Abstract
Many organisms harbor circadian clocks with periods close to 24 h. These cellular clocks allow organisms to anticipate the environmental cycles of day and night by synchronizing circadian rhythms with the rising and setting of the sun. These rhythms originate from the oscillator components of circadian clocks and control global gene expression and various cellular processes. The oscillator of photosynthetic cyanobacteria is composed of three proteins, KaiA, KaiB, and KaiC, linked to a complex regulatory network. Synechocystis sp. strain PCC 6803 possesses the standard cyanobacterial kaiABC gene cluster plus multiple kaiB and kaiC gene copies and antisense RNAs for almost every kai transcript. However, there is no clear evidence of circadian rhythms in Synechocystis sp. PCC 6803 under various experimental conditions. It is also still unknown if and to what extent the multiple kai gene copies and kai antisense RNAs affect circadian timing. Moreover, a large number of small noncoding RNAs whose accumulation dynamics over time have not yet been monitored are known for Synechocystis sp. PCC 6803. Here we performed a 48-h time series transcriptome analysis of Synechocystis sp. PCC 6803, taking into account periodic light-dark phases, continuous light, and continuous darkness. We found that expression of functionally related genes occurred in different phases of day and night. Moreover, we found day-peaking and night-peaking transcripts among the small RNAs; in particular, the amounts of kai antisense RNAs correlated or anticorrelated with those of their respective kai target mRNAs, pointing toward the regulatory relevance of these antisense RNAs. Surprisingly, we observed that the amounts of 16S and 23S rRNAs in this cyanobacterium fluctuated in light-dark periods, showing maximum accumulation in the dark phase. Importantly, the amounts of all transcripts, including small noncoding RNAs, did not show any rhythm under continuous light or darkness, indicating the absence of circadian rhythms in Synechocystis.
INTRODUCTION
Cyanobacteria must regulate many metabolic reactions in synchrony with the environment, such as day-night cycles. Every metabolic reaction has to be controlled temporally and/or spatially. Cyanobacteria perform photosynthesis during the day and respiration during the night (1, 2), as well as cell division only in certain phases (3). Enzymes (e.g., cytochrome b6f) shared by two incompatible biochemical processes (e.g., photosynthesis and respiration) are not fed into both pathways at the same time (4), and other disparate cellular processes, such as photosynthesis (produces oxygen as a by-product) and oxygen-sensitive nitrogen fixation, are separated in time in unicellular diazotrophic cyanobacterial species (5–7). In fact, the temporal separation of incompatible metabolic processes appears to be one of the driving forces for evolving an endogenous timing mechanism—the circadian clock (8).
At the heart of the circadian clock is an oscillator that integrates a series of biochemical events and stepwise modifications to its components to produce a 24-h timing loop (9). A true circadian clock keeps time of day. In consequence, rhythms in gene expression driven by a circadian clock proceed even under constant (free-running) conditions. In contrast, oscillations are expected to stop in constant light or darkness if the gene expression is regulated only in response to day-night cycles.
Circadian rhythms have been found in some cyanobacteria (10). The core circadian clock of cyanobacteria consists of solely three proteins (KaiA, KaiB, and KaiC) and is well described for the model strain Synechococcus elongatus PCC 7942 (referred to here as simply Synechococcus). In this strain, interactions among the three Kai proteins and cyclic KaiC phosphorylation are thought to set the timing signal for almost every cellular process. Examples include amino acid uptake (11), cell division (3, 12), global gene expression (10), chromosomal compaction (13), and DNA supercoiling (14–17).
In Synechococcus, the transcript amounts of 30 to 100% of all genes oscillate in a circadian fashion under free-running conditions, depending on the experimental setup (10, 15, 18, 19). For example, a promoter trap analysis detected oscillations in transcript abundance for almost all genes (10), while more recent microarray studies indicated that transcript abundance levels oscillate only for 30% of all genes (15, 19).
Synechocystis sp. strain PCC 6803 (referred to here as Synechocystis) possesses the standard kaiABC gene cluster (kaiAB1C1) plus multiple kaiB and kaiC gene copies (kaiC2B2 cluster, kaiB3, and kaiC3). KaiB3 and KaiC3 might fine-tune the oscillator encoded by kaiAB1C1 as to period length and phase of the circadian rhythm (20). While transcript abundance levels of at least 36% of all genes in the Synechocystis genome oscillate linked to diurnal cycles (21), the percentage of circadian-regulated genes is small, and the oscillations in the abundances of transcripts display small amplitudes in this cyanobacterium (19, 22–24). A microarray survey by Kucho et al. (22) reported that the amounts of transcripts of 2 to 9% of all genes in Synechocystis oscillate under constant light, indicating the involvement of a circadian clock system. A reanalysis of the same data supported the small percentage of circadian-regulated genes (19).
In many bacteria, including cyanobacteria, small regulatory RNAs (sRNAs) have been reported as important regulators of gene expression (25–29). By base pairing with their target mRNA(s), small RNAs can interfere with the ribosome binding site or other sequence stretches and, consequently, alter mRNA translation and stability. The small regulatory antisense RNA (asRNA) IsrR from Synechocystis was one of the first examples reported. We previously demonstrated that this antisense RNA causes a pronounced delay in induction of isiA (the target mRNA) under iron depletion conditions. Moreover, it ensures that the isiA mRNA is degraded rapidly once the external stress is removed (30, 31).
Recent global screenings using high-density microarrays and RNA sequencing approaches showed that 65% of all individual transcripts in Synechocystis represent small noncoding RNAs (ncRNAs) and that at least 26% of all transcripts are influenced by antisense RNAs (32, 33). In addition, there are antisense RNAs for almost every Synechocystis kai gene, encoded in cis on the respective noncoding strand (33). The regulatory relevance of small RNAs in diurnal gene expression in general, and of antisense RNAs in the stability of kai mRNAs in particular, has not been discussed so far.
In this study, we used microarrays to address the contributions of the circadian clock and light to the expression of protein-encoding genes and small noncoding RNAs in Synechocystis. We found that the transcript abundance levels of at least 27% of the genes analyzed oscillated under light-dark (LD) conditions but stopped being rhythmic in constant light or darkness. Under 12-h–12-h light-dark cycles, transcripts of functionally related genes accumulated in the same time phases, and small noncoding RNAs were upregulated at day or night. Furthermore, the diurnal accumulation dynamics of the antisense RNAs, including the kai antisense RNAs, correlated or anticorrelated with those of the target mRNAs. Surprisingly, the amounts of 16S and 23S rRNAs increased strongly during the night.
MATERIALS AND METHODS
Strains and growth conditions.
The glucose-tolerant and motile wild-type strain “PCC-M” of Synechocystis sp. PCC 6803, obtained from S. Shestakov (Moscow State University, Russia), was grown photoautotrophically in BG11-medium (34) at 30°C under continuous illumination with white light at 80 μmol of photons/m2-s1 (Versatile environmental test chamber; Sanyo) and with a continuous stream of air. Cell concentration was determined by measuring the optical density at 750 nm (OD750) of the culture (Specord200 Plus; Analytik Jena). Additionally, the cell number per ml was determined by manual counting (C-Chip, Neubauer improved; Biochrome). The culture was kept in log growth phase (OD750 of up to 1.0) by regular dilution to a specific volume.
Three days prior to the time series experiments, cultures grown in identical Schlenk tubes were diluted to a certain volume and an OD750 of ca. 0.4 and transferred to a 12-h–12-h LD cycle for synchronization. The time point of the transition from the third dark period to light was denoted time zero. All stated times are relative to this time point. The sampling series started at 5.5 h.
While culture 1 (four replicates) was maintained under a 12-h–12-h LD cycle, culture 2 (four replicates) and culture 3 (four replicates) were shifted to continuous light (LL) and continuous darkness (DD), respectively, at 24 h. During the time series experiments, 15 ml of each culture was sampled for RNA extraction every 2 h and, additionally, around the dark-to-light transition, at intervals of 10 and 30 min.
Monitoring the culture fitness throughout the whole experiment was realized by recording absorption spectra in the range of 400 to 750 nm (Specord200 Plus; Analytik Jena). Spectra were corrected for light scattering at 750 nm, and the ratio of phycocyanin to chlorophyll a was estimated according to the method of Myers et al. (35). Chlorophyll measurements were done according to a modified protocol by Houmard and de Marsac (36), as follows. A total of 0.5 ml of culture was centrifuged at 15,000 × g for 10 min. Four hundred microliters of the supernatant was removed, and the resuspended cell pellet was frozen in liquid nitrogen prior to storage at −20°C. Nine hundred microliters of 100% methanol was added and incubated in the dark on ice for 30 min. Samples were then centrifuged at 15,000 × g for 10 min, and absorption was measured at 665 nm. The chlorophyll concentration (in μg/ml) was calculated using the following formula: chlorophyll concentration = (A665 × 13.9) × 2.
RNA extraction and amount of total RNA per cell.
