Abstract
Despite the existence of 10 avian paramyxovirus (APMV) serotypes, very little is known about the distribution, host species, and ecological factors affecting virus transmission. To better understand the relationship among these factors, we conducted APMV wild bird surveillance in regions of Ukraine suspected of being intercontinental (north to south and east to west) flyways. Surveillance for APMV was conducted in 6,735 wild birds representing 86 species and 8 different orders during 2006 to 2011 through different seasons. Twenty viruses were isolated and subsequently identified as APMV-1 (n = 9), APMV-4 (n = 4), APMV-6 (n = 3), and APMV-7 (n = 4). The highest isolation rate occurred during the autumn migration (0.61%), with viruses isolated from mallards, teals, dunlins, and a wigeon. The rate of isolation was lower during winter (December to March) (0.32%), with viruses isolated from ruddy shelducks, mallards, white-fronted geese, and a starling. During spring migration, nesting, and postnesting (April to August) no APMV strains were isolated out of 1,984 samples tested. Sequencing and phylogenetic analysis of four APMV-1 and two APMV-4 viruses showed that one APMV-1 virus belonging to class 1 was epidemiologically linked to viruses from China, three class II APMV-1 viruses were epidemiologically connected with viruses from Nigeria and Luxembourg, and one APMV-4 virus was related to goose viruses from Egypt. In summary, we have identified the wild bird species most likely to be infected with APMV, and our data support possible intercontinental transmission of APMVs by wild birds.
INTRODUCTION
Over the past 40 years, a large number of different paramyxoviruses have been isolated from animals and birds (1–17). Paramyxoviruses belong to the Paramyxoviridae family. The subfamily Paramyxovirinae is divided into 5 genera: Respirovirus, Morbillivirus, Rubulavirus, Henipavirus, and Avulavirus (18). Until recently, the genus Avulavirus included 9 avian paramyxovirus serotypes (APMV-1 to -9), but recently the existence of avian paramyxovirus serotype 10 (APMV-10) was established, which was isolated from penguins (19), as well as avian paramyxovirus serotype 11 (APMV-11), found in snipes (20), and serotype 12 (APMV-12), detected in a wigeon (21).
The avian paramyxovirus host range is very large, and the natural reservoirs of paramyxoviruses of different serotypes are wild birds of aquatic, shore, and terrestrial ecosystems (22, 23). Most of the countries with developed industrial poultry production monitor APMVs among both poultry and wild birds. The National Reference Laboratories of the European Union (EU) report a large number of different serotypes of APMV in poultry every year. In most cases, these are APMV-2, APMV-3, APMV-4, APMV-6, and APMV-9. In 2010, in 18 EU countries, 199 isolates of APMV-1, APMV-2 from chickens, turkeys, ducks, and geese, APMV-3 from turkeys, APMV-4 from wild ducks, and APMV-6 were reported (24).
Antibodies to APMV-1, APMV-2, APMV-3, APMV-4, APMV-6, APMV-8, and APMV-9 have been found in wild waterfowl in Spain (25). APMV-2 was detected in wild birds, mainly passerines, in countries of Europe, Asia, Africa, and the Americas (26–28). APMV-3 were isolated from exotic birds and turkeys in the United States, Canada, Britain, France, and Germany (29, 30). APMV-4 strains have been isolated from ducks (31). APMV-5 strains have been isolated only from parrots, and some of APMV-5's properties are significantly different from those of other avian paramyxoviruses (32). The main hosts of APMV-6 are ducks and geese, but APMV-6 strains also have been found in turkeys (33, 34). Viruses of this serotype were isolated in Russia, the United States, and other countries. The main carriers of APMV-7 are pigeons and doves (35). The main carriers of APMV-8 are geese and ducks, and the main carriers of APMV-9 are ducks (25–27, 34).
All avian paramyxoviruses of serotype 1 (APMV-1), including Newcastle disease virus (NDV), as well as some APMV-2, APMV-3, APMV-6, and APMV-7 viruses, are important for the poultry industry, as they can cause disease in poultry (23). Newcastle disease virus is the most studied APMV-1 virus and is of great agricultural importance, and in spite of vaccination, Newcastle disease virus continues to be one of the most widespread infections in poultry, causing significant economic losses (23). Newcastle disease virus has been reported in domestic and wild birds since 1926 in many countries around the world. Every year, the number of countries reporting infections ranges from 50 to 89, and the disease has been detected on all continents. Also there are multiple reports of APMV-1 isolated from pigeons, so-called “pigeon PMV-1” (PPMV-1) (36). In most cases, it is virulent and a source of pathogenic viruses for poultry. The other known natural reservoirs of virulent APMV-1 are cormorants, which have been reported to maintain viruses of genotype V (37).
Ukraine was considered free of Newcastle disease from 1992 to 2006. The last officially reported cases of the disease were in 2006 in the Kharkiv and Rivne regions. Since then, poultry in Ukraine has been free of Newcastle disease. However, the circulation of various paramyxoviruses has not been excluded among wild and synanthropic birds, and Newcastle disease virus was isolated from pigeons in the Kiev and Donetsk regions in 2001 to 2005 (38).
This study presents the results of a large-scale monitoring of wild birds for ortho-and paramyxoviruses in the Azov-Black Sea region of Ukraine. Part of the research findings on orthomyxoviruses has already been published (39). The aim of our research was to study the circulation of serotypes APMV-1 to APMV-9 of avian paramyxovirus in wild birds of different ecological groups in the Azov-Black Sea region of Ukraine.
MATERIALS AND METHODS
Sample collection.
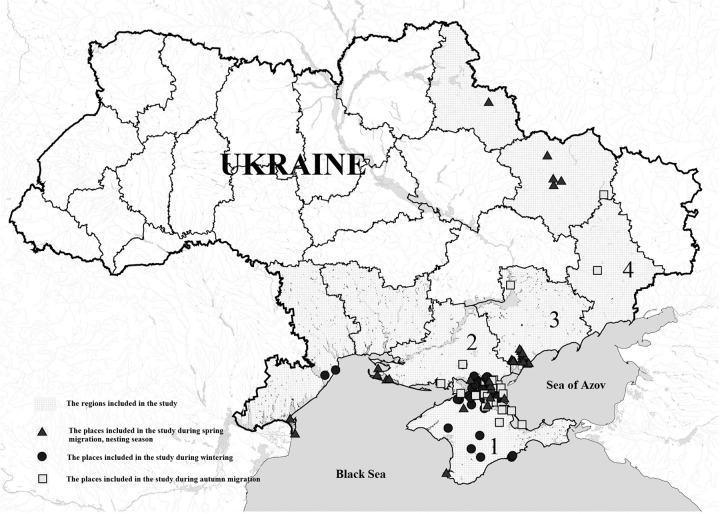
Collection of biological material for virological investigation was carried out in areas of wild bird gatherings in the wetlands of the Azov-Black Sea region of Ukraine (Kherson, Odesa, Mukalayv, and Zaporizhia regions and Autonomous Republic of Crimea [AR Crimea]) and the eastern region of Ukraine (Donetsk, Kharkiv, and Sumy regions) during 2006 to 2011. These collections were conducted during different life cycles of birds, including migration, wintering, and nesting. A total of 6,735 samples were collected from wild birds of 86 species. During the autumn migration (months of September to November), 1,628 samples were collected from 38 species of birds in the Zaporizhia, Kherson (northern coast of Sivash Bay, Arabatsky Strelka), and Donetsk regions and AR Crimea (hunting regions of Krasnoperekopsky, Djankoiskiy, and Nizhnegorskiy districts and the Southern part of the coast of Sivash Bay) (Fig. 1). During wintering months (December to March), 3,123 samples were collected from birds of 22 species in the central part of Kherson region, as well as in wetlands of Sivash Bay (Kherson region and AR Crimea). During the spring migration, nesting season, and after the breeding movements (April to August), 1,984 biological samples were collected from 59 species of birds in the wetlands of the Kherson and Zaporizhia regions (Sivash Bay).
