Abstract
An increase in the proportion of ambiguous base calls in HIV-1 pol population sequences during the course of infection has been demonstrated in different study populations, and sequence ambiguity thresholds to classify infections as recent or nonrecent have been suggested. The aim of our study was to evaluate sequence ambiguities as a candidate biomarker for use in an HIV-1 incidence assay using samples from antiretroviral treatment-naive seroconverters with known durations of infection (German HIV-1 Seroconverter Study). We used 2,203 HIV-1 pol population sequences derived from 1,334 seroconverters to assess the sequence ambiguity method (SAM). We then compared the serological incidence BED capture enzyme immunoassay (BED-CEIA) with the SAM for a subset of 723 samples from 495 seroconverters and evaluated a multianalyte algorithm that includes BED-CEIA results, SAM results, viral loads, and CD4 cell counts for 453 samples from 325 seroconverters. We observed a significant increase in the proportion of sequence ambiguities with the duration of infection. A sequence ambiguity threshold of 0.5% best identified recent infections with 76.7% accuracy. The mean duration of recency was determined to be 208 (95% confidence interval, 196 to 221) days. In the subset analysis, BED-CEIA achieved a significantly higher accuracy than the SAM (84.6 versus 75.5%, P < 0.001) and results were concordant for 64.2% (464/723) of the samples. Also, the multianalyte algorithm did not show better accuracy than the BED-CEIA (83.4 versus 84.3%, P = 0.786). In conclusion, the SAM and the multianalyte algorithm including SAM were inferior to the BED-CEIA, and the proportion of sequence ambiguities is therefore not a preferable biomarker for HIV-1 incidence testing.
INTRODUCTION
Estimation of the HIV-1 incidence in populations is important for developing specific prevention strategies. Methods to identify recently acquired infections are needed for estimation of the incidence of HIV-1 from cross-sectional surveys. Whether or not an HIV-1 infection was acquired recently can only be determined within a short period of approximately 14 weeks after infection by routine HIV-1 diagnostic methods. Early HIV-1 infection is characterized by an eclipse phase of approximately 10 days before viral RNA becomes detectable, followed by the orderly appearance of other laboratory markers until seroconversion is completed, as indicated by a positive immunoblot assay (1). However, the level and avidity of HIV-specific antibodies increase during the months following seroconversion. This allows recent and nonrecent infections to be distinguished by serology-based HIV-1 incidence assays in which the increase in the proportion of HIV-specific antibodies (2), the avidity of HIV-specific antibodies (3, 4), or a combination of HIV-specific antibody level and avidity (5) is measured. Unfortunately, samples from individuals infected with HIV-1 non-B subtypes (6, 7), from elite controllers (8), from individuals treated with antiretroviral drugs (8, 9), and from individuals with advanced stages of disease (7, 9) can be misclassified on the basis of serological criteria because of delayed or reduced production of HIV-specific antibodies. Nonserological HIV-1 incidence assays (10, 11) and algorithms combining serological and nonserological biomarkers have therefore been developed (12, 13). HIV-1 diversity is considered to be a candidate nonserological biomarker, as it increases during the course of infection in antiretroviral treatment (ART)-naive individuals. Viral diversity can be determined by single-genome sequencing (11), next-generation sequencing (10), and high-resolution melting assays (12), but direct Sanger population sequencing can also reflect HIV-1 polymorphisms via ambiguous base calls (14–16).
The aim of this study was to evaluate ambiguous base calls in HIV-1 pol population sequences (sequence ambiguities) as a biomarker for use in an HIV-1 incidence assay (sequence ambiguity method, SAM) or as part of a multianalyte algorithm. To achieve this, we used samples from ART-naive study patients within the German HIV-1 Seroconverter Study with known durations of infection (17–20).
MATERIALS AND METHODS
Seroconverter study population.
The German HIV-1 Seroconverter Study of the Robert Koch Institute is a long-term, open, prospective, nationwide, multicenter cohort study of HIV-1-infected patients with documented dates of infection (17–20). Newly diagnosed HIV-1 seroconverters are recruited by clinical centers and private practitioners. The study was approved (EA 2/105/05) by the ethical committee of the Charité University Medicine (Berlin, Germany). All study patients sign an informed consent form prior to enrollment. Primary and ideally yearly follow-up blood samples are collected along with demographic, laboratory, and clinical data. HIV-1 genotyping is performed at the Robert Koch Institute on the basis of plasma from ART-naive seroconverters with viral loads of ≥1,000 copies/ml in order to monitor trends in transmitted drug resistance (TDR) and the spread of HIV-1 subtypes.
In this study, we included only seroconverters from whom at least one viral sequence had been determined and whose test results conformed to one of the following criteria: (i) detectable HIV-1 RNA or p24 antigen combined with a negative or indeterminate enzyme-linked immunosorbent assay (ELISA) result, (ii) reactive HIV-1 ELISA result combined with a negative or indeterminate immunoblot result, or (iii) last negative and first confirmed positive HIV-1 antibody test within a maximum of 365 days. Either the blood sampling date for the first reactive test (i, ii) or the mean of the blood sampling dates for the last negative and first confirmed positive HIV-1 antibody tests (iii) was used as the date of infection. The duration of infection was calculated as the difference between the date of blood sampling and the date of infection.
