Abstract
The enzyme triphenylmethane reductase (TMR) reduces toxic triphenylmethane dyes into colorless, nontoxic derivatives, and TMR-producing microorganisms have been proposed as bioremediation tools. Analysis of the genome of Listeria monocytogenes H7858 (1998-1999 hot dog outbreak) revealed that the plasmid (pLM80) of this strain harboring a gene cassette (bcrABC) conferring resistance to benzalkonium chloride (BC) and other quaternary ammonium disinfectants also harbored a gene (tmr) highly homologous to TMR-encoding genes from diverse Gram-negative bacteria. The pLM80-associated tmr was located two genes downstream of bcrABC as part of a putative IS1216 composite transposon. To confirm the role of tmr in triphenylmethane dye detoxification, we introduced various tmr-harboring fragments of pLM80 in a pLM80-cured derivative of strain H7550, from the same outbreak as H7858, and assessed the resistance of the constructs to the triphenylmethane dyes crystal violet (CV) and malachite green. Transcriptional and subcloning data suggest that the regulation of TMR is complex. Constructs harboring fragments spanning bcrABC and tmr were CV resistant, and in such constructs tmr transcription was induced by sublethal levels of either BC or CV. However, constructs harboring only tmr and its upstream intergenic region could also confer resistance to CV, albeit at lower levels. Screening a panel of BC-resistant L. monocytogenes strains revealed that all those harboring bcrABC and adjacent pLM80 sequences, including tmr, were resistant to CV and decolorized this dye. The findings suggest a potential role of TMR as a previously unknown adaptive attribute for environmental persistence of L. monocytogenes.
INTRODUCTION
Listeria monocytogenes is a food-borne facultative intracellular pathogen that is able to contaminate a variety of ready-to-eat food products and food processing facilities (1–4). It has been hypothesized that biofilm formation, resistance to disinfectants, resistance to Listeria-specific viruses, and the ability to replicate at low temperatures are among the major attributes that contribute to the prevalence and persistence of L. monocytogenes in food-related environments (5–10). The current list of adaptive attributes of L. monocytogenes is not exhaustive. Substantial effort has been directed toward the identification of new adaptations and niches that might lead to persistence of L. monocytogenes in the environment.
Triphenylmethane dyes such as crystal violet (CV) and malachite green have had numerous industrial applications, e.g., as dyes in the textile industry or antifungal agents in aquaculture. Such uses have been accompanied with release into the environment as industrial effluents (11, 12). Due to increasing interest in bioremediation tools for these dyes, a number of studies have utilized the ability of microbes, including fungi and bacteria, to enzymatically decolorize and detoxify the dyes (13). Triphenylmethane reductase (TMR) is one such enzyme produced by diverse Gram-negative bacteria such as Citrobacter (accession number AY756172), Aeromonas (accession number EF010984), and Pseudomonas (accession number FJ649187) spp. (14–17). A TMR produced by Citrobacter sp. strain MY-5 (accession number AY756172) and by Pseudomonas sp. has been characterized in a heterologous host (Escherichia coli) (14, 16–19); however, limited information is currently available on the ecological importance and role of TMR in its native hosts or on genes encoding similar TMRs in other bacteria.
Genome analysis of L. monocytogenes strain H7858 from the 1998-1999 listeriosis outbreak involving contaminated hot dogs revealed a large (ca. 80 kb) plasmid, pLM80, which harbored a gene cassette (bcrABC) conferring resistance to benzalkonium chloride (BC) and other quaternary ammonium disinfectants (20–22). Analysis of bcrABC from strain H7550, from the same 1998-1999 outbreak as strain H7858, revealed that bcrABC (1.2 kb) was transcribed from a canonical promoter and was part of a larger, 3.8-kb transcript. Interestingly, the last gene in this operon, two genes downstream from bcrC, was annotated as a putative TMR (21, 22). However, functional confirmation of the involvement of this gene (tmr) in dye detoxification in L. monocytogenes has been lacking. In the present study, we characterized tmr in L. monocytogenes H7550 and assessed tmr prevalence and dye detoxification potential among L. monocytogenes isolates.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The L. monocytogenes strains used for subcloning of tmr-harboring fragments of pLM80 are listed in Table 1 and were grown in brain heart infusion broth (BHI; Becton Dickinson) or on BHI agar (BHI with 1.2% Bacto-agar [Becton Dickinson]). Other L. monocytogenes strains used in the present study were from our laboratory's Listeria strain collection at North Carolina State University. These included 18 strains (14 environmental and 4 from human listeriosis) of serotype 1/2a or 3a, 12 environmental strains of serotype 1/2b or 3b, 3 environmental strains of serotype 1/2c or 3c, and 14 strains (2 environmental and 12 from human listeriosis) of serotype 4b. Environmental strains were from turkey processing plants (8). Escherichia coli strains were grown at 30°C in Luria-Bertani (LB) broth (Becton Dickinson) or on LB broth supplemented with 1.2% Bacto agar. When indicated, the antibiotics used for Listeria were nalidixic acid (20 μg/ml) and chloramphenicol (6 μg/ml), while chloramphenicol (25 μg/ml) was used for E. coli. The antibiotics were purchased from Sigma-Aldrich (St. Louis, MO).
TABLE 1.
