Abstract
Carbapenem-resistant Acinetobacter baumannii (CRAb) is emerging worldwide as a public health problem in various settings. The aim of this study was to investigate the prevalence of CRAb isolates in Italy and to characterize their resistance mechanisms and genetic relatedness. A countrywide cross-sectional survey was carried out at 25 centers in mid-2011. CRAb isolates were reported from all participating centers, with overall proportions of 45.7% and 22.2% among consecutive nonreplicate clinical isolates of A. baumannii from inpatients (n = 508) and outpatients (n = 63), respectively. Most of them were resistant to multiple antibiotics, whereas all remained susceptible to colistin, with MIC50 and MIC90 values of ≤0.5 mg/liter. The genes coding for carbapenemase production were identified by PCR and sequencing. OXA-23 enzymes (found in all centers) were by far the most common carbapenemases (81.7%), followed by OXA-58 oxacillinases (4.5%), which were found in 7 of the 25 centers. In 6 cases, CRAb isolates carried both blaOXA-23-like and blaOXA-58-like genes. A repetitive extragenic palindromic (REP)-PCR technique, multiplex PCRs for group identification, and multilocus sequence typing (MLST) were used to determine the genetic relationships among representative isolates (n = 55). Two different clonal lineages were identified, including a dominant clone of sequence type 2 (ST2) related to the international clone II (sequence group 1 [SG1], SG4, and SG5) and a clone of ST78 (SG6) previously described in Italy. Overall, our results demonstrate that OXA-23 enzymes have become the most prevalent carbapenemases and are now endemic in Italy. In addition, molecular typing profiles showed the presence of international and national clonal lineages in Italy.
INTRODUCTION
Acinetobacter baumannii has emerged in recent years as a leading cause of nosocomial infections, especially in intensive care units (ICUs), becoming a public health problem of major concern in several countries (1). Of note, cases of community-acquired infections are also increasingly reported (2). The ability of the organism to survive in the environment during prolonged periods of time, combined with its innate resistance to desiccation and disinfectants, make A. baumannii difficult to eradicate in the clinical setting (3).
The extensive use of antimicrobial chemotherapy, particularly carbapenems, has contributed to the emergence of carbapenem-resistant A. baumannii (CRAb), which usually exhibits a multidrug-resistant (MDR) phenotype (4). For critically ill patients with MDR infections, therapeutic options are limited, and colistin remains the last resource for treatment (5). Carbapenem resistance is mostly associated with the production of carbapenem-hydrolyzing class D β-lactamases, including the acquired OXA-23, OXA-24, OXA-58, OXA-143, OXA-235, and the intrinsic OXA-51 enzyme (6, 7). The significant contribution of OXA-type carbapenemases in A. baumannii has been emphasized, particularly when blaOXA genes are associated with ISAba sequences, which provide strong promoters for their expression (3). Comparative typing of the outbreak strains of A. baumannii from geographically scattered European hospitals has demonstrated the occurrence of three successful clones originally named European clones I to III, which were renamed as international clones (ICs) I to III, with worldwide distribution. In addition to these major clones, a wide geographic distribution of some other clones has been reported (1). Multilocus sequence typing (MLST) analysis conducted on 496 A. baumannii strains isolated from around the world identified 17 clones, considering both clonal complexes (CCs) and sequence types (STs), distributed also in European countries, with six clones apparently restricted to Europe (8).
In Italy, hospital outbreaks caused by CRAb isolates have been repeatedly reported during the last years. CRAb isolates were found to be mostly related to the production of OXA-58 carbapenemases and belong to IC-II, while strains genetically related to IC-I and IC-III were found to be less common (9–12). More recently, CRAb isolates from Italy were characterized by different STs (e.g., ST1, ST2, ST4, ST20, ST78, ST95, ST109, ST196, and ST197) and sequence groups (SGs) (including SG1, SG2, SG5, and SG6), thus highlighting the presence of international and national clonal lineages in Italy (13–15). A high proportion of carbapenem-resistant Acinetobacter species among the bloodstream isolates in Italy was recently reported by the EARS-Net surveillance system, which has been monitoring Acinetobacter resistance in Europe since 2012 (16).
In this work, we report the results of the first countrywide cross-sectional survey, promoted by the Italian Society of Clinical Microbiologist (AMCLI) and carried out in mid-2011, to investigate the prevalence of CRAb isolates in Italy and to characterize the most common resistance mechanisms. The genetic relatedness of CRAb isolates was also studied in order to trace the epidemiologic evolution of these strains.
MATERIALS AND METHODS
Study design.
