Abstract
In this study, we developed a new rapid, economic, and automated microarray-based genotyping test for the standardized subtyping of Shiga toxins 1 and 2 of Escherichia coli. The microarrays from Alere Technologies can be used in two different formats, the ArrayTube and the ArrayStrip (which enables high-throughput testing in a 96-well format). One microarray chip harbors all the gene sequences necessary to distinguish between all Stx subtypes, facilitating the identification of single and multiple subtypes within a single isolate in one experiment. Specific software was developed to automatically analyze all data obtained from the microarray. The assay was validated with 21 Shiga toxin-producing E. coli (STEC) reference strains that were previously tested by the complete set of conventional subtyping PCRs. The microarray results showed 100% concordance with the PCR results. Essentially identical results were detected when the standard DNA extraction method was replaced by a time-saving heat lysis protocol. For further validation of the microarray, we identified the Stx subtypes or combinations of the subtypes in 446 STEC field isolates of human and animal origin. In summary, this oligonucleotide array represents an excellent diagnostic tool that provides some advantages over standard PCR-based subtyping. The number of the spotted probes on the microarrays can be increased by additional probes, such as for novel alleles, species markers, or resistance genes, should the need arise.
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) organisms are an important causative agent of diarrhea, hemorrhagic colitis (HC), and hemolytic-uremic syndrome (HUS), and they are the most common cause of acute renal failure in children (1–5). Clinical STEC isolates that resemble (with regard to clinical, epidemiological, and pathogenetic characteristics) the prototype strain E. coli O157:H7 are also designated enterohemorrhagic E. coli (EHEC) (6). EHEC O157:H7 is one of the predominant STEC serovars responsible for several food-transmitted outbreaks and epidemics all over the world (7, 8). However, non-O157 STEC serogroups, such as O26, O103, O111, and O145, contribute significantly to cases of HC and HUS (4). In early summer 2011, a massive outbreak in Germany caused by a hitherto rare EHEC serotype O104:H4 (9–12) was reported. This outbreak led to >3,800 known cases, including 855 patients with HUS and 53 deaths (13), and it was the largest outbreak of HUS in recorded history. The most likely source for the outbreak was contaminated fenugreek sprouts (9).
Major virulence factors of STEC that contribute to pathogenesis include the phage-encoded Shiga toxins (Stx), also known as Vero toxin or verocytotoxins (VT). The Stx toxins are typical AB5 toxins with an operon structure (i.e., the StxA subunit is immediately upstream of the StxB subunit, with a short intergenic sequence). The enzymatic N-glycosidase activity of the StxA subunit is responsible for the cytotoxicity by cleavage of adenine residues in the rRNA of host cell ribosomes, causing the inhibition of protein biosynthesis and cell death (14, 15). The five StxB subunits are able to bind to specific glycolipid receptors. Shiga toxins show a remarkable degree of sequence variation. Within the Stx family, two major types are distinguished based on their antigenic differences: Stx1, which is nearly identical to Stx from Shigella dysenteriae type 1, and Stx2. The Stx1 and Stx2 alleles do not display DNA-DNA cross hybridization under conditions of high stringency. In the past, many subtypes and toxin variants have been described in either branch. Several authors (16–18) have shown that some subtypes or variants of Stx2 appear to be clearly associated with serious sequelae of EHEC infection, namely, with HC or HUS, while other subtypes or variants of Stx1 and Stx2 are mainly associated with a milder course of disease. Nevertheless, a previous lack of uniform guidelines for defining and naming subtypes and the significant diversity among sequences within the main families has caused much confusion (19). Consistent nomenclature and subtyping strategies are essential for surveillance and for predicting the risks associated with particular STEC infections. Scheutz et al. (19) developed a molecular protocol for the detection and subtyping of both Stx1 and Stx2 using a set of defined PCRs and created a standardized nomenclature for the Shiga toxins and their subtypes. Thus, Stx1 is divided into three subtypes, Stx1a, Stx1c, and Stx1d. The more heterogeneous Stx2 is grouped into seven subtypes, designated Stx2a, Stx2b, Stx2c, Stx2d, Stx2e, Stx2f, and Stx2g (19).
The aim of the study was to develop a new, rapid, economic, and automated microarray-based genotyping test for standardized subtyping of Shiga toxins 1 and 2 of E. coli, based on the nomenclature described by Scheutz et al. (19). The primers and probes were designed and processed by biomathematical methods. The microarray was established based on the ArrayStrip format by Alere Technologies GmbH (Jena, Germany), which allows high-throughput tests in a 96-well format as well as fully automated microarray imaging analysis and Stx subgroup assignment. Moreover, the test protocol was optimized. Subsequently, we verified the performance of the microarray by testing STEC reference and field strains of human and animal origin.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and genomic DNA extraction.
