Abstract
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker Biotyper) was able to accurately identify 98.6% (142/144) of Acinetobacter baumannii isolates, 72.4% (63/87) of A. nosocomialis isolates, and 97.6% (41/42) of A. pittii isolates. All Acinetobacter junii, A. ursingii, A. johnsonii, and A. radioresistens isolates (n = 28) could also be identified correctly by Bruker Biotyper.
TEXT
Isolates of Acinetobacter species causing human infections predominantly belong to Acinetobacter calcoaceticus (genospecies 1), A. baumannii (genospecies 2), A. pittii (genospecies 3), and A. nosocomialis (genospecies 13TU) (1–4). Because these isolates are phenotypically similar and difficult to distinguish using traditional microbiological and biochemical tests, they are frequently reported as A. calcoaceticus-A. baumannii complex (ACB complex) (1–4). Although several commercially available matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) systems are rapidly emerging novel technology widely used in clinical microbiology laboratories for rapid identification of commonly encountered bacteria and fungi (5–8), few studies have compared the accuracy of different MALDI-TOF MS systems in identifying Acinetobacter species to the genospecies level (9–12).
A total of 286 blood isolates of ACB complex and 39 isolates of non-ACB complex that had been recovered from patients treated at the National Taiwan University Hospital from 2010 to 2012 were obtained. Conventional biochemical identification methods as well as the Phoenix bacterial identification system (NMIC/ID-72 cards; Becton, Dickinson Diagnostic Instrument Systems, Sparks, MD) and Vitek 2 (GN cards; bioMérieux, Marcy l'Etoile, France) were used for the identification of the isolates (4). We sequenced the 16S-23S rRNA gene intergenic spacer (ITS) region and a 350-bp highly variable zone of the rpoB gene to identify the isolates to the genospecies level (2, 4). The sequences obtained were compared with published sequences in the GenBank database using the BLASTN algorithm (http://www.ncbi.nlm.nih.gov/blast).
Bacteria were prepared for analysis by the MALDI Bruker Biotyper system as previously described (8, 10). Identification scores of ≥2.000 indicated species-level identification, scores of 1.700 to 1.999 indicated genus-level identification, and scores of <1.700 indicated no reliable identification (8, 10). Clustering analysis of four genetically identified genospecies of ACB complex was performed using ClincProtools 3.0 (Bruker Daltonics GmbH, Bremen, Germany) (8). Characteristic peaks of the spectra of the four genospecies of ACB complex were identified. Cluster analysis of half of the A. baumannii isolates identified by the Bruker Biotyper system was also performed (8). The remaining half of the A. baumannii isolates identified by the Bruker Biotyper system were used as external validation isolates.
Tables 1 and 2 show the results of two molecular methods for identification of the 286 isolates of ACB complex and 39 non-ACB complex isolates. No discrepant results were found for ACB complex isolates by the two molecular identification methods (Table 1). The Phoenix and Vitek 2 systems identified all isolates of ACB complex as A. baumannii (complex). The overall accuracy rate of identifying Acinetobacter species other than ACB complex was higher in the Vitek 2 system (61.5% [24/39]) than in the Phoenix system (0%). Vitek 2 had good identification results for A. junii (91.7%), A. ursingii (100%), and A. radioresistens (100%). Of the 14 species (genospecies) of Acinetobacter isolates evaluated in the present study, Acinetobacter genospecies “close to 13TU,” Acinetobacter genospecies 14TU, Acinetobacter genospecies 16BJ, A. bereziniae (Acinetobacter genospecies 10), and A. gyllenbergii are not included in the Bruker Biotyper database (DB 5627). Of the 286 isolates of ACB complex, the accuracy rate of genospecies identification by the Bruker Biotyper system was 98.6% (142/144) for A. baumannii, 72.4% (63/87) for A. nosocomialis, and 97.6% (41/42) for A. pittii. Twelve of the 13 isolates of Acinetobacter genospecies “close to 13TU” were identified as A. baumannii, and 10 of these isolates had score values of ≥2.000 (Table 1).
