Abstract
Bifidobacterium bifidum MIMBb75 is a human intestinal isolate demonstrated to be interactive with the host and efficacious as a probiotic. However, the molecular biology of this microorganism is yet largely unknown. For this reason, we undertook whole-genome sequencing of B. bifidum MIMBb75 to identify potential genetic factors that would explain the metabolic and probiotic attributes of this bacterium. Comparative genomic analysis revealed a 45-kb chromosomal region that comprises 19 putative genes coding for a potential type IV secretion system (T4SS). Thus, we undertook the initial characterization of this genetic region by studying the putative virB1-like gene, named tgaA. Gene tgaA encodes a peptidoglycan lytic enzyme containing two active domains: lytic murein transglycosylase (LT, cd00254.3) and cysteine- and histidine-dependent amidohydrolase/peptidase (CHAP, pfam05257.4). By means of several in vitro assays, we experimentally confirmed that protein TgaA, consistent with its computationally assigned role, has peptidoglycan lytic activity, which is principally associated to the LT domain. Furthermore, immunofluorescence and immunogold labeling showed that the protein TgaA is abundantly expressed on the cell surface of B. bifidum MIMBb75. According to the literature, the T4SSs, which have not been characterized before in bifidobacteria, can have important implications for bacterial cell-to-cell communication as well as cross talk with host cells, justifying the interest for further studies aimed at the investigation of this genetic region.
INTRODUCTION
Bifidobacteria are Gram-positive members of the human gut microbiota that include species typically representing the dominant colonic bacteria in breast-fed infants (1, 2). Together with lactobacilli, bifidobacteria are the microorganisms most widely exploited in probiotic formulations because of their various health-promoting properties. In this regard, the ability of Bifidobacterium to interact with the host has been demonstrated to be species and strain specific (3–5). Notably, members of the species Bifidobacterium bifidum are capable of interaction with the host, including degradation of mucin (6, 7), metabolism of human milk oligosaccharides (8), and modulation of host immune responses (9–11). Therefore, the species B. bifidum represents a good source of bacterial strains to be employed for the development of novel effective probiotic (12) or paraprobiotic (13) products.
B. bifidum MIMBb75 is a human intestinal isolate with a marked ability to adhere to intestinal mucosae (6, 8), transiently colonize murine intestine, and impact the resident microbial communities (14). Furthermore, the probiotic potential of this strain was evidenced in a clinical trial in which daily consumption of B. bifidum MIMBb75 significantly alleviated the global symptoms of irritable bowel syndrome and improved the quality of life (15). Though MIMBb75 is efficacious as a probiotic, the molecular mechanisms underlying such interaction are yet largely unknown. For this reason, we undertook whole-genome sequencing of MIMBb75 to identify potential genetic factors useful to elucidate the metabolic and probiotic potentialities of this bacterium.
Type IV secretion systems (T4SSs) are large bacterial protein complexes that transport macromolecules across the cell envelope (16). Reportedly, T4SSs are involved in naked DNA uptake and release (17), conjugation (18), and delivery of effector molecules (DNA and/or protein) into eukaryotic host cells (19). To support these activities, the proteins constituting T4SSs include (i) peptidoglycan lytic enzymes, (ii) energizing (motor) components, (ii) scaffold/putative core components, and (iv) surface factors/adhesins (20). T4SSs have been studied in detail in certain Gram-negative bacteria, which use these conjugation apparatuses for the translocation of genetic material and effector proteins into other bacteria or eukaryotic host cells (19). In contrast, T4SSs described so far in Gram-positive bacteria appear to be exclusively dedicated to conjugative DNA transfer, as demonstrated by the presence of members of an additional family of DNA processing enzymes not present in T4SSs participating in protein effector transport (20). Furthermore, Gram-positive conjugative T4SSs do not produce pilus filaments but contain surface proteins that operate as adhesins (21). It is plausible that T4SSs in Gram-positive cells serve additional and yet unknown purposes.
Although identified in the genome of numerous Gram-positive bacteria, T4SSs have not been characterized in bifidobacteria. In this regard, here we show the identification by comparative genomics of a genetic region putatively coding for a type IV secretion system in B. bifidum MIMBb75. As a first step in the characterization of the putative T4SS of strain MIMBb75, in this study, we describe the molecular analysis of one of its constitutive elements, namely, the putative VirB1-like peptidoglycan-lytic component encoded by gene tgaA. In addition, gene tgaA attracted our attention because its product probably is located on the outer surface of the bacterial cells and therefore has major chances to interact with host cells.
MATERIALS AND METHODS
Bifidobacterial culture conditions.
Bifidobacterium strains were grown under anaerobic condition (Anaerocult A System; Merck, Darmstadt, Germany) at 37°C in prereduced DeMan-Rogosa-Sharpe (MRS) broth (Difco Laboratories, Inc., Detroit, MI) supplemented with 0.05% l-cysteine hydrochloride (cMRS; Sigma-Aldrich, St. Louis, MO).
Genome sequencing, sequence annotation, and comparative analysis.
The genome sequence of B. bifidum MIMBb75 was determined by the use of an Illumina HiSeq 2000 system with paired-end and shotgun libraries. A total of 5,722,028 reads with an average length of 35 bp were assembled into 133 contigs using SOAPde novo, version 1.04, software (22). Protein-encoding open reading frames (ORFs) were predicted using a combination of Prodigal (23) and BLASTX (24) for comparative analysis. Results of the gene-finder program were combined manually with data from BLASTP (25) analysis against a nonredundant protein database provided by the National Center for Biotechnology Information. The combined results were inspected by Artemis (26), which was used for a manual editing effort to verify and, if necessary, to redefine the start of each predicted coding region or to remove or add coding regions within the 45-kb region containing the tgaA gene (i.e., the putative type IV secretion system region [T4SS]). The TMHMM server, version 2.0 (27), was used to predict transmembrane sequences, and SignalP (28) was used for the prediction of signal peptides. Whole-genome sequence alignments for similarity were performed at the DNA level using LAST (http://last.cbrc.jp/).
tgaA gene expression analysis.
Total RNA was extracted from B. bifidum MIMBb75 collected at the early logarithmic (optical density at 600 nm [OD600] of 0.5 to 1), late logarithmic (OD600 of 2 to 2.5), and stationary (OD600 of 2.5 to 3) growth phases as previously described (29). In brief, a volume of cMRS broth culture containing 109 cells was collected and centrifuged (at 4°C for 1 min at 8,000 × g). The pellet was resuspended in 1 ml of PureZol (Bio-Rad Laboratories, Milan, Italy) and subjected to a mechanical disruption using a Precellys bead beater (Advanced Biotech Italia s.r.l., Seveso, Italy). Total RNA extraction was performed with an Aurum Total RNA Fatty and Fibrous Tissue kit (Bio-Rad Laboratories), and samples were additionally treated with DNase I (Sigma-Aldrich) for 30 min at room temperature. Subsequently, iScriptTM cDNA Synthesis (Bio-Rad Laboratories) was used to obtain cDNA from 1 μg of RNA. Finally, PCR was carried out using a volume of cDNA solution corresponding to 15 ng of initial RNA in a total reaction volume of 25 μl using DreamTaq DNA polymerase (Fermentas, Vilnius, Lithuania). The expression level of the gene tgaA was assessed with primers Cha-F (5′-CGGCGACGTGATCTGCTTCGA-3′) and Cha-R (5′-CCGGATTGGCTGATGAGGATC-3′), which allow the amplification of a 115-bp tgaA internal fragment. Internal normalization was obtained through the amplification of an approximately 500-bp fragment from the 16S rRNA gene with primers g-Bifid-F (5′-CTCCTGGAAACGGGTGG-3′) and g-Bifid-R (5′-GGTGTTCTTCCCGATATCTACA-3′). Annealing temperatures in the PCR were 60°C and 55°C for the Cha and g-Bifid primers, respectively.
