Abstract
Nonduplicate blood cultures that were positive for Gram-negative bacilli (n = 125) were tested by the Verigene Gram-negative blood culture (BC-GN) assay; 117 (90.7%) isolates were members of the panel. For identification and resistance markers, the agreements with routine methods were 97.4% (114/117) and 92.3% (12/13). The BC-GN assay is a rapid and accurate tool for the detection of pathogens from blood cultures and could be integrated alongside conventional systems to enable faster patient management, but the clinical benefits should be further evaluated.
TEXT
The Verigene Gram-positive blood culture (BC-GP) and Gram-negative blood culture (BC-GN) nucleic acid tests (Nanosphere, Inc., Northbrook, IL) are microarray-based assays designed to rapidly identify multiple bacterial species and their associated resistance markers directly from positive blood cultures. Two separate panels dedicated for Gram-positive (1–3) and Gram-negative (4, 5) organisms are currently available. The system instrumentation supplies random access test processing that is easy to perform and requires no personnel trained in molecular approaches. The major advantages of these tests are the short turnaround time (TAT) following positivity and the ease of use, which allows testing 24 hours a day. These assays provide results in ≤3 h and may significantly impact patient management by reducing the time needed for laboratory processing.
The targets of the BC-GN assay are Klebsiella pneumoniae, Klebsiella oxytoca, Escherichia coli, Serratia marcescens, Proteus spp., Citrobacter spp., Enterobacter spp., Pseudomonas aeruginosa, and Acinetobacter spp. In addition, the BC-GN assay detects resistance markers, including the extended-spectrum β-lactamase (ESBL) CTX-M and the carbapenemases KPC, NDM, VIM, IMP, and OXA groups. Several studies on the BC-GP assay were published, but so far, only a few studies evaluated the BC-GN test, particularly in a European epidemiology context (4, 5). The aim of this study was to evaluate the diagnostic performance of the BC-GN assay and the TAT for positive blood cultures, with the results of culture and antimicrobial susceptibility testing (AST) considered the gold standard.
A total of 125 nonduplicate blood cultures that were positive for Gram-negative bacilli (Bactec FX, Becton, Dickinson) were consecutively enrolled from patients admitted to Erasme Hospital from March 2013 to September 2013. An aliquot of 700 μl was used for the testing of each blood culture flagging positive within 12 h, as recommended by the manufacturer. All samples which were not tested within 12 h were stored at −20°C to be analyzed retrospectively with the BC-GN assay. Verigene provides a qualitative result for the presence (“detected”) or absence (“not detected”) of the bacterial targets and resistance markers. Tests generating invalid results (reported as “no call”) were repeated. Identification was performed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics, Bremen, Germany) and AST by disk diffusion for nonfermenting Gram-negative rods and by Vitek2 (bioMérieux, Lyon, France) for Enterobacteriaceae according to CLSI guidelines (6). The presence of ESBLs was confirmed by PCR and sequencing for the blaTEM, blaSHV, and blaCTX-M genes (7). The presence of the blaNDM, blaIMP, blaVIM, blaKPC, or blaOXA-48 allelic genes was tested by a multiplex PCR (8). The TAT corresponding to the elapsed time between the positivity of blood cultures and the results of identification/AST was calculated considering the phenotypic methods and the BC-GN results for only the samples analyzed within 12 h of the positive blood culture signal (n = 71).