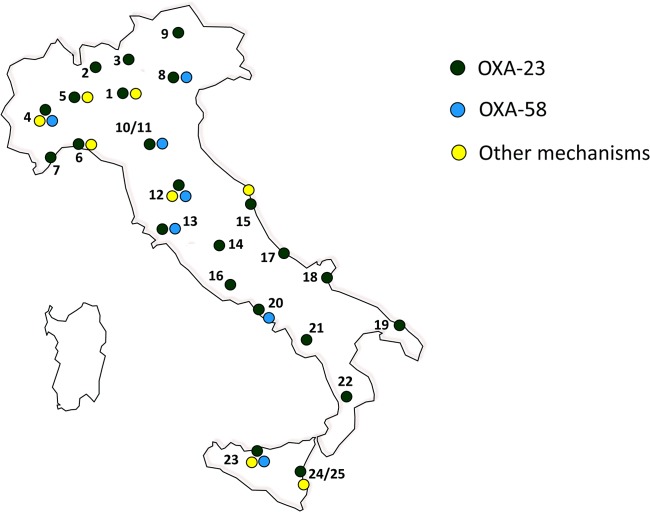
Twenty-five clinical microbiology laboratories from 23 Italian cities, distributed across the national territory, participated in the study (Fig. 1). Each laboratory collected consecutive nonreplicate clinical isolates of A. baumannii that exhibited MICs for imipenem and/or meropenem of ≥8 mg/liter from any type of clinical specimen, with the exception of surface and rectal swabs, during the period of 15 May to 30 June 2011. Both inpatients and outpatients were included in the study. Outpatients were defined as patients not hospitalized at the time of specimen collection. Isolates from patients living in nursing homes were excluded from the analysis. The collected isolates were sent to reference laboratories for confirmation of both species identification and carbapenem MICs, and for characterization of the carbapenem resistance mechanisms and analysis of clonal relatedness. For each isolate, information on the clinical specimen and type of ward (for isolates from inpatients) were provided. Moreover, each participating laboratory provided information on the total number of consecutive nonreplicate clinical isolates of A. baumannii observed during the collection period.
FIG 1.
Location of the laboratories participating in the survey (n = 25). 1, Milan; 2, Varese; 3, Lecco; 4, Turin; 5, Novara; 6, Genoa; 7, Sanremo; 8, Verona; 9, Bolzano; 10 to 11, Modena; 12, Florence; 13, Siena; 14, Perugia; 15, Ancona; 16, Rome; 17, Pescara; 18, San Giovanni Rotondo; 19, Lecce; 20, Naples; 21, Avellino; 22, Cosenza; 23, Palermo; and 24 and 25, Catania. The presence of OXA-23 and/or OXA-58 determinants (as well as other mechanisms) is also indicated. The map was generated using Magic Maps version 1.4.6.
Characterization of bacterial isolates.
Bacterial identification and antimicrobial susceptibility testing were carried out by the collecting laboratories using either the Phoenix automated microbiology system (Becton Dickinson Diagnostic Systems, Sparks, MD, USA) or the Vitek 2 system (bioMérieux, Marcy l'Etoile, France). Confirmatory identification was carried out by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Vitek MS; bioMérieux), followed by the detection of blaOXA-51-like alleles (17). Confirmatory MIC testing for imipenem and meropenem was carried out by Etest (bioMérieux). All collected isolates confirmed to be resistant to imipenem and/or meropenem according to the EUCAST breakpoints (18) were considered to be CRAb isolates for the purpose of this study and were evaluated for the presence of carbapenem resistance mechanisms. For the purpose of antimicrobial susceptibility evaluation, the CRAb MICs of ampicillin-sulbactam, trimethoprim-sulfamethoxazole, ceftazidime, cefepime, imipenem, meropenem, doripenem, amikacin, gentamicin, levofloxacin, colistin, and tigecycline were determined by a reference broth microdilution method using a TREK Sensititre custom panel (TREK Diagnostic Systems, Cleveland, OH). When available, the MIC data were interpreted according to the current EUCAST breakpoints (18). In the case of cefepime, ceftazidime, and ampicillin-sulbactam, the results were evaluated according to CLSI criteria (19). Statistical differences were determined using as the chi-square test, and confidence intervals (CI) were calculated by the Stata statistical software (release 13; College Station, TX, USA) as parameters.
PCR analysis for carbapenem resistance determinants.
PCR assays were carried out by using specific primers to identify blaKPC, blaIMP, blaVIM, blaGIM, blaSIM, blaDIM, blaAIM, blaNDM, blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like, and blaOXA-143-like genes (20–22). The presence of ISAba1 elements adjacent to blaOXA-51-like genes was also investigated, as previously described (23).
Molecular typing by REP-PCR, multiplex PCRs, and MLST.
REP-PCR was carried out using the Acinetobacter kit of the DiversiLab microbial typing system (bioMérieux). The extraction of DNA was performed by the UltraClean microbial DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA). DNA fragment separation and detection were obtained by the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and the results were analyzed and interpreted using the 2100 expert software. Pearson's correlation coefficient for measuring similarity and the unweighted-pair group method using average linkages (UPGMA) for clustering were used. Isolates were defined as genetically related when ≥90% similarity was found between them (24). Representatives of the major European clones I (A. baumannii strain RUH 875) and II (A. baumannii strain RUH 134) were used as controls.
Two multiplex PCRs designed to selectively amplify group 1 or group 2 alleles of the ompA, csuE, and blaOXA-51-like genes were performed. The allelic profiles were determined as described previously (25, 26). Multilocus sequence typing (MLST) of A. baumannii isolates was performed using the primers and conditions described on the Institut Pasteur website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html). Sequence types (STs) were assigned using the same MLST website.
RESULTS
Proportions of CRAb isolates from inpatients and outpatients.