The microarray was validated with a set of 21 reference strains from 7 different reference laboratories that were previously tested by the complete set of conventional subtyping PCRs described by Scheutz et al. (19) (Table 1). Additionally, 446 STEC strains of human and animal origin were analyzed to determine their Stx subtype using the newly developed test.
TABLE 1.
Comparison of the results of the PCR described by Scheutz et al. (19) and the microarray results of selected reference strains
| Reference strain | Results of microarray | Results of PCR for stx1 |
Results of PCR for stx2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Stx1a | Stx1c | Stx1d | Stx2a | Stx2b | Stx2c | Stx2d | Stx2e | Stx2f | Stx2g | ||
| CB168_120310a | Stx1a | + | − | − | − | − | − | − | − | − | − |
| PT199_120310a | Stx1c | − | + | − | − | − | − | − | − | − | − |
| DSM15856b | Stx1d | − | − | + | − | − | − | − | − | − | − |
| EDL933 (ATCC 700927)c | Stx1a, Stx2a | + | − | − | + | − | − | − | − | − | − |
| 1760/98 (O129:H-)c | Stx1a, Stx2b | + | − | − | − | + | − | − | − | − | − |
| WH-01/26/021-5d | Stx1a, Stx2c | + | − | − | − | − | + | − | − | − | − |
| DD_1380/99_156_#311c | Stx1a, Stx2d | + | − | − | − | − | − | + | − | − | − |
| 12 (O128:H2)e | Stx1c, Stx2b | − | + | − | − | + | − | − | − | − | − |
| 13 (O172:H-)e | Stx1a, Stx2a, Stx2c | + | − | − | + | − | + | − | − | − | − |
| 17 (O8:H20)e | Stx1a, Stx2a, Stx2d | + | − | − | + | − | − | + | − | − | − |
| CB5805a | Stx2a | − | − | − | + | − | − | − | − | − | − |
| 20 (O146:H28)e | Stx2b | − | − | − | − | + | − | − | − | − | − |
| 02_23877 (O157)f | Stx2c | − | − | − | − | − | + | − | − | − | − |
| CB12533a | Stx2d | − | − | − | − | − | − | + | − | − | − |
| 11 (O101:H-)e | Stx2e | − | − | − | − | − | − | − | + | − | − |
| T4-97g | Stx2f | − | − | − | − | − | − | − | − | + | − |
| 7Vg | Stx2g | − | − | − | − | − | − | − | − | − | + |
| 01_23209 (O157)f | Stx2a, Stx2c | − | − | − | + | − | + | − | − | − | − |
| 45_28154 (O174)f | Stx2a, Stx2d | − | − | − | + | − | − | + | − | − | − |
| 43 (O116:H-)e | Stx2a, Stx2g | − | − | − | + | − | − | − | − | − | + |
| 21 (O2:H29)e | Stx2b, Stx2c | − | − | − | − | + | + | − | − | − | − |
L. Beutin, Federal Institute for Risk Assessment, Berlin, Germany.
Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany.
F. Gunzer, Institute for Medical Microbiology and Hygiene, Dresden, Germany.
L. Geue, Friedrich Loeffler Institute, Jena, Germany.
H. Hächler, Institute for Food Safety and Food Hygiene, Zürich, Switzerland.
E. Bingen, Service de Microbiology, Paris, France.
H. Karch/M. Bielaszewska, Institute for Hygiene, Münster, Germany.
The strains were cultivated on tryptone yeast agar or blood agar (VWR International GmbH, Darmstadt, Germany). A full 1-μl loop (diameter, 1 mm) of the clonal colony material of each strain was picked from solid medium, resuspended in 200 μl lysis reagents (lysis enhancer A2 dissolved in 200 μl lysis buffer A1; Alere Technologies GmbH, Jena, Germany), and incubated for 30 to 60 min at 37°C and 550 rpm in a thermomixing device (Eppendorf GmbH, Hamburg, Germany). RNA-free unfragmented genomic DNA was extracted with the Qiagen DNeasy blood and tissue kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's instructions. The DNA concentration was determined spectrophotometrically at 260 nm and finally analyzed for fragmentation by use of electrophoresis in a 1% nondenaturing agarose gel.
Heat lysis protocol for DNA extraction.
In order to reduce costs and save time, a fast and robust heat lysis protocol was developed. Fifty microliters of molecular-grade water was used to homogenize a loop full (1-μl loop size) of fresh clonal bacterial culture directly harvested from a tryptone yeast agar or blood agar plate (VWR International GmbH, Darmstadt, Germany). The mixture was heated in an Eppendorf Safe-Lock Tube for 15 min at 99°C in an Eppendorf ThermoShaker (Eppendorf, Hamburg, Germany). After centrifugation (for 5 min at 16,000 × g), 25 μl supernatant was used for subsequent RNase A treatment (for 5 min at 37°C), with a final concentration of 200 μg/μl RNase (Qiagen GmbH, Hilden, Germany). Five microliters of recovered genomic DNA was used directly for the internal biotin-labeling procedure and subsequently for hybridization.