As of January 2013, 2,203 HIV-1 pol sequences (1,334 primary, 869 follow-up) derived from 1,334 seroconverters were included in the analysis. The majority of the study patients were male (1,265/1,334, 94.8%), and the most common (1,164/1,334, 87.3%) risk factor for transmission was being a man who has sex with men (MSM). The median age of the study patients at primary blood sampling was 33 years (interquartile range, 27 to 39). Study patients were infected between 1996 and 2012. A total of 49.8% (1,096/2,203) of the samples were taken within 182 days after infection, 10.1% (222/2,203) were taken between 183 and 365 days after infection, 16.3% (359/2,203) were taken between 366 and 730 days after infection, 10.6% (234/2,203) were taken between 731 and 1,095 days after infection, 5.9% (130/2,203) were taken between 1,096 and 1,460 days after infection, 2.9% (65/2,203) were taken between 1,461 and 1,825 days after infection, and 4.4% (97/2,203) were taken >1,825 days after infection. A detailed description of the study population is provided in Table 1.
TABLE 1.
Characteristics of the study population
| Characteristic | No. (%) of patients |
|---|---|
| Gender | |
| Both | 1,334 (100.0) |
| Male | 1,265 (94.8) |
| Female | 69 (5.2) |
| Risk factor for transmission | |
| MSM | 1,164 (87.3) |
| Heterosexual | 114 (8.5) |
| Injection drug user | 18 (1.4) |
| High-prevalence country | 7 (0.5) |
| Occupational exposure | 4 (0.3) |
| Unknown | 27 (2.0) |
| HIV-1 subtype | |
| B | 1,212 (90.8) |
| CRF02_AG | 32 (2.4) |
| C | 25 (1.9) |
| A | 23 (1.7) |
| CRF01_AE | 21 (1.6) |
| Other | 21 (1.6) |
| TDR | |
| Sensitive | 1,176 (88.2) |
| Resistant | 158 (11.8) |
| Origin of study patient | |
| Germany | 1,129 (84.6) |
| Other | 205 (15.4) |
HIV-1 genotyping.
HIV-1 from 500 μl plasma stored frozen at −70°C was pelleted by centrifugation (20,800 × g, 90 min, 4°C), and viral RNA was isolated with the Viral RNA minikit (Qiagen) according to the manufacturer's instructions. Reverse transcription (RT) with 75 μl plasma equivalents and PCR was performed either with the Viroseq HIV-1 Genotyping System (Abbott Molecular; n = 1,688) according to the manufacturer's instructions or with a previously described in-house pol RT-PCR (n = 515) (17, 21). Direct Sanger population sequencing was performed with the Viroseq HIV-1 Genotyping System (Abbott Molecular; n = 1,688) according to the manufacturer's instructions or BigDye terminator cycle sequencing (Life Technologies) with in-house primers as described previously (n = 515) (17, 21). SeqMan Pro (Lasergene version 10.0.1; DNASTAR; n = 1,866) or AutoAssembler (version 2.0; Applied Biosystems; n = 337) was used to assemble the sequence contigs. Ambiguous base calls were identified at a threshold of 20% if the minor peak was three times as high as the background and evident in at least two differently primed sequences. The consensus sequence was used to determine TDR according to the surveillance drug resistance mutation (SDRM) list (22) and the HIV-1 subtype (REGA HIV Subtyping Tool, version 2.0). If the viral subtype was not assigned by REGA, distance-based neighbor-joining phylogeny (Kimura two-parameter model, bootstrapping with 1,000 replicates) of an end-trimmed sequence alignment including the Los Alamos HIV subtype reference sequences of 2010 (www.hiv.lanl.gov) was performed (PHYLIP version 3.6; Joe Felsenstein). To confirm the HIV-1 sequences of longitudinal samples, the resistance-associated positions according to the SDRM list (22) were deleted prior to neighbor-joining phylogeny. For this study, sequences were trimmed to a length of 1,170 nucleotides spanning amino acids 1 to 99 of the protease and 1 to 291 of the reverse transcriptase, thus covering all of the positions mediating resistance to protease and reverse transcriptase inhibitors according to the SDRM list (22). The number and proportion of ambiguous base calls in each Sanger population sequence were determined with Microsoft Excel 2010 (Microsoft).
Definition of a sequence ambiguity threshold.
Sensitivity (classified as recent among truly recent samples), specificity (classified as nonrecent among truly nonrecent samples), and accuracy (classified correctly among all samples) were calculated with a custom Visual Basic for Applications (VBA) program within Microsoft Excel 2010 (Microsoft). Thresholds for the proportion of sequence ambiguities ranging from 0.1 to 1.5% in increments of 0.1% were analyzed for durations of infection ranging from 180 to 540 days in increments of 1 day. The best threshold was defined as the highest accuracy with sensitivity and specificity both set to ≥75.0%.
Serological HIV-1 incidence assay.
The Aware BED capture enzyme immunoassay (BED-CEIA) HIV-1 incidence assay (Calypte Biomedical Corporation) was performed according to the manufacturer's instructions. The results were reported as normalized optical density units (ODn) with a threshold ≤0.8 ODn to identify recent infections. A subset of 723 samples from 495 seroconverters was analyzed with the BED-CEIA. Only samples with durations of infection of ≤400 days from study patients whose seroconversion had been documented according to criterion i or ii described above were included.