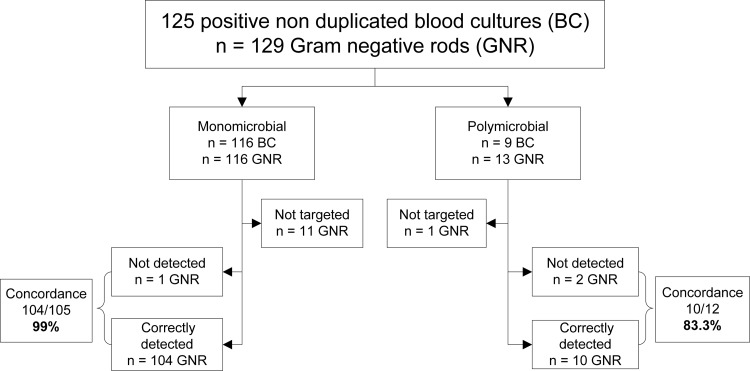
Out of the 125 positive blood cultures, 116 were monomicrobial, and 9 were polymicrobial, which made a total of 129 Gram-negative rods (Fig. 1). Among the polymicrobial blood cultures, 5 were positive for Gram-negative and Gram-positive organisms. Out of the 10 samples (8%) with a “no call” result, 5 were available for a retest. Four of them were correctly identified after repeat testing, and one remained with a “no call” result. The samples were divided into two distinct groups of organisms. The first group consisted of 12 “non-BC-GN” panel organisms (9.3%) not targeted on the BC-GN assay, including Pseudomonas stutzeri, Stenotrophomonas maltophilia, Elizabethkingia meningoseptica, Haemophilus influenzae, Moraxella osloensis, Capnocytophaga spp., Porphyromonas spp., and Bacteroides spp. None of these isolates were detected by the BC-GN assay, as expected, but four of them were reported as “no call” instead of “not detected.” The second group consisted of 117 “BC-GN panel” organisms (90.7%) (Table 1). For the organisms targeted by the BC-GN assay, the agreement for identification to the species or genus level with routine laboratory methods was 97.4% (114/117). The Verigene result was “not detected” for one isolate identified as K. pneumoniae by the reference method. Two distinct positive blood cultures containing K. pneumoniae and Proteus mirabilis were reported as “no call.”
FIG 1.
BC-GN performance for identification of Gram-negative rods from positive blood cultures.
TABLE 1.
Distribution and numbers of isolates correctly identified or not detected by BC-GN compared to those of conventional methods
| Organism | No. of isolates: |
Agreement (%) | ||
|---|---|---|---|---|
| Tested | Correctly identified | Not detected | ||
| E. coli | 69 | 69 | 100 | |
| K. pneumoniae | 14 | 12 | 2 | 85.7 |
| K. oxytoca | 4 | 4 | 100 | |
| S. marcescens | 0 | NAa | ||
| Enterobacter spp. | 9 | 9 | 100 | |
| Citrobacter spp. | 2 | 2 | 100 | |
| Proteus spp. | 7 | 6 | 1 | 85.7 |
| P. aeruginosa | 9 | 9 | 100 | |
| Acinetobacter spp. | 3 | 3 | 100 | |
| Total no. of isolates | 117 | 114 | 3 | 97.4 |
NA, not applicable.
Considering the genetic resistance determinants, the overall concordance was 92.3% (12/13). Out of the nine ESBLs, the BC-GN assay correctly detected CTX-M in two K. pneumoniae isolates and five E. coli isolates. Among carbapenemases, OXA-48 was recovered from two Enterobacter aerogenes isolates, and metallo-β-lactamases, including VIM (n = 2) and IMP (n = 1), were recovered from three P. aeruginosa isolates. Two ESBLs not detected in K. pneumoniae and E. coli isolates were confirmed by molecular analysis to be blaSHV-12 and blaCTX-M-1. Agreement for the identification of Gram-negative organisms and resistance determinants was more accurate in monomicrobial (104/105, 99.0%) than in polymicrobial cultures (10/12, 83.3%). The median times to bacterial identification and to AST results were 21 h and 43 h, respectively. The median TAT for the BC-GN assay results was 10 h. Consequently, the organism's identification and susceptibility could have been available approximately 11 h and 33 h earlier, respectively, than with conventional methods.
