Abstract
What defines central carbon metabolism? The classic textbook scheme of central metabolism includes the Embden-Meyerhof-Parnas (EMP) pathway of glycolysis, the pentose phosphate pathway, and the citric acid cycle. The prevalence of this definition of central metabolism is, however, equivocal without experimental validation. We address this issue using a general experimental approach that combines the monitoring of transcriptional and metabolic flux changes between steady states on alternative carbon sources. This approach is investigated by using the model bacterium Pseudomonas putida with glucose, fructose, and benzoate as carbon sources. The catabolic reactions involved in the initial uptake and metabolism of these substrates are expected to show a correlated change in gene expressions and metabolic fluxes. However, there was no correlation for the reactions linking the 12 biomass precursor molecules, indicating a regulation mechanism other than mRNA synthesis for central metabolism. This result substantiates evidence for a (re)definition of central carbon metabolism including all reactions that are bound to tight regulation and transcriptional invariance. Contrary to expectations, the canonical Entner-Doudoroff and EMP pathways sensu stricto are not a part of central carbon metabolism in P. putida, as they are not regulated differently from the aromatic degradation pathway. The regulatory analyses presented here provide leads on a qualitative basis to address the use of alternative carbon sources by deregulation and overexpression at the transcriptional level, while rate improvements in central carbon metabolism require careful adjustment of metabolite concentrations, as regulation resides to a large extent in posttranslational and/or metabolic regulation.
INTRODUCTION
Understanding the relationship between the structure and function of microbial metabolic networks is of prime interest in systems biology (1, 2), with a major impact on research areas such as biochemistry, metabolic engineering, and synthetic biology. To this end, significant progress has been made in elucidating the global structure properties of microbial networks by using graph theory (3, 4), demonstrating that bacterial metabolic networks are highly organized in the form of a “bow tie” (5), in which different nutrients are catabolized (fan-in) to produce precursor metabolites and activated carriers required for the synthesis of larger building blocks (fan-out) (6). A striking example of the bow tie framework in nature can be observed with many soil bacteria (e.g., pseudomonads), which follow an unbiased trait called the “catabolic funnel” to mineralize complex aromatic molecules (7, 8).
The versatile carbon metabolism of soil bacteria is currently being explored and exploited in several biotechnological applications, including biofuel and fine chemical synthesis (9, 10), the design of new catabolic routes for mineralizing toxic compounds (11), or protecting plants from pathogenic interactions and supporting their growth (12). All these diverse aspects of bacterial metabolism are linked to the functional organization of the carbon metabolic network, with the majority of control directed toward anabolic reactions (13), to ensure a balanced provision of energy equivalents and building blocks for cell growth. Microorganisms, especially the soil generalists, have evolved enormous flexibility in fueling central carbon metabolism. Central metabolism, according to the classical textbook definition, includes the Embden-Meyerhof-Parnas (EMP) pathway of glycolysis, the pentose phosphate pathway, and the citric acid cycle (14, 15), with individual variations depending on the ecological niche in which the organism lives; e.g., Pseudomonas has an additional central pathway, such as the Entner-Doudoroff (ED) pathway, replacing the EMP pathway. In some species of saccharolytic Archaea, carbohydrates are known to be assimilated through modified nonphosphorylated ED pathways because of the absence of the conventional EMP pathway (16). All these definitions are the outcome of extensive investigations with carbohydrates, and a comprehensive experimental elucidation of the functional structure of the metabolic network and central carbon metabolism with multiple carbon sources is missing to date.
As conceived notions from textbook chemistry and carbohydrate-centered metabolism mask the views on central carbon metabolism, it is not only interesting but also important to ask, what defines central carbon metabolism? This is true because environmental pressure and needs may indeed and finally determine pathway usage (17). Explicitly, answers to this question can be found by exploring the response of the stimulons (i.e., all the operons responding together to an environmental stimulus) in the alternate routes of carbon metabolism toward the synthesis of the 12 biomass precursor metabolites. Such knowledge may provide clues towards an understanding of the regulatory mechanisms of the metabolic pathways in redirecting carbon flux to biomass precursors and industrially relevant metabolite synthesis (18–20) and have indirect implications, for example, in the field of metabolic engineering. In a recent study, Noor et al. showed that the central carbon metabolism of Escherichia coli follows an optimality principle: it uses a minimal number of enzymes to traverse between 12 key precursor metabolites and between input sugars and precursor metabolites (21). However, the extent to which an optimality principle can describe the operation of a metabolic network may depend on the bacterium's growth conditions and evolutionary selection (22). Heterotrophic microbes are known to efficiently adapt and modulate flux to environmental changes by the concerted action of different regulatory mechanisms (23). These mechanisms are collectively represented as hierarchical (modulation of enzyme concentrations by processes such as transcription, translation, and posttranscriptional modifications) and/or metabolic (alteration of enzyme activity through a change in substrate, product, or effector concentration). The classic example of hierarchical regulation via gene expression is the induction of the lac operon in response to lactose availability (24) or the induction of the histidine biosynthesis pathway in response to histidine limitation in the medium (25). ter Kuile and Westerhoff proposed “regulation analysis” to quantitatively resolve hierarchical from metabolic regulation (26, 27). In this study, the different modes of regulation in the metabolic network of Pseudomonas putida are examined by analyzing the required metabolic and transcriptional responses to sustain growth on limiting alternative carbon sources (i.e., glucose, fructose, and benzoate). Glucose, fructose, and benzoate were chosen to trigger the innate response of the three classical biochemical pathways, namely, the Entner-Doudoroff pathway, the Embden-Meyerhof-Parnas pathway, and the β-ketoadipate (ortho-cleavage) pathway (Fig. 1). By comprehensive analyses of steady-state gene expression and in vivo flux data, the existence of hierarchical and metabolic regulation in the P. putida metabolic network is qualitatively identified. The results are discussed in the context of revisiting the definition of central carbon metabolism based on regulatory analyses and the application thereof in metabolic engineering using alternate carbon sources derived, e.g., from glucose.
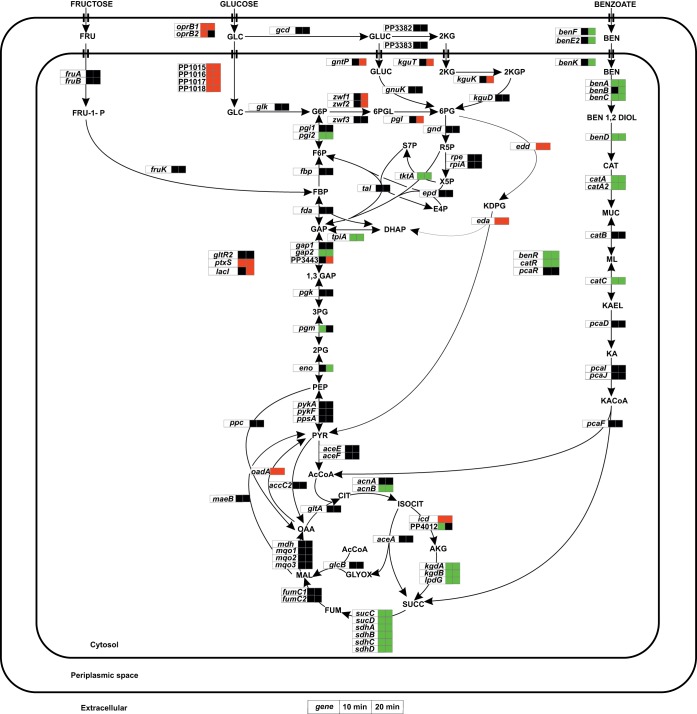
FIG 1.

