Abstract
Human bocavirus 1 (HBoV1) is associated with respiratory infections worldwide, mainly in children. Similar to other parvoviruses, it is believed that HBoV1 can persist for long periods of time in humans, probably through maintaining concatemers of the virus single-stranded DNA genome in the nuclei of infected cells. Recently, HBoV-1 was detected in high rates in adenoid and palatine tonsils samples from patients with chronic adenotonsillar diseases, but nothing is known about the virus replication levels in those tissues. A 3-year prospective hospital-based study was conducted to detect and quantify HBoV1 DNA and mRNAs in samples of the adenoids (AD), palatine tonsils (PT), nasopharyngeal secretions (NPS), and peripheral blood (PB) from patients undergoing tonsillectomy for tonsillar hypertrophy or recurrent tonsillitis. HBoV1 was detected in 25.3% of the AD samples, while the rates of detection in the PT, NPS, and PB samples were 7.2%, 10.5%, and 1.7%, respectively. The viral loads were higher in AD samples, and 27.3% of the patients with HBoV had mRNA detectable in this tissue. High viral loads and detectable mRNA in the AD were associated with HBoV1 detection in the other sample sites. The adenoids are an important site of HBoV1 replication and persistence in children with tonsillar hypertrophy. The adenoids contain high HBoV1 loads and are frequently positive for HBoV mRNA, and this is associated with the detection of HBoV1 in secretions.
INTRODUCTION
Human bocavirus (HBoV) is a small nonenveloped single-stranded DNA virus of the family Parvoviridae (1). HBoV is highly prevalent in different human populations and has been detected in association with acute respiratory and gastrointestinal diseases worldwide (2). Four different species of HBoV are currently known, designated HBoV1, HBoV2, HBoV3, and HBoV4 (3–5). HBoV1 is mainly detected in respiratory secretions, while HBoV2 to HBoV4 are frequently found in stool samples from children.
Koch's postulates have not yet been fulfilled for HBoV and, therefore, an association of the agent with human diseases has not been established, which is further hampered by the high frequency of simultaneous codetection with other respiratory and enteric viruses (6, 7). However, there has been increasing evidence that HBoV, especially HBoV1, is a pathogen rather than an innocent bystander.
Acute infections by HBoV1, evidenced by methods, such as the detection of high viral loads in respiratory samples, the presence of HBoV1 mRNA in nasopharyngeal aspirate samples, and the detection of HBoV1-specific IgM and/or viremia, have frequently been associated with respiratory illness (6, 8–11). In keeping with this, in vitro HBoV1 infection of polarized primary human airway epithelia causes remarkable airway epithelial damage, with a disruption of the tight junction barrier, loss of cilia, and epithelial cell hypertrophy (12).
In addition to acute infections, recent studies have shown that HBoV can establish persistence in humans, mainly by maintaining covalently closed circular viral genomic DNA in infected cells in several sites, such as respiratory and gut epithelia (13–15). It has been speculated that chronic HBoV infection might lead to ongoing inflammation, which may be subclinical, eventually leading to damage in chronically infected tissues. In line with this, HBoV1 DNA has been detected in bronchoalveolar lavage fluid samples and in different tissues from patients with idiopathic lung fibrosis, either in association with acute exacerbations of interstitial pneumonia or as a finding in postmortem investigations (16).
Despite the evidence for HBoV persistence, not all sites that harbor such persistence are currently known. The adenoids and palatine tonsils are prominent secondary lymphoepithelial organs associated with the upper respiratory tract, where inhaled or ingested antigens may come in contact with host defense cells (17). These tissues have been recognized as sites of persistence of viruses, such as adenovirus and Epstein-Barr virus (18–20). HBoV1 has been detected in high rates in adenotonsillar tissue (21, 22), but whether HBoV1 can persist in the adenoids and tonsils remains unknown. The present study was done to investigate HBoV1 persistence and replication in tonsillar tissues and to check for its possible association with the pathogenesis of tonsillar hypertrophy.
MATERIALS AND METHODS
Patients and sample collection.
Detection of the HBoV1 genomic DNA and mRNAs and a determination of viral loads were done in samples of the adenoids (AD), palatine tonsils (PT), nasopharyngeal secretions (NPS), and peripheral blood (PB) from patients of the Division of Otorhinolaryngology of the Clinical Hospital of the Ribeirão Preto School of Medicine, University of São Paulo, from May 2010 to July 2012. The enrolled patients were divided in two groups: one composed of 180 patients (93 males) 1 to 18 years of age (median, 5 years) who underwent surgical removal of the adenoids and tonsils to treat adenotonsillar hypertrophy with clinical evidence of obstructive sleep apnea, or who had recurrent adenotonsillitis according to the criteria of Paradise et al. (23), and a second group with 12 control patients (7 males) 1 to 12 years of age (median, 3 years) without chronic adenotonsillitis who underwent surgery for cochlear implant placement. An adenoidectomy was not performed in 6 patients; therefore, only 174 adenoid samples were analyzed. In the control patients, only small biopsy specimens were collected from both the adenoids and palatine tonsils. The exclusion criteria were the presence of symptoms of acute respiratory infection or the use of antibiotic treatment 1 month prior to surgery. The study was conducted in accordance with the principles expressed in the Declaration of Helsinki and was approved by the clinic's hospital research ethics committee (file 10466/2008). Written informed consent was obtained from all parents and guardians prior to enrollment. A detailed description of the criteria for disease classification and the methods for clinical sample processing were previously published (22).
DNA and RNA extraction.