During the study period (15 May to 30 June 2011), a total of 571 consecutive nonreplicate clinical isolates of A. baumannii were isolated at the 25 Italian laboratories participating in the survey (inpatients, n = 508; outpatients, n = 63) (Table 1). Overall, 246 isolates (43.1%) were confirmed as CRAb (inpatients, n = 232; outpatients, n = 14), with the proportion of CRAb organisms being significantly higher among isolates from inpatients (45.7%; 95% CI, 41.4 to 50%; P = 0.38) than among those from outpatients (22.2%; 95% CI, 13.7 to 33.9%; P = 0.0014). CRAb organisms were found in all the 25 participating centers (Fig. 1), although the overall proportions ranged from 9.1 to 100%. The isolates were mostly obtained from the lower respiratory tract, followed by skin and soft tissue, urine, and blood. Of note, blood isolates from inpatients showed the highest proportion of CRAb (54%; 95% CI, 40.4 to 67%; P = 0.17). Concerning the intrahospital distribution, most isolates were obtained from ICUs (109 [47.0%]), whereas the remaining isolates were from medical (86 [37.1%]) and surgical wards (37 [15.9%]) (Table 2). Focusing on CRAb organisms obtained from the lower respiratory tracts of ICU patients, these isolates showed the highest proportion versus other specimen types (56.0%) compared to those obtained from medical and surgical wards (34.9 and 37.8%, respectively).
TABLE 1.
Numbers and proportions of carbapenem-resistant Acinetobacter baumannii isolates obtained from different clinical sources
| Source | Isolates from inpatients |
Isolates from outpatients |
||
|---|---|---|---|---|
| No. | CRAb (no. [%]) | No. | CRAb (no. [%]) | |
| Blood | 50 | 27 (54.0) | 1 | 0 (0.0) |
| Lower respiratory tract | 214 | 105 (49.1) | 10 | 0 (0.0) |
| Urine | 72 | 35 (48.6) | 21 | 7 (33.3) |
| Skin and soft tissue | 112 | 40 (35.7) | 31 | 7 (22.6) |
| Othera | 60 | 25 (41.7) | 0 | 0 (0.0) |
| Total | 508 | 232 (45.7) | 63 | 14 (22.2) |
Other includes vascular catheters, various biological fluids (e.g., pleural, peritoneal, and cerebrospinal fluid), and the upper respiratory tract.
TABLE 2.
Numbers and proportions of carbapenem-resistant A. baumannii isolates obtained from different wardsa
| Source | Medical ward | Surgical ward | ICU |
|---|---|---|---|
| Blood | 13 (15.1/48.2) | 5 (13.5/18.5) | 9 (8.2/33.3) |
| Lower respiratory tract | 30 (34.9/28.6) | 14 (37.8/13.3) | 61 (56.0/58.1) |
| Urine | 20 (23.3/57.1) | 3 (8.1/8.6) | 12 (11.0/34.3) |
| Skin and soft tissue | 21 (24.4/52.5) | 9 (24.3/22.5) | 10 (9.2/25.0) |
| Otherb | 2 (2.3/8.0) | 6 (16.2/24.0) | 17 (15.6/68.0) |
| Total (no. [%]) | 86 (37.1) | 37 (15.9) | 109 (47.0) |
Data are no. (proportion [%] within the specimen type/proportion [%] within the hospitalization ward) (i.e., the ward in which the patients were hospitalized at the time the culture was obtained), unless otherwise stated.
Others includes vascular catheters, various biological fluids (e.g., pleural, peritoneal, and cerebrospinal fluid), and the upper respiratory tract.
Antimicrobial susceptibility of CRAb.
The susceptibility results, including MIC range, MIC50, and MIC90, are summarized in Table 3. Most isolates showed combined resistance to carbapenems, fluoroquinolones, and aminoglycosides (amikacin and gentamicin), while all of them were susceptible to colistin. Of note, a small proportion of CRAb isolates showed an MIC value lower than the CLSI resistance breakpoint for ceftazidime, cefepime, and ampicillin-sulbactam (4.1, 18.3, and 19.9%, respectively). In the case of tigecycline, for which EUCAST or CLSI breakpoints are not available, the MIC50 and MIC90 values were ≤0.5 mg/liter and 1 mg/liter, respectively.
TABLE 3.
Susceptibility results obtained from carbapenem-resistant A. baumannii isolates (n = 246)
| Antibiotic | MIC data (mg/liter)b |
Interpretationa |
|||
|---|---|---|---|---|---|
| Range | MIC50 | MIC90 | % S | % R | |
| Imipenem | 4 to >16 | >16 | >16 | 0 | 95.1 |
| Meropenem | 2 to >64 | 64 | >64 | 1.2 | 96.4 |
| Doripenem | 2 to >8 | >8 | >8 | 0 | 98.0 |
| Amikacin | ≤4 to >16 | >16 | >16 | 2.9 | 97.1 |
| Gentamicin | ≤1 to >4 | >4 | >4 | 5.3 | 94.7 |
| Colistin | ≤0.5 to >2 | ≤0.5 | ≤0.5 | 100 | 0 |
| Levofloxacin | 4 to >4 | >4 | >4 | 0 | 100 |
| Trimethoprim-sulfamethoxazole | ≤0.5/9.5 to >4/76 | >4/76 | >4/76 | 4.1 | 95.1 |
| Ampicillin-sulbactam | ≤8/4 to >32/16 | 32/16 | >32/16 | ||
| Ceftazidime | 4 to >128 | 64 | 128 | ||
| Cefepime | 8 to >32 | 32 | >32 | ||
| Tigecycline | ≤0.12 to 4 | 0.5 | 1 | ||
MICs interpreted according to EUCAST criteria (18). S, susceptible; R, resistant.
