Abstract
We investigated three nosocomial Candida quercitrusa candidemia cases occurring within 2 months in a Chinese hospital. Isolates were identifiable only by DNA sequencing of the rRNA genes. Genetic (via random amplified polymorphic DNA [RAPD]) and protein mass spectral (via matrix-assisted laser desorption ionization–time of flight mass spectrometry [MALDI-TOF MS]) analyses yielded identical profiles suggesting an outbreak. The fluconazole MICs of all the strains were 16 to 32 μg/ml.
TEXT
Candidemia in hospitalized patients causes substantial mortality and has high costs (1, 2). Some studies have suggested that up to one-third of cases may occur as nosocomial clusters (3, 4). Furthermore, uncommon or novel Candida species are increasingly recognized to cause candidemia (5–8). Here, we describe the clinical characteristics of three candidemia cases caused by the novel species Candida quercitrusa, which occurred within a 2-month period in a single hospital's intensive care unit (ICU) in Harbin, China (9).
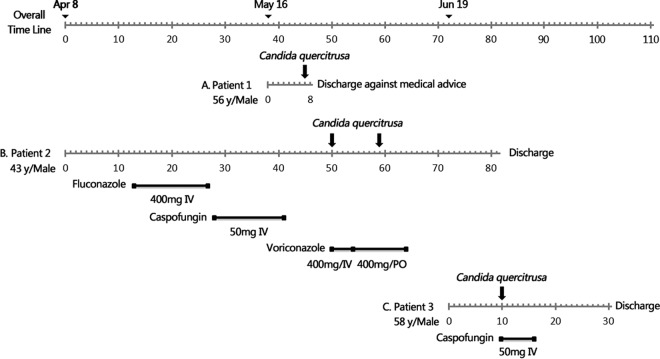
Patient 1 was admitted to the hospital's ICU in May 2010 with acute upper gastrointestinal bleeding. On day 7 of the admission, peripheral blood cultures (Bactec Myco/F Lytic; Becton, Dickinson, Sparks, MD, USA) grew a strain identified as Candida pulcherrima (strain 10H1064), as identified by the Vitek 2 YST system (bioMérieux, Marcy l'Etoile, France) (identification accuracy, 89%) (Fig. 1A). Patient 2 presented to the same ICU in April 2010 with major trauma. On days 50 and 59 of the admission, blood cultures from a central venous catheter (CVC) grew a strain identified as C. pulcherrima (strain 10H1067, initially identified as for patient 1) (Fig. 1B). Patient 3 was admitted to the ICU in July 2010 with fever, seizures, and obtundation. A strain identified as Candida lusitaniae (strain 10H1076) by the API 20C AUX system (bioMérieux) (identification accuracy, 86.2%) was recovered from peripheral blood cultures on day 10 (Fig. 1C). Table 1 summarizes the clinical details of the three cases. Patients 2 and 3 had central lines in situ and were receiving total parenteral nutrition (TPN).
FIG 1.
Clinical features, treatment regimens, and outcomes of patients with Candida quercitrusa candidemia. Time axis in each case (A, B, and C, respectively) indicates period of hospital stay, with numbers of days since the beginning of hospitalization. Lines with black squares, period and dose per day of antifungal treatment; down-facing black arrows, dates of positive C. quercitrusa blood culture.
TABLE 1.