Fifteen milliliters of cyanobacterial culture was filtered through Supor 0.45-μm membrane filters (Pall), immediately mixed with TRIzol reagent (Invitrogen), and frozen in liquid nitrogen. Samples stored at −20°C for no more than 2 days were directly heated to 65°C for 5 min, mixed with 0.2 ml chloroform per ml of TRIzol, and incubated for another 15 min. The dissolving of the membrane and lysis of the cells were supported by vortexing. Centrifugation at maximum speed for 10 min at 4°C was performed to separate the phases. The RNA in the supernatant was precipitated by adding 1 volume of isopropanol. Finally, extracted total RNA was treated with RNase-free Turbo DNase (Ambion) by following the manufacturer's instructions, resulting in 40 μl purified RNA solution. For quantification and a quality check, a NanoDrop ND-1000 spectrophotometer (peqLab Biotechnology) and separation by electrophoresis on 1.3% agarose-formaldehyde gels were used.
The total RNA concentration value (total RNA in 1 ml culture) at each time point was derived from measurements on NanoDrop and Bioanalyzer instruments, which were normalized to the optical density of the culture at that time to account for different growth rates and then averaged. A total of 7.85 × 107 cells in 1 ml of cyanobacterial culture with an OD of 1 were counted. Given this cell number, the amount of total RNA per cell was calculated.
RNA analysis with an Agilent 2100 Bioanalyzer instrument.
The composition of the total RNA preparations was analyzed with an Agilent 2100 Bioanalyzer instrument, using an RNA 6000 nanokit according to the manufacturer's instructions (37). The unaligned raw data provided by Agilent 2100 Expert software were used to reanalyze the composition of the total RNA preparations with our own method, as follows. The unaligned raw data were divided by the migration time to compensate for higher fluorescence intensities of larger RNAs due to a longer pass by the detector. The ladder data were used to calculate the fluorescence of a given RNA amount. The migration times for all RNA sizes were estimated, and the rRNA peaks were identified. The background was estimated by linear regression between the left and right boundaries of the peak. The RNA concentrations were determined by calculating the respective peak areas over the background and relating the values to the ladder data. The RNA values calculated with our method were in good agreement with the concentrations obtained via 2100 Expert software. For analysis of the accumulation patterns of total RNA and rRNAs, the RNA samples from at least three biological replicates were used (i.e., at least three biological replicates each for LD, LL, and DD conditions).
Microarray design and hybridization.
Single custom-made 44K Agilent RNA microarrays, containing 20,443 probes for 3,352 protein-encoding genes, 1,931 antisense RNAs, and 620 other noncoding RNAs of the Synechocystis sp. PCC 6803 chromosome (NC_000911.1), as well as plasmid pSYSA (NC_005230), identified by Mitschke et al. (33), were used. Each probe was spotted two times on each chip. The second spots served as technical replicates. Full information on the microarray design is stored in the GEO database (accession number GPL15867).
For the microarray experiments, the RNA samples for one biological replicate each from LD, LL, and DD conditions were used. Two micrograms of RNA was directly labeled with Cy5 by using a Kreatech ULS labeling kit for Agilent gene expression arrays. A sample of 1.5 μg of the labeled RNA was hybridized to each microarray by following the Agilent protocol for single-color microarrays (32). RNAs from time points 11.5 and 23.5 (h) of the LD time series were hybridized twice to test the reproducibility of the results. RNAs from 10 time points of the constant-light time series (LL; 7.5, 9.5, 13.5, 15.5, 19.5, 21.5, 25.5, 37.5, 31.5, and 33.5 h) and from 4 time points of the constant-dark time series (DD; 7.5, 9.5, 13.5, and 15.5 h) were not hybridized (see File S2 in the supplemental material). These missing data were interpolated with the corresponding time points from the LD time series by using the raw chip data.
The microarrays were digitalized with an Agilent G2505B microarray scanner, using Agilent Feature extraction software 10.7.3.1 and protocol GE1_107_Sep09. The raw data for each spot were extracted with the R-package limma (38). Computational analysis was done with R (http://www.r-project.org/) and MATLAB (The MathWorks Inc.). The corresponding median background value was subtracted from the median intensity of each spot.
Data normalization and analysis.
Synechocystis sp. PCC 6803 exhibits strong oscillations in the total amount and composition of RNA, which adds strong bias when standard normalization methods, such as quantile normalization, are applied to the data. Moreover, the intensities of rRNAs could not be detected reliably because of saturation effects. These factors were considered in a previous study where several normalization approaches and data analyses were compared (39). Therefore, a least-oscillating-gene-set (LOS) method classified as best for analyzing Synechocystis time series data was used. A discrete Fourier transformation was applied to the time course of each probe. The same was applied to 500,000 random permutations. The combined power of the 24-h oscillations and their two harmonics, with periods of 12 and 8 h, was then calculated. The estimated likelihood (E value) was computed as the ratio of permutations with oscillatory power above the measured data. Probes with an E value of >0.7 (around 17% of all probes) were selected as the LOS. For each microarray, a LOWESS (locally weighted scatterplot smoothing) (40) polynome with a smoothing parameter of 0.2 was fitted to the raw data from LOS probes and their average expression over the whole time series, similar to a procedure described previously (41). All probe values were normalized regarding these polynomes. After that, intensities of multiple probes corresponding to the same gene were averaged. The E values for the normalized data were recalculated, and transcripts with ratios of <0.01 were classified as significantly oscillating. Several clustering methods were also tested by Lehmann et al. (39). Again, the method rated best for our data was chosen. All protein-encoding genes were grouped based on the accumulation profiles of their transcripts under LD conditions, using the model-based clustering algorithm flowClust (42) with standard parameters. Visual inspections of clusterings with 2 to 15 groups revealed a most promising separation when the data were divided into 10 groups. A gene ontology (GO) analysis was performed; all GO terms for Synechocystis were downloaded from the gene ontology database (www.geneontology.org; accessed 5 January 2011). Fisher's exact test was applied to each GO term-cluster combination to test for statistical significance.
Comparison with Synechococcus data.
Our results were compared with the existing transcript accumulation profiles of homologous genes in Synechococcus elongatus PCC 7942, taken from the supplemental material provided by two studies (15, 19). Only transcripts classified as cyclic in both studies and with peak times of transcripts differing by less than 8 h were considered oscillating in Synechococcus. Gene homology between both species was investigated in a previous study (43).
Microarray data accession number.
The microarray data are accessible in the GEO database under accession number GSE47482.
RESULTS
We used a microarray technique that allowed us to screen Synechocystis for mRNAs and small regulatory RNAs. So far, compared with previous investigations, this microarray technique has the highest time and transcript resolution, comprising 3,352 protein-encoding genes and 2,251 noncoding transcripts. The microarray data are accessible in the GEO database under accession number GSE47482.
To analyze how many genes and small noncoding RNAs are circadian or light regulated in Synechocystis, we performed one time series experiment where we kept synchronized Synechocystis cultures under a 12-h–12-h light-dark (LD) cycle, whereas in a second time series experiment, we transferred cultures to continuous light (LL), and in a third time series, we used continuous darkness (DD). We chose these three light regimens to overcome the various experimental setups used in different studies and to reliably answer the question of whether gene transcripts and small noncoding RNAs are regulated by circadian rhythms in Synechocystis.
The cultures were sampled for RNA extraction every 2 h or within 10- and 30-min intervals (around the dark-to-light transition) over a time course of 48 h. The sample times indicated here refer to the time point of transition from darkness to light. This event marked the beginning of the time series experiments and is defined as time zero. The sampling started at 5.5 h (see Materials and Methods). Growth, chlorophyll concentration, and the ratio of phycocyanin to chlorophyll were monitored during sampling. In summary, the physiological data indicated that all cultures grew under unstressed conditions (see Fig. S1 and File S1 in the supplemental material). For the microarray analysis, the RNA samples from one biological replicate each from LD, LL, and DD conditions were used; for the analysis of the accumulation patterns of total RNA and rRNAs, the RNA samples from at least three biological replicates were employed.
Rhythms in transcript amounts are found only during cyclic light-dark periods.