L. monocytogenes strains used in this study
| L. monocytogenes straina | CV MICb (μg/ml) | Source or reference |
|---|---|---|
| H7550 | 20 | 22 |
| H7550-CdS | 2.5 | 22 |
| CdS-pPL81 | 15 | This study |
| CdS-pPL82 | 10 | This study |
| CdS-pPL83 | 17.5 | This study |
| CdS-pPL84 | 10 | This study |
| CdS-pPL2 | 2.5 | This study |
| CdS-pPLBC | 2.5 | This study |
Strains harboring various integrated constructs are described in Materials and Methods.
MICs were determined in two independent trials as described in Materials and Methods. Identical values were obtained from the two trials for all strains with the exception of CdS-pPL83 (MIC = 20 and 15 μg/ml in trials 1 and 2, respectively).
BC and crystal violet (CV) resistance and MIC determinations.
BC resistance was determined as described before (8). For CV susceptibility determination, a single colony from a blood agar plate (Remel) was inoculated in 100 μl of BHI broth, and 5 μl was spotted in duplicate onto BHI agar supplemented with 15 μg/ml CV (Fisher, Fair Lawn, NJ). The plates were incubated at 37°C for 48 h. Strains H7550 and its pLM80-cured derivative H7550-CdS (22) were used as resistant and susceptible controls, respectively. For CV MIC determinations, overnight cultures were grown at 37°C (ca. 109 CFU/ml), and 3 μl was spotted in duplicate onto the surfaces of BHI agar plates supplemented with variable concentrations of CV (0, 2.5, 5, 10, 15, and 20 μg/ml). The plates were incubated at 37°C for 48 h, and the MIC was defined as the lowest assessed concentration of CV that prevented confluent growth on the spots.
PCR and recombinant plasmid constructs.
The locations of the primers used to construct the recombinant plasmids are indicated in Fig. 1. PCR analyses employed the TaKaRa ExTaq kit (TaKaRa, Madison, WI) and a T1 thermal cycler (Biometra, Gottingen, Germany). Primers p1/p2 and 0067F/0067R were used for bcrABC and for tmr, respectively (22, 23).
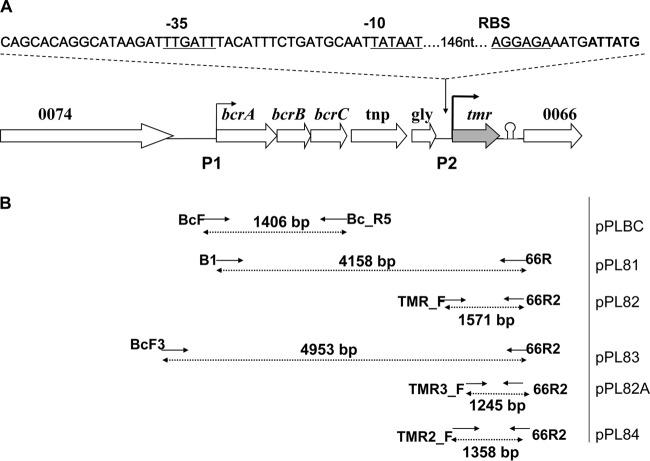
FIG 1.
Genetic characterization of tmr in L. monocytogenes. (A) Schematic diagram of the tmr-harboring region of pLM80 (20). P1 and P2 represent the two putative promoters where P1 is a canonical promoter upstream of bcrABC described before (22), and P2 is immediately upstream of tmr. (B) Recombinant plasmids harboring different tmr segments. Solid arrows indicate the position and orientation of the primers used for recombinant plasmid construction. The plasmids were introduced in the pLM80-cured derivative H7550-CdS and integrated into the chromosome, as described in Materials and Methods.
For recombinant constructs, the primers B1 (5′-GACTCCCGGGGATTCTGGAACATCCCTATC-3′) and 66R (5′-GGTTCCCGGGGGCAATGCAATTGTCTATATCAAC-3′; the XmaI sites are underlined) were used to amplify a 4,158-bp fragment extending from 105 nucleotides (nt) upstream of bcrA, along with the canonical promoter (P1), to 281 nt downstream of the tmr stop codon. This fragment was digested with XmaI (New England BioLabs, Beverly, MA) and ligated with T4 DNA ligase (Promega, Madison, WI) into similarly digested pPL2 (24) to produce pPL81. The primers TMR_F (5′-GACTGGTACCCAATCAATTACGCATTTGTACTTG-3′) and 66R2 (5′-GACTGGTACCGGCAATGCAATTGTCTATATCAAC-3′; the KpnI sites are underlined) were used to amplify a 1,571-bp fragment harboring the entire intergenic space (426 bp) upstream of the start codon of tmr (including P2, located 175 nt upstream of the tmr start codon), along with tmr and 281 nt downstream of the tmr stop codon. This fragment was restricted with KpnI (New England BioLabs) and then ligated into similarly digested pPL2, resulting in pPL82 (Fig. 1). The primers BcF3 (5′-GACTGGTACC TTCAATTAGATCGAGGCACG-3′; the KpnI site is underlined) and 66R2 were used to amplify a 4,953-bp fragment that harbored the entire (850 nt) intergenic sequence upstream of bcrA and 257 nt downstream of the tmr intergenic region. This fragment was restricted with KpnI and was ligated into similarly digested pPL2, resulting in pPL83 (Fig. 1). The primers TMR2_F (5′-GACTGGTACC TCCAGTCTTCAATTGCGGCC-3′; the KpnI site is underlined) and 66R2 were used to amplify a 1,358-bp fragment harboring tmr, together with its upstream (213 bp, including P2) and downstream (281 bp) intergenic regions. The fragment was restricted with KpnI (New England BioLabs) and ligated into similarly digested pPL2, resulting in pPL84. The primers TMR3_F (5′-GACTGGTACCGGAGGCATTCACCTTGTTTAG-3′; the KpnI site is underlined) and 66R2 were used to amplify a fragment (1,245 bp) with a shorter (100-bp) intergenic region upstream of tmr, thus lacking P2. This fragment was restricted with KpnI (New England BioLabs) and ligated into similarly digested pPL2, resulting in pPL82A (Fig. 1). Recombinant plasmids were electroporated first into E. coli DH5α, followed by selection on LB medium supplemented with 