FIG 1.
Map of Ukraine indicating the region and the place included in the APMV wild bird surveillance study. Numerals indicate the regions where APMVs were isolated from wild birds: 1, AR Crimea; 2, Kherson region; 3, Zaporizhia region; 4, Donetsk region.
Sampling from wild birds was carried out in cooperation with ornithologists, who helped determine the identity of the bird species. Cloacal and tracheal swabs were collected from captured birds and from birds shot by hunters. Fresh feces were collected from certain species of birds in places of mass bird accumulations. Feces were collected only if the origin and type of bird had been established. Samples of feces were taken in a checkerboard pattern at a distance of at least 1.5 to 2 m from each other, to avoid selecting feces from the same bird. Cloacal and tracheal swabs and feces were taken from adult birds regardless of gender. The sample size depended on the size of the flock and was at least 25 samples if the flock was up to 500 birds, at least 35 samples if the flock was from 500 to 1,000 birds, and at least 50 samples per 1,000 birds if the number of birds in the flock was more than 1,000. Estimations of the numbers of birds were conducted by the ornithologists.
Swab samples were collected in cryotubes containing 1.0 ml of transport medium (phosphate-buffered saline [PBS]-glycerin at 1:1) with antibiotics (penicillin, 2,000 U/ml; streptomycin, 2 mg/ml; gentamicin, 50 μg/ml; and nystatin, 1,000 U/ml). A 5-fold concentration of antibiotics was used for the fecal samples and cloacal swabs (40–42). Samples were stored at −196°C in liquid nitrogen, where they were kept until processing.
Virus isolation and identification.
Virus isolation was conducted in accordance with the World Organisation for Animal Health procedures. Cloacal, tracheal, and fecal swab samples were inoculated into the allantoic cavity of 9- to 10-day-old specific-pathogen-free (SPF) chicken embryonated eggs. Every sample was passaged three times. The presence of hemagglutinating viruses in allantoic fluid was determined by the hemagglutination test with a 1% suspension of chicken red blood cells (40–42).
The hemagglutinin (HA) virus subtype was determined by hemagglutination inhibition tests as previously described (40–42). Avian influenza viruses were identified as previously described and reported (39). For identification of APMVs, antisera against APMV-1 to -9 produced by either the Veterinary Laboratories Agency, Weybridge, United Kingdom, or by the Instituto Zooprofilattio Sperimentale delle Venezie, Padua, Italy were used.
Nucleic acid extraction, PCR, and sequencing.
Molecular characterization of four APMV-1 and two APMV-4 viruses isolated was conducted. The nucleic acid extraction was carried out by affinity adsorption using Rybo-sorb-50 weighted sorbents (FDUN Central Research Institute of Epidemiology, Moscow, Russian Federation). The resulting RNA samples were used for production of cDNA using a commercial kit for reverse transcription of RNA, Reverte-L (FDUN Central Research Institute of Epidemiology, Moscow, Russian Federation) or RT-Core (Isogene, Moscow, Russian Federation). The concentration of cDNA was determined using the NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) at a wavelength of 260 nm in a volume of 1 ml.
Sequencing and phylogenetic analysis of the fusion (F) gene was conducted on 4 isolates of avian paramyxovirus serotype 1 (APMV-1/Ruddy Shelduck/AN/37-15-02/2011, APMV-1/Ruddy Shelduck/AN/38-15-02/2011, APMV-1/Teal/Krasnooskilsky/5-11/2009, and APMV-1/Mallard/Krasnoperekopsk/18-23-10/2010) and two isolates of the avian paramyxovirus serotype 4 (APMV-4/teal/Dzhankoy/9-17-11/10 and APMV-4/starling/Medvedkovo/5-24-12/10).
To amplify the complete coding region for the F gene of APMV-1, reverse transcriptase PCR (RT-PCR) was conducted as previously described (43).
To amplify the F gene of the APMV-4 isolates, the following primers were used with the following sequences: AGAAAGAAAAGGCTCGACTCAACC for APMV4-4157F, CCCTGATAACCAACAGCTGATACT for APMV4-5253R, ATGGGGAATCGCCTTGGTGTAT for APMV4-5009F, and CAATGGGCAGGAATTGGCTACCTT for APMV4-6257R. The thermal reaction parameters consisted of 45 min of reverse transcriptase, 5 min of an initial denaturation, and 35 amplification cycles with 30 s of denaturation at 94°C, 30 s at 57°C, and 1 min of elongation at 72°C, followed by a final elongation at 72°C for 5 min.
Amplicons of 679, 1,096, and 1,248 nucleotide residues were purified and sequenced. Electrophoretic analysis was performed using 0.8% and 1.5% agarose gel. Sequencing was performed using a commercial kit, the ABI Prism Terminator kit (Applied Biosystems), and an ABI-3000 DNA analyzer (ABI Prism). The resulting sequence was assembled using the software package DNAStar LaserGene (DNAStar Inc., Madison, WI).
Phylogenetic analysis.
The construction of multiple alignments of the homologous region of the fusion gene, using genes published in the GenBank database, was carried out using the alignment program Muscle as implemented in MEGA5. Phylogenetic analysis of the nucleotide sequences was conducted using the maximum likelihood method based on the general time-reversible model. The trees with the highest log likelihood are shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial trees for the heuristic search were obtained automatically by applying the maximum parsimony method. A discrete gamma distribution was used to model evolutionary rate differences among sites (4 categories [+G {gamma}, parameter = 200.0000]). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 14 nucleotide sequences. The codon positions included were 1st + 2nd + 3rd + noncoding. There were a total of 1,762 positions in the final data set in the serotype 4 data set, 1,649 in the APMV-1 full fusion data set, and 5xx in the class I data set. Evolutionary analyses were conducted in MEGA5.
Nucleotide sequence accession numbers.
Nucleotide sequences were submitted to GenBank under accession no. KF851266, KF851267, KF851268, KF851269, and KF851270.
RESULTS
The main goal of this research is to better understand the ecology of APMV in wild birds. In order to obtain a more accurate estimate of the transmission potential of each bird species, virus isolation from swabs or from feces was conducted instead of the more sensitive but less reliable nucleic acid-based methods. During the years 2006 to 2011, the virological examination of biological material collected from 6,735 wild birds belonging to 86 species and 8 different orders was conducted. The largest number of samples was collected from birds from the orders Anseriformes (4,106 samples), followed by Charadriiformes (2,039 samples), and Passeriformes (247 samples). These monitoring studies covered the Azov-Black Sea region of Ukraine, where there are massive gatherings of wild birds from various ecological groups throughout the year (Fig. 1). In addition, this region is the meeting point of the transcontinental migration routes of different wild birds from Siberia, Africa, Europe, and Asia. Ninety percent of the samples were collected in this region. The rest of the biological samples (∼10%) were collected in the eastern region of Ukraine. The results of the number of biological samples collected and the virological findings are shown in Table 1. All viruses were isolated from cloacal swabs and fecal samples collected in the AR Crimea, Kherson, and Donetsk regions.
TABLE 1.
