Abstract
Human cytomegalovirus (CMV) has historically been the major infectious cause of morbidity and mortality among patients receiving hematopoietic cell or organ transplant. Standard care in a transplant setting involves frequent monitoring of CMV viral load over weeks to months to determine when antiviral treatment may be required. Quantitative PCR (qPCR) is the standard molecular diagnostic method for monitoring. Recently, digital PCR (dPCR) has shown promise in viral diagnostics, although current dPCR systems have lower throughput than qPCR systems. Here, we compare qPCR and droplet digital PCR (ddPCR) for CMV detection in patient plasma samples. Droplet digital PCR exhibits increased precision over qPCR at viral loads of ≥4 log10 with equivalent sensitivity. However, retrospective analysis of longitudinal samples from transplant patients with CMV viral loads near therapeutic thresholds did not provide evidence that the improved precision of ddPCR would be of clinical benefit. Given the throughput advantages of current qPCR systems, a widespread switch to dPCR for CMV monitoring would appear premature.
INTRODUCTION
Clinical viral diagnostic approaches rely heavily on quantitative PCR (qPCR) as a method to detect and quantify viral load in patient samples. However, accuracy and precision of this testing is limited by the lack of universal standard material for many pathogens and reliance upon a standard curve for quantitation (1). Digital PCR (dPCR) offers potential improvement over current testing methods through absolute quantitation of viral load without the need for a calibration curve (2, 3).
Human cytomegalovirus (CMV) is a major contributor to morbidity and mortality of immunocompromised patients, including transplant and HIV-infected patients. Tracking CMV viral load in transplant patients helps predict disease development and inform antiviral treatment decisions (4). The World Health Organization (WHO) has recently released a CMV standard material (IU/ml) for the calibration of qPCR assays run in different labs, which should improve commutability of viral load measurements between labs (5, 6). However, digital PCR has the potential to further improve clinical viral diagnostics by providing absolute viral load measurements, abrogating the need for standardization of calibration curves between different laboratory-developed and commercial assays. Digital PCR has also shown increased precision over qPCR in certain applications (7). This precision advantage could aid in monitoring viral disease progression, particularly at low viral load ranges where therapeutic decisions are made.
Several studies have previously investigated the potential of droplet digital PCR (ddPCR) in molecular diagnostics for pathogens, including assays for chlamydia (8), HIV (9, 10), CMV (11), and chromosomally integrated human herpesvirus 6 (HHV-6) (12). In the previous CMV study, investigators concluded that quantitative PCR showed greater sensitivity and less variability than ddPCR in clinical samples (11). However, in that study, the DNA input volumes were not equivalent in the qPCR and ddPCR assays, and therefore we hypothesized that the sensitivity of ddPCR might be improved by increasing the input volume of DNA. Here, we compare the sensitivity and precision of optimized ddPCR and qPCR assays on clinical CMV samples. We then evaluated whether ddPCR could be of clinical benefit to transplant patients, by asking if ddPCR provides greater precision than qPCR in samples with viral loads near therapeutic thresholds critical in making antiviral treatment decisions.
MATERIALS AND METHODS
CMV standards and patient specimens.
An AcroMetrix CMVtc panel (Life Technologies, Benicia, CA) composed of known dilutions (in IU/ml) of AD169 whole virus in EDTA plasma was extracted using the protocol of 1 ml plasma to 80 μl DNA extraction on the Roche MagnaPure LC (Basel, Switzerland) with the large-volume total nucleic acid extraction kit. The WHO standard material (NIBSC, South Mimms, Potters Bar, Herts, United Kingdom) was reconstituted according to the manufacturer's instructions and then diluted 1:1 in negative serum control (Bio-Rad Laboratories; Lyphochek immunoassay plus control level 3 no. 373) before 1:10 dilutions were made. Negative serum control is our laboratory's standard diluent, as it is economical, consistent, and performs well in all plasma PCR assays. Dilutions were extracted using the protocol of 200 μl plasma to 100 μl DNA extraction on the Roche MagnaPure 96 with the DNA and viral NA small-volume kit. The NIST CMV standard reference material (2366), prepared from a bacterial artificial chromosome of CMV TowneΔ147 (13), was purchased from the National Institute of Standards and Technology (Gaithersburg, MD), and component C (19,641 copies/μl) was diluted 10-fold in 10 mM Tris (pH 8), while components A (420 copies/μl) and B (1,702 copies/μl) were run neat; all components were run without extraction.