Glucose, fructose, and benzoate metabolism in P. putida KT2440 through the Entner-Doudoroff (ED) pathway, the Embden-Meyerhof-Parnas (EMP) pathway, and the β-ketoadipate pathway. Glucose transport into the cytoplasm is mediated through an ABC uptake system and phosphorylated to glucose-6-phosphate (G6P) and further to 6-phosphogluconate (6PG). Alternatively, glucose diffuses into the periplasm and is converted into gluconate and 2-ketogluconate (39). These intermediates are transported to the cytoplasm and are subsequently phosphorylated and reduced to 6-phosphogluconate, which is further metabolized by Entner-Doudoroff pathway enzymes or the pentose phosphate pathway enzymes. Fructose is transported into the cytoplasm by a PTS permease system and converted first to fructose-1-phosphate (F1P) and then to fructose-1,6-bisphosphate (FBP). FBP can then follow a reverse/anabolic EMP route and then enter the ED pathway or be processed by the standard glycolytic EMP route. Benzoate uptake into the cytoplasm is mediated by a permease (65) and is then processed by the β-ketoadipate pathway enzymes, resulting in the central intermediates acetyl coenzyme A (AcCoA) and succinate (SUCC). The metabolic intermediates involved in the corresponding pathways are abbreviated as follows: F6P, fructose-6-phosphate; R5P, ribulose-5-phosphate; GAP, glyceraldehyde-3-phosphate; DHAP, dihydroxyacetone phosphate; PYR, pyruvate.
MATERIALS AND METHODS
Strain, medium, and preculture conditions.
Pseudomonas putida KT2440, a plasmid-free derivative of P. putida mt-2 (28), was used throughout this study. Cells from glycerol stocks were recovered on LB agar plates. Liquid cultures were started on LB medium, followed by two subsequent cultivations on modified M9 minimal medium. Erlenmeyer flasks (1 liter) were used in a rotary shaker at 30°C and 150 rpm, ensuring fully aerated growth conditions.
The composition of the modified M9 medium for chemostat cultivations was calculated based on elementary balancing for the production of 5 g biomass (dry weight) per liter, with the following composition per liter: 15 g Na-benzoate, 20 g glucose, or 15 g d-fructose; 2 g Na2SO4 · 10 H2O; 2.68 g (NH4)2SO4; 1 g NH4Cl; 1.46 g K2HPO4; 0.4 g NaH2PO4 · 2H2O; 0.25 g MgSO4 · 7H2O; 22 mg CaCl2 · 2H2O; 0.27 mg ZnSO4 · 7H2O; 0.15 mg MnSO4 · H2O; 30.2 mg Na-EDTA; 0.24 mg CuSO4 · 5H2O; 24.1 mg FeCl3 · 6H2O; and 0.27 mg CoCl2 · 6H2O. The final pH of the medium was 7.1. For all precultures, sodium benzoate (1 g liter−1) was used as the sole carbon source. To maintain a constant biomass concentration during the substrate shift experiments, the concentration of the carbon source was modified according to the biomass growth yield on the respective substrates, with the remaining components of the medium being unchanged.
For the determination of the biomass concentration, 10 ml of the cell suspension was centrifuged (Heraeus Megafuge 1.0 R with an 8030 rotor for 15 min at 3,220 × g at 4°C) in preweighed glass tubes. The biomass pellet was washed twice with a 0.9% NaCl solution and dried to a constant weight at 105°C, followed by weighing.
Shake flask cultivations.
P. putida KT2440 was grown in 200 ml modified M9 minimal medium in 1-liter baffled Erlenmeyer flasks at 30°C in a rotary shaker (200 rpm). The medium was supplemented with benzoate (0.10%) together with glucose (0.25%) or d-fructose (0.63%). Samples were withdrawn at regular intervals and centrifuged (17,700 × g at 4°C for 10 min). The supernatant was analyzed for substrate concentrations by using high-performance liquid chromatography (HPLC) or enzymatic kits. Growth was monitored by measurements of the optical density at 450 nm (OD450).
Startup of continuous cultivation and criteria for steady state.
Cultivation of P. putida KT2440 was initiated with a batch mode of operation with benzoate minimal medium at 30°C in a stirred-tank bioreactor (KLF 2000; Bioengineering, Wald, Switzerland), with a working volume of 1.5 liters. The airflow was kept at 2 liters min−1, and the pH was maintained at 7.0 by the addition of 20% H3PO4 and 2 M NaOH. Batch cultivation was continuously monitored by detecting changes in the dissolved oxygen (DO) concentration and increases in the OD450. Fed-batch operation was initiated with exponential feeding of a benzoate solution (50 g/liter), initiated after the complete consumption of benzoate in the growth medium (marked by a steep increase in the DO concentration). Exponential feeding was performed at a growth rate of 0.65 h−1, controlled by a computer-aided feed-forward control. After the desired biomass concentration of 5 g cell dry weight (gCDW) liter−1 (corresponding to an OD450 of 30) was reached, the continuous mode of operation was started with a dilution rate of 0.1 h−1. Steady-state conditions were confirmed when the biomass concentration as well as the exhaust analysis remained constant over a period of five mean residence times.
Substrate shift experiments.
Carbon source shift experiments were performed with a well-established benzoate-limited chemostat. Three shifts were introduced; i.e., the carbon source limitations were changed to glucose and fructose and finally back to benzoate. The chemostat culture was maintained on each carbon source for no more than two residence times. To monitor the transient changes during each shift, samples were withdrawn from the reactor simultaneously for transcriptome analysis and for analysis of substrate concentrations. For transcriptome analysis, 1 ml of the biosuspension from the reactor was withdrawn into 4 ml of RNAprotect solution (RNAprotect Bacteria reagent; Qiagen) in 15-ml falcon tubes. The diluted sample was vortexed immediately and incubated at room temperature for 5 min, followed by centrifugation at room temperature (Heraeus Megafuge 1.0 R with an 8030 rotor for 10 min at 3,220 × g). The supernatant was discarded, and the pellet was frozen at −70°C until analysis. For analysis of substrate concentrations, 2 ml of the biosuspension was filtered directly through a 0.22-μm cellulose filter, and the filtrate was stored at −20°C until further analysis.
Analytical method.
The filtrate obtained from the biosuspension was used for the analysis of the metabolites benzoate, glucose, and fructose. The benzoate concentration was quantified by HPLC (Elite LaChrom; Merck-Hitachi, Germany) on a CC Nucleosil C18 HD column (100-Å pore size, 5-μm particle size, and 25-cm by 4-mm inner diameter; Macharey-Nagel, Oensingen, Switzerland). The conditions for the analysis were as follows: solvent A was 100 mM ortho-phosphoric acid (pH 2), and solvent B was methanol. The starting concentration was 10% solvent B in solvent A. After 5 min, a gradient was established by increasing the concentration of solvent B by 3% per minute at a flow rate of 1 ml min−1. Analysis was followed by UV absorption studies using a diode array detector (DAD) with wavelength of 230 nm for benzoate. Glucose and fructose concentrations were determined enzymatically with commercial kits (Enzytec; R-Biopharm). Irrespective of the method of choice, 10 μl of the filtrate was used for the analysis.
Net flux analysis.