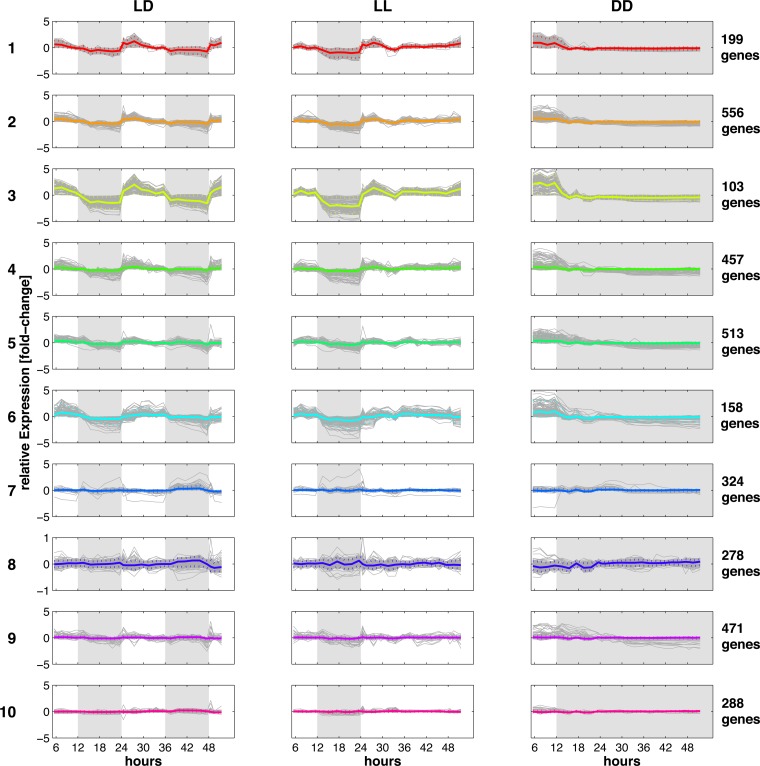
We first investigated if there are genes in Synechocystis whose transcript amounts show circadian rhythm. We clustered the transcripts of all protein-encoding genes of the LD time series with respect to their abundance to obtain a day-and-night pattern of coaccumulated transcripts. The left column of Fig. 1 shows the gene transcript abundance profile and the mean abundance pattern of each cluster. The clusters are sorted by phase, as indicated by colors, ranging from transcripts upregulated early in the light phase (cluster 1; red) to transcripts most abundant in the late light phase (cluster 6; turquoise) and transcripts of genes active at night (clusters 7 and 8; blue). Transcripts showing no oscillation are grouped in clusters 9 (purple) and 10 (pink). We estimated the percentage of genes whose transcripts were oscillating by performing a Fourier transformation and comparing the power of the 24-h oscillation as well as its harmonic with the power in randomly permutated expression profiles (see Materials and Methods).
FIG 1.
Clustering of the microarray data. The 48-h sampling of Synechocystis cultures started 5.5 h after the onset of light. During the 48-h period, culture 1 was kept in a 12-h–12-h light-dark cycle (LD), culture 2 was transferred to constant light at 24 h (LL), and culture 3 was kept in darkness at 12 h (DD). For LD, LL, and DD conditions, the transcripts of 3,352 protein-encoding genes were each grouped into 10 clusters according to their accumulation phases seen in LD, starting with transcripts of genes peaking in the early morning (cluster 1) and ending with gene transcripts peaking in darkness (cluster 8). Clusters 9 and 10 consist of transcripts whose amounts were almost unchanged. For each cluster, the relative transcript amounts throughout the 48 h are given on a logarithmic scale. Solid colored lines depict the averaged transcript accumulation profiles. Dashed colored lines show the 5% and 95% quantiles contouring the accumulation pattern for the majority of the gene transcripts. Dark phases are indicated in gray. The number of genes in each cluster is shown on the right.
According to our analysis, transcripts of at least 27% of all genes in Synechocystis cycled with a light-dark rhythm. However, the rhythms of these transcript levels were damped immediately after the shift from LD to LL or DD (Fig. 1, middle and right columns). No transcripts were found to be oscillating significantly in LL and DD (E values of <0.01). In particular, the level of day-peaking gene transcripts in LD (clusters 1 to 6) reached a steady high level in constant light and a steady low level in constant darkness. The microarray results from LD and LL conditions were confirmed for a set of genes by using quantitative real-time PCR (see File S1 and Fig. S2 in the supplemental material). In summary, these findings indicate an absence of circadian rhythm at the transcriptome level.
Day-night accumulation pattern of functionally related transcripts.
Even though we did not observe circadian rhythms in Synechocystis, we discovered that functionally related genes were coregulated under LD conditions.
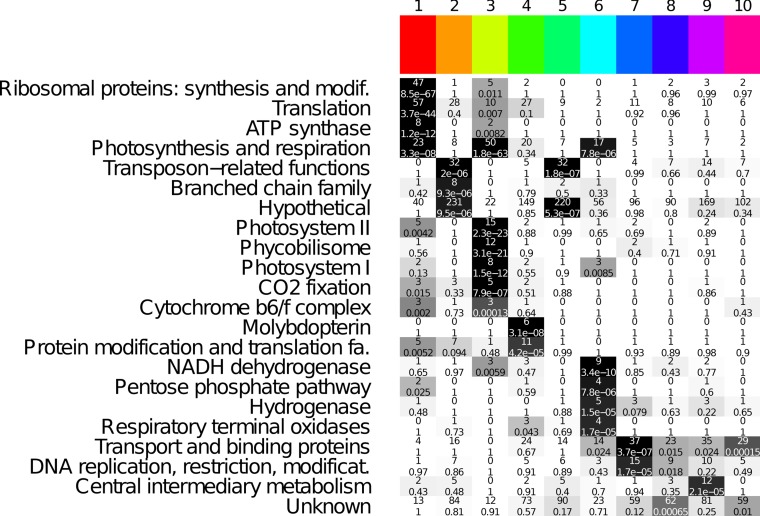
For the LD transcriptome analysis, we determined the associated gene ontology (GO) term for every clustered gene transcript and computed the GO enrichment within each cluster (see Materials and Methods for details). The most significant enrichments are listed in Fig. 2. Our GO enrichment analysis revealed that various cellular processes were already upregulated early in the morning—but not before the transition to light—such as genes encoding ribosomal proteins involved in translation, genes necessary for energy production (ATP synthase subunit genes) and amino acid biosynthesis (branched-chain family amino acid metabolism), genes associated with photosynthesis, including photosystems I and II (PSI and PSII), genes for carbon fixation, and genes encoding cytochrome and phycobilisome complexes (clusters 1 to 3). Later in the day, the molybdopterin gene cluster and genes encoding several translation factors (e.g., ribosome releasing factor and elongation factor EF-G) were activated (cluster 4). The transcript profile of cluster 5, comprising mainly genes with unknown function, was similar to that of cluster 4, though the amplitudes of the transcript abundance levels were lower. Toward the end of the day (cluster 6), processes such as glycolysis and catabolic metabolism (pentose phosphate pathway), as well as respiration, seemed to be activated to prepare for the upcoming night. The upregulation included genes coding for NADH dehydrogenase subunits, cytochrome c oxidase, pyridine nucleotide transhydrogenase, 6-phosphogluconate dehydrogenase (6PGD), glucose-6-phosphate dehydrogenase (G6PD), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and the enzyme transaldolase. During the night (clusters 7 and 8), the mRNA levels for genes coding for several transport processes, as well as DNA repair (e.g., mutL and recN) and replication (e.g., the gyrase gene, dnaX, mutT, and the heat shock gene dnaK2), were enhanced. Besides gene groups whose transcript amounts were increased at specific times, there also existed groups of transcripts of functionally related genes distributed over various clusters. These groups included transcripts of genes essential for cell division (minC and ftsZ [cluster 4], minD [cluster 1], minE [cluster 10], and zipN [cluster 5]) and mRNAs of sigma factor genes, such as sigA (slr0653 [cluster 1]), sigB, sigC (sll0306 and sll0184 [cluster 5]), sigD (sll2012 [cluster 10]), and sigE (sll1689 [cluster 6]). Overall, our analysis revealed that the schedule within which the transcript amounts of genes are regulated is consistent with the times at which cellular processes are expected to occur. This indicates that Synechocystis possesses an endogenous timing mechanism even when it does not generate circadian rhythms under the conditions applied here.
FIG 2.
GO term enrichment analysis. A tabular overview of the cluster-wise functional enrichment is shown. Each row corresponds to one biological function (GO term). Each column corresponds to a single cluster of coaccumulated gene transcripts. Each cell shows the absolute number of genes with the respective biological function in the cluster, as well as an enrichment P value calculated with Fisher's exact test. For ease of presentation, the background of each cell is shaded according to its enrichment. More significant entries appear darker. Genes with maximum transcript accumulation during the day are in clusters 1 to 6 (early to late day). Genes with maximum transcript accumulation during the night are in clusters 7 and 8. Genes showing constant transcript accumulation are in clusters 9 and 10. The P values were corrected using the Bonferroni method.
Diurnal changes in abundance of long (ribosomal) RNAs.
