Abstract
Sake (Japanese rice wine) production is a complex, multistage process in which fermentation is performed by a succession of mixed fungi and bacteria. This study employed high-throughput rRNA marker gene sequencing, quantitative PCR, and terminal restriction fragment length polymorphism to characterize the bacterial and fungal communities of spontaneous sake production from koji to product as well as brewery equipment surfaces. Results demonstrate a dynamic microbial succession, with koji and early moto fermentations dominated by Bacillus, Staphylococcus, and Aspergillus flavus var. oryzae, succeeded by Lactobacillus spp. and Saccharomyces cerevisiae later in the fermentations. The microbiota driving these fermentations were also prevalent in the production environment, illustrating the reservoirs and routes for microbial contact in this traditional food fermentation. Interrogating the microbial consortia of production environments in parallel with food products is a valuable approach for understanding the complete ecology of food production systems and can be applied to any food system, leading to enlightened perspectives for process control and food safety.
INTRODUCTION
Humans have employed food fermentation since time immemorial to improve the safety, stability, flavor, nutrition, and value of their agricultural products. Traditionally, these processes have been driven by indigenous fungi and bacteria originating in raw materials, in autochthonous starter cultures, or in the processing environment itself (1), organisms that are responsible for these beneficial transformative processes as well as for product spoilage (2, 3). While most modern fermented foods are inoculated with defined starter cultures, traditional, uninoculated products remain celebrated for their historical and cultural significances (4), and indigenous microbial activity is often considered to increase the flavor complexity of these foods (5). The advent of high-throughput sequencing technologies has enhanced our ability to investigate the role of microbial communities in food systems with greater scale and sensitivity than ever possible (4), connecting the transmission of microbial communities in food production and food processing environments to their impact on food products.
Sake is the traditional, national alcoholic beverage of Japan. Sake is produced from rice through the saccharification of starch by Aspergillus flavus var. oryzae and subsequent alcoholic fermentation by Saccharomyces cerevisiae. Sake brewing involves a serial propagation process, beginning with koji, a solid culture consisting of rice and A. flavus var. oryzae (6) (Fig. 1). Polished, steamed rice is mixed with the dried spores of A. flavus var. oryzae and incubated for approximately 2 days. Koji is then pitched with more steamed rice, water, and yeast into the moto (seed mash) tank, an open mashing vessel, wherein fermentation occurs for 10 to 25 days. Next, the moto is moved to a larger vessel and mixed with increasing amounts of water, rice, and koji in three additions to form moromi, the main fermentation. Moromi fermentation occurs for 20 to 30 days, after which it is pressed, filtered, and typically pasteurized to become finished sake. Originally, sake brewing was performed entirely by autochthonous microorganisms. However, as sake fermentations are conducted in open fermenters, such methods are prone to microbial contamination. Thus, most modern sake production is inoculated with pure yeast and acidified with lactic acid in the moto to inhibit the growth of undesirable organisms. In contrast, in traditional moto fermentations, the growth of undesirable bacteria and wild yeast is inhibited by several factors (low pH, high concentration of sugar and nitrite, and low temperature). In particular, lactic acid and nitrite produced by specific bacteria play an important role for inhibition of undesirable bacteria (7). After a decrease of undesirable microorganisms, the indigenous yeast that is suitable for sake fermentation grows spontaneously or pure culture yeast is added. This traditional method of moto process is called “kimoto.”
FIG 1.

Generalized sake production schematic. Times and temperatures listed at each stage represent those typically used in the sake brewery featured in this study. A. oryzae, Aspergillus flavus var. oryzae.
Many studies have been conducted to reveal the microbial transitions that occur during kimoto-style sake production using culture-based techniques (7–9), but few have employed culture-independent techniques (10). In the early stages, bacteria (Micrococcus, Escherichia, Pseudomonas, Enterobacter, Aerobacter, and Achromobacter) and non-Saccharomyces yeasts (Pichia spp., Candida spp., Zygosaccharomyces spp.) have been detected (7–9). Among these, Gram-negative bacteria, including Escherichia and Pseudomonas, initially increase. Then, lactic acid bacteria such as Leuconostoc mesenteroides subsp. sake and Lactobacillus sakei grow and produce lactic acid, leading to decreased pH (6, 7, 11, 12). In parallel with these microbial community changes, rice starches are saccharified by A. flavus var. oryzae amylase activity in the moto, and wild yeasts and bacteria are inhibited by the low pH, high sugar concentration, and high concentration of nitrite (7). Subsequently, sake yeast (Saccharomyces cerevisiae) increases in the moto and conducts the main alcoholic fermentation (7).