15 μg/ml CV for 24 to 48 h at 30°C, until the medium was decolorized. The primers BcF and BC_R5 (22) were used to amplify a 1,406-bp fragment harboring the 1,231-bp bcrABC, along with 105 bp upstream of bcrA and 70 bp downstream of bcrC. This fragment was digested with SacI/XmaI (restriction sites harbored on the primers) and ligated into similarly digested pPL2, resulting in pPLBC (Fig. 1). Like the other plasmids, pPLBC was first electroporated in E. coli DH5α, followed by selection on LB agar supplemented with chloramphenicol at 25 μg/ml (24). Plasmids from the E. coli DH5α transformants were confirmed with PCR and restriction analysis, followed by electroporation into E. coli S17-1. The recombinant plasmids pPL81, pPL82, pPL83, pPL84, and pPLBC and the empty vector (pPL2) were then mobilized into H7550-CdS via conjugation and integrated into the chromosome, as described previously (22, 24). Transconjugants were selected on BHI agar plates supplemented with chloramphenicol (6 μg/ml) and nalidixic acid (20 μg/ml) at 30°C for 2 to 3 days and confirmed using PCR as described previously (24).
Decolorization curves.
Overnight cultures (37°C in BHI) of L. monocytogenes strains H7550, H7550-CdS harboring pPL2 (Cds-pPL2), and derivatives of H7550-CdS, i.e., pPL81, pPL82, pPL83, and pPL84 were diluted 1:20 in BHI supplemented with 15 μg/ml CV in separate wells of 24-well plates (Corning, NY). The inoculated plates were incubated at 37°C and the decrease in A590 of the cultures was measured at several intervals using a spectrophotometer (SmartSpec 3000; Bio-Rad, Hercules, CA) until the CV was completely decolorized. Experiments were done in at least two independent trials. For growth assessments, overnight cultures were diluted 1:20 in BHI, followed by incubation at 37°C. The A600 was recorded using a spectrophotometer (SmartSpec 3000) at hourly intervals for up to 8 h. To assess the impact of CV on growth of H7550 and Cds-pPL2, cultures (1:20 dilutions in BHI, as described above) were incubated at 37°C in the presence of CV (7.5 μg/ml), and CFU were determined by plating dilutions at 4.5, 24 and 48 h. The assay was done in two independent trials.
Transcriptional analyses.
Bacteria were grown in BHI at 37°C until mid-stationary phase (A600 ∼ 0.7 to 0.9, measured using a spectrophotometer [SmartSpec 3000]). The cultures were then divided into two equal portions, one of which was supplemented with 15 μg/ml CV, while the other was left untreated and incubated at 37°C for 30 min. The total RNA was isolated from the CV-treated and untreated cultures as described previously (22, 23, 25). RNA was isolated in three independent trials and reverse transcribed as described before (22, 23, 25). The gene-specific cDNA was reverse transcribed using 0067-R for tmr (23) and s2 for the housekeeping gene spoVG, which was used as a reference (22, 25, 26).
Quantitative real-time PCR (qPCR) was used to assess the expression of tmr in the presence or absence of CV using the Applied Biosystems 7500 Fast real-time PCR system according to the manufacturer's instructions (Applied Biosystems, Foster City, CA) and as described before (25). The primers qTMR_F (5′-GAGTCTGGAGCAATTGTTAC-3′), qTMR_R (5′-CAAGGTTATACGTTTTGTTTTCATG-3′), and TaqMan probe qTMR_P (5′FAM-TTCCTCTGTAAGAACCGTTGCG3-′BHQ1) were used for tmr; the primers and probe used for spoVG were as described before (25). Assays were performed in triplicate and in at least three independent trials.
Statistics.
Expression levels for tmr were reported relative to those of the housekeeping gene spoVG, used as an internal control (26). The quantification of tmr transcript levels for each strain and treatment (with or without CV) was done by normalizing the tmr CT values to those of spoVG for the same strain and treatment. The resulting values (ΔCT) were used (2−ΔCT) to express the fold change between tmr and spoVG transcript levels, assuming a gene amplification efficiency for both tmr and spoVG of 2 (25, 27).
A mixed linear model (SAS, Cary, NC) was fit to the ΔCT measurements in order to test hypotheses regarding factorial effects of strain and CV treatment. In particular, random effects were included for trial and trial-by-treatment interaction so that appropriate error terms were used in tests for these hypotheses. Residual diagnostics did not reveal any violation of the assumptions of normally distributed ΔCT measurements and homogeneity of variance necessary for statistical inference. To test for equality of gene expression across strains separately with or without the CV treatment, a nested model was fit in which the strain effect was nested within the CV factor.
RESULTS AND DISCUSSION
Distribution of tmr suggests recent transfer from Gram-negative bacteria to Listeria, possibly in triphenylmethane dye-contaminated environments.