For the present analysis, small-scale metabolic models for benzoate, glucose, and fructose metabolism were compiled individually from the genome-scale metabolic model iJN746 for P. putida (29) (see Table S2 in the supplemental material). The biomass composition was assumed to be similar to that of E. coli (30). Intracellular net fluxes were estimated by using (i) the stoichiometric reaction matrix, (ii) physiological data (substrate uptake rates and growth rates), (iii) flux ratios, and (iv) precursor requirements for biomass synthesis, as described previously (31). The benzoate metabolic network is well determined without the use of flux ratios. Appropriate choices for flux ratios were made to constrain the metabolic network for glucose and fructose and to ensure the continued supply of tricarboxylic acid cycle intermediates. The following flux ratios were used: the fraction of pyruvate derived through the Entner-Doudoroff pathway, the fraction of oxaloacetate (OAA) originating from phosphoenolpyruvate (PEP) or pyruvate, the fraction of PEP originating from OAA, and the upper bound of the PEP fraction derived through the pentose phosphate pathway. We used the NETTO module of the FiatFlux software to estimate net fluxes by minimizing the sum of weighted square residuals of the constraints from both metabolite balances and flux ratios (32). The estimated net fluxes for benzoate, glucose, and fructose metabolic networks are given in Table S2 in the supplemental material.
Transcriptome analysis.
RNA isolation was performed by using the protocol provided by Qiagen (RNeasy minikit). RNA integrity and purity were checked by agarose gel electrophoresis. cDNA generation was performed with a statistically distributed mixture of hexanucleotides as primers (random priming). cDNA was generated from ∼10 μg total RNA pooled from three independent RNA samples by using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's protocols. After cDNA purification, samples for P. putida genome oligonucleotide microarrays were labeled according to a protocol provided by Progenika Biopharma (Spain) (33). Since the Progenika microarray is a two-dye array, by using the fluorescent dyes cyanine-3 (Cy3) and cyanine-5 (Cy5) (GE Healthcare), both samples (reference and test samples) are hybridized to one array. In this study, four arrays per experiment were used in parallel, and each sample was labeled twice with each dye, resulting in eight cDNA samples per experiment. A minimum of 5 μg of total cDNA per sample was used for the labeling reaction. Labeled cDNA samples were purified by using PCR purification columns provided by Qiagen. An aliquot of eluted DNA was used for quantity and quality control of cDNA and measuring labeling efficiency. Equal amounts of cDNA of the samples to be compared on one array were mixed and dried in a vacuum concentrator at 40°C for several hours. Labeled cDNA was sent to the array facility at the Helmholtz Centre for Infection Research, Braunschweig, Germany, for array hybridization and data evaluation. For Progenika genomic DNA microarrays, array slides were incubated in preheated blocking buffer at 42°C, and after 45 min, the slides were washed several times by dipping them into double-distilled H2O, followed by a final washing step with isopropanol (33, 34). Slides were dried by centrifugation (2,000 × g for 5 min) and subsequently used for hybridization. The desiccated cDNA samples were resuspended in 110 μl hybridization buffer and denatured at 95°C for 2 min. Meanwhile, the array slide was placed into the hybridization chamber and preheated to 42°C. A coverslip was placed on top of the slide, with white spacers facing down, and the cDNA-solution was added slowly in between the slide and cover until the array area beneath was completely covered. Samples were hybridized at 42°C for 16 h. Hybridized arrays were treated with several washing steps, with subsequent centrifugation at 2,000 × g for 5 min (33, 34).
For evaluation of signal intensities, Agilent feature extraction software was used. Scan pictures had a resolution of 5 μm per pixel. The two-channel gene expression feature was used. The signal intensity per spot was calculated by subtracting the background from the foreground and is provided as the median value of all measured pixels per spot. Progenika array data were normalized by using the LOWESS (locally weighted linear regression) function to remove intensity-dependent effects that can appear by low signal intensities and to avoid bias due to the different label efficiencies of the fluorescent dyes (35). Raw data were provided by the array facility and analyzed with the freely available software BRB ArrayTools (http://linus.nci.nih.gov/pilot/index.htm). Signal intensities were log2 transformed and median normalized over all arrays of one experiment, and an output file providing mean signal intensities, P values (two-sample t test with a random-variance model), FDRs (false discovery rates), and fold changes for all represented probe sets was generated. Genes were considered to be significantly differentially expressed if they showed a combination of a P value of ≤0.05 and an FDR of ≤0.1.
Microarray data accession number.
The transcriptome data discussed in this publication have been deposited in accordance with MIAME standards at the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) and can be accessed through GEO series accession number GSE26785.
RESULTS
Metabolic network operation depends on cultivation modes.
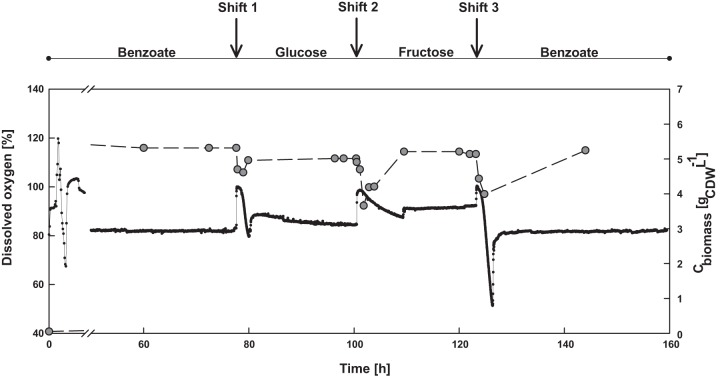
Careful choice of the experimental approach was essential for isolating the carbon source-specific response of P. putida. The experiments were thus designed to satisfy two different criteria: first to maintain substrate supply and demand of the cell at a constant level and second to transiently disturb the supply and demand by changing the carbon source. A chemostat mode of cultivation (at a constant growth rate and a constant biomass concentration) was chosen to satisfy the above-described criteria and also to alleviate the effects of carbon catabolite repression and the synthesis of storage polymers, which are otherwise observed under carbon excess or other nutrient-limiting conditions (36, 37). The experimental approach was favorable in introducing sequential carbon limitations, i.e., from (i) benzoate to glucose, (ii) glucose to fructose, and, finally, (iii) fructose to benzoate (Fig. 2). Notably, with this experimental approach, the initial steady-state conditions on benzoate were replicated after 130 h, confirming the validity of the experimental setup to monitor the flexible response of the metabolic network and to experimentally elucidate the “bow tie” structure in bacterial metabolism.
FIG 2.
Continuous cultivation of P. putida KT2440 on benzoate, glucose, and fructose. Gray circles and dots represent the measured biomass and dissolved oxygen concentrations, respectively.
The network response to the change in carbon source was characterized by a physiological adaptation time, which is defined as the time taken by the bacteria to attain a new steady state after the shift to another carbon source. At a physiological level, the biomass concentration and the percentage of dissolved oxygen were considered indicators. The adaptation time for the shift from glucose to fructose was rather long, 9 h, while only 3 h was required for the shift from benzoate to glucose or fructose to benzoate (Fig. 2). Alternatively, an additional cofeed experiment with benzoate-fructose and benzoate-glucose (see Fig. S1 in the supplemental material) gave us good proof that benzoate is the preferred carbon source among the three carbon sources tested. In this experiment with benzoate-fructose as mixed substrates (see Fig. S1B in the supplemental material), a diauxic lag of 2 to 3 h also indicated that the induction times required for the fructose-metabolizing enzymes are longer than those for the enzymes of the ED and the β-ketoadipate (ortho-cleavage) pathway, catabolizing glucose and benzoate, respectively. To disclose the regulation of these metabolic pathways in detail, gene expression and metabolic flux analyses were performed.
Transcriptional response of Pseudomonas putida to changes in substrates.
The three different shifts in carbon sources (i.e., from benzoate to glucose, from glucose to fructose, and from fructose to benzoate) resulted in a remarkable rearrangement of the gene expression levels in steady state and under conditions of transience. In comparison to the other parts of the metabolic network, only a few changes (i.e., in the fruB and fruK genes) were observed between the steady states on fructose and glucose (Fig. 3). Hence, the changes observed during the alternative shifts (i.e., between benzoate and glucose and between fructose and benzoate) are presented here.