Values separated by slashes are the respective concentrations of drugs in the indicated combination.
Carbapenem resistance mechanisms in A. baumannii isolates.
Carbapenemase determinants were detected in the vast majority of CRAb (227/246 [92.3%]) isolates. OXA-23 enzymes (found in all centers) were by far the most common acquired carbapenemases (201/246 [81.7%]), followed by OXA-58 oxacillinases (11/246 [4.5%]), which were detected in 7 of 25 centers (Fig. 1). With respect to the different hospital areas, the proportions of OXA-23 enzymes were 73% in surgical wards, 82.6% in medicine, and 84.5% in ICUs. Overall, no significant differences were observed depending on the ward of hospitalization or specimen type (data not shown). Of note, 6 CRAb isolates carried both blaOXA-23-like and blaOXA-58-like genes. Twenty-one isolates (found in 10 centers equally distributed across the country) were positive for the presence of an ISAba1 genetic element upstream from the resident blaOXA-51-like gene. The mechanism(s) of carbapenem resistance was not identified in 19 of 246 (7.7%) isolates. The carbapenem MIC values for these isolates were as follows: imipenem MIC50 and MIC90, >16 mg/liter (MIC range, 16 to >16 mg/liter), and meropenem MIC50 and MIC90, 32 and 64 mg/liter, respectively (MIC range, 16 to >64 mg/liter).
Genetic relationships among carbapenem-resistant A. baumannii isolates.
Based on different antimicrobial susceptibility profiles, carbapenem resistance determinants, and geographic distribution, 47 CRAb isolates from inpatients were chosen as representatives and characterized by REP-PCR, as well as multiplex PCRs for group identification, together with 8 representative CRAb isolates obtained from outpatients. A subset of isolates was also characterized by MLST.
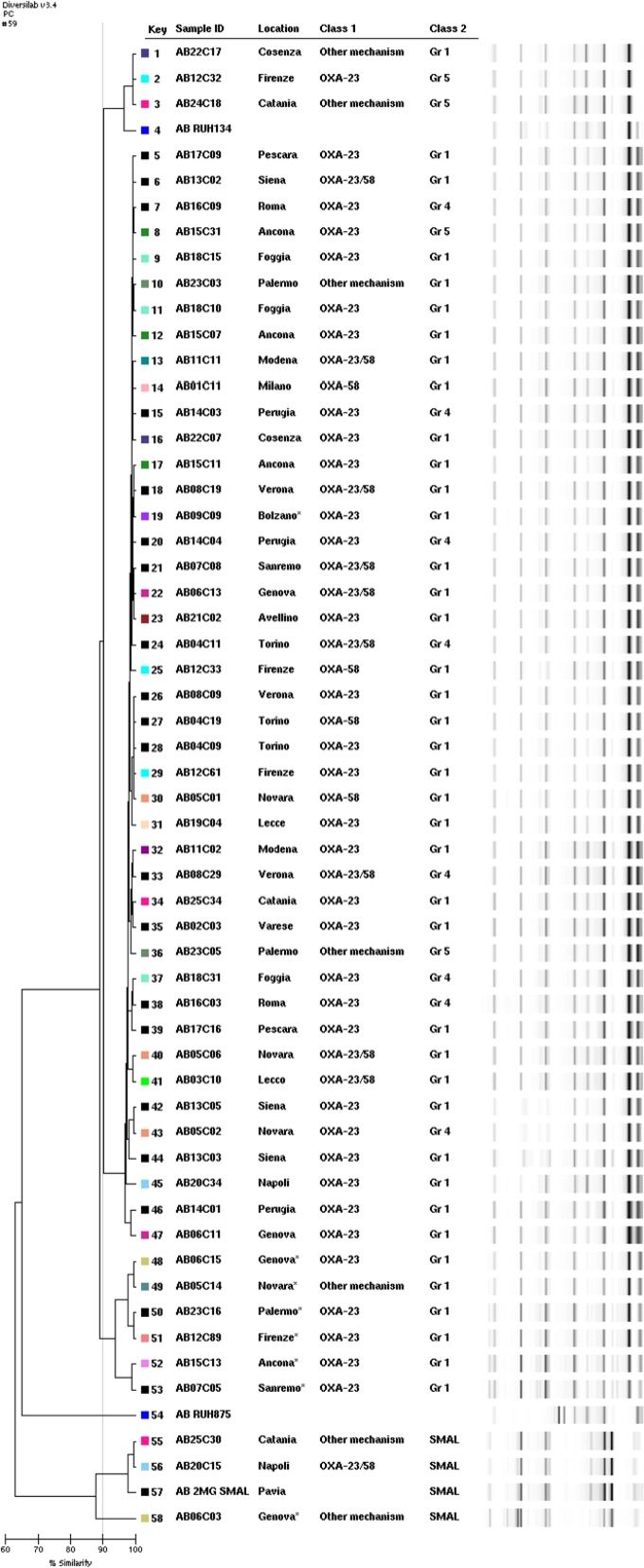
As shown in Fig. 2, molecular typing by REP-PCR revealed the presence of two different clonal lineages, with the dominant clone being related to IC-II. Three major subgroups, however, were distinguishable within the dominant clone when setting the cutoff value to 95% similarity. Interestingly, most outpatients were grouped into a single subgroup within the dominant clone. Multiplex PCRs identified three different sequence groups (SGs), including SG1, SG4, and SG5. MLST revealed that these isolates were consistently from ST2.