A residual high-viral-load CMV plasma patient sample (6 log10 copies/ml) was used to create a 10-fold dilution series in negative serum control. Low-viral-load (3 log10 copies/ml) residual patient samples were used to create 2-fold dilutions in a negative serum control at low viral quantities (2 log10 to 1 log10) to determine linearity and 95% cutoff for limit of detection (Probit Analysis; SPSS 15.0). For longitudinal analysis of patient specimens, we selected samples left over after routine CMV clinical testing at the University of Washington Molecular Virology Laboratory. Inclusion criteria for the 19 patients in the study are as follows: testing performed between October 2012 and January 2013, 2 or more CMV-positive samples remaining with ≥1 ml of plasma, and at least one specimen with a viral load of ≤4 log10. All DNA extractions were performed with the 1-ml protocol detailed above. Use of these specimens was approved by the University of Washington Institutional Review Board.
qPCR.
The quantitative PCR (qPCR) assay targets two genes in CMV simultaneously, the UL55 and UL123 genes, to increase the likelihood of accurately detecting all possible clinical variants. Both targets are detected using the same fluorophore (6-carboxyfluorescein [FAM]), and the standard numbers are set to account for the dual amplification of each CMV template copy. The following primer and TaqMan probe sets (14) were mixed with 2× ABI fast mix and 15 μl of template DNA for a final reaction volume of 30 μl: UL55 forward, TGG GCG AGG ACA ACG AA; UL55 reverse, TGA GGC TGG GAA GCT GAC AT; UL55probe, FAM-TGG GCA ACC ACC GCA CTG AGG-TAM (tetramethylrhodamine azide); UL123exon-4F, TCC CGC TTA TCC TCR GGT ACA; UL123exon-4R, TGA GCC TTT CGA GGA SAT GAA; UL123exon4, FAM-TCT CAT ACA TGC TCT GCA TAG TTA GCC CAA TAC A-TAM. The reactions were run on a StepOnePlus system (Applied Biosystems, Benicia, CA) using the following thermocycler parameters: 95°C for 20 s followed by 45 cycles of 95°C for 3 s and 60°C for 30 s. A three-point standard curve was included in all runs. Data were analyzed with StepOnePlus (version 2.1) analysis software, and quantitation of virus was presented as copies/ml of plasma.
ddPCR.
Droplet digital PCR (ddPCR) has been reported to be less affected by target sequence polymorphisms than qPCR (10), and therefore the assay used a single primer-probe set targeting UL55. Both UL55 and UL123 primer/probe sets were tested in ddPCR, and the UL55 set was selected as the most efficient. Additionally, on the standard materials tested in Fig. 1, the UL55 and UL123 primer-probe sets behaved identically. The UL55 primer-probe set was the same as utilized for qPCR, but HEX (6-carboxy-2,4,4,5,7,7 hexachlorofluorescein succinimidyl ester) was substituted for FAM and black hole quencher 1 (BHQ1) was substituted for TAM (Sigma-Aldrich, Saint Louis, MO) to accommodate detection capabilities of the droplet reader. The ddPCR reaction mixture consisted of 12.5 μl of a 2× or 6.25 μl of a 4× ddPCR supermix for probes (Bio-Rad Laboratories, Pleasanton, CA), 1.25 μl of each 20× primer-probe mix, and 10 μl or 16.25 μl of template DNA in a final volume of 25 μl. A total of 20 μl of each reaction mixture was loaded onto a disposable plastic cartridge (Bio-Rad) with 70 μl of droplet generation oil (Bio-Rad) and placed in the droplet generator (Bio-Rad), resulting in 8 μl (2× mix) or 13 μl (4× mix) template DNA in the final PCR. The droplets generated from each sample were transferred to a 96-well PCR plate, and PCR amplification was performed on a 2720 Thermal Cycler (Applied Biosystems) with the following conditions: 94°C for 10 min, 40 cycles of 94°C for 10 min and 60°C for 1 min, followed by 98°C for 10 min and ending at 4°C. After amplification, the plate was loaded onto the droplet reader (Bio-Rad), and the droplets from each well of the plate were automatically read at a rate of 32 wells/hour. Data were analyzed with QuantaSoft analysis (version 1.3.2.0) software, and quantitation of target molecules was presented as copies/μl of PCR, which was then calculated to reflect copies/ml of plasma.