Clinical features of three patients with Candida quercitrusa candidemia
| Clinical feature | Patient 1 | Patient 2 | Patient 3 |
|---|---|---|---|
| Age (yr) | 56 | 43 | 58 |
| Gender | Male | Male | Male |
| Reason for hospital admission | Acute upper gastrointestinal bleeding, high fever | Fracture dislocation caused by a falling | Clonic spasm, high fever, meningitis |
| Underlying disease | Hypertension, coronary heart disease, diabetes | Aspiration pneumonia | Hypertension |
| Total no. of blood cultures taken | 1 | 9 | 4 |
| Date(s) (day/mo/yr) of C. quercitrusa isolationa | 23/05/2010 (7) | 06/06/2010 (50), 28/05/2010 (59) | 29/06/2010 (10) |
| Clinical status at time of positive culture | |||
| Immunosuppressive state | No | No | No |
| Neutropenia (<109 per liter) | No | No | No |
| Presence of CVC | No | Yes | Yes |
| Broad-spectrum antibiotics | Yes | Yes | Yes |
| Total parenteral nutrition | No | Yes | Yes |
| Surgery within 30 days | No | Yes | No |
| Intensive care | Yes | Yes | Yes |
| Previous antifungal agents within 30 days | No | Yes | No |
| Concomitant bacteremia | No | No | No |
| Concomitant candidemia | No | Yes (Candida lipolytica) | No |
| Indwelling urinary catheter | No | Yes | Yes |
| Therapy | |||
| Antifungal | No antifungal therapy commenced | Voriconazole 400 mg daily, 14 days | Caspofungin 50 mg daily, 8 days |
| CVC removala | Not applicable | Yes (64) | Yes (10) |
| Antifungal, after culture | Yes | Yes | Yes |
| Outcome | Unknownb | Recovered | Recovered |
Numbers in parentheses indicate days since beginning of hospitalization.
Patient discharged against medical advice and was lost to further follow-up.
The isolates identified as C. pulcherrima and C. lusitaniae were forwarded to the reference mycology laboratory at the Peking Union Medical College Hospital for further mycological studies, including observing their appearance on chromogenic media and species identification by molecular-based approaches (see below). This practice was in place for all suspected yeast strains causing invasive infection, per protocol, for a national survey of invasive fungal infections in China (the China Hospital Invasive Fungal Surveillance Net [CHIF-NET] study) (9).
Specifically, DNA sequencing of the isolates was performed by amplifying the internal transcribed spacer (ITS) region and the D1/D2 domain of the rRNA gene as previously described (9, 10). A comparison of all available C. quercitrusa sequences, including those of the isolates from the current study, was then performed using the maximum-parsimony (MP) method (11). The isolates were also analyzed by matrix-assisted laser desorption ionization–time of flight mass spectroscopy (MALDI-TOF MS) using Bruker Biotyper version 3.1 software (Bruker Daltonics, Bremen, Germany), and protein profiles were further studied by use of main spectra projections (12). The primers RAPD24 and RAPD1283 were then used for random amplified polymorphic DNA (RAPD) analysis (13). In vitro susceptibility to amphotericin B, fluconazole, voriconazole, itraconazole, and caspofungin was determined by Clinical and Laboratory Standards Institute M27–A3 methodology (14).
On Sabouraud dextrose agar and chromogenic media, the isolates grew well at 25°C and 30°C. They grew slowly at 37°C over 5 days (cf. with the type strain C. quercitrusa CBS 4412, which failed to grow at 37°C) but did not grow at 42°C. The colonies were dark blue on CHROMagar Candida medium (CHROMagar Company, Paris, France) (Fig. 2A) and dark green on Brilliance Candida agar (Oxoid Ltd., Hampshire, United Kingdom) (Fig. 2B).
FIG 2.
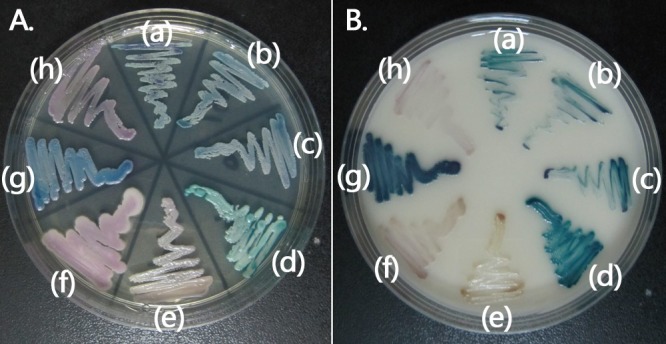
Phenotypic characteristics of C. quercitrusa isolates on CHROMagar Candida medium (A) and Brilliance Candida agar (B). (a) C. quercitrusa 10H1064; (b) C. quercitrusa 10H1067; (c) C. quercitrusa 10H1076; (d) C. albicans ATCC 90028; (e) Candida parapsilosis sensu stricto ATCC 22019; (f) C. krusei ATCC 6258; (g) C. tropicalis 10H1048; and (h) C. glabrata sensu stricto 10H1043.