In L. monocytogenes H7858 the putative tmr (864 bp) was harbored on the large plasmid pLM80 (LMOh7858_plm80_0067, accession number NZ_AADR00000000), two genes downstream of the BC resistance cassette bcrABC (20–22; Fig. 1). The gene had unusually high GC content (45%) in comparison to the genome average (38%) of L. monocytogenes. BLAST analysis of other Listeria genomes revealed conserved (100% identity) tmr sequences in a pLM80-like (∼80-kb) plasmid harbored by L. monocytogenes J0161 (serotype 1/2a, implicated in a 2000 outbreak involving turkey deli meats) (21) and a ∼149-kb plasmid from strain L. monocytogenes N1-011A; sequences with 100% identity were also detected in L. monocytogenes 11GZL18 (serotype 1/2c strain from raw pork), L. monocytogenes Lm1986, and L. seeligeri N1-067, isolated from trout brine (28). However, various homologs with 99 to 100% nucleotide identity over the entire length of the gene were also detected among high-GC-content Gram-negative bacteria from wastewater treatment systems, aquaculture water, and soil in a textile effluent treatment facility. These included Citrobacter sp. strain MY-5 [66% GC], accession number AY756172; Pseudomonas sp. [63% GC], accession number FJ649187; Aeromonas hydrophila subsp. decolorationis [61% GC], accession number EF010984; Comamonas sp. [52% GC], accession number JN648092; and Delftia sp. [66% GC], accession number NC_019312. With the exception of Listeria, tmr homologs were under-represented in Gram-positive bacteria, being reported only in Geobacillus sp. (43% GC; 98 to 99% identity over 86% of the gene).
Analysis of the intergenic sequence upstream of tmr in pLM80 revealed a putative promoter (P2: −10 TATAAT; −35 TTGATT) 146 nt upstream of the tmr start codon (Fig. 1). The upstream 395-nt intergenic region, including P2, was conserved in several of the Gram-negative tmr homologs. The promoter sequence for P2 has not been characterized before since the Gram-negative tmr homologs were expressed in a heterologous host under a non-native promoter (14, 16, 17). Analysis of the 239-nt intergenic sequence downstream of tmr in pLM80 also revealed that this sequence was conserved (100% identity) in the Gram-negative tmr homologs, including the previously identified rho-independent terminator 110 nt downstream of the tmr stop codon (22). In certain cases (Comamonas sp. strain KV11 and KV36, Delftia sp. strain KV29 and uncultured bacterium pGNB1) the downstream conserved sequences extended beyond the 239-nt intergenic region downstream of tmr to include a hypothetical gene, LMOh7858_plm80_0066 (34% GC) (Fig. 1).
The high-level conservation of tmr and its upstream and downstream sequences between L. monocytogenes and the Gram-negative homologs, together with the unusually high GC content of tmr and its over-representation among Gram-negative bacteria, suggest recent transfer of these sequences from Gram-negative bacteria to Listeria. The Gram-negative bacteria themselves likely acquired tmr from other sources, since their genome GC content (52 to 66%, as discussed above) was significantly higher than that for tmr (45%) (14, 16). It is tempting to speculate that transfer of tmr took place and conferred selective advantages to Listeria in environments contaminated with triphenylmethane dyes, such as those associated with aquaculture, wastewater treatment, and textile industry effluents.
tmr confers CV decolorization and CV resistance to L. monocytogenes strain H7550.
Preliminary evidence for the role of tmr in CV resistance and decolorization was obtained from comparisons between strain H7550 (harboring tmr on pLM80) and its pLM80-cured derivative, H7550-CdS. After overnight incubation at 37°C on BHI agar with 15 μg/ml CV (CV-15), confluent growth with a zone of decolorized CV was seen for H7550, while no growth or decolorization could be noted for H7550-CdS (data not shown). The CV MICs for H7550 and H7550-CdS were 20 and 2.5 μg/ml, respectively. These strains were also tested with a different triphenylmethane dye, malachite green. Strain H7550 grew noticeably better than H7550-CdS on agar with this dye (data not shown). Differences were more pronounced and consistent with CV, and the latter was used for the remainder of the study.
Cloning of tmr with different upstream and downstream sequences resulted in the chromosomally integrated plasmids shown in Fig. 1. Strain CdS-pPL2 (H7550-CdS harboring the integrated empty vector, pPL2) was susceptible to CV (MIC = 2.5 μg/ml) and unable to decolorize CV-15 (Table 1 and Fig. 2). However, the genetically complemented strains CdS-pPL81 and CdS-pPL83, harboring tmr and upstream genes (Fig. 1), recovered the ability to grow on CV (MIC ≥ 15 μg/ml) and to decolorize the dye (Table 1 and Fig. 2). Resistance to CV was also restored (MIC = 10 μg/ml; Table 1) in the complemented strains that harbored only tmr, together with either the entire (426 nt) or a portion (213 nt) of the upstream intergenic sequence (CdS-pPL82 and CdS-pPL84, respectively) (Fig. 1). However, pPL82A harboring only tmr with 100 nt upstream of tmr start codon and thus lacking P2 failed to yield E. coli DH5α recombinants on CV-15. These data suggest that tmr can be expressed from the P1 promoter, together with bcrABC and other genes in the bcrABC multicistronic unit, as previously reported (22). Further studies are needed to determine whether tmr in pLM80 and constructs such as pPL81 and pPL83 (which harbor both P1 and P2) is expressed from P1 only or from both P1 and P2. Furthermore, it is currently not known whether certain environmental signals or conditions impact the relative contribution of P1 and P2 in pLM80 and constructs harboring both P1 and P2. However, CV resistance and decolorization conferred by pPL82 and pPL84 (which harbor only P2) suggest that tmr can also be expressed from its own promoter, P2.