FIG 2.
Molecular typing of representative CRAb isolates by REP-PCR (DiversiLab microbial typing system; bioMérieux). The participating centers, sequence groups, and carbapenem resistance determinants are also indicated. Isolates AB09C09, AB06C15, AB05C14, AB23C16, AB12C89, AB15C13, AB07C05, and AB06C03 were obtained from outpatients.
Three isolates obtained from centers located in Naples, Catania, and Genoa (the Genoan isolate was from an outpatient) belonged to a previously described national clonal lineage (27). MLST and SG analyses revealed that these isolates belong to ST78 and SG6. Notably, different carbapenem resistance determinants were detected in these isolates, including the overexpression of blaOXA-51 associated with the ISAba1 genetic element (Catania and Genoa) and the simultaneous presence of blaOXA-23 and blaOXA-58 genes (Naples).
DISCUSSION
This survey was conducted in 25 Italian centers distributed across the country in order to take an overall picture of A. baumannii in both hospitalized and community patients. All clinical isolates identified as A. baumannii during the survey were included in the study and investigated to evaluate both epidemiologic features and antibiotic resistance phenotypes; however, for specimens from nonsterile sites, we could not distinguish between infection and colonization based on the clinical data provided by participating centers. Focusing on carbapenems, CRAb isolates comprised 45.7% of all A. baumannii isolates from Italian hospitals during the survey. Similarly, a recent study (2008 to 2009) in 16 countries (14 of which are in Europe) reported an overall rate of imipenem-resistant strains of 48.9% (28). A lower rate (34%) was observed from the National Healthcare Safety Network (NHSN) surveillance system from U.S. hospitals between 2006 and 2008 (29). In another U.S. survey, however, nonsusceptibility to carbapenems increased from 22% in 2002 to 52% in 2008 (30). In 2012, data on Acinetobacter spp. were included for the first time in the EARS-Net surveillance report (16), showing a high percentage of CRAb (83.3%; 95% CI, 78 to 88%) isolates, as well as combined resistance to multiple antibiotics (78%) for the Italian centers. That report, however, analyzed only invasive isolates recovered from hospitalized patients. In our survey, carried out during 2011 in different centers, the proportion of CRAb organisms among blood isolates was lower (54%) than that reported in the EARS-Net data. Furthermore, due to a limited number of invasive isolates, our data showed a broad confidence interval (95% CI, 40.4 to 67%) that might account for this difference. It is to be noted that colistin fully maintained its activity against CRAb isolates, and tigecycline was characterized by low MIC50 and MIC90 values (0.5 and 1 mg/liter, respectively).
From an epidemiological point of view, our data demonstrate that similarly to carbapenem-resistant Enterobacteriaceae (CRE), CRAb organisms are commonly found in Italy, with a widespread distribution across the national territory. To date, no guidelines have been issued in Italy to prevent Acinetobacter infections, but the present findings suggest to implement them as soon as possible. Compared with the results obtained by the CRE nationwide surveillance (carried out simultaneously in the same Italian centers), CRAb organisms appear to have a hospital diffusion comparable to that of carbapenem-nonsusceptible Klebsiella pneumoniae isolates (232 versus 234, respectively) (31). Notably, a high number of CRAb isolates were from the lower respiratory tracts of ICU patients, whereas blood isolates accounted for a number lower than the CREs (27 versus 40, respectively). Similarly to CREs, however, CRAb isolates were not restricted to ICU settings but affected all major hospital areas, a finding that should be carefully considered when planning infection control strategies. Overall, similar numbers of carbapenem-resistant isolates were found among outpatients (14 CRAb versus 21 CRE isolates). However, A. baumannii obtained from outpatients was mainly from skin and soft tissue specimens, but K. pneumoniae was mostly from urine specimens.
In this nationwide survey, carbapenem resistance in A. baumannii was primarily associated with the OXA-23 enzyme (found in all centers), which was the most commonly acquired carbapenemase. On the contrary, OXA-58 oxacillinases, previously reported to be prevalent in Italy (10, 13, 14), were detected in few centers even though they were distributed across the country. Taken together, our results agree with the described worldwide dissemination of the blaOXA-23-like genes (32) and highlight the recent evolution of CRAb organisms in Italy, with isolates belonging to IC-II now almost exclusively carrying the blaOXA-23 determinant. A progressive change from blaOXA-58 to blaOXA-23 gene carriage had been already observed among IC-II CRAb isolates responsible for ICU outbreaks in the main hospitals in central Italy, with A. baumannii strains producing OXA-23 appearing in 2007 (33).
In our survey, the coexistence of blaOXA-23-like and blaOXA-58-like genes was found in 6 isolates obtained from inpatients hospitalized at 5 different centers. A similar finding was previously reported (14, 34), but the potential clinical implications remain unknown, and further investigations are needed to clarify this point.