Number of samples of biological material taken from wild birds of different ecological groups in the central part of the Azov-Black Sea region from 2006 to 2011 and the results of APMV isolation
| Bird | No. of samples duringa: |
Total no. of samplesa | ||
|---|---|---|---|---|
| Autumn migration | Wintering | Spring migration, nesting, and postnesting movements | ||
| Pelecaniformes | ||||
| Cormorant (Phalacrocorax carbo) | 10 | 50 | 60 | |
| Ciconiiformes | ||||
| Gray heron (Ardea cinerea) | 4 | 35 | 39 | |
| Purple heron (Ardea purpurea) | 1 | 1 | ||
| Great white egret (Egretta alba) | 12 | 12 | ||
| Night heron (Nycticorax nycticorax) | 27 | 27 | ||
| Little egret (Egretta garzetta) | 21 | 21 | ||
| Anseriformes | ||||
| Mute swan (Cygnus olor) | 3 | 21 | 24 | |
| Whooper swan (Cygnus cygnus) | 6 | 6 | ||
| White-fronted goose (Anser albifrons) | 419 | 1,014/2 (APMV-7 [2]) | 20 | 1,453/2 (0.13%) |
| Greylag goose (Anser anser) | 18 | 20 | 38 | |
| Branta ruficollis (Rufibrenta ruficollis) | 295 | 295 | ||
| Shelduck (Tadorna tadorna) | 23 | 316 | 33 | 372 |
| Ruddy shelduck (Tadorna ferruginea) | 170 | 360/4 (APMV-1 [2], APMV-1/7 [2]) | 530/4 (0.75%) | |
| Mallard duck (Anas platyrhynchos) | 469/3 (APMV-1/7 [1], APMV-4[1], APMV-6 [1]) | 668/3 (APMV-4 [1], APMV-6 [1], APMV-7 [1]) | 1,137/6 (0.52%) | |
| Red-crested pochard (Netta rufina) | 3 | 3 | ||
| Wigeon (Anas penelope) | 22/1 (APMV-6 [1]) | 120 | 142/1 (0.70%) | |
| Pochard (Aythya ferina) | 1 | 1 | ||
| Pintail (Anas acuta) | 3 | 3 | ||
| Garganey (Anas querquedula) | 13 | 13 | ||
| Teal (Anas crecca) | 58/2 (APMV-1 [1], APMV-4 [1]) | 20 | 2 | 80/2 (2.5%) |
| Shoveler (Anas clypeata) | 6 | 6 | ||
| Gadwall (Anas strepera) | 3 | 3 | ||
| Galliformes | ||||
| Gray partridge (Perdix perdix) | 3 | 1 | 4 | |
| Gruiformes | ||||
| Crane (Grus grus) | 40 | 68 | 108 | |
| Coot (Fulica atra) | 35 | 3 | 25 | 63 |
| Water rail (Rallus aquaticus) | 1 | 1 | ||
| Moorhen (Gallinula chloropus) | 2 | 2 | ||
| Little crake (Porzana parva) | 2 | 2 | ||
| Charadriiformes | ||||
| Yellow-legged gull (Larus cachinnans) | 9 | 97 | 165 | 271 |
| Slender-billed gull (Larus genei) | 12 | 76 | 88 | |
| Mediterranean gull (Larus melanocephalus) | 2 | 397 | 399 | |
| Common gull (Larus canus) | 4 | 13 | 17 | |
| Black-headed gull (Larus ridibundus) | 180 | 180 | ||
| Sandwich tern (Thalasseus sandvicensis) | 2 | 61 | 63 | |
| Gull-billed tern (Gelochelidon nilotica) | 85 | 85 | ||
| Gray plover (Pluvialis squatarola) | 21 | 58 | 79 | |
| Kentish plover (Charadrius alexandrinus) | 1 | 1 | ||
| Sanderling (Calidris alba) | 3 | 3 | ||
| Dunlin (Calidris alpina) | 231/4 (APMV-1 [4]) | 37 | 268/4 (1.49%) | |
| Little stint (Calidris minuta) | 3 | 14 | 17 | |
| Temminck's stint (Calidris temminckii) | 3 | 3 | ||
| Common sandpiper (Actitis hypoleucos) | 5 | 5 | ||
| Wood sandpiper (Tringa glareola) | 38 | 38 | ||
| Green sandpiper (Tringa ochropus) | 7 | 7 | ||
| Marsh sandpiper (Tringa stagnatilis) | 1 | 1 | ||
| Greenshank (Tringa nebularia) | 16 | 6 | 22 | |
| Black-winged stilt (Himantopus himantopus) | 4 | 4 | ||
| Oystercatcher (Hematopus ostralegus) | 4 | 4 | ||
| Collared pratincole (Glareola pratincola) | 1 | 1 | ||
| Snipe (Gallinago gallinago) | 1 | 1 | 2 | |
| Ruff (Phylomachus pugnax) | 3 | 210 | 213 | |
| Curlew sandpiper (Calidris ferruginea) | 1 | 141 | 142 | |
| Redshank (Tringa totanus) | 12 | 3 | 15 | |
| Jack snipe (Lymnocryptes minimus) | 1 | 1 | ||
| Spotted redshank (Tringa erythropus) | 2 | 2 | ||
| Common tern (Sterna hirundo) | 3 | 3 | ||
| Curlew (Numenius arquata) | 3 | 3 | ||
| Bar-tailed godwit (Limosa lapponica) | 1 | 1 | ||
| Broad-billed sandpiper (Limicola falcinellus) | 15 | 15 | ||
| Little ringed plover (Charadrius dubius) | 10 | 10 | ||
| Little tern (Sterna albifrons) | 27 | 27 | ||
| Gull-billed tern (Gelochelion nilotica) | 24 | 24 | ||
| Avocet (Recurvirostra avosetta) | 22 | 22 | ||
| Glossy ibis (Plegadis falcinellus) | 1 | 1 | ||
| Sociable plover (Chettusia gregaria) | 2 | 2 | ||
| Coraciiformes | ||||
| Kingfisher (Alcedo atthis) | 3 | 3 | ||
| Passeriformes | ||||
| Sand martin (Riparia riparia) | 3 | 3 | ||
| Swallow (Hirundo rustica) | 14 | 14 | ||
| Jackdaw (Corvus monedula) | 10 | 10 | ||
| Calandra lark (Melanocorypha calandra) | 60 | 60 | ||
| Pied wagtail (Motacilla alba) | 1 | 1 | ||
| Yellow wagtail (Motacilla flava) | 2 | 2 | ||
| Magpie (Pica pica) | 35 | 35 | ||
| Rook (Corvus frugilegus) | 30 | 30 | ||
| Chaffinch (Fringilla coelebs) | 2 | 2 | ||
| Reed bunting (Emberiza schoeniclus) | 20 | 1 | 21 | |
| Starling (Sturnus vulgaris) | 36/1 (APMV-4 [1]) | 2 | 38/1 (2.63%) | |
| Reed warbler (Acrocephalus scirpaceus) | 5 | 5 | ||
| Great reed warbler (Acrocephalus arundinaceus) | 11 | 11 | ||
| Sedge warbler (Acrocephalus schoenobaenus) | 1 | 1 | ||
| Savis̀ warbler (Locustella luscinioides) | 1 | 1 | ||
| Bearded tit (Panurus biarmicus) | 8 | 8 | ||
| Icterine warbler (Hippolais icterina) | 1 | 1 | ||
| Olivaceous warbler (Hippolais pallida) | 1 | 1 | ||
| Song thrush (Turdus philomelos) | 1 | 1 | ||
| Blackbird (Turdus merula) | 2 | 2 | ||
| Total | 1,628/10 (0.61%) | 3,123/10 (0.32%) | 1,984 | 6,735/20 (0.29%) |
Results are presented as the no. of samples alone or the total no./no. of isolated viruses (serotype [no. of viruses of this serotype]). The percentages given in parentheses represent the percentage of positive samples.
Twenty different APMVs were isolated from the samples. During the fall migration, 10 viruses were isolated. Based on serology, they were identified as APMV-1, APMV-4, and APMV-6 (Table 2). All of these viruses were isolated from representatives of wild waterfowl and shorebirds of the Charadriiformes (4 isolates) and Anseriformes (6 isolates).
TABLE 2.