Multianalyte algorithm.
Viral loads and CD4 cell counts determined within 7 days of the blood collection dates were available for 453 samples from 325 seroconverters tested with the BED-CEIA. A multianalyte algorithm that integrates BED-CEIA results, SAM results, viral loads, and CD4 cell counts was developed to analyze these data. With the Excel VBA program, the highest accuracy (with sensitivity and specificity both set to ≥70.0%) for discriminating recent from nonrecent infections was defined by comparing 2,069,550 combinations of thresholds for BED-CEIA results (0.6 to 1.4 ODn, increments of 0.1 ODn), SAM results (0.2 to 1.0%, increments of 0.1%), viral loads (100 to 5,000 copies/ml, increments of 100 copies/ml), CD4 cell counts (100 to 400 cells/μl, increments of 50 cells/μl), and duration of infection (180 to 540 days, increments of 5 days).
Determination of the mean duration of recency and the false recency rate.
The mean duration of recency is the average duration of infection for which individuals in a population are classified as recently infected by an HIV-1 incidence assay (13, 23). In our study, the mean duration of recency was computed by using an adaption of the method described by Brookmeyer et al. (13). We defined the function ϕ(t) as the probability that an individual who has been infected for at most τ days is classified as recent. The probability ϕ(t) was computed by using the empirical cumulative frequency of being classified as recent by the respective biomarker PR(τ > t), i.e., ϕ(t) = 1 − PR(τ > t) = PR(τ ≤ t). The mean duration of recency, μ, is then given by μ = ∫ϕ(t)dt, which was computed by standard integration. In order to account for the uncertainty in the duration of infection τ for samples derived from study patients whose seroconversion had been documented according to criterion iii described above, we conducted a multiple imputation by assuming a uniform distribution of actual infection dates within the interval defined by the last negative and first confirmed positive HIV-1 antibody tests. Furthermore, to estimate confidence ranges for μ and to account for multiple samples of the same study patient, we performed bootstrapping with 1,000 replicates of the data set. Computations were conducted with the statistics toolbox of MATLAB 8 (MathWorks).
The false recency rate is the proportion of individuals in a population who have been infected for longer than the duration of a recency cutoff (T) but classified as recent by an HIV-1 incidence assay (23). For calculation of the false recency rate, we defined T as ≤540 days.
Statistical analyses.
The relationship between the proportion of sequence ambiguities and the duration of infection was analyzed by Pearson correlation and linear regression. Partial Pearson correlation was used to examine confounding factors. Because the data did not show normal distribution and multiple samples from 478 seroconverters were included, 95% confidence intervals (CIs) were generated by bootstrapping with 1,000 replicates. The Mann-Whitney U test and the Fisher exact test were used to compare continuous and categorical variables, respectively. Statistical analysis was performed with SPSS Statistics 20 (IBM) or GraphPad QuickCalcs (GraphPad). Two-sided probability values of type I errors (P values) of ≤0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
The sequences used in this study (n = 2,203) have been deposited in the GenBank database and assigned accession numbers KJ769682 to KJ771884.
RESULTS
Correlation between the proportion of sequence ambiguities and the duration of infection.
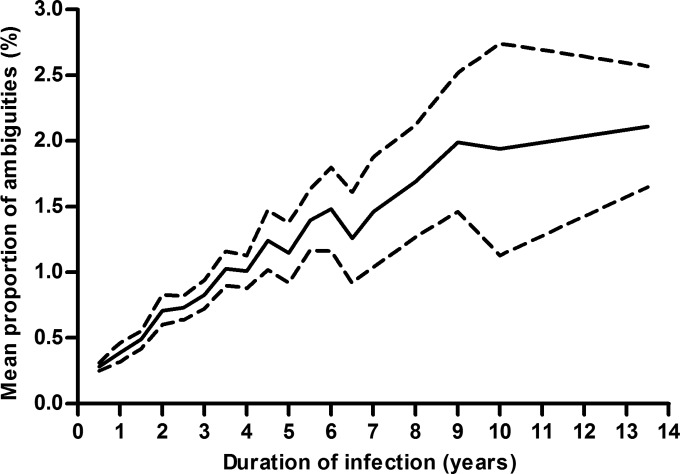
Using Pearson correlation, we identified a significant increase in the proportion of sequence ambiguities as the duration of infection increased (Fig. 1) that was not affected by the HIV-1 subtype (B versus non-B), TDR (sensitive versus resistant), risk factor for transmission (MSM versus non-MSM), gender (male versus female), origin of the study patient (Germany versus other countries), age, and year of infection (Table 2). The inclusion of multiple samples from 478 seroconverters did not bias the correlation (Pearson coefficient of correlation, 0.54; 95% CI, 0.50 to 0.58; P < 0.001). Linear regression (coefficient of determination, 0.29; P < 0.001) indicated an overall rate of 0.2% (95% CI, 0.19 to 0.22%) sequence ambiguities per year of infection with an intercept point of 0.26% (95% CI, 0.23 to 0.28%) sequence ambiguities.
FIG 1.
Mean proportion of sequence ambiguities (solid line) and 95% CI (dashed lines) stratified by duration of infection.
TABLE 2.