All samples tested in this study were maintained in a preservative solution (RNAlater; Invitrogen, Carlsbad, CA, USA) at −70°C until nucleic acid extraction. DNA from the tonsils was extracted from 30 mg of tissue sample with the AllPrep DNA minikit (Qiagen GmbH, Hilden, Germany), while RNA was obtained using the AllPrep RNA minikit (Qiagen). Total nucleic acids were extracted from 200 μl of NPS with the QIAamp MinElute virus spin kit (Qiagen) and from 1 ml of peripheral blood with the QIAamp RNA and DNA blood minikits (Qiagen). All procedures were performed according to the manufacturer's instructions.
Detection and quantification of HBoV1 viral load.
HBoV1 was detected by TaqMan real-time PCR, according to a previously published protocol (22). Briefly, the reactions were assembled in a final volume of 10 μl with 50 ng of DNA, 10 mM forward and reverse primers (HBoV-F, 5′-GCACAGCCACGTGACGAA-3′ and HBoV-R, 5′-TGGACTCCCTTTTCTTTTGTAGGA-3′), 5 mM probe (HBoV-P, 5′-FAM-TGAGCTCAGGGAATATGAAAGACAAGCATCG-TAMRA-3′ [FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine]), and 5.0 μl of TaqMan master mix (Applied Biosystems, Foster City, CA, USA), with the following parameters: 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. All TaqMan PCR assays were done on the StepOnePlus thermocycler (Applied Biosystems), and tests for the housekeeping control genes β-actin and RNase P were done in all samples (22). Applicable measures to prevent contamination of the PCRs were taken, including sample handling and reaction mix preparation in separate rooms. In addition, all TaqMan PCR plates included appropriate negative controls matched to every step of clinical sample processing.
Quantitative PCR (qPCR) for HBoV1 was targeted to the viral nucleoprotein 1 (NP1) gene according to previously published procedures (24). Viral quantification was done in comparison with a standard curve produced using serial decimal dilutions of a plasmid in which a fragment of the HBoV1 NP1 gene (nucleotides [nt] 2270 to 3280) had been cloned. HBoV1 qPCR was considered positive when the threshold was reached before the 40th cycle. The detection limit was 1 copy of HBoV1 plasmid. All qPCR assays for HBoV1 were performed in triplicate, and the results were normalized by amplification of the β-actin gene, tested in duplicate in all batches. The viral loads were determined as the number of copies of HBoV1 DNA per g of tissue or per ml of NPS or blood sample.
Detection of VP1 and NP1 mRNAs of HBoV1.
HBoV1 VP1 and NP1 mRNAs were detected by real-time reverse transcription-PCR (RT-PCR) in NPS, peripheral blood, and tonsil samples to ascertain the presence of active viral replication in the patients. To ensure target-specific amplification of cDNA without genomic DNA contamination, total RNA extraction products were treated with DNase I (Invitrogen, Carlsbad, CA, USA) for 2 h prior to PCR and, as a control, all PCRs were carried out in parallel using RNA extraction products without previous reverse transcription (RT). In addition, to ensure RNA quality after DNase treatment, all samples with cDNAs and DNase-treated RNAs were tested by for β-actin by qPCR (6). All RT reactions were performed with 1 mg of RNA, using 10 pmol of oligo(dT) primer, according to the manufacturer's protocol. PCR was carried out with 150 ng of cDNA and 300 nmol/liter each primer (VP1-F, 5′-CTCACTTTTCAGACAAATATGTGGTTACT-3′ and VP1-R, 5′-TGTTGTTTCAATGGCCTCTGTT-3′), 150 nmol/liter probe (VP1-P, 5′-FAM-TTCAGAATGGTCACCTCT-AMGBNFQ-3′), and 5 μl of TaqMan universal PCR master mix (Applied Biosystems), according to the same cycling parameters used for the detection of the HBoV1 genomic DNA. The detection of NP1 mRNA was done by the same protocol used for VP1 mRNA, except for the primers targeting the NP1 gene, mentioned above in the description of the detection of HBoV1 and quantification of viral loads.
Detection of other respiratory viruses.
All samples were tested by qualitative real-time PCR for human rhinovirus (HRV), human respiratory syncytial virus (HRSV), human metapneumovirus (HMPV), influenza A and B (FLU), human parainfluenza (HPIV), human coronaviruses (HCoV) 229E and OC43, human adenovirus (HAdV), and human enterovirus (HEV), according to previously published procedures (22).
Statistical analysis.
Comparisons between the patient groups were done using chi-square and Fisher's exact tests; comparisons of the viral loads between two groups were done by Mann-Whitney or unpaired t test for groups of less than four individuals; comparisons involving three or more groups were done by one-way analysis of variance with Bonferroni's correction as a posttest. All tests were done using GraphPad Prism version 5.00 for Mac (GraphPad Software, San Diego, CA, USA). A P value of ≤0.05 was chosen for significance.
RESULTS
Frequency of HBoV1 DNA detection.