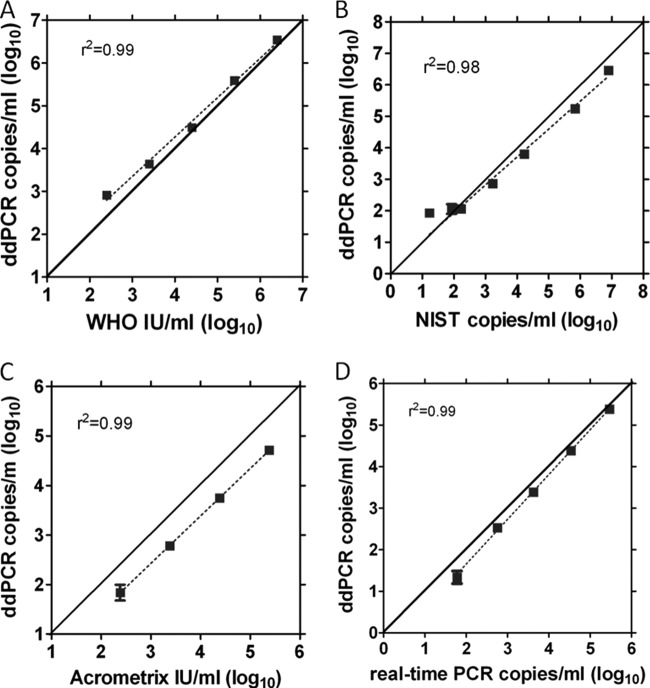
FIG 1.
Analysis of CMV standard materials by ddPCR for CMV UL55. (A) WHO dilution series, run in singlet; (B) NIST CMV DNA standard dilution series. Eight replicate PCRs were performed per dilution. (C) Acrometrix CMVtc dilution panel. Eight replicate PCRs were performed per dilution. (D) CMV patient plasma sample dilution series. Each dilution was assayed in 4 replicate reactions for ddPCR and duplicate reactions for real-time PCR.
Assessment of contamination potential.
A checkerboard experiment consisting of alternating wells containing CMV plasmid standard or water as the template in ddPCR reactions was used to determine potential for well-to-well contamination while performing the assay in a clinical setting. Three 96-well plates were run with CMV-positive (∼300 copies/μl PCR) and -negative wells alternating. The PCRs were mixed in a 96-well plate, covered and sealed with PCR optical tape, vortexed for 5 s, and centrifuged for 30 s at ∼3,000 × g, and the optical tape was removed for transfer of the PCRs to the droplet generation cartridge. After droplet generation and thermocycling, the number of negative-control wells resulting in positive droplets was determined.
Statistical analysis.
Linear mixed-effects models were used to assess differences in log10-transformed quantity between qPCR and ddPCR within persons and to assess the potential for differences in precision as well. The function “lme” within the software package “R,” version 3.0.0, was used, and the potential for variability to differ by method of quantitation was assessed by comparing models with and without the incorporation of heteroskedasticity parameters using a likelihood ratio test (15).
RESULTS
Evaluation of CMV standard material by ddPCR.
To assess the accuracy of the CMV ddPCR assay, CMV standards and a patient plasma sample were evaluated by ddPCR (Fig. 1A to D). Digital PCR values correlated well with WHO, NIST, and our laboratory's real-time qPCR values with marked linearity (r2 = 0.98 to 0.99). However, for the Acrometrix panel, the ddPCR results were approximately 4-fold lower than would have been expected based on information provided by the manufacturer, despite the fact that both the AcroMetrix and WHO materials both are calibrated in IU/ml. Utilizing the WHO standard, the conversion factor for digital copies/ml to IU/ml was 1.5 copies/IU.
Comparison of precision and sensitivity between ddPCR and qPCR.