Multiplex linear DNA amplification and labeling.
For multiplex linear DNA amplification, a set of 46 primers was used. The primers are shown in Table 2. These oligonucleotides are located in a nonoverlapping but as close as possible fashion on the complementary strand, upstream of the position of the oligonucleotide probe with which it is covalently coupled on the microarray. For labeling and biotinylation of the genomic DNA, a site-specific labeling approach was used (20) (see Fig. S1 in the supplemental material). The primer elongation reaction was performed using the primer mixture, the HybPlus kit (Alere Technologies, Jena, Germany), and 0.5 to 1.5 μg unfragmented RNA-free genomic DNA of the E. coli isolates, according to the manufacturer's instructions. The reaction was started with a denaturation step (for 5 min at 96°C). Next, 50 cycles of 60 s at 96°C, 20 s at 50°C, and 40 s at 72°C were performed. The sample was finally cooled down to 4°C.
TABLE 2.
Primers for internal labeling with biotin-dUTP
| Primer no. | Primer name | Species | GenBank accession no. (defined DNA sequence positions within the reference sequences) |
|---|---|---|---|
| 1 | gad_11 | Escherichia coli | AAJT02000001.1 (76948–76969-r) |
| 2 | ipaH9.8_21 | E. coli | AAKB02000001.1 (5059278–5059295-r) |
| 3 | lacY_04 | E. coli | AAMK02000012.1 (123534–123550-r) |
| 4 | lb_basC_721 | Acinetobacter baumannii | ACYR02000085.1 (5172–5192) |
| 5 | lb_basC_722 | A. baumannii | ABXK01000012.1 (117503–117524-r) |
| 6 | lb_cfa_741 | Citrobacter freundii | ANAV01000025.1 (14988–15008) |
| 7 | lb_cfa_742 | C. freundii | U09771.1 (65–84-r) |
| 8 | lb_dnaE_651_rv | E. coli | AAJW02000003.1 (64127–64144-r) |
| 9 | lb_dnaE_652_rv | E. coli | AAMK02000042.1 (8271–8287-r) |
| 10 | lb_ecfX_371 | Pseudomonas aeruginosa | DQ996551.1 (44–60-r) |
| 11 | lb_ecfX_372 | P. aeruginosa | DQ996551.1 (511–527-r) |
| 12 | lb_efp_301 | A. baumannii | CU468230.2 (1055312–1055330-r) |
| 13 | lb_efp_302 | A. baumannii | AMST01000070.1 (255183–255206-r) |
| 14 | lb_khe_431 | Klebsiella pneumoniae | AF293352.1 (323–343-r) |
| 15 | lb_pld_381 | A. baumannii | CP000863.1 (3397120–3397142-r) |
| 16 | lb_pld_382 | A. baumannii | CP000521.1 (3478048–3478071-r) |
| 17 | lb_rrs_651 | E. coli | U70214.1 (55203–55222-r) |
| 18 | lb_stx1_g10n52 | E. coli | AY170851.1 (274–294-r) |
| 19 | lb_stx1_g20n51 | E. coli | AM230663.1 (566–584-r) |
| 20 | lb_stx1_g30n53 | E. coli | Z36901.1 (996–1014-r) |
| 21 | lb_stx1_g30n54 | E. coli | AM230663.1 (962–980-r) |
| 22 | lb_stx1_g30n55 | E. coli | AY170851.1 (851–869-r) |
| 23 | lb_stx1_g40n56 | E. coli | Z36901.1 (1128–1152-r) |
| 24 | lb_stx1_g40n57 | E. coli | AM230663.1 (1093–1114-r) |
| 25 | lb_stx1_g40n58 | E. coli | AY170851.1 (988–1011-r) |
| 26 | lb_stx1_g50n59 | E. coli | AM230663.1 (1150–1169-r) |
| 27 | lb_stx2_g11n01 | E. coli | FM998838.1 (230–250-r) |
| 28 | lb_stx2_g11n02 | E. coli | AB048236.1 (222–241-r) |
| 29 | lb_stx2_g21n01 | E. coli | AB472687.1 (468–488-r) |
| 30 | lb_stx2_g22n01 | E. coli | FM998851.1 (471–491-r) |
| 31 | lb_stx2_g23n01 | E. coli | AB472687.1 (523–541-r) |
| 32 | lb_stx2_g23n02 | E. coli | AY443052.1 (659–678-r) |
| 33 | lb_stx2_g44n01 | E. coli | AY443052.1 (1101–1124-r) |
| 34 | lb_stx2_g44n02 | E. coli | L11078.1 (1156–1179-r) |
| 35 | lb_stx2_g46n01 | E. coli | AB472687.1 (1030–1047-r) |
| 36 | lb_stx2_g51n01 | E. coli | AY443052.1 (1235–1250-r) |
| 37 | lb_stx2_g51n02 | E. coli | L11078.1 (1288–1304-r) |
| 38 | lb_stx2_g51n03 | E. coli | FM998838.1 (1111–1129-r) |
| 39 | lb_stx2_g63n01 | E. coli | AY443052.1 (1331–1349-r) |
| 40 | lb-2156-manC | Salmonella enterica | AE014613.1 (857318–857336-r) |
| 41 | lb-2278-galF | S. enterica | AM933173.1 (2174904–2174924) |
| 42 | lb-2291-invA | S. enterica | CP000880.1 (74624–74646-r) |
| 43 | lb-3317-invA | Salmonella bongori | FR877557.1 (2759648–2759674) |
| 44 | lb-3319-invA | S. enterica | CP000880.1 (74907–74932-r) |
| 45 | prim_gapA_651_rv | E. coli | CU928164.2 (1341195–1341215-r) |
| 46 | prim_ihfA_651_rv | E. coli | CU928164.2 (1407630–1407654-r) |
Hybridization of the ArrayStrips.