HBoV1 DNA was detected in at least one sample in 56 (31.1%) patients with chronic tonsillar disease. The detection rate was higher in samples from the adenoids (AD) (25.3%) than in palatine tonsils (PT) (7.2%), nasopharyngeal secretions (NPS) (10.5%), and peripheral blood (1.7%). Of 180 patients with chronic tonsillar diseases, 49 (27.2%) had HBoV1 DNA detected in tissue fragments from AD and/or PT. Among the 56 HBoV1-positive patients, HBoV1 DNA was detected exclusively in AD in 29 (51.8%) patients, while HBoV1 was detected solely in PT tissue in only 5 (8.9%) patients and solely in NPS tissue in only 7 (12.5%) patients. In 5 (8.9%), 3 (5.3%), and 7 (12.5%) patients, HBoV1 genomic DNA was simultaneously detected in AD, PT, and NS samples, AD and PT samples, and AD and NS samples, respectively, while in the vast majority of HBoV1-positive patients, the viral genomic DNA was detected in only one of the tested sample sites. In 3 (5.3%) patients, HBoV1 DNA was detected in the peripheral blood and simultaneously in AD samples, and in 2 of those patients, the virus was detected also in PT and NPS samples.
The detection of HBoV1 DNA in the adenoids was not associated with particular clinical signs/symptoms, and virus genomic DNA was detected in similar rates in patients with chronic tonsillar diseases and in controls without chronic or acute respiratory symptoms (Table 1). Positive associations of HBoV1 detection in AD samples were noted with the presence of viral coinfections (P < 0.001), male gender (P = 0.03), and younger age (P = 0.04) (Table 1).
TABLE 1.
Clinical and demographic data of patients with HBoV1 detected in adenoidsa
| Patient demographic or condition | Hypertrophic patients |
Control patients |
||
|---|---|---|---|---|
| HBoV1 positive | HBoV1 negative | HBoV1 positive | HBoV1 negative | |
| Patients | 44 | 130 | 4 | 8 |
| Male genderb | 29 (65.9) | 62 (47.7) | 3 (75.0) | 4 (50.0) |
| Median age (yr)b | 4.0 | 6.0 | 2.0 | 4.0 |
| Viral coinfectionb | 38 (86.4) | 69 (53.1) | 2 (50.0) | 4 (50.0) |
| Airway obstruction | ||||
| 0% to 50% | 3 (6.8) | 14 (10.8) | ||
| 50% to 75% | 17 (38.6) | 60 (46.1) | ||
| 75% to 100% | 24 (54.6) | 56 (43.1) | ||
| Sleep apnea | 29 (65.9) | 75 (57.7) | ||
| Secretory otitis media | 5 (11.4) | 25 (19.2) | ||
| Allergies | 12 (27.3) | 36 (27.7) | ||
Data are presented as no. (%), unless otherwise indicated.
P < 0.05.
The detection of HBoV1 in PT samples was also associated with younger age (P = 0.04), presence of viral coinfections (P < 0.001), and sleep apnea (P = 0.04). However, these associations should be approached with caution, since the number of patients with HBoV detected in the PT samples was small (Table 2). HBoV1 genomic DNA was not detected in PT samples from the control patients (Table 2).
TABLE 2.
Clinical and demographic data of patients with HBoV1 detected in palatine tonsilsa
| Patient demographic or condition | Hypertrophic patients |
Control patients |
||
|---|---|---|---|---|
| HBoV1 positive | HBoV1 negative | HBoV1 positive | HBoV1 negative | |
| Patients | 13 | 167 | 0 | 12 |
| Male gender | 6 (46.1) | 87 (52.1) | 0 (0.0) | 7 (58.3) |
| Median age (yr)b | 4.0 | 6.0 | 3.0 | |
| Viral coinfectionb | 10 (76.9) | 38 (22.7) | 0 (0.0) | 0 (00.0) |
| Recurrent tonsillitis | 6 (46.1) | 84 (50.3) | ||
| Tonsillar hypertrophy | 11 (84.6) | 137 (82.0) | ||
| Sleep apneab | 11 (84.6) | 93 (55.6) | ||
| Secretory otitis media | 1 (7.7) | 29 (17.3) | ||
| Allergies | 2 (15.4) | 46 (27.5) | ||
Data are presented as no. (%), unless otherwise indicated.
P < 0.05.
The high rates of HBoV1 genomic DNA detection in AD and PT samples, both from patients with tonsillar hypertrophy and controls, as well as the high frequency of viral coinfections are indicative of a lack of a causative role for respiratory viruses in chronic tonsillar hypertrophy. In keeping with this, the number of different virus types codetected with HBoV1 showed no apparent correlation with the clinical or demographic data of patients with chronic tonsillar diseases (see Tables S1 and S2 in the supplemental material). In addition, the HBoV1 detection rates were not higher in the presence of any specific virus. In order to verify the replicative status of HBoV1 in the AD and PT and its potential relevance for chronic disease development, we determined viral loads and checked for the presence of mRNAs for two key HBoV1 proteins in all tissues obtained in the study.
The viral load of HBoV1 in tonsillar tissue.