Pearson correlations of the proportion of sequence ambiguities and the duration of infection
| Adjustment | Coefficient of correlation (95% CI) | P value |
|---|---|---|
| None | 0.54 (0.50–0.58) | <0.001 |
| HIV-1 subtype | 0.54 (0.50–0.58) | <0.001 |
| TDR | 0.54 (0.50–0.58) | <0.001 |
| Risk factor for transmission | 0.54 (0.50–0.58) | <0.001 |
| Gender | 0.54 (0.50–0.58) | <0.001 |
| Origin of study patient | 0.54 (0.50–0.58) | <0.001 |
| Age | 0.54 (0.50–0.58) | <0.001 |
| Yr of infection | 0.54 (0.50–0.58) | <0.001 |
Identification of recent infections by using a sequence ambiguity threshold.
A sequence ambiguity threshold of 0.5% was found to best discriminate recent from nonrecent infections in our study population with an accuracy of 76.7%. The false recency rate accounted for 23.3%. The mean duration of recency was computed as 208 (95% CI, 196 to 221) days. Samples classified as recent were derived from study patients with a significantly shorter duration of infection than study patients whose samples were classified as nonrecent (median, 101 days versus 729 days; P < 0.001). This finding was independent of the HIV-1 subtype (B, P < 0.001; non-B, P < 0.001), TDR (sensitive, P < 0.001; resistant, P < 0.001), risk factor for transmission (MSM, P < 0.001; non-MSM, P < 0.001), gender (male, P < 0.001; female, P < 0.001), and origin of the study patient (Germany, P < 0.001; other countries, P < 0.001).
Comparison of the BED-CEIA and the SAM.
The BED-CEIA achieved a significantly higher accuracy than the SAM (84.6 versus 75.5%; P < 0.001) in the subset of 723 samples with available BED-CEIA results. The false recency rate of the BED-CEIA was significantly lower than that of the SAM (6.6 versus 28.5%; P < 0.001). The mean durations of recency calculated for the BED-CEIA and the SAM were 98 (95% CI, 76 to 121) days and 236 (95% CI, 209 to 263) days, respectively. The concordance of BED-CEIA and SAM results reached 64.2% (464/723) and did not significantly differ between subgroups regarding the HIV-1 subtype, TDR, risk factor for transmission, gender, or origin of the study patient (Table 3). Of discordant results, 74.5% (193/259) were caused by classification as recent by the SAM but nonrecent by the BED-CEIA.
TABLE 3.
Concordance of the SAM and the BED-CEIA in subgroups
| Subgroup | % Concordance (no. of samples concordant/total) | P value |
|---|---|---|
| Subtype | ||
| B | 64.0 (406/634) | |
| Non-B | 65.2 (58/89) | 0.906 |
| TDR | ||
| Sensitive | 64.0 (405/633) | |
| Resistant | 65.6 (59/90) | 0.815 |
| Risk | ||
| MSM | 64.2 (396/617) | |
| Non-MSM | 64.2 (68/106) | 1.000 |
| Gender | ||
| Male | 63.8 (434/680) | |
| Female | 69.8 (30/43) | 0.513 |
| Origin of study patient | ||
| Germany | 63.8 (383/600) | |
| Other | 65.9 (81/123) | 0.757 |
Multianalyte algorithm.
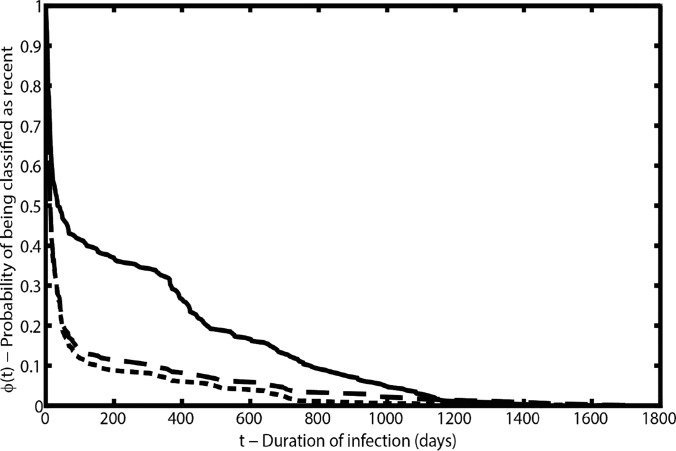
Combination of biomarkers was based on the subset of 453 samples with data for BED-CEIA, viral loads, and CD4 cell counts. The thresholds for the best classification of recent infections with the SAM in combination with viral loads or CD4 cell counts were a SAM result of ≤0.3%, a viral load of ≥100 copies/ml, and a CD4 cell count of ≥100 cells/μl. Combination of the SAM result with the viral load or the CD4 cell count did not significantly improve accuracy but did improve the false recency rate (Table 4). The mean duration of recency was shorter with both combinations than with the SAM alone (Table 4). The thresholds that best identified recent infections through the combination of the BED-CEIA result with the SAM result, the viral load, or the CD4 cell count were a BED-CEIA result of ≤0.8 ODn, a SAM result of ≤1.0%, a viral load of ≥300 copies/ml, and a CD4 cell count of ≥100/μl. Identical thresholds were determined for the combination of all four biomarkers (multianalyte algorithm). Neither combination of the BED-CEIA result with the SAM result, the viral load, or the CD4 cell count nor the multianalyte algorithm achieved significantly higher accuracy or a lower false recency rate than the BED-CEIA alone (Table 4). All biomarker combinations exhibited shorter mean durations of recency than the BED-CEIA (Table 4). Compared to the ϕ(t) of the BED-CEIA or the multianalyte algorithm, the decay of ϕ(t) of the SAM was much slower in the infection duration range of 50 to 500 days (Fig. 2), indicating that fewer samples within this interval were classified as recent and that the individual durations of recency were more broadly distributed.