TABLE 1.
Comparison of the Bruker Biotyper MALDI TOF MS system with molecular methods in identifying 286 blood isolates of Acinetobacter calcoaceticus-Acinetobacter baumannii complex
| Acinetobacter species identification with identical results by molecular methods (no. of isolates) | Acinetobacter species identified by Bruker Biotyper system (no. of isolates) | No. (%) of isolates with the following range of scores by the Bruker Biotyper system: |
||
|---|---|---|---|---|
| ≥2.0 | 1.7–1.999 | <1.7 | ||
| A. baumannii (144) | A. baumannii (142) | 142 (100) | 0 (0) | 0 (0) |
| A. nosocomialis (2) | 2 (100) | 0 (0) | 0 (0) | |
| A. nosocomialis (87) | A. nosocomialis (63) | 62 (98.4) | 1 (1.6) | 0 (0) |
| A. baumannii (22) | 17 (77.3) | 5 (22.7) | 0 (0) | |
| A. pittii (2) | 0 (0) | 2 (100) | 0 (0) | |
| A. pittii (42) | A. pittii (41) | 40 (97.6) | 1 (2.4) | 0 (0) |
| A. nosocomialis (1) | 1 (100) | 0 (0) | 0 (0) | |
| Acinetobacter genospecies “close to 13TU” (13)a | A. baumannii (12) | 10 (83.3) | 2 (16.7) | 0 (0) |
| A. pittii (1) | 1 (100) | 0 (0) | 0 (0) | |
Not included in Bruker Biotyper database (DB 5627).
TABLE 2.
Results of identification of 39 Acinetobacter species other than Acinetobacter calcoaceticus-Acinetobacter baumannii complex isolates by different methods
| Species no. | Species identified by gene sequencinga |
MALDI Bruker Biotyper resultb |
Vitek 2 result |
Phoenix result |
||||
|---|---|---|---|---|---|---|---|---|
| ITS gene | rpoB gene | Species identified | Score | Species identified | Identity (%)c | Species identified | Identity (%)c | |
| 1 | A. junii | A. junii | A. junii | 2.351 | A. junii | 99 | Acinetobacter sp. | 90 |
| 2 | A. junii | A. junii | A. junii | 2.42 | A. junii | 99 | A. lwoffii | 90 |
| 3 | A. junii | A. junii | A. junii | 2.268 | A. junii | 99 | A. lwoffii | 99 |
| 4 | A. junii | A. junii | A. junii | 2.34 | A. junii | 98 | A. haemolyticus | 91 |
| 5 | A. junii | A. junii | A. junii | 2.462 | A. junii | 99 | A. lwoffii | LD |
| 6 | A. junii | A. junii | A. junii | 2.369 | A. junii | 99 | A. lwoffii | 90 |
| 7 | A. junii | A. junii | A. junii | 2.397 | A. junii | 99 | Acinetobacter sp. | 90 |
| 8 | A. junii | A. junii | A. junii | 2.346 | A. junii | 94 | Acinetobacter sp. | 92 |
| 9 | A. junii | A. junii | A. junii | 2.322 | A. junii | 98 | A. haemolyticus | 91 |
| 10 | A. junii | A. junii | A. junii | 2.315 | A. junii | 99 | A. lwoffii | 90 |
| 11 | A. junii | A. junii | A. junii | 2.323 | A. ursingii | 97 | A. lwoffii | LD |
| 12 | A. junii | A. junii | A. junii | 2.420 | A. junii | 98 | Acinetobacter sp. | 90 |
| 13 | A. ursingii | A. ursingii | A. ursingii | 2.041 | A. ursingii | 96 | Acinetobacter sp. | 98 |
| 14 | A. ursingii | A. ursingii | A. ursingii | 2.018 | A. ursingii | 97 | A. lwoffii | 90 |