APMV isolates from wild birds during the fall migration periods from 2006 to 2010
| Isolate | APMV type |
|---|---|
| Mallard/Krasnoperekopsk/18-23-10/2010 | APMV-1 |
| Dunlin/Solone Ozero/19/2006 | APMV-1 |
| Dunlin/Solone Ozero/20/2006 | APMV-1 |
| Dunlin/Solone Ozero/22/2006 | APMV-1 |
| Dunlin/Solone Ozero/23/2006 | APMV-1 |
| Mallard/Krasnoperekopsk/9-10-10/2010 | APMV-4 |
| Mallard/Dzhankoy/3-17-11/2010 | APMV-6 |
| Teal/Dzhankoy/9-17-11/2010 | APMV-4 |
| Wigeon/Nyjnigirskiy/2-20-11/2010 | APMV-6 |
| Teal/Krasnooskilsky/5-11/2009 | APMV-1 |
The rate of APMV isolation varied, depending on the season and wild bird species sampled. During the autumn migration, the rate of isolation ranged from 1.92 to 25% in wild birds of the orders Charadriiformes (dunlins) and Anseriformes (mallards, wigeons, and teals). Among dunlins, isolation was the lowest at 1.92% of the captured birds. Among the mallards sampled during the fall migration in the different locations, isolation averaged 4.34 to 10.00%. It should be noted that several populations (in different hunting locations) of mallards were sampled, and the rate of infection was different in each population. In 2010, the rates of APMV isolation in mallards at 3 different locations were 4.34, 4.54, and 5.26%. In 2011, the rate of APMV isolation in mallards was 10.0%. The paramyxovirus isolation rates of teals in two different populations were 11.10 and 14.28%. The highest isolation rate was among wigeons (25%), but in our studies, we had few samples from this species.
During wintering, 10 viruses were isolated from wild birds and subsequently identified as APMV-1, APMV-4, APMV-6, and APMV-7 (Table 3). During this season, all viruses were only isolated from wild birds in the Azov-Black Sea region (AR Crimea, Kherson region). It should be noted that among them there was a mixed sample of paramyxovirus and orthomyxovirus, H10/APMV-7, underlining the possibility of coinfection of an individual bird with more than one virus. It is noteworthy that during wintering, paramyxoviruses were isolated from the members of Anseriformes (9 isolates) and Passeriformes (1 isolate). An APMV-4 virus was isolated from a starling (Sturnus vulgaris), which is not typical for the birds of this ecological group. The virus isolation rate in starlings was 1.66%, while the isolation rate in wild waterfowl ranged from 0.93 to 1.33%. The rate of APMV isolation for ruddy shelducks from different populations ranged from 1.11 to 1.66%, and that for mallards ranged from 1.07 to 1.33%. The lowest isolation rate was in white-fronted geese: 0.93%. During the periods of spring migration, nesting, and postnesting movements, no paramyxoviruses were isolated in all regions studied.
TABLE 3.
APMV viruses isolated from wild birds during wintering periods in 2008 to 2011
| Isolate | APMV type |
|---|---|
| Ruddy Shelduck/AN/3-20-11/2010 | APMV-1 |
| Mallard/Novomychalivka/9-23-12/2010 | APMV-4 |
| Starling/Medvedkovo/5-24-12/2010 | APMV-4 |
| Mallard/Ermakovo/6-7-02/2011 | APMV-6 |
| Mallard/Ermakovo/9-7-02/2011 | H10/APMV-7 |
| White-fronted Goose/AN/48-15-02/2011 | APMV-1 |
| White-fronted Goose/AN/50-15-02/2011 | APMV-7 |
| Ruddy Shelduck/AN/36-15-02/2011 | APMV-7 |
| Ruddy Shelduck/AN/37-15-02/2011 | APMV-7 |
| Ruddy Shelduck/AN/38-15-02/2011 | APMV-1 |
To determine the genetic characteristics of the APMV-1 isolated from wild birds in the Azov-Black Sea region, we carried out sequencing and phylogenetic analysis of four isolates from wild waterfowl of different species and isolated in different seasons: APMV-1/Mallard/Krasnoperekopsk/18-23-10/2010, isolated from a cloacal swab of a clinically healthy mallard during the fall migration in 2010 in the Crimea; APMV-1/Teal/Krasnooskilsky/5-11/2009, isolated from the cloacal swab of a clinically healthy teal collected by fall hunters in 2009 in the Donetsk region; and APMV-1/Ruddy Shelduck/AN/37-15-02/2011 and APMV-1/Ruddy Shelduck/AN/38-15-02/2011, isolated from fecal samples from ruddy shelduck during wintering in the Kherson region in 2011. The pathotype and genotype of the viruses are shown in Table 4.
TABLE 4.
Genotype and pathotype of APMV-1 isolates
| Isolate | Class (genotype) | Cutting site |
|---|---|---|
| APMV-1/Ruddy Shelduck/AN/37-15-02/2011 | II (1b) | GKQGRL |
| APMV-1/Ruddy Shelduck/AN/38-15-02/2011 | II (1b) | GKQGRL |
| APMV-1/Teal/Krasnooskilsky/5-11/2009 | II (1b) | GKQGRL |
| APMV-1/Mallard/Krasnoperekopsk/18-23-10/2010 | I | ERQERL |
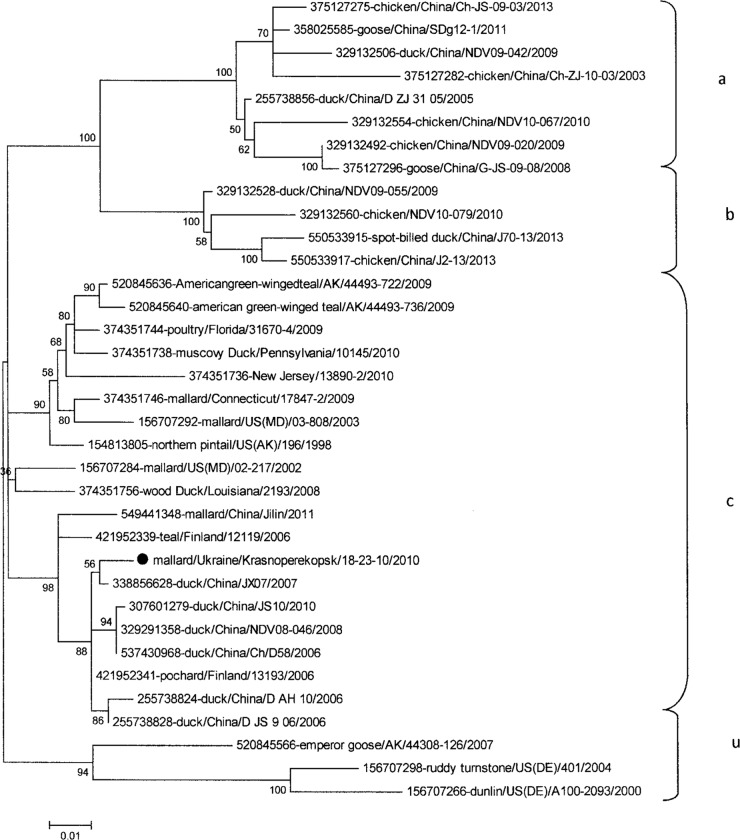
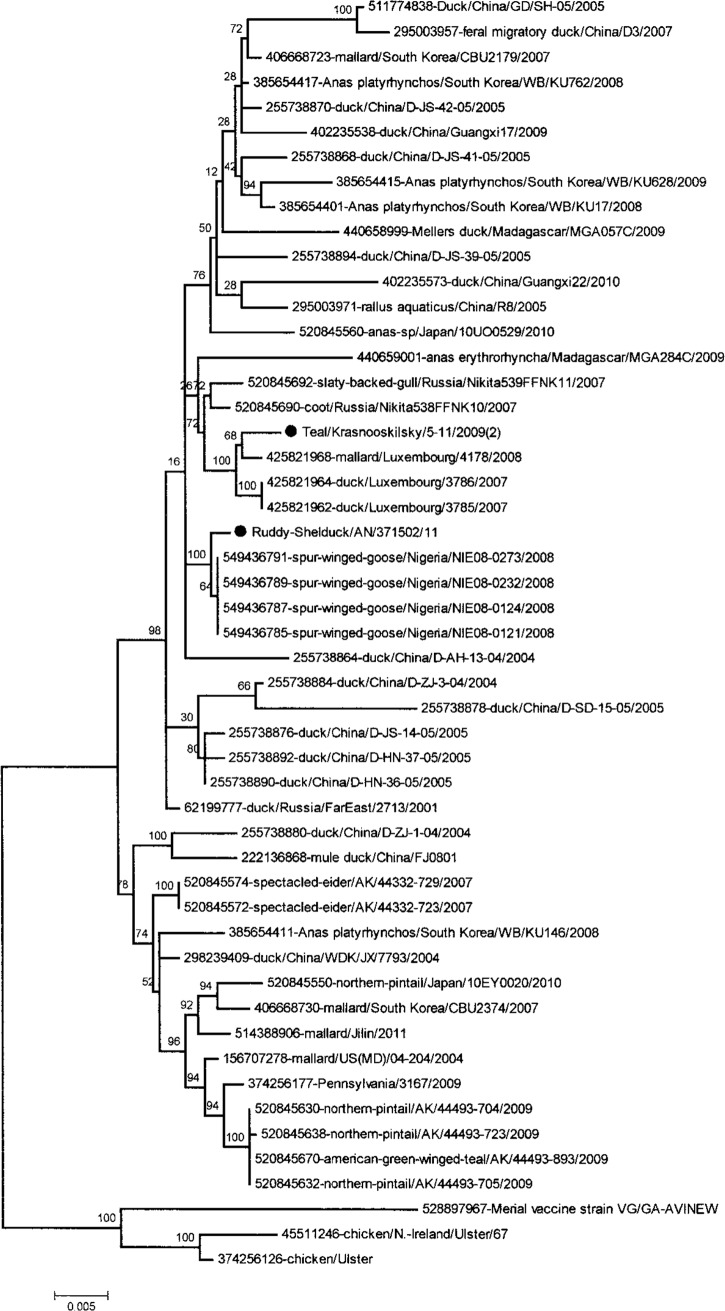
According to the results of the phylogenetic analysis, we found that the APMV-1/mallard/Krasnoperekopsk/18-23-10/2010 virus, isolated in the Crimea, belongs to class I of Newcastle disease viruses (Fig. 2). The other three viruses isolated in Kherson and Donetsk regions, APMV-1/Ruddy Shelduck/AN/37-15-02/2011, APMV-1/Ruddy Shelduck/AN/38-15-02/2011, and APMV-1/Teal/Krasnooskilsky/5-11/2009, belong to genotype 1b of class II (Fig. 3). The viruses (APMV-1/Ruddy Shelduck/AN/37-15-02/2011, APMV-1/Ruddy Shelduck/AN/38-15-02/2011) are nearly identical, and because of it, we included only APMV-1/Ruddy Shelduck/AN/37-15-02/2011 on the tree.
FIG 2.