Expression of TgaA-derived recombinant proteins in Escherichia coli.
All enzymes and reagents for molecular biology reactions (polymerases, T4 ligase, and restriction enzymes) were from Fermentas or New England BioLabs (EuroClone S.p.A., Pero, Italy). Purification of PCR fragments was performed with UltraClean PCR Cleanup (MoBio Laboratories/Cabru s.a.s., Arcore, Italy). Plasmid extraction from E. coli was carried out with a QIAprep Spin miniprep kit (Qiagen s.r.l., Milan, Italy). DNA fragments in agarose gel were extracted and purified with a QIAquick gel extraction kit (Qiagen). Antibiotics for the selection of plasmids were from Sigma-Aldrich (St. Louis, MO). For the preparation of vector pAK-tga, the gene tgaA (without the original signal peptide [SP]) was amplified by PCR from B. bifidum MIMBb75 chromosomal DNA with primers tgaFOR-MA (5′-TTACTCGCGGCCCAGCCGGCCATGGCGACGGAATCCACGGTC-3′) and tgaREV (5′-ATGGTGATCGGCCCCCGAGGCCGTGTGAATGAAGCTGACGCT-3′). The amplicon was digested with SfiI restriction enzyme (SfiI restriction sites in the primers are underlined) and ligated in the same restriction site of vector pAK400 (30), yielding a transcriptional fusion of the tgaA amplicon with an E. coli pelB signal sequence (SPpelB) that was present in vector pAK400. The ligation mix was used to transform E. coli JM109 by electroporation according to standard protocols. Recombinants of E. coli JM109 were cultured at 37°C in Super Optimal Broth (SOB) medium supplemented with 25 μg ml−1 chloramphenicol. Once expressed, the signal peptide SPPelB allows the export of the recombinant protein to the periplasm of E. coli cells. For the overexpression of TgaA recombinant proteins, we prepared three plasmids based on expression vector pET26b(+) (Novagen/Merck KGaA, Darmstadt, Germany) and the expression host E. coli BL21(DE3) pLysS (Promega, Madison, WI, USA). In detail, gene tgaA (without the original signal peptide) was amplified by PCR from B. bifidum MIMBb75 chromosomal DNA with primers TgaNdeF (5′-CCTTGGAACATATGAGCGAATCCACGGTCG-3′) and TgaHindR (5′-TCTAGGAAAAGCTTGTGAATGAAGCTGACG-3′), which include restriction sites NdeI and BamHI, respectively (underlined). The resulting amplicon was digested and ligated in the same restriction sites of vector pET26b(+), yielding a transcriptional fusion with a six-His tag at the 3′ terminus of the gene. The resulting vector, named pET-Tga, was subsequently digested with Bsp19I, yielding two DNA fragments. The larger fragment (containing the vector replicon) was separated in and purified from agarose gel and finally self-ligated, yielding vector pET-ΔCHAP (where CHAP is cysteine- and histidine-dependent amidohydrolase/peptidase, pfam05257.4). Afterwards, we prepared vector pET-ΔLT (where LT is lytic murein transglycosylase, cd00254.3) by inverse PCR amplification of plasmid pET-Tga with primers dLT-F (5′-ATACCGGAATTCACGGAGACCACCA-3′) and dLT-R (5′-TTCCAATTGAATTCCTGGTCGATCT-3′), followed by digestion of the amplicon with EcoRI (the restriction site is underlined in primer sequences), and self-ligation. The recombinants of E. coli BL21(DE3) pLysS were cultured at 37°C in the presence of 25 μg ml−1 chloramphenicol (selection for pLysS plasmid) and 25 μg ml−1 kanamycin (selection for pET vectors). The absence of mutations or recombinations in the obtained plasmids was confirmed by sequencing analysis. Vectors pAK-Tga, pET-ΔCHAP, and pET-ΔLT were used for the overexpression of the TgaA-derived recombinant proteins.
Analysis of intracellular accumulation of cell wall breakdown products.
In order to determine the accumulation of fragments deriving from cell wall lysis in E. coli cells, we used the bioluminescence reporter system based on vector pBlaLux1 and E. coli SNO301 (31). Briefly, we transformed by electroporation the cell reporter system with vector pAK-Tga. The expression of TgaA recombinant protein from pAK-Tga was obtained by adding isopropyl β-d-1-thiogalactopyranoside (IPTG; Sigma-Aldrich) to the cell culture. To assess cell wall lysis, immediately after IPTG addition, the broth cultures were aliquoted in a microtiter plate and incubated at 37°C in a luminometer (Chameleon Multilabel Detection Platform; Hidex OY, Turku, Finland) that was set to automatically measure light emission every 5 min for several hours.
Protein overproduction.
Vectors pAK-Tga, pET-ΔCHAP, and pET-ΔLT were used to overexpress in E. coli the recombinant proteins ΔSP-TgaA-His (containing both domains of TgaA protein; also named SPPelB-TgaA here and rTgaA for recombinant TgaA in an accompanying paper [32]), protein ΔSP-TgaA-ΔCHAP-His (containing only the LT domain; rLT), and ΔSP-TgaA-ΔLT-His (containing only the CHAP domain; rCHAP), respectively. To this aim, recombinant E. coli strains were grown over night in 2× YT (yeast extract, tryptone) broth at 37°C and then inoculated in 200 ml of the same medium (1:100 inoculum). After 2 h of incubation under strong agitation at 37°C, 1 mM IPTG was added, and incubation was prolonged for 4 h. Afterwards, cells were recovered by centrifugation, weighed, and treated for protein extraction under denaturing conditions with PerfectPro Ni-nitrilotriacetic acid (NTA) agarose (5 Prime; Eppendorf Italia s.r.l., Milan, Italy) according to the manufacturer's instructions.
Detection of peptidoglycan lytic activity by renaturing SDS-PAGE.
Renaturing SDS-PAGE was performed as described by Mora et al. (33). In brief, autoclaved cells of Micrococcus luteus ATCC 4698 (Sigma-Aldrich) were polymerized using a 12% SDS-PAGE gel at a concentration of 0.2% (wt/vol). Sample preparation was carried out by mixing 5 μg of recombinant protein with 30 μl of SDS-PAGE denaturing loading buffer. After electrophoresis, gels were soaked in 200 ml of sterilized, distilled water for 30 min and then in 200 ml of 25 mmol liter−1 Tris-HCl (pH 7) containing 1% (wt/vol) Triton X-100 for 12 to 36 h. Bands of lytic activity were visualized by staining the gel with 1% (wt/vol) methylene blue (Sigma) in 0.1% (wt/vol) KOH and subsequent destaining in sterilized distilled water.