To improve our understanding of kimoto fermentations, we analyzed the bacterial and fungal communities of koji and kimoto production in parallel with the processing environment of a North American sake brewery, using high-throughput marker gene sequencing, quantitative PCR (qPCR), and terminal restriction fragment length polymorphism (TRFLP). Results suggest that microbial transfer from the processing environment is responsible for driving microbial successions throughout sake fermentations.
MATERIALS AND METHODS
Sample collection and DNA extraction.
All samples were collected from a single sake brewery located in North America. This facility produces sake using the kimoto method, using no starter cultures except for Aspergillus flavus var. oryzae in their koji preparations. Koji samples were collected before inoculation with A. flavus var. oryzae, at mixing times, and at harvest across the 48-h preparation time of two separate batches. Moto and moromi samples from two separate production batches were collected in duplicate every other day for the first 2 weeks and then weekly thereafter. Biological replicates were collected from the moromi, which was fermented in two separate fermentation tanks. Equipment and environmental surfaces were sampled as previously described (1). Sterile cotton-tipped swabs (Puritan Medical, Guilford, ME) were moistened with sterile phosphate-buffered saline and streaked across a 100-cm2 area of the target surface in two perpendicular series of firm, overlapping S strokes, rotating the swab to ensure full contact of all parts of the swab tip and the surface. Samples were placed on ice and frozen immediately in a −20°C freezer for storage. Fermentation samples were centrifuged at 4,000 × g prior to DNA extraction. DNA was extracted using the standard protocol for the ZR fecal DNA MiniPrep kit (Zymo Research, Irvine, CA), with bead beating in a FastPrep-24 bead beater (MP Bio, Solon, OH), and stored at −20°C until further processing.
Sequencing library construction.
Amplification and sequencing was performed as described previously for bacterial (13) and fungal (14) communities. Briefly, the V4 domain of bacterial 16S rRNA genes was amplified using primers F515 (5′-NNNNNNNNGTGTGCCAGCMGCCGCGGTAA-3′) and R806 (5′-GGACTACHVGGGTWTCTAAT-3′) (15), with the forward primer modified to contain a unique 8-nucleotide (nt) barcode (poly-N section of primer is italicized) and 2-nt linker sequence (underlined portion) at the 5′ terminus. PCR mixtures contained 5 to 100 ng DNA template, 1× GoTaq Green master mix (Promega), 1 mM MgCl2, and 2 pmol of each primer. Reaction conditions consisted of an initial 94°C for 3 min followed by 40 cycles of 94°C for 45 s, 50°C for 60 s, and 72°C for 90 s and a final extension of 72°C for 10 min. Fungal internal transcribed spacer (ITS) 1 loci were amplified with primers BITS (5′-NNNNNNNNCTACCTGCGGARGGATCA-3′) and B58S3 (5′-GAGATCCRTTGYTRAAAGTT-3′) (14), with a unique 8-nt barcode and linker sequence incorporated in each forward primer. PCR mixtures contained 5 to 100 ng DNA template, 1× GoTaq Green master mix (Promega, Madison, WI), 1 mM MgCl2, and 2 pmol of each primer. Reaction conditions consisted of an initial 95°C for 2 min followed by 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 60 s and a final extension of 72°C for 5 min. Amplicons were combined into two separate pooled samples (keeping bacterial and fungal amplicons separate) at roughly equal amplification intensity ratios, purified using the QIAquick spin kit (Qiagen), and submitted to the UC Davis Genome Center DNA Technologies Core for Illumina paired-end library preparation, cluster generation, and 250-bp paired-end sequencing on an Illumina MiSeq instrument in two separate runs.
Data analysis.
Raw fastq files were demultiplexed, quality filtered, and analyzed using QIIME version 1.7.0 (16). The 250-bp reads were truncated at any site of more than three sequential bases receiving a quality score of <Q20, and any read containing ambiguous base calls or barcode/primer errors were discarded, as were reads with <75% (of total read length) consecutive high-quality base calls (17). Reverse primer sequences were trimmed from the ends of ITS sequences following demultiplexing. Operational taxonomic units (OTUs) were clustered at 97% identity using the QIIME subsampled reference OTU-picking pipeline using UCLUST Reference (18) against either the Greengenes 16S rRNA database (May 2013 release) (19) or the UNITE fungal ITS database (20), modified as described previously (14). OTUs were classified taxonomically against these same databases using the QIIME-based wrappers of RDP classifier (21) (16S sequences) or a UCLUST-based classifier (18). Any OTUs comprising less than 0.001% of total sequences for each run were removed prior to further analysis (17). Environmental surveillance heatmaps based on taxonomic abundances were visualized using SitePainter 1.1 (22).