| 15 | A. ursingii | A. ursingii | A. ursingii | 2.037 | A. ursingii | 97 | Acinetobacter sp. | 90 |
| 16 | A. ursingii | A. ursingii | A. ursingii | 2.09 | A. ursingii | 98 | Acinetobacter sp. | 90 |
| 17 | A. ursingii | A. ursingii | A. ursingii | 2.058 | A. ursingii | 97 | A. lwoffii | LD |
| 18 | A. ursingii | A. ursingii | A. ursingii | 2.116 | A. ursingii | LD | Acinetobacter sp. | 90 |
| 19 | A. ursingii | A. ursingii | A. ursingii | 2.101 | A. ursingii | 98 | Acinetobacter sp. | 90 |
| 20 | A. ursingii | A. ursingii | A. ursingii | 2.099 | A. ursingii | 97 | Acinetobacter sp. | 95 |
| 21 | A. ursingii | A. ursingii | A. ursingii | 2.107 | A. ursingii | 97 | Acinetobacter sp. | 90 |
| 22 | A. ursingii | A. ursingii | A. ursingii | 2.098 | A. ursingii | 93 | Acinetobacter sp. | 90 |
| 23 | A. johnsonii | A. johnsonii | A. johnsonii | 2.242 | Sphingomonas paucimobilis | 86 | A. lwoffii | 90 |
| 24 | A. johnsonii | A. johnsonii | A. johnsonii | 2.373 | A. lwoffii | 97 | A. lwoffii | 90 |
| 25 | A. johnsonii | A. johnsonii | A. johnsonii | 2.390 | A. lwoffii | LD | A. lwoffii | 99 |
| 26 | A. radioresistens | A. radioresistens | A. radioresistens | 2.359 | A. radioresistens | 99 | A. lwoffii | 99 |
| 27 | A. radioresistens | A. radioresistens | A. radioresistens | 2.305 | A. radioresistens | LD | A. lwoffii | 99 |
| 28 | A. radioresistens | A. radioresistens | A. radioresistens | 2.327 | A. radioresistens | 99 | A. lwoffii | 99 |
| 29 | Acinetobacter genospecies 14TU | A. gyllenbergii | A. junii | 1.866 | A. baumannii | 99 | A. haemolyticus | 90 |
| 30 | Acinetobacter genospecies 14TU | Acinetobacter genospecies 16BJ | A. junii | 1.958 | Alcaligenes faecalis subsp. faecalis | 98 | A. haemolyticus | 97 |
| 31 | Acinetobacter genospecies 14TU | A. gyllenbergii | A. parvus | 1.743 | A. baumannii | 97 | Burkholderia cepacia | 98 |
| 32 | Acinetobacter genospecies 16BJ | Acinetobacter genospecies 16BJ | A. junii | 1.857 | A. junii | 99 | A. haemolyticus | LD |
| 33 | Acinetobacter genospecies 16BJ | Acinetobacter genospecies 16BJ | A. haemolyticus | 1.858 | Pseudomonas pseudoalcaligenes | 97 | A. haemolyticus | 95 |
| 34 | Acinetobacter genospecies 16BJ | Acinetobacter genospecies 16BJ | A. junii | 2.035 | A. ursingii | LD | A. haemolyticus | 98 |
| 35 | A. tjernbergiae | Acinetobacter genospecies 14TU | A. junii | 1.914 | A. baumannii | 99 | A. haemolyticus | 90 |
| 36 | A. tjernbergiae | A. parvus | A. junii | 1.921 | A. ursingii | 98 | A. lwoffii | 90 |
| 37 | A. bereziniae | A. bereziniae | Acinetobacter sp. | 1.741 | A. lwoffii | 99 | ACB complex | 99 |
| 38 | A. guillouiae | A. radioresistens | A. guillouiae | 1.914 | A. baumannii | 99 | ACB complex | 92 |
| 39 | A. haemolyticus | A. junii | A. junii | 2.279 | A. junii | 99 | A. lwoffii | 90 |
Species are identified by maximal identification of gene sequencing.