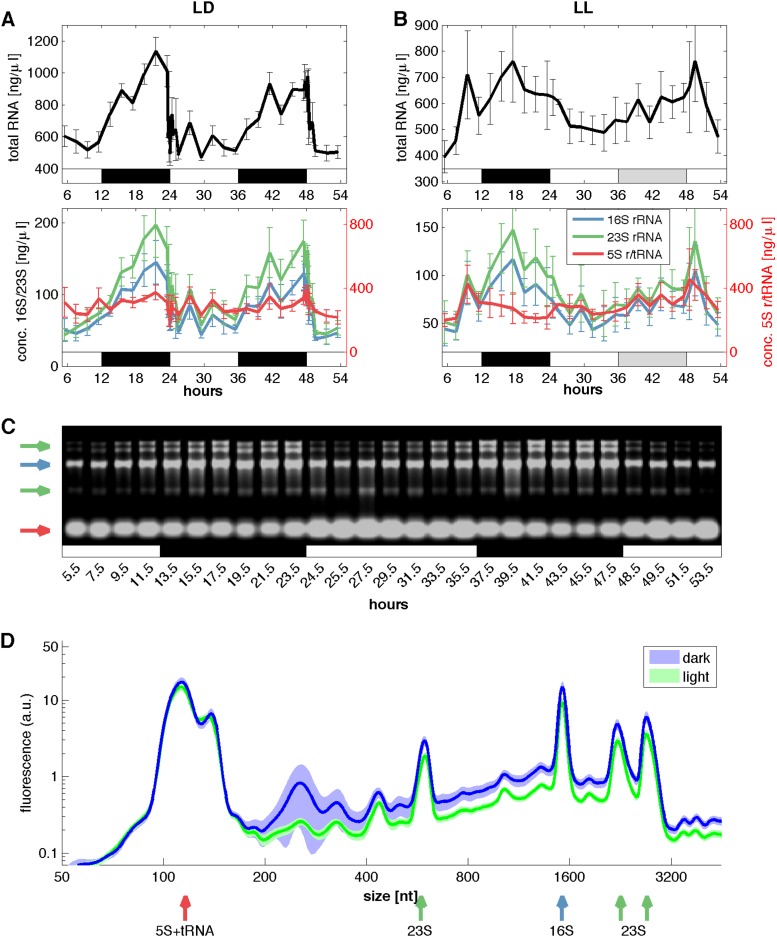
When we surveyed the total RNA accumulation dynamics of the LD and LL cultures (hours 5 to 24), we discovered that the amount of total RNA per cell increased during the dark periods (Fig. 3A and B, upper panels) and thus was anticorrelated with cell growth (see Fig. S1 in the supplemental material). In particular, total RNA accumulated steadily over the night phase, until the level suddenly dropped when the light was switched on at 24 or 48 h (Fig. 3A). The increase in the amount of total RNA during the night phase was also quantified numerically. For the LD time series, we calculated the absolute amount of RNA per cell and found about 1.76 × 10−14 g RNA per cell during the day. During the night, the absolute RNA level rose to more than twice that amount, i.e., 3.76 × 10−14 g RNA per cell. This oscillatory accumulation pattern was lost in LL at subjective night (hours 36 to 48) (Fig. 3B).
FIG 3.
Total amount and composition of RNA in Synechocystis sp. PCC 6803. Synechocystis shows massive diurnal changes in the total amount of RNA as well as in the composition of RNA. (A) Concentrations of total RNA and rRNAs in LD. The total RNA data are means from four biological replicates measured with an Agilent 2100 Bioanalyzer and a NanoDrop spectrometer. rRNA data are means from four biological replicates measured with an Agilent 2100 Bioanalyzer. (B) Concentrations of total RNA and rRNAs in LL. The RNA data are means from four measurements on an Agilent 2100 Bioanalyzer, plus two additional measurements on a NanoDrop spectrometer for the total RNA. The data in panels A and B are concentrations of RNA in 1 ml cell culture with an OD750 of 1. Error bars indicate the standard errors of the means. The concentrations of 5S rRNA in both lower panels are shown by the red y axes on the right. Please note the different scaling of the LD and LL axes. (C) Agarose gel showing the relative abundances of rRNAs at selected time points. Arrows indicate the bands of 23S (cleaved and uncleaved), 16S, and 5S rRNAs, using the same color code as in panels A and B. White and black bars indicate light and dark phases, while gray bars indicate the subjective night of LL. (D) Averaged RNA data from the light phase (green) and dark phase (blue) of the LD conditions from panel A were translated into electropherogram traces, in which the different RNA fragments are separated by size (given in nucleotides [nt]). The fluorescence signal of an RNA fragment is related to its concentration, represented by the area under the curve. Shaded areas indicate the 95% confidence interval of the mean (assuming a normal distribution of the measurements). RNAs shorter than 500 nt were present in similar amounts at day and night, while the amount of longer RNAs (>500 nt) increased significantly during the dark phase. Arrows indicate the rRNA peaks. a.u., arbitrary units.
We observed that long transcripts (>500 nucleotides), such as 16S rRNA and 23S rRNA, were up to three and four times, respectively, more abundant during the night than in the day phase. However, the amounts of RNAs shorter than 500 nucleotides, such as 5S rRNA and tRNAs, remained close to constant (Fig. 3A, lower panel). These findings were confirmed by an agarose gel analysis (Fig. 3C). During the subjective night of the LL time series, the increases in the total RNA amount and, in particular, in 16S and 23S rRNA levels were much smaller (Fig. 3B). This indicated that the diurnal oscillation of the RNA abundance is driven by cyclic light-dark periods. The electropherograms from the Agilent Bioanalyzer system showed increases of not only the long rRNA amounts during the night phase but also the amounts of all RNAs longer than 500 nucleotides. However, the long 23S and 16S rRNAs are most abundant compared to other RNAs (together with the less-abundant tRNAs, between 60 and 82% of total RNA). It can therefore be expected that the long rRNAs are mainly responsible for the increase of total RNA during the night (Fig. 3D). RNA analysis of the DD time series between 5 h and 24 h confirmed these findings.
In contrast to the long rRNAs, the amounts of ribosomal mRNAs rose in the early light hours. Also, the abundances of 50S ribosomal protein L1 (Rpl1) and 30S ribosomal protein S1 tended to decrease during the dark phase and increase during the light phase (see Fig. S3 in the supplemental material). Thus, the 23S and 16S rRNAs seem to be upregulated at different phases compared with ribosomal mRNAs and, likely, ribosomal proteins. The amounts of total protein under LD, LL, and DD conditions were nearly constant (see File S1).
Diurnal accumulation dynamics of small noncoding RNAs.
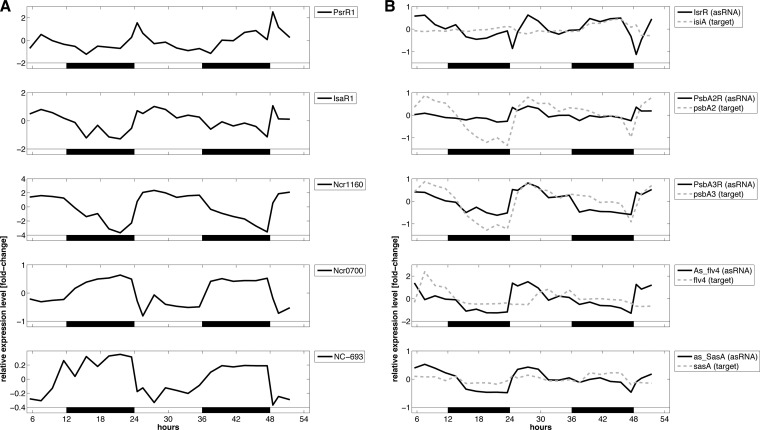
A plethora of small RNAs (sRNAs), such as antisense RNAs (asRNAs) and other noncoding RNAs (ncRNAs), has been identified with recent global screening methods. In this study, we found the amounts of many sRNAs to be rhythmic in LD and grouped these into day-peaking and night-peaking transcripts (Fig. 4), in a way comparable to that for the mRNAs (Fig. 1). For instance, the amount of photosynthesis regulatory RNA 1 (PsrR1) rose rapidly toward the end of the night phase, and the level peaked with the onset of light (Fig. 4A, first panel), in accordance with its function (J. Georg, D. Dienst, N. Schürgers, E. Kuchmina, T. Wallner, S. Klähn, D. Koop, H. Lokstein, W. R. Hess, and A. Wilde, submitted for publication). IsaR1, the single most strongly activated sRNA under iron limitation conditions (44, 45), was upregulated in the early part of and throughout the light period. During darkness, the amount of IsaR1 decreased (Fig. 4A, second panel). Ncr1160 (unknown function) did likewise (Fig. 4A, third panel). The group of noncoding RNAs showing maximum expression during the night phase included Ncr0700, a very abundant small RNA, and NC-693 (Fig. 4A, fourth and fifth panels).
FIG 4.
Temporal transcript profiles of small RNAs of Synechocystis sp. PCC 6803, derived from microarray analysis, under LD conditions. (A) Examples of accumulation of noncoding RNAs. There were day-peaking noncoding transcripts, such as PsrR1 (photosynthesis regulatory RNA 1; also called SyR1), IsaR1 (iron-stress-activated RNA 1; also called NC-181), and Ncr1160, as well as night-peaking noncoding RNAs, including Ncr0700 and NC-693. (B) Examples of accumulation of antisense RNAs and target mRNAs. The isiA mRNA and the antisense RNA IsrR occurred simultaneously under the unstressed conditions applied here. The psbA2 and psbA3 mRNAs and their antisense RNAs showed a correlated coaccumulation pattern. The antisense transcript amounts of As_flv4 and as_SasA increased sharply with the onset of illumination, whereas the accumulation of the corresponding flv4 and sasA mRNAs was delayed or showed no correlation. White and black bars indicate light and dark phases, respectively.
