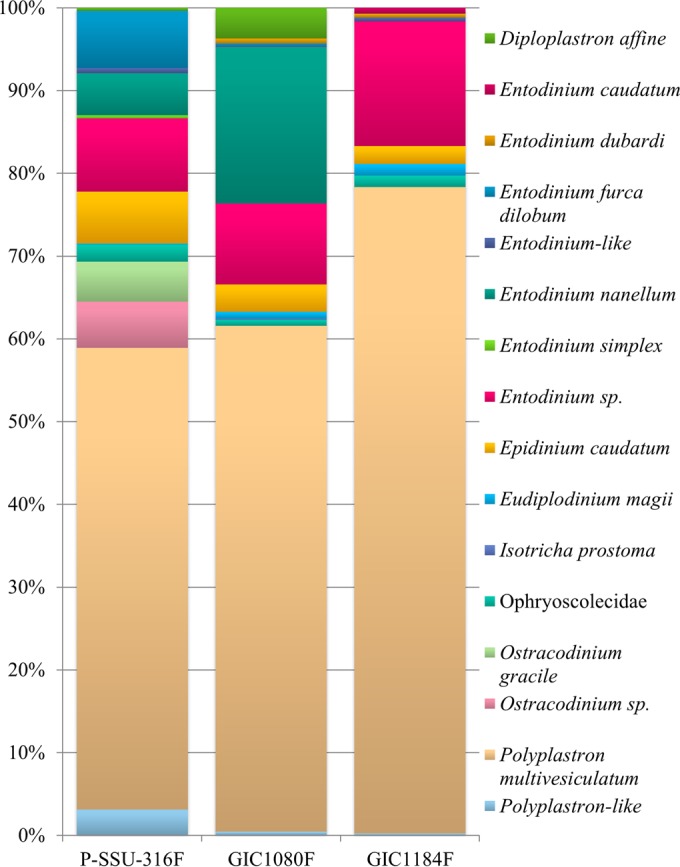
Taxonomy and proportions of unique pyrosequences by NCBI BLAST using forward primers P-SSU-316F (12), GIC1080F (present study), and GIC1184F (present study). All sequences used passed all quality assurance steps outlined in Materials and Methods.
FIG 3.
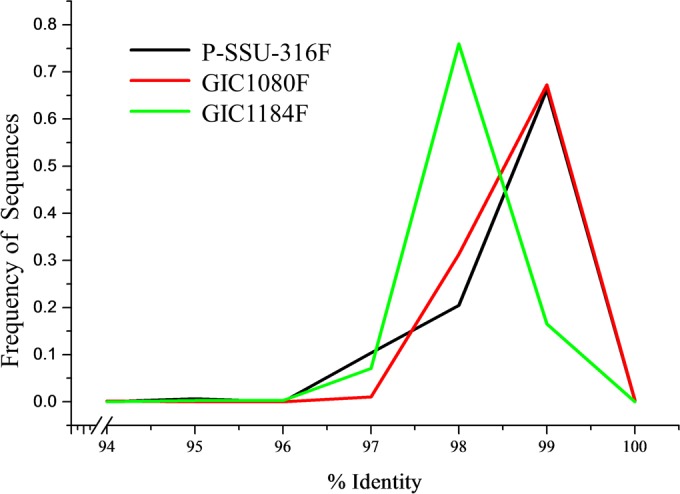
Distribution of percentages of sequence identity to known sequences, based on BLAST, for the sequencing primers.
During the calculation of genetic distances using pairwise comparisons, it was noted that the amplicon created by primer set 1 could not differentiate between Blepharocorys microcorys (AB794975) and Blepharocorys uncinata (AB530162), between Hemiprorodon gymnoprosthium (AB795028) and Prorodonopsis coli (AB795029), between Tetratoxum excavatum (AB794971) and Tetratoxum parvum (AB794969), between Bandia deveneyi (AY380823) and Bandia smalesae (AF298822), and between Polycosta roundi (AF298819) and Polycosta turniae (AF298818).
Primer set 2, GIC1080F and GIC1578R.