Generation of ΔSP-TgaA-ΔLT-His-specific antibodies.
An antibody against the recombinant protein rCHAP was raised in rabbits by Primm s.r.l. (Milan, Italy). Briefly, after preimmune serum collection, an initial immunization was followed by a second immunization at day 21, a third immunization at day 28, a bleed step and enzyme-linked immunosorbent assay (ELISA) at day 35, and a fourth immunization at day 37. Then, a final serum sampling and affinity purification of antibodies were carried out.
Preparation and analysis of immunofluorescent B. bifidum MIMBb75.
B. bifidum MIMBb75 cells were cultivated until early stationary growth phase, harvested by centrifugation, washed once with deionized sterile water, and incubated at room temperature for 10 min with ΔSP-TgaA-ΔLT-His antiserum (1:100 diluted in deionized water). Subsequently, bacterial cells were centrifuged, resuspended in a solution of Cy5-conjugated goat anti-rabbit secondary antibody (Molecular Probes, Eugene, OR) (1:10 diluted in deionized water), and incubated for 10 min at room temperature in the dark. Afterwards, stained cells were visualized with a fluorescence optical digital microscope (Leica DM1000; Leica Micro-Systems, Wetzlar, Germany) at a magnification of ×1,000.
Electron microscopy and immunogold labeling.
Bifidobacterial cells were cultivated in broth to stationary phase (15 to 18 h of cultivation) or on agar plates of cMRS medium. Bacterial cells were collected, and the resulting pellet was processed for transmission electron microscopy. Sample preparation was performed according to Arioli et al. (34). In brief, cells were fixed in 2.5% glutaraldehyde and later postfixed with 1% osmium tetroxide (in 0.1 M cacodylate buffer, pH 7.2) for 2 h at room temperature. After the remaining osmium tetroxide was eliminated, the samples were dehydrated in a graduated cold ethanol series (35 to 100%); each step was performed for about 10 to 15 min at room temperature. The fixed cells were embedded in Epon 812. Blocks were cut with an ultramicrotome and collected on nickel grids. Sections were poststained with 5% uranyl acetate for 5 min at room temperature and treated with lead citrate for 1 min. Sections were observed and photographed with a Philips CM 12 electron microscope and a Zeiss 900 (Carl Zeiss S.p.A., Arese, Milan, Italy). Bacterial cells were also observed after direct staining (intact-cell analysis). For immunogold electron microscopy analysis, samples were treated with anti-ΔSP-TgaA-ΔLT-His rabbit immune serum (or preimmune serum), and the antibodies were then labeled with protein A-conjugated gold particles (10 nm). Finally, the samples were negatively stained with uranyl acetate and lead citrate as indicated above and examined under an electron microscope.
Nucleotide sequence accession number.
The DNA sequence of the T4SS region from B. bifidum MIMBb75 has been deposited in the EMBL database under the accession number HG794715.
RESULTS AND DISCUSSION
In silico analysis of the B. bifidum MIMBb75 genome reveals a chromosomal region putatively coding for a type IV secretion system.
We generated a draft genome sequence of Bifidobacterium bifidum MIMBb75 consisting of 133 contigs, which in total contained 2.25 Mbp. The guanine and cytosine (G+C) content was calculated to be 62.76%, which is consistent with that of other B. bifidum genomes (7).
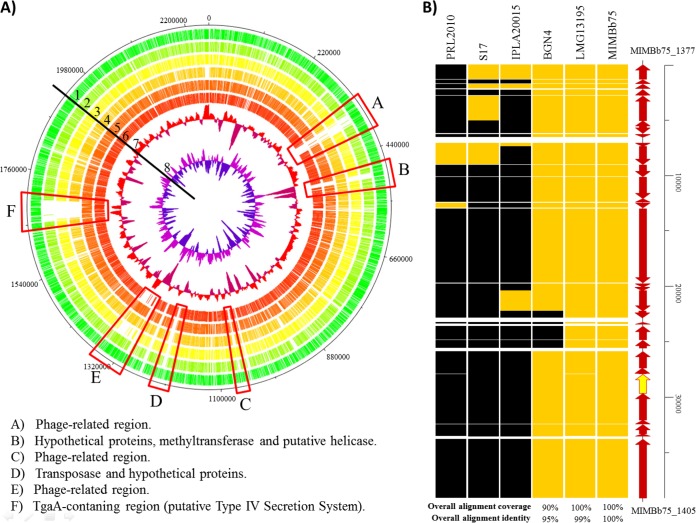
Comparative genomics revealed that more than 90% of the putative coding sequences identified in MIMBb75's contigs share high sequence homology with all of the B. bifidum genomes available so far (GenBank accession numbers: strain BGN4, NC_017999; PRL2010, NC_014638; S17, NC_014616; IPLA20015, from AMPM01000001 to AMPM01000050; LMG13195, from AMPL01000001 to AMPL01000050). The only exceptions were six chromosomal regions, three of which are related to putative integrated phages or phage remnants (Fig. 1A, regions A, C, and E) and two of which are related to DNA modification enzymes (regions B and D); the other nonconserved genetic locus (region F) consists of a 45-kb region, which includes the putative genes of a type IV secretion system (T4SS).
FIG 1.
Comparative genomic analysis of B. bifidum MIMBb75 with other complete genome sequences of B. bifidum strains. (A) Circular genome atlas of B. bifidum MIMBb75 (circle 1) with mapped orthologues (defined as reciprocal best BLASTP hits with more than 30% identity over at least 80% of both protein lengths) in six publicly available B. bifidum genomes: B. bifidum PRL2010 (circle 2), B. bifidum S17 (circle 3), B. bifidum IPLA20015 (circle 4), B. bifidum LMG13195 (circle 5), and B. bifidum BGN4 (circle 6). Circle 6 shows the GC plot, and circle 8 shows the GC skew. Highlighted in the atlas are the main regions of difference between the genomes. (B) Heatmap showing genes encompassing the tgaA-containing region and conserved among the other sequenced B. bifidum genomes. Yellow indicates the chromosomal regions homologous to the putative MIMBb75 T4SS, whereas black indicates the absence of homology.
Even though T4SSs are widespread among all bacterial phyla, their molecular component functions have been studied in depth only within Gram-negative bacteria, such as the vir region of Agrobacterium tumefaciens (35) and the cag type IV secretion apparatus of Helicobacter pylori (36). The gene nomenclature of the 43-kbp Agrobacterium vir T4SS region has been used to refer also to T4SSs identified in Gram-positive bacteria (37, 38).