All patient isolates were identified as C. quercitrusa by querying their ITS region and D1/D2 domain sequences against the GenBank database (accessed 31 December 2013) (Table 2). The ITS and D1/D2 sequences of all three study isolates were identical and shared 98.9 and 99.8% sequence similarity, respectively, to those of C. quercitrusa type strain CBS 4412 (Table 2). The GenBank sequences of the remaining C. quercitrusa isolates also shared high sequence similarity (95.8 to 100%) with those of C. quercitrusa CBS 4412 (Table 2), but at least moderate intraspecies sequence variation was indicated within the ITS region and D1/D2 domain for this species. MP analysis of the ITS region revealed genetic heterogeneity among the C. quercitrusa strains analyzed; there was clustering of the three patient isolates (strains 10H1064, 10H1967, and 10H1076), which were clearly separated from the nonclinical strains (Fig. 3). MALDI-TOF MS assigned no identification to the patient isolates (spectral score, <1.5). Their RAPD profiles were identical, as were their MALDI-TOF MS spectra (see Fig. S1A and S1B in the supplemental material).
TABLE 2.
Summary of C. quercitrusa isolates from published literature or GenBank
| Strain | Country | Origin | Reference or source | ITS |
D1/D2a |
||
|---|---|---|---|---|---|---|---|
| Accession no. | Identityb | Accession no. | Identityb | ||||
| Type strain CBS 4412 | Australia | Insect | 22 | AM158924 | Reference | U45831 | Reference |
| Clinical isolates | |||||||
| 10H1064 | Mainland China | Human blood | This study | KF220648 | 625/632 (98.9) | KF220651 | 564/565 (99.8) |
| 10H1067 | Mainland China | Human blood | This study | KF220649 | 625/632 (98.9) | KF220652 | 564/565 (99.8) |
| 10H1076 | Mainland China | Human blood | This study | KF220650 | 625/632 (98.9) | KF220653 | 564/565 (99.8) |
| Isolates from other sources | |||||||
| BJ50 | Mainland China | Apple orchard | Unpublished | NAc | NA | JQ219335 | 565/565 (100) |
| JM36 | Mainland China | Coffee | Unpublished | NA | NA | KC510074 | 548/551 (99.4) |
| LYSJLFL-1 | Mainland China | Food | Unpublished | NA | NA | JX049432 | 554/560 (98.9) |
| JHSb | Mainland China | Marine | Unpublished | DQ665264 | 566/591 (95.8) | EF375703 | 555/566 (98.1) |
| PH-M32 | Mainland China | Wastewater | Unpublished | NA | NA | GU373802 | 548/551 (99.4) |
| TA256 | Mainland China | Apple orchard | Unpublished | NA | NA | JQ219336 | 565/565 (100) |
| WG1 | Mainland China | Wine grape | 17 | GU237045 | 537/558 (96.2) | NA | NA |
| 339 | Taiwan | Plant | Unpublished | NA | NA | JN544056 | 565/565 (100) |