Bruker Biotyper database (DB 5627) does not include Acinetobacter genospecies 14TU, Acinetobacter genospecies 16BJ, A. bereziniae (Acinetobacter genospecies 10), and A. gyllenbergi.
LD, low discrimination.
Of the 286 ACB complex isolates, 176 were identified as A. baumannii by the Bruker Biotyper system, and 142 (80.7%) were genetically confirmed to be A. baumannii, 22 (12.5%) were confirmed to be A. nosocomialis, and 12 (6.8%) were confirmed to be Acinetobacter genospecies “close to 13TU.” Of the 66 isolates of A. nosocomialis identified by the Bruker Biotyper, 63 (95.5%) were identified correctly. Of the 44 isolates of A. pittii identified by the Bruker Biotyper, three (6.8%) were not correctly identified by molecular methods as being A. pittii. Of the 39 isolates of Acinetobacter species other than ACB complex, accurate identification by the Bruker Biotyper was obtained for all A. junii, A. ursingii, A. johnsonii, and A. radioresistens isolates (Table 2).
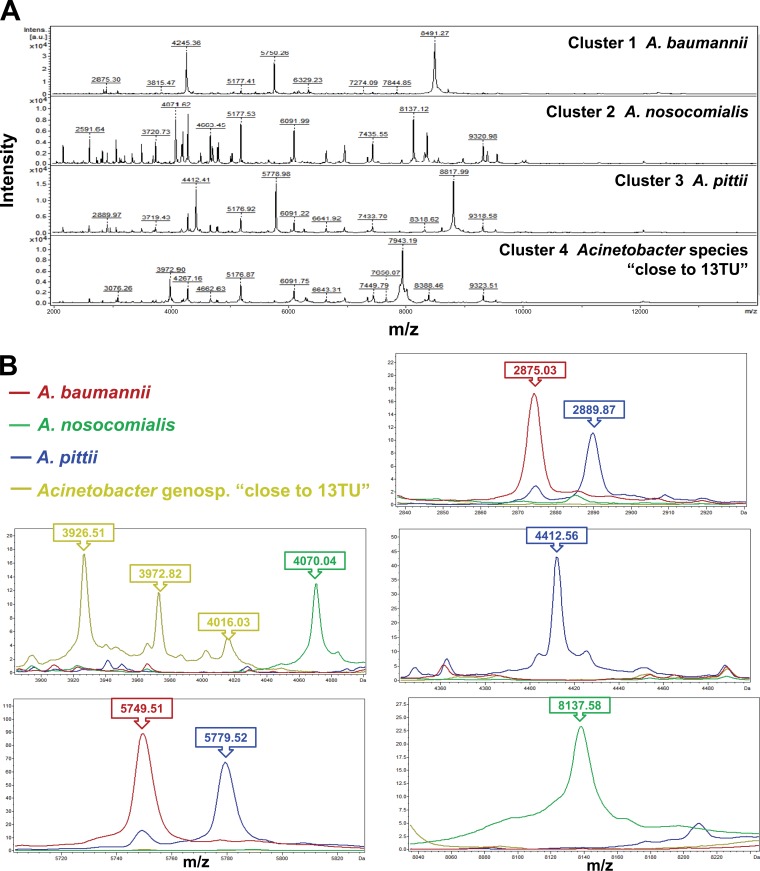
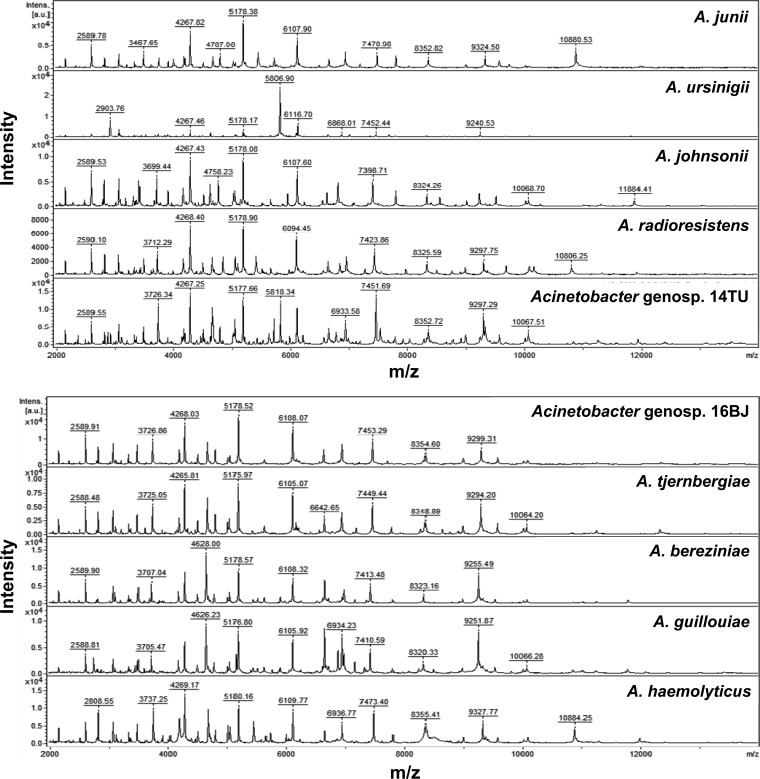
Cluster analysis of spectra generated by the Bruker Biotyper for 286 isolates of four genospecies of ACB complex isolates was performed. Four clusters of spectra for ACB complex isolates, i.e., cluster 1 (A. baumannii), cluster 2 (A. nosocomialis), cluster 3 (A. pittii), and cluster 4 (Acinetobacter genospecies “close to 13TU”) were identified based on the 10 peaks generated by ClinProTools with the genetic algorithm, i.e., 2875.03 m/z and 5749.51 m/z (A. baumannii), 4070.04 