FIG 3.
Differential expression of genes under steady-state conditions on three different substrates. The gene expression level is relative to that under the previous steady-state conditions. SSglu represents the glucose/benzoate transcript ratio at steady state, SSfru represents fructose/glucose transcript ratio at steady state, and SSben represents the benzoate/fructose transcript ratio at steady state. Green and red boxes indicate down- and upregulation of genes, respectively. Black boxes indicate no differential regulation. Abbreviations: GLC, glucose; GLUC, gluconate; 6PGL, 6-phospho-glucono-1,5-lactone; S7P, sedoheptulose-7-phosphate; X5P, xylulose-5-phosphate; 2KG, 2-ketogluconate; KDPG, 2-keto-3-deoxy-6-phosphogluconate; CIT, citrate; GLYOX, glyoxylate; FUM, fumarate; CAT, catechol; MUC, cis,cis-muconate; ML, muconolactone; KAEL, beta-ketoadipate enol lactone; KA, beta-ketoadipate; KACoA, beta-ketoadipyl coenzyme A; 3PG, phosphoglyceric acid; AKG, alpha-ketoglutarate; E4P, erythrose-4-phosphate; PYR, pyruvate.
The transient transcriptional response (determined with samples taken at 10 and 20 min) (Fig. 4) during the shift from benzoate to glucose consisted of prompt downregulation of the benR and catR genes, encoding the positive regulators of the β-ketoadipate (ortho-cleavage) pathway, and upregulation of the ptxS and lacI genes, encoding the major regulators of glucose metabolism. PtxS and LacI control the utilization of glucose via the ketogluconate and gluconate pathways, whereas GltR controls the glucokinase pathway (38). Transient upregulation of the regulatory genes (ptxS and lacI) induced the expression of genes for glucose uptake (oprB1, oprB2, and PP1015 to PP1018; sample taken at 10 min sample) (Fig. 4) and the genes involved in gluconate and ketogluconate uptake and conversion (gntP, kguT, and kguK; sample at 20 min) (Fig. 4). Subsequently, the genes responsible for the formation of the central ED pathway intermediate 6-phosphogluconate (zwf1, zwf2, and pgl; sample at 20 min) (Fig. 4) were also upregulated. During steady state, the genes encoding pyruvate carboxylase (accC2) and pyruvate dehydrogenase (aceE) were reciprocally regulated with the upregulation of the pyruvate carboxylase-encoding gene accC2 (Fig. 3), favoring CO2 fixation. Induction of the oadA (Fig. 3 and 4) gene, encoding the oxaloacetate decarboxylase, is consistent with the results of a previous study comparing growth on citrate to growth on glucose (39), supporting the hypothesis of scavenging excess oxaloacetate produced in the cycle via oxaloacetate decarboxylase during growth on glucose.
FIG 4.
Transcriptome responses during the shift from benzoate toward the glucose steady state. Data are from transcript analyses of samples taken at 10 and 20 min after the shift to glucose from the benzoate steady state. The gene expression level is relative to that under the previous steady-state conditions on benzoate (i.e., glucose/benzoate transcript ratio). Green and red boxes indicate down- and upregulation of genes, respectively. Black boxes indicate no differential regulation.
Differential expression of genes during the shift from fructose to benzoate is indicative of reciprocal regulation compared to the shift from benzoate to glucose. The transcriptional response of benzoate-activated regulators (benR, catR, and pcaR) (Fig. 5) triggered the consecutive expression of the genes involved in the β-ketoadipate (ortho-cleavage) pathway (benF, benK, benABC, benD, catA, catA2, catB, pcaD, pcaJ, and pcaF), with benzoate, cis-cis-muconate, and β-ketoadipate as the effectors (40, 41). As reported previously for growth on acetogenic carbon sources (42, 43), we observed dynamic induction of the malate synthase-encoding gene glcB and, thus, activation of the glyoxylate shunt, which functions as an anaplerotic route to satisfy requirements of biomass synthesis by fixing the acetyl moiety formed during benzoate degradation (Fig. 5).
FIG 5.
Transcriptome responses during the shift from fructose to the benzoate steady state. Data are from transcript analyses of samples taken at 10 and 120 min after the shift to benzoate from the fructose steady state. The gene expression level is relative to that under the previous steady-state conditions on fructose (i.e., benzoate/fructose transcript ratio). Green and red boxes indicate down- and upregulation of genes, respectively. Black boxes indicate no differential regulation.
Metabolic flux responses to changing carbon sources.
To estimate net flux distributions, substrate-specific metabolic models for growth on benzoate, glucose, and fructose were derived from the genome-scale metabolic model iJN746 (29). In absolute numbers, the fluxes and their absolute changes through the ED, EMP, and pentose phosphate pathways and tricarboxylic acid cycle are by far higher than those in any of the peripheral pathways. Hence, analysis was carried out with small-scale models only (see Materials and Methods). In P. putida KT2440, glucose is assimilated through three initial pathways (39), which converge at the level of 6-phosphogluconate. Although their simultaneous operation is required to achieve maximal growth rates, for flux analysis, these three pathways were lumped together, and a glucose uptake rate of 2.21 mmol gCDW−1 h−1 and a growth rate of 0.1 h−1 were used for flux calculation. Flux distributions on glucose indicated that the flow of carbon was directed toward the ED pathway, generating most of the pyruvate, and a minor fraction is directed toward the anabolic route for synthesis of pentose phosphates (Fig. 6A), as reported previously (39, 44). Flux data also revealed a highly active tricarboxylic acid cycle during growth on glucose, with no carbon flux through the glyoxylate cycle.
FIG 6.

In vivo flux distribution of P. putida KT2440 during growth on glucose (A), fructose (B), and benzoate (C). Molar net fluxes (mmol gCDW−1 h−1) are obtained from the best fit with substrate utilization rates on the respective substrates with a growth rate of 0.1 h−1. The arrows are drawn in proportion to the flux. Metabolites in a gray-shaded background represent the biomass precursor molecules. Broken lines indicate zero flux or nonconsidered reactions. Abbreviations: T3P, triose-3-phosphate; P5P, pentose-5-phosphates; PGA, 3-phosphoglycerate; PO, phosphate/oxygen ratio.
The flux distribution on fructose was, however, different from that on glucose. In Pseudomonas, the uptake of fructose is known to proceed via the phosphoenolpyruvate-fructose phosphotransferase (PTSFru) system (45–47), generating a pool of fructose-1-phosphate and pyruvate. Catabolism of fructose-1-phosphate proceeds via the fructose-1-phosphate kinase instead of the fructose-6-phosphate kinase and is thus converted to fructose-1,6-bisphosphate (FBP). Notably, flux analysis indicated that the flux from FBP was directed completely through the gluconeogenic fructose-bisphosphatase and further directed via the Entner-Doudoroff pathway (Fig. 6B). This concurrent operation of the reverse/anabolic EMP and the ED pathways might serve to sustain the additional need for phosphoenolpyruvate in fructose transport during growth on this carbon source (46) and for the generation of NADPH to endure oxidative conditions (48). Flux data revealed that the majority of the glyceraldehyde-3-phosphate generated from the ED pathway is directed to lower glycolysis (from glyceraldehyde-3-phosphate to phosphoenolpyruvate), and no flux through the reaction catalyzed by the enzymes triose-phosphate isomerase and fructose-1,6-bisphosphate aldolase was observed. In P. putida, this sugar is metabolized through a less energetically favorable route, where there is no net synthesis of ATP until the formation of pyruvate (46, 47). To our surprise, such modified forms of EMP pathway (reverse/anabolic) operation in addition to the ED pathway are observed in some species of archaea (16), indicating that in evolution, the EMP pathway was probably an anabolic pathway to begin with. An examination of the further metabolism of pyruvate revealed the presence of an active tricarboxylic acid cycle, with a smaller fraction of flux (∼4%) from isocitrate diverted through the glyoxylate cycle (Fig. 6B). In the cases of both glucose and fructose metabolism, approximately 11 to 13% of the flux from oxaloacetate in comparison to citrate synthase flux is diverted through the gluconeogenic reaction catalyzed by phosphoenolpyruvate carboxykinase to form phosphoenolpyruvate. Finally, a fraction of pyruvate formed through the ED pathway is converted to oxaloacetate through the anaplerotic reaction catalyzed by pyruvate carboxylase.