CRAb isolates in most cases were found to belong to IC-II, whereas other clonal lineages were sporadically detected. This picture appears to be the most common in both the United States and Europe (8, 35). Molecular epidemiology surveys conducted in the United States on CRAb isolates obtained from 1995 to 2008 showed that IC-II isolates had spread to many centers as early as 1996 and became the dominant lineage (35, 36). Interestingly, strains assigned to IC-II are known to form biofilms and adhere to abiotic surfaces more efficiently than strains assigned to IC-I (mainly characterized by elevated resistance to desiccation). The association of biofilm formation with the ability to acquire different antimicrobial resistance determinants might have favored their spread and persistence in the hospital environment (37).
In Italy, the IC-II appears to be widely distributed since 2004, even though different clones and STs have often been described (13–15). Noteworthy, a single clone of CRAb belonging to the IC-II/ST2 lineage was most recently reported in an ICU in Palermo, Sicily (11). Our results demonstrate that this evolution occurred at a countrywide level, with the ST2 clone able to disseminate in Italian hospitals. Similarly, a high predominance of the ST2 clone was recently reported in several European countries, including Poland, Romania, Latvia, and the Czech Republic (38–41). In these cases, however, interesting differences were observed among countries with respect to the associated carbapenem resistance determinants, thus demonstrating a multidirectional evolution of the IC-II/ST2 clonal lineage. It is worth noting that most outpatients were grouped into a single subgroup within the dominant clone. This finding, together with the low rate of carbapenem resistance, seems to confirm the different origins of these isolates.
Three A. baumannii isolates belonged to the ST78 clone, previously known as the SMAL clone or the Italian clone, and are now assigned to the worldwide clone 6 (1, 42). The ST78 genotype was first reported from Naples in 2006 and then detected in several Italian hospitals, as well as in other Mediterranean countries (34). Similarly to ST2, ST78 strains have been shown to have a higher adherence to both biotic and antibiotic surfaces, with ST78 also showing an elevated resistance to desiccation (37, 42). Our survey demonstrates that ST78 strains acquired different resistance determinants, which may contribute to the persistence of this genotype in Italian hospitals.
In conclusion, our data indicate that resistance to carbapenems is more and more common among A. baumannii isolates, and they demonstrate that the OXA-23 enzyme has become the most prevalent carbapenemase, which is now endemic in Italy. This further evolution of carbapenem-resistant A. baumannii strains confirms the need to continuously monitor this worrisome bacterial pathogen.
ACKNOWLEDGMENTS
We thank bioMérieux and Becton Dickinson for their contributions in supporting the transport of isolates from the collecting laboratories to the reference laboratories.
The participants in the AMCLI-CRAb Survey (the first Italian survey on carbapenem-resistant A. baumannii, promoted by the Italian Society of Clinical Microbiologists) were G. Gesu (Niguarda Ca' Granda Hospital, Milan), A. Toniolo (Circolo and Fondazione Macchi University Hospital, Varese), R. Serra (San Giovanni Battista [Molinette] Hospital, Turin), B. Pini (Alessandro Manzoni Hospital, Lecco), G. Fortina (Maggiore Hospital, Novara), M. Mori (Galliera Hospital, Genoa), P. A. Dusi (Azienda Sanitaria Imperiese Hospital, Sanremo), R. Fontana (Borgo Roma University Hospital, Verona), R. Aschbacher (Azienda Sanitaria dell'Alto Adige, Bolzano), M. Sarti (Sant'Agostino Estense Hospital, Modena), F. Rumpianesi (Modena University Hospital, Modena), P. Pecile (Careggi University Hospital, Florence), G. M. Rossolini (Santa Maria alle Scotte University Hospital, Siena), A. Mencacci (Santa Maria della Misericordia University Hospital, Perugia), E. Manso (Torrette University Hospital, Ancona), M. Tronci (San Camillo Forlanini Hospital, Rome), P. Fazii (Santo Spirito Hospital, Pescara), M. Labonia (IRCCS Casa Sollievo della Sofferenza Hospital, San Giovanni Rotondo), M. Pizzolante (V. Fazzi Hospital, Lecce), G. Amato (A. Cardarelli Hospital, Naples), P. Buonopane (San Giuseppe Moscati Hospital, Avellino), C. Giraldi (Annunziata Hospital, Cosenza), P. G. Conaldi (Mediterranean Institute of Transplantation [ISMETT], Palermo), and S. Stefani (Policlinico and Vittorio Emanuele University Hospitals, Catania).