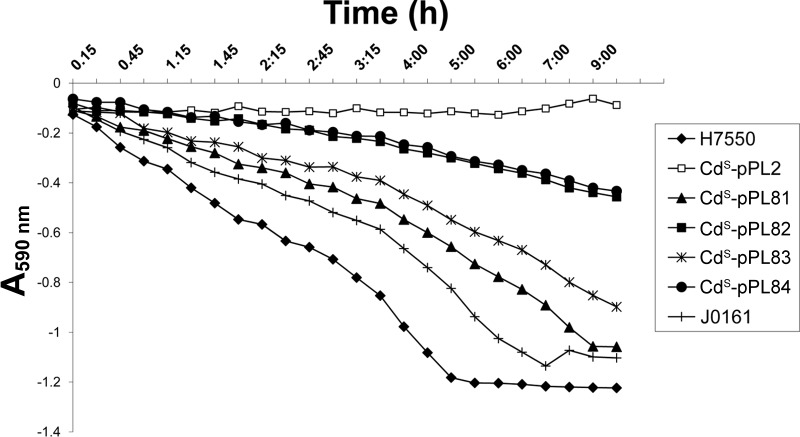
FIG 2.
L. monocytogenes strain H7550 and the derivatives of H7550-CdS harboring different tmr segments decolorize CV (15 μg/ml) at different rates. The experimental details are provided in Materials and Methods. This graph is a representative experiment from three independent trials.
Among the complemented integrated derivatives, CdS-pPL81 exhibited a consistently higher CV decolorization efficiency than CdS-pPL83 (Fig. 2). This was surprising, since CdS-pPL81 and CdS-pPL83 had similar CV MICs (Table 1). CdS-pPL83 harbored the entire (795-nt) intergenic region upstream of bcrA, whereas CdS-pPL81 only included 105 nt upstream of bcrA; both constructs included the promoter P1 (Fig. 1). The higher decolorization efficiency of CdS-pPL81 suggests the possibility of negative regulatory controls based on sequences within portions of the intergenic region present in CdS-pPL83 but lacking in CdS-pPL81. Indeed, this intergenic region is unusually long (795 nt), suggesting potential for regulatory involvement, e.g., via small noncoding RNAs (10). The decolorization potential of CdS-pPL82 and CdS-pPL84, although identical, was lowest among the complemented strains (Fig. 2). Together, these data suggest that, even though tmr can indeed be expressed on its own likely via P2 (as in pPL82 and pPL84), such expression is at lower levels than those observed with the multicistronic message that includes bcrABC and the other genes (as in pPL81 and pPL83).
Previous analysis of the bcrABC region indicated that bcrABC was transcribed together with several downstream genes, of which tmr was the last in the operon (22). It is tempting to speculate that the current location of tmr in the bcrABC region, and under control by P1, was selected upon due to the higher levels of expression observed in the presence of both P1 and P2.
Even though CV resistance and decolorization potential was restored in CdS-pPL81, CdS-pPL82, CdS-pPL83, and CdS-pPL84, both the MICs and the decolorization potential remained below those observed with the wild-type strain H7550 harboring pLM80 (Table 1 and Fig. 2). Copy number differences may account for this finding, since in the complemented strains the genes were integrated as single copies in the chromosome. It is also possible that pLM80 harbors additional, as-yet-uncharacterized determinants that contribute to CV resistance and decolorization.
As expected by the documented role of bcrABC in BC resistance, only CdS-pPL81 and CdS-pPL83 acquired BC resistance (BC 20 μg/ml), whereas CdS-pPL82 and CdS-pPL84 remained BC susceptible (data not shown). Strain CdS-pPLBC harboring just P1 and bcrABC restored resistance to BC but not to CV and did not decolorize CV-15 or confer resistance to the dye (data not shown).
No differences were found in the growth rates of H7550, H7550-CdS, and the H7550-CdS complemented derivatives in BHI without CV (data not shown). After a 4.5-h exposure of L. monocytogenes H7550 and CdS-pPL2 to 7.5 μg/ml CV, both strains exhibited a decrease in viability (∼2-log10 reduction in CFU/ml) (Fig. 3). During this period, L. monocytogenes H7550 completely decolorized CV and resumed growth, as evidenced by higher CFU at 24 h; in contrast, no decolorization was detected for CdS-pPL2, and the survival of this strain continued to decline, with an ∼4.5-log10 reduction by 24 h (Fig. 2 and 3). Assessing the CFU/ml at 48 h indicated further reductions (∼6-log10 reduction) in CdS-pPL2 but not in strain H7550 (data not shown). Together with the recovery of CV tolerance and decolorization potential by the genetically complemented strains, these findings indicate that tmr-mediated decolorization of CV results in detoxification of this dye for L. monocytogenes.
FIG 3.