m/z and 8137.58 m/z (A. nosocomialis); 2889.87 m/z, 4412.56 m/z and 5779.52 m/z (A. pittii); and 3926.51 m/z, 3972.82 m/z, and 4016.03 m/z (Acinetobacter genospecies “close to 13TU”) (Fig. 1A and B). The spectra of 10 species (genospecies) of Acinetobacter species other than ACB complex generated by the MALDI-TOF Bruker Biotyper system are shown in Fig. 2.
FIG 1.
Cluster analysis of spectra generated by the MALDI-TOF Bruker Biotyper system for 286 A. baumannii complex (ACB complex) isolates. (A) Four clusters of spectra for ACB complex isolates, i.e., cluster 1 (A. baumannii), cluster 2 (A. nosocomialis), cluster 3 (A. pittii), and cluster 4 (Acinetobacter genospecies “close to 13TU”) were identified. (B) Clusters were based on the 10 peaks generated by ClinProTools with a genetic algorithm, i.e., 2875.03 m/z and 5749.51 m/z (A. baumannii)), 4070.04 m/z and 8137.58 m/z (A. nosocomialis); 2889.87 m/z, 4412.56 m/z, and 5779.52 m/z (A. pittii); and 3926.51 m/z, 3972.82 m/z, and 4016.03 m/z (Acinetobacter genospecies “close to 13TU”). The absolute intensities of the ions are shown on the y axis, and the masses (m/z) of the ions are shown on the x axis. The m/z values represent the mass-to-charge ratios.
FIG 2.
Spectra of 10 species (genospecies) of Acinetobacter species other than ACB complex generated by the MALDI-TOF Bruker Biotyper system. The absolute intensities of the ions are shown on the y axis, and the masses (m/z) of the ions are shown on the x axis. The m/z values represent the mass-to-charge ratios.