| NU9L75 | Taiwan | Leaf | Unpublished | NA | NA | HM461728 | 537/538 (99.8) |
| NRRL Y-27941 | America | Insect | 18 | NA | NA | DQ655691 | 543/544 (99.8) |
| B176 | America | Sugar beet root | Unpublished | NA | NA | EU196383 | 294/294 (100) |
| UNC MB27 | America | Flower | 15 | NA | NA | JN642539 | 473/474 (99.8) |
| HA 1669 | Australia | Insect | Unpublished | AM160627 | 606/606 (100) | AM160627 | 565/565 (100) |
| N17 | Brazil | Grape | 16 | GQ999840 | 457/465 (98.3) | NA | NA |
| 17a/3 | Germany | Root tip | Unpublished | HQ680959 | 443/445 (99.6) | NA | NA |
| 129 | Ghana | Cocoa bean | 23 | NA | NA | AY529522 | 561/563 (99.6) |
| G4 | Ghana | Cocoa bean | 24 | NA | NA | DQ466526 | 555/556 (99.8) |
| NCL 6 | India | Grape | 25 | FJ231428 | 538/549 (98.0) | NA | NA |
| 3.1 | Italy | Beverage | Unpublished | NA | NA | JN417623 | 503/503 (100) |
| EGV28 | Mexico | Zea mays | Unpublished | JX455761 | 566/582 (97.2) | NA | NA |
| EGV70 | Mexico | Zea mays | Unpublished | JX455759 | 518/523 (99.0) | NA | NA |
| DMKU-RK1 | Thailand | Rice/Corn/Sugarcane | Unpublished | NA | NA | AB773291 | 562/567 (99.1) |
| DMKU-RK14 | Thailand | Rice/Corn/Sugarcane | Unpublished | NA | NA | AB772038 | 564/565 (99.8) |
| DMKU-RK2 | Thailand | Rice/Corn/Sugarcane | Unpublished | NA | NA | AB773292 | 564/565 (99.8) |
| DMKU-RK4 | Thailand | Rice/Corn/Sugarcane | Unpublished | NA | NA | AB773294 | 562/567 (99.1) |
| DMKU-RK5 | Thailand | Rice/Corn/Sugarcane | Unpublished | NA | NA | AB773295 | 563/565 (99.6) |
| DMKU-RK504 | Thailand | Rice/Corn/Sugarcane | Unpublished | NA | NA | AB773375 | 562/567 (99.1) |
| DMKU-RK506 | Thailand | Rice/Corn/Sugarcane | Unpublished | NA | NA | AB773377 | 539/546 (98.7) |
| DMKU-RK516 | Thailand | Rice/Corn/Sugarcane | Unpublished | NA | NA | AB773384 | 562/567 (99.1) |
| EC4 | Thailand | Water | Unpublished | NA | NA | AB436403 | 564/565 (99.8) |
D1/D2, D1/D2 domain of the 26S ribosomal DNA.
Number of nucleotides identical/number of nucleotides compared (%) between the indicated C. quercitrusa strain and type strain CBS 4412.
NA, not available.
FIG 3.
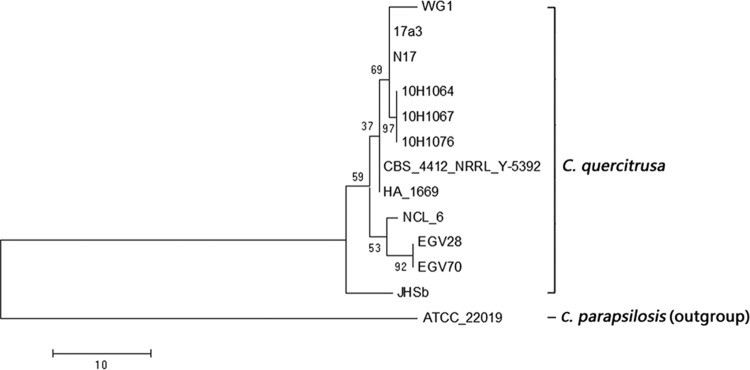
The maximum-parsimony tree generated from C. quercitrusa ITS sequences available in GenBank (see Table 2), with C. parapsilosis ATCC 22019 (GenBank accession no. FJ872015) as outgroup.
All the isolates had fluconazole MICs of 16 to 32 μg/ml. The MIC ranges for itraconazole and voriconazole were 0.25 to 0.5 and 0.125 to 0.25 μg/ml, respectively. The MICs for caspofungin and amphotericin B were low (MIC ranges, 0.5 to 1 and ≤0.5 μg/ml, respectively).