We found 19 putative genes (Fig. 2A, ORFs 1 to 19) in the predicted T4SS genetic locus of B. bifidum MIMBb75. These putative genes include the minimal components of the secretory system in Gram-positive bacteria (VirB1-, VirB4-, VirB6-, and VirD4-like proteins) and the proteins needed to support the conjugative transfer of DNA (37, 38). Specifically, we found in MIMBb75 seven genes whose products contain conserved domains associated to vir molecular components of the Agrobacterium tumefaciens T4SS (Fig. 2A): ORF4 putatively codes for a VirD2-like relaxase/mobilization protein (conserved domain pfam03432) (Fig. 3, D2), whereas the polypeptide deduced from ORF8 shows the conserved domains cd01126 and pfam12696, which characterize TraG/TraD/VirD4-like proteins (Fig. 3, D4); ORF11 shares homology with a VirB1-type lytic transglycosylase, functioning to degrade peptidoglycan to facilitate insertion of the Gram-positive bacterial type IV secretion machinery into the cell envelope (38–40); the ORF12-encoded peptide contains a sequence homologous to the conserved domains pfam12846, TIGR02746, and TIGR00929, which are typical of TrbE/VirB4-like proteins (Fig. 3, B4). The VirB4- and VirD4-like enzymes of the T4SSs are coupling protein NTPases that play essential roles in supplying the energy for substrate translocation and apparatus assembly (31, 36, 41). ORF13 putatively codes for VirB6-like proteins (Fig. 3, B6), which constitute a component of the transmembrane channel (16), whereas ORF15 is the potential gene for VirC1 protein, a factor required for efficient DNA transfer (42). Therefore, the 45-kb genetic locus of B. bifidum MIMBb75 seems to have the main elements necessary for a functional type IV secretion system (Fig. 3 describes the potential mechanism of DNA transfer by the putative T4SS of B. bifidum MIMBb75).
FIG 2.
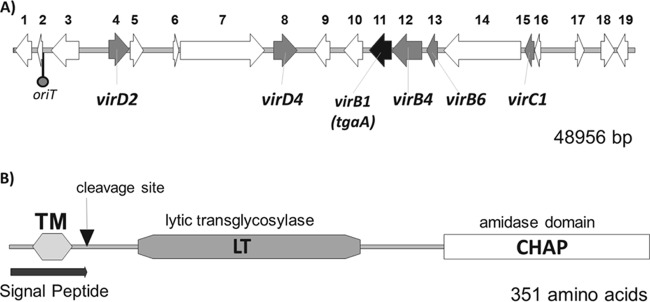
Organization of the putative type IV secretion system genetic region of Bifidobacterium bifidum MIMBb75 (A) and structural organization of the TgaA protein (B). ORFs indicated in the map in panel A putatively code for the following proteins (according to a BLASTP search): 1, HipA-like domain-containing protein (PF07805); 2, helix-turn-helix XRE family-like protein (HipB); 3, DNA topoisomerase III; 4, relaxase (VirD2); 5, mobilization protein (MobC); 6, conserved transposon-encoded protein (TnpV); 7, methyl transferase type 11; 8, VirD4 ATPase (TraG/TraD family protein); 9, HipB transcriptional regulator (HTH XRE); 10, molecular chaperone (containing the pfam02459 motif); 11, TgaA (VirB1); 12, transfer complex protein (VirB4); 13, plasmid conjugal transfer protein (VirB6/TrbL); 14, aggregation substance (containing domains pfam10698 and TIGR04228); 15, ParA ATPase (VirC1); 16, transcription regulator WhiB-related protein; 17, acetyltransferase (GNAT) family; 18, HTH XRE (HipB, containing the domain cd00093); 19, HAD-type hydrolase (COG0561). ORFs putatively coding for hypothetical proteins have not been included. oriT, putative origin of transfer. TM, transmembrane region.
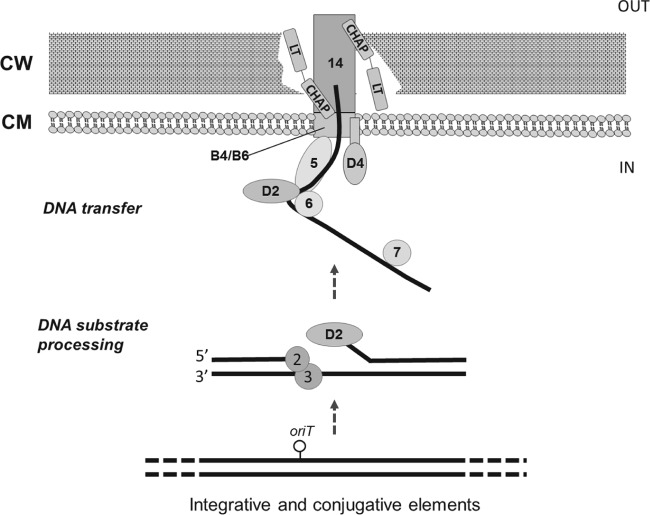
FIG 3.
Proposed mechanism of the putative type IV secretion system (T4SS) of Bifidobacterium bifidum MIMBb75. (The illustration is based on the scheme in reference 51.) The numbers and codes of genes and proteins are according to Fig. 2A. CW, cell wall; CM, cell membrane. The T4SS is comprised of conjugation, effector translocator, and DNA release/uptake subfamilies of proteins. For conjugative transfer, DNA substrates are processed through (i) excision from the chromosome by excisionase/integrase enzymes or transposases (e.g., VirD2), (ii) processing of the transfer intermediate at the origin-of-transfer sequence (oriT) by the relaxosome (e.g., ORF5 and ORF6), (iii) recruitment of the relaxase T-strand intermediate to the type IV coupling proteins (e.g., VirB4/D4), and (iv) translocation through the T4SS channel (e.g., ORF14).
ORF11 encodes TgaA, a putative VirB1-like enzyme.
With the aim of characterizing the putative T4SS of B. bifidum MIMBb75, in this study we initially investigated ORF11, which potentially corresponds to a VirB1-like protein. In the T4SS, VirB1 is a protein that harbors a lysozyme-like structural fold. Consistently, we found that the protein encoded by ORF11 contains two conserved domains involved in the metabolism of the bacterial cell wall, lytic murein transglycosylase (LT, cd00254.3) and cysteine- and histidine-dependent amidohydrolase/peptidase (CHAP, pfam05257.4) (Fig. 2B), according to an organization that resembles the structure of autolytic enzymes (43, 44). Bacterial lytic transglycosylases degrade murein by nonhydrolytic cleavage of the β-1,4-glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine and are part of the autolytic enzymes involved in controlled turnover of the bacterial cell wall (45, 46). The CHAP domain is found in a wide range of protein architectures, often in association with other domains that cleave peptidoglycan (41, 47), and presumably shows N-acetylmuramoyl-l-alanine amidase activity (48). Consequently, the putative gene corresponding to ORF11 was named tgaA for transglycosylase amidase gene A.
Distribution of TgaA protein in bifidobacteria.