Of the 286 ACB complex isolates, 176 were identified as A. baumannii by Bruker Biotyper. Of these 176 isolates, 142 (80.7%) were genetically confirmed to be A. baumannii, 22 (12.5%) were confirmed to be A. nosocomialis, and 12 (6.8%) were confirmed to be Acinetobacter genospecies “close to 13TU”. Cluster analysis of spectra generated by the Bruker Biotyper system for model establishment was performed for nearly half (n = 89) of the 176 isolates of A. baumannii identified by the Bruker Biotyper system. Genetically, 72 were A. baumannii, 11 were A. nosocomialis, and 6 were Acinetobacter genospecies “close to 13TU”. The characteristic signals generated using clustering analysis by ClinProTools with the genetic algorithm specific for separating the three genospecies of ACB complex were 3091.35 m/z, 3926.18 m/z, 4070.04 m/z, 5749.51 m/z, and 8137.58 m/z. Of the remaining 87 BioTyper spectra of A. baumannii isolates used for external validation, 70 were genotypically confirmed A. baumannii isolates, 11 were A. nosocomialis isolates, and 6 were Acinetobacter genospecies “close to 13TU.” The overall correct validation rates by the established model were 98.8% for the three genospecies of the ACB complex (Table 3).
TABLE 3.
Results of external validation of 87 spectra of the three genospecies of the A. baumannii complex (ACB complex) isolates by the Bruker Biotyper system
| Acinetobacter species | No. of isolates | No. (%) of isolates with the following genospecies of ACB complex by external validationa: |
||
|---|---|---|---|---|
| A. baumannii | A. nosocomialis | Acinetobacter genospecies 13TU | ||
| A. baumannii | 70 | 69 (98.6) | 1 (1.4) | 0 |
| A. nosocomialis | 11 | 0 | 11 (100) | 0 |
| Acinetobacter genospecies “close to 13TU” | 6 | 1 (3.3) | 6 (100) | |
The rates of correct identification by external validation results are given in boldface type.
Several important findings were demonstrated. First, the overall rate of the Bruker Biotyper in correctly identifying ACB complex isolates to the species level (score values ≥2.0) was only 85.3%. This is partly because Acinetobacter species 13TU is not included in the current database. Using cluster analysis of spectra from the 286 isolates of the ACB complex, 10 peaks were identified to be unique to the four genospecies and might be useful to generate a local database. Second, although A. nosocomialis is included in the current database of the Bruker Biotyper system, the correct identification rate was only 72.4%. Third, of the 176 A. baumannii isolates identified by the Bruker Biotyper system (score values of ≥1.700), about 80% were correctly identified to the species level (score values of ≥2.000). Interestingly, using six characteristic markers created by cluster analysis of spectra of the 176 isolates, nearly all (98.8%) could be reidentified correctly. Finally, compared with the Vitek identification system, the Phoenix identification system is not suitable for identification of Acinetobacter species, particularly for species other than ACB complex isolates.
Alvarez-Buylla et al. reported that the MALDI-TOF Bruker Biotyper system is useful for identifying A. baumannii and A. pittii but not for identifying A. calcoaceticus, A. nosocomialis, A. berezinae, or A. genospecies 14BJ (10). The total misidentification rate by the MALDI-TOF Bruker Biotyper system was 16.4% for all Acinetobacter species tested compared with the identification results by rpoB gene sequencing analysis. Espinal et al. analyzed 60 genotypically characterized ACB complex isolates, including 18 A. nosocomialis isolates, 17 A. pittii strains, 18 A. baumanni isolates, and 7 other Acinetobacter species isolates against a local database (inclusion of specific signature profiles for A. nosocomialis within the Bruker database) and found that 98.3% of all isolates were identified at the species level (log score > 2.0), and only 1.7% (one of seven other Acinetobacter species) were identified at the probable genus level with a log score between 1.7 and 2.0 (9).
Our work documented high identification rates of A. baumannii, A. pittii, and non-ACB complex by the MALDI-TOF Bruker Biotyper system. However, further expansion and improvement of the database, including A. nosocomialis and Acinetobacter genospecies “close to 13TU,” is mandatory to make MALDI-TOF MS an efficient method for identification of all Acinetobacter species.