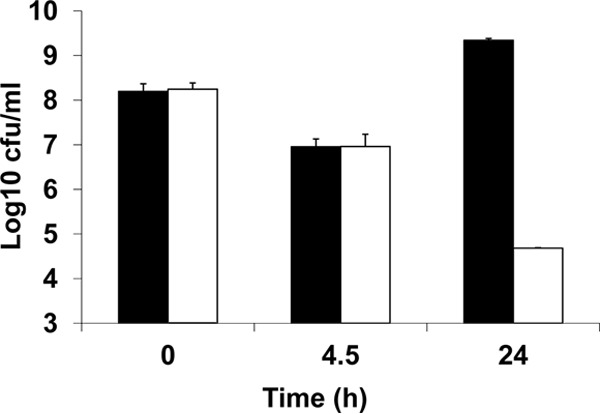
Decolorization of crystal violet renders the dye nontoxic for L. monocytogenes. Black bars represent CFU of H7550 upon exposure to CV (15 μg/ml), whereas white bars represent CFU of CdS-pPL2 upon exposure to CV (15 μg/ml). These data are based on two independent trials.
The transcript levels of bcrABC and tmr increased in the presence of CV.
Previous data indicated that tmr was part of a 3.8-kb transcript that also included bcrABC (mediating resistance to BC). The amounts of this polycistronic transcript increased in the presence of sublethal amounts of BC (22, 23). To determine whether the exposure to CV could also result in enhanced amounts of this transcript, we used reverse transcription-PCR (RT-PCR) to assess the impact of CV treatment on the expression of bcrABC. The exposure of H7550 to CV-15 indeed resulted in enhanced amounts of the bcrABC transcript, even though these amounts were lower than observed by exposure to BC at 10 μg/ml (Fig. 4) (22). These findings suggest that bcrABC expression could be induced not only by BC but also by CV.
FIG 4.
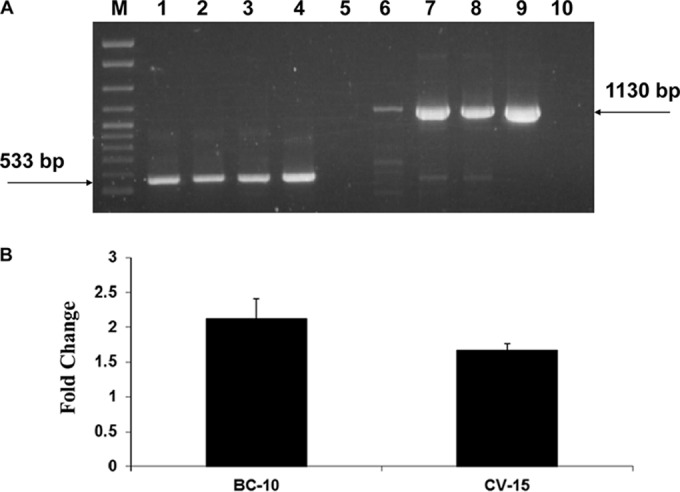
Transcription of bcrABC is induced by CV (15 μg/ml). (A) RT-PCR of spoVG and bcrABC. Lanes 1 to 5, RT-PCR of spoVG using primer s2 (for cDNA) and primers s1 and s2 for PCR (22, 23) as follows: lane 1, H7550 in the absence of BC or CV; lane 2, H7550 exposed to BC-10 μg/ml; lane 3, H7550 exposed to CV (15 μg/ml); lanes 4 and 5, H7550 genomic DNA and total RNA, respectively, used as a positive and negative controls for RT-PCR. An arrow points to the expected spoVG PCR product of 533 bp. Lanes 6 to 10, RT-PCR of bcrABC using primer p2 (for cDNA) and primers p1 and p2 for PCR (22, 23), as follows: lane 6, H7550 in the absence of BC or CV; 7, H7550 exposed to BC (10 μg/ml); 8, H7550 exposed to CV (15 μg/ml); 9 and 10, H7550 genomic DNA and total RNA, respectively, used as a positive and negative control for RT-PCR. Arrow points to the expected bcrABC PCR product of 1130 bp. M, 100- to 2,686-bp DNA molecular marker XIV (Roche). The CV and BC exposures were for 30 min, as described in Materials and Methods. The product in lane 6 represents the baseline levels of bcrABC, as previously reported (22). (B) Fold change in transcript level of bcrABC upon exposure to BC (10 μg/ml) and CV (15 μg/ml). The fold change was calculated using the ImageJ software (rsb.info.nih.gov/ij) as described previously (22).
qPCR analysis with H7550 revealed that tmr was expressed at low levels in the absence of CV, but expression increased significantly (2.6-fold; P = 0.012) upon CV exposure. Increases were also noted for the integrated constructs CdS-pPL81 (4.3-fold, P = 0.003) and CdS-pPL83 (3-fold, P = 0.005) (Fig. 5). In spite of repeated efforts, we were unable to obtain RNA of sufficient quantity and quality from CdS-pPL82 and CdS-pPL84 upon exposure to CV-15, possibly due to increased cell death due to CV exposure even during the 30-min incubation. Thus, the impact of CV on expression of tmr under its own promoter could not be assessed. The baseline levels of tmr in the absence of CV did not differ significantly between H7550 and any of the complemented derivatives (data not shown).
FIG 5.