The estimation of intracellular flux for growth on benzoate (Fig. 6C) was based on the fact that benzoate degradation via the β-ketoadipate (ortho-cleavage) pathway yields equimolar quantities of CO2, succinate, and acetyl coenzyme A (acetyl-CoA) (49). Succinate and acetyl-CoA serve as fuels for anabolic reactions. The carbon skeletons forming glucose-6-phosphate originate from gluconeogenesis. Due to the reported frameshifts in the A subunit of the pyruvate carboxykinase-encoding pckA gene (44), oxaloacetate decarboxylase (encoded by oadA) was considered the first enzyme unique to gluconeogenesis. Pyruvate enters a series of predominantly gluconeogenic reactions, leading to the formation of fructose-6-phosphate and glucose-6-phosphate by fructose-1,6-bisphosphatase and phosphoglucoisomerase. A major metabolic shift between the aromatic (benzoate) and the sugar (glucose and fructose) substrates is reflected at the branch point of isocitrate. Nearly 85% of the flux (in comparison to isocitrate dehydrogenase flux) was diverted through the glyoxylate cycle (Fig. 6C). Posttranslational modifications of isocitrate dehydrogenase by phosphorylation might play a vital role in regulating the flux at this branch point (50, 51).
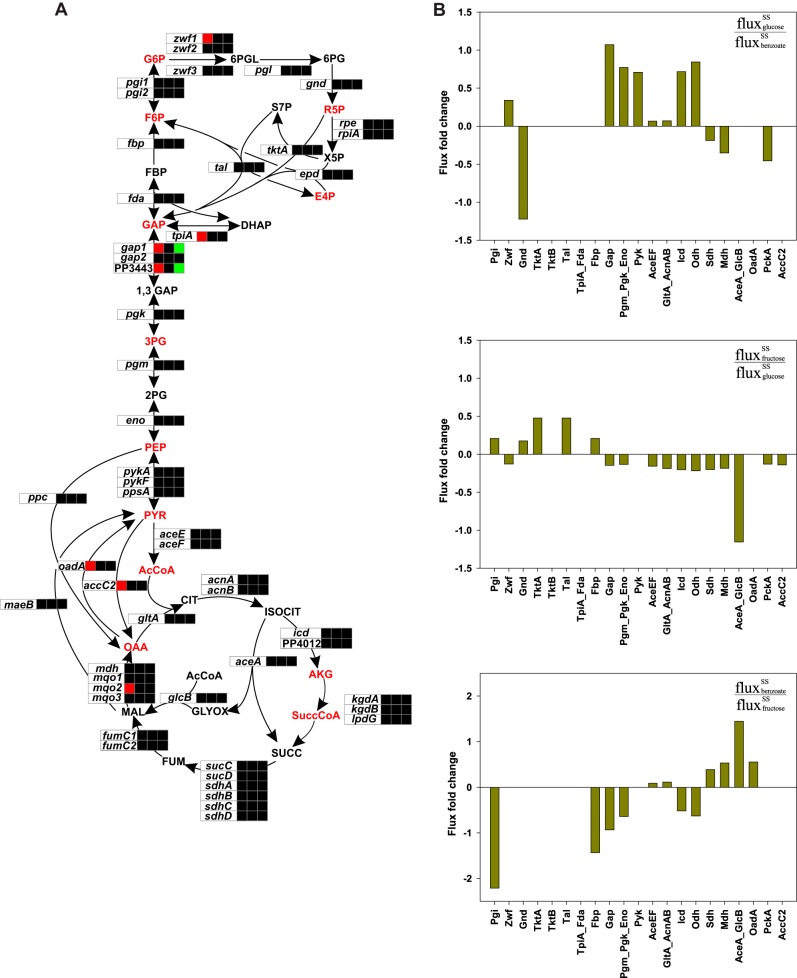
Correlation between flux and transcriptome analyses.
From a combined analysis of transcriptional and metabolic responses of the metabolic network to the imposed changes in the carbon source, we were able to identify the mode of regulation in the reactions involved in the interconversion of the 12 biomass precursor metabolites. Steady-state gene expression on three different substrates revealed no differential expression for a majority of the genes involved in the interconversion of the 12 biomass precursor metabolites (Fig. 7A). However, a few genes (zwf1, tpiA, gap1, PP3443, oadA, accC2, and mqo2) are subject to differential expression, the mechanism of which is uncertain. In contrast to the transcriptional silence observed by gene expression analysis, the steady-state flux responses showed a massive reorganization of carbon flux in the central metabolic reactions under the three growth conditions investigated (Fig. 6). From the set of observed fluxes, fold changes in fluxes on each of three substrates were calculated relative to the flux on the alternate substrate (i.e., the substrate at the previous steady state) (Fig. 7B). The shift in the substrate from benzoate to glucose resulted in an increased flux in the enzymes of the lower part of glycolysis (i.e., from glyceraldehyde-3-phosphate to acetyl-CoA) and also in the tricarboxylic acid cycle enzymes (citrate synthase, isocitrate dehydrogenase, and alpha-ketoglutarate dehydrogenase). However, the shift from glucose to fructose resulted in an increase in flux through the gluconeogenic enzyme fructose-1,6-bisphosphatase due to an increased pool of fructose-1,6-bisphosphate from fructose assimilation. Although the major flux from 6-phosphogluconate is diverted through the ED pathway, a considerable fraction of flux is diverted through the pentose phosphate pathway. This finding is in agreement with data from a previous analysis by Chavarria et al., where ∼14% of the flux from fructose was channeled through the pentose phosphate pathway (47). During the last shift, i.e., from fructose to benzoate, the majority of the flux from benzoate is diverted through the glyoxylate cycle, which is evident from the ∼1.5-fold increase in the flux through the enzymes isocitrate lyase (AceA) and malate synthase (GlcB) (Fig. 7B). Furthermore, the flow of carbon from oxaloacetate is diverted through a series of gluconeogenic reactions toward the synthesis of the remaining biomass precursor metabolites.
FIG 7.
Transcriptional (A) and flux (B) fold changes of the central metabolic reactions between steady states (SS) on glucose, fructose, and benzoate. (A) The three boxes adjacent to the genes denote the gene expression levels at steady state on glucose (glucose/benzoate transcript ratio), fructose (fructose/glucose transcript ratio), and benzoate (benzoate/fructose transcript ratio), in sequential order. Black boxes indicate no differential regulation. Metabolic intermediates in red type represent the 12 biomass precursor metabolites, which are abbreviated as follows: G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; R5P, ribulose-5-phosphate; E4P, erythrose-4-phosphate; GAP, glyceraldehyde-3-phosphate; 3PG, phosphoglyceric acid; PEP, phosphoenolpyruvate; PYR, pyruvate; AcCoA, acetyl coenzyme A; SuccCoA, succinyl coenzyme A; OAA, oxaloacetate; AKG, alpha-ketoglutarate. (B) The fold changes in fluxes were calculated from the net fluxes with the following formula: log10(fluxC source/fluxprevious C source).