The ArrayStrips were manufactured using the probes listed in Table 3. For hybridization, the HybPlus kit was used with an adapted and optimized protocol. Each ArrayStrip was initially washed with 200 μl double-distilled water and then with 150 μl of buffer C1 using a thermomixing device, the BioShake iQ (QInstruments GmbH, Jena, Germany) or the Eppendorf ThermoShaker (Eppendorf GmbH, Hamburg, Germany), for 5 min each at 50°C and 550 rpm. For the hybridization, 10 μl labeled amplification product and 90 μl buffer C1 were mixed, transferred into the ArrayStrip, and incubated for 60 min at 50°C and 550 rpm. The sample was then removed from the tube, and the array was washed twice with buffer C2 for 10 min on a shaker at 550 rpm. With the BioShake iQ, washing was performed at 40°C, while for the Eppendorf ThermoShaker, a washing temperature of 45°C was used. Afterwards, 100 μl conjugate solution (1 μl C3 horseradish peroxidase [HRP] conjugate plus 99 μl C4 conjugation buffer) was added and incubated for 10 min at 30°C and 550 rpm, followed by a washing step with 200 μl buffer C5 for 5 min at 30°C and 550 rpm. The ArrayStrip was then stained with 100 μl substrate solution D1 (at 25°C for 10 min, with no agitation). After complete removal of the liquid, the microarrays were photographed using the ArrayMate instrument (Alere Technologies GmbH, Jena, Germany) and automatically analyzed with an assay-specific software script (see Fig. S1 in the supplemental material). The hybridization signals were processed using the IconoClust software version 3.2r1. All spots were recognized and subsequently normalized automatically by the software according to the equation NI = 1 − (M/BG), where NI is the normalized intensity, M is the average intensity of the automatically recognized spot, and BG is the intensity of the local background (21–23). The output range of the signals was from 0 to 1, with 0 being negative and 1 being the maximum possible signal value. Resulting values of <0.1 were considered to be negative and >0.3 to be positive. Values between 0.1 and 0.3 were considered inconclusive. The calculated data were combined in a gray value table for all probes. Finally, an HTML report is provided by the ArrayMate instrument (see Fig. S1 in the supplemental material), giving information on the presence of stx genes and the affiliation to one of the more common species.
TABLE 3.