A total of 6,070 sequences passed quality assurance measures and were used for sequence analysis with primer set 2, GIC1080F and GIC1578R. Of these, 697 sequences were unique (Table 3). Using aligned sequences of valid protozoa to generate pairwise genetic distances (see Fig. S2 in the supplemental material), the species and genus-level cutoffs were determined to be 0.039 and 0.079, respectively. For statistical analysis, sequences were trimmed to a uniform length of 450 bases. At a 4% species-level genetic distance cutoff, 25 species-level OTUs were observed, and at an 8% genus-level cutoff, 14 genus-level OTUs were observed (Table 3). ACE, CHAO, Shannon-Weaver, and inverse Simpson values can be found in Table 3.
Over 60% of sequences from this primer set represented Polyplastron multivesiculatum (Fig. 2). The next most predominant genus was Entodinium, with 30% of the unique sequences, and of those, Entodinium nanellum represented approximately two-thirds of the genus (20% of the unique sequences) (Fig. 2). Diploplastron affine and Epidinium caudatum were the third most prevalent taxa, with 5% of the unique sequences each. The percent identity to known sequences ranged from 94 to 100% for primer set 2, with 67% of sequences having a 99% identity to a known sequence in BLAST (Fig. 3). Only 1 unique (9 total) sequence had <96% identity to known sequences.
Primer set 3.
A total of 8,265 total sequences passed quality assurance measures and were used for sequence analysis with primer set 3, GIC1184F and GIC1578R. Of these, 424 sequences were unique and used for BLAST (Table 3). Using aligned valid protozoal sequences to generate pairwise genetic distances (see Fig. S3 in the supplemental material), the species and genus-level cutoffs were determined to be 0.042 and 0.096, respectively. For statistical analysis, sequences were trimmed to a uniform length of 375 bases. At a 4% species-level genetic distance cutoff, 20 species-level OTUs were observed, and at 10% genus-level cutoff, 12 genus-level OTUs were observed (Table 3). ACE, CHAO, Shannon-Weaver, and inverse Simpson values can be found in Table 3.
Over 75% of the unique sequences in the primer set 3 data set were classified as Polyplastron multivesiculatum using BLAST (Fig. 2). The next most predominant genus was Entodinium, representing approximately 15% of the unique sequences (Fig. 2). Epidinium was the third most prevalent genus, with <5% of the unique sequences. The percent identity to known sequences ranged from 95 to 99% for primer set 3, with 76% of sequences having a 98% identity to a known sequence in GenBank (Fig. 3). There were 2 unique sequences, which had <96% identity to a publically available sequence.
Comparison of the three primer sets.
Primer set 1 (P-SSU-316F and GIC758R) had the highest number of observed OTUs, as well as the highest ACE, CHAO, Shannon-Weaver and inverse Simpson values of all three primer sets, indicating the largest amount of diversity of the three amplicon libraries (Table 3). Primer set 1 had the lowest Good's coverage (0.95). Primer set 2 (GIC1080F and GIC1578R) had the second largest number of observed OTUs and the second largest CHAO index. However, primer set 2 had the lowest ACE, Shannon-Weaver, and inverse Simpson values, indicating a small amount of diversity (Table 3). Primer set 3 had the lowest number of observed OTUs, as well as the lowest CHAO estimate (Table 3). However, primer set 3 had the second highest ACE, Shannon-Weaver, and inverse Simpson values (Table 3). Weighted and unweighted UniFrac analyses were also run, using MOTHUR. The primer sets were not significantly different based on weighted (0.96 to 1.0) or unweighted (0.98 to 1.0) UniFrac (P < 0.001). There was no correlation between sequence length and percent identity to known sequences in BLAST for any of the three data sets.
The genera of rumen ciliates were confirmed using light microscopy. Various species of Entodinium, as well as Polyplastron multivesiculatum and Epidinium cattanei, were found in abundance in all three samples. Isotricha were found in two samples, and Ostracodinium was found in one sample.
DISCUSSION
This study validated three primer sets for the amplification of gastrointestinal tract ciliate protozoa for use in high-throughput sequencing, as well as determining the diversity of rumen protozoa of moose from Alaska, USA. All three test primer sets were able to amplify 18S rRNA protozoal sequences. 18S rRNA genes are highly conserved among eukaryotes, and finding potential primer sites that are specific to certain taxa can be challenging. Using the new primers reported in the present study under the same amplification parameters (i.e., PCR annealing temperature and removal of short amplicons), only 18S rRNA genes from gastrointestinal tract (rumen, forestomach, cecum, and colon) ciliate protozoa should be targeted for amplification.