The overall structure of the TgaA protein (i.e., the LT conserved domain cd00254.3, followed by the CHAP domain pfam05257.4 at the C terminus) was found in eight Bifidobacterium coding sequences available in GenBank, including three putative proteins belonging to the species B. breve and two belonging to B. bifidum. Besides the above-mentioned eight sequences, the LT cassette was found in only one additional putative protein (YP_004209100 from B. longum subsp. infantis 157F) (see Fig. S4 in the supplemental material), whereas a conserved domain of the CHAP superfamily was identified in numerous bifidobacterial putative proteins and particularly in proteins where no other conserved domains are identified (see Fig. S4). The CHAP domain is, therefore, widespread among bifidobacteria (18 species), indicating that its enzymatic activity could be involved in cellular functions of fundamental importance for cell wall metabolism (44). Interestingly, about 14 protein spots in the extracellular proteome of B. animalis subsp. lactis BB-12 were attributed to BIF_00825 and BIF_01398, two proteins containing a CHAP amidase domain (49). In contrast, in our study we failed to detect any significant positive signal when we tested an MIMBb75 cell-free cMRS broth culture using Western blot analysis with anti-CHAP polyclonal antibodies (data not shown).
TgaA is a peptidoglycan lytic enzyme.
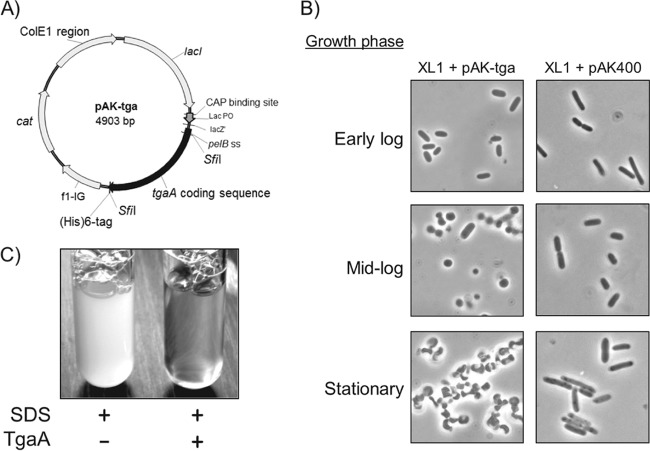
We initially verified by reverse transcription-PCR (RT-PCR) that the tgaA gene is expressed by B. bifidum MIMBb75 during the initial exponential growth phase (see Fig. S1 in the supplemental material). Subsequently, we prepared an engineered E. coli construct expressing a recombinant form of TgaA (SPPelB-TgaA) on IPTG induction in order to biologically confirm the enzymatic activity computationally assigned to the protein TgaA. In SPPelB-TgaA, the original signal peptide of TgaA was replaced by a pelB signal sequence for export at the periplasmic level in E. coli (Fig. 4A). As an initial confirmation of TgaA lytic activity, we observed a drastic morphological modification of recombinant E. coli cells at the end of the exponential growth phase (Fig. 4B), which was not detected in the same E. coli strain harboring the empty vector pAK400. In addition, incubation for a few hours of a suspension of wild-type E. coli XL1 with purified SPPelB-TgaA protein resulted in a complete lysis of the bacterial cells (Fig. 4C).
FIG 4.
Determination of the cell wall lytic activity of TgaA protein. A recombinant form of TgaA (SPPelB-TgaA) was expressed in E. coli XL1 through vector pAK-tga (A), resulting in altered cell morphology after 24 h of growth at 37°C in 2× YT medium (B). After wild-type XL1 cells were incubated at 37°C for 4 h in the presence of 50 μg of purified SPPelB-TgaA, the addition of 10% SDS to a final concentration of 0.2% determined instantaneous complete cell lysis (C). Vector pAK-tga was obtained by cloning gene tgaA in the empty vector pAK400.
In a following experiment, we confirmed the lytic activity of TgaA by means of a luminescence reporter system based on vector pBlaLux1 (31). In pBlaLux1, the luxCDABE operon of Photorhabdus luminescens is under the control of the regulatory element ampR-ampC of Citrobacter freundii (PampC), which was demonstrated to be activated by peptidoglycan fragments (see Fig. S2 in the supplemental material). The introduction of the vector expressing SPPelB-TgaA in the microbial biosensor resulted in a double expression system in which the addition of IPTG produced a dose-dependent increase in bioluminescence (Fig. 5). This experiment showed, therefore, that the intracellular expression of SPPelB-TgaA can cause the accumulation in E. coli cells of peptidoglycan breakdown products, possibly originating from cell wall lysis by TgaA.
FIG 5.
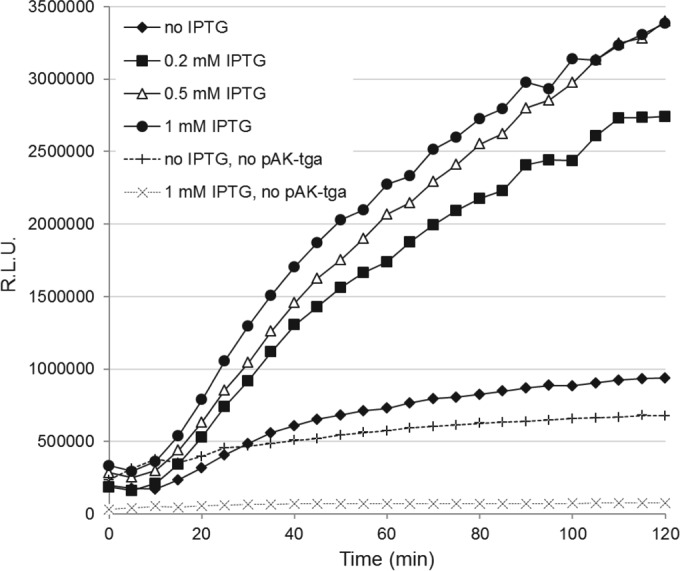
Double expression reporter system for the determination of the peptidoglycan lytic activity of TgaA. A dose-dependent increase of bioluminescence by IPTG induction in E. coli SNO301 containing pAK-tga and pBlaLux1 was observed. The same strain containing only vector pBlaLux1 was used as a control. The addition of IPTG to E. coli SNO301 containing only pBlaLux1 (and not pAK-tga) determined a reduction of the light emission (dotted lines). RLU, relative luminescence units. Each point in the plot represents the mean of the values obtained in one representative experiment conducted in quadruplicates. The variation between the replicates was less than 10%.
Finally, purified SPPelB-TgaA displayed peptidoglycan lytic activity in a renaturing SDS-PAGE assay with cell wall substrates from Micrococcus luteus (see Fig. S3 in the supplemental material). We ran the same experiment also with the two TgaA protein domains (LT and CHAP), expressed separately by the preparation of recombinant forms of TgaA through overexpression in E. coli. Recombinant proteins lacked the original TgaA signal peptide and were named as follows: protein ΔSP-TgaA-ΔCHAP-His (containing only the LT domain; rLT) and ΔSP-TgaA-ΔLT-His (containing only the CHAP domain; rCHAP). Renaturing SDS-PAGE analyses clearly showed TgaA peptidoglycan lytic activity predominantly linked with the transglycosylase domain LT, as confirmed by a distinct band corresponding to the position to which rLT had migrated on SDS-PAGE (Fig. 6). In addition, the clarification determined by the rCHAP, although faint, was detectable (Fig. 6). These results agree with those of Arends et al., who found that both the LT and CHAP domains can degrade peptidoglycan (40).