Probes spotted on the microarray
| Probe no. | Probe name | Species | GenBank accession no. (defined DNA sequence positions within the reference sequences) |
|---|---|---|---|
| 1 | gad_10 | E. coli | U00039.1 (81049–81071-r) |
| 2 | hp_basC-391 | A. baumannii | CU459141.1 (1165074–1165104-r) |
| 3 | hp_basC-392 | A. baumannii | AY571146.1 (7569–7596-r) |
| 4 | hp_cfa-491 | C. freundii | ANAV01000025.1 (15036–15062-r) |
| 5 | hp_cfa-492 | C. freundii | CACD01000086.1 (28997–29021-r) |
| 6 | hp_dnaE_612 | E. coli | U70214.1 (38283–38310) |
| 7 | hp_dnaE_613 | E. coli | U70214.1 (39602–39623) |
| 8 | hp_ecfX-231 | P. aeruginosa | FM209186.1 (4293685–4293709-r) |
| 9 | hp_ecfX-232 | P. aeruginosa | FM209186.1 (4293251–4293276-r) |
| 10 | hp_efp-321 | A. baumannii | CP003856.1 (3111621–3111648-r) |
| 11 | hp_efp-323 | A. baumannii | AMST01000070.1 (255118–255148) |
| 12 | hp_khe-511 | K. pneumoniae | JH930438.1 (5083526–5083553-r) |
| 13 | hp_lacY_04 | E. coli | X56095.1 (1348–1373-r) |
| 14 | hp_pld-281 | A. baumannii | CU459141.1 (515551–515582-r) |
| 15 | hp_pld-282 | A. baumannii | CU459141.1 (515134–515164-r) |
| 16 | hp_rrs_611 | Vibrio splendidus | FM954972.2 (3252371–3252397-r) |
| 17 | hp_rrs_612 | E. coli | U18997.1 (209197–209222-r) |
| 18 | hp_stx1_g10n01 | E. coli | AY170851.1 (229–253) |
| 19 | hp_stx1_g20n02 | E. coli | CP001925.1 (2853382–2853407-r) |
| 20 | hp_stx1_g30n03 | E. coli | Z36901.1 (968–994) |
| 21 | hp_stx1_g30n04 | E. coli | AE005174.2 (2996142–2996166-r) |
| 22 | hp_stx1_g40n06 | E. coli | AY170851.1 (927–956) |
| 23 | hp_stx1_g40n07 | E. coli | AB083044.1 (922–945) |
| 24 | hp_stx1_g40n08 | E. coli | Z36901.1 (1078–1100) |
| 25 | hp_stx2_g11n01 | E. coli | AY286000.1 (371–397) |
| 26 | hp_stx2_g11n02 | E. coli | FN182286.1 (181–207) |
| 27 | hp_stx2_g12n01 | E. coli | AY286000.1 (383–411) |
| 28 | hp_stx2_g12n02 | E. coli | AJ313016.1 (425–451) |
| 29 | hp_stx2_g21n01 | E. coli | AB472687.1 (417–437) |
| 30 | hp_stx2_g23n03 | E. coli | AB472687.1 (495–517) |
| 31 | hp_stx2_g43n01_aa291–297_SE_neu | E. coli | X61283.1 (1094–1124) |
| 32 | hp_stx2_g43n02_aa291–297_FK | E. coli | L11079.1 (1104–1138) |
| 33 | hp_stx2_g44n01_aa297_E | E. coli | X61283.1 (1109–1137) |
| 34 | hp_stx2_g44n03_aa297_K | E. coli | CP003301.1 (3342104–3342130-r) |
| 35 | hp_stx2_g44n04 | E. coli | X81418.1 (948–977) |
| 36 | hp_stx2_g44n05 | E. coli | AY286000.1 (1141–1167) |
| 37 | hp_stx2_g46n01 | E. coli | M29153.1 (1254–1279) |
| 38 | hp_stx2_g51n04_aa34–36 | E. coli | EF441605.1 (1060–1090) |
| 39 | hp_stx2_g51n05_aa34–36 | E. coli | AB472687.1 (1061–1093) |
| 40 | hp_stx2_g51n06_aa34–36 | E. coli | M29153.1 (1315–1346) |
| 41 | hp_stx2_g51n07_aa34–36 | E. coli | Z37725.1 (1237–1267) |
| 42 | hp_stx2_g51n08_aa34–36 | E. coli | EF441618.1 (1064–1092) |
| 43 | hp_stx2_g51n11_aa34–36 | E. coli | X65949.1 (1632–1661) |
| 44 | hp_stx2_g63n03 | E. coli | X65949.1 (1748–1775) |
| 45 | hp_stxA2_611 | E. coli | M29153.1 (670–692) |
| 46 | hp_stxA2_613 | E. coli | M29153.1 (747–770) |
| 47 | hp_stxA2_616 | E. coli | X81418.1 (198–225) |
| 48 | hp_stxA2_617 | E. coli | GU244510.1 (198–224) |
| 49 | hp-3165-manC | S. enterica | FQ312003.1 (2186375–2186399-r) |
| 50 | hp-3300-galF | S. enterica | FQ312003.1 (2176996–2177023-r) |
| 51 | hp-3314-invA | S. bongori | FR877557.1 (2759714–2759739-r) |
| 52 | hp-3315-invA | S. enterica | CP000880.1 (74841–74867) |
| 53 | hp-3316-invA | S. bongori | FR877557.1 (2759962–2759995-r) |
| 54 | ipaH9.8_20 | Shigella flexneri | M76445.1 (1148–1170) |
| 55 | prob_gapA_611 | E. coli | X02662.1 (495–523-r) |
| 56 | prob_ihfA_611 | E. coli | U00096.3 (1795548–1795575-r) |
RESULTS
Target gene selection and assay design.