Previously used primers for high-throughput sequencing were often universal eukaryotic primers (15–19) or primers which targeted only one signature region for the ciliate protozoa (18, 20). In several studies, this resulted in sequences which were not ciliate or protozoan in nature (17, 19) or which were too short (<200 bases), thereby increasing the risk for misidentification (18) given the overall high degree of conservation of 18S genes across eukaryotic taxa. In the present study, no nonprotozoal eukaryotic sequences (i.e., plant, fungal, or host DNA) were amplified.
Previously, using classical microbiology, Dehority (6) identified just five species of protozoa from the rumen of Alaskan moose, Sládeček (4) identified four species from moose, and Westerling (34) identified just two species from moose in Finland. In the present study, 12 species representing 7 genera were found across the three Alaskan moose from which samples were obtained. Previously, between 16 and 24 rumen ciliate species, represented by 1 to 9 genera, were found in in studies of wild reindeer (9, 35–39), wild musk ox (Ovibos moschatus) (6), wildebeest (Connochaetes spp.) (40), Kafue lechwe antelope (Kobus leche kafuensis) (41), Sassaby antelope (Damaliscus lunatus) (10), and tsessebe antelope (Damaliscus lunatus lunatus) (42).
Only one study exists which investigated the rumen protozoa from three bull moose in Alaska, USA, using culturing and microscopy techniques (6). Previously, Entodinium alces and Entodinium exiguum were reported to be the two dominant species in moose rumen contents, whereas Entodinium dubardi, Entodinium simplex, and Entodinium longinucleatum were present in one moose (6). Additionally, Entodinium dubardi and Epidinium caudatum were identified in moose rumen samples from Finnish Lapland (34) and Entodinium dubardi, Entodinium simplex, Ostracodinium obtusum, and Epidinium ecaudatum were isolated from moose in Slovakia (4). Eudiplodinium neglectum was also first identified in a moose from Canada (5).
Based on the previous studies that identified the rumen ciliate protozoa in the moose, it was surprising that Polyplastron species are the dominant species in the present study. Polyplastron produce xylanases, carboxymethylcellulases, and various other endoglucanases which digest fiber in the rumen. While the presence of Polyplastron was validated using light microscopy in the present study, a large number of sequences related to Polyplastron may be explained by the rRNA copy numbers in ciliates, which could be highly variable from one species to another. Also, there is a lack of publically available sequences for the rumen ciliates, especially for genera closely related to Polyplastron, such as Elytroplastron and Eudiplodinium. This is also true of Entodinium alces, Entodinium exiguum, Ostracodinium obtusum, Epidinium ecaudatum, and Eudiplodinium neglectum, all of which were previously found in moose (4–6) but for none of which do representative sequences exist. This makes identification at a species level very difficult until additional sequences from all described species are elucidated.
There were 74 total sequences (24 unique sequences) that had <96% identity to publically available sequences. Given that the genetic distance between Epidinium caudatum and Epidinium ecaudatum is 1.3%, some sequences in the present analysis with genetic distances of between 1.0 and 1.5% from E. caudatum may represent other species of Epidinium, such as Epidinium cattenei, which was observed under light microscopy. Similarly, given that Polyplastron multivesiculatum has 98% sequence identity to Ostracodinium gracile and Ostracodinium clipoleum, the large number of sequences that have <98% identity to P. multivesiculatum may in fact represent other closely related species or genera. As we stated previously, more representative sequences from other rumen ciliates, such as E. cattenei, are needed to confirm these interpretations of the data.
Based on the analysis in the present study, the primer set which gave the best representation of diversity using BLAST was primer set 1, P-SSU-316F and GIC758R. Primer set 1 amplified Entodinium simplex and Ostracodinium spp., which were not amplified by either of the other two primer sets. However, primer set 2 had a slightly higher percentage of sequences classified at a higher percent identity in BLAST than primer set 1. Both primer sets produced an amplicon of more than 400 bases. Primer set 1 (P-SSU-316F and GIC758R) had the highest number of observed OTUs, as well as the highest ACE, CHAO, Shannon-Weaver, and inverse Simpson values of all three primer sets, indicating the largest amount of diversity of the three amplicon libraries. Primer set 3 had the second highest ACE, Shannon-Weaver, and inverse Simpson values. While primer sets 2 and 3 target the same variable and signature regions, primer set 2 spans a larger area of the conserved region, which may account for the lower estimated diversity than was obtained with primer set 3.