The absolute abundance of individual bacterial and fungal taxa detected by marker gene sequencing was estimated as the product of their relative abundances (number of sequences identified as that taxon divided by total number of sequences observed) multiplied by the observed copy number of the corresponding gene detected by qPCR (16S rRNA gene copies for bacteria, ITS copies for fungi).
TRFLP.
Lactic acid bacterium-specific terminal restriction fragment length polymorphism (LAB-TRFLP) was performed as described previously using the primers NLAB2F (5′-[HEX]-GGCGGCGTGCCTAATACATGCAAGT-3′, where HEX is 6-carboxy-2′,4,4′,5′,7,7′-hexachlorofluorescein) and WLAB1R (5′-TCGCTTTACGCCCAATAAATCCGGA-3′) (23). PCR conditions consisted of an initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 45 s, annealing at 66°C for 30 s, and extension at 72°C for 45 s, with a final extension at 72°C for 5 min. Samples were purified using a QIAquick PCR purification kit (Qiagen), digested using enzymes MseI and Hpy188I according to the manufacturer's instructions, and submitted to the UC Davis College of Biological Sciences Sequencing Facility for capillary electrophoresis fragment separation. Electropherogram traces were visualized using the program Peak Scanner v1.0 (Applied Biosystems, Carlsbad, CA) using a baseline detection value of 10 fluorescence units. Peak filtration and clustering were performed with R software using TRFLP-STATS (24). OTUs were identified based on an empirical TRFLP database (23) and an in silico digest database generated with MiCA (25) of good-quality 16S rRNA gene sequences from RDP (26), allowing up to 3 nucleotide mismatches within 15 bp of the 5′ terminus of the forward primer. OTUs detected by TRFLP are reported as relative abundance, or the peak area of the corresponding terminal restriction fragment(s) divided by the total peak area observed for each sample.
Quantitative PCR.
In order to quantify net microbial biomass in sake samples and on equipment surfaces, qPCR was used to enumerate total fungi and bacteria. qPCR was performed in 20-μl reaction mixtures containing 2 μl of DNA template, 8 pmol of each respective primer, and 10 μl of TaKaRa SYBR 2× Perfect real-time master mix (TaKaRa Bio Inc.). Total fungi were quantified using the primers BITS (5′-ACCTGCGGARGGATCA-3′) and B58S3 (5′-GAGATCCRTTGYTRAAAGTT-3′) (14). Reaction conditions involved an initial step at 95°C for 30 s, followed by 40 cycles of 5 s at 95°C, 1 min at 55°C, and 1 min at 72°C. For amplification of total bacteria, the primers Uni334F (5′-ACTCCTACGGGAGGCAGCAGT-3′) and Uni514R (5′-ATTACCGCGGCTGCTGGC-3′) (27) were used. Reaction conditions consisted of an initial hold at 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. All reactions were performed in triplicate in optical-grade 96-well plates on an ABI Prism 7500 Fast real-time PCR system (Applied Biosystems). The instrument automatically calculated cycle threshold (CT), efficiency (E), confidence intervals, and Saccharomyces cerevisiae cell equivalents (fungi) or 16S rRNA gene copy number (bacteria) by comparing sample CT values to a standard curve of serially diluted genomic DNA extracted from a known concentration of S. cerevisiae or Escherichia coli cells.
Nucleotide sequence accession numbers.
Raw marker gene sequencing data were deposited in QIIME-DB (www.microbio.me/qiime) under the accession numbers 2278 (16S rRNA gene sequences) and 2279 (fungal ITS sequences).
RESULTS
Koji and sake preparation involve multistage microbial succession.