We compared the intraassay and interassay precision of ddPCR and qPCR CMV assays using CMV-positive patient plasma samples. Utilizing 2× ddPCR master mix with a concentrating DNA extraction (1 ml of plasma to 80 μl of DNA) and 8 replicate PCRs, at 4 log10 copies/ml CMV the average intraassay coefficient of variation was 4-fold less for ddPCR than for qPCR (P = 0.004). Likewise, intraassay variation at viral loads of ≤1,500 copies/ml appeared less for ddPCR than for qPCR (see Table 2). The same was true of interassay variation (see Table 2), with average coefficient of variations appearing consistently lower for ddPCR than for qPCR (P = 0.2 to 0.9). However, these results failed to reach statistical significance, likely due to the limited sample size (n = 12 per dilution). Power calculations indicate 25 to 65 samples would be needed to reach significance when evaluating differences of this magnitude. Unexpectedly, the interassay variation appears slightly lower than the intraassay variation, which may also be an artifact of limited sample size.
TABLE 2.
Comparison of within-run (intraassay) variability and between-run (interassay) variability for ddPCR and qPCRa
| Dilution (copies/ml) | CV (%) |
|||
|---|---|---|---|---|
| Interassay |
Intraassay |
|||
| ddPCR | qPCR | ddPCR | qPCR | |
| 150 | 24.1 | 44.0 | 33.5 | 37.7 |
| 300 | 19.8 | 26.4 | 19.0 | 28.1 |
| 1,500 | 8.6 | 13.2 | 11.7 | 14.9 |
Utilizing 3 dilutions of CMV patient plasma sample, each run in triplicate and repeated in four separate runs. CV, coefficient of variation.
We assessed the sensitivity of the ddPCR assay compared to that of the qPCR assay, utilizing the standard 2× ddPCR master mix commercially available from Bio-Rad, as well as an experimental 4× ddPCR master mix. The 4× master mix was utilized to increase the volume of template DNA that could be included in a reaction, in order to increase sensitivity. Analyzing a 2-fold plasma dilution series from a CMV-positive patient plasma sample yielded a 95% cutoff for limit of detection of 9 copies/ml for qPCR, 32 copies/ml for ddPCR using the 2× master mix, and 11 copies/ml for ddPCR using the 4× master mix. Table 1 shows the number of replicates positive at each dilution of this patient plasma sample. The ddPCR assay utilizing the 4× master mix approached the sensitivity of the standard clinical qPCR assay; the slight difference in sensitivity between the two assays is likely due to the addition of 15 μl of template DNA in the qPCR assay versus only 13 μl of template DNA in the 4× ddPCR assay. It should also be noted that the qPCR assay utilized two primer/probe sets to detect one viral template which could also account for a slight sensitivity advantage.
TABLE 1.
Analysis of the limit of detection of the ddPCR CMV assay using 2× master mix or 4× master mix compared with the CMV qPCR assay
| Dilution (copies/ml) | No. of samples | No. of positive samples |
||
|---|---|---|---|---|
| 2× ddPCR | 4× ddPCR | qPCR | ||
| 110 | 8 | 8 | 8 | 8 |
| 55 | 8 | 8 | 8 | 8 |
| 27.5 | 8 | 7 | 8 | 8 |
| 13.75 | 8 | 6 | 8 | 8 |
| 6.875 | 8 | 3 | 3 | 7 |
Evaluation of contamination potential.
A standard checkerboard experiment was performed to assess the likelihood of well-to-well contamination when performing the digital assay clinically. Out of 236 wells on three 96-well plates, 118 were set up to be positive, and 118 were set up to be negative. All expected positive wells were positive. A single negative well was positive with a single positive droplet.
Analysis of longitudinal CMV plasma samples.