It is important to note that primer set 1 (P-SSU-316F and GIC758R) produced amplicons which could not differentiate between the pairs Blepharocorys microcorys and Blepharocorys uncinata, Hemiprorodon gymnoprosthium and Prorodonopsis coli, Tetratoxum excavatum and Tetratoxum parvum, Bandia deveneyi and Bandia smalesae, and Polycosta roundi and Polycosta turniae. The aforementioned Blepharocorys spp. (S. Imai, A. Ito, Y. Miyazaki, M. Ishihara, and K. Nataami, 2009, unpublished data), Hemiprorodon gymnoprosthium (43), Prorodonopsis coli (44), and Tetratoxum spp. (45) have previously been found in the hind gut of the horse, and Bandia deveneyi, B. smalesae, Polycosta roundi, and P. turniae in Australian marsupials (46). Since these ciliate species mainly occur in nonruminants, either P-SSU-316F and GIC758R or GIC1080F and GIC1578R could be used in ruminants, but only GIC1080F and GIC1578R should be used in nonruminants. However, in order to bring about standardization of the amplification of rumen ciliates and to better enable comparison across studies, GIC1080F and GIC1578R seem to be the better choice for a general gut ciliate primer set.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge John Crouse and Kimberlee Beckmen of the Alaskan Department of Fish and Game for their assistance in rumen sample collection.
Footnotes
Published ahead of print 27 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01644-14.
REFERENCES
- 1.Gijzen HJ, Lubberding HJ, Gerhardus MJT, Vogels GD. 1988. Contribution of rumen protozoa to fibre degradation and cellulase activity in vitro. FEMS Microbiol. Lett. 53:35–43. 10.1111/j.1574-6968.1988.tb02645.x [DOI] [Google Scholar]
- 2.Varel VH, Dehority BA. 1989. Ruminal cellulolytic bacteria and protozoa from bison, cattle-bison hybrids, and cattle fed three alfalfa-corn diets. Appl. Environ. Microbiol. 55:148–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams AG, Withers SE. 1993. Changes in the rumen microbial population and its activities during the refaunation period after the reintroduction of ciliate protozoa into the rumen of defaunated sheep. Can. J. Microbiol. 39:61–69. 10.1139/m93-009 [DOI] [PubMed] [Google Scholar]
- 4.Sládeček F. 1946. Ophryoscolecidae from the stomach of Cervus elaphus L., Dama dama L., and Capreolus capreolus L. Vestn. Cesk. Zool. Spol. 10:201–231 (In Czech) [Google Scholar]
- 5.Krascheninnikow S. 1955. Observations on the morphology and division of Eudiplodinium neglectum Dogiel (Ciliata Entodiniomorpha) from the stomach of a moose (Alces americana). J. Protozool. 2:124–134. 10.1111/j.1550-7408.1955.tb02412.x [DOI] [Google Scholar]
- 6.Dehority BA. 1974. Rumen ciliate fauna of Alaskan moose (Alces americana), musk-ox (Ovibos moschatus) and Dall mountain sheep (Ovis dalli). J. Eukaryot. Microbiol. 21:26–32 [DOI] [PubMed] [Google Scholar]
- 7.Coleman GS, Laurie JI, Bailey JE, Holdgate SA. 1976. The cultivation of cellulolytic protozoa isolated from the rumen. J. Gen. Microbiol. 95:144–150. 10.1099/00221287-95-1-144 [DOI] [PubMed] [Google Scholar]
- 8.Krumholz LR, Forsberg CW, Veira DM. 1983. Association of methanogenic bacteria with rumen protozoa. Can. J. Microbiol. 29:676–680. 10.1139/m83-110 [DOI] [PubMed] [Google Scholar]
- 9.Imai S, Oku Y, Morita T, Ike K, Guirong 2004. Rumen ciliate protozoal fauna of reindeer in Inner Mongolia, China. J. Vet. Med. Sci. 66:209–212. 10.1292/jvms.66.209 [DOI] [PubMed] [Google Scholar]
- 10.Ito A, Arai N, Tsutsumi Y, Imai S. 2007. Ciliate protozoa in the rumen of Sassaby antelope, Damaliscus lunatus lunatus, including the description of a new species and form. J. Eukaryot. Microbiol. 44:586–591. 10.1111/j.1550-7408.1997.tb05964.x [DOI] [PubMed] [Google Scholar]