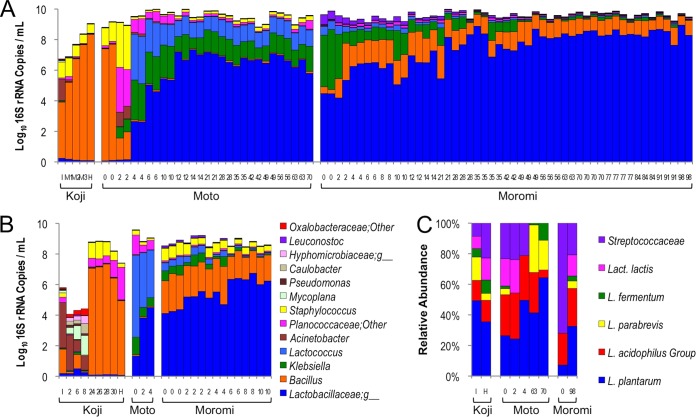
To elucidate the microbial processes involved in traditional kimoto sake fermentations, a combined culture-independent approach of marker gene sequencing, qPCR, and LAB-TRFLP (23) was used to profile two separate fermentations and koji preparations in a single North American sake brewery. Results demonstrated large changes in bacterial community composition and abundance over time (Fig. 2). Koji preparations were characterized by several-log bacterial growth over 48 h, reaching a maximum of close to 109 16S rRNA gene copies/ml (Fig. 2A and B). For the first 24 h following inoculation, the bacterial communities appear unpredictable and differed between batches but primarily consisted of Acinetobacter, Bacillus, and Staphylococcus. After 24 h, both koji batches were dominated by Bacillus with secondary populations of Staphylococcus and Planococcaceae. After being mixed with steamed rice and water to initiate moto production, the bacterial communities quickly changed from a koji-like profile to become dominated by Lactobacillaceae, Klebsiella, and Lactococcus within 4 days, accompanied by another 1-log increase in bacterial abundance to between 109 and 1010 16S rRNA gene copies/ml (Fig. 2A). As moto fermentation proceeded, Lactococcus and Klebsiella gradually declined, replaced by increasing populations of Lactobacillaceae. The onset of moromi, during which more steamed rice is added as the fermentation is mixed and transferred to a larger vessel, is characterized by another drastic change in bacterial community composition. Bacillus and Leuconostoc suddenly emerged in both batches before gradually decreasing during the course of fermentation. Lactococcus, Staphylococcus, and Klebsiella continued to decrease over the course of moromi fermentation, yielding to increasing Lactobacillaceae populations. However, no appreciable change in bacterial abundance occurred during the moto-to-moromi transition or during the remainder of the fermentation, through which it hovered around 108 to 109 16S rRNA copies/ml (Fig. 2A and B). Although some of the taxonomic groups detected in these fermentations include potentially pathogenic organisms (e.g., Klebsiella), both batches tested finished at 10% alcohol and pH 3.7, effectively preventing growth or survival of pathogenic organisms and explaining the decreased abundance of these groups as moromi fermentation progressed.
FIG 2.
Kimoto fermentations involve multistage bacterial succession. Bacterial community abundance and structure across time for batch A (A) and batch B (B). Column height indicates qPCR 16S rRNA gene copy number/ml. Relative abundance of each bacterial taxon (sequence count/total sequence count) derived from marker gene sequencing (key in panel B) is superimposed on each bar and does not correspond to the y axis. Only taxa detected at ≥1% maximum relative abundance are shown. (C) Relative abundance (OTU peak area/total peak area) of Lactobacillales detected in batch A by LAB-TRFLP (23). Units along x axis indicate hours (koji) or days (moto and moromi) since initiation of stage. I, inoculation with Aspergillus flavus var. oryzae; M, mixing; H, koji harvest.
Due to the limited heterogeneity of some bacterial taxonomic groups within the 16S rRNA gene V4 domain with the short read lengths achievable by marker gene sequencing methods, the dominant bacterial taxon in sake fermentations could be confidently identified only to the family level: Lactobacillaceae. Therefore, LAB-TRFLP (23) was used to identify which Lactobacillaceae were present during the course of sake fermentations. Results identified Lactobacillus plantarum as the most abundant species during the course of fermentation, with large populations of Lactobacillus parabrevis, Lactobacillus fermentum, and a group identified as either Lactobacillus acidophilus, Lactobacillus helveticus, or Lactobacillus amylolyticus (Fig. 2C). L. plantarum, L. acidophilus, and L. fermentum have all been described in sake previously (12). Consistent with the marker gene sequencing results, Lactococcus lactis and other Streptococcaceae (most likely other lactococci) were also detected during the fermentation course.
The fungal communities of kimoto fermentations exhibited comparatively less complexity, consisting primarily of S. cerevisiae and A. flavus var. oryzae throughout the course of fermentation (Fig. 3). Rapid fungal growth was observed over the 48-h course of koji preparation, from around 106 to 1010 S. cerevisiae cell equivalents/ml, consisting almost entirely of A. flavus var. oryzae with minor populations of S. cerevisiae. Populations continued to increase and stabilized around 1010 cell equivalents/ml through moto and moromi stages, during which time A. flavus var. oryzae dramatically decreases in favor of S. cerevisiae. Minor populations of Wickerhamomyces anomalus were observed sporadically in both batches in these later stages. In the initial days of batch 1 moromi, several other fungi were observed, including Phoma, Aspergillus, and an unknown Nectriaceae, disappearing within the first week (“Other” in Fig. 3).
FIG 3.