While remarkable changes in fluxes in central carbon metabolism are evident from the use of glucose, fructose, and benzoate, the changes are not reflected in the level of transcription (Fig. 7A). In contrast, the genes of the catabolic pathways, including the ED, the reverse EMP, and the β-ketoadipate (ortho-cleavage) pathways, were differentially expressed (Fig. 3), having a direct correlation with the observed change in flux (Fig. 6). With no differential expression of genes of central carbon metabolism but strong flux rerouting, P. putida requires additional means for flux regulation, including presumably allosteric and posttranslational mechanisms.
DISCUSSION
The systematic interpretation of steady-state transcript and flux levels revealed two modes of regulation, i.e., hierarchical and metabolic regulation, in the P. putida metabolic network. We qualitatively identified the modes of regulation in distinct parts of the metabolic network (i.e., central carbon metabolism and catabolic pathways). The rate-controlling steps (the ED, the EMP, and the β-ketoadipate pathways) are optimized to modify fluxes effectively according to the change in carbon sources, which is reflected by their correlated change in transcript and flux levels. Interestingly, the changes in metabolic fluxes of central carbon metabolism are not mirrored at the transcript level under the three growth conditions investigated (Fig. 7). This is in line with previously reported findings for the yeast Saccharomyces cerevisiae (52, 53), the freshwater cyanobacterium Synechocystis (54), and Lactococcus lactis (55), where, depending on the energy source availability, only a few reactions of central carbon metabolism were transcriptionally regulated. For example, the drastic metabolic readjustment of flux at the branch point of isocitrate, i.e., decreased flux through isocitrate dehydrogenase on benzoate and increased flux on glucose or fructose, was not reflected transcriptionally. Studies of E. coli demonstrated that during growth on acetogenic or gluconeogenic carbon sources, ∼80% of the total isocitrate dehydrogenase is in a phosphorylated form (43, 56), thus leading to a spontaneous inactivation state of the enzyme and a subsequent decrease of the flux through the tricarboxylic acid cycle. In yeast, a negative correlation between metabolite and enzyme concentration levels was found (57), and metabolic regulation was found to play a vital role in significantly altering the fluxes through glycolytic enzymes (58). By combining flux analysis with proteome and phosphoproteome analyses, Oliveira et al. (59) demonstrated in yeast the role of phosphorylation and posttranslational modifications in modulating the flux through nine key enzymes of central metabolism under five nutritional conditions. A different study, dealing with multiple knockout mutants of E. coli, revealed the robustness of central carbon metabolism in handling deletions. The possible modifications are counteracted by a change in metabolite levels linked to the deleted enzyme (60). The results support the hypothesis that central carbon metabolism is the part of a metabolic network where tight regulation dominates. The majority of flux modifications are effectively regulated at metabolic and posttranslational levels and are not regulated through mRNA synthesis, which is evident from the transcriptional silence of the reactions involved in the interconversion of the 12 biomass precursor metabolites despite changes in flux (Fig. 7). This observation invites reformulation of genetic engineering approaches based on transcriptional modifications to increase the flux through central metabolic reactions, because regulation of central carbon metabolism is not (only) exerted by gene expression but resides to a large extent in posttranscriptional and posttranslational modifications.
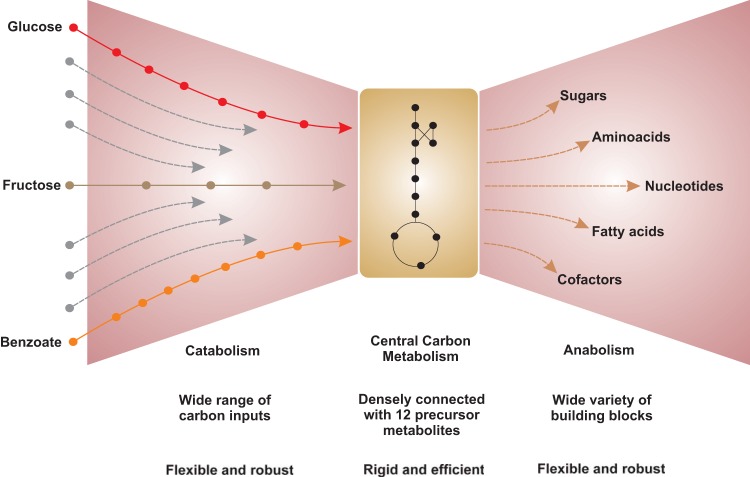
Using P. putida as an example, we provided a dedicated experimental design to elucidate responses and define central carbon metabolism. By our definition, the ED and the EMP pathways are not a part of central carbon metabolism, as they are transcriptionally regulated, allowing activation and repression of the corresponding genes by demand. The functional operations of these sugar-driven (ED and EMP) pathways are similar to those of the β-ketoadipate pathway, a classical biodegradation pathway, and hence are part of the fan-in reactions where the catabolic pathways are organized (Fig. 8). These results support the classical/theoretical view of a bacterial metabolic network as a bow tie with functional modules (5, 6, 61, 62), where nutrients are catabolized to produce energy, redox cofactors, and the 12 biomass precursor metabolites, which are then synthesized into larger building blocks.
FIG 8.
Functional structure and organization of carbon metabolism in Pseudomonas putida. The metabolic network structure is organized as a “bow tie.” The bow tie is decomposed into three modules; i.e., the catabolic and anabolic reactions are organized as fan-in and fan-out parts of the bow tie, and the knot part of the bow tie is comprised of the 12 biomass precursor metabolites that form the central carbon metabolism.
However, the reactions included in central carbon metabolism most likely differ in individual organisms depending on the ecological niche occupied by the organism. In several Archaea, peripheral modules like the ED and EMP pathways may have diverse structures and functionalities, such as gluconeogenesis, or are not present at all, such as the pentose phosphate pathway (63, 64). The present study might therefore provide new leads for how, through structure, changes in operation can be efficiently introduced into rational strain engineering approaches to utilize the abundant plant-derived aromatic material lignin, which today has the potential to regain its central position as a renewable feedstock.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matthias Reuss, Ralf Takors, and Burkhard Tümmler for laboratory support and fruitful discussions. We also thank Alexander Dietrich, Ilona Grimm, and Lara Bogner for their skillful assistance in fermentation and analytical techniques.
This project was supported by the International Max Planck Research School in Chemical Biology (IMPRS-CB) and was partially supported by the German Ministry of Science and Education (BMBF project ERA-NET SysMO, no. 0313980A) (VAPMdS) and by the Ministry of Innovation, Science, Research and Technology of North Rhine-Westphalia (Bio.NRW, Technology Platform Biocatalysis, RedoxCell).
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 20 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01643-14.