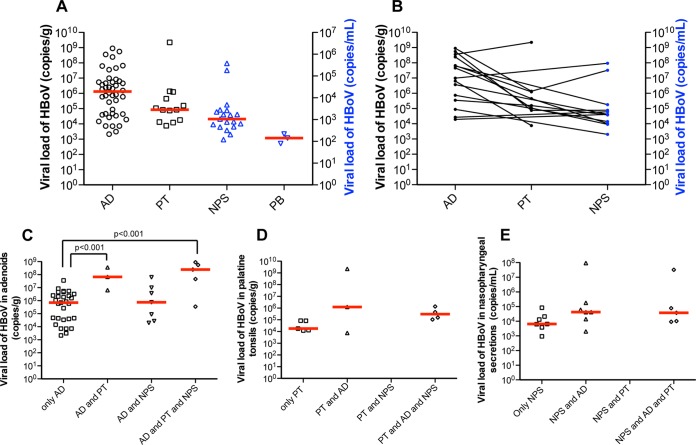
The median viral load in AD samples as determined by qPCR for HBoV1 genomic DNA was 1.32 × 106 copies of genomic DNA/g of tissue, which was not significantly different (P = 0.38) from the median of 4.1 × 105 copies/g obtained in the control patients. In PT samples, HBoV1 was found only in samples from patients with chronic adenotonsillitis, with a median viral load of 8.4 × 104 copies/g. In NPS samples, the median viral load was 2.1 × 104 copies/ml in samples from patients with chronic tonsillar disease, while in peripheral blood samples, it was 1.1 × 103 copies/ml. Although the median viral loads were not significantly different between the four sample sites (Fig. 1A), in most patients, there was a pattern of increased HBoV1 viral load in AD samples compared to those in the other sites (Fig. 1B). The median HBoV1 viral loads in AD samples were significantly higher in patients in whom HBoV1 was detected in several sites than in those with HBoV1 detected in a single site (Fig. 1C to E), as shown by analysis of variance with Bonferroni's correction as a posttest. Although HBoV1 DNA was detected in the peripheral blood samples from only 3 patients, all of them had high viral load in their AD samples (>107 copies/g), and HBoV was simultaneously detected in PT and NPS samples, reinforcing that loads of HBoV1 in AD tissue are likely associated with virus detection in other sites, such as PT, NPS, and peripheral blood.
FIG 1.
HBoV1 viral loads in the different sample sites in patients with hypertrophic tonsillar diseases. (A) Median viral loads (red lines) of HBoV1 per gram of tissue were higher in adenoid (AD) than in palatine tonsil (PT) samples, and the viral loads per ml were higher in nasopharyngeal secretion (NPS) samples than in peripheral blood (PB) samples. (B) Median HBoV1 viral loads in the different sample sites of the same patients are connected by lines; (C to E) Median HBoV1 viral loads (red lines) in AD, PT, and NPS samples from patients with HBoV1 detected in these respective sites only, as well as from patients with HBoV1 detected in more than one site.
The HBoV1 genomic loads were higher in AD samples from patients with higher degrees of adenoidal hypertrophy and with sleep apnea than in those with lesser airway obstruction and without sleep apnea (Fig. 2A). Such differences in the HBoV1 viral loads were not apparent in PT or in NPS samples (Fig. 2B and C). A firm association between the genomic DNA loads of HBoV1 and more severe AD hypertrophy was hard to establish, considering the high frequency of codetection of other respiratory viruses by qPCR (see Tables S1 and S2 in the supplemental material). No significant differences were noted in HBoV1 viral loads in AD, PT, or NPS samples when stratified by gender or age.
FIG 2.
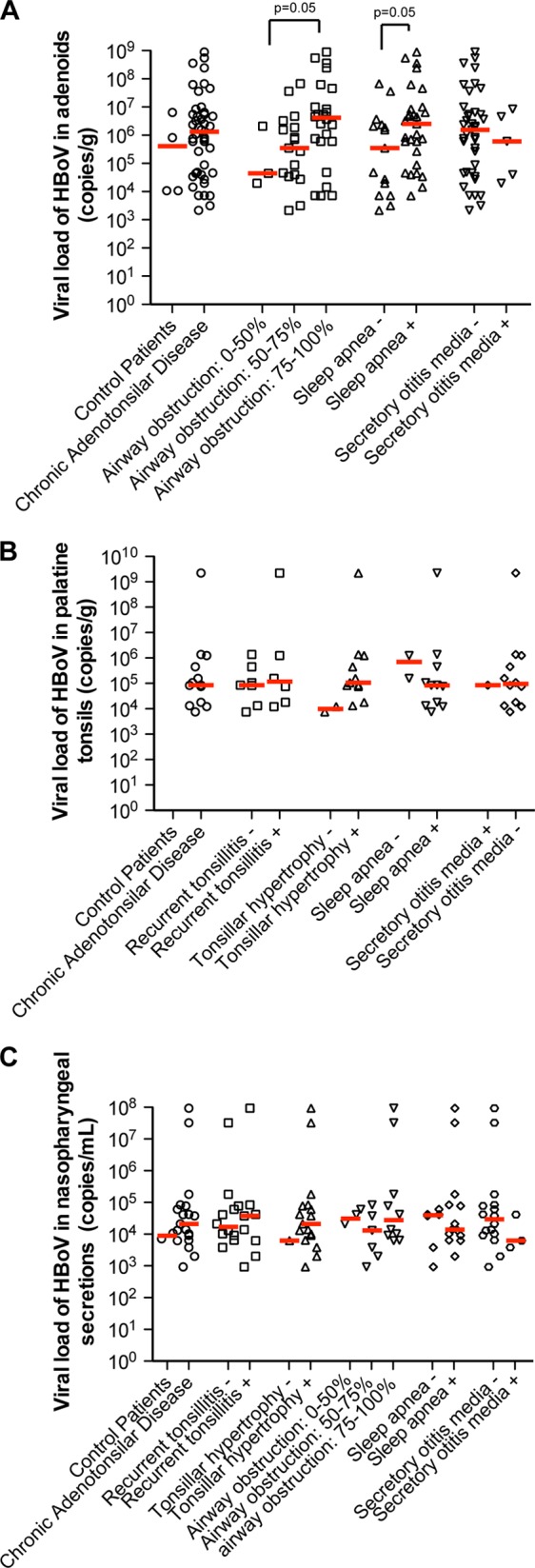
HBoV viral loads in adenoid (A), palatine tonsils (B), and nasopharyngeal secretion (C) samples from patients with chronic tonsillar diseases and different clinical features. Median HBoV viral loads (red lines) were significantly higher in patients with more severe adenoidal hypertrophy and with sleep apnea (A), whereas differences were not present in palatine tonsils (B) or in nasopharyngeal secretions (C).