Phylogenetic tree of the class I isolates of APMV-1 based on the 374-nucleotide variable region of the F gene. Shown are results from the maximum likelihood method with a bootstrap of 1,000. ●, Ukrainian designation of isolates.
FIG 3.
Phylogenetic tree of the class II isolates of APMV-1 based on the F gene full-length sequences. Shown are results from the maximum likelihood method with a bootstrap of 1,000. ●, Ukrainian designation of isolates.
We also conducted full F gene sequencing and phylogenetic analysis of the nucleotide sequences of two isolates of the APMV serotype 4, APMV-4/Teal/Dzhankoy/9-17-11/2010 and APMV-4/Starling/Medvedkovo/5-24-12/2010, which were isolated from a teal and a starling in the Azov-Black Sea region in 2010 during the fall migration and wintering. The phylogenetic tree is shown in Fig. 4. The reference strain for APMV serotype 4, APMV-4/Duck/Hong Kong/D3/75, was used as a polarizer sequence. Sequence divergence between the two Ukrainian isolates was 0.7%. In general, based on the distribution of F gene sequences, two clusters of viruses were observed. The Ukrainian isolates belong to the first one and are closely related to virus isolated from an Egyptian goose in 2010 in North Africa. The degree of nucleotide identity between these viruses was 97%. The second cluster was formed by isolates from Western Europe of Belgian and Italian descent, and the level of nucleotide differences ranged from 2 to 5%.
FIG 4.
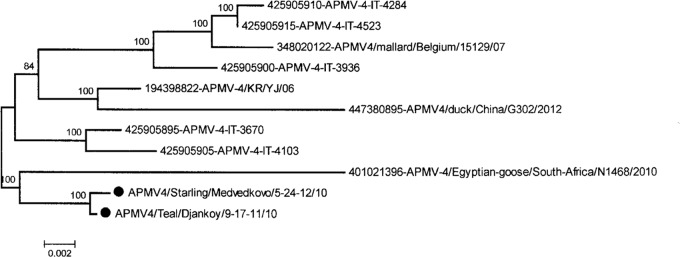
Phylogenetic tree of APMV-4 isolates based on the F gene full-length sequences. Shown are results from neighbor joining with a bootstrap of 1,000. ●, Ukrainian designation of isolates.
DISCUSSION
Overall, the results of our studies have shown the circulation of different APMV serotypes among wild birds in the Azov-Black Sea region of Ukraine. During the period from 2006 to 2011, viruses from 4 of the 12 known APMV serotypes were isolated from wild birds of 3 different orders: Charadriiformes, Anseriformes, and Passeriformes. Most viruses (19 isolates) were obtained from waterfowl and shorebirds, and only one virus was obtained from a land bird (starling), which generally reflects the existing view of reservoirs of APMV in nature. From the 20 isolated viruses, 9 were identified as APMV-1, 4 were APMV-4, 4 were APMV-7, and 3 were APMV-6. Similar data were obtained by other researchers in monitoring studies of waterfowl who identified viruses of serotypes APMV-1, APMV-4, and APMV-6 (22, 34, 44, 45). In our studies, we obtained only 4 APMV-1 isolates from Charadriiformes. All of these isolates were obtained from dunlins (Calidris alpina). The rate of isolation among them was 1.7%. It should be noted that we observed such a high rate only in 2007; in other years, no viruses were isolated from Charadriiformes. Additionally, all 4 viruses were isolated in the same geographic location and in a short period of time according to our observations of the same bird populations. Other authors have reported APMV-2 isolation from dunlins (46). In North America, the prevalence of APMV-1 infections among dunlins was 0.5%. In Europe, apart from APMV-1, several APMV-6 strains have been isolated as well from waders (47). In those cases, the prevalence of APMV-1 infection was 2.4%, and that of APMV-6 was 1.7%.
Virus isolation was conducted to determine levels of APMV infection in the wild birds sampled because other molecular techniques, such as RT-PCR, that are more sensitive for virus detection have severe limitations. For example, if a viable isolate is not recovered, one cannot infer that an RT-PCR-positive strain would contribute to the basic reproduction number of the APMV strains in the sites at the times of sampling. RT-PCR might be detecting dead virus or such a small amount of virus that it may be irrelevant for transmission. In addition, real-time PCR or PCR-based methods are highly sequence specific, and viruses that display mutations in primers or probe regions may produce a negative result. In conclusion, because of this more realistic assessment of the potential for transmission and because virus isolation also allows to conduct the biological characterization of the isolates, this approach was chosen.
In our study, most viruses were isolated from members of the order Anseriformes. In total, 15 APMV isolates of different serotypes (APMV-1, -4, -6, and -7) were obtained from waterfowl. This serotype distribution is similar to what other research groups have reported in other countries (45, 48). In other studies, APMV serotypes 1, 2, and 3 were mostly isolated from the members of the order Passeriformes (23, 49–51).
In agreement with previous studies, other APMV serotypes were not frequently detected in our surveillance. APMV-6 viruses have been previously isolated from meadow pipit (Anthus pratensis) by others, but here we only have been able to detect this serotype in mallards (47, 50). In our studies, one isolate from a starling (Sturnus vulgaris) was identified as APMV-4. No other viruses were isolated from this order. It is also important to note the fact of simultaneous isolation of two avian influenza viruses and an avian paramyxovirus from an individual bird during our investigation. These coinfections have also been reported by others (44).