TABLE 4.
Accuracy, false recency rate, and mean duration of recency for different biomarker combinations
| Assay(s) | % Accuracy | P valuea | False recency rate (%) | P valuea | Mean duration of recency in days (95% CI) |
|---|---|---|---|---|---|
| SAM | 75.5 | R | 28.2 | R | 243 (210–277) |
| SAM + viral load | 78.1 | 0.387 | 13.2 | 0.001 | 169 (137–200) |
| SAM + CD4 cell count | 77.5 | 0.531 | 13.8 | 0.002 | 173 (141–205) |
| BED-CEIA | 84.3 | R | 6.3 | R | 94 (63–124) |
| BED-CEIA + SAM | 83.0 | 0.653 | 5.2 | 0.819 | 85 (57–112) |
| BED-CEIA + viral load | 85.0 | 0.854 | 5.2 | 0.819 | 80 (53–106) |
| BED-CEIA + CD4 cell count | 84.1 | 1.000 | 6.3 | 1.000 | 93 (62–123) |
| Multianalyte algorithm | 83.4 | 0.786 | 4.0 | 0.469 | 69 (46–91) |
Compared to respective referent (R).
FIG 2.
Probability of being classified as recent by duration of infection for the SAM (solid line), the BED-CEIA (long-dashed line), and the multianalyte algorithm (short-dashed line).
DISCUSSION
In this study, we evaluated ambiguous base calls in HIV-1 pol population sequences as a candidate biomarker for use in an HIV-1 incidence assay or as part of a multianalyte algorithm. By using a large sample set derived from ART-naive study patients of the German HIV-1 Seroconverter Study that have well-documented dates of infection, it was possible to confirm previous reports based on study populations from Switzerland (14), Canada (15), and Sweden (16).
The concordance between the SAM and the BED-CEIA reached 64.2%, which is in agreement with the analysis by Ragonnet-Cronin et al. (15), which yielded a 67.1% concordance between their “mixed base classifier” and the BED-CEIA. The discordance between the SAM and the BED-CEIA was caused mainly by a recent result with the SAM and a nonrecent result with the BED-CEIA, which is also in agreement with the related study (15). Referring to previously published data (2, 24, 25) and our subset analyses, the BED-CEIA yields significantly higher accuracy and a lower false recency rate than the SAM and the multianalyte algorithm that includes the SAM. Moreover, with the SAM, the probability of a sample being classified as recent showed a broad distribution over the duration of infection. This considerably reduces the utility of the proportion of sequence ambiguities as a biomarker for use in an HIV-1 incidence assay. Nevertheless, it should be noted that our data set, comprising a high proportion of recent samples of HIV-1 subtype B, was highly suitable for testing with the BED-CEIA. In populations with predominantly nonrecent or HIV-1 non-B subtype infections, less accurate results with the BED-CEIA have been reported (26, 27).
In line with the analysis by Kouyos et al. (14), we detected a significant increase in the proportion of sequence ambiguities as the duration of infection increased, giving an overall sequence ambiguity rate of 0.2% (95% CI, 0.19 to 0.22%) per year. In the previous studies, sequence ambiguity thresholds of 0.45% (15), 0.47% (16), and 0.5% (14) were described to best identify infections acquired within ≤155 days (15) or ≤365 days (14, 16) with accuracies of 79.9% (15), 79.7% (16), and 70.9% (14). Our analysis is in accordance with these reports, yielding a sequence ambiguity threshold of 0.5%, a mean duration of recency of 208 days, and an accuracy of 76.7%. The slightly different study outcomes may be attributed to the following issues.
First, technical differences between the studies exist. The detection limit of ambiguous base calls depends on the plasma virus load and the method of HIV-1 genotyping, including RNA extraction, cDNA synthesis, PCR, sequencing, and sequence analysis. For example, primer mismatches, which may especially occur among different HIV-1 subtypes, can introduce a bias to specific virus variants. Because of the retrospective nature of our analysis, two different PCR systems, sequencing systems, and software packages to call ambiguous bases were used over time, which is an inherent technical limitation of the SAM. However, the methodological diversity in our study was still lower than in related studies (14–16). Since Ragonnet-Cronin et al. (15) demonstrated that differences in ambiguous base calling were at least negligible in the commonly used threshold range of 15 to 25%, we consider the sequencing software to have only a minor influence on the overall frequency of ambiguous base calling. Second, durations of infection were assessed in different ways. Our analysis involved samples of seroconverters with well-documented dates of infection and therefore reliably calculated durations of infection for each blood sampling date. On the contrary, approaches used to determine the duration of infection in previous studies have been less precise (14, 16). Third, the size and composition of the study panels, as far as documented, differed between the studies. The analysis of Andersson et al. included 57% HIV-1 non-B subtypes (16), whereas our analysis, as well as related studies, focused mainly (14) or only on HIV-1 subtype B (15). However, we found that the HIV-1 subtype did not affect the SAM, which is in agreement with the two related studies evaluating HIV-1 non-B subtypes (14, 16). In comparison with previous studies (14, 16), our study panel comprised more recent than nonrecent infections and more samples derived from MSM. Recent infections with multiple virus variants exhibit a higher viral diversity than those founded by a single virus variant and will therefore be misclassified as nonrecent. Furthermore, transmission of multiple virus variants is more frequently observed in injection drug users (IDU) and MSM than in heterosexuals (28, 29). Indeed, we detected a proportion of sequence ambiguities higher than 0.5% in 17.7% of the samples collected within 30 days of infection. Nevertheless, potential transmissions of multiple virus variants in recent infections were also noticed in previous studies (14, 16). Finally, Kouyos et al. (14) found an overall higher proportion of sequence ambiguities in samples derived from IDU, which was not observed in our analysis or in related studies (15, 16).