Footnotes
Published ahead of print 11 June 2014
REFERENCES
- 1.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents 41:11–19. 10.1016/j.ijantimicag.2012.09.008 [DOI] [PubMed] [Google Scholar]
- 2.Eveillard M, Kempf M, Belmonte O, Pailhoriès H, Joly-Guillou ML. 2013. Reservoirs of Acinetobacter baumannii outside the hospital and potential involvement in emerging human community-acquired infections. Int. J. Infect. Dis. 17:802–805. 10.1016/j.ijid.2013.03.021 [DOI] [PubMed] [Google Scholar]
- 3.Roca I, Espinal P, Vila-Farrés X, Vila J. 2012. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 3:148. 10.3389/fmicb.2012.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3471–3484. 10.1128/AAC.01464-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kempf M, Rolain J-M. 2012. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int. J. Antimicrob. Agents 39:105–114. 10.1016/j.ijantimicag.2011.10.004 [DOI] [PubMed] [Google Scholar]
- 6.Patel G, Bonomo RA. 2013. “Stormy waters ahead”: global emergence of carbapenemases. Front. Microbiol. 4:48. 10.3389/fmicb.2013.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins PG, Pérez-Llarena FJ, Zander E, Fernández A, Bou G, Seifert H. 2013. OXA-235, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 57:2121–2126. 10.1128/AAC.02413-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karah N, Sundsfjord A, Towner K, Samuelsen Ø. 2012. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updat. 15:237–247. 10.1016/j.drup.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 9.Mendes RE, Spanu T, Deshpande L, Castanheira M, Jones RN, Fadda G. 2009. Clonal dissemination of two clusters of Acinetobacter baumannii producing OXA-23 or OXA-58 in Rome, Italy. Clin. Microbiol. Infect. 15:588–592. 10.1111/j.1469-0691.2009.02770.x [DOI] [PubMed] [Google Scholar]
- 10.D'Arezzo S, Principe L, Capone A, Petrosillo N, Petrucca A, Visca P. 2011. Changing carbapenemase gene pattern in an epidemic multidrug-resistant Acinetobacter baumannii lineage causing multiple outbreaks in central Italy. J. Antimicrob. Chemother. 66:54–61. 10.1093/jac/dkq407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mammina C, Palma DM, Bonura C, Aleo A, Fasciana T, Sodano C, Saporito MA, Verde MS, Calà C, Cracchiolo AN, Tetamo R. 2012. Epidemiology and clonality of carbapenem-resistant Acinetobacter baumannii from an intensive care unit in Palermo, Italy. BMC Res. Notes 5:365. 10.1186/1756-0500-5-365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brigante G, Migliavacca R, Bramati S, Motta E, Nucleo E, Manenti M, Migliorino G, Pagani L, Luzzaro F, Viganò FE. 2012. Emergence and spread of a multidrug-resistant Acinetobacter baumannii clone producing both the carbapenemase OXA-23 and the 16S rRNA methylase ArmA. J. Med. Microbiol. 61:653–661. 10.1099/jmm.0.040980-0 [DOI] [PubMed] [Google Scholar]
- 13.Migliavacca R, Espinal P, Principe L, Drago M, Fugazza G, Roca I, Nucleo E, Bracco S, Vila J, Pagani L, Luzzaro F. 2013. Characterization of resistance mechanisms and genetic relatedness of carbapenem-resistant Acinetobacter baumannii isolated from blood, Italy. Diagn. Microbiol. Infect. Dis. 75:180–186. 10.1016/j.diagmicrobio.2012.11.002 [DOI] [PubMed] [Google Scholar]
- 14.Mezzatesta ML, D'Andrea MM, Migliavacca R, Giani T, Gona F, Nucleo E, Fugazza G, Pagani L, Rossolini GM, Stefani S. 2012. Epidemiological characterization and distribution of carbapenem-resistant Acinetobacter baumannii clinical isolates in Italy. Clin. Microbiol. Infect. 18:160–166. 10.1111/j.1469-0691.2011.03527.x [DOI] [PubMed] [Google Scholar]
- 15.Carretto E, Barbarini D, Dijkshoorn L, van der Reijden TJ, Brisse S, Passet V, Farina C, ASPI Acinetobacter Study Group 2011. Widespread carbapenem resistant Acinetobacter baumannii clones in Italian hospitals revealed by a multicenter study. Infect. Genet. Evol. 11:1319–1326. 10.1016/j.meegid.2011.04.024 [DOI] [PubMed] [Google Scholar]
- 16.European Centre for Disease Prevention and Control. 2013. Surveillance report: antimicrobial resistance surveillance in Europe 2012.Annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net)ECDC, Stockholm, Sweden: http://ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-surveillance-europe-2012.pdf [Google Scholar]
- 17.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44:2974–2976. 10.1128/JCM.01021-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.European Committee on Antimicrobial Susceptibility Testing. 2014. Breakpoint tables for interpretation of MICs and zone diameters, version 4.0. EUCAST, Basel, Switzerland: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_4.0.pdf [Google Scholar]
- 19.CLSI. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI document M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 20.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 21.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351–353. 10.1016/j.ijantimicag.2006.01.004 [DOI] [PubMed] [Google Scholar]
- 22.Higgins PG, Lehmann M, Seifert H. 2010. Inclusion of OXA-143 primers in a multiplex polymerase chain reaction (PCR) for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 35:305. 10.1016/j.ijantimicag.2009.10.014 [DOI] [PubMed] [Google Scholar]
- 23.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, Pitt TL. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72–77. 10.1111/j.1574-6968.2006.00195.x [DOI] [PubMed] [Google Scholar]