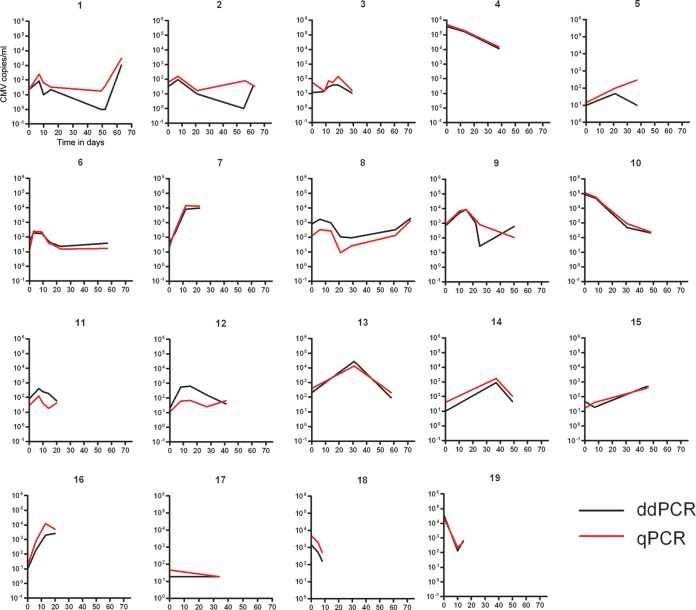
We sought to determine if ddPCR's observed increased precision might offer a clinical advantage over qPCR in following CMV viral load in transplant patients. We performed ddPCR analysis (utilizing 4× master mix) of plasma samples from 19 patients with repeated CMV monitoring and compared ddPCR results to previous clinical qPCR results. Results from qPCR and ddPCR correlated well in all patients, as evidenced by the matching trends in viral load over time in each patient (Fig. 2). Statistical analysis of these data (total of 85 time points, median of 4 per patient, range of 2 to 7) assessed differences in precision between qPCR and ddPCR within persons. Precision within persons was not found to differ between qPCR and ddPCR (P = 0.26).
FIG 2.
Comparison of ddPCR and qPCR results for longitudinal plasma samples in 19 patients. The y axis is CMV copies/ml and the x axis is time in days.
DISCUSSION
We developed a ddPCR assay for CMV that performs with increased precision at viral loads of ≥4 log10 (Table 2) and equivalent sensitivity (Table 1) compared to our laboratory's qPCR assay for CMV. The ddPCR assay has improved intraassay and interassay precision compared to that of qPCR, particularly at viral loads above 4 log10. These data are consistent with previous studies comparing ddPCR and qPCR (7, 10, 11).
This CMV ddPCR assay accurately quantitates CMV without the use of a traditional standard curve utilized in qPCR (Fig. 1A to D). As noted, the ddPCR assay underquantified the Acrometrix standard but very closely matched the WHO standard, both of which are calibrated in IU/ml. This discrepancy may be due to the fact that the Acrometrix panel is a secondary calibrant produced from the WHO standard and may reflect differences in how the calibrants were originally produced and quantitated. A different lot of the Acrometrix panel also might have improved concordance. The ddPCR assay also closely replicated NIST values, showing concordance of two different digital PCR measurements, as the NIST standard was originally quantitated by a chip-based digital PCR system (13).
We demonstrated that the process of ddPCR is not susceptible to cross-contamination of samples during droplet generation or reading. We did observe a single positive droplet in one negative well, but this is likely due to low-level noise in the assay, which has been observed in other ddPCR assays (10).
By optimizing DNA extraction and utilizing a concentrated digital PCR master mix, we show the potential for ddPCR to be used in a clinical diagnostic setting. We hypothesized that the increased precision afforded by ddPCR might contribute to more accurate monitoring of CMV disease progression, particularly in transplant patients with continuous, often weekly, CMV monitoring. Changes in CMV plasma quantities at levels below around 250 IU/ml (1,000 copies/ml) are critical in making decisions about antiviral treatments that can be toxic and expensive (4). Our data do not show a statistically significant difference in precision of the two assays on longitudinal samples from patients with continuous CMV monitoring and viral loads below 4 log10 (Fig. 2). Given the analytical precision of the ddPCR assay at viral loads above 4 log10, we conclude that ddPCR offers a precision advantage over qPCR, but this may not be clinically relevant in the transplant setting, where CMV viral loads below 3 log10 are critical for making treatment decisions. It should be noted that this study was conducted on a relatively low number of clinical samples, and ddPCR was not compared to an FDA-cleared CMV qPCR assay, so the results of this study might not directly apply in other settings. Similarly, different results might be obtained using a different digital PCR platform. Taken together, our results suggest that since the current dPCR systems require more hands-on time and have lower throughput than current qPCR systems, a widespread switch to dPCR for CMV monitoring would be premature.