Among mRNA-asRNA pairs, we observed that the amounts of the isiA transcript (upregulated under conditions of iron limitation [46]) and its regulatory antisense RNA, IsrR (30), occurred simultaneously during the light-dark period, consistent with the unstressed conditions applied here (Fig. 4B, first panel). The mRNAs of the photosynthesis genes psbA2 and psbA3 and their antisense RNAs (PsbA2R and PsbA3R) showed a tightly correlated accumulation pattern under LD conditions (Fig. 4B, second and third panels). This is consistent with the protective function described for these antisense RNAs, which bind to the 5′ untranslated regions (5′-UTRs) of their targets and thereby preventing degradation of the psbA2 and psbA3 mRNAs (47). The amounts of the flv4 mRNA and the as_flv4 antisense RNA were negatively correlated (Fig. 4B, fourth panel). The flv4 gene encodes the flavodiiron protein Flv4. Flv4 and Flv2 are suggested to mediate the electron transfer from PSII, very likely having a photoprotective role that comes into play around noon (48). The antisense RNA as_flv4 regulates the gene expression of flv4 and flv2, carried in the flv4-sll0218-flv2 operon (49). In this study, the amount of as_flv4 rose rapidly as soon as the light was turned on, whereas the abundance of flv4 target mRNA was anticorrelated with this rise, indicating a negative role of as_flv4 in flv4 mRNA stability. The sasA gene (also called hik8, sarA, or sll0750) encodes the histidine kinase SasA, which very likely plays a key role in gene expression regulation in Synechocystis (50). In this study, the amount of the antisense RNA as_SasA was enhanced during the light phase and correlated marginally with the amount of sasA mRNA (Fig. 4B, fifth panel).
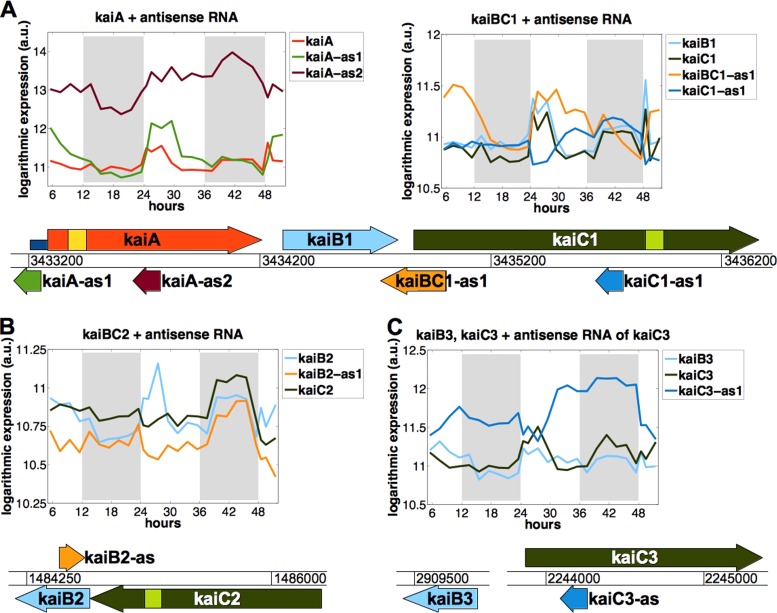
Diurnal accumulation dynamics of kai antisense RNAs.
There is an antisense RNA for almost every Synechocystis kai gene, encoded in cis on the respective noncoding strand (Fig. 5). We confirmed the existence of two kai antisense RNAs by Northern blot hybridization (see Fig. S4 in the supplemental material). We found that a kaiA antisense RNA, kaiA-as1, and the kaiA mRNA showed very similar accumulation patterns under LD conditions, with a correlation coefficient of 0.37 (Fig. 5A; see Fig. S5). Immediately after the shift from dark to light, the amounts of both RNAs started out in parallel, peaking toward the middle of the light phase. Afterwards, both RNA amounts declined likewise, and they remained at low levels in darkness. This observation indicates that kaiA-as1, which is encoded in antisense orientation relative to the 5′-UTR of kaiA, might have a positive effect on kaiA mRNA stability. kaiA-as1 may even regulate the entire kaiAB1C1 gene cluster at the pre-mRNA level, as the three kai mRNAs showed similar accumulation patterns and therefore very likely form an operon (Fig. 5A).
FIG 5.
Temporal transcript profiles of homologous kai clock genes and their antisense RNAs derived from microarray analysis. Synechocystis harbors multiple gene copies of the standard cyanobacterial clock genes kaiB1 and kaiC1. There are cis-encoded antisense RNAs for the majority of the kai genes, except for kaiC2 and kaiB3 (33). (A) The kaiA gene possesses two antisense RNAs, kaiA-as1 and kaiA-as2, whose abundances are correlated and noncorrelated, respectively, with that of the target mRNA. The kaiC1 gene possesses one antisense RNA, kaiC1-as1. Both RNAs accumulate in an inverse fashion. In addition, kaiBC-as1 overlaps the 3′-UTR of kaiB1 and the 5′-UTR of kaiC1. kaiBC-as1 possibly harbors an additional internal transcription start site. Data for the shorter version of this antisense transcript are shown (solid orange line). (B) For the kaiB2C2 gene cluster, there exists only one antisense RNA, kaiB2-as. Due to the different accumulation patterns, kaiB2 and kaiC2 likely have separate promoters. (C) The kaiB3 and kaiC3 genes are carried at different loci of the genome, and no antisense RNA for kaiB3 was found. The transcript amounts of kaiC3 and kaiC3-as are strictly inverse to each other. Gray areas indicate the dark phases. The RNA abundance data can be found in File S3 in the supplemental material. Results from the analysis of the correlation between the accumulation dynamics of the kai mRNAs and that of their cis-encoded antisense RNAs are shown in Fig. S5.
We also found examples where antisense RNAs might negatively control the accumulation of kai mRNAs, as follows. (i) While the kaiC1 mRNA amount increased after the onset of light, the abundance of kaiC1-as decreased in an inverse fashion (correlation coefficient of −0.22) (Fig. 5B; see Fig. S5 in the supplemental material). (ii) The kaiB2 mRNA and its antisense RNA (kaiB2-as) showed an opposite accumulation pattern over the daytime (correlation coefficient of −0.47) (Fig. 5B; see Fig. S5). Given that the amounts of mRNA for the kaiB2 and kaiC2 genes are not correlated with each other, we inferred that these clustered genes have separate promoters (Fig. 5B). In accordance with this, the kaiC2 transcript amount did not seem to be affected by kaiB2-as. However, we cannot exclude the possibility that regulation of kaiB2 mRNA occurred after kaiB2C2 mRNA maturation. (iii) The amount of kaiC3 mRNA peaked in the first half of the light phase. The increase/decrease of the amount of kaiC3-as was always converse to that of its target mRNA (Fig. 5C; see Fig. S5). It appeared as if kaiC3-as downregulated kaiC3 mRNA accumulation from the second half of the day onwards and during the night. The absolute value of the correlation coefficient (−0.68) for kaiC3 and kaiC3-as was the highest among the kai mRNA-asRNA pairs (see Fig. S5). Despite these examples of correlation, the kai antisense RNAs seemed to cause only subtle amplitude changes in the amounts of kai mRNAs under our growth and light conditions.
DISCUSSION
In this study, we analyzed the transcriptome of Synechocystis, including the small noncoding RNAs, under periodic LD conditions, constant light (LL), and constant darkness (DD). In contrast to previous transcriptome analyses of Synechocystis, we used a microarray technique where total RNA is directly labeled and hybridized with microarrays so that small noncoding RNAs can be analyzed as well. In addition, this approach guarantees a very high detection sensitivity and avoids primer-caused sequence bias, as well as reverse transcriptase-introduced artifacts such as those found with DNA microarrays. The RNA microarray used here has the highest existing time and transcript resolution so far, comprising 3,352 protein-encoding genes and 2,251 noncoding RNAs.