The BC-GN assay showed excellent performance for the identification of organisms with no misidentification observed, as previously reported (4, 5). Among the three false-negative results, two bacteria were part of mixed blood cultures. The BG-GN panel covers a broad range of Gram-negative bacteria, including the most frequently isolated pathogens, allowing for inference of the natural profile of the organisms. In this study, the BC-GN panel identified the microorganism in 90% of the positive blood cultures. The accurate identification of pathogens together with the patient's history and the hospital epidemiology may help to institute empirical antibiotic treatment. However, besides intrinsic resistance, the rising of acquired resistance mechanisms, including in ESBLs and carbapenemases, makes it difficult to predict resistance phenotypes. Worldwide resistance to carbapenems increases constantly due to the dissemination of carbapenemase-producing Gram-negative bacteria (9). The BC-GN assay is able to detect the most frequently acquired resistance determinants, including the genes coding for the different carbapenemases (4, 5). All the carbapenemases encountered during this study were correctly detected by the BC-GN assay. Along with the detection of carbapenemase, the BC-GN assay also detects the presence of the CTX-M type, which is rapidly spreading among Enterobacteriaceae worldwide, replacing the TEM and SHV types as the predominant ESBLs in many countries (10, 11). Out of the nine ESBLs detected by the phenotypic method, all but one were the CTX-M type. The assay failed to detect one CTX-M gene confirmed by our triplex ESBL PCR. This discrepancy may be attributed to the known mixed infection limitation for the BC-GN test. As expected, one SHV-12 ESBL was not detected by the BC-GN assay. The negative signal for the CTX-M target may not exclude the presence of another type of ESBL, making it difficult to shift to a narrow-spectrum antibiotic.
Our workflow was not optimized to perform the Verigene assays directly after blood cultures had become positive, but even in our suboptimal laboratory workflow conditions, the Verigene assay enabled same-day reporting of positive blood cultures to physicians. This major diminution in TAT (33 h shorter) warrants further study to determine the positive impacts on patient care improvement and reduction of length of stay.
Using the BC-GN assay to identify blood culture pathogens provides several advantages; the assay is simple (short hands-on time), is easy to perform (minimal technician training), has a short TAT (<3 h), is comprehensive (15 targets), and shows excellent performance compared to conventional methods. Another promising rapid test is the FilmArray blood culture identification (BCID) panel (BioFire Diagnostic), which simultaneously tests 24 organisms (Gram-positive and -negative bacteria and yeasts) and 3 resistance genes (KPC, mecA, and vanA/B) and provides results within 1 h (12, 13). Compared to Verigene, the BCID test does not report resistance genes for the CTX-M, VIM, IMP, NDM, and OXA groups, recognized as major emerging concerns worldwide. In contrast, the BCID test offers the targets for Neisseria meningitidis, Haemophilus influenzae, and Acinetobacter baumannii, which require prompt and adequate therapy. Moreover, A. baumannii is recognized as a major drug-resistant agent of nosocomial infections. Potential limitations of the Verigene assays include the need to initially obtain a positive blood culture and a Gram stain before the appropriate panel is chosen. Also, the assay is limited by the number of targets included on the panel.
In conclusion, the BC-GN assay provided rapid and accurate organism identification and detection of resistance genes compared to routine laboratory methods. The Verigene assay could enable earlier evidence-based management for bacteremic patients, but it cannot replace the phenotypic methods. Isolation on solid medium is still needed to differentiate mixed cultures, to identify organisms yielding a “not detected” or “no call” result, and to obtain a complete antimicrobial susceptibility pattern. Cost-effectiveness analyses on clinical outcomes and overall health care costs are needed to confirm the impact that a 1.5-day improvement in identification and resistance reporting will have.
ACKNOWLEDGMENTS
We thank Soultana Pavlov, Angélique Fourmeaux, Sandra Cuvellier, and Anne-Laurence Debyttere for their technical assistance with blood culture processing and Nanosphere for kindly providing the kits used in this study.
We declare no conflicts of interest in relation to this work.