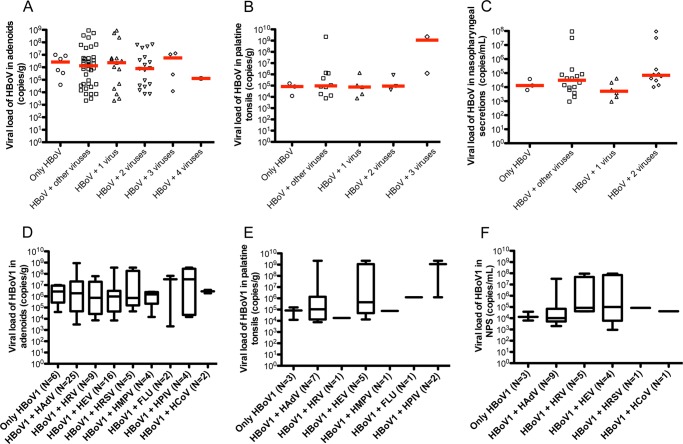
The presence of other respiratory viruses, which conceivably might stimulate or restrict the replication of HBoV1, apparently did not affect HBoV1 viral loads in AD, PT, and NPS samples (Fig. 3). Dual infections of HBoV1 with certain specific respiratory viruses were not significantly associated with higher HBoV1 viral loads (Fig. 3D to Fig. 3F).
FIG 3.
HBoV viral loads in tissues (copies of genomic DNA/g) and nasopharyngeal secretion samples (copies of genomic DNA/ml) from patients with coinfections by other respiratory viruses. (A to C) HBoV1 viral loads in adenoid, palatine tonsils, and nasopharyngeal secretion samples from patients with HBoV1 as a single agent or simultaneously detected with 1 to 4 other respiratory viruses. (D to F) HBoV1 viral loads in adenoid, palatine tonsils, and nasopharyngeal secretion samples from patients with HBoV1 as a single agent or in dual infection with other respiratory viruses: human adenovirus (HAdV), human rhinovirus (HRV), human enterovirus (HEV), human respiratory syncytial virus (HRSV), human metapneumovirus (HMPV), influenza virus (FLU), human parainfluenza virus (HPIV) and human coronavirus (HCoV). Boxes extend from 25th to 75th percentiles, middle whiskers mark median values, and upper and lower whiskers, respectively, mark the highest and the lowest values.
Productive infection by HBoV1 in tonsillar tissues.
The high rates of HBoV1 genomic DNA detection and high viral loads are indirect evidence that the adenoids are an important site for HBoV1 replication. In order to ascertain the presence of HBoV1 replication in the AD, PT, and NPS, we performed an assay to detect HBoV1 VP1 and NP1 mRNAs by real-time RT-PCR in total RNA extracts previously treated with DNase I. Of 56 patients with chronic adenotonsillar disease positive for HBoV1, 12 (21.4%) were positive for VP1 or NP1 mRNAs. Of those, 9 patients (75%) had levels of VP1 and NP1 mRNA detectable in AD samples only, while 3 were positive in AD plus NPS samples and 1 in AD plus PT plus NPS samples. The frequency of detection of HBoV genomic DNA in several sites was significantly higher in patients positive for HBoV mRNA in their AD samples than in those in whom mRNA was not detected (66.6% versus 21.8%; P = 0.02) (see Table S3 in the supplemental material). All 3 patients with HBoV1 in the peripheral blood had viral mRNA detectable in their AD samples. Thus, the adenoids appear to be the main site of HBoV1 replication in patients with chronic tonsillar diseases.
The detection of HBoV1 mRNA significantly correlated with high viral load (Fig. 4). The median HBoV1 viral load in patients with detectable viral mRNA in their AD samples was 2.3 × 107 copies/g, while in AD samples without detectable viral mRNA, the median viral load was 6.6 × 105 copies/g (see Table S3 in the supplemental material). Such a comparison could not be done in PT samples because mRNA was detected in hypertrophic PT samples from only one patient who had an HBoV1 viral load of 2.2 × 109 copies/g (see Table S4 in the supplemental material). It is noteworthy that the HBoV1 viral load in that one patient was much higher than the median load of 8.4 × 104 obtained in PT samples from patients without detectable mRNA (Fig. 4; see also Table S4 in the supplemental material).
FIG 4.
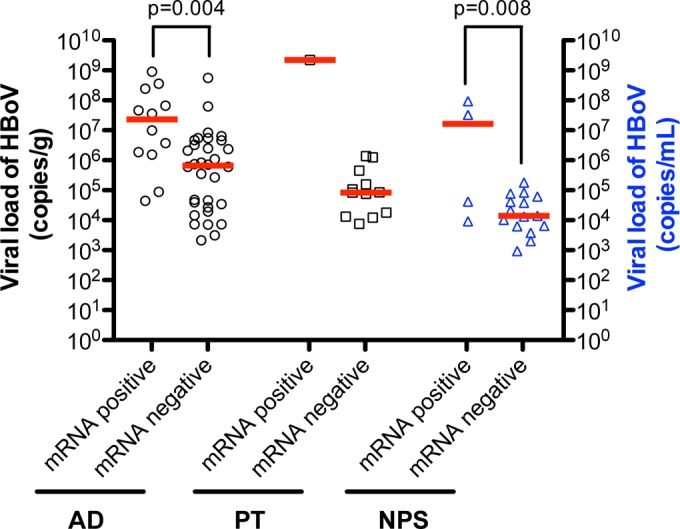
Quantification of HBoV viral loads (in copies of genomic DNA/g of tissue or ml, as appropriate) and detection of HBoV VP1 or NP1 mRNAs in adenoid (AD), palatine tonsils (PT) and nasopharyngeal secretion (NPS) samples collected from patients with chronic tonsillar hypertrophy. The median loads (red lines) of HBoV were significantly higher in AD and NPS samples in patients positive for HBoV mRNA. Since HBoV1 mRNA was detected in a PT samples from only 1 patient, statistical analysis was not feasible.