This report details, for the first time, candidemia due to the novel species C. quercitrusa in three Chinese patients identified through surveillance (9). Although C. quercitrusa isolates have been recovered from plant, water, and insect material in a number of countries (15–18), the species has not been reported to cause human infection (Table 2). The cases herein are also notable for their clustering in the same hospital within only a 2-month period.
Of interest, an analysis of the ITS and D1/D2 region sequences of the available C. quercitrusa isolates in the GenBank database to date suggests that this species may be genetically diverse, with intraspecies sequence heterogeneity in these gene regions (Table 2). However, the ITS and D1/D2 sequences of the patient isolates in the present report were identical. Taken together with their identical RAPD and MALDI-TOF MS protein profiles, the findings suggest that the isolates may have originated from a common source. Furthermore, MP analysis of their ITS sequences indicated that they are genetically distinct from previously isolated strains from nonclinical sources. A limitation of the present study is that it was performed retrospectively. Given the initial misidentification of the isolates and the resultant delay in accurate species identification, we were consequently unable to investigate for a possible environmental source or the potential for human-to-human transmission, including that through hospital staff. Although definite clustering of cases cannot be confirmed, the proximity in time and close similarity of the genetic and protein profiles suggest a minioutbreak.
The transmission of infection through the contamination of blood culture vials or of intravenous preparations such as TPN bags remains another possibility, suggesting a pseudo outbreak (19). The TPN bags were not available for culture. However, as not all the patients received TPN, this was unlikely to be the source. The blood culture bottles were of two different batches (lot 9320384 for patients 1 and 2, lot 0055634 for patient 3). There were no other patients with Candida in their blood at the time that our patients were ill with candidemia.
Note that phenotypic identification methods, including the use of chromogenic media, failed to correctly identify this species. MALDI-TOF MS, while not being able to assign species, did not misidentify the organism. Analyses of spectra from other clinical isolates are important in creating a robust spectral library for this species (20, 21). The fact that DNA sequencing of the ITS region and/or the D1/D2 domain remains necessary for species identification emphasizes the need to be vigilant for unusual species assignments by standard identification methods.
Curiously, all three C. quercitrusa strains grew very slowly at 37°C yet were able to cause candidemia in humans. The fungal load necessary to result in clinical infection is unknown. If infection is hypothesized to originate from contaminated intravenous fluids/materials, it is possible that a relatively large fungal load was inoculated into the bloodstream and resulted in candidemia.
The relatively high MICs (16 to 32 μg/ml) to fluconazole suggest that this antifungal agent should not be used to treat C. quercitrusa infections. However, the species was more susceptible to amphotericin B, the newer azoles, and caspofungin. Patients 2 and 3 recovered with no relapse after receiving voriconazole and caspofungin therapy, respectively.
In conclusion, we detail C. quercitrusa as a human pathogen, for the first time, with a clustering of three C. quercitrusa candidemia cases in a single ICU. Molecular methods are needed for the accurate identification of this species. The species is less susceptible to fluconazole.
Nucleotide sequence accession numbers.
Nucleotide sequences for the ITS regions and D1/D2 domains of C. quercitrusa 10H1064, C. quercitrusa 10H1067, and C. quercitrusa 10H1076 were deposited in GenBank under accession numbers KF220648 to KF220653.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Special Research Foundation for Capital Medical Development: Epidemiology and in Vitro Antifungal Susceptibility of Yeast Species Causing Invasive Fungal Infections in Beijing (grant 2011-4001-09) and the Youth Research Foundation in Peking Union Medical College: Novel Identification Assays of Clinical Important Invasive Fungal Species (grant 2012X03).
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 2 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00246-14.