We found that transcripts of functionally related genes were upregulated at the same times (Fig. 1). With the onset of light, the cyanobacterial cells met the demand for proteins by enhancing the transcript amounts of translation-associated genes, such as those for ribosomal proteins, and of genes related to energy production, in particular ATP synthase subunits (cluster 1). Transcripts of genes coding for components of amino acid biosynthesis appeared in the first hour of the day (cluster 2). Moreover, most genes associated with photosynthesis were highly upregulated during the day, with a sharp increase in their transcript amounts in the early morning hours (cluster 3). Toward the end of the light phase, the transcript amounts of genes encoding proteins involved in nonphotosynthetic and catabolic metabolism processes, as well as respiration, increased (cluster 6). A previous transcriptome study performed under quite different experimental conditions arrived at similar results (21). In addition, the temporal expression patterns observed here are in good agreement with data from Synechococcus (15, 19). The GO term enrichment analysis confirmed the cluster analysis, suggesting a tight day-night regulation of the cellular activities (Fig. 2). However, in contrast to the case for Synechococcus, our transcriptome analysis of Synechocystis showed that the diurnal rhythms of RNA accumulation vanished under LL and DD conditions, indicating that transcript amounts and cellular processes are regulated by an endogenous mechanism that itself is driven by light only. Such a mechanism works rather like an hourglass that is turned around at the first glimpse of the day. There is support in the literature for this hypothesis: Synechocystis does not grow on glucose under complete darkness unless a daily light pulse is given (51). The requirement for a light pulse appeared to be enigmatic for a long time, as it is insufficient for photoautotrophy. An hourglass-like mechanism would explain the phenomenon of light-activated heterotrophic growth. However, a previous study reported circadian rhythms (22), and indeed, the Kai proteins of Synechocystis should be able to drive circadian rhythms. Synechocystis possesses the standard KaiABC protein set, which is known to constitute the core circadian clockwork in cyanobacteria such as Synechococcus (52). Our very recent in vitro analysis clearly demonstrated that the KaiC1 protein of Synechocystis exhibits autophosphorylation activity and that KaiC1's kinase activity is positively affected by KaiA (53). On the other hand, laboratory substrains of Synechocystis have evolved over the course of the last 2 decades, with different phenotypes and single nucleotide polymorphisms (SNPs), but mutations in known circadian clock-related genes were not found in the Synechocystis substrain “PCC-M” used here (54). However, one has to consider that the number of previously reported circadian clock-regulated genes is very small for Synechocystis. Transcripts of only 2 to 9% of the genes oscillated in the study of Kucho et al. (22). Ito et al. (19) found even fewer, illustrating the challenge in detecting circadian clock-regulated genes in Synechocystis with certainty. The real number of circadian-regulated genes might be even smaller than the number proposed by Ito et al. (19), or these genes might have escaped detection by the methods used here. Alternatively, we cannot exclude the possibility that circadian rhythms appear if growth conditions are different. A recent study showed that circadian clockworks are not constitutively active. The circadian clock in Synechococcus was found to be suppressed at low temperatures, at which cyanobacteria without a functional clock had a higher fitness level (55). Thus, one might speculate that Synechocystis contains a circadian clock system which is solely repressed under the conditions tested here. Under this premise, other conditions would provoke circadian rhythms. On the other hand, the multiple Kai proteins might not fine-tune the circadian clock (20) but may compromise a robust circadian timing. Alternatively, the amount of the phosphorylated form of KaiC might not oscillate in Synechocystis. In contrast to the two different promoters for kaiA and kaiBC in Synechococcus (52), the Synechocystis kaiA, kaiB1, and kaiC1 genes are transcribed from a single promoter, located upstream of kaiA (45). This situation further explains why their mRNAs accumulated in the same phase, to roughly equal levels (Fig. 5). If the Kai proteins accumulate in the cytosol at similar levels as well, sustained oscillation of the KaiC phosphorylation rhythm would be impossible (56–58). Furthermore, if transcription is not regulated by a particular kai promoter sensitive to the KaiC phospho-state, like the kaiBC promoter is in Synechococcus (59, 60), kai gene transcription and the Kai protein oscillator are decoupled from each other, making synchronous oscillation in a population of cells infeasible (61).
Surprisingly, we observed a doubling in the total RNA amount during the night, which suddenly dropped at the transition from darkness to light (Fig. 3A). The percentage of rRNAs in a cellular RNA population of Synechocystis is considerably high (60 to 82% of the total RNA), similar to the case in Escherichia coli (62, 63). A closer look revealed that, in fact, long RNAs (>500 nucleotides), such as 16S and 23S rRNAs, oscillated with high amplitudes in LD and accumulated during the night phase (Fig. 3). This disproves the belief in constant amounts of 16S and 23S rRNAs under any experimental conditions. These observations (an increase in RNA quantity in the dark and a sudden drop of the RNA amount at the onset of light) have not been made before or reported for other cyanobacteria in this way. Accumulations of total RNA and 16S rRNA during the night phase have been observed in Synechococcus sp. PCC 6301 but were explained by de novo synthesis of cellular RNA and a lack of cell division at night (64). In this study, this explanation cannot hold, because the cyanobacterial cells did not double during the 48-h experiment (see Fig. S1 in the supplemental material). It remains puzzling why shorter RNAs, such as 5S rRNA, did not oscillate and why long rRNAs accumulated at night, whereas genes encoding ribosomal proteins were induced during the early morning hours.
The huge light-induced drop in the amount of long rRNAs contradicts what is known about RNAs. For example, the amount of RNAs correlates with the growth rate (64–68), and the abundance of rRNAs is proportional to the number of ribosomes (69). However, the increase in the amount of rRNAs (Fig. 3), as well as the slight decrease in abundance of the two ribosomal proteins at night (see Fig. S3 in the supplemental material), indicates that rRNAs have different functions at day and night. A counterintuitive but possible explanation could be that during the day, rRNAs are the RNA components of ribosomes, whereas at night, long rRNAs serve as storage and are degraded at the transition from darkness to light in order to induce phototrophic growth very early in the morning. It was recently observed in yeast that carbon starvation triggers degradation of RNA, ultimately leading to ribulose-5-phosphate production (70). Going one step further, Synechocystis might degrade RNA in the morning to kick-start the Calvin cycle. Whether or not such an explanation holds can be ascertained only as more metabolome data become available.
Like the amounts of mRNAs, the amounts of many antisense RNAs and other noncoding RNAs were rhythmic in LD and peaked at day or night. Examples are shown in Fig. 4. The abundance of the antisense RNAs was positively (e.g., PsbAR2) or negatively (e.g., as_flv4) correlated with that of their target mRNAs, consistent with their known role. The positive correlation between the sasA mRNA and as_SasA amounts was very weak. Both RNAs are particularly interesting. The sasA gene encodes the histidine kinase SasA, which very likely plays a key role in gene expression regulation in Synechocystis (50). The Synechococcus homolog SasA is directly involved in the clock output pathway (71–74). It remains to be seen whether the very low positive correlation in the accumulation of the two RNAs has a functional relevance and, if so, how it influences the timing mechanism in Synechocystis. A regulatory relevance remains to be found for all these examples, including for the kai antisense RNAs. The kai antisense RNAs and the kai mRNAs also showed different types of correlation (Fig. 5). In particular, the accumulation dynamics of kaiA-as1 was always positively correlated with that of the kaiA mRNA. Moreover, the transcription initiation site of the antisense RNA kaiA-as1 does not interfere with the ribosome binding site of the kaiA mRNA (33). Theoretically, kaiA-as1 is able to bind within the 5′-UTR of kaiA mRNA to prevent mRNA degradation, as in the case of the antisense RNA PsbAR2 and the psbA2 mRNA (47). We find it interesting that in the phylogenetically quite remote marine organism Synechococcus sp. WH 7803, a similar short antisense RNA (i1642) also overlaps the 5′-UTR of the kaiA mRNA (75), suggesting a possible conserved role for this antisense RNA. Also, the internal transcription initiation site of the antisense RNA kaiC3-as and the converse increase/decrease in its amount compared with the amount of its target mRNA make it very likely that this antisense RNA plays a negative role in the expression of the kaiC3 gene.
The amplitudes of the cycling kai gene transcripts and the kai antisense RNAs are fairly low. Stress applied to Synechocystis might provoke major changes in accumulation. Otherwise, the presence of kai antisense RNAs might be the reason for the overall low kai mRNA amplitude rhythms and the absence of circadian rhythms in Synechocystis. In Synechococcus, kai transcripts cycle with high amplitudes (19). It would be interesting to know whether Synechococcus species have kai antisense RNAs as well.
Conclusions.
In this paper, we used microarrays to globally analyze the temporal transcript amounts of protein-encoding genes and small noncoding RNAs in Synechocystis under periodic LD, LL, and DD conditions. We found that transcripts of at least 27% of the genes analyzed oscillated in their amounts under LD conditions. Furthermore, transcripts of functionally related genes accumulated at the same specific times of day or night, in accordance with the assumed times at which cellular processes would take place.
Like the mRNA amounts, the sRNA levels increased at day or at night, and antisense RNA amounts correlated consistently with those of their target mRNAs, indicating that posttranscriptional regulation is crucial in Synechocystis. Two kai antisense RNAs (kaiA-as1 and kaiC3-as1) are promising candidates for playing a regulatory role in kai gene expression. Moreover, the increase in long rRNAs (e.g., 16S rRNA and 23S rRNA) during the night phase indicates an alternative function of these RNAs. However, mRNA, rRNA, tRNA, and sRNA amounts stopped being rhythmic under conditions of constant light or darkness, indicating that the RNA accumulation rhythms seen in LD are not circadian rhythms. The reasons for why we did not observe circadian rhythms in Synechocystis remain unclear. It is possible that the few gene transcripts regulated by the circadian clock were not detected in this study or that multiple Kai proteins, posttranscriptional regulation by antisense RNAs, or a lack of feedback regulation of kai gene expression indeed interferes with circadian rhythms in this cyanobacterium. Finally, a conditionality of the circadian clock cannot be excluded.