The presence of HBoV1 VP1 and/or NP1 mRNAs in AD samples showed no association with age, gender, or any specific clinical features, such as the severity of airway obstruction, presence of sleep apnea, or secretory otitis media (see Table S3 in the supplemental material). Since the detection of HBoV1 mRNA in PT samples occurred in only one patient (see Table S4 in the supplemental material), no conclusions can be made about its relevance. As observed with HBoV viral loads, the detection of viral coinfections was not associated with the detection of HBoV mRNA, suggesting that infection by other respiratory viruses does not increase HBoV production in the tonsils (see Table S5 in the supplemental material).
DISCUSSION
The results of this prospective study of patients with chronic tonsillar diseases seen at a reference hospital in southeast Brazil indicate that the adenoids are an important site for the maintenance and replication activity of HBoV1. Roughly one-third of these patients, more frequently males around 4 years of age, were positive for HBoV1 in at least one sample site.
Since its discovery in 2005, HBoV1 has been reported worldwide in the respiratory secretions of patients with acute respiratory infections (2), and recent studies have shown that it can be detected with a high frequency in palatine tonsils and adenoids from patients with chronic tonsillar disease (21, 22). Lu et al. (21) reported a rate of detection of HBoV1 DNA of approximately 32% in the adenoids of young children (median age, 3.7 years) undergoing adenoidectomy, an observation similar to ours.
Recently, using a reverse genetics model developed by Huang et al. (25), Deng et al. (12) showed that HBoV1 replicates in well-differentiated primary cultures of human airway epithelium (HAE). In addition, those authors reported that HBoV1 infects commercially available primary cultures of human airway epithelium (26). Therefore, it is reasonable to speculate that the higher frequency of HBoV genomic DNA detected in AD (25.1%) than in PT (7.2%) samples may be related to the fact that in the adenoids, the epithelium is largely of the pseudostratified ciliated respiratory kind, while the palatine tonsils are covered by nonkeratinized stratified squamous epithelia. Moreover, the adenoids are located in the nasopharynx, where they are exposed to inhaled pathogens, while the palatine tonsils are located lower in the pharynx, more distant from inhaled pathogens. Interestingly, the rate of HBoV1 detection in AD tissue was also higher than the rate of detection in NPS (10.5%), which suggests that the AD are an important site of HBoV1 replication in the upper airways of young children.
HBoV1 was detected in adenoid samples from control patients without tonsillar hypertrophy who received cochlear implants at a frequency similar to that observed in adenoid samples from patients with tonsillar hypertrophy. In addition, HBoV1 is prone to the establishment of prolonged shedding (27) and to be detected along with other pathogenic viruses (6, 10), a finding that was also confirmed in the present study. Taken together, these findings indicate that HBoV1 is not the primary etiology of tonsillar hypertrophy.
HBoV1 can persist in the nuclei of infected cells, either as single episomal genomes or as viral DNA concatemers, consistent with the rolling-hairpin replication of parvoviruses. In fact, several published results corroborate this hypothesis, including the detection of viral DNA in the nuclei of infected cells by in situ hybridization (28) and the characterization of head-to-tail parvoviral DNA sequences in human samples (13–15). In this regard, the PCR results obtained in respiratory secretions from children in Norway indicate that upon acute infection, HBoV1 DNA can persist in detectable levels for several months (9).
It is reasonable to hypothesize that the prolonged persistence of a DNA virus might trigger the development of chronic inflammation and tonsillar hyperplasia/hypertrophy, facilitating recurrent infections and the maintenance of clinical symptoms seen in the patients in our study. In fact, DNA sensors that are part of the innate immune system, such as IFI16 and cyclic GMP-AMP synthase (cGAS), might trigger chronic inflammation and damage stimulated by prolonged HBoV1 infection, similar to that observed with HIV (29) and herpes simplex virus (HSV) (30). In addition, HBoV1 DNA has been found in the nuclei of lung cancer cells, suggesting that this virus may contribute to the development of tissue hyperplasias, as also observed with other DNA viruses, such as hepatitis B virus (HBV) (28).
Therefore, in order to try to understand the association between HBoV1 persistence in the adenoids and palatine tonsils and the development of chronic tonsillar diseases, we determined the viral load and checked for the presence of two viral mRNAs, VP1 and NP1. To the best of our knowledge, this is the first report on HBoV1 viral loads and the presence of HBoV1 mRNAs in tissue samples from chronically ill patients without acute respiratory symptoms. There was no apparent correlation of the detection of HBoV mRNA with demographic or clinical features. In fact, in the present study, a significant proportion of patients had undetectable levels of viral mRNA and low viral loads, consistent with viral DNA persistence.