FIG 6.
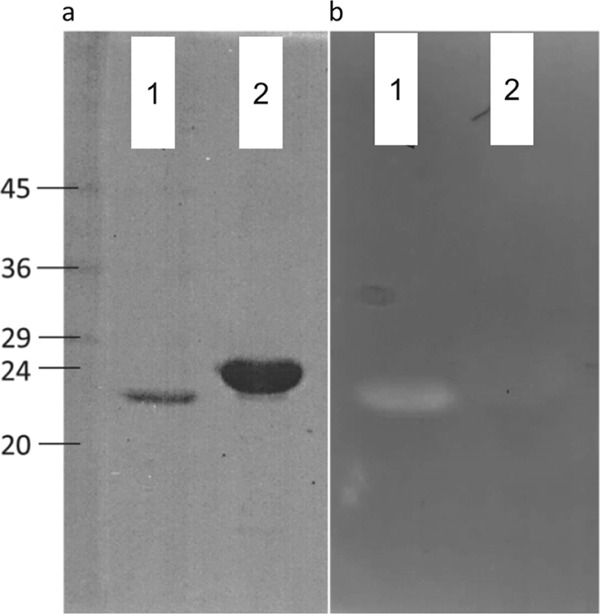
SDS-PAGE (a) and renaturing SDS-PAGE containing 0.2% autoclaved M. luteus cells (b) of the purified six-His-tagged TgaA-derived recombinant proteins. Lane 1, ΔSP-TgaA-ΔCHAP-His (rLT); lane 2, ΔSP-TgaA-ΔLT-His (rCHAP). The relative molecular masses (in kDa) of standard proteins are indicated on the left.
In conclusion, our results show unambiguously that the TgaA protein displays peptidoglycan lytic enzymatic activity, which is consistent with the supposed VirB1-like role of this enzyme.
Protein TgaA is expressed on the B. bifidum MIMBb75 cell surface.
The enzymatic activity of TgaA protein and its potential role in the putative T4SS suggest its location at cell wall level. Therefore, to confirm the presence and localization of the TgaA protein in B. bifidum cells, we prepared rabbit polyclonal antibodies against a purified recombinant form of TgaA protein. We first studied the presence of TgaA in MIMBb75 cells by fluorescence immunostaining with a goat anti-rabbit phycoerythrin (PE)-Cy5.5 secondary antibody via fluorescence microscopy, which revealed evident fluorescence emission by many cells, particularly by cell aggregates (Fig. 7A).
FIG 7.
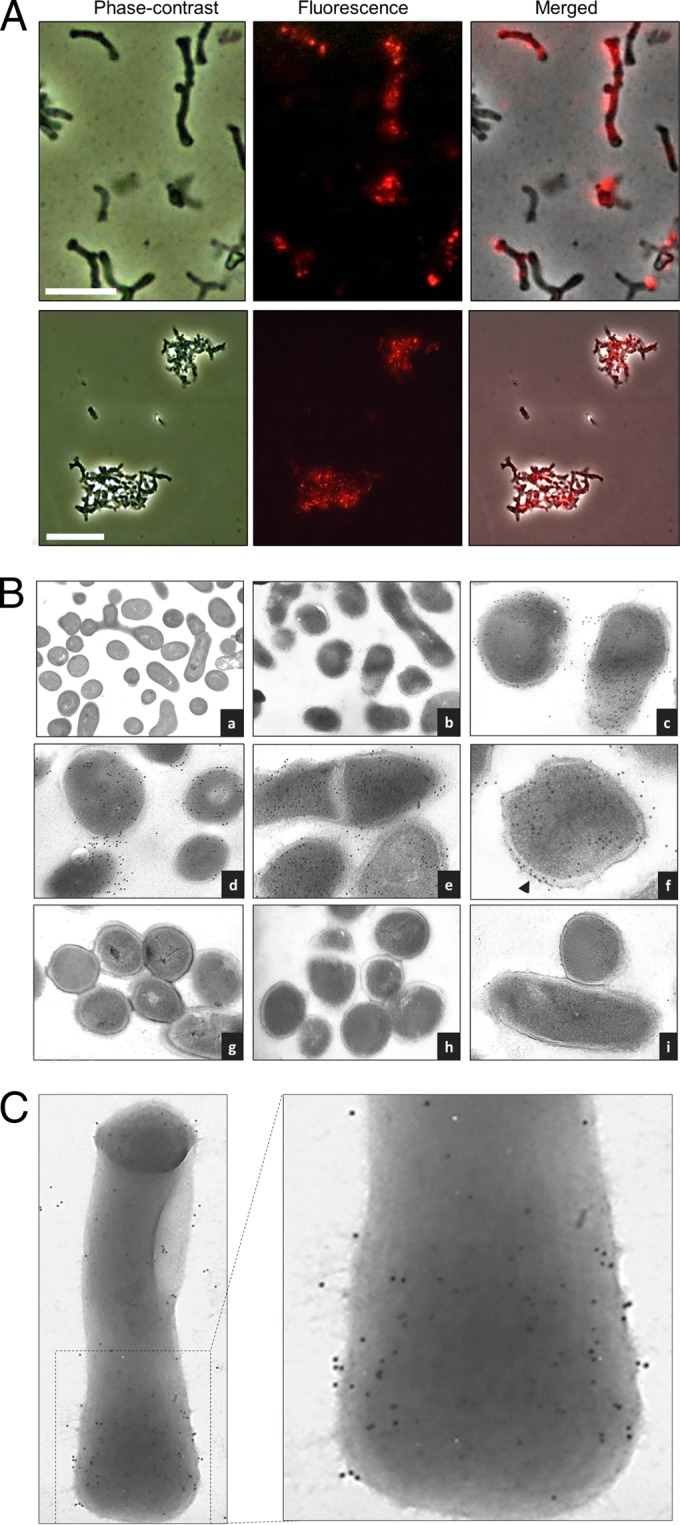
Localization of the TgaA protein in B. bifidum MIMBb75 cells by immunofluorescence staining (A) and immunogold labeling (B and C). (A) Bacterial cells were probed with ΔSP-TgaA-ΔLT-His antiserum, followed by a reaction with a Cy5-conjugated goat anti-rabbit secondary antibody. Cells were visualized with a fluorescence microscope. Fluorescence, phase-contrast, and merged images of the bacterial cells in the same field are shown. The upper pictures show free bacterial cells (scale bar, 5 μm), whereas lower panels show cell aggregates (scale bar, 10 μm). (B and C) Bacterial cells were probed with the ΔSP-TgaA-ΔLT-His antiserum, followed by interaction with gold (12-nm)-conjugated goat anti-rabbit IgG, and negatively stained. We tested thin sections of cell-embedded Lowicryl HM20 resin (B) and intact cells (C). Iden- tifications for panel B are as follows: frame a, nonlabeled MIMBb75 cells; frames b to f, B. bifidum MIMBb75; frames g to i, B. bifidum PRL2010. The arrow in frame f indicates abundant staining of the MIMBb75 cell wall. Bacterial cells were visualized with a transmission electron microscope at a magnification of 23,000 to 60,000.