Footnotes
Published ahead of print 4 June 2014
REFERENCES
- 1.Samuel LP, Tibbetts RJ, Agotesku A, Fey M, Hensley R, Meier FA. 2013. Evaluation of a microarray-based assay for rapid identification of Gram-positive organisms and resistance markers in positive blood cultures. J. Clin. Microbiol. 51:1188–1192. 10.1128/JCM.02982-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott LJ. 2013. Verigene Gram-positive blood culture nucleic acid test. Mol. Diagn. Ther. 17:117–122. 10.1007/s40291-013-0021-z [DOI] [PubMed] [Google Scholar]
- 3.Wojewoda CM, Sercia L, Navas M, Tuohy M, Wilson D, Hall GS, Procop GW, Richter SS. 2013. Evaluation of the Verigene Gram-positive blood culture nucleic acid test for rapid detection of bacteria and resistance determinants. J. Clin. Microbiol. 51:2072–2076. 10.1128/JCM.00831-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tojo M, Fujita T, Ainoda Y, Nagamatsu M, Hayakawa K, Mezaki K, Sakurai A, Masui Y, Yazaki H, Takahashi H, Miyoshi-Akiyama T, Totsuka K, Ohmagari N. 2014. Evaluation of an automated rapid diagnostic assay for detection of Gram-negative bacteria and their drug-resistance genes in positive blood cultures. PLoS One. 9:e94064. 10.1371/journal.pone.0094064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mancini N, Infurnari L, Ghidoli N, Valzano G, Clementi N, Burioni R, Clementi M. 2014. Potential impact of a microarray-based nucleic acid assay for rapid detection of Gram-negative bacteria and resistance markers in positive blood cultures. J. Clin. Microbiol. 52:1242–1245. 10.1128/JCM.00142-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7.Rodriguez-Villalobos H, Malaviolle V, Frankard J, de Mendonça R, Nonhoff C, Struelens MJ. 2006. In vitro activity of temocillin against extended spectrum beta-lactamase-producing Escherichia coli. J. Antimicrob. Chemother. 57:771–774. 10.1093/jac/dkl046 [DOI] [PubMed] [Google Scholar]
- 8.Bogaerts P, Rezende de Castrol R, de Mendonça R, Huang TD, Denis O, Glupczynski Y. 2013. Validation of carbapenemase and extended-spectrum beta-lactamase multiplex endpoint PCR assays according to ISO 15189. J. Antimicrob. Chemother. 68:1576–1582. 10.1093/jac/dkt065 [DOI] [PubMed] [Google Scholar]
- 9.Nordmann P. 2013. Carbapenemase-producing Enterobacteriaceae: overview of a major public health challenge. Med. Mal. Infect. 44:51–56. 10.1016/j.medmal.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 10.Carrer A, Nordmann P. 2011. CTX-M-15-producing Klebsiella pneumoniae: a change in the epidemiology of ESBL. Pathol. Biol. (Paris) 59:e133–e135. 10.1016/j.patbio.2009.06.003 (In French) [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez-Villalobos H, Bogaerts P, Berhin C, Bauraing C, Deplano A, Montesinos I, Jans B, Glupczynski Y. 2011. Trends in production of extended-spectrum beta-lactamases among Enterobacteriaceae of clinical interest: results of a nationwide survey in Belgian hospitals. J. Antimicrob. Chemother. 66:37–47. 10.1093/jac/dkq388 [DOI] [PubMed] [Google Scholar]
- 12.Altun O, Almuhayawi M, Ullberg M, Ozenci V. 2013. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J. Clin. Microbiol. 51:4130–4136. 10.1128/JCM.01835-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blaschke AJ, Heyrend C, Byington CL, Fisher MA, Barker E, Garrone NF, Tatcher SA, Pavia AT, Barney T, Alger GD, Daly JA, Ririe KM, Ota I, Poritz MA. 2012. Rapid identification of pathogens from positive blood cultures by multiplex polymerase chain reaction using the FilmArray system. Diagn. Microbiol. Infect. Dis. 74:349–355. 10.1016/j.diagmicrobio.2012.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]