In patients with acute respiratory illnesses, there is a strong correlation between high HBoV1 DNA load (>107 copies/ml) in nasopharyngeal secretions and the detection of mRNA as a marker of viral replication, the absence of viral coinfections, and the presence of HBoV1 DNAemia (6, 9). In the present study, we also observed a correlation between higher viral loads and the detection of HBoV1 mRNAs, both in adenoid and nasopharyngeal secretion samples. These results indicate that for clinical purposes, the quantification of HBoV1 genomic DNA loads or the detection of mRNA may be useful for diagnosing acute HBoV1 infections. In contrast to previous studies of patients with symptoms of acute respiratory infections, in the present study, the viral loads of HBoV1 were not higher in patients without viral coinfections.
Higher HBoV1 viral loads but not the presence of HBoV mRNAs were associated with more severe adenoidal hypertrophy and sleep apnea. This may mean that there is a higher abundance of HBoV1 episomal DNA in proliferating cells of hypertrophic tonsils rather than more intense viral replication. It should also be kept in mind, however, that the persistence of HBoV1 DNA may be a contributing factor in helping to maintain chronic inflammation in already hypertrophic tonsils. In this regard, a recent study reported an association between HBoV1 and another chronic ailment, otitis media with effusion, suggesting that persisting viruses can cause lymphoid hyperplasia, leading to ventilation disorders and impaired immunoreactivity of the middle ear (31). The current lack of practical assays for markers of replication for all respiratory viruses makes it hard to conclusively assess how their replicative status may influence infection by the others.
The detection of HBoV1 mRNA and high viral DNA levels in some palatine tonsils that lack ciliated epithelia suggests that nonepithelial cells in that tissue can harbor HBoV1 viral replication. Based on previous evidence (21), such cells are probably lymphocytes, but definitive in situ studies are lacking.
In the present study, we documented that the adenoids are a main site of HBoV1 replication in patients with chronic adenotonsillar diseases. The adenoids of these patients contained the highest HBoV1 loads and yielded high rates of HBoV1 mRNA, which was significantly associated with viral DNA detection in other sites. Additional studies will be required to provide definitive localization of HBoV1 replication and persistence in the adenoids and palatine tonsils.
Supplementary Material
ACKNOWLEDGMENTS
We thank Maria Cecília Onofre and Helder G. de Souza for secretarial assistance and Lúcia Lopes and Jamila Mendonça de Souza for expert technical support.
This study was supported by the São Paulo Research Foundation–FAPESP (grant 2009/51818-8). E.A., W.T.A.-L., and F.C.P.V. are recipients of longstanding research scholarships from the National Research Council (CNPq). The funders had no role in study design, data collection, or analysis, the decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 11 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00870-14.
REFERENCES
- 1.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 102:12891–12896. 10.1073/pnas.0504666102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schildgen O, Müller A, Allander T, Mackay IM, Völz S, Kupfer B, Simon A. 2008. Human bocavirus: passenger or pathogen in acute respiratory tract infections? Clin. Microbiol. Rev. 21:291–304, table of contents. 10.1128/CMR.00030-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur JL, Higgins GD, Davidson GP, Givney RC, Ratcliff RM. 2009. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog. 5:e1000391. 10.1371/journal.ppat.1000391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapoor A, Simmonds P, Slikas E, Li L, Bodhidatta L, Sethabutr O, Triki H, Bahri O, Oderinde BS, Baba MM, Bukbuk DN, Besser J, Bartkus J, Delwart E. 2010. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J. Infect. Dis. 201:1633–1643. 10.1086/652416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapoor A, Slikas E, Simmonds P, Chieochansin T, Naeem A, Shaukat S, Alam MM, Sharif S, Angez M, Zaidi S, Delwart E. 2009. A newly identified bocavirus species in human stool. J. Infect. Dis. 199:196–200. 10.1086/595831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Proença-Modena JL, Gagliardi TB, da Paula FE, Iwamoto MA, Criado MF, Camara AA, Acrani GO, Cintra OA, Cervi MC, Arruda LK, Arruda E. 2011. Detection of human bocavirus mRNA in respiratory secretions correlates with high viral load and concurrent diarrhea. PLoS One 6:e21083. 10.1371/journal.pone.0021083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Proenca-Modena JL, Martinez M, Amarilla AA, Espínola EE, Galeano ME, Fariña N, Russomando G, Aquino VH, Parra GI, Arruda E. 2013. Viral load of human bocavirus-1 in stools from children with viral diarrhoea in Paraguay. Epidemiol. Infect. 141:2576–2580. 10.1017/S095026881300023X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allander T, Jartti T, Gupta S, Niesters HG, Lehtinen P, Osterback R, Vuorinen T, Waris M, Bjerkner A, Tiveljung-Lindell A, van den Hoogen BG, Hyypiä T, Ruuskanen O. 2007. Human bocavirus and acute wheezing in children. Clin. Infect. Dis. 44:904–910. 10.1086/512196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen A, Døllner H, Skake LH, Krokstad S, Moe N, Nordbø SA. 2013. Detection of spliced mRNA from human bocavirus 1 in clinical samples from children with respiratory tract infections. Emerg. Infect. Dis. 19:574–580. 10.3201/eid1904.121775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen A, Nordbø SA, Krokstad S, Rognlien AG, Døllner H. 2010. Human bocavirus in children: mono-detection, high viral load and viraemia are associated with respiratory tract infection. J. Clin. Virol. 49:158–162. 10.1016/j.jcv.2010.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nascimento-Carvalho CM, Cardoso MR, Meriluoto M, Kemppainen K, Kantola K, Ruuskanen O, Hedman K, Söderlund-Venermo M. 2012. Human bocavirus infection diagnosed serologically among children admitted to hospital with community-acquired pneumonia in a tropical region. J. Med. Virol. 84:253–258. 10.1002/jmv.22268 [DOI] [PubMed] [Google Scholar]