- 11.Karnati SKR, Yu Z, Sylvester JT, Dehority BA, Morrison M, Firkins JL. 2003. Specific PCR amplification of protozoal 18S rDNA sequences from DNA extracted from ruminal samples of cows. J. Anim. Sci. 81:812–815 [DOI] [PubMed] [Google Scholar]
- 12.Sylvester JT, Karnati SKR, Yu Z, Morrison M, Firkins JL. 2004. Development of an assay to quantify rumen ciliate protozoal biomass in cows using real-time PCR. J. Nutr. 134:3378–3384 [DOI] [PubMed] [Google Scholar]
- 13.Kittelmann S, Janssen PH. 2011. Characterization of rumen ciliate community composition in domestic sheep, deer, and cattle, feeding on varying diets, by means of PCR-DGGE and clone libraries. FEMS Microbiol. Ecol. 75:468–481. 10.1111/j.1574-6941.2010.01022.x [DOI] [PubMed] [Google Scholar]
- 14.Shin EC, Cho KM, Lim WJ, Hong SY, An CL, Kim EJ, Kim YK, Choi BR, An JM, Kang JM, Kim H, Yun HD. 2004. Phylogenetic analysis of protozoa in the rumen contents of cow based on the 18S rDNA sequences. J. Appl. Microbiol. 97:378–383. 10.1111/j.1365-2672.2004.02304.x [DOI] [PubMed] [Google Scholar]
- 15.Moon-van der Staay SY, De Wachter R, Vaulot D. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607–610. 10.1038/35054541 [DOI] [PubMed] [Google Scholar]
- 16.Lara E, Berney C, Harms H, Chatzinotas A. 2007. Cultivation-independent analysis reveals a shift in ciliate 18S rRNA gene diversity in a polycyclic aromatic hydrocarbon-polluted soil. FEMS Microbiol. Ecol. 62:365–373. 10.1111/j.1574-6941.2007.00387.x [DOI] [PubMed] [Google Scholar]
- 17.Dopheide A, Lear G, Stott R, Lewis G. 2008. Molecular characterization of ciliate diversity in stream biofilms. Appl. Environ. Microbiol. 74:1740–1747. 10.1128/AEM.01438-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. 2009. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One 4:e6372. 10.1371/journal.pone.0006372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brulc JM, Antonopoulos DA, Miller MEB, Wilson MK, Yannarell AC, Dinsdale EA, Edwards RE, Frank ED, Emerson JB, Wacklin P, Coutinho PM, Henrissat B, Nelson KE, White BA. 2009. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Natl. Acad. Sci. U. S. A. 106:1948–1953. 10.1073/pnas.0806191105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kittelmann S, Seedorf H, Walters WA, Clemente JC, Knight R. 2013. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS One 8:e47879. 10.1371/journal.pone.0047879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Q, Lin J, Qin Y, Zhou J, Bu W. 2011. Structural diversity of eukaryotic 18S rRNA and its impact on alignment and phylogenetic reconstruction. Protein Cell 2:161–170. 10.1007/s13238-011-1017-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright A-DG, Dehority BA, Lynn DH. 1997. Phylogeny of the rumen ciliates Entodinium, Epidinium and Polyplastron (Litostomatea: Entodiniomorphida) inferred from small subunit ribosomal RNA sequences. J. Eukaryot. 44:61–67. 10.1111/j.1550-7408.1997.tb05693.x [DOI] [PubMed] [Google Scholar]
- 23.Wright A-DG, Lynn DH. 1997. Maximum ages of ciliate lineages estimated using a small subunit rRNA molecular clock: crown eukaryotes that date back to the Paleoproterozoic. Arch. Protist. 148:329–341. 10.1016/S0003-9365(97)80013-9 [DOI] [Google Scholar]
- 24.Yu Z, Morrison M. 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36:808–812 [DOI] [PubMed] [Google Scholar]
- 25.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods 6:639–641. 10.1038/nmeth.1361 [DOI] [PubMed] [Google Scholar]