Our study confirms that the prevalence of APMV-1 infection in wild birds varies within biological cycles in different years, with the highest rates of infection detected in autumn. During the autumn season migration, the rate of isolation in wild waterfowl and birds of different species ranged from 1.92 to 25.0%. Isolation from wild birds during wintering was 0.93 to 1.66%. In contrast to this, we did not isolate any APMV during the spring migration, nesting, and after nesting movements. In general, these rates are similar to data of other authors (45–47) and might be associated with environmental and biological characteristics of the life cycle of birds, especially migratory and wintering strategies. As is known, autumn migration of wild waterfowl occurs more slowly, when the local moving is gradually turning into migration (52–56). During this process, the concentration of wild birds significantly increases in the locations of wild bird accumulations. An important factor that might increase concentrations of wild waterfowl in the beginning of autumn migration is when birds fully or partially lose their ability to fly and for safety localize in limited areas, mostly in shallow water. This leads to the increase in bird concentration in a limited area and increases the probability of direct contact between birds of different species from different geographical regions. As for the wintering season, this period of the biological cycle of birds is also characterized by significant concentrations of birds. However, the main factor that determines the number of birds during the wintering is weather (temperature, presence of snow cover, food availability, etc.). Rapid temperature change contributes the formation of large groups of birds in a limited area (food area of the ponds that do not freeze, as well as other factors), which also significantly increases the probability of direct contact of wild birds of different species and from different geographic regions.
It should be noted the possible significant role of fecal-oral route transmission of APMV especially during the autumn and winter seasons, when low temperatures contribute to long-term storage of paramyxoviruses in the environment, particularly water contaminated by feces. In contrast, during spring migration, high concentrations of birds are usually not observed (excluding breeding colonies). During this period, the birds migrate rapidly and have little contact with each other. Additionally, environmental conditions (temperature and solar radiation) do not contribute to the preservation of pathogens in the environment.
The results of the phylogenetic analysis of selected APMVs showed that some Ukrainian viruses have a high level of identity to viruses from other geographical regions in the globe. This might be explained in terms of migration of wild birds into the Azov-Black Sea region. For example, isolate APMV-1/Mallard/Krasnoperekopsk/18-23-10/10, which was obtained from a mallard (Anas platyrhynchos) in the Crimea region, belongs to class I of APMV1 and has a high degree of similarity to the Chinese lentogenic strains NDV08-046 and JX07, which were isolated in 2007 to 2008 from ducks. Such similarity of the viruses can be explained by the migratory characteristics of mallards. Mallards are very numerous, and their species are widely distributed on the Azov-Black Sea region of Ukraine. As with the other species of wild ducks, there are no clear boundaries between mallard and other species populations, and usually mixing of birds from different population groups occurs in the molting area and migration routes, especially in winter. Mallards stay in the winter, mostly in the south and southeast Russia. At the same time, these regions are the intersection of migration routes of wild birds from Asia (52–56); thus, the viruses could possibly be carried by wild ducks from China through the Russian Federation into the Azov-Black Sea region.
The isolate APMV-1/Teal/Krasnooskilsky/5-11/2009, which belongs to the class II genotype 1b, is most related to nonpathogenic strains of NDV, such as those circulating among populations of wild birds in the territory of Luxembourg in 2007 and 2008. The other APMV-1 isolates, APMV-1/Ruddy Shelduck/AN/37-15-02/11 and APMV-1/Ruddy Shelduck/AN/38-15-02/11, that belong to class II genotype 1b are also related to nonpathogenic strains of NDV. Similar viruses circulated among a population of wild birds in Nigeria in 2008. A nonpathogenic APMV-1 strain from mallards isolated in Sweden in 2010 clustered with genotype Ib and was closely related to the viruses from Luxembourg (57).
Eurasian teals (Anas crecca) are wild ducks characterized by extensive intermixing with birds from the same species from different geographical populations. There are mixed populations of teals on the vast territory from Britain to the Yenisei, especially on migration routes and wintering areas. Birds of this species are associated with migration routes in Western Europe, northern and southwestern Russia, Asia, and Africa (52–56). The fact that some viruses have been isolated from ruddy shelducks (Tadorna ferrugine), which are a population of birds originally from the southern part of Ukraine, can be explained by interpopulation and interspecific exchanges of pathogens, which occur on the Azov-Black Sea region. It is most likely that ruddy shelduck populations were infected by these viruses during contacts with other bird species, which carry pathogens in long-distance migration.
Additional evidence of viruses spreading among wild bird populations in different geographic regions is the presence of the two APMV-4 viruses isolated from a starling and a teal in 2010 during fall migration and wintering, which were related to a virus of the same subtype isolated from an African goose from Nigeria during 2010.
Other species of wild birds from which APMV strains were isolated deserve attention because they travel long distances and play an important role in interpopulation and interspecies exchange of infectious avian diseases. Dunlins, from which 4 isolates of APMV-4 were obtained during the autumn migration, transmigrate for significant distances. Dunlins nest in the tundra area throughout the Palearctic ecozone. The Azov-Black Sea coast is used as one of the migratory routes and locations of stops to restore energy reserves. According to results from experts from the Azov Black Sea Ornithological Station (Melitopol, Ukraine), seasonal migration connects considerable territory from the British Isles to South Africa. Widgeons (Anas penelope) nest in northeastern Ukraine, and during migration might be found throughout the country, but they winter in the Azov-Black Sea region. During migration and wintering, they can join in large flocks.
Birds that are found in the Azov-Black Sea region geographically relate to the west Siberian populations (a bird of western and partly southern central Siberia and Kazakhstan). Starlings are nesting, migrating, and wintering birds in Ukraine. They nest throughout of the country, always migrate, and during the winter season stay in the southern regions. In the Azov-Black Sea region, starlings are ordinary birds that nest, but the number of birds nesting is unknown. During severe winters, the birds disappear from this area and fly for wintering to different regions of the Mediterranean and the northern part of Africa.
Particular attention during monitoring studies should be paid to white-fronted geese. Analysis of migration, wintering strategies, and other biological characteristics of these birds as a migratory and wintering species in the Azov-Black Sea region indicates their potential role in virus transfer in new geographic regions.
The results of our study confirm the widespread circulation of avian paramyxoviruses in wild bird populations in the Azov-Black Sea region, as well as their high level of genetic variability. Our study clearly shows the possibility of interpopulation and interspecific exchange of infectious agents. The APMV isolates are related to viruses from other geographical regions, and their presence suggests the potential risk of pathogens being carried into the country and possibly infecting poultry. It also underlines the importance of monitoring for viruses the wild birds in an area like the Azov-Black Sea region to explore possible introduction of new genetic variants from other geographic regions.
ACKNOWLEDGMENTS
We gratefully acknowledge ornithologists from Azov-Black Sea Ornithological Station, and especially Raisa Chernychko for support during identification of bird species and for providing quality advice and information concerning the biological and ecological characteristics of wild birds from the Azov-Black Sea region of Ukraine.
Part of the research was funded by USDA project P444, through the Ukrainian Science and Technology Center.