In addition, to directly pinpoint TgaA in B. bifidum MIMBb75 cells, we ran experiments using immunogold transmission electron microscopy. The specificity of TgaA recognition by the antibodies was demonstrated by the fact that cell staining was insignificant when we immunolabeled B. bifidum PRL2010, a bifidobacterial strain with no gene in its genome sharing significant sequence similarity with tgaA. In contrast, the electron micrographs showed numerous positive signals in the cytoplasm and, particularly, associated with the cell wall of MIMBb75 (Fig. 7B and C). Gold particles were also partially distributed in an extracellular matrix (Fig. 7B, frames c to f), which could indicate either a molecule naturally produced by the bacterium (e.g., exopolysaccharides) or cytoplasm extruded from bacterial cells during sectioning. These data confirm that the TgaA protein is abundantly expressed in B. bifidum MIMBb75. In particular, experiments with intact cells established unequivocally that TgaA was present on the outer surface of the bacterial cells (Fig. 7B), consistent with the role proposed for VirB1-like proteins in T4SS (50).
Conclusions.
The results of this study can be summarized as follows: (i) the genome of Bifidobacterium bifidum MIMBb75 contains a 45-kb region that includes 19 putative genes coding for a potential type IV secretion system; (ii) one gene of this region, named tgaA, putatively encodes the VirB1-like protein of the system, containing two enzymatic domains (lytic transglycosylase LT and amidase CHAP) reportedly associated with peptidoglycan lysis; (iii) we experimentally confirmed that protein TgaA, consistent with its putative VirB1 role, has peptidoglycan lytic activity and is abundantly expressed on the cell surface of B. bifidum MIMBb75.
T4SSs have not been characterized before in bifidobacteria. However, a database search revealed that a genetic region significantly similar to MIMBb75's putative T4SS is also present in the genome of Bifidobacterium bifidum BGN4 (93% nucleotide identity) and Bifidobacterium longum subsp. infantis 157F (91% nucleotide identity). In this study, we undertook the initial characterization of this genetic region, which, according to the literature, can have important implications for bacterial cell-to-cell communication as well as cross talk with host cells (19). In addition, the localization and abundance of TgaA on the B. bifidum cell surface support the hypothesis that this protein may potentially act as a molecular pattern recognized by host cell receptors at the intestinal level, justifying the interest for further studies aimed at the investigation of the direct interaction of TgaA protein with host cells. Accordingly, in a companion paper (32), we describe the ability of TgaA protein domains to activate dendritic cell maturation.
Supplementary Material
ACKNOWLEDGMENT
This work was financially supported by Fondazione Cariplo (grant 2010-0678).
Footnotes
Published ahead of print 20 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01413-14.
REFERENCES
- 1.Zivkovic AM, German JB, Lebrilla CB, Mills DA. 2011. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci. U. S. A. 108:4653–4658. 10.1073/pnas.1000083107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. 10.1371/journal.pone.0036957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young SL, Simon MA, Baird MA, Tannock GW, Bibiloni R, Spencely K, Lane JM, Fitzharris P, Crane J, Town I, Addo-Yobo E, Murray CS, Woodcock A. 2004. Bifidobacterial species differentially affect expression of cell surface markers and cytokines of dendritic cells harvested. Clin. Diagn. Lab. Immunol. 11:686–690. 10.1128/CDLI.11.4.686-690.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menard O, Butel MJ, Gaboriau-Routhiau V, Waligora-Dupriet AJ. 2008. Gnotobiotic mouse immune response induced by Bifidobacterium sp. strains isolated from infants. Appl. Environ. Microbiol. 74:660–666. 10.1128/AEM.01261-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guglielmetti S, Tamagnini I, Mora D, Minuzzo M, Scarafoni A, Arioli S, Hellman J, Karp M, Parini C. 2008. Implication of an outer surface lipoprotein in adhesion of Bifidobacterium bifidum to Caco-2 cells. Appl. Environ. Microbiol. 74:4695–4702. 10.1128/AEM.00124-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guglielmetti S, Tamagnini I, Minuzzo M, Arioli S, Parini C, Comelli E, Mora D. 2009. Study of the adhesion of Bifidobacterium bifidum MIMBb75 to human intestinal cell lines. Curr. Microbiol. 59:167–172. 10.1007/s00284-009-9415-x [DOI] [PubMed] [Google Scholar]
- 7.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. U. S. A. 107:19514–19519. 10.1073/pnas.1011100107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, Yamaguchi M, Kumagai H, Katayama T, Yamamoto K. 2008. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl. Environ. Microbiol. 74:3996–4004. 10.1128/AEM.00149-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss G, Rasmussen S, Fink LN, Jarmer H, Nielsen BN, Frokiaer H. 2010. Bifidobacterium bifidum actively changes the gene expression profile induced by Lactobacillus acidophilus in murine dendritic cells. PLoS One 5:e11065. 10.1371/journal.pone.0011065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez P, Gonzalez-Rodriguez I, Gueimonde M, Margolles A, Suarez A. 2011. Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PLoS One 6:e24776. 10.1371/journal.pone.0024776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turroni F, Serafini F, Foroni E, Duranti S, Motherway MO, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, Sanchez B, Santoni A, Gioiosa L, Ferrarini A, Delledonne M, Margolles A, Piazza L, Palanza P, Bolchi A, Guglielmetti S, van Sinderen D, Ventura M. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc. Natl. Acad. Sci. U. S. A. 110:11151–11156. 10.1073/pnas.1303897110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.FAO/WHO. 2002. Report of a joint FAO/WHO expert consultation on guidelines for the evaluation of probiotics in food. World Health Organization and Food and Agriculture Organization of the United Nations, London, Ontario, Canada [Google Scholar]
- 13.Taverniti V, Guglielmetti S. 2011. The immunomodulatory properties of probiotic microorganisms beyond their viability (ghost probiotics: proposal of paraprobiotic concept). Genes Nutr. 6:261–274. 10.1007/s12263-011-0218-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh N, Arioli S, Wang A, Villa CR, Jahani R, Song YS, Mora D, Guglielmetti S, Comelli EM. 2013. Impact of Bifidobacterium bifidum MIMBb75 on mouse intestinal microorganisms. FEMS Microbiol. Ecol. 85:369–375. 10.1111/1574-6941.12124 [DOI] [PubMed] [Google Scholar]
- 15.Guglielmetti S, Mora D, Gschwender M, Popp K. 2011. Randomised clinical trial: Bifidobacterium bifidum MIMBb75 significantly alleviates irritable bowel syndrome and improves quality of life—a double-blind, placebo-controlled study. Aliment. Pharmacol. Therap. 33:1123–1132. 10.1111/j.1365-2036.2011.04633.x [DOI] [PubMed] [Google Scholar]
- 16.Juhas M, Crook DW, Hood DW. 2008. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 10:2377–2386. 10.1111/j.1462-5822.2008.01187.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704–1721. 10.1111/j.1365-2958.2005.04521.x [DOI] [PubMed] [Google Scholar]