- 24.Saeed S, Fakih MG, Riederer K, Shah AR, Khatib R. 2006. Interinstitutional and intrainstitutional transmission of a strain of Acinetobacter baumannii detected by molecular analysis: comparison of pulsed-field gel electrophoresis and repetitive sequence-based polymerase chain reaction. Infect. Control Hosp. Epidemiol. 27:981–983. 10.1086/507286 [DOI] [PubMed] [Google Scholar]
- 25.Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807–815. 10.1111/j.1469-0691.2007.01759.x [DOI] [PubMed] [Google Scholar]
- 26.Towner KJ, Levi K, Vlassiadi M, ARPAC Steering Group 2008. Genetic diversity of carbapenem-resistant isolates of Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 14:161–167. 10.1111/j.1469-0691.2007.01911.x [DOI] [PubMed] [Google Scholar]
- 27.Giannouli M, Cuccurullo S, Crivaro V, Di Popolo A, Bernardo M, Tomasone F, Amato G, Brisse S, Triassi M, Utili R, Zarrilli R. 2010. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii in a tertiary care hospital in Naples Italy, shows the emergence of a novel epidemic clone. J. Clin. Microbiol. 48:1223–1230. 10.1128/JCM.02263-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nordmann P, Picazo JJ, Mutters R, Korten V, Quintana A, Laeuffer JM, Seak JC, Flamm RK, Morrissey I, COMPACT Study Group 2011. Comparative activity of carbapenem testing: the COMPACT Study. J. Antimicrob. Chemother. 66:1070–1078. 10.1093/jac/dkr056 [DOI] [PubMed] [Google Scholar]
- 29.Kallen AJ, Srinivasan A. 2010. Current epidemiology of multidrug-resistant Gram-negative bacilli in the United States. Infect. Control Hosp. Epidemiol. 31(Suppl 1):S51–S54. 10.1086/655996 [DOI] [PubMed] [Google Scholar]
- 30.Mera RM, Miller LA, Amrine-Madsen EH, Sahm DF. 2010. Acinetobacter baumannii 2002–2008: increase of carbapenem-associated multiclass resistance in the United States. Microb. Drug Resist. 16:209–215. 10.1089/mdr.2010.0052 [DOI] [PubMed] [Google Scholar]
- 31.Giani T, Pini B, Arena F, Conte V, Bracco S, Migliavacca R, AMCLI-CRE Survey Participants. Pantosti A, Pagani L, Luzzaro F, Rossolini GM. 2013. Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro Surveill. 18:pii=20489 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20489 [PubMed] [Google Scholar]
- 32.Mugnier PD, Poirel L, Naas T, Nordmann P. 2010. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 16:35–40. 10.3201/eid1601.090852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Popolo A, Giannouli M, Triassi M, Brisse S, Zarrilli R. 2011. Molecular epidemiological investigation of multidrug-resistant Acinetobacter baumannii strains in four Mediterranean countries with a multilocus sequence typing scheme. Clin. Microbiol. Infect. 17:197–201. 10.1111/j.1469-0691.2010.03254.x [DOI] [PubMed] [Google Scholar]
- 34.Higgins PG, Dammhayn C, Hackel M, Seifert H. 2010. Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65:233–238. 10.1093/jac/dkp428 [DOI] [PubMed] [Google Scholar]
- 35.Adams-Haduch JM, Onuoha EO, Bogdanovich T, Tian GB, Marschall J, Urban CM, Spellberg BJ, Rhee D, Halstead DC, Pasculle AW, Doi Y. 2011. Molecular epidemiology of carbapenem-nonsusceptible Acinetobacter baumannii in the United States. J. Clin. Microbiol. 49:3849-3854. 10.1128/JCM.00619-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higgins PG, Janßen K, Fresen MM, Wisplinghoff H, Seifert H. 2012. Molecular epidemiology of Acinetobacter baumannii bloodstream isolates obtained in the United States from 1995 to 2004 using rep-PCR and multilocus sequence typing. J. Clin. Microbiol. 50:3493–3500. 10.1128/JCM.01759-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giannouli M, Antunes LCS, Marchetti V, Triassi M, Visca P, Zarrilli R. 2013. Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I–III and to emerging phenotypes ST25 and ST78. BMC Infect. Dis. 13:282. 10.1186/1471-2334-13-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izdebski R, Fiett J, Hryniewicz W, Gniadkowski M. 2012. Molecular analysis of Acinetobacter baumannii isolates from invasive infections in 2009 in Poland. J. Clin. Microbiol. 50:3813–3815. 10.1128/JCM.02271-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonnin RA, Poirel L, Licker M, Nordmann P. 2011. Genetic diversity of carbapenem-hydrolysing β-lactamases in Acinetobacter baumannii from Romanian hospitals. Clin. Microbiol. Infect. 17:1514–1530. 10.1111/j.1469-0691.2011.03539.x [DOI] [PubMed] [Google Scholar]
- 40.Saule M, Samuelsen Ø, Dumpis U, Sundsfjord A, Karlsone A, Balode A, Miklasevics E, Karah N. 2012. Dissemination of a carbapenem-resistant Acinetobacter baumannii strain belonging to international clone II/sequence type 2 and harboring a novel AbaR4-like resistance island in Latvia. Antimicrob. Agents Chemother. 57:1069–1072. 10.1128/AAC.01783-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nemec A, Krízová L, Maixnerová M, Diancourt L, van der Reijden TJ, Brisse S, van der Broek P, Dijkshoorn L. 2008. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug-resistant strains of European clone II. J. Antimicrob. Chemother. 62:484–489. 10.1093/jac/dkn205 [DOI] [PubMed] [Google Scholar]
- 42.Nucleo E, Steffanoni L, Fugazza G, Migliavacca R, Giacobone E, Navarra A, Pagani L, Landini P. 2009. Growth in glucose-based medium and exposure to subinhibitory concentrations of imipenem induce biofilm formation in a multidrug-resistant clinical isolate of Acinetobacter baumannii. BMC Microbiol. 9:270–274. 10.1186/1471-2180-9-270 [DOI] [PMC free article] [PubMed] [Google Scholar]