Altogether, the remarkable concordance of the yearly increase in sequence ambiguities (15) and the suggested sequence ambiguity thresholds (15–17) argues against a major impact of technical differences and major inaccuracies in the determination of the durations of infection. Instead, the diverse compositions of the four study panels most likely account for the slightly different accuracies and mean durations of recency determined in the four analyses. This underscores the need to adjust the mean duration of recency and the false recency rate for any biomarker-based estimation of HIV-1 incidence to the population being investigated (6, 23). We consider it important to also determine the accuracy of HIV-1 incidence assays since the accuracy provides information as to whether both the false recency and false nonrecency rates will impact the estimation of HIV-1 incidence. This information is particularly crucial in populations with a low HIV-1 prevalence. A uniform sequence ambiguity threshold to identify recent infections could be reasonably reduced to one decimal place since our analysis (data not shown) and a previous analysis (16) found that this has a negligible impact on accuracy.
One limitation common to our analysis and others (14–16) is that only samples from ART-naive individuals can be analyzed because ART impacts viral diversity by selecting for replicating virus variants. Obviously, it is not possible to derive a viral sequence from plasma if virus replication is successfully suppressed by ART. This selection inevitably introduces a bias to individuals with slower disease progression. Furthermore, similar to other studies evaluating sequence-based methods (14–16), we used the same data set to define the sequence ambiguity threshold and to determine the accuracy of the SAM. As this could result in an overestimation of accuracy, it would be preferable for training and validation to be performed with two different study populations. Alternatively, randomized splitting of a single study population into two for training and validation could be performed. However, this was precluded by the unequal proportions of recent and nonrecent infections in our data set.
In summary, in our population level analysis, the proportion of sequence ambiguities increased with the duration of infection and a proportion of sequence ambiguities larger than 0.5% indicated nonrecent or multiple virus variant infections in our study population, corroborating previous reports (14–16). However, the SAM achieved a lower accuracy than serological HIV-1 incidence assays (2, 3, 24, 25) and demonstrated an inconveniently broad distribution of individual durations of recency. Moreover, the SAM would not be cost-effective if HIV-1 genotyping was performed solely for cross-sectional surveys to estimate HIV-1 incidence. In conclusion, compared to HIV-1 incidence assays using more sensitive methods to determine HIV-1 diversity (10–12), the SAM is not preferable for HIV-1 incidence studies whether alone or as part of a multianalyte algorithm.
ACKNOWLEDGMENTS
We are very grateful to all of the study patients and medical doctors who have participated in the German HIV-1 Seroconverter Study since 1997. We thank Sabrina Neumann, Hanno von Spreckelsen, and Katrin Arndt for their enduring excellent technical assistance; Julia Tesch, Silvia Muschter, and Julia Hinzmann for excellent sequencing service; and Parvinolsadat Ghassim for careful data documentation. Special thanks to Matthias an der Heiden and Ramona Scheufele for statistical advice.
The German Seroconverter Study was partially funded by the Federal Ministry of Health (BMG) and the Federal Ministry of Education and Research (BMBF). K.P.Y. and M.V.K acknowledge funding through the DFG Research Center MATHEON (project A21) and the BMBF JRG Meth4SysPharm (grant 031A307).