REFERENCES
- 1.Arnold HM, Micek ST, Shorr AF, Zilberberg MD, Labelle AJ, Kothari S, Kollef MH. 2010. Hospital resource utilization and costs of inappropriate treatment of candidemia. Pharmacotherapy 30:361–368. 10.1592/phco.30.4.361 [DOI] [PubMed] [Google Scholar]
- 2.Falagas ME, Apostolou KE, Pappas VD. 2006. Attributable mortality of candidemia: a systematic review of matched cohort and case-control studies. Eur. J. Clin. Microbiol. Infect. Dis. 25:419–425. 10.1007/s10096-006-0159-2 [DOI] [PubMed] [Google Scholar]
- 3.Asmundsdottir LR, Erlendsdottir H, Haraldsson G, Guo H, Xu J, Gottfredsson M. 2008. Molecular epidemiology of candidemia: evidence of clusters of smoldering nosocomial infections. Clin. Infect. Dis. 47:e17–e24. 10.1086/589298 [DOI] [PubMed] [Google Scholar]
- 4.Chou HH, Lo HJ, Chen KW, Liao MH, Li SY. 2007. Multilocus sequence typing of Candida tropicalis shows clonal cluster enriched in isolates with resistance or trailing growth of fluconazole. Diagn. Microbiol. Infect. Dis. 58:427–433. 10.1016/j.diagmicrobio.2007.03.014 [DOI] [PubMed] [Google Scholar]
- 5.Chen SC, Marriott D, Playford EG, Nguyen Q, Ellis D, Meyer W, Sorrell TC, Slavin M. 2009. Candidaemia with uncommon Candida species: predisposing factors, outcome, antifungal susceptibility, and implications for management. Clin. Microbiol. Infect. 15:662–669. 10.1111/j.1469-0691.2009.02821.x [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez GM, Trevino-Rangel Rde J, Palma-Nicolas JP, Martinez C, Gonzalez JG, Ayala J, Caballero A, Morfin-Otero R, Rodriguez-Noriega E, Velarde F, Ascencio EP, Tinoco JC, Vazquez JA, Cano MA, Leon-Sicairos N, Gonzalez R, Rincon J, Elias MA, Bonifaz A. 2013. Species distribution and antifungal susceptibility of bloodstream fungal isolates in paediatric patients in Mexico: a nationwide surveillance study. J. Antimicrob. Chemother. 68:2847–2851. 10.1093/jac/dkt283 [DOI] [PubMed] [Google Scholar]
- 7.Pfaller MA, Diekema DJ, Colombo AL, Kibbler C, Ng KP, Gibbs DL, Newell VA. 2006. Candida rugosa, an emerging fungal pathogen with resistance to azoles: geographic and temporal trends from the ARTEMIS DISK antifungal surveillance program. J. Clin. Microbiol. 44:3578–3582. 10.1128/JCM.00863-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma C, Wankhede S, Muralidhar S, Prakash A, Singh PK, Kathuria S, Kumar DA, Khan N, Randhawa HS, Meis JF, Chowdhary A. 2013. Candida nivariensis as an etiologic agent of vulvovaginal candidiasis in a tertiary care hospital of New Delhi, India. Diagn. Microbiol. Infect. Dis. 76:46–50. 10.1016/j.diagmicrobio.2013.02.023 [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Xiao M, Chen SC, Kong F, Sun ZY, Liao K, Lu J, Shao HF, Yan Y, Fan H, Hu ZD, Chu YZ, Hu TS, Ni YX, Zou GL, Xu YC. 2012. In vitro susceptibilities of yeast species to fluconazole and voriconazole as determined by the 2010 National China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study. J. Clin. Microbiol. 50:3952–3959. 10.1128/JCM.01130-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taverna CG, Bosco-Borgeat ME, Murisengo OA, Davel G, Boite MC, Cupolillo E, Canteros CE. 2013. Comparative analyses of classical phenotypic method and ribosomal RNA gene sequencing for identification of medically relevant Candida species. Mem. Inst. Oswaldo Cruz 108:178–185. 10.1590/0074-0276108022013009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall BG. 2013. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30:1229–1235. 10.1093/molbev/mst012 [DOI] [PubMed] [Google Scholar]