Footnotes
Published ahead of print 27 June 2014
REFERENCES
- 1.Barrett T, Blixenkrone-Moller M, Di GG, Domingo M, Duignan P, Hall A, Mamaev L, Osterhaus AD. 1995. Morbilliviruses in aquatic mammals: report on round table discussion. Vet. Microbiol. 44:261–265. 10.1016/0378-1135(95)00019-7 [DOI] [PubMed] [Google Scholar]
- 2.Chua KB, Wang LF, Lam SK, Crameri G, Yu M, Wise T, Boyle D, Hyatt AD, Eaton BT. 2001. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology 283:215–229. 10.1006/viro.2000.0882 [DOI] [PubMed] [Google Scholar]
- 3.Clark HF, Lief FS, Lunger PD, Waters D, Leloup P, Foelsch DW, Wyler RW. 1979. Fer de Lance virus (FDLV): a probable paramyxovirus isolated from a reptile. J. Gen. Virol. 44:405–418. 10.1099/0022-1317-44-2-405 [DOI] [PubMed] [Google Scholar]
- 4.Jack PJ, Boyle DB, Eaton BT, Wang LF. 2005. The complete genome sequence of J virus reveals a unique genome structure in the family Paramyxoviridae. J. Virol. 79:10690–10700. 10.1128/JVI.79.16.10690-10700.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kusagawa S, Komada H, Mao X, Kawano M, Nishikawa F, Tsurudome M, Matsumura H, Ohta H, Yuasa T, Nishio M. 1993. Antigenic and molecular properties of Murayama virus isolated from cynomolgus monkeys: the virus is closely related to avian paramyxovirus type 2. Virology 194:828–832. 10.1006/viro.1993.1325 [DOI] [PubMed] [Google Scholar]
- 6.Lee KE, Umapathi T, Tan CB, Tjia HT, Chua TS, Oh HM, Fock KM, Kurup A, Das A, Tan AK, Lee WL. 1999. The neurological manifestations of Nipah virus encephalitis, a novel paramyxovirus. Ann. Neurol. 46:428–432 [PubMed] [Google Scholar]
- 7.Li Z, Yu M, Zhang H, Magoffin DE, Jack PJ, Hyatt A, Wang HY, Wang LF. 2006. Beilong virus, a novel paramyxovirus with the largest genome of non-segmented negative-stranded RNA viruses. Virology 346:219–228. 10.1016/j.virol.2005.10.039 [DOI] [PubMed] [Google Scholar]
- 8.Mamaev LV, Denikina NN, Belikov SI, Volchkov VE, Visser IK, Fleming M, Kai C, Harder TC, Liess B, Osterhaus AD. 1995. Characterisation of morbilliviruses isolated from Lake Baikal seals (Phoca sibirica). Vet. Microbiol. 44:251–259. 10.1016/0378-1135(95)00018-6 [DOI] [PubMed] [Google Scholar]
- 9.Miller PJ, Boyle DB, Eaton BT, Wang LF. 2003. Full-length genome sequence of Mossman virus, a novel paramyxovirus isolated from rodents in Australia. Virology 317:330–344. 10.1016/j.virol.2003.08.013 [DOI] [PubMed] [Google Scholar]
- 10.Osterhaus AD, de Swart RL, Vos HW, Ross PS, Kenter MJ, Barrett T. 1995. Morbillivirus infections of aquatic mammals: newly identified members of the genus. Vet. Microbiol. 44:219–227. 10.1016/0378-1135(95)00015-3 [DOI] [PubMed] [Google Scholar]
- 11.Philbey AW, Kirkland PD, Ross AD, Davis RJ, Gleeson AB, Love RJ, Daniels PW, Gould AR, Hyatt AD. 1998. An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerg. Infect. Dis. 4:269–271. 10.3201/eid0402.980214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Renshaw RW, Glaser AL, Van CH, Weiland F, Dubovi EJ. 2000. Identification and phylogenetic comparison of Salem virus, a novel paramyxovirus of horses. Virology 270:417–429. 10.1006/viro.2000.0305 [DOI] [PubMed] [Google Scholar]
- 13.Shi LY, Li M, Yuan LJ, Wang Q, Li XM. 2008. A new paramyxovirus, Tianjin strain, isolated from common cotton-eared marmoset: genome characterization and structural protein sequence analysis. Arch. Virol. 153:1715–1723. 10.1007/s00705-008-0184-9 [DOI] [PubMed] [Google Scholar]
- 14.Tidona CA, Kurz HW, Gelderblom HR, Darai G. 1999. Isolation and molecular characterization of a novel cytopathogenic paramyxovirus from tree shrews. Virology 258:425–434. 10.1006/viro.1999.9693 [DOI] [PubMed] [Google Scholar]
- 15.Tikasingh ES, Jonkers AH, Spence L, Aitken TH. 1966. Nariva virus, a hitherto undescribed agent isolated from the Trinidadian rat, Zygodontomys b. brevicauda (J. A. Allen & Chapman). Am. J. Trop. Med. Hyg. 15:235–238 [DOI] [PubMed] [Google Scholar]
- 16.van den Hoogen BG, de Jong JC, Groen J, Kuiken T, Fouchier RA, Osterhaus AD. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719–724. 10.1038/89098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wild TF. 2009. Henipaviruses: a new family of emerging paramyxoviruses. Pathol. Biol. (Paris) 57:188–196. 10.1016/j.patbio.2008.04.006 [DOI] [PubMed] [Google Scholar]
- 18.Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed). 2007. Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 19.Miller PJ, Afonso CL, Spackman E, Scott MA, Pedersen JC, Senne DA, Brown JD, Fuller CM, Uhart MM, Karesh WB, Brown IH, Alexander DJ, Swayne DE. 2010. Evidence for a new avian paramyxovirus serotype 10 detected in rockhopper penguins from the Falkland Islands. J. Virol. 84:11496–11504. 10.1128/JVI.00822-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briand FX, Henry A, Massin P, Jestin V. 2012. Complete genome sequence of a novel avian paramyxovirus. J. Virol. 86:7710. 10.1128/JVI.00946-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terregino C, Aldous EW, Heidari A, Fuller CM, De NR, Manvell RJ, Beato MS, Shell WM, Monne I, Brown IH, Alexander DJ, Capua I. 2013. Antigenic and genetic analyses of isolate APMV/wigeon/Italy/3920-1/2005 indicate that it represents a new avian paramyxovirus (APMV-12). Arch. Virol. 158:2233–2243. 10.1007/s00705-013-1735-2 [DOI] [PubMed] [Google Scholar]
- 22.Camenisch G, Bandli R, Hoop R. 2008. Monitoring of wild birds for Newcastle disease virus in Switzerland using real time RT-PCR. J. Wildl. Dis. 44:772–776. 10.7589/0090-3558-44.3.772 [DOI] [PubMed] [Google Scholar]
- 23.Saif YM, Fadly AM, Glisson JR, McDougald LR, Nolan LK, Swayne DE. (ed). 2008. Diseases of poultry, 12th ed. Blackwell Publishing, Ames, IA [Google Scholar]
- 24.Brown IH. 2011. Country reports for avian paramyxoviruses 2010. Joint 17th Annu. Meet. Natl. Lab. Avian Influenza Newcastle Dis. Eur. Union Member States. http://ec.europa.eu/food/animal/diseases/controlmeasures/avian/docs/programme17th_2011_en.pdf [Google Scholar]
- 25.Maldonado A, Arenas A, Tarradas MC, Luque I, Astorga R, Perea JA, Miranda A. 1995. Serological survey for avian paramyxoviruses from wildfowl in aquatic habitats in Andalusia. J. Wildl. Dis. 31:66–69. 10.7589/0090-3558-31.1.66 [DOI] [PubMed] [Google Scholar]
- 26.Goodman BB, Hanson RP. 1988. Isolation of avian paramyxovirus-2 from domestic and wild birds in Costa Rica. Avian Dis. 32:713–717. 10.2307/1590989 [DOI] [PubMed] [Google Scholar]
- 27.Lipkind M, Alexander D, Shihmanter E, Weisman Y, Collins M. 1995. Antigenic heterogeneity of avian paramyxoviruses of serotype 2 (“Yucaipa-like”) isolated from domestic and wild birds in Israel. Comp. Immunol. Microbiol. Infect. Dis. 18:189–207. 10.1016/0147-9571(95)00003-Q [DOI] [PubMed] [Google Scholar]