Footnotes
Published ahead of print 4 June 2014
REFERENCES
- 1.Chang HC, Wei YF, Dijkshoorn L, Vaneechoutte M, Tang CT, Chang TC. 2005. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J. Clin. Microbiol. 43:1632–1639. 10.1128/JCM.43.4.1632-1639.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gundi VA, Dijkshoorn L, Burignat S, Raoult D, La Scola B. 2009. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology 155:2333–2341. 10.1099/mic.0.026054-0 [DOI] [PubMed] [Google Scholar]
- 3.Tien N, You BJ, Chang HL, Lin HS, Lee CY, Chung TC, Lu JJ, Chang CC. 2012. Comparison of genospecies and antimicrobial resistance profiles of isolates in the Acinetobacter calcoaceticus-Acinetobacter baumannii complex from various clinical specimens. Antimicrob. Agents Chemother. 56:6267–6271. 10.1128/AAC.01304-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee YC, Huang YT, Tan CK, Kuo YW, Liao CH, Lee PI, Hsueh PR. 2011. Acinetobacter baumannii and Acinetobacter genospecies 13TU and 3 bacteraemia: comparison of clinical features, prognostic factors and outcomes. J. Antimicrob. Chemother. 66:1839–1846. 10.1093/jac/dkr200 [DOI] [PubMed] [Google Scholar]
- 5.Dubois D, Grare M, Prere MF, Segonds C, Marty N, Oswald E. 2012. Performances of the Vitek MS matrix-assisted laser desorption ionization–time of flight mass spectrometry system for rapid identification of bacteria in routine clinical microbiology. J. Clin. Microbiol. 50:2568–2576. 10.1128/JCM.00343-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seng P, Abat C, Rolain JM, Colson P, Lagier JC, Gouriet F, Fournier PE, Drancourt M, La Scola B, Raoult D. 2013. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 51:2182–2194. 10.1128/JCM.00492-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray PR. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry: usefulness for taxonomy and epidemiology. Clin. Microbiol. Infect. 16:1626–1630. 10.1111/j.1469-0691.2010.03364.x [DOI] [PubMed] [Google Scholar]
- 8.Teng SH, Chen CM, Lee MR, Lee TF, Chien KY, Teng LJ, Hsueh PR. 2013. Matrix-assisted laser desorption ionization–time of flight mass spectrometry can accurately differentiate between Mycobacterium masilliense (M. abscessus subspecies bolletti) and M. abscessus (sensu stricto). J. Clin. Microbiol. 51:3113–3116. 10.1128/JCM.01239-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espinal P, Seifert H, Dijkshoorn L, Vila J, Roca I. 2012. Rapid and accurate identification of genomic species from the Acinetobacter baumannii (Ab) group by MALDI-TOF MS. Clin. Microbiol. Infect. 18:1097–1103. 10.1111/j.1469-0691.2011.03696.x [DOI] [PubMed] [Google Scholar]
- 10.Alvarez-Buylla A, Culebras E, Picazo JJ. 2012. Identification of Acinetobacter species: is Bruker biotyper MALDI-TOF mass spectrometry a good alternative to molecular techniques? Infect. Genet. Evol. 12:345–349. 10.1016/j.meegid.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 11.Kishii K, Kikuchi K, Matsuda N, Yoshida A, Okuzumi K, Uetera Y, Yasuhara H, Moriya K. 28 August 2013. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for species identification of Acinetobacter strains isolated from blood cultures. Clin. Microbiol. Infect. 10.1111/1469-0691.12376 [DOI] [PubMed] [Google Scholar]
- 12.Sedo O, Nemec A, Krizova L, Kacalova M, Zdrahal Z. 2013. Improvement of MALDI-TOF MS profiling for the differentiation of species within the Acinetobacter calcoaceticus-Acinetobacter baumannii complex. Syst. Appl. Microbiol. 36:572–578. 10.1016/j.syapm.2013.08.001 [DOI] [PubMed] [Google Scholar]