Supplementary Material
ACKNOWLEDGMENTS
We thank Reimo Zoschke and Michael Tillich for their help in collecting samples, Monika Reissmann (Tierphysiologie, HU Berlin) for help with RNA measurements using the Agilent Bioanalyzer system, Gudrun Krüger (Genetics and Experimental Bioinformatics, ALU Freiburg) for technical assistance with microarray hybridizations, and Annegret Wilde (Molekulare Genetik, ALU Freiburg), Ralf Steuer, and Henning Knoop (Institute for Theoretical Biology, HU Berlin) for their valuable comments on the manuscript.
This work was financially supported by the German Ministry of Education and Research (BMBF), FORSYS partner program (grant 0315294 to A.R., B.H., S.H., and I.M.A.), the Einstein Foundation Berlin (to R.L. and A.K.), the Deutsche Forschungsgemeinschaft (DFG) (to C.B. and A.W.), and BMBF grant e:bio RNAsys 0316165 (to W.R.H.).
Footnotes
Published ahead of print 13 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01086-14.
REFERENCES
- 1.Whitton BA. 1992. Diversity, ecology and taxonomy of the cyanobacteria, p 1–51 In Mann NH, Carr NG. (ed), Photosynthetic prokaryotes. Plenum Press, New York, NY [Google Scholar]
- 2.Ormerod JG. 1992. Physiology of the photosynthetic prokaryotes, p 93–120 In Mann NH, Carr NG. (ed), Photosynthetic prokaryotes. Plenum Press, New York, NY [Google Scholar]
- 3.Mori T, Binder B, Johnson CH. 1996. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc. Natl. Acad. Sci. U. S. A. 93:10183–10188. 10.1073/pnas.93.19.10183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vermaas WF. 2001. Photosynthesis and respiration in cyanobacteria, p 245–251 In Encyclopedia of life sciences. Nature Publishing Group, London, United Kingdom [Google Scholar]
- 5.Gallon JR, LaRue TA, Kurz WG. 1974. Photosynthesis and nitrogenase activity in the blue-green alga Gloeocapsa. Can. J. Microbiol. 20:1633–1637. 10.1139/m74-254 [DOI] [PubMed] [Google Scholar]
- 6.Postgate J. 1998. Nitrogen fixation. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 7.Millineaux PM, Gallon JR, Chaplin AE. 1981. Acetylene reduction (nitrogen fixation) by cyanobacteria grown under alternating light-dark cycles. FEMS Microbiol. Lett. 10:245–247. 10.1111/j.1574-6968.1981.tb06249.x [DOI] [Google Scholar]
- 8.Tu BP, McKnight SL. 2006. Metabolic cycles as an underlying basis of biological oscillations. Nat. Rev. Mol. Cell Biol. 7:696–701. 10.1038/nrm1980 [DOI] [PubMed] [Google Scholar]
- 9.Golden SS, Canales SR. 2003. Cyanobacterial circadian clocks—timing is everything. Nat. Rev. Microbiol. 1:191–199. 10.1038/nrmicro774 [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, Ishiura M, Kondo T. 1995. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 9:1469–1478. 10.1101/gad.9.12.1469 [DOI] [PubMed] [Google Scholar]
- 11.Chen TH, Chen TL, Hung LM, Huang TC. 1991. Circadian rhythm in amino acid uptake by Synechococcus RF-1. Plant Physiol. 97:55–59. 10.1104/pp.97.1.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sweeney BM, Borgese MB. 1989. A circadian-rhythm in cell-division in a prokaryote, the cyanobacterium Synechococcus WH 7803. J. Phycol. 25:183–186. 10.1111/j.0022-3646.1989.00183.x [DOI] [Google Scholar]
- 13.Smith RM, Williams SB. 2006. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. U. S. A. 103:8564–8569. 10.1073/pnas.0508696103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori T, Johnson CH. 2001. Circadian programming in cyanobacteria. Semin. Cell Dev. Biol. 12:271–278. 10.1006/scdb.2001.0254 [DOI] [PubMed] [Google Scholar]
- 15.Vijayan V, Zuzow R, O'Shea EK. 2009. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 106:22564–22568. 10.1073/pnas.0912673106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woelfle MA, Xu Y, Qin X, Johnson CH. 2007. Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 104:18819–18824. 10.1073/pnas.0706069104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennebaker K, Mackey KR, Smith RM, Williams SB, Zehr JP. 2010. Diel cycling of DNA staining and nifH gene regulation in the unicellular cyanobacterium Crocosphaera watsonii strain WH 8501 (Cyanophyta). Environ. Microbiol. 12:1001–1010. 10.1111/j.1462-2920.2010.02144.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakahira Y, Katayama M, Miyashita H, Kutsuna S, Iwasaki H, Oyama T, Kondo T. 2004. Global gene repression by KaiC as a master process of prokaryotic circadian system. Proc. Natl. Acad. Sci. U. S. A. 101:881–885. 10.1073/pnas.0307411100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito H, Mutsuda M, Murayama Y, Tomita J, Hosokawa N, Terauchi K, Sugita C, Sugita M, Kondo T, Iwasaki H. 2009. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc. Natl. Acad. Sci. U. S. A. 106:14168–14173. 10.1073/pnas.0902587106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoki S, Onai K. 2009. Circadian clocks of Synechocystis sp. strain PCC 6803, Thermosynechococcus elongatus, Prochlorococcus spp., Trichodesmium spp. and other species, p 259–282 In Ditty JL, Mackey SR, Johnson CH. (ed), Bacterial circadian programs. Springer, Berlin, Germany [Google Scholar]
- 21.Labiosa RG, Arrigo KR, Tu CJ, Bhaya D, Bay S, Grossman AR, Shrager J. 2006. Examination of diel changes in global transcript accumulation in Synechocystis (cyanobacteria). J. Phycol. 42:622–636. 10.1111/j.1529-8817.2006.00217.x [DOI] [Google Scholar]
- 22.Kucho K, Okamoto K, Tsuchiya Y, Nomura S, Nango M, Kanehisa M, Ishiura M. 2005. Global analysis of circadian expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 187:2190–2199. 10.1128/JB.187.6.2190-2199.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aoki S, Kondo T, Ishiura M. 1995. Circadian expression of the dnaK gene in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 177:5606–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoki S, Kondo T, Wada H, Ishiura M. 1997. Circadian rhythm of the cyanobacterium Synechocystis sp. strain PCC 6803 in the dark. J. Bacteriol. 179:5751–5755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottesman S, Storz G. 2011. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3:a003798. 10.1101/cshperspect.a003798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koo JT, Alleyne TM, Schiano CA, Jafari N, Lathem WW. 2011. Global discovery of small RNAs in Yersinia pseudotuberculosis identifies Yersinia-specific small, noncoding RNAs required for virulence. Proc. Natl. Acad. Sci. U. S. A. 108:E709–E717. 10.1073/pnas.1101655108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Georg J, Hess WR. 2011. cis-Antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 75:286–300. 10.1128/MMBR.00032-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madhugiri R, Pessi G, Voss B, Hahn J, Sharma CM, Reinhardt R, Vogel J, Hess WR, Fischer HM, Evguenieva-Hackenberg E. 2012. Small RNAs of the Bradyrhizobium/Rhodopseudomonas lineage and their analysis. RNA Biol. 9:47–58. 10.4161/rna.9.1.18008 [DOI] [PubMed] [Google Scholar]
- 29.Sharma UK, Chatterji D. 2010. Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol. Rev. 34:646–657. 10.1111/j.1574-6976.2010.00223.x [DOI] [PubMed] [Google Scholar]
- 30.Dühring U, Axmann IM, Hess WR, Wilde A. 2006. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl. Acad. Sci. U. S. A. 103:7054–7058. 10.1073/pnas.0600927103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legewie S, Dienst D, Wilde A, Herzel H, Axmann IM. 2008. Small RNAs establish delays and temporal thresholds in gene expression. Biophys. J. 95:3232–3238. 10.1529/biophysj.108.133819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Georg J, Voss B, Scholz I, Mitschke J, Wilde A, Hess WR. 2009. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 5:305. 10.1038/msb.2009.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voss B, Steglich C, Wilde A, Vogel J, Hess WR. 2011. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. U. S. A. 108:2124–2129. 10.1073/pnas.1015154108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1–61. 10.1099/00221287-111-1-1 [DOI] [Google Scholar]
- 35.Myers J, Graham JR, Wang RT. 1980. Light harvesting in Anacystis nidulans studied in pigment mutants. Plant Physiol. 66:1144–1149. 10.1104/pp.66.6.1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houmard J, de Marsac NT. 1988. Cyanobacterial genetic tools: current status. Methods Enzymol. 167:808–847. 10.1016/0076-6879(88)67092-3 [DOI] [PubMed] [Google Scholar]
- 37.Panaro NJ, Yuen PK, Sakazume T, Fortina P, Kricka LJ, Wilding P. 2000. Evaluation of DNA fragment sizing and quantification by the Agilent 2100 Bioanalyzer. Clin. Chem. 46:1851–1853 [PubMed] [Google Scholar]