Fungal succession of kimoto fermentations. Fungal community abundance and structure across time for batch A (A) and batch B (B). Column height indicates qPCR S. cerevisiae cell equivalents/ml. Relative abundance of each fungal taxon (sequence count/total sequence count) derived from marker gene sequencing; key in panel B) is superimposed on each bar and does not correspond to the y axis. Only taxa detected at ≥1% average relative abundance are shown.
The processing environment is the source of adventitious microbiota in sake fermentations.
Marker gene sequencing and qPCR were also both applied to characterize the bacterial and fungal communities on equipment and surfaces within the sake brewery environment in order to observe sites of microbial transfer between the processing environment and these autochthonous sake fermentations. The adventitious microbiota detected during these sake fermentations were observed frequently throughout the brewery environment (Fig. 4). The greatest abundance of bacteria and fungi was detected within the main fermentation cellar, particularly in and around the moto, fermentation, and aging tanks. S. cerevisiae (99.9% maximum relative abundance) and the Lactobacillaceae OTUs (70.7%) detected in the fermentations were highly abundant at these sites, as were Bacillus (40.8%), Klebsiella (10.7%), Lactococcus (10.3%), Leuconostoc (1.8%), Staphylococcus (3.1%), and W. anomalus (38.5%), all organisms detected in the moto and moromi fermentations. The koji room and rice steaming room both displayed lower bacterial and fungal abundance than the main cellar. Microbes detected in the fermentations were less prevalent here, but Bacillus and Staphylococcus were detected at higher abundances on equipment surfaces within these rooms, corresponding to their detection in the koji preparations. While A. flavus var. oryzae was the dominant fungus in koji preparations, it was detected less frequently in the environment (73.5% maximum relative abundance inside koji room, 6.5% maximum elsewhere).
FIG 4.
Microbial drivers of kimoto fermentations are residents of the processing environment. Floor plan key (top) depicts all environmental surfaces analyzed. Microbial heatmaps (below) indicate estimated absolute abundance of select microbial taxa detected in high abundance in kimoto fermentations. Total bacteria and total fungi are results of actual qPCR data; estimated abundances of other taxa are the products of marker gene sequencing relative abundance (sequence count/total sequence count) multiplied by absolute abundance of the appropriate qPCR target. Color gradient logarithmic scale is indicated in the key (top right). White surfaces (fungal plots) were below the limit of detection.
DISCUSSION
Kimoto sake fermentations are a unique and increasingly rare fermentation tradition, employing indigenous microbiota to perform a multistage food fermentation. The several stages of production apparently involve a parallel succession of bacteria and fungi responsible for the fermentation. The initial stage, koji preparation, is a semiaerobic, stirred, solid fermentation, dominated by A. flavus var. oryzae (the only organism inoculated in the koji), Bacillus, and Staphylococcus, accompanied by a complex, variable consortium of adventitious bacteria (Fig. 2 and 3). Some of these groups, e.g., Pseudomonas, have been detected in early sake fermentations previously but do not persist (6, 7), consistent with our observations. A. flavus var. oryzae was the dominant fungus detected in the koji and the koji room environment, reflecting its use as an inoculum here. The marker gene sequencing method could not distinguish varieties of this fungus, but this OTU presumably represents Aspergillus flavus var. oryzae, the pure commercial inoculum used in this facility (and traditionally in sake and other food fermentations), and not other phytopathogenic, aflatoxin-producing varieties of A. flavus (28).
The moto, or seed mash, is the next propagation stage involved in sake production, during which prepared koji is mixed with steamed rice and water, precipitating a dramatic shift in the microbial communities and initiating alcoholic fermentation (Fig. 2 and 3). A. flavus var. oryzae, Bacillus, and other koji organisms rapidly declined, most likely because of decreased aerobiosis following hydration, and were replaced by S. cerevisiae, Lactobacillus spp., Lactococcus, and Klebsiella. This consortium bears considerable similarity to another autochthonous beverage fermentation, lambic-style coolship beers (29), providing a similar niche as a grain-based sugar substrate with relatively high pH and low alcohol prior to fermentation. While the roles of Saccharomyces and lactic acid bacteria in sake fermentations are well characterized—alcohol production and acidification, respectively—those of several other microbiota that appear in the moto are unclear. Klebsiella may play a similar role as in lambic-style beers, in which enterobacteria produce short-chain fatty acids and organic acids that contribute to product complexity (30). Consequently, they may be responsible for some of the more pungent aromas of kimoto compared to modern sake production.