- 27.Needleman S, Wunsch C. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443–453. 10.1016/0022-2836(70)90057-4 [DOI] [PubMed] [Google Scholar]
- 28.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon CE, Weaver W. 1949. The mathematical theory of communication. University of Illinois Press, Urbana, IL [Google Scholar]
- 30.Good IJ. 1953. On population frequencies of species and the estimation of population parameters. Biometrika 40:237–264. 10.2307/2333344 [DOI] [Google Scholar]
- 31.Jost L. 2006. Entropy and diversity. Oikos 113:363. 10.1111/j.2006.0030-1299.14714.x [DOI] [Google Scholar]
- 32.Chao A, Shen T-J. 2003. Nonparametric estimation of Shannon's index of diversity when there are unseen species in sample. Environ. Ecol. Stat. 10:429–443. 10.1023/A:1026096204727 [DOI] [Google Scholar]
- 33.Chao A, Shen T-J. 2010. User's guide for program SPADE (Species Prediction And Diversity Estimation). http://chao.stat.nthu.edu.tw
- 34.Westerling B. 1969. The rumen ciliate fauna of cattle and sheep in Finnish Lapland, with special reference to the species regarded as specific to reindeer. Nord Vet. Med. 21:14–19 [Google Scholar]
- 35.Dogiel VA. 1925. Neue parasitische Infusorien aus dem Magen des Renntieres (Rangifer tarandus). Arch. Rus. Protistol. 4:43–65 [Google Scholar]
- 36.Dogiel VA. 1927. Monographie der Familie Ophryoscolecidae. Archiv fur Protistenkunde, vol 59 Fischer, Jena, Germany [Google Scholar]
- 37.Lubinsky G. 1957. Ophryoscolecidae (ciliata: entodiniomorphida) of the reindeer (Rangifer tarandus l.) from the Canadian arctic. ii. Diplodiniinae. Can. J. Zool. 36:927–959 [Google Scholar]
- 38.Westerling B. 1970. Rumen ciliate fauna of semi-domestic reindeer (Rangifer tarandus t.) in Finland: composition, volume and some seasonal variations. Acta Zool. Fenn. 127:1–76 [Google Scholar]
- 39.Dehority BA. 1975. Characterization studies on rumen bacteria isolated from Alaskan reindeer (Rangifer rarandus L.), p 228–240 In Luick JR. (ed), Proc. 1st Int. Reindeer Caribou Symp. Institute of Arctic Biology, Fairbanks, AK [Google Scholar]
- 40.Booyse DG, Dehority BA. 2012. Protozoa and digestive tract parameters in blue wildebeest (Connochaetes taurinus) and black wildebeest (Connochaetes gnou), with description of Entodinium taurinus n. sp. Eur. J. Protistol. 48:283–289. 10.1016/j.ejop.2012.04.004 [DOI] [PubMed] [Google Scholar]
- 41.Imai S, Tsutsumi Y, Yumara S, Mulenga A. 1992. Ciliate protozoa in the rumen of Kafue lechwe, Kobus leche kafuensis, in Zambia, with the description of four new species. J. Protozool. 39:564–572. 10.1111/j.1550-7408.1992.tb04852.x [DOI] [PubMed] [Google Scholar]
- 42.Van Hoven W. 1975. Rumen ciliates of the Tsessebe (Damaliscus lunatus lunatus) in South Africa. J. Eukaryot. Microbiol. 22:457–462 [DOI] [PubMed] [Google Scholar]
- 43.Gürelli G, Göçmen B. 2010. The occurrence of the hindgut ciliate Hemiprorodon gymnoposthium (Ciliophora: Buetschliidae) from domestic horses in Cyprus. Turkiye Parazitol. Derg. 34:206–208 (In Turkish) [PubMed] [Google Scholar]
- 44.Gürelli G, Göçmen B. 2012. Intestinal ciliate composition found in the feces of racing horses from Izmir, Turkey. Eur. J. Protistol. 48:215–226. 10.1016/j.ejop.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 45.Imai S, Ozeki K, Fujita J. 1979. Scanning electron microscopy of ciliary zones of the ciliate protozoa in the large intestine of the horse. J. Parasitol. 65:434–440. 10.2307/3280291 [DOI] [PubMed] [Google Scholar]
- 46.Cameron SL, O'Donoghue PJ. 2004. Phylogeny and biogeography of the “Australian” trichostomes (Ciliophora: Litostomata). Protist 155:215–235. 10.1078/143446104774199600 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