- 28.Zhang GZ, Zhao JX, Wang M. 2007. Serological survey on prevalence of antibodies to avian paramyxovirus serotype 2 in China. Avian Dis. 51:137–139. 10.1637/0005-2086(2007)051[0137:SSOPOA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 29.Alexander DJ, Pattison M, Macpherson I. 1983. Avian paramyxoviruses of PMV-3 serotype in British turkeys. Avian Pathol. 12:469–482. 10.1080/03079458308436192 [DOI] [PubMed] [Google Scholar]
- 30.Tumova B, Robinson JH, Easterday BC. 1979. A hitherto unreported paramyxovirus of turkeys. Res. Vet. Sci. 27:135–140 [PubMed] [Google Scholar]
- 31.Parthiban M, Kaliyaperumal M, Xiao S, Nayak B, Paldurai A, Kim SH, Ladman BS, Preskenis LA, Gelb J, Collins PL, Samal SK. 2013. Complete genome sequence of an avian paramyxovirus type 4 from North America reveals a shorter genome and new genotype. Genome Announc. 1:e00075-12. 10.1128/genomeA.00075-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nerome K, Nakayama M, Ishida M, Fukumi H. 1978. Isolation of a new avian paramyxovirus from budgerigar (Melopsittacus undulatus). J. Gen. Virol. 38:293–301. 10.1099/0022-1317-38-2-293 [DOI] [PubMed] [Google Scholar]
- 33.Chang PC, Hsieh ML, Shien JH, Graham DA, Lee MS, Shieh HK. 2001. Complete nucleotide sequence of avian paramyxovirus type 6 isolated from ducks. J. Gen. Virol. 82:2157–2168 [DOI] [PubMed] [Google Scholar]
- 34.Kelleher CJ, Halvorson DA, Newman JA, Senne DA. 1985. Isolation of avian paramyxoviruses from sentinel ducks and turkeys in Minnesota. Avian Dis. 29:400–407. 10.2307/1590501 [DOI] [PubMed] [Google Scholar]
- 35.Saif YM, Mohan R, Ward L, Senne DA, Panigrahy B, Dearth RN. 1997. Natural and experimental infection of turkeys with avian paramyxovirus-7. Avian Dis. 41:326–329. 10.2307/1592185 [DOI] [PubMed] [Google Scholar]
- 36.Collins MS, Strong I, Alexander DJ. 1996. Pathogenicity and phylogenetic evaluation of the variant Newcastle disease viruses termed “pigeon PMV-1 viruses” based on the nucleotide sequence of the fusion protein gene. Arch. Virol. 141:635–647. 10.1007/BF01718322 [DOI] [PubMed] [Google Scholar]
- 37.Susta L, Miller PJ, Afonso CL, Brown CC. 2011. Clinicopathological characterization in poultry of three strains of Newcastle disease virus isolated from recent outbreaks. Vet. Pathol. 48:349–360. 10.1177/0300985810375806 [DOI] [PubMed] [Google Scholar]
- 38.Maxymchuk SI. 2008. Defining virulence isolates of Newcastle disease virus isolated in 2001–2005 from pigeons. Vet. Biotechnol. 13:309–317 (In Ukrainian.) [Google Scholar]
- 39.Muzyka D, Pantin-Jackwood M, Spackman E, Stegniy B, Rula O, Shutchenko P. 2012. Avian influenza virus wild bird surveillance in the Azov and Black Sea regions of Ukraine (2010–2011). Avian Dis. 56:1010–1016. 10.1637/10157-040912-ResNote.1 [DOI] [PubMed] [Google Scholar]
- 40.Alexander DJ, Capua I. 2009. Avian influenza and Newcastle disease: a field and laboratory manual. Springer, Milan, Italy [Google Scholar]
- 41.International Office of Epizootics. 2008. Manual of diagnostic tests and vaccines for terrestrial animals: mammals, birds and bees, 6th ed. International Office of Epizootics, World Organisation for Animal Health, Paris, France: [PubMed] [Google Scholar]
- 42.Spackman E. 2008. Avian influenza virus. Humana Press, Totowa, NJ [Google Scholar]
- 43.Perozo F, Merino R, Afonso CL, Villegas P, Calderon N. 2008. Biological and phylogenetic characterization of virulent Newcastle disease virus circulating in Mexico. Avian Dis. 52:472–479. 10.1637/8276-022908-Reg.1 [DOI] [PubMed] [Google Scholar]
- 44.Dormitorio TV, Giambrone JJ, Guo K, Hepp GR. 2009. Detection and characterization of avian influenza and other avian paramyxoviruses from wild waterfowl in parts of the southeastern United States. Poult. Sci. 88:851–855. 10.3382/ps.2008-00337 [DOI] [PubMed] [Google Scholar]
- 45.Goekjian VH, Smith JT, Howell DL, Senne DA, Swayne DE, Stallknecht DE. 2011. Avian influenza viruses and avian paramyxoviruses in wintering and breeding waterfowl populations in North Carolina, U.S.A. J. Wildl. Dis. 47:240–245. 10.7589/0090-3558-47.1.240 [DOI] [PubMed] [Google Scholar]
- 46.Coffee LL, Hanson BA, Luttrell MP, Swayne DE, Senne DA, Goekjian VH, Niles LJ, Stallknecht DE. 2010. Avian paramyxoviruses in shorebirds and gulls. J. Wildl. Dis. 46:481–487. 10.7589/0090-3558-46.2.481 [DOI] [PubMed] [Google Scholar]
- 47.Hlinak A, Muhle RU, Werner O, Globig A, Starick E, Schirrmeier H, Hoffmann B, Engelhardt A, Hubner D, Conraths FJ, Wallschlager D, Kruckenberg H, Muller T. 2006. A virological survey in migrating waders and other waterfowl in one of the most important resting sites of Germany. J. Vet. Med. B Infect. Dis. Vet. Public Health 53:105–110. 10.1111/j.1439-0450.2006.00935.x [DOI] [PubMed] [Google Scholar]
- 48.Tian Z, Chai H, Li F, Sun J, Chen G, Hu X, Hua Y, Xiang W. 2012. Complete nucleotide sequence of avian paramyxovirus type 6 strain JL isolated from mallard ducks in China. J. Virol. 86:13112. 10.1128/JVI.02317-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schnebel B, Dierschke V, Rautenschlein S, Ryll M. 2005. No detection of avian influenza A viruses of the subtypes H5 and H7 and isolation of lentogenic avian paramyxovirus serotype 1 in passerine birds during stopover in the year 2001 on the island Helgoland (North Sea). Dtsch. Tierarztl. Wochenschr. 112:456–460 [PubMed] [Google Scholar]
- 50.Shihmanter E, Weisman Y, Lublin A, Mahani S, Panshin A, Lipkind M. 1998. Isolation of avian serotype 3 paramyxoviruses from imported caged birds in Israel. Avian Dis. 42:829–831. 10.2307/1592725 [DOI] [PubMed] [Google Scholar]
- 51.Zhang GZ, Zhao JX, Wang HW, Yang AM, Bu CY, Wang M. 2006. Isolation, identification, and comparison of four isolates of avian paramyxovirus serotype 2 in China. Avian Dis. 50:386–390. 10.1637/7502-010906R1.1 [DOI] [PubMed] [Google Scholar]
- 52.Delany S, Scott D. 2002. Waterbird population estimates, 3rd ed. Wetlands International, Wageningen, The Netherlands [Google Scholar]
- 53.Gilissen N. 2002. Numbers and distribution of wintering waterbirds in the Western Palearctic and Southwest Asia in 1997, 1998 and 1999: results from the International Waterbird Census. Wetlands International, Wageningen, The Netherlands [Google Scholar]
- 54.Madsen J, Cracknell G, Fox AD. 1999. Goose populations of the Western Palearctic: a review of status and distribution. J. Appl. Ecol. 36:843–844 [Google Scholar]
- 55.Madsen J. 1991. Status and trends of goose populations in the Western Palearctic in the 1980s. Ardea 79:113–122 [Google Scholar]
- 56.Scott DA, Rose PM. 1996. Atlas of Anatidae populations in Africa and western Eurasia. Wetlands International, Wageningen, The Netherlands [Google Scholar]
- 57.Tolf C, Wille M, Haidar AK, Avril A, Zohari S, Waldenstrom J. 2013. Prevalence of avian paramyxovirus type 1 in mallards during autumn migration in the western Baltic Sea region. Virol. J. 10:285. 10.1186/1743-422X-10-285 [DOI] [PMC free article] [PubMed] [Google Scholar]