- 12.Deng X, Yan Z, Luo Y, Xu J, Cheng F, Li Y, Engelhardt F, Qiu J. 2013. In vitro modeling of human bocavirus 1 infection of polarized primary human airway epithelia. J. Virol. 87:4097–4102. 10.1128/JVI.03132-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lüsebrink J, Schildgen V, Tillmann RL, Wittleben F, Böhmer A, Müller A, Schildgen O. 2011. Detection of head-to-tail DNA sequences of human bocavirus in clinical samples. PLoS One 6:e19457. 10.1371/journal.pone.0019457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapoor A, Hornig M, Asokan A, Williams B, Henriquez JA, Lipkin WI. 2011. Bocavirus episome in infected human tissue contains non-identical termini. PLoS One 6:e21362. 10.1371/journal.pone.0021362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H, Zhao L, Sun Y, Qian Y, Liu L, Jia L, Zhang Y, Dong H. 2012. Detection of a bocavirus circular genome in fecal specimens from children with acute diarrhea in Beijing, China. PLoS One 7:e48980. 10.1371/journal.pone.0048980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Windisch W, Schildgen V, Malecki M, Lenz J, Brockmann M, Karagiannidis C, Schildgen O. 2013. Detection of HBoV DNA in idiopathic lung fibrosis, Cologne, Germany. J. Clin. Virol. 58:325–327. 10.1016/j.jcv.2013.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nave H, Gebert A, Pabst R. 2001. Morphology and immunology of the human palatine tonsil. Anat. Embryol. (Berl.) 204:367–373. 10.1007/s004290100210 [DOI] [PubMed] [Google Scholar]
- 18.Garnett CT, Erdman D, Xu W, Gooding LR. 2002. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J. Virol. 76:10608–10616. 10.1128/JVI.76.21.10608-10616.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garnett CT, Talekar G, Mahr JA, Huang W, Zhang Y, Ornelles DA, Gooding LR. 2009. Latent species C adenoviruses in human tonsil tissues. J. Virol. 83:2417–2428. 10.1128/JVI.02392-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hislop AD, Kuo M, Drake-Lee AB, Akbar AN, Bergler W, Hammerschmitt N, Khan N, Palendira U, Leese AM, Timms JM, Bell AI, Buckley CD, Rickinson AB. 2005. Tonsillar homing of Epstein-Barr virus-specific CD8+ T cells and the virus-host balance. J. Clin. Invest. 115:2546–2555. 10.1172/JCI24810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu X, Gooding LR, Erdman DD. 2008. Human bocavirus in tonsillar lymphocytes. Emerg. Infect. Dis. 14:1332–1334. 10.3201/eid1408.080300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proenca-Modena JL, Valera FCP, Jacob MG, Buzatto GP, Saturno TH, Lopes L, Souza JM, Paula FE, Silva ML, Carenzi LR, Tamashiro E, Arruda E, Anselmo-Lima WT. 2012. High rates of detection of respiratory viruses in tonsillar tissues from children with chronic adenotonsillar disease. PLoS One 7:e42136. 10.1371/journal.pone.0042136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paradise JL, Bluestone CD, Bachman RZ, Colborn DK, Bernard BS, Taylor FH, Rogers KD, Schwarzbach RH, Stoll SE, Friday GA, Smith IH, Saez CA. 1984. Efficacy of tonsillectomy for recurrent throat infection in severely affected children. Results of parallel randomized and nonrandomized clinical trials. N. Engl. J. Med. 310:674–683 [DOI] [PubMed] [Google Scholar]
- 24.Neske F, Blessing K, Tollmann F, Schubert J, Rethwilm A, Kreth HW, Weissbrich B. 2007. Real-time PCR for diagnosis of human bocavirus infections and phylogenetic analysis. J. Clin. Microbiol. 45:2116–2122. 10.1128/JCM.00027-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Q, Deng X, Yan Z, Cheng F, Luo Y, Shen W, Lei-Butters DC, Chen AY, Li Y, Tang L, Söderlund-Venermo M, Engelhardt JF, Qiu J. 2012. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog. 8:e1002899. 10.1371/journal.ppat.1002899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng X, Li Y, Qiu J. 2014. Human bocavirus 1 infects commercially available primary human airway epithelium cultures productively. J. Virol. Methods 195:112–119. 10.1016/j.jviromet.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin ET, Fairchok MP, Kuypers J, Magaret A, Zerr DM, Wald A, Englund JA. 2010. Frequent and prolonged shedding of bocavirus in young children attending daycare. J. Infect. Dis. 201:1625–1632. 10.1086/652405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schildgen V, Malecki M, Tillmann RL, Brockmann M, Schildgen O. 2013. The human bocavirus is associated with some lung and colorectal cancers and persists in solid tumors. PLoS One 8:e68020. 10.1371/journal.pone.0068020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, Greene WC. 2014. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343:428-432. 10.1126/science.1243640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, Knipe DM. 2013. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc. Natl. Acad. Sci. U. S. A. 110:E4492–E4501. 10.1073/pnas.1316194110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szalmás A, Papp Z, Csomor P, Kónya J, Sziklai I, Szekanecz Z, Karosi T. 2013. Microbiological profile of adenoid hypertrophy correlates to clinical diagnosis in children. Biomed. Res. Int. 2013:629607. 10.1155/2013/629607 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.