We have constructed multisequence alignments of the DNA sequences derived from the 62 reference isolates, as defined by Scheutz et al. (19). The sequences were clustered into groups corresponding to the subtypes. The exceptions were Stx2a, Stx2c, and Stx2d, which fell into a single group named Stx2acd in our study. Next, we identified sites that are highly conserved within a given group but that are divergent from those in other groups. We placed hybridization probes onto these characteristic sites. The length of the probes in the initial design varied between 25 and 30 nucleotides in order to have probes with very similar melting temperature (Tm) values. Tm values were calculated applying the method of SantaLucia (24). The biological activities of mature Stx complexes encoded by gene sequences of the Stx2acd group differ widely, and Scheutz et al. (19) have related this to the amino acid constitution at two sites, the StxA activatable tail and the END motif within StxB (see Table S2 in the supplemental material). In order to discriminate subtypes Stx2a, Stx2c, and Stx2d, we have further constructed a set of probes to detect single nucleotide polymorphisms at these two sites. Our search for conserved sites has also revealed that GenBank accession no. AM904726.1, classified as Stx2e by Scheutz et al. (19), is in fact a chimera between stx2e (5′ end, nucleotide positions 1 to 400) and stx2a (3′ end, nucleotide positions 400 to 1242). Instead of assigning it to subtype Stx2e, we designated it by an additional subtype Stx2exa, with ‘x' being a mnemonic for its chimeric character. Another notable finding is that the 3′ end (nucleotide positions 1050 to 1240) of regular stx2e is identical to that of stx2f. Thus, either stx2e or stx2f originated from a recombination event.
The DNA microarray was manufactured using 56 probes. Each probe was spotted in triplicate. A staining control (a biotinylated marker), specific controls for the family Enterobacteriaceae (hp_dnaE_613, prob_gapA_611, and prob_ihfA_611), genus-specific probes for Citrobacter (hp_cfa-491 and hp_cfa-492), Salmonella (hp-3314-invA, hp-3315-invA, and hp-3316-invA), Shigella (ipaH9.8_20), and species-specific probes for Acinetobacter baumannii (hp_basC-391, hp_basC-392, hp_efp-321, hp_efp-323, hp_pld-281, and hp_pld-282), Klebsiella pneumoniae (hp_khe-511), Pseudomonas aeruginosa (hp_ecfX-231 and hp_ecfX-232), as well as a species control for E. coli (gad_10 and hp_dnaE_612) completed the array. DNA-free spotting buffer was used as the negative control.
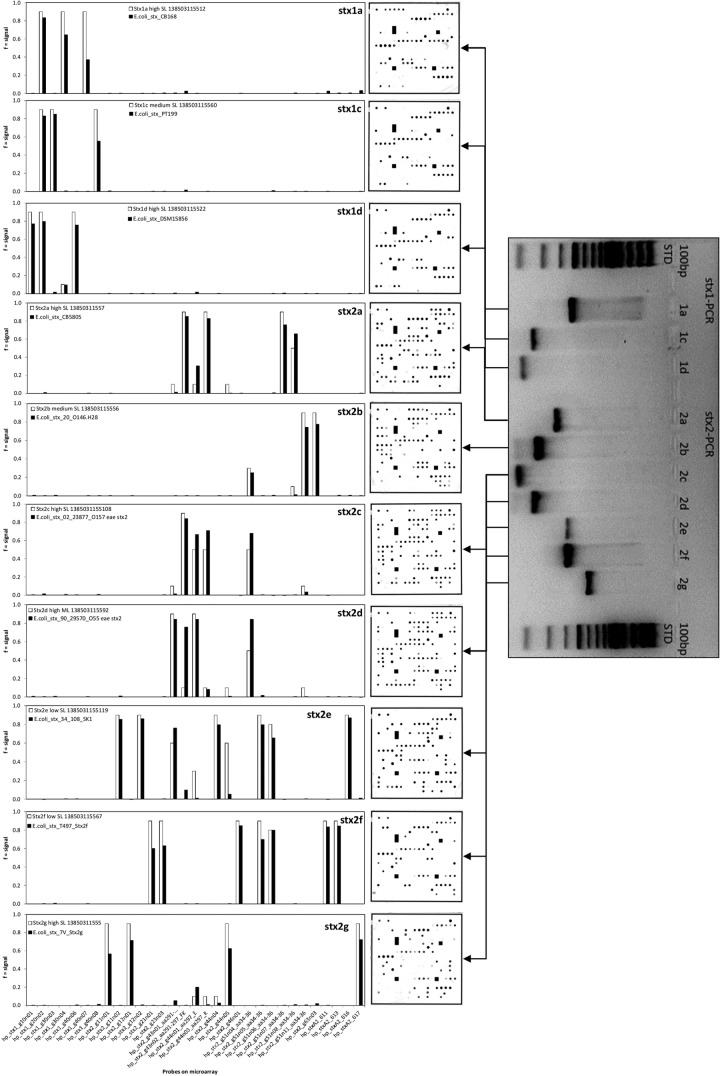
A probe-matching matrix was used to construct theoretical hybridization patterns of the fully sequenced strains listed in the NCBI database (GenBank accession no. Z36900, Z36901.1, AY170851, EF441578, AB048224, AF291819, X61283, AJ313016, M29153, and AY286000). The definition of the theoretical signal intensity was 0.9 for a perfect match, 0.6 for 1 mismatch, 0.3 for 2 mismatches, ≤0.1 for 3 mismatches, and no signal for more mismatches. For each of these sequenced strains, at least one reference strain was used to assign the expected pattern with the pattern of the real hybridization experiments (Fig. 1). The final hybridization protocols for the BioShake iQ and the Eppendorf ThermoShaker were optimized by a comparison of the theoretical and real hybridization patterns at different hybridization temperatures (40°C to 50°C) and washing temperatures (40°C to 45°C).