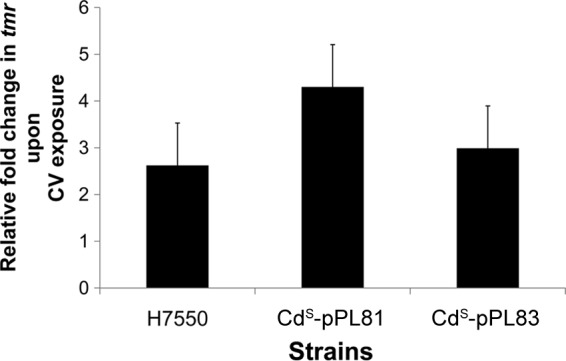
Relative increase in tmr transcript levels upon exposure to CV (15 μg/ml) in H7550 and H7550-CdS derivatives. Quantification of tmr transcripts was performed using qRT-PCR as described in Materials and Methods. The tmr transcript levels were first normalized to those of spoVG from the same treatments. These normalized tmr transcript levels were used to calculate the relative fold increase in tmr upon exposure to CV (15 μg/ml). These data were computed from three independent trials.
Presence of tmr is correlated with bcrABC and resistance to CV decolorization in L. monocytogenes strains of diverse serotypes.
PCR analysis of 47 strains of diverse serotypes and of environmental or clinical origins, as described in Materials and Methods (including 34 strains resistant to both CV and BC, 9 strains resistant only to BC, and 4 strains susceptible to both BC and CV), confirmed that all 34 CV-resistant strains harbored tmr, as well as bcrABC, whereas CV-susceptible strains were negative for tmr, regardless of whether they were BC resistant or not (data not shown). Analysis of numerous other L. monocytogenes strains has failed to identify any that are CV resistant without also being resistant to BC (data not shown). Previous data have indeed shown that strains harboring conserved bcrABC and adjacent sequences, including tmr, had a pLM80-like bcrABC region and were of diverse serotypes and sources (23). The findings are in agreement with BLAST analysis of L. monocytogenes genomes discussed above, which revealed tmr only when bcrABC was also present. Additional analysis of recently sequenced L. monocytogenes genomes (n = 100) from the Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/sra) further supports these findings (data not shown).
In conclusion, our findings suggest that in L. monocytogenes CV resistance is associated with tmr and that this gene is typically encountered as a member of a multisubstrate resistance region on pLM80 and related plasmids. In pLM80 this region confers resistance to quaternary ammonium disinfectants, triphenylmethane dyes, and the heavy metal cadmium and appears to be a component of a composite transposon (20–22). Such genetic attributes associated with mobility likely account for the observed dissemination of tmr and the other resistance determinants among strains of diverse serotypes and sources.
It is intriguing that tmr appears to have been introduced into Listeria from a Gram-negative source and that it was incorporated into the resistance region in a way that allows its transcription not only from its own promoter, conserved in numerous triphenylmethane dye-resistant Gram-negative bacteria, but also utilizing the bcrABC promoter for expression. This genetic arrangement appears to have resulted in higher levels of tmr expression and was likely selected in environments with high triphenylmethane dye burdens. Findings from the current study will be useful in elucidating adaptations and evolution of Listeria in such ecosystems and in future assessments of the potential role of tmr in additional adaptations, including virulence.
ACKNOWLEDGMENTS
This study was partially supported by a grant from the American Meat Institute Foundation, USDA-AFRI grant 2011-2012-67017-30218, and USDA grant 2006-35201-17377.
We thank all of the members of our laboratory, especially Robin M. Siletzky, for their support and encouragement.
Footnotes
Published ahead of print 20 June 2014
REFERENCES
- 1.Lawrence LM, Gilmour A. 1995. Characterization of Listeria monocytogenes isolated from poultry products and from the poultry-processing environment by random amplification of polymorphic DNA and multilocus enzyme electrophoresis. Appl. Environ. Microbiol. 61:2139–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ojeniyi B, Wegener HC, Jensen NE, Bisgaard M. 1996. Listeria monocytogenes in poultry and poultry products: epidemiological investigations in seven Danish abattoirs. J. Appl. Bacteriol. 80:395–401. 10.1111/j.1365-2672.1996.tb03234.x [DOI] [PubMed] [Google Scholar]
- 3.Soumet C, Ragimbeau C, Maris P. 2005. Screening of benzalkonium chloride resistance in Listeria monocytogenes strains isolated during cold smoked fish production. Lett. Appl. Microbiol. 41:291–296. 10.1111/j.1472-765X.2005.01763.x [DOI] [PubMed] [Google Scholar]
- 4.Thevenot D, Delignette-Muller ML, Christieans S, Vernozy-Rozand C. 2005. Prevalence of Listeria monocytogenes in 13 dried sausage processing plants and their products. Int. J. Food Microbiol. 102:85–94. 10.1016/j.ijfoodmicro.2004.12.008 [DOI] [PubMed] [Google Scholar]
- 5.Norwood DE, Gilmour A. 1999. Adherence of Listeria monocytogenes strains to stainless steel coupons. J. Appl. Microbiol. 86:576–582. 10.1046/j.1365-2672.1999.00694.x [DOI] [PubMed] [Google Scholar]
- 6.Gandhi M, Chikindas ML. 2007. Listeria: a food-borne pathogen that knows how to survive. Int. J. Food Microbiol. 113:1–15. 10.1016/j.ijfoodmicro.2006.07.008 [DOI] [PubMed] [Google Scholar]