ACKNOWLEDGMENTS
Bio-Rad Laboratories provided reagents and materials toward the completion of this work.
Footnotes
Published ahead of print 28 May 2014
REFERENCES
- 1.Wolff DJ, Heaney DL, Neuwald PD, Stellrecht KA, Press RD. 2009. Multi-site PCR-based CMV viral load assessment-assays demonstrate linearity and precision, but lack numeric standardization: a report of the association for molecular pathology. J. Mol. Diagn. 11:87–92. 10.2353/jmoldx.2009.080097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sedlak RH, Jerome KR. 2013. Viral diagnostics in the era of digital polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 75:1–4. 10.1016/j.diagmicrobio.2012.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker M. 2012. Digital PCR hits its stride. Nat. Meth. 9:541–544. 10.1038/nmeth.2027 [DOI] [Google Scholar]
- 4.Green ML, Leisenring W, Stachel D, Pergam SA, Sandmaier BM, Wald A, Corey L, Boeckh M. 2012. Efficacy of a viral load-based, risk-adapted, preemptive treatment strategy for prevention of cytomegalovirus disease after hematopoietic cell transplantation. Biol. Blood Marrow Transplant. 18:1687–1699. 10.1016/j.bbmt.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsch HH, Lautenschlager I, Pinsky BA, Cardeñoso L, Aslam S, Cobb B, Vilchez RA, Valsamakis A. 2013. An international multicenter performance analysis of cytomegalovirus load tests. Clin. Infect. Dis. 56:367–373. 10.1093/cid/cis900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Razonable RR, Åsberg A, Rollag H, Duncan J, Boisvert D, Yao JD, Caliendo AM, Humar A, Do TD. 2013. Virologic suppression measured by a cytomegalovirus (CMV) DNA test calibrated to the World Health Organization international standard is predictive of CMV disease resolution in transplant recipients. Clin. Infect. Dis. 56:1546–1553. 10.1093/cid/cit096 [DOI] [PubMed] [Google Scholar]
- 7.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. 2013. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 10:1003–1005. 10.1038/nmeth.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts CH, Last A, Molina-Gonzalez S, Cassama E, Butcher R, Nabicassa M, McCarthy E, Burr SE, Mabey DC, Bailey RL, Holland MJ. 2013. Development and evaluation of a next-generation digital PCR diagnostic assay for ocular Chlamydia trachomatis infections. J. Clin. Microbiol. 51:2195–2203. 10.1128/JCM.00622-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O'Doherty U, Palmer S, Deeks SG, Siliciano JD. 2013. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 9:e1003174. 10.1371/journal.ppat.1003174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. 2013. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 8:e55943. 10.1371/journal.pone.0055943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden RT, Gu Z, Ingersoll J, Abdul-Ali D, Shi L, Pounds S, Caliendo AM. 2013. Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. J. Clin. Microbiol. 51:540–546. 10.1128/JCM.02620-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sedlak RH, Cook L, Huang M-L, Magaret A, Zerr D, Boeckh M, Jerome KR. 2014. Identification of chromosomally integrated human herpesvirus 6 by droplet digital PCR. Clin. Chem. 60:765–772. 10.1373/clinchem.2013.217240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haynes RJ, Kline MC, Toman B, Scott C, Wallace P, Butler JM, Holden MJ. 2013. Standard reference material 2366 for measurement of human cytomegalovirus DNA. J. Mol. Diagn. 15:177–185. 10.1016/j.jmoldx.2012.09.007 [DOI] [PubMed] [Google Scholar]
- 14.Boeckh M, Huang M, Ferrenberg J, Stevens-Ayers T, Stensland L, Nichols WG, Corey L. 2004. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J. Clin. Microbiol. 42:1142–1148. 10.1128/JCM.42.3.1142-1148.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinheiro J, Bates D. 2000. Mixed-effects models in S and S-PLUS, p 177 Springer-Verlag, New York, NY [Google Scholar]