The moromi, or main mash, involves mixing the moto with increasing quantities of water and steamed rice to start the main fermentation. Interestingly, Bacillus and Leuconostoc emerged at this stage in both batches analyzed, as well as Staphylococcus in batch B, reminiscent of the bacterial composition of the koji (Fig. 2). Though these taxa were detected throughout the main cellar, their sudden emergence in the moromi may suggest that the severalfold dilution of the moto with water and steamed rice introduces this microbial influx and encourages their growth until conditions restabilize. Spore-forming bacilli may survive rice steaming (31) and grow on the surface before alcohol increases and oxygen decreases, yielding the large populations observed in all moromi tanks and batches. Bacillus spp. are commonly reported in other rice wines and solid rice fermentations (32–38) in which the amylolytic activity of these bacteria may be an important contributor to saccharification (33). The role of bacilli in sake flavor development is unknown, but they can produce an array of ketones, acids, esters, and other compounds important to soybean fermentations (39) and may play a similar role here. Staphylococcus is frequently detected on human skin (40) and in food fermentations, including other Asian beverage fermentations that employ semisolid stages similar to koji preparation (34–37). The common pattern observed in rice fermentations of early dominance by Staphylococcus and Bacillus species succeeded by lactic acid bacteria has led other authors to speculate that these bacteria may produce growth factors conducive to lactic acid bacteria growth later during the fermentation (38), but this relationship has yet to be demonstrated. Leuconostoc species have also been frequently isolated from sake fermentations, in which it produces lactic acid (11, 12). In some kimoto fermentations, Leuconostoc mesenteroides can directly compete with L. sakei, providing the opportunity for growth of wild yeasts (9).
Surprisingly, no yeasts other than S. cerevisiae were detected in appreciable quantities throughout any of the sake fermentations or within the processing environment. Non-Saccharomyces yeasts have been reported in other sakes previously (8), as well as in other food processing environments (1, 41). Unlike wine and some cheese production, sake production involves raw material sterilization, rice steaming, prior to any production stage. This may limit the carryover of microbiota associated with the raw materials into the fermentation and into the processing areas. The low fungal biomass observed in the rice steaming and koji preparation rooms may be further evidence of this theory. The one non-Saccharomyces yeast detected in sakes and in the cellar was W. anomalus. This yeast is commonly detected in fermented beverages and other food products (42). It is considered typical in some food fermentations, including other rice wines (32), but causes spoilage in many foods through excessive ethyl acetate production (42). It was detected at low abundances in these kimoto fermentations, likely inhibited by the high alcohol concentration (42), but may be a typical member of these types of fermentations.
Most of the organisms commonly detected in these kimoto fermentations were also detected on equipment and other surfaces throughout the main cellar, particularly on processing equipment and fermentation tanks (Fig. 4). As these fermentations rely entirely on the growth of adventitious microbiota, their presence within the cellar demonstrates the importance of surface contact for possible bidirectional transfer of these organisms between fermentations. Similarly to artisan cheesemaking facilities (1), individual sake breweries may harbor unique, resident microbiota, potentially leading to regional differences in kimoto characteristics. However, the resident populations may not necessarily be stable and likely fluctuate seasonally as previously observed in wineries (41), altering the propensity for flavor development and spoilage by indigenous microbiota on a seasonal basis in response to changing environmental conditions. This may reflect the practice of performing traditional sake fermentations only during winter months, when cooler conditions would dampen spoilage potential. Further studies across multiple sake breweries and seasons will be necessary to establish the stability and regionality of sake brewery microbiota.
This study illuminates the role of brewery-resident, adventitious microbiota in spontaneous sake fermentations. The microbial succession of these fermentations closely corresponds to the microbial consortia inhabiting the production environment, illustrating the reservoirs and routes for microbial contact in traditional food fermentations. Interrogating the microbial consortia of production environments in parallel with food products is a valuable approach for understanding the complete ecology of food production systems. Using this model, a similar approach could—and should—be applied to any food production system, leading to enlightened perspectives for process control, spoilage prevention, and food safety.
ACKNOWLEDGMENTS
This study was supported in part by the National Tax Agency of Japan. N.A.B. was supported by the Samuel Adams Scholarship Fund (awarded by the American Society of Brewing Chemists Foundation), the American Wine Society Educational Foundation Endowment Fund scholarship, an American Society for Enology and Viticulture scholarship, and grant number T32-GM008799 from NIGMS-NIH during the completion of this work. M.O. is a visiting scholar from the National Tax Agency Japan.
We thank Masayuki Takahashi, Kazuhiro Iwashita, and Nami Goto-Yamamoto from National Research Institute of Brewing (Japan) for their insightful comments and suggestions. We also thank Chad Masarweh for technical support.