Footnotes
Published ahead of print 11 June 2014
REFERENCES
- 1.Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, Heldebrant C, Smith R, Conrad A, Kleinman SH, Busch MP. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871–1879. 10.1097/00002030-200309050-00005 [DOI] [PubMed] [Google Scholar]
- 2.Parekh BS, Kennedy MS, Dobbs T, Pau CP, Byers R, Green T, Hu DJ, Vanichseni S, Young NL, Choopanya K, Mastro TD, McDougal JS. 2002. Quantitative detection of increasing HIV type 1 antibodies after seroconversion: a simple assay for detecting recent HIV infection and estimating incidence. AIDS Res. Hum. Retroviruses 18:295–307. 10.1089/088922202753472874 [DOI] [PubMed] [Google Scholar]
- 3.Suligoi B, Rodella A, Raimondo M, Regine V, Terlenghi L, Manca N, Casari S, Camoni L, Salfa MC, Galli C. 2011. Avidity index for anti-HIV antibodies: comparison between third- and fourth-generation automated immunoassays. J. Clin. Microbiol. 49:2610–2613. 10.1128/JCM.02115-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duong YT, Qiu M, De AK, Jackson K, Dobbs T, Kim AA, Nkengasong JN, Parekh BS. 2012. Detection of recent HIV-1 infection using a new limiting-antigen avidity assay: potential for HIV-1 incidence estimates and avidity maturation studies. PLoS One 7(3):e33328. 10.1371/journal.pone.0033328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis KA, Hanson DL, Kennedy MS, Owen SM. 2013. Evaluation of a multiplex assay for estimation of HIV-1 incidence. PLoS One 8(5):e64201. 10.1371/journal.pone.0064201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parekh BS, Hanson DL, Hargrove J, Branson B, Green T, Dobbs T, Constantine N, Overbaugh J, McDougal JS. 2011. Determination of mean recency period for estimation of HIV type 1 Incidence with the BED-capture EIA in persons infected with diverse subtypes. AIDS Res. Hum. Retroviruses 27:265–273. 10.1089/aid.2010.0159 [DOI] [PubMed] [Google Scholar]
- 7.Longosz AF, Serwadda D, Nalugoda F, Kigozi G, Franco V, Gray RH, Quinn TC, Eshleman SH, Laeyendecker O. 2014. Impact of HIV subtype on performance of the limiting antigen-avidity enzyme immunoassay, the Bio-Rad avidity assay, and the BED capture immunoassay in Rakai, Uganda. AIDS Res. Hum. Retroviruses 30:339–344. 10.1089/aid.2013.0169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wendel SK, Mullis CE, Eshleman SH, Blankson JN, Moore RD, Keruly JC, Brookmeyer R, Quinn TC, Laeyendecker O. 2013. Effect of natural and ARV-induced viral suppression and viral breakthrough on anti-HIV antibody proportion and avidity in patients with HIV-1 subtype B infection. PLoS One 8(2):e55525. 10.1371/journal.pone.0055525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laeyendecker O, Brookmeyer R, Oliver AE, Mullis CE, Eaton KP, Mueller AC, Jacobson LP, Margolick JB, Brown J, Rinaldo CR, Quinn TC, Eshleman SH, Multicenter AIDS Cohort Study (MACS) 2012. Factors associated with incorrect identification of recent HIV infection using the BED capture immunoassay. AIDS Res. Hum. Retroviruses 28:816–822. 10.1089/aid.2011.0258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park SY, Goeken N, Lee HJ, Bolan R, Dube MP, Lee HY. 2014. Developing high-throughput HIV incidence assay with pyrosequencing platform. J. Virol. 88:2977–2990. 10.1128/JVI.03128-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park SY, Love TM, Nelson J, Thurston SW, Perelson AS, Lee HY. 2011. Designing a genome-based HIV incidence assay with high sensitivity and specificity. AIDS 25:F13–F19. 10.1097/QAD.0b013e328349f089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cousins MM, Konikoff J, Laeyendecker O, Celum C, Buchbinder SP, Seage GR, Kirk GD, Moore RD, Mehta SH, Margolick JB, Brown J, Mayer KH, Koblin BA, Wheeler D, Justman JE, Hodder SL, Quinn TC, Brookmeyer R, Eshleman SH. 2014. HIV diversity as a biomarker for HIV incidence estimation: including a high-resolution melting diversity assay in a multiassay algorithm. J. Clin. Microbiol. 52:115–121. 10.1128/JCM.02040-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brookmeyer R, Konikoff J, Laeyendecker O, Eshleman SH. 2013. Estimation of HIV incidence using multiple biomarkers. Am. J. Epidemiol. 177:264–272. 10.1093/aje/kws436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouyos RD, von Wyl V, Yerly S, Boni J, Rieder P, Joos B, Taffe P, Shah C, Burgisser P, Klimkait T, Weber R, Hirschel B, Cavassini M, Rauch A, Battegay M, Vernazza PL, Bernasconi E, Ledergerber B, Bonhoeffer S, Gunthard HF, Swiss HIV Cohort Study 2011. Ambiguous nucleotide calls from population-based sequencing of HIV-1 are a marker for viral diversity and the age of infection. Clin. Infect. Dis. 52:532–539. 10.1093/cid/ciq164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ragonnet-Cronin M, Aris-Brosou S, Joanisse I, Merks H, Vallee D, Caminiti K, Rekart M, Krajden M, Cook D, Kim J, Malloch L, Sandstrom P, Brooks J. 2012. Genetic diversity as a marker for timing infection in HIV-infected patients: evaluation of a 6-month window and comparison with BED. J. Infect. Dis. 206:756–764. 10.1093/infdis/jis411 [DOI] [PubMed] [Google Scholar]