- 38.Smyth GK. 2005. limma: linear models for microarray data, p 397–420 In Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S. (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY [Google Scholar]
- 39.Lehmann R, Machné R, Georg J, Benary M, Axmann I, Steuer R. 2013. How cyanobacteria pose new problems to old methods: challenges in microarray time series analysis. BMC Bioinformatics 14:133. 10.1186/1471-2105-14-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cleveland WS, Devlin SJ. 1988. Locally weighted regression: an approach to regression analysis by local fitting. J. Am. Stat. Assoc. 83:596–610. 10.1080/01621459.1988.10478639 [DOI] [Google Scholar]
- 41.Calza S, Valentini D, Pawitan Y. 2008. Normalization of oligonucleotide arrays based on the least-variant set of genes. BMC Bioinformatics 9:140. 10.1186/1471-2105-9-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo K, Hahne F, Brinkman RR, Gottardo R. 2009. flowClust: a Bioconductor package for automated gating of flow cytometry data. BMC Bioinformatics 10:145. 10.1186/1471-2105-10-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beck C, Knoop H, Axmann IM, Steuer R. 2012. The diversity of cyanobacterial metabolism: genome analysis of multiple phototrophic microorganisms. BMC Genomics 13:56. 10.1186/1471-2164-13-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernández-Prieto MA, Schon V, Georg J, Barreira L, Varela J, Hess WR, Futschik ME. 2012. Iron deprivation in Synechocystis: inference of pathways, non-coding RNAs, and regulatory elements from comprehensive expression profiling. G3 (Bethesda) 2:1475–1495. 10.1534/g3.112.003863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kopf M, Klähn S, Scholz I, Matthiessen J, Hess WR, Voss B. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res., in press. 10.1093/dnares/dsu018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park Y-I, Sandström S, Gustafsson P, Öquist G. 1999. Expression of the isiA gene is essential for the survival of the cyanobacterium Synechococcus sp. PCC 7942 by protecting photosystem II from excess light under iron limitation. Mol. Microbiol. 32:123–129 [DOI] [PubMed] [Google Scholar]
- 47.Sakurai I, Stazic D, Eisenhut M, Vuorio E, Steglich C, Hess WR, Aro EM. 2012. Positive regulation of psbA gene expression by cis-encoded antisense RNAs in Synechocystis sp. PCC 6803. Plant Physiol. 160:1000–1010. 10.1104/pp.112.202127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang P, Eisenhut M, Brandt AM, Carmel D, Silen HM, Vass I, Allahverdiyeva Y, Salminen TA, Aro EM. 2012. Operon flv4-flv2 provides cyanobacterial photosystem II with flexibility of electron transfer. Plant Cell 24:1952–1971. 10.1105/tpc.111.094417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eisenhut M, Georg J, Klahn S, Sakurai I, Mustila H, Zhang P, Hess WR, Aro EM. 2012. The antisense RNA As1_flv4 in the cyanobacterium Synechocystis sp. PCC 6803 prevents premature expression of the flv4-2 operon upon shift in inorganic carbon supply. J. Biol. Chem. 287:33153–33162. 10.1074/jbc.M112.391755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Axmann IM, Hertel S, Wiegard A, Dorrich AK, Wilde A. Diversity of KaiC-based timing systems in marine Cyanobacteria. Mar. Genomics 14:3–16. 10.1016/j.margen.2013.12.006 [DOI] [PubMed] [Google Scholar]
- 51.Anderson SL, McIntosh L. 1991. Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: a blue-light-requiring process. J. Bacteriol. 173:2761–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. 1998. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281:1519–1523. 10.1126/science.281.5382.1519 [DOI] [PubMed] [Google Scholar]
- 53.Wiegard A, Dörrich AK, Deinzer HT, Beck C, Wilde A, Holtzendorff J, Axmann IM. 2013. Biochemical analysis of three putative KaiC clock proteins from Synechocystis sp. PCC 6803 suggests their functional divergence. Microbiology 159:948–958. 10.1099/mic.0.065425-0 [DOI] [PubMed] [Google Scholar]
- 54.Trautmann D, Voss B, Wilde A, Al-Babili S, Hess WR. 2012. Microevolution in cyanobacteria: re-sequencing a motile substrain of Synechocystis sp. PCC 6803. DNA Res. 19:435–448. 10.1093/dnares/dss024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y, Ma PJ, Shah P, Rokas A, Liu Y, Johnson CH. 2013. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature 495:116–120. 10.1038/nature11942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. 2005. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308:414–415. 10.1126/science.1108451 [DOI] [PubMed] [Google Scholar]
- 57.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. 2003. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 22:2127–2134. 10.1093/emboj/cdg212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayashi F, Ito H, Fujita M, Iwase R, Uzumaki T, Ishiura M. 2004. Stoichiometric interactions between cyanobacterial clock proteins KaiA and KaiC. Biochem. Biophys. Res. Commun. 316:195–202. 10.1016/j.bbrc.2004.02.034 [DOI] [PubMed] [Google Scholar]
- 59.Murayama Y, Oyama T, Kondo T. 2008. Regulation of circadian clock gene expression by phosphorylation states of KaiC in cyanobacteria. J. Bacteriol. 190:1691–1698. 10.1128/JB.01693-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hertel S, Brettschneider C, Axmann IM. 2013. Revealing a two-loop transcriptional feedback mechanism in the cyanobacterial circadian clock. PLoS Comput. Biol. 9:e1002966. 10.1371/journal.pcbi.1002966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teng SW, Mukherji S, Moffitt JR, de Buyl S, O'Shea EK. 2013. Robust circadian oscillations in growing cyanobacteria require transcriptional feedback. Science 340:737–740. 10.1126/science.1230996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delong EF, Wickham GS, Pace NR. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360–1363. 10.1126/science.2466341 [DOI] [PubMed] [Google Scholar]
- 63.Poulsen LK, Ballard G, Stahl DA. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59:1354–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lepp PW, Schmidt TM. 1998. Nucleic acid content of Synechococcus spp. during growth in continuous light and light/dark cycles. Arch. Microbiol. 170:201–207. 10.1007/s002030050634 [DOI] [PubMed] [Google Scholar]
- 65.Kjeldgaard NO, Kurland CG. 1963. Distribution of soluble and ribosomal RNA as a function of growth rate. J. Mol. Biol. 6:341. 10.1016/S0022-2836(63)80093-5 [DOI] [Google Scholar]
- 66.Rosset R, Julien J, Monier R. 1966. Ribonucleic acid composition of bacteria as a function of growth rate. J. Mol. Biol. 18:308–320. 10.1016/S0022-2836(66)80248-6 [DOI] [PubMed] [Google Scholar]
- 67.Parrott LM, Slater JH. 1980. The DNA, RNA and protein composition of the cyanobacterium Anacystis nidulans grown in light- and carbon dioxide-limited chemostats. Arch. Microbiol. 127:53–58. 10.1007/BF00414355 [DOI] [PubMed] [Google Scholar]
- 68.Kerkhof L, Kemp P. 1999. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol. Ecol. 30:253–260. 10.1111/j.1574-6941.1999.tb00653.x [DOI] [PubMed] [Google Scholar]
- 69.Gourse RL, Gaal T, Bartlett MS, Appleman JA, Ross W. 1996. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 50:645–677. 10.1146/annurev.micro.50.1.645 [DOI] [PubMed] [Google Scholar]
- 70.Xu YF, Letisse F, Absalan F, Lu W, Kuznetsova E, Brown G, Caudy AA, Yakunin AF, Broach JR, Rabinowitz JD. 2013. Nucleotide degradation and ribose salvage in yeast. Mol. Syst. Biol. 9:665. 10.1038/msb.2013.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iwasaki H, Williams SB, Kitayama Y, Ishiura M, Golden SS, Kondo T. 2000. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell 101:223–233. 10.1016/S0092-8674(00)80832-6 [DOI] [PubMed] [Google Scholar]
- 72.Takai N, Nakajima M, Oyama T, Kito R, Sugita C, Sugita M, Kondo T, Iwasaki H. 2006. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 103:12109–12114. 10.1073/pnas.0602955103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang YG, Tseng R, Kuo NW, LiWang A. 2012. Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc. Natl. Acad. Sci. U. S. A. 109:16847–16851. 10.1073/pnas.1211508109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pattanayek R, Yadagiri KK, Ohi MD, Egli M. 2013. Nature of KaiB-KaiC binding in the cyanobacterial circadian oscillator. Cell Cycle 12:810–817. 10.4161/cc.23757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gierga G, Voss B, Hess WR. 2012. Non-coding RNAs in marine Synechococcus and their regulation under environmentally relevant stress conditions. ISME J. 6:1544–1557. 10.1038/ismej.2011.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.