FIG 1.
Comparison between PCR and microarray results (microarray image and bar graph) of selected STEC reference strains. The white bars represent the expected hybridization values from theoretical experiments obtained from the matching of all array probes to the known sequence of all Stx alleles. The black bars represent experimental data under optimized and stringent hybridization conditions.
Development of a specific software.
To automatically analyze all data obtained from the microarray, a software tool was compiled in which specific probes for different Stx subtypes were summarized. For instance, to identify Stx subtype 2g, the probes hp_stx2_g11n01, hp_stx2_g12n01, hp_stx2_g44n05, and hp_stxA2_617 were to be positive. To identify Stx subtype 2e, the probes hp_stx2_g11n02, hp_stx2_g12n02, hp_stx2_g44n04, and hp_stxA2_616 were to be positive. The software summarized each set of probes with one value and compared these values with each other. When one of these values (Stx2g or Stx2e) was above the threshold of 0.3, a positive was given for either Stx2g or Stx2e in a final result sheet in html format.
Validation of the test with reference strains.
A set of 21 Shiga toxin-producing E. coli reference strains representing all known Stx1 and Stx2 subtypes (Table 1) were used to evaluate the probes printed on the array and the primers in the labeling mixture. All reference strains were first screened by applying the complete set of conventional subtyping PCRs described by Scheutz et al. (19). The microarray results showed 100% identity with the PCR results (Table 1). Ten E. coli reference strains harbor only one Stx subtype, while nine and two E. coli reference strains simultaneously were found to carry two and three different Stx subtypes, respectively (Table 1). A comparison of the results of the PCR described by Scheutz et al. (19) and the microarray results (microarray image and bar graph) of selected STEC reference strains is shown in Fig. 1. Two examples of microarray images and bar graphs for STEC strains that produce two Shiga toxin subtypes are shown in Fig. S2 in the supplemental material.
Field study of the test with Stx-positive strains.
In total, 446 Stx-positive E. coli field isolates were analyzed to determine their Stx subtype using the newly developed microarray. Stx2-positive isolates were detected more often (n = 255) than Stx1-positive isolates (n = 121) or isolates positive for both Stx1 and Stx2 (n = 70). The most frequently detected subtypes were Stx2a (n = 180), followed by Stx1a (n = 117) and the combination of Stx1a-Stx2a (n = 32). While the subtypes Stx2c (n = 26), especially the combinations Stx2a-Stx2c (n = 24), Stx1a-Stx2c (n = 19), and Stx1c-Stx2b (n = 15) were also found frequently, the subtypes Stx1c (n = 4), Stx2b (n = 4), Stx2d (n = 7), Stx2e (n = 11), and the combinations Stx1a-Stx2b (n = 4), Stx2a-Stx2d (n = 2), and Stx2b-Stx2c (n = 1) were rarely detected. The subtypes Stx1d, Stx2f, and Stx2g did not occur in the field strain population (see Table S3 in the supplemental material). A number of the 47 selected field isolates with usually less frequent Stx subtypes or combinations of Stx subtypes detected by the microarray were tested in parallel using the PCRs described by Scheutz et al. (19) (5 with Stx1a, 1 with Stx1c, 2 with Stx2a, 2 with Stx2b, 8 with Stx2c, 6 with Stx2d, 5 with Stx2e, 1 with Stx1a/Stx2a, 2 with the combination Stx1a-Stx2c, 4 with the combination Stx1c-Stx2b, 10 with the combination Stx2a-Stx2c, and 1 with the combination Stx2a-Stx2d). The PCR results were completely in concordance with the microarray results (data not shown).
Comparison of the heat lysis protocol with the standard DNA isolation protocol.
A set of 20 reference strains (all strains listed in Table 1, except DD_1380/99) was used to compare a commercial DNA extraction kit (Qiagen, Hilden, Germany) and a new developed heat lysis protocol in different laboratories and with different technical assistants. The signals and the local background of the microarrays were highly similar between the methods, and no significant differences were observed. In all cases, the same Stx subtype was detected (data not shown).