REFERENCES
- 1.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. 1999. From molecular to modular cell biology. Nature 402:C47–C52. 10.1038/35011540 [DOI] [PubMed] [Google Scholar]
- 2.Sauer U, Heinemann M, Zamboni N. 2007. Getting closer to the whole picture. Science 316:550–551. 10.1126/science.1142502 [DOI] [PubMed] [Google Scholar]
- 3.Barabási A-L, Oltvai ZN. 2004. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 5:101–113. 10.1038/nrg1272 [DOI] [PubMed] [Google Scholar]
- 4.Jeong H, Tombor B, Albert R, Oltval ZN, Barabasi A-L. 2000. The large-scale organization of metabolic networks. Nature 407:651–654. 10.1038/35036627 [DOI] [PubMed] [Google Scholar]
- 5.Ma H-W, Zeng A-P. 2003. The connectivity structure, giant strong component and centrality of metabolic networks. Bioinformatics 19:1423–1430. 10.1093/bioinformatics/btg177 [DOI] [PubMed] [Google Scholar]
- 6.Csete M, Doyle J. 2004. Bow ties, metabolism and disease. Trends Biotechnol. 22:446–450. 10.1016/j.tibtech.2004.07.007 [DOI] [PubMed] [Google Scholar]
- 7.Wackett LP. 2003. Pseudomonas putida—a versatile biocatalyst. Nat. Biotechnol. 21:136–137. 10.1038/nbt0203-136 [DOI] [PubMed] [Google Scholar]
- 8.Díaz E. 2004. Bacterial degradation of aromatic pollutants: a paradigm of metabolic versatility. Int. Microbiol. 7:173–180 [PubMed] [Google Scholar]
- 9.Kiener A. April 1992. Microbiological oxidation of methyl groups in heterocycles. US patent 5,104,798. [Google Scholar]
- 10.Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B. 2001. Industrial biocatalysis today and tomorrow. Nature 409:258–268. 10.1038/35051736 [DOI] [PubMed] [Google Scholar]
- 11.Erb RW, Eichner CA, Wagner-Döbler I, Timmis KN. 1997. Bioprotection of microbial communities from toxic phenol mixtures by a genetically designed pseudomonad. Nat. Biotechnol. 15:378–382. 10.1038/nbt0497-378 [DOI] [PubMed] [Google Scholar]
- 12.Espinosa-Urgel M, Kolter R, Ramos J-L. 2002. Root colonization by Pseudomonas putida: love at first sight. Microbiology 148:341–343 [DOI] [PubMed] [Google Scholar]
- 13.Koebmann BJ, Westerhoff HV, Snoep JL, Nilsson D, Jensen PR. 2002. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J. Bacteriol. 184:3909–3916. 10.1128/JB.184.14.3909-3916.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingraham JL, Maaløe O, Neidhardt FC. 1983. Growth of the bacterial cell, p 87–173. Sinauer Associates, Inc, Sunderland, MA [Google Scholar]
- 15.Neidhardt FC, Ingraham JL, Schaechter M. 1992. Physiology of the bacterial cell. A molecular approach, p 133–173 Sinauer Associates, Inc, Sunderland, MA. [Google Scholar]
- 16.Romano AH, Conway T. 1996. Evolution of carbohydrate metabolic pathways. Res. Microbiol. 147:448–455. 10.1016/0923-2508(96)83998-2 [DOI] [PubMed] [Google Scholar]
- 17.Stettner AI, Segre D. 2013. The cost of efficiency in energy metabolism. Proc. Natl. Acad. Sci. U. S. A. 110:9629–9630. 10.1073/pnas.1307485110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keasling JD. 2010. Manufacturing molecules through metabolic engineering. Science 330:1355–1358. 10.1126/science.1193990 [DOI] [PubMed] [Google Scholar]
- 19.Nielsen J. 2011. Transcriptional control of metabolic fluxes. Mol. Syst. Biol. 7:478. 10.1038/msb.2011.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vickers CE, Blank LM, Krömer JO. 2010. Chassis cells for industrial biochemical production. Nat. Chem. Biol. 6:875–877. 10.1038/nchembio.484 [DOI] [PubMed] [Google Scholar]
- 21.Noor E, Eden E, Milo R, Alon U. 2010. Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol. Cell 39:809–820. 10.1016/j.molcel.2010.08.031 [DOI] [PubMed] [Google Scholar]
- 22.Schuetz R, Kuepfer L, Sauer U. 2007. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol. Syst. Biol. 3:119. 10.1038/msb4100162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerosa L, Sauer U. 2011. Regulation and control of metabolic fluxes in microbes. Curr. Opin. Biotechnol. 22:566–575. 10.1016/j.copbio.2011.04.016 [DOI] [PubMed] [Google Scholar]
- 24.Jacob F, Monod J. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3:318–356. 10.1016/S0022-2836(61)80072-7 [DOI] [PubMed] [Google Scholar]
- 25.Alifano P, Fani R, Lio P, Lazcano A, Bazzicalupo M, Carlomagno MS, Bruni CB. 1996. Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol. Rev. 60:44–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blasi F, Bruni CB. 1981. Regulation of the histidine operon: translation-controlled transcription termination (a mechanism common to several biosynthetic operons). Curr. Top. Cell. Regul. 19:1–45. 10.1016/B978-0-12-152819-5.50018-X [DOI] [PubMed] [Google Scholar]
- 27.ter Kuile BH, Westerhoff HV. 2001. Transcriptome meets metabolome: hierarchical and metabolic regulation of the glycolytic pathway. FEBS Lett. 500:169–171. 10.1016/S0014-5793(01)02613-8 [DOI] [PubMed] [Google Scholar]
- 28.Bagdasarian M, Lurz R, Riickert B, Bagdasarian MM, Frey J, Timmis KN. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSFlOlO-derived, and a host-vector system for gene cloning in Pseudomonas. Gene 16(1–3):237–247 [DOI] [PubMed] [Google Scholar]
- 29.Nogales J, Palsson BØ, Thiele I. 2008. A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC Syst. Biol. 2:79. 10.1186/1752-0509-2-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emmerling M, Dauner M, Ponti A, Fiaux J, Hochuli M, Szyperski T, Wu K, Bailey JE, Sauer U. 2002. Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J. Bacteriol. 184:152–164. 10.1128/JB.184.1.152-164.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer E, Zamboni N, Sauer U. 2004. High-throughput metabolic flux analysis based on gas chromatography-mass spectrometry derived 13C constraints. Anal. Biochem. 325:308–316. 10.1016/j.ab.2003.10.036 [DOI] [PubMed] [Google Scholar]
- 32.Zamboni N, Fischer E, Sauer U. 2005. FiatFlux—a software for metabolic flux analysis from 13C-glucose experiments. BMC Bioinformatics 6:209. 10.1186/1471-2105-6-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuste L, Hervás AB, Canosa I, Tobes R, Jiménez JI, Nogales J, Pérez-Pérez MM, Santero E, Díaz E, Ramos J-L, de Lorenzo V, Rojo F. 2006. Growth phase-dependent expression of the Pseudomonas putida KT2440 transcriptional machinery analysed with a genome-wide DNA microarray. Environ. Microbiol. 8:165–177. 10.1111/j.1462-2920.2005.00890.x [DOI] [PubMed] [Google Scholar]
- 34.Frank S, Klockgether J, Hagendorf P, Geffers R, Schock U, Pohl T, Davenport CF, Tummler B. 2011. Pseudomonas putida KT2440 genome update by cDNA sequencing and microarray transcriptomics. Environ. Microbiol. 13:1309–1326. 10.1111/j.1462-2920.2011.02430.x [DOI] [PubMed] [Google Scholar]
- 35.Quackenbush J. 2002. Microarray data normalization and transformation. Nat. Genet. 32:496–501. 10.1038/ng1032 [DOI] [PubMed] [Google Scholar]
- 36.Preiss J. 1984. Bacterial glycogen synthesis and its regulation. Annu. Rev. Microbiol. 38:419–458. 10.1146/annurev.mi.38.100184.002223 [DOI] [PubMed] [Google Scholar]
- 37.Kessler B, Witholt B. 2001. Factors involved in the regulatory network of polyhydroxyalkanoate metabolism. J. Biotechnol. 86:97–104. 10.1016/S0168-1656(00)00404-1 [DOI] [PubMed] [Google Scholar]
- 38.del Castillo T, Duque E, Ramos JL. 2008. A set of activators and repressors control peripheral glucose pathways in Pseudomonas putida to yield a common central intermediate. J. Bacteriol. 190:2331–2339. 10.1128/JB.01726-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.del Castillo T, Ramos JL, Rodríguez-Herva JJ, Fuhrer T, Sauer U, Duque E. 2007. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. J. Bacteriol. 189:5142–5152. 10.1128/JB.00203-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiménez JI, Miñambres B, Luis J, Díaz E. 2002. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4:824–841. 10.1046/j.1462-2920.2002.00370.x [DOI] [PubMed] [Google Scholar]
- 41.Moreno R, Rojo F. 2008. The target for the Pseudomonas putida Crc global regulator in the benzoate degradation pathway is the BenR transcriptional regulator. J. Bacteriol. 190:1539–1545. 10.1128/JB.01604-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerstmeir R, Wendisch VF, Schnicke S, Ruan H, Farwick M, Reinscheid D, Eikmanns BJ. 2003. Acetate metabolism and its regulation in Corynebacterium glutamicum. J. Biotechnol. 104:99–122. 10.1016/S0168-1656(03)00167-6 [DOI] [PubMed] [Google Scholar]
- 43.Laporte DC, Koshland DE., Jr 1983. Phosphorylation of isocitrate dehydrogenase as a demonstration of enhanced sensitivity in covalent regulation. Nature 305:286–290. 10.1038/305286a0 [DOI] [PubMed] [Google Scholar]
- 44.Blank LM, Ionidis G, Ebert BE, Bühler B, Schmid A. 2008. Metabolic response of Pseudomonas putida during redox biocatalysis in the presence of a second octanol phase. FEBS J. 275:5173–5190. 10.1111/j.1742-4658.2008.06648.x [DOI] [PubMed] [Google Scholar]
- 45.Sawyer MH, Baumann P, Baumann L, Berman SM, Cánovas JL, Berman RH. 1977. Pathways of D-fructose catabolism in species of Pseudomonas. Arch. Microbiol. 112:49–55. 10.1007/BF00446653 [DOI] [PubMed] [Google Scholar]
- 46.Van Dijken JP, Quayle JR. 1977. Fructose metabolism in four Pseudomonas species. Arch. Microbiol. 114:281–286. 10.1007/BF00446874 [DOI] [PubMed] [Google Scholar]
- 47.Chavarria M, Kleijn RJ, Sauer U, Pfluger-Grau K, de Lorenzo V. 2012. Regulatory tasks of the phosphoenolpyruvate-phosphotransferase system of Pseudomonas putida in central carbon metabolism. mBio 3(2):e00028-12. 10.1128/mBio.00028-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chavarria M, Nikel PI, Perez-Pantoja D, de Lorenzo V. 2013. The Entner-Doudoroff pathway empowers Pseudomonas putida KT2440 with a high tolerance to oxidative stress. Environ. Microbiol. 15:1772–1785. 10.1111/1462-2920.12069 [DOI] [PubMed] [Google Scholar]
- 49.Ampe F, Lindley ND. 1996. Flux limitations in the ortho pathway of benzoate degradation of Alcaligenes eutrophus: metabolite overflow and induction of the meta pathway at high substrate concentrations. Microbiology 142:1807–1817. 10.1099/13500872-142-7-1807 [DOI] [PubMed] [Google Scholar]
- 50.Laporte DC, Walsh K, Koshland DE. 1984. The branch point effect. Ultrasensitivity and subsensitivity to metabolic control. J. Biol. Chem. 259:14068–14075 [PubMed] [Google Scholar]
- 51.Walsh K, Koshland DE. 1985. Branch point control by the phosphorylation state of isocitrate dehydrogenase. A quantitative examination of fluxes during a regulatory transition. J. Biol. Chem. 260:8430–8437 [PubMed] [Google Scholar]
- 52.Bordel S, Agren R, Nielsen J. 2010. Sampling the solution space in genome-scale metabolic networks reveals transcriptional regulation in key enzymes. PLoS Comput. Biol. 6:e1000859. 10.1371/journal.pcbi.1000859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daran-Lapujade P, Jansen ML, Daran J-M, van Gulik W, de Winde JH, Pronk JT. 2004. Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiae. A chemostat culture study. J. Biol. Chem. 279:9125–9138. 10.1074/jbc.M309578200 [DOI] [PubMed] [Google Scholar]
- 54.Yang C, Hua Q, Shimizu K. 2002. Integration of the information from gene expression and metabolic fluxes for the analysis of the regulatory mechanisms in Synechocystis. Appl. Microbiol. Biotechnol. 58:813–822. 10.1007/s00253-002-0949-0 [DOI] [PubMed] [Google Scholar]
- 55.Even S, Lindley ND, Cocaign-Bousquet M. 2003. Transcriptional, translational and metabolic regulation of glycolysis in Lactococcus lactis subsp. cremoris MG 1363 grown in continuous acidic cultures. Microbiology 149:1935–1944. 10.1099/mic.0.26146-0 [DOI] [PubMed] [Google Scholar]
- 56.Edlin JD, Sundaram TK. 1989. Regulation of isocitrate dehydrogenase by phosphorylation in Escherichia coli K-12 and a simple method for determining the amount of inactive phosphoenzyme. J. Bacteriol. 171:2634–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fendt S-M, Buescher JM, Rudroff F, Picotti P, Zamboni N, Sauer U. 2010. Tradeoff between enzyme and metabolite efficiency maintains metabolic homeostasis upon perturbations in enzyme capacity. Mol. Syst. Biol. 6:356. 10.1038/msb.2010.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daran-Lapujade P, Rossell S, van Gulik WM, Luttik MA, de Groot MJ, Slijper M, Heck AJR, Daran J-M, de Winde JH, Westerhoff HV, Pronk JT, Bakker BM. 2007. The fluxes through glycolytic enzymes in Saccharomyces cerevisiae are predominantly regulated at posttranscriptional levels. Proc. Natl. Acad. Sci. U. S. A. 104:15753–15758. 10.1073/pnas.0707476104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oliveira AP, Ludwig C, Picotti P, Kogadeeva M, Aebersold R, Sauer U. 2012. Regulation of yeast central metabolism by enzyme phosphorylation. Mol. Syst. Biol. 8:623. 10.1038/msb.2012.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakahigashi K, Toya Y, Ishii N, Soga T, Hasegawa M, Watanabe H, Takai Y, Honma M, Mori H, Tomita M. 2009. Systematic phenome analysis of Escherichia coli multiple-knockout mutants reveals hidden reactions in central carbon metabolism. Mol. Syst. Biol. 5:306. 10.1038/msb.2009.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma H-W, Zhao X-M, Yuan Y-J, Zeng A-P. 2004. Decomposition of metabolic network into functional modules based on the global connectivity structure of reaction graph. Bioinformatics 20:1870–1876. 10.1093/bioinformatics/bth167 [DOI] [PubMed] [Google Scholar]
- 62.Zhao J, Yu H, Luo J-H, Cao Z-W, Li Y-X. 2006. Hierarchical modularity of nested bow-ties in metabolic networks. BMC Bioinformatics 7:386. 10.1186/1471-2105-7-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siebers B, Schonheit P. 2005. Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr. Opin. Microbiol. 8:695–705. 10.1016/j.mib.2005.10.014 [DOI] [PubMed] [Google Scholar]
- 64.Zaparty M, Siebers B. 2011. Physiology, metabolism, and enzymology of thermoacidophiles, p 601–639 In Horikoshi K. (ed), Extremophiles handbook. Springer, Tokyo, Japan [Google Scholar]
- 65.Thayer JR, Wheelis ML. 1982. Active transport of benzoate in Pseudomonas putida. J. Gen. Microbiol. 128:1749–1753 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.