- 7.Kim JW, Kathariou S. 2009. Temperature-dependent phage resistance of Listeria monocytogenes epidemic clone II. Appl. Environ. Microbiol. 75:2433–2438. 10.1128/AEM.02480-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullapudi S, Siletzky RM, Kathariou S. 2008. Heavy-metal and benzalkonium chloride resistance of Listeria monocytogenes isolates from the environment of turkey-processing plants. Appl. Environ. Microbiol. 74:1464–1468. 10.1128/AEM.02426-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freitag NE, Port GC, Miner MD. 2009. Listeria monocytogenes—from saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 7:623–628. 10.1038/nrmicro2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Régnault B, Coppée JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. 10.1038/nature08080 [DOI] [PubMed] [Google Scholar]
- 11.Gregory P. 1993. Dyes and dyes intermediates, p 544–545 In Kroschwitz JI. (ed), Encyclopedia of chemical technology, vol 8 John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 12.Hessel A, Allegre C, Maisseu M, Charbit F, Moulin P. 2007. Guidelines and legislation for dye house effluents. J. Environ. Manage. 83:171–180. 10.1016/j.jenvman.2006.02.012 [DOI] [PubMed] [Google Scholar]
- 13.Azmi W, Sani RK, Banerjee UC. 1998. Biodegradation of triphenylmethane dyes. Enzyme Microb. Technol. 22:185–191. 10.1016/S0141-0229(97)00159-2 [DOI] [PubMed] [Google Scholar]
- 14.Jang MS, Lee YM, Kim CH, Lee JH, Kang DW, Kim SJ, Lee YC. 2005. Triphenylmethane reductase from Citrobacter sp. strain KCTC 18061P: purification, characterization, gene cloning, and overexpression of a functional protein in Escherichia coli. Appl. Environ. Microbiol. 71:7955–7960. 10.1128/AEM.71.12.7955-7960.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlüter A, Krahn I, Kollin F, Bönemann G, Stiens M, Szczepanowski R, Schneiker S, Pühler A. 2007. IncP-1-beta plasmid pGNB1 isolated from a bacterial community from a wastewater treatment plant mediates decolorization of triphenylmethane dyes. Appl. Environ. Microbiol. 73:6345–6350. 10.1128/AEM.01177-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huan M, Tai LL, Fang YC, Jin SJ, Gao Y, Hong Q, Peng LS. 2011. Biodegradation of malachite green by strain Pseudomonas sp. K9 and cloning of the tmr2 gene associated with an ISPpu12. World J. Microbiol. Biotechnol. 27:1323–1329 [DOI] [PubMed] [Google Scholar]
- 17.Li LT, Hong Q, Yan X, Fang GH, Ali SW, Li SP. 2009. Isolation of a malachite green-degrading Pseudomonas sp. MDB-1 strain and cloning of the tmr2 gene. Biodegradation 20:769–776 [DOI] [PubMed] [Google Scholar]
- 18.Jang MS, Kang NY, Kim KS, Kim CH, Lee JH, Lee YC. 2007. Mutational analysis of NADH-binding residues in triphenylmethane reductase from Citrobacter sp. strain KCTC 18061P. FEMS Microbiol. Lett. 271:78–82. 10.1111/j.1574-6968.2007.00709.x [DOI] [PubMed] [Google Scholar]
- 19.Kim MH, Kim Y, Park HJ, Lee JS, Kwak SN, Jung WH, Lee SG, Kim D, Lee YC, Oh TK. 2008. Structural insight into bioremediation of triphenylmethane dyes by Citrobacter sp. triphenylmethane reductase. J. Biol. Chem. 283:31981–31990. 10.1074/jbc.M804092200 [DOI] [PubMed] [Google Scholar]
- 20.Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT, Kolonay JF, Rasko DA, Angiuoli SV, Gill SR, Paulsen IT, Peterson J, White O, Nelson WC, Nierman W, Beanan MJ, Brinkac LM, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Haft DH, Selengut J, Van Aken S, Khouri H, Fedorova N, Forberger H, Tran B, Kathariou S, Wonderling LD, Uhlich GA, Bayles DO. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386–2395. 10.1093/nar/gkh562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuenne C, Voget S, Pischimarov J, Oehm S, Goesmann A, Daniel R, Hain T, Chakraborty T. 2010. Comparative analysis of plasmids in the genus Listeria. PLoS One 5:e12511. 10.1371/journal.pone.0012511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elhanafi D, Dutta V, Kathariou S. 2010. Genetic characterization of plasmid-associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998-1999 outbreak. Appl. Environ. Microbiol. 76:8231–8238. 10.1128/AEM.02056-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutta V, Elhanafi D, Kathariou S. 2013. Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl. Environ. Microbiol. 79:6067–6074. 10.1128/AEM.01751-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauer P, Chow MYN, Loessner MJ, Portnoy DA, Calendar R. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177–4186. 10.1128/JB.184.15.4177-4186.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JW, Dutta V, Elhanafi D, Lee S, Osborne JA, Kathariou S. 2012. A novel restriction-modification system is responsible for temperature-dependent phage resistance in Listeria monocytogenes ECII. Appl. Environ. Microbiol. 78:1995–2004. 10.1128/AEM.07086-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Ream A. 2008. Gene expression profiling of Listeria monocytogenes strain F2365 during growth in ultrahigh-temperature-processed skim milk. Appl. Environ. Microbiol. 74:6859–6866. 10.1128/AEM.00356-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.den Bakker HC, Cummings CA, Ferreira V, Vatta P, Orsi RH, Degoricija L, Barker M, Petrauskene O, Furtado MR, Wiedmann M. 2010. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics 11:688. 10.1186/1471-2164-11-688 [DOI] [PMC free article] [PubMed] [Google Scholar]