DISCUSSION
The commonly used nomenclature to discern variants of Stx proteins has evolved historically over several decades. Sequence variants differing in only a few amino acid positions may show altered biological activity. Scheutz et al. (19) recently reviewed and unified the nomenclature. They assigned their unified subtypes to >110 DNA sequences publically available in GenBank. The assignment of types and subtypes is based on two criteria: the overall sequence similarity of the concatenated StxA and StxB subunit sequences and the presence of certain amino acids in two protein motifs, the StxA activatable tail and the END motif within StxB. Most Stx subtypes, except Stx2a, Stx2c, and Stx2d, can be classified based solely on their overall sequence similarities. These three exceptions are defined by their amino acid constitution in the StxA activatable tail and the END motif within StxB. To match these definitions, we have developed a set of DNA primer-probe combinations for a microarray-based assay. The assay was validated on a panel of STEC reference strains that had been tested previously applying the complete set of conventional subtyping PCRs described by Scheutz et al. (19). The results of the tests matched 100%. This was also true when we detected more than one Stx subtype per reference strain. Additionally, we determined the subtypes of 446 STEC-positive field strains. All subtypes or combinations of subtypes were identified correctly. No uncertain reactions were observed.
The multiplex primer extension reaction used for labeling is highly specific, but the assay has a lower sensitivity based on the linear (nonexponential) amplification of single genes than that of the exponential DNA amplification performed in classic standard PCRs. However, this is not an issue for the typing of colony material of quickly growing organisms, such as E. coli. The use of colony material instead of original field samples allows one to obtain the necessary amount of DNA and to ensure the pureness and clonality of the cultures to be genotyped. Besides, the limited amplification can prove to be an advantage under routine conditions, as the assay becomes less susceptible to contamination. The advantage of the linear DNA amplification is the possibility of amplifying and detecting virtually any number of target genes simultaneously in a single-tube reaction.
The microarray-based technique described here provides a couple of advantages over standard PCR based subtyping. Alere Technologies provides two platforms, ArrayTubes and ArrayStrips, for use with these special DNA microarrays. ArrayStrips have the advantage of facilitating the screening of up to 96 samples in parallel in a standard 96-well format. The reading device for both platforms analyzes the microarrays by taking images and automatically assigning the observed pattern to a Stx subtype using a designated software. One microarray chip harbors all gene sequences needed to distinguish between all Stx subtypes, and even the identification of multiple subtypes within one isolate can be performed in a single experiment. It requires significantly less hands-on time compared with the PCRs described by Scheutz et al. (19), because with those methods, almost every subtype must be determined in a single PCR. In order to save more time, a heat lysis protocol was developed that provides ready-to-use genomic DNA in <30 min. In contrast, a standard commercial kit for DNA isolation takes >1 h. We observed no significant differences in the signal and background quality of the microarray between the methods, which allows the conclusion that the DNA isolated by heat lysis is sufficiently pure and highly concentrated (0.5 to 1.5 μg) in order to be used in the standard protocol for labeling and hybridization. A further advantage of the microarray is that the number of spotted probes on the array can be extended by additional probes, e.g., for resistance genes or markers for further characterization of E. coli or the identification of other Gram-negative bacteria that we have described before (12, 25–27).
In summary, the microarray is an excellent tool for fast and accurate identification of all Shiga toxin subtypes based on the standardized nomenclature. Furthermore, this diagnostic tool is user-friendly and can be used in almost every STEC diagnostic routine lab. Moreover, it may be easily expanded and standardized.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Bingen, H. Hächler, L. Beutin, H. Karch, and M. Bielaszewska for kindly providing the reference strains. We also thank E. Jacobs (Technical University of Dresden), InfectoGnostics, and E. Ermantraut (Alere Technologies, Jena, Germany) for supporting this work, as well as S. Proft and K. Clack for critical proofreading.
Footnotes
Published ahead of print 4 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01048-14.
REFERENCES
- 1.Griffin PM, Tauxe RV. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60–98 [DOI] [PubMed] [Google Scholar]
- 2.Remuzzi G, Ruggenenti P. 1998. The hemolytic uremic syndrome. Kidney Int. 53(Suppl 66):S54–S57 [PubMed] [Google Scholar]
- 3.Su C, Brandt LJ. 1995. Escherichia coli O157:H7 infection in humans. Ann. Intern. Med. 123:698–714. 10.7326/0003-4819-123-9-199511010-00009 [DOI] [PubMed] [Google Scholar]
- 4.Karch H, Tarr PI, Bielaszewska M. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295:405–418. 10.1016/j.ijmm.2005.06.009 [DOI] [PubMed] [Google Scholar]
- 5.Tarr PI, Gordon CA, Chandