Abstract
Bifidobacteria are Gram-positive inhabitants of the human gastrointestinal tract that have evolved close interaction with their host and especially with the host's immune system. The molecular mechanisms underlying such interactions, however, are largely unidentified. In this study, we investigated the immunomodulatory potential of Bifidobacterium bifidum MIMBb75, a bacterium of human intestinal origin commercially used as a probiotic. Particularly, we focused our attention on TgaA, a protein expressed on the outer surface of MIMBb75's cells and homologous to other known bacterial immunoactive proteins. TgaA is a peptidoglycan lytic enzyme containing two active domains: lytic murein transglycosylase (LT) and cysteine- and histidine-dependent amidohydrolase/peptidase (CHAP). We ran immunological experiments stimulating dendritic cells (DCs) with the B. bifidum MIMBb75 and TgaA, with the result that both the bacterium and the protein activated DCs and triggered interleukin-2 (IL-2) production. In addition, we observed that the heterologous expression of TgaA in Bifidobacterium longum transferred to the bacterium the ability to induce IL-2. Subsequently, immunological experiments performed using two purified recombinant proteins corresponding to the single domains LT and CHAP demonstrated that the CHAP domain is the immune-reactive region of TgaA. Finally, we also showed that TgaA-dependent activation of DCs requires the protein CD14, marginally involves TRIF, and is independent of Toll-like receptor 4 (TLR4) and MyD88. In conclusion, our study suggests that the bacterial CHAP domain is a novel microbe-associated molecular pattern actively participating in the cross talk mechanisms between bifidobacteria and the host's immune system.
INTRODUCTION
The human intestinal microbiota comprises more than 1,000 microbial taxa, which through evolution have adopted different strategies to interact with the host, from commensalism or symbiosis to parasitism (1). A well-established and rapidly growing body of literature demonstrates how deeply the intestinal microbiota is involved in several host physiologic dysfunctions such as obesity, diabetes, autoimmune diseases, and cancer (1, 2). Furthermore, there is scientific evidence that certain members of the microbiota more than others play a crucial role in maintaining a physiological homeostasis in the host (3, 4). For instance, bifidobacteria are Gram-positive inhabitants of the human gastrointestinal tract that have evolved a deep interaction with the host (5, 6, 7, 8). In particular, bifidobacteria, which colonize the human intestine immediately after birth, are the predominant taxon of the microbiota of breast-fed infants and affect the maturation of the host's immune system during the neonatal period, especially the TH1/TH2 balance (9, 10). Accordingly, a relation has been established between allergic diseases and bifidobacterial colonization (11, 12, 13, 14). In fact, allergic patients have exhibited lower Bifidobacterium counts than healthy control subjects. In addition, the species Bifidobacterium adolescentis and Bifidobacterium longum subsp. longum have been isolated from allergic infants as the predominant bifidobacteria, whereas the predominant ones isolated from age-matched healthy infants have been Bifidobacterium breve, Bifidobacterium longum subsp. infantis, and Bifidobacterium bifidum (13). These findings testify to a plausible link between bifidobacteria and atopy/tolerance balance (15, 16), with certain species such as B. bifidum being potentially crucial in limiting the development of a long-term TH2-skewed immunological memory in infants and, therefore, in preventing allergies. The ability of B. bifidum strains to interact with the host immune system has been reported in several studies (17, 18), but very little information is available concerning the molecular components that support the host-B. bifidum cross talk (8, 19).
Whereas adaptive immunity recognizes a microorganism by its specific microbial molecular components known as antigens, innate immunity relies on recognizing microbe-associated molecular patters (MAMPs), that is, conserved structures present within a class of microorganisms but not in the host tissue (20). Several MAMPs have been identified, such as lipopolysaccharide (LPS, also referred to as endotoxin), peptidoglycan, lipoteichoic acid, and bacterial CpG DNA motifs. However, many other yet uncharacterized MAMPs govern the intricate mechanisms of host-microbiota interactions and either help maintain or compromise immunological homeostasis. Consequently, it is critically important to study the immunological role of microbial molecular cell components to decipher the operating principles of the immune system. In this regard, our use of a reductionist molecular approach allowed us to define the immunological role of an intramolecular region belonging to an outer surface enzyme, TgaA, identified in an accompanying paper (21) by comparative genomic analysis in strain B. bifidum MIMBb75.
MATERIALS AND METHODS
Bifidobacterial culture conditions.
Bifidobacteria were grown under anaerobic conditions (Anaerocult A System; Merck, Darmstadt, Germany) at 37°C in prereduced DeMan-Rogosa-Sharpe (MRS) broth (Difco Laboratories, Inc., Detroit, MI) supplemented with 0.05% l-cysteine hydrochloride (cMRS).
Overproduction and purification of TgaA-derived recombinant proteins.
All enzymes and reagents for molecular biology reactions were from Fermentas (Vilnius, Lithuania) or New England BioLabs (EuroClone S.p.A., Pero, Italy). Commercial kits for the extraction and purification of nucleic acids were from MoBio Laboratories (Cabru s.a.s., Arcore, Italy) or Qiagen s.r.l. (Milan, Italy). Vectors pET-Tga, pET-ΔCHAP (where CHAP is cysteine- and histidine-dependent amidohydrolase/peptidase) and pET-ΔLT (where LT is lytic murein transglycosylase) (prepared as described in the accompanying paper [21]) were used for the isopropyl-β-d-thiogalactopyranoside (IPTG)-dependent overexpression in Escherichia coli of the recombinant proteins ΔSP-TgaA-His (containing both domains of TgaA protein; also named here recombinant TgaA, or rTgaA, and SPPelB-TgaA in the accompanying paper [21]), protein ΔSP-TgaA-ΔCHAP-His (containing only the LT domain; rLT), and ΔSP-TgaA-ΔLT-His (containing only the CHAP domain; rCHAP), respectively. In brief, mutant E. coli strains were grown in 2× YT (yeast extract, tryptone) broth at 37°C for 2 h before 1 mM IPTG was added, and incubation was prolonged for 4 h. Afterwards, proteins were extracted under denaturing conditions with PerfectPro Ni-nitrilotriacetic acid (NTA) agarose (5 Prime; Eppendorf Italia, s.r.l., Milan, Italy) according to the manufacturer's instructions. The molecular mass of recombinant ΔSP-TgaA-His, ΔSP-TgaA-ΔCHAP-His, and ΔSP-TgaA-ΔLT-His proteins was confirmed by reverse-phase high-pressure liquid chromatography (RP-HPLC)/electrospray ionization mass spectrometry (ESI-MS) analysis under conditions reported by Taverniti et al. (22). A further purification and removal of residue LPS from recombinant proteins was carried out by analytical or preparative RP-HPLC. After acidification to pH 2 by addition of 0.1% of trifluoroacetic acid (TFA), aliquots of 0.1 ml of Ni-NTA-purified protein were injected into a Waters 600 HPLC system fitted with a VYDAC C4 (4.6 by 250 mm; 300-Å pore size; 5-μm particle size) column (Grace, Deerfield, IL). The eluents used for the separation were solvent A, consisting of 0.1% trifluoroacetic acid (TFA) in MilliQ-treated water, and solvent B, consisting of 0.1% TFA in acetonitrile. The protein separation was performed at room temperature by using a linear elution gradient (60% to 30% of solvent A in 25 min). Proteins were eluted at a flow rate of 0.8 ml min−1 and monitored at 220 and 280 nm. Protein-containing fractions were collected, and the degree of purification was checked by SDS-PAGE. Finally, eluted fractions were lyophilized and stored at −20°C.
Generation of ΔSP-TgaA-ΔLT-His-specific antibodies and immunogold labeling.
The preparation of an antibody against the recombinant protein ΔSP-TgaA-ΔLT-His (rCHAP) was raised in rabbits by Primm s.r.l. (Milan, Italy). Immunogold labeling was performed as described in the companion paper (21).
Expression of TgaA in Bifidobacterium longum NCC2705.
All cloning steps for the preparation of the vectors used to transform B. longum NCC2705 were carried out in E. coli XL1-Blue (see Fig. S1 in the supplemental material). In detail, the promoter region from phage T5 (PT5) was obtained from vector pGBL8b (23) through digestion with restriction enzymes BamHI and HindIII. The resulting DNA fragment was introduced in vector pUC19 in the same restriction sites, yielding vector pUC-T5. At the same time, the DNA region containing the gene coding for the chloramphenicol acetyltransferase of pC194 (a natural plasmid of Staphylococcus aureus) with its original promoter was obtained through PCR with primers catBC-f (5′-CATGGATCCATCGATCTGCA-3′; restriction sites BamHI and ClaI are underlined) and catS-r 5′-TACCCGGAGCTCCTCTAGA-3′; the restriction site SalI is underlined) from vector pGBL8b. The resulting amplicon was digested with BamHI and SalI and cloned in pUC-T5 digested with the same restriction enzymes, yielding vector pUT5cat. Subsequently, the gene tgaA was amplified from the chromosomal DNA of B. bifidum MIMBb75 with primers tgaDie-f (5′-AACTTCGTGAGATCTGCCGCGTCCCGCGCCAT-3′; restriction site BglII is underlined) and tgaC-r (5′-GCTCATCGATTTTACTTTCCTT-3′; restriction site ClaI is underlined); the amplicon was digested with enzymes BglII and ClaI and cloned in sites BamHI and ClaI of vector pUT5cat, yielding vector pUTgaCat. Plasmid pUTgaCat was then digested with HindIII and XbaI; then, the restriction fragment containing the gene tgaA was extracted from the agarose gel, purified, and cloned in the same restriction sites of vector pGOSBif33 (24), yielding the final vector pTgaBif5, which was employed for the expression of tgaA in B. longum. The control vector pT5CatBif21 (empty vector) was prepared according to the same protocol, with the exception of the cloning step for the introduction of gene tgaA, which was not carried out. Finally, vectors pTgaBif5 and pT5CatBif21 were introduced in B. longum NCC2705 (courteously provided by Nestlé Research Center, Lausanne, Switzerland) as previously described (23, 24).
RT-PCR.
The expression of the gene tgaA from plasmid pTgaBif5 was verified in B. longum NCC2705 by reverse transcription-PCR (RT-PCR). For the extraction of RNA, 20 ml of exponentially growing cultures of B. bifidum MIMBb75 were centrifuged for 10 min at 4,000 × g at 4°C in the presence of RNA-Later (Ambion). The pellet was then immediately frozen in liquid nitrogen and subjected to RNA extraction using a previously described method, which includes a DNase treatment (25). The quality and integrity of the RNA were checked by Experion (Bio-Rad) analysis. cDNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad), according to the supplier's instructions. PCR was performed with primers B-7x (5′-CACAGTTCATACCGTCCACA-3′) and Bext-f-II (5′-GTAGTTGGTGGTCTCCGTGA-3′) according to the following PCR protocol: 1 cycle of 95°C for 3 min, 39 cycles consisting of 95°C for 30 s, 58°C for 40 s and 72°C for 30 s, and a final elongation step of 72°C for 7 min. The obtainment of an amplicon with the expected size of 260 bp demonstrated the presence of tgaA mRNA in the recombinant NCC2705 strain.
Preparation and analysis of immunofluorescent B. bifidum MIMBb75.
B. bifidum MIMBb75 cells were cultivated until early stationary growth phase, harvested by centrifugation, washed once with deionized sterile water, and incubated at room temperature for 10 min with ΔSP-TgaA-ΔLT-His antiserum (1:100 diluted in deionized water). Subsequently, bacterial cells were centrifuged, resuspended in a solution of Cy5-conjugated goat anti-rabbit secondary antibody (Molecular Probes, Eugene, OR) (1:10 diluted in deionized water), and incubated for 10 min at room temperature in the dark. Afterwards, stained cells were visualized with a fluorescence optical digital microscope (Leica DM1000; Leica Microsystems, Wetzlar, Germany) at a magnification of ×1,000.
Study of BMDC activation.
Dendritic cells (DCs) were obtained in vitro from bone marrow hematopoietic precursors isolated from C57BL/6 mouse femurs as described previously (26). In brief, hematopoietic precursors were recovered from mouse femoral bone marrow and resuspended in conditioned medium (complete medium supplemented with 10% of the growth supernatant of granulocyte-macrophage colony-stimulating factor [GM-CSF]-transduced B16 cells). About 7 × 106 cells were plated in 100-mm suspension plates. The proportion of CD11c+ cells (corresponding to dendritic cells) was monitored periodically by flow cytometry until it reached 90% (ca. 8 days). The bone marrow-derived dendritic cells (BMDCs) were then used for bacterial activation assays. On the day of bacterial infection, BMDCs were plated at a concentration of 0.5 million per ml in 96-well plates (105 cells/well). After 1 h, BMDCs were incubated with four different concentrations of bacterial cells for 2 h, washed with saline, resuspended in a culture medium containing penicillin G, streptomycin, tetracycline, and gentamicin, and incubated overnight. Finally, interleukin-2 (IL-2) and tumor necrosis factor alpha (TNF-α) in the supernatant were quantified by enzyme-linked immunosorbent assay (ELISA) using DuoSet kits (R&D Systems, Minneapolis, MN). The same procedure was followed to prepare BMDCs from four mutant mouse lines. Wild-type animals were supplied by Harlan Italy. Ticam1Lps2 (Trif−/−) mice were purchased from The Jackson Laboratory. Myd88−/− and Tlr4−/− mice were provided by S. Akira (IFReC, Japan). Cd14−/− mice were from CNRS d'Orléans (Orléans, France). D1 cell line was cultured in Iscove's modified Dulbecco's medium (IMDM; Sigma, St. Louis, MO) containing 10% heat-inactivated fetal bovine serum (Gibco-BRL, Gaithersburg, MD), 100 IU of penicillin, 100 μg ml−1 of streptomycin, 2 mM l-glutamine (all from Sigma), and 50 μM β-mercaptoethanol (in complete IMDM) with 30% supernatant from R1 medium (supernatant from NIH 3T3 fibroblasts transfected with GM-CSF). Unpaired Student's t tests were run to determine statistically significant differences.
RESULTS AND DISCUSSION
Recently, we generated a draft genome sequence of Bifidobacterium bifidum MIMBb75, a probiotic strain with demonstrated ability to interact with the host (19, 27–29). Comparative genomics revealed in MIMBb75's genome the presence of a gene encoding TgaA, a peptidoglycan-lytic enzyme which contains two conserved domains: lytic murein transglycosylase (LT; cd00254.3) and cysteine- and histidine-dependent amidohydrolase/peptidase (CHAP; pfam05257.4) (21).
The biomolecular composition of a bacterium's cell wall mainly determines the cross talk processes with the host. In fact, most known bacterial molecules governing bacterial adhesion to epithelia or interaction with immune cells are cell wall constituents (lipopolysaccharides, teichoic and lipoteichoic acids, murein, and proteinaceous adhesins) or cell wall appendages (flagella, pili, and fimbriae) (6, 8, 19, 22). Interestingly, TgaA protein was found to be abundantly expressed and located on the outer surface of MIMBb75 bacterial cells (21). Furthermore, notably, a search of the Conserved Domain Database (30) for the TgaA protein produced a significant match (E value of 8.30e-23) between the CHAP module and the conserved domain COG3942, which has been annotated as a surface antigen. The description of COG3942 originated from two studies that identified the CHAP domain-containing protein as an immune-reactive molecule of Listeria monocytogenes (31) and Streptococcus pyogenes (32). Stimulated by the above data, we evaluated the potential contribution of the TgaA protein to the immunomodulatory activity of the strain MIMBb75.
We assessed the immunological properties of B. bifidum MIMBb75 in parallel and independently of the TgaA protein study and discovered through our immunological model a peculiar immunomodulatory capacity of this strain. In particular, we focused on dendritic cells (DCs), the sentinels of the immune system present at various mucosal sites and especially in the intestinal mucosa. At this site, they may sense the intestinal lumen content (including bacteria) via transepithelial processes by responding to the ligation of the DCs' specific microbe recognition receptors (MRRs) to MAMPs (such as LPS, peptidoglycan, and bacterial and viral nucleic acids) to trigger immune responses (33, 34). Particularly, upon activation and migration to secondary lymphoid organs, DCs induce immunocompetence in lymphocytes through antigen presentation and cytokine production. Hence, DCs form a link between innate and adaptive immunity and are considered arbiters of immunological tolerance (34).
In DCs, a major outcome of the ligation of pattern recognition receptors (PRRs) may be the production of interleukin-2 (IL-2), which has several effects. In fact, DC-derived IL-2 increases gamma interferon (IFN-γ) production by the natural killer (NK) cells (35), enhances T cell responses in both mice (26) and humans (36), regulates thymic development (37), and promotes regulatory T cell (Treg) expansion and function (38). Furthermore, it has been linked with TH1-skewing stimuli (35). Inducing IL-2 production in DCs can thus steer the adaptive immune system toward preventing TH2 cell-dominated immune responses, such as those associated with allergy (39, 40).
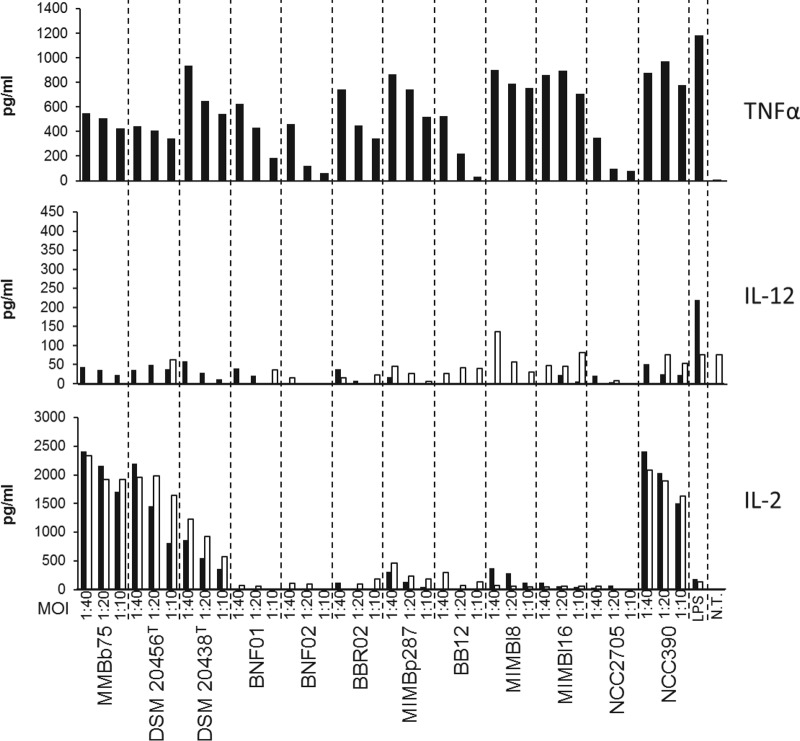
Our experiments showed that out of 12 bifidobacterial strains belonging to six different species and subspecies, the strain B. bifidum MIMBb75 was among the strongest inducers of IL-2 production by murine bone marrow-derived DCs (BMDCs) (Fig. 1).
FIG 1.
ELISA quantification of cytokines produced by dendritic cells after stimulation with 12 different strains belonging to the genus Bifidobacterium and 10 μg/ml of lipopolysaccharides (LPS) from E. coli. NT, sample not treated. One representative experiment out of three performed is shown. Black bars, bone marrow-derived dendritic cells (BMDCs); white bars, D1 cell line. All strains were tested at four different multiplicities of induction (MOI; DC/bacteria ratio). Strains belong to the following bifidobacterial species: B. bifidum (MIMBb75, DMS 20456T, NCC390); B. longum subsp. infantis (BNF01, BNF02); B. breve (BBR02); B. pseudocatenulatum (DSM20438T, MIMBp287); B. longum subsp. longum (MIMBl8, MIMBl16, NCC2705); B. animalis subsp. lactis (BB12).
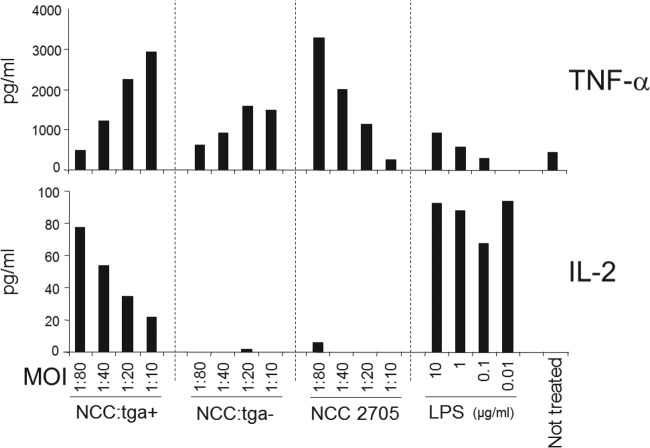
Subsequently, we studied the potential immunomodulatory activity of TgaA with the same immunological model we used for bifidobacterial cells. Initially, we cloned a complete tgaA gene (including the original signal sequence for exporting the protein at a cell wall level) in Bifidobacterium longum NCC2705 using a shuttle vector previously developed and used for successful expression of heterologous genes in B. longum (23, 24). We followed this strategy because all of our previous efforts to knock out tgaA had been unsuccessful since this species has been proved to be recalcitrant to genetic manipulation (41, 42). We selected B. longum NCC2705 as the host for tgaA expression after analysis of its whole genome revealed that the tgaA gene as absent in this human intestinal bacterium. Furthermore, our immunological data (Fig. 1) showed that the strain NCC2705, unlike MIMBb75, is incapable of inducing IL-2 expression by BMDCs. We named the recombinant strain we obtained NCC:tga+. After verifying by reverse transcription-PCR (RT-PCR) and immunogold labeling (see Fig. S2A and B in the supplemental material) that NCC:tga+ expressed the recombinant tgaA gene, we incubated BMDCs with supernatant-free wild-type live cells and recombinant NCC2705 strains and then measured by ELISA the levels of the secreted tumor necrosis factor alpha (TNF-α) and IL-2. Secretion by BMDCs of the TNF-α cytokine, the principal marker of DC activation, was induced by all bacterial preparations (Fig. 2). In contrast, our experiments showed a significant secretion of IL-2 only when DCs were stimulated with the recombinant strain NCC:tga+ (i.e., NCC2705 transformed with the vector containing the gene tgaA) and not with control preparations (wild-type NCC2705 and NCC2705 transformed with the empty vector) (Fig. 2). Therefore, our data suggest that the TgaA protein may be directly involved in inducing the cytokine IL-2 in dendritic cells.
FIG 2.
ELISA quantification of cytokine production by BMDCs after stimulation with E. coli lipopolysaccharides (LPS), Bifidobacterium longum NCC2705, and its recombinant strains NCC:tga+ (expressing TgaA protein) and NCC:tga− (harboring the empty vector). All samples were tested at four different multiplicities of infection (MOI; BMDC/bacteria ratio). One representative experiment out of two performed is shown.
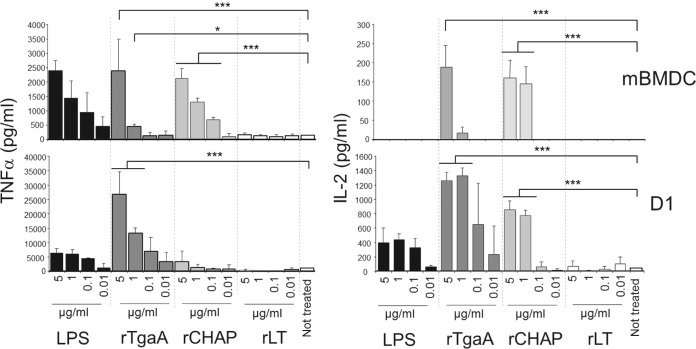
To further test our hypothesis, we thoroughly purified from E. coli cells the recombinant proteins rTgaA, rLT, and rCHAP by Ni-NTA affinity chromatography and subsequent RP-HPLC to remove any residual LPS and extraneous proteins (Fig. 3). Purified proteins were then used to stimulate two dendritic cell models: BMDCs and the D1 cell line, which is a long-term growth factor-dependent immature myeloid (CD11c+ CD8α−) DC line of splenic origin. Our results show clearly that the rTgaA protein can in vitro activate dendritic cells and induce dose-dependent secretion of IL-2 (Fig. 4). A similar stimulatory effect was found with the recombinant protein rCHAP, whereas rLT activated both dendritic cell models only marginally but could not markedly trigger IL-2 expression (Fig. 4). Therefore, the TgaA protein activates DCs and triggers IL-2 secretion by DCs by means of the CHAP domain.
FIG 3.
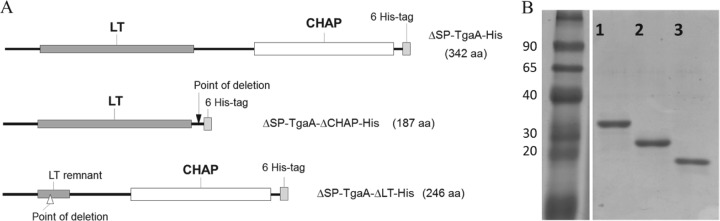
Preparation of TgaA-derived recombinant proteins. (A) Functional map of the recombinant proteins originated from a tgaA gene sequence and produced by overexpression in E. coli BL21(DE3) pLysS using the pET26b(+) expression vector. (B) SDS-PAGE of the purified six-His-tagged TgaA-derived recombinant proteins. Lane 1, ΔSP-TgaA-His (containing both TgaA domains; rTgaA); lane 2, ΔSP-TgaA-ΔLT-His (containing the CHAP domain; rCHAP), lane 3, ΔSP-TgaA-ΔCHAP-His (containing the LT domain; rLT). The relative molecular masses (in kDa) of standard proteins are indicated on the left. aa, amino acids.
FIG 4.
ELISA quantification of cytokine production by BMDCs and the D1 cell line stimulated with LPS or TgaA-derived recombinant proteins: rTgaA corresponds to ΔSP-TgaA-His, which contains both TgaA domains; rLT corresponds to ΔSP-TgaA-ΔCHAP-His, which contains only the LT domain; rCHAP corresponds to ΔSP-TgaA-ΔLT-His, which contains only the CHAP domain. Statistically significant differences were determined by an unpaired Student's t test (*, P < 0.05; ***, P < 0.001). mBMDC, murine BMDCs.
Finally, we studied the pathway induced by the protein TgaA in DCs. Since TgaA can induce TNF-α production by DCs and since the cluster of differentiation 14 (CD14) protein is essential in inducing such cytokines in LPS-stimulated DCs (43), we first tested if CD14 might have a role also in recognizing TgaA. As shown in Fig. 5, BMDC activation was totally abolished in the absence of CD14. Since CD14 is the best characterized as a coreceptor for Toll-like receptor 4 (TLR4), we then tested if BMDCs isolated from knockout mice lacking the expression of TLR4 can be activated by TgaA. According to our data, the receptor TLR4 was not necessary to induce TNF-α in BMDCs by TgaA (Fig. 5). Therefore, whereas LPS required TLR4 to induce TNF-α (43), the protein TgaA directly activated the downstream pathways through CD14 or involved CD14 for its presentation to a different unknown receptor, independently of TLR4.
FIG 5.
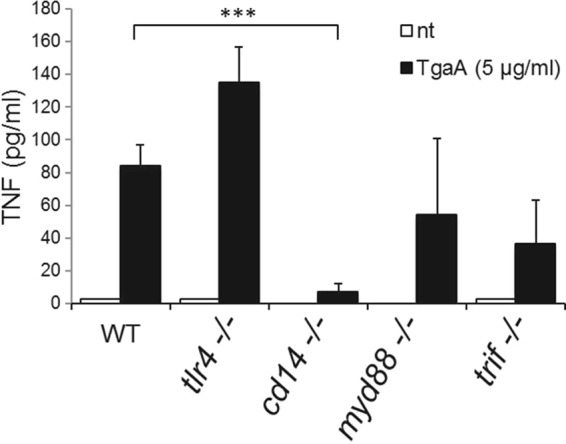
ELISA quantification of TNF-α produced by rTgaA-stimulated BMDCs isolated from wild-type (WT) and knockout mice. tlr4−/−, Toll-like receptor 4 (TLR4) KO mutant mouse; cd14−/−, CD14 protein KO mutant mouse; myd88−/−, myeloid differentiation primary response gene 88 (MYD88) KO mutant mouse; trif−/−, Toll IL-1 receptor (TIR) domain-containing adaptor-inducing IFN-β (TRIF) KO mutant mouse. Statistically significant differences were determined by an unpaired Student's t test (***, P < 0.001). nt, not treated.
To test if other TLRs might be important to recognizing TgaA by DCs, we used BMDCs derived from mice lacking the transducer of TLR signaling. In particular, TLR downstream signaling can occur via two pathways, which depend on two proteins: (i) the myeloid differentiation primary response gene 88 (MyD88) and (ii) the Toll IL-1 receptor (TIR) domain-containing adaptor-inducing IFN-β (TRIF) (44). Surprisingly, we found that without a functional TRIF or MyD88, production of TNF-α by TgaA-stimulated BMDCs was largely preserved (Fig. 5), suggesting that, in dendritic cells, the downstream signaling induced by TgaA could follow a CD14-dependent, TLR-independent signaling pathway.
Collectively, the above data show that TgaA is a cell wall protein of B. bifidum MIMBb75 and contains a C-terminal domain that initiates a CD14-dependent and TLR4-independent signaling pathway, which only partially involves the TRIF adapter protein.
A putative CHAP coding sequence is present in many bacterial genomes and is included in a number of different deduced protein architectures (45). Interestingly, several proteins from Gram-positive bacteria that include the CHAP domain actively interact with the host's immune system. For instance, the Streptococcus pyogenes SibA protein binds all immunoglobulin G (IgG) subclasses, the Fc and Fab fragments, and also IgA and IgM (32). PcsB, another CHAP-containing protein of Streptococcus spp. and a known antigen of Streptococcus pneumoniae, is capable of activating T cells in humans by inducing strong IL-17A responses (46). Furthermore, P40, a CHAP-containing protein produced by Lactobacillus rhamnosus GG (perhaps the most studied probiotic strain so far), can inhibit cytokine-induced epithelial cell apoptosis, reduce TNF-induced colon epithelial damage, and promote cell growth in human and mouse colon epithelial cells and cultured mouse colon explants (47, 48). In addition, our study showed that TgaA from B. bifidum MIMBb75 is a cell surface-exposed molecule capable alone through its C-terminal CHAP domain of inducing DC activation and IL-2 production. Based on observed induction of IL-2 secretion, the TgaA stimulatory capacity here described may stimulate dendritic cells to acquire TH1 stimulatory capacity (35). Furthermore, the ability of B. bifidum MIMBb75 and its protein TgaA to induce IL-2 by DCs, together with the previous observation that B. bifidum strains induce an immune response affecting Treg/TH17 plasticity (49), supports the hypothesis that these commensal bacteria have a key role in mucosal tolerance. The presence of specific microbial stimuli, such as the CHAP-containing TgaA protein, may then have served as a way for commensals to coevolve with the host and produce mechanisms for maintaining homeostasis and controlled reactions in the adaptive immune system (i.e., prevent an imbalance in T-helper cell subsets).
Interestingly, a BLASTP search revealed the presence of a gene closely similar to the CHAP domain of TgaA (with more than 80% identity in the amino acid sequence) not only in B. bifidum but also in the genome of B. breve (e.g., GenBank accession number WP_016462813), which is another Bifidobacterium species typically associated with healthy breast-fed infants (13). Therefore, the presence in B. bifidum and other bifidobacteria of molecules similar to the TgaA protein and capable of stimulating the immune system could help explain the ability of these bacteria to protect the host and nurture the immune system in early life (15, 16).
Conclusions.
The results of this study can be summarized as follows: (i) both the cells of B. bifidum MIMBb75 and its surface protein TgaA can activate dendritic cells and trigger IL-2 production; (ii) TgaA activates dendritic cells through its CHAP amidase domain; and (iii) the TgaA-dependent activation of dendritic cells requires the protein CD14, only marginally involves TRIF, and is independent of TLR4 and MyD88.
The bacterium B. bifidum MIMBb75, which is of human origin, has probiotic properties (28), and its ability to interact with the host may partly depend on the presence of the CHAP domain of its TgaA surface protein. In conclusion, the bacterial CHAP domain may well be considered a novel, potentially widespread MAMP that may participate in the cross talk mechanisms among Gram-positive bacteria and their mammalian host. The possible physiological consequences of the ability of B. bifidum MIMBb75 and its TgaA protein to activate DCs are schematically summarized in Fig. 6.
FIG 6.
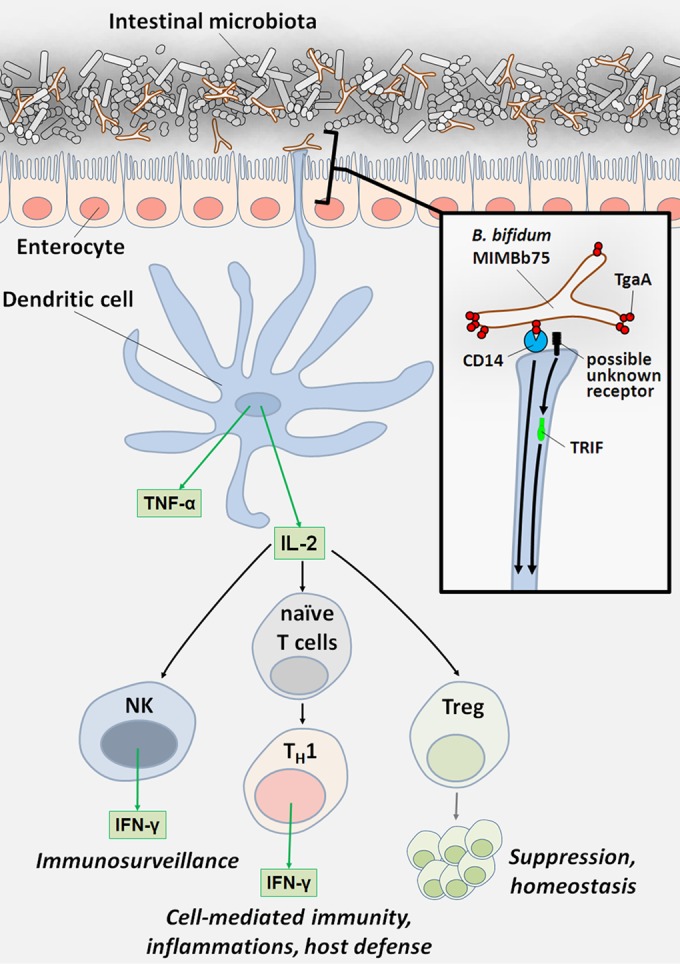
DC-mediated host immune response to TgaA protein from Bifidobacterium bifidum MIMBb75: possible consequences of IL-2 production. The contact between B. bifidum MIMBb75 and dendritic cells (DCs) determines DC activation and IL-2 secretion. Reportedly, stimuli able to induce IL-2 production by DCs are associated with a Th1-skewing of the immune responses, Treg lymphocyte proliferation, and natural killer (NK) cell activation (for a review, see reference 50).
Supplementary Material
ACKNOWLEDGMENT
This work was financially supported by Fondazione Cariplo (grant 2010-0678).
Footnotes
Published ahead of print 9 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00761-14.
REFERENCES
- 1.Tremaroli V, Backhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- 2.Tlaskalova-Hogenova H, Stepankova R, Kozakova H, Hudcovic T, Vannucci L, Tuckova L, Rossmann P, Hrncir T, Kverka M, Zakostelska Z, Klimesova K, Pribylova J, Bartova J, Sanchez D, Fundova P, Borovska D, Srutkova D, Zidek Z, Schwarzer M, Drastich P, Funda DP. 2011. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell. Mol. Immunol. 8:110–120. 10.1038/cmi.2010.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottiere HM, Dore J, Marteau P, Seksik P, Langella P. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 105:16731–16736. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belzer C, de Vos WM. 2012. Microbes inside-from diversity to function: the case of Akkermansia. ISME J. 6:1449–1458. 10.1038/ismej.2012.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U. S. A. 105:18964–18969. 10.1073/pnas.0809584105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, Motherway MO, Shanahan F, Nally K, Dougan G, van Sinderen D. 2012. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. U. S. A. 109:2108–2113. 10.1073/pnas.1115621109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. 2012. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 20:467–476. 10.1016/j.tim.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 8.Turroni F, Serafini F, Foroni E, Duranti S, Motherway MO, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, Sanchez B, Santoni A, Gioiosa L, Ferrarini A, Delledonne M, Margolles A, Piazza L, Palanza P, Bolchi A, Guglielmetti S, van Sinderen D, Ventura M. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc. Natl. Acad. Sci. U. S. A. 110:11151–11156. 10.1073/pnas.1303897110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaboriau-Routhiau V, Raibaud P, Dubuquoy C, Moreau MC. 2003. Colonization of gnotobiotic mice with human gut microflora at birth protects against Escherichia coli heat-labile enterotoxin-mediated abrogation of oral tolerance. Pediatr. Res. 54:739–746. 10.1203/01.PDR.0000086902.52137.C9 [DOI] [PubMed] [Google Scholar]
- 10.Dong P, Yang Y, Wang WP. 2010. The role of intestinal bifidobacteria on immune system development in young rats. Early Hum. Dev. 86:51–58. 10.1016/j.earlhumdev.2010.01.002 [DOI] [PubMed] [Google Scholar]
- 11.Bjorksten B, Naaber P, Sepp E, Mikelsaar M. 1999. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin. Exp. Allergy 29:342–346. 10.1046/j.1365-2222.1999.00560.x [DOI] [PubMed] [Google Scholar]
- 12.He F, Ouwehand AC, Isolauri E, Hashimoto H, Benno Y, Salminen S. 2001. Comparison of mucosal adhesion and species identification of bifidobacteria isolated from healthy and allergic infants. FEMS Immunol. Med. Microbiol. 30:43–47. 10.1111/j.1574-695X.2001.tb01548.x [DOI] [PubMed] [Google Scholar]
- 13.Ouwehand AC, Isolauri E, He F, Hashimoto H, Benno Y, Salminen S. 2001. Differences in Bifidobacterium flora composition in allergic and healthy infants. J. Allergy Clin. Immunol. 108:144–145. 10.1067/mai.2001.115754 [DOI] [PubMed] [Google Scholar]
- 14.Watanabe S, Narisawa Y, Arase S, Okamatsu H, Ikenaga T, Tajiri Y, Kumemura M. 2003. Differences in fecal microflora between patients with atopic dermatitis and healthy control subjects. J. Allergy Clin. Immunol. 111:587–591. 10.1067/mai.2003.105 [DOI] [PubMed] [Google Scholar]
- 15.Young SL, Simon MA, Baird MA, Tannock GW, Bibiloni R, Spencely K, Lane JM, Fitzharris P, Crane J, Town I, Addo-Yobo E, Murray CS, Woodcock A. 2004. Bifidobacterial species differentially affect expression of cell surface markers and cytokines of dendritic cells harvested. Clin. Diagn. Lab. Immunol. 11:686–690. 10.1128/CDLI.11.4.686-690.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menard O, Butel MJ, Gaboriau-Routhiau V, Waligora-Dupriet AJ. 2008. Gnotobiotic mouse immune response induced by Bifidobacterium sp. strains isolated from infants. Appl. Environ. Microbiol. 74:660–666. 10.1128/AEM.01261-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss G, Rasmussen S, Nielsen Fink L, Jarmer H, Nøhr Nielsen B, Frøkiaer H. 2010. Bifidobacterium bifidum actively changes the gene expression profile induced by Lactobacillus acidophilus in murine dendritic cells. PLoS One 5:e11065. 10.1371/journal.pone.0011065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turroni F, Taverniti V, Ruas-Madiedo P, Duranti S, Guglielmetti S, Lugli GA, Gioiosa L, Palanza P, Margolles A, van Sinderen D, Ventura M. 2014. Bifidobacterium bifidum PRL2010 modulates the host innate immune response. Appl. Environ. Microbiol. 80:730–740. 10.1128/AEM.03313-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guglielmetti S, Tamagnini I, Mora D, Minuzzo M, Scarafoni A, Arioli S, Hellman J, Karp M, Parini C. 2008. Implication of an outer surface lipoprotein in adhesion of Bifidobacterium bifidum to Caco-2 cells. Appl. Environ. Microbiol. 74:4695–4702. 10.1128/AEM.00124-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janeway CA., Jr 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor Symp. Quant. Biol. 54:1–13. 10.1101/SQB.1989.054.01.003 [DOI] [PubMed] [Google Scholar]
- 21.Guglielmetti S, Balzaretti S, Taverniti V, Miriani M, Milani C, Scarafoni A, Corona S, Arioli S, Santala V, Iametti S, Bonomi F, Ventura M, Mora D, Karp M. 2014. TgaA, a VirB1-like component belonging to a putative type IV secretion system of Bifidobacterium bifidum MIMBb75. Appl. Environ. Microbiol. 80:5161–5169. 10.1128/AEM.01413-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taverniti V, Stuknyte M, Minuzzo M, Arioli S, De Noni I, Scabiosi C, Cordova ZM, Junttila I, Hämäläinen S, Turpeinen H, Mora D, Karp M, Pesu M, Guglielmetti S. 2013. S-layer protein mediates the stimulatory effect of Lactobacillus helveticus MIMLh5 on innate immunity. Appl. Environ. Microbiol. 79:1221–1231. 10.1128/AEM.03056-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guglielmetti S, Ciranna A, Mora D, Parini C, Karp M. 2008. Construction, characterization and exemplificative application of bioluminescent Bifidobacterium longum biovar longum. Int. J. Food Microbiol. 124:285–290. 10.1016/j.ijfoodmicro.2008.03.033 [DOI] [PubMed] [Google Scholar]
- 24.Guglielmetti S, Karp M, Mora D, Tamagnini I, Parini C. 2007. Molecular characterization of Bifidobacterium longum biovar longum NAL8 plasmids and construction of a novel replicon screening system. Appl. Microbiol. Biotechnol. 74:1053–1061. 10.1007/s00253-006-0755-1 [DOI] [PubMed] [Google Scholar]
- 25.Ventura M, Zhang Z, Cronin M, Canchaya C, Kenny JG, Fitzgerald GF, van Sinderen D. 2005. The ClgR protein regulates transcription of the clpP operon in Bifidobacterium breve UCC 2003. J. Bacteriol. 187:8411–8426. 10.1128/JB.187.24.8411-8426.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Granucci F, Vizzardelli C, Pavelka N, Feau S, Persico M, Virzi E, Rescigno M, Moro G, Ricciardi-Castagnoli P. 2001. Inducible IL-2 production by dendritic cells revealed by global gene expression analysis. Nat. Immunol. 2:882–888. 10.1038/ni0901-882 [DOI] [PubMed] [Google Scholar]
- 27.Guglielmetti S, Tamagnini I, Minuzzo M, Arioli S, Parini C, Comelli E, Mora D. 2009. Study of the adhesion of Bifidobacterium bifidum MIMBb75 to human intestinal cell lines. Curr. Microbiol. 59:167–172. 10.1007/s00284-009-9415-x [DOI] [PubMed] [Google Scholar]
- 28.Guglielmetti S, Mora D, Gschwender M, Popp K. 2011. Randomised clinical trial: Bifidobacterium bifidum MIMBb75 significantly alleviates irritable bowel syndrome and improves quality of life—a double-blind, placebo-controlled study. Aliment. Pharmacol. Therap. 33:1123–1132. 10.1111/j.1365-2036.2011.04633.x [DOI] [PubMed] [Google Scholar]
- 29.Singh N, Arioli S, Wang A, Villa CR, Jahani R, Song YS, Mora D, Guglielmetti S, Comelli EM. 2013. Impact of Bifidobacterium bifidum MIMBb75 on mouse intestinal microorganisms. FEMS Microbiol. Ecol. 85:369–375. 10.1111/1574-6941.12124 [DOI] [PubMed] [Google Scholar]
- 30.Marchler-Bauer A, Lu SN, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke ZX, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang DC, Zhang NG, Zheng CJ, Bryant SH. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229. 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schubert K, Bichlmaier AM, Mager E, Wolff K, Ruhland G, Fiedler F. 2000. P45, an extracellular 45 kDa protein of Listeria monocytogenes with similarity to protein p60 and exhibiting peptidoglycan lytic activity. Arch. Microbiol. 173:21–28. 10.1007/s002030050003 [DOI] [PubMed] [Google Scholar]
- 32.Fagan PK, Reinscheid D, Gottschalk B, Chhatwal GS. 2001. Identification and characterization of a novel secreted immunoglobulin binding protein from group A Streptococcus. Infect. Immun. 69:4851–4857. 10.1128/IAI.69.8.4851-4857.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Granucci F, Feau S, Zanoni I, Pavelka N, Vizzardelli C, Raimondi G, Ricciardi-Castagnoli P. 2003. The immune response is initiated by dendritic cells via interaction with microorganisms and interleukin-2 production. J. Infect. Dis. 187:S346–S350. 10.1086/374748 [DOI] [PubMed] [Google Scholar]
- 34.Lewis KL, Reizis B. 2012. Dendritic cells: arbiters of immunity and immunological tolerance. Cold Spring Harb. Perspect. Biol. 4:a007401. 10.1101/cshperspect.a007401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zanoni I, Foti M, Ricciardi-Castagnoli P, Granucci F. 2005. TLR-dependent activation stimuli associated with Th1 responses confer NK cell stimulatory capacity to mouse dendritic cells. J. Immunol. 175:286–292. 10.4049/jimmunol.175.1.286 [DOI] [PubMed] [Google Scholar]
- 36.Wuest SC, Edwan JH, Martin JF, Han S, Perry JSA, Cartagena CM, Matsuura E, Maric D, Waldmann TA, Bielekova B. 2011. A role for interleukin-2 trans-presentation in dendritic cell-mediated T cell activation in humans, as revealed by daclizumab therapy. Nat. Med. 17:604–U125. 10.1038/nm.2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng GY, Yu AX, Malek TR. 2011. T-cell tolerance and the multi-functional role of IL-2R signaling in T-regulatory cells. Immunol. Rev. 241:63–76. 10.1111/j.1600-065X.2011.01004.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guiducci C, Valzasina B, Dislich H, Colombo MP. 2005. CD40/CD40L interaction regulates CD4+ CD25+ Treg homeostasis through dendritic cell-produced IL-2. Eur. J. Immunol. 35:557–567. 10.1002/eji.200425810 [DOI] [PubMed] [Google Scholar]
- 39.Lampinen M, Hakansson L, Venge P. 2001. Interleukin-2 inhibits eosinophil migration but is counteracted by IL-5 priming. Clin. Exp. Allergy 31:249–258. 10.1046/j.1365-2222.2001.00968.x [DOI] [PubMed] [Google Scholar]
- 40.Palm NW, Rosenstein RK, Medzhitov R. 2012. Allergic host defences. Nature 484:465–472. 10.1038/nature11047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serafini F, Turroni F, Guglielmetti S, Gioiosa L, Foroni E, Sanghez V, Bartolomucci A, Motherway MO, Palanza P, van Sinderen D, Ventura M. 2012. An efficient and reproducible method for transformation of genetically recalcitrant bifidobacteria. FEMS Microbiol. Lett. 333:146–152. 10.1111/j.1574-6968.2012.02605.x [DOI] [PubMed] [Google Scholar]
- 42.Guglielmetti S, Mayo B, Alvarez-Martin P. 2013. Mobilome and genetic modification of bifidobacteria. Benef. Microbes 4:143–166. 10.3920/BM2012.0031 [DOI] [PubMed] [Google Scholar]
- 43.Zanoni I, Ostuni R, Capuano G, Collini M, Caccia M, Ronchi AE, Rocchetti M, Mingozzi F, Foti M, Chirico G, Costa B, Zaza A, Ricciardi-Castagnoli P, Granucci F. 2009. CD14 regulates the dendritic cell life cycle after LPS exposure through NFAT activation. Nature 460:264–U130. 10.1038/nature08118 [DOI] [PubMed] [Google Scholar]
- 44.Takeda K, Akira S. 2005. Toll-like receptors in innate immunity. Int. Immunol. 17:1–14. 10.1093/intimm/dxh186 [DOI] [PubMed] [Google Scholar]
- 45.Bateman A, Rawlings ND. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234–237. 10.1016/S0968-0004(03)00061-6 [DOI] [PubMed] [Google Scholar]
- 46.Lundgren A, Bhuiyan TR, Novak D, Kaim J, Reske A, Lu YJ, Qadri F, Malley R. 2012. Characterization of Th17 responses to Streptococcus pneumoniae in humans: Comparisons between adults and children in a developed and a developing country. Vaccine 30:3897–3907. 10.1016/j.vaccine.2012.03.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan F, Cao HW, Cover TL, Whitehead R, Washington MK, Polk DB. 2007. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 132:562–575. 10.1053/j.gastro.2006.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan F, Cao HW, Cover TL, Washington MK, Shi Y, Liu LS, Chaturvedi R, Peek RM, Wilson KT, Polk DB. 2011. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J. Clin. Invest. 121:2242–2253. 10.1172/JCI44031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez P, Gonzalez-Rodriguez I, Gueimonde M, Margolles A, Suarez A. 2011. Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PLoS One 6:e24776. 10.1371/journal.pone.0024776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zanoni I, Granucci F. 2012. Regulation and dysregulation of innate immunity by NFAT signaling downstream of pattern recognition receptors (PRRs). Eur. J. Immunol. 42:1924–1931. 10.1002/eji.201242580 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.