Footnotes
Published ahead of print 27 June 2014
REFERENCES
- 1.Bokulich NA, Mills DA. 2013. Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Appl. Environ. Microbiol. 79:5214–5123. 10.1128/AEM.00934-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bokulich NA, Bamforth CW. 2013. The microbiology of malting and brewing. Microbiol. Mol. Biol. Rev. 77:157–172. 10.1128/Microbiol.Mol.Biol.Rev.00060-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bokulich NA, Bamforth CW, Mills DA. 2012. A review of molecular methods for microbial community profiling of beer and wine. J. Am. Soc. Brew. Chem. 70:150–162. 10.1094/asbcj-2012-0709-01 [DOI] [Google Scholar]
- 4.Bokulich NA, Mills DA. 2012. Next-generation approaches to the microbial ecology of food fermentations. BMB Rep. 45:377–389. 10.5483/BMBRep.2012.45.7.148 [DOI] [PubMed] [Google Scholar]
- 5.Capozzi V, Spano G. 2011. Food microbial biodiversity and “microbes of protected origin.” Front. Microbiol. 2:237. 10.3389/fmicb.2011.00237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshizawa K. 1999. Sake: production and flavor. Food Rev. Int. 15:83–107. 10.1080/87559129909541178 [DOI] [Google Scholar]
- 7.Kitagaki H, Kitamoto K. 2013. Breeding research on sake yeasts in Japan: history, recent technological advances, and future perspectives. Annu. Rev. Food Sci. Technol. 4:215–235. 10.1146/annurev-food-030212-182545 [DOI] [PubMed] [Google Scholar]
- 8.Akiyama H. 1978. A microbiological control of sake brewing from the standpoint of ecology of yeasts. J. Ferment. Technol. Japan. 56:618–629 [Google Scholar]
- 9.Ashizawa T. 1976. The mystery of Japanese sake brewing focused on kimoto. J. Brew. Soc. Japan 71:424–427. 10.6013/jbrewsocjapan1915.71.424 [DOI] [Google Scholar]
- 10.Masuda Y, Noguchi T, Takahashi T, Iguchi A, Osawa R, Mizoguchi H. 2012. DGGE and PFGE analysis of lactic acid bacterial succession during kimoto making. Seibutsu-kogaku 90:684–690 [Google Scholar]
- 11.Katagiri H, Kitahara K, Fukami K. 1934. The characteristics of the lactic acid bacteria isolated from moto, yeast mashes for sake manufacture. Bull. Agr. Chem. Soc. Japan 10:156–157. 10.1271/bbb1924.10.156 [DOI] [Google Scholar]
- 12.Kitahara K, Kaneko T, Goto O. 1957. Taxonomic studies on the hoichi-bacteria, specific saprophytes of sake. J. Gen. Appl. Microbiol. 3:111–120. 10.2323/jgam.3.111 [DOI] [Google Scholar]
- 13.Bokulich NA, Joseph CML, Allen GR, Benson A, Mills DA. 2012. Next-generation sequencing reveals significant bacterial diversity of botrytized wine. PLoS One 7(5):e36357. 10.1371/journal.pone.0036357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bokulich NA, Mills DA. 2013. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 79:2519–2526. 10.1128/AEM.03870-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108:4516–4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez Pena A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. Qiime allows analysis of high-throughput community sequence data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10:57–59. 10.1038/nmeth.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 19.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME 6:610–618. 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abarenkov K, Nilsson RH, Larsson K-H, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Ursing BM, Vrålstad T, Liimatainen K, Peintner U, Kõljalg U. 2010. The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol. 186:1447–1452. 10.1111/j.1469-8137.2009.03160.x [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez A, Stombaugh J, Lauber CL, Fierer N, Knight R. 2012. SitePainter: a tool for exploring biogeographical patterns. Bioinformatics 28:436–438. 10.1093/bioinformatics/btr685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bokulich NA, Mills DA. 2012. Differentiation of mixed lactic acid bacteria communities in beverage fermentations using targeted terminal restriction fragment length polymorphism. Food Microbiol. 31:126–132. 10.1016/j.fm.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 24.Abdo Z, Schuette UME, Bent SJ, Williams CJ, Forney LJ, Joyce P. 2006. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ. Microbiol. 8:929–938. 10.1111/j.1462-2920.2005.00959.x [DOI] [PubMed] [Google Scholar]