- 12.Pulcrano G, Roscetto E, Iula VD, Panellis D, Rossano F, Catania MR. 2012. MALDI-TOF mass spectrometry and microsatellite markers to evaluate Candida parapsilosis transmission in neonatal intensive care units. Eur. J. Clin. Microbiol. Infect. Dis. 31:2919–2928. 10.1007/s10096-012-1642-6 [DOI] [PubMed] [Google Scholar]
- 13.Pfliegler WP, Horvath E, Kallai Z, Sipiczki M. 2014. Diversity of Candida zemplinina isolates inferred from RAPD, micro/minisatellite and physiological analysis. Microbiol. Res. 169:402–410. 10.1016/j.micres.2013.09.006 [DOI] [PubMed] [Google Scholar]
- 14.CLSI. 2009. Reference method for broth dilution antifungal susceptibility testing of yeasts, approved standard, 3rd edition M27–A3. CLSI, Wayne, PA [Google Scholar]
- 15.Belisle M, Peay KG, Fukami T. 2012. Flowers as islands: spatial distribution of nectar-inhabiting microfungi among plants of Mimulus aurantiacus, a hummingbird-pollinated shrub. Microb. Ecol. 63:711–718. 10.1007/s00248-011-9975-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bezerra-Bussoli C, Baffi MA, Gomes E, Da-Silva R. 2013. Yeast diversity isolated from grape musts during spontaneous fermentation from a Brazilian winery. Curr. Microbiol. 67:356–361. 10.1007/s00284-013-0375-9 [DOI] [PubMed] [Google Scholar]
- 17.Li SS, Cheng C, Li Z, Chen JY, Yan B, Han BZ, Reeves M. 2010. Yeast species associated with wine grapes in China. Int. J. Food. Microbiol. 138:85–90. 10.1016/j.ijfoodmicro.2010.01.009 [DOI] [PubMed] [Google Scholar]
- 18.Nguyen NH, Suh SO, Blackwell M. 2007. Five novel Candida species in insect-associated yeast clades isolated from Neuroptera and other insects. Mycologia 99:842–858. 10.3852/mycologia.99.6.842 [DOI] [PubMed] [Google Scholar]
- 19.Medeiros EA, Lott TJ, Colombo AL, Godoy P, Coutinho AP, Braga MS, Nucci M, Brandt ME. 2007. Evidence for a pseudo-outbreak of Candida guilliermondii fungemia in a university hospital in Brazil. J. Clin. Microbiol. 45:942–947. 10.1128/JCM.01878-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castanheira M, Woosley LN, Diekema DJ, Jones RN, Pfaller MA. 2013. Candida guilliermondii and other species of Candida misidentified as Candida famata: assessment by Vitek 2, DNA sequencing analysis, and matrix-assisted laser desorption ionization–time of flight mass spectrometry in two global antifungal surveillance programs. J. Clin. Microbiol. 51:117–124. 10.1128/JCM.01686-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohmann C, Sabou M, Moussaoui W, Prevost G, Delarbre JM, Candolfi E, Gravet A, Letscher-Bru V. 2013. Comparison between the Biflex III-Biotyper and the Axima-SARAMIS systems for yeast identification by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 51:1231–1236. 10.1128/JCM.03268-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtzman CP, Robnett CJ. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jespersen L, Nielsen DS, Honholt S, Jakobsen M. 2005. Occurrence and diversity of yeasts involved in fermentation of West African cocoa beans. FEMS Yeast Res. 5:441–453. 10.1016/j.femsyr.2004.11.002 [DOI] [PubMed] [Google Scholar]
- 24.Nielsen DS, Teniola OD, Ban-Koffi L, Owusu M, Andersson TS, Holzapfel WH. 2007. The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int. J. Food Microbiol. 114:168–186. 10.1016/j.ijfoodmicro.2006.09.010 [DOI] [PubMed] [Google Scholar]
- 25.Chavan P, Mane S, Kulkarni G, Shaikh S, Ghormade V, Nerkar DP, Shouche Y, Deshpande MV. 2009. Natural yeast flora of different varieties of grapes used for wine making in India. Food Microbiol. 26:801–808. 10.1016/j.fm.2009.05.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.