- 18.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1–15. 10.1016/S0378-1097(03)00430-0 [DOI] [PubMed] [Google Scholar]
- 19.Cascales E, Christie PJ. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137–149. 10.1038/nrmicro753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goessweiner-Mohr N, Arends K, Keller W, Grohmann E. 2013. Conjugative type IV secretion systems in Gram-positive bacteria. Plasmid 70:289–302. 10.1016/j.plasmid.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451–485. 10.1146/annurev.micro.58.030603.123630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zerbino DR, Birney E. 2008. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyatt D, Chen GL, Lo Cascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gish W, States DJ. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266–272. 10.1038/ng0393-266 [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 26.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- 27.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 28.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795. 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- 29.Arioli S, Ragg E, Scaglioni L, Fessas D, Signorelli M, Karp M, Daffonchio D, De Noni I, Mulas L, Oggioni M, Guglielmetti S, Mora D. 2010. Alkalizing reactions streamline cellular metabolism in acidogenic microorganisms. PLoS One 5:e15520. 10.1371/journal.pone.0015520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krebber A, Bornhauser S, Burmester J, Honegger A, Willuda J, Bosshard HR, Pluckthun A. 1997. Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. J. Immunol. Methods 201:35–55. 10.1016/S0022-1759(96)00208-6 [DOI] [PubMed] [Google Scholar]
- 31.Valtonen SJ, Kurittu JS, Karp MT. 2002. A luminescent Escherichia coli biosensor for the high throughput detection of beta-lactams. J. Biomol. Screen. 7:127–134. 10.1177/108705710200700205 [DOI] [PubMed] [Google Scholar]
- 32.Guglielmetti S, Zanoni I, Balzaretti S, Miriani M, Taverniti V, De Noni I, Presti I, Stuknyte M, Scarafoni A, Arioli S, Iametti S, Bonomi F, Mora D, Karp M, Granucci F. 2014. Murein lytic enzyme TgaA of Bifidobacterium bifidum MIMBb75 modulates dendritic cell maturation through its cysteine- and histidine-dependent amidohydrolase/peptidase (CHAP) amidase domain. Appl. Environ. Microbiol. 80:5170–5177. 10.1128/AEM.00761-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mora D, Musacchio F, Fortina MG, Senini L, Manachini PL. 2003. Autolytic activity and pediocin-induced lysis in Pediococcus acidilactici and Pediococcus pentosaceus strains. J. Appl. Microbiol. 94:561–570. 10.1046/j.1365-2672.2003.01868.x [DOI] [PubMed] [Google Scholar]
- 34.Arioli S, Roncada P, Salzano AM, Deriu F, Corona S, Guglielmetti S, Bonizzi L, Scaloni A, Mora D. 2009. The relevance of carbon dioxide metabolism in Streptococcus thermophilus. Microbiology 155:1953–1965. 10.1099/mic.0.024737-0 [DOI] [PubMed] [Google Scholar]
- 35.Stachel SE, Nester EW. 1986. The genetic and transcriptional organization of the Vir region of the A6-Ti plasmid of Agrobacterium tumefaciens. EMBO J. 5:1445–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terradot L, Waksman G. 2011. Architecture of the Helicobacter pylori Cag-type IV secretion system. FEBS J. 278:1213–1222. 10.1111/j.1742-4658.2011.08037.x [DOI] [PubMed] [Google Scholar]
- 37.Abajy MY, Kopec J, Schiwon K, Burzynski M, Doring M, Bohn C, Grohmann E. 2007. A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in gram-positive bacteria. J. Bacteriol. 189:2487–2496. 10.1128/JB.01491-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Rong CB, Chen C, Gao GF. 2012. Type-IVC secretion system: a novel subclass of type IV secretion system (T4SS) common existing in Gram-positive genus Streptococcus. PLoS One 7:e46390. 10.1371/journal.pone.0046390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fronzes R, Schäfer E, Wang L, Saibil HR, Orlova EV, Waksman G. 2009. Structure of a type IV secretion system core complex. Science 323:266–268. 10.1126/science.1166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arends K, Celik EK, Probst I, Goessweiner-Mohr N, Fercher C, Grumet L, Soellue C, Abajy MY, Sakinc T, Broszat M, Schiwon K, Koraimann G, Keller W, Grohmann E. 2013. TraG encoded by the pIP501 type IV secretion system is a two-domain peptidoglycan-degrading enzyme essential for conjugative transfer. J. Bacteriol. 195:4436–4444. 10.1128/JB.02263-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pritchard DG, Dong SL, Baker JR, Engler JA. 2004. The bifunctional peptidoglycan lysin of Streptococcus agalactiae bacteriophage B30. Microbiology 150:2079–2087. 10.1099/mic.0.27063-0 [DOI] [PubMed] [Google Scholar]
- 42.Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. 2007. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J. 26:2540–2451. 10.1038/sj.emboj.7601696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rigden DJ, Jedrzejas MJ, Galperin MY. 2003. Amidase domains from bacterial and phage autolysins define a family of gamma-D,L-glutamate-specific amidohydrolases. Trends Biochem. Sci. 28:230–234. 10.1016/S0968-0004(03)00062-8 [DOI] [PubMed] [Google Scholar]
- 44.Bateman A, Rawlings ND. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234–237. 10.1016/S0968-0004(03)00061-6 [DOI] [PubMed] [Google Scholar]
- 45.Engel H, Kazemier B, Keck W. 1991. Murein-metabolizing enzymes from Escherichia coli—sequence-analysis and controlled overexpression of the Slt gene, which encodes the soluble lytic transglycosylase. J. Bacteriol. 173:6773–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Asselt EJ, Dijkstra AJ, Kalk KH, Takacs B, Keck W, Dijkstra BW. 1999. Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand. Structure 7:1167–1180. 10.1016/S0969-2126(00)80051-9 [DOI] [PubMed] [Google Scholar]
- 47.Kausmally L, Johnsborg O, Lunde M, Knutsen E, Havarstein LS. 2005. Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J. Bacteriol. 187:4338–4345. 10.1128/JB.187.13.4338-4345.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loessner MJ, Gaeng S, Wendlinger G, Maier SK, Scherer S. 1998. The two-component lysis system of Staphylococcus aureus bacteriophage Twort: a large TTG-start holin and an associated amidase endolysin. FEMS Microbiol. Lett. 162:265–274. 10.1111/j.1574-6968.1998.tb13008.x [DOI] [PubMed] [Google Scholar]
- 49.Gilad O, Svensson B, Viborg AH, Stuer-Lauridsen B, Jacobsen S. 2011. The extracellular proteome of Bifidobacterium animalis subsp. lactis BB-12 reveals proteins with putative roles in probiotic effects. Proteomics 11:2503–2514. 10.1002/pmic.201000716 [DOI] [PubMed] [Google Scholar]
- 50.Aguilar J, Cameron TA, Zupan J, Zambryski P. 2011. Membrane and core periplasmic Agrobacterium tumefaciens virulence Type IV secretion system components localize to multiple sites around the bacterial perimeter during lateral attachment to plant cells. mBio 2(6):e00218-11. 10.1128/mBio.00218-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73:775–808. 10.1128/MMBR.00023-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.