- 25.Shyu C, Soule SJ, Bent SJ, Forster JA, Forney LJ. 2007. MiCA: a Web-based tool for the analysis of microbial communities based on terminal-restriction fragment length polymorphisms of 16S and 18S rRNA genes J. Microb. Ecol. 53:562–570. 10.1007/s00248-006-9106-0 [DOI] [PubMed] [Google Scholar]
- 26.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrel DM, Marsh TL, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartmann AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, Eisen JA. 2009. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc. Natl. Acad. Sci. U. S. A. 106:17187–17192. 10.1073/pnas.0904847106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurtzman C, Smiley MJ, Robnett CJ, Wicklow DT. 1986. DNA relatedness among wild and domesticated species in the Aspergillus flavus group. Mycologia 78:955–959. 10.2307/3807436 [DOI] [Google Scholar]
- 29.Bokulich NA, Bamforth CW, Mills DA. 2012. Brewhouse-resident microbiota are responsible for multistage fermentation of American coolship ale. PLoS One 7(4):e35507. 10.1371/journal.pone.0035507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martens H, Dawoud E, Verachtert H. 1992. Synthesis of aroma compounds by wort enterobacteria during the first stage of lambic fermentation. J. Inst. Brew. 98:421–425. 10.1002/j.2050-0416.1992.tb01126.x [DOI] [Google Scholar]
- 31.Lee SY, Chun HJ, Shin JH, Dougherty RH, Kang DH. 2006. Survival and growth of foodborne pathogens during cooking and storage of oriental-style rice cakes. J. Food Prot. 12:3037–3042 [DOI] [PubMed] [Google Scholar]
- 32.Thanh VN, Mai le, Tuan TDA. 2008. Microbial diversity of traditional Vietnamese alcohol fermentation starters (banh men) as determined by PCR-mediated DGGE. Int. J. Food Microbiol. 128:268–273. 10.1016/j.ijfoodmicro.2008.08.020 [DOI] [PubMed] [Google Scholar]
- 33.Ramos CL, de Almeida EG, Freire AL, Freitas Schwan R. 2011. Diversity of bacteria and yeast in the naturally fermented cotton seed and rice beverage produced by Brazilian Amerindians. Food Microbiol. 28:1380–1386. 10.1016/j.fm.2011.06.012 [DOI] [PubMed] [Google Scholar]
- 34.Li XR, Ma EB, Yan LZ, Meng H, Du XW, Zhang SW, Quan ZX. 2011. Bacterial and fungal diversity in the traditional Chinese liquor fermentation process. Int. J. Food Microbiol. 146:31–37. 10.1016/j.ijfoodmicro.2011.01.030 [DOI] [PubMed] [Google Scholar]
- 35.Zheng XW, Yan Z, Han BZ, Zwietering MH, Samson RA, Boekhout T, Robert Nout MJ. 2012. Complex microbiota of a Chinese “Fen” liquor fermentation starter (Fen-Daqu), revealed by culture-dependent and culture-independent methods. Food Microbiol. 31:293–300. 10.1016/j.fm.2012.03.008 [DOI] [PubMed] [Google Scholar]
- 36.Lv X-C, Weng X, Zhang W, Rao P-F, Ni L. 2012. Microbial diversity of traditional fermentation starters for Hong Qu glutinous rice wine as determined by PCR-mediated DGGE. Food Control 28:426–434. 10.1016/j.foodcont.2012.05.025 [DOI] [Google Scholar]
- 37.Jung MJ, Nam YD, Roh SW, Bae JW. 2012. Unexpected convergence of fungal and bacterial communities during fermentation of traditional Korean alcoholic beverages inoculated with various natural starters. Food Microbiol. 30:112–23. 10.1016/j.fm.2011.09.008 [DOI] [PubMed] [Google Scholar]
- 38.Koyanagi T, Nakagawa A, Kiyohara M, Matsui H, Yamamoto K, Barla F, Take H, Katsuyama Y, Tsuji A, Shijimaya M, Nakamura S, Minami H, Enomoto T, Katayama T, Kumagai H. 2013. Pyrosequencing analysis of microbiota in kaburazushi, a traditional medieval sushi in Japan. Biosci. Biotechnol. Biochem. 77:2125–2130. 10.1271/bbb.130550 [DOI] [PubMed] [Google Scholar]
- 39.Owens JD, Allagheny N, Kipping G, Ames JM. 1997. Formation of volatile compounds during Bacillus subtilis fermentation of soya beans. J. Sci. Food Agric. 74:132–140. [DOI] [Google Scholar]
- 40.Sanford JA, Gallo RL. 2013. Functions of the skin microbiota in health and disease. Semin. Immunol. 25:370–377. 10.1016/j.smim.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bokulich NA, Ohta M, Richardson P, Mills DA. 2013. Monitoring seasonal changes in winery-resident microbiota. PLoS One 8(6):e66437. 10.1371/journal.pone.0066437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Passoth V, Fredlund E, Druvefors UA, Schnurer J. 2006. Biotechnology, physiology and genetics of the yeast Pichia anomala. FEMS Yeast Res. 6:3–13. 10.1111/j.1567-1364.2005.00004.x [DOI] [PubMed] [Google Scholar]