- 16.Andersson E, Shao W, Bontell I, Cham F, Cuong DD, Wondwossen A, Morris L, Hunt G, Sonnerborg A, Bertagnolio S, Maldarelli F, Jordan MR. 2013. Evaluation of sequence ambiguities of the HIV-1 pol gene as a method to identify recent HIV-1 infection in transmitted drug resistance surveys. Infect. Genet. Evol. 18:125–131. 10.1016/j.meegid.2013.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duwe S, Brunn M, Altmann D, Hamouda O, Schmidt B, Walter H, Pauli G, Kucherer C. 2001. Frequency of genotypic and phenotypic drug-resistant HIV-1 among therapy-naive patients of the German Seroconverter Study. J. Acquir. Immune Defic. Syndr. 26:266–273. 10.1097/00126334-200103010-00010 [DOI] [PubMed] [Google Scholar]
- 18.Poggensee G, Kucherer C, Werning J, Somogyi S, Bieniek B, Dupke S, Jessen H, Hamouda O, HIV-1 Seroconverter Study Group 2007. Impact of transmission of drug-resistant HIV on the course of infection and the treatment success. Data from the German HIV-1 Seroconverter Study. HIV Med. 8:511–519. 10.1111/j.1468-1293.2007.00504.x [DOI] [PubMed] [Google Scholar]
- 19.Bartmeyer B, Kuecherer C, Houareau C, Werning J, Keeren K, Somogyi S, Kollan C, Jessen H, Dupke S, Hamouda O, German HIV-1 Seroconverter Study Group 2010. Prevalence of transmitted drug resistance and impact of transmitted resistance on treatment success in the German HIV-1 Seroconverter Cohort. PLoS One 5(10):e12718. 10.1371/journal.pone.0012718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zu Knyphausen F, Scheufele R, Kucherer C, Jansen K, Somogyi S, Dupke S, Jessen H, Schurmann D, Hamouda O, Meixenberger K, Bartmeyer B. 2014. First line treatment response in patients with transmitted HIV drug resistance and well defined time point of HIV infection: updated results from the German HIV-1 Seroconverter Study. PLoS One 9(5):e95956. 10.1371/journal.pone.0095956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter H, Schmidt B, Korn K, Vandamme AM, Harrer T, Uberla K. 1999. Rapid, phenotypic HIV-1 drug sensitivity assay for protease and reverse transcriptase inhibitors. J. Clin. Virol. 13:71–80. 10.1016/S1386-6532(99)00010-4 [DOI] [PubMed] [Google Scholar]
- 22.Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, Heneine W, Kantor R, Jordan MR, Schapiro JM, Vandamme AM, Sandstrom P, Boucher CA, van de Vijver D, Rhee SY, Liu TF, Pillay D, Shafer RW. 2009. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 4(3):e4724. 10.1371/journal.pone.0004724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kassanjee R, McWalter TA, Welte A. 2014. Short communication: defining optimality of a test for recent infection for HIV incidence surveillance. AIDS Res. Hum. Retroviruses 30:45–49. 10.1089/aid.2013.0113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loschen S, Batzing-Feigenbaum J, Poggensee G, Cordes C, Hintsche B, Rausch M, Dupke S, Gohlke-Micknis S, Rodig J, Hamouda O, Kucherer C. 2008. Comparison of the human immunodeficiency virus (HIV) type 1-specific immunoglobulin G capture enzyme-linked immunosorbent assay and the avidity index method for identification of recent HIV infections. J. Clin. Microbiol. 46:341–345. 10.1128/JCM.01055-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hauser A, Santos-Hoevener C, Meixenberger K, Zimmermann R, Somogyi S, Fiedler S, Hoffmann A, Bartmeyer B, Jansen K, Hamouda O, Bannert N, Kuecherer C. 2014. Improved testing of recent HIV-1 infections with the BioRad Avidity Assay compared to the Limiting Antigen Avidity Assay and BED Capture Enzyme Immunoassay: evaluation using reference sample panels from the German Seroconverter Cohort. PLoS One 9(6):e98038. 10.1371/journal.pone.0098038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laeyendecker O, Brookmeyer R, Mullis CE, Donnell D, Lingappa J, Celum C, Baeten JM, Campbell MS, Essex M, de Bruyn G, Farquhar C, Quinn TC, Eshleman SH, Partners in Prevention HSV/HIV Transmission Study Team 2012. Specificity of four laboratory approaches for cross-sectional HIV incidence determination: analysis of samples from adults with known nonrecent HIV infection from five African countries. AIDS Res. Hum. Retroviruses 28:1177–1183. 10.1089/aid.2011.0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karita E, Price M, Hunter E, Chomba E, Allen S, Fei L, Kamali A, Sanders EJ, Anzala O, Katende M, Ketter N, IAVI Collaborative Seroprevalence and Incidence Study T 2007. Investigating the utility of the HIV-1 BED capture enzyme immunoassay using cross-sectional and longitudinal seroconverter specimens from Africa. AIDS 21:403–408. 10.1097/QAD.0b013e32801481b7 [DOI] [PubMed] [Google Scholar]
- 28.Li H, Bar KJ, Wang S, Decker JM, Chen Y, Sun C, Salazar-Gonzalez JF, Salazar MG, Learn GH, Morgan CJ, Schumacher JE, Hraber P, Giorgi EE, Bhattacharya T, Korber BT, Perelson AS, Eron JJ, Cohen MS, Hicks CB, Haynes BF, Markowitz M, Keele BF, Hahn BH, Shaw GM. 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 6(5):e1000890. 10.1371/journal.ppat.1000890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bar KJ, Li H, Chamberland A, Tremblay C, Routy JP, Grayson T, Sun C, Wang S, Learn GH, Morgan CJ, Schumacher JE, Haynes BF, Keele BF, Hahn BH, Shaw GM. 2010. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J. Virol. 84:6241–6247. 10.1128/JVI.00077-10 [DOI] [PMC free article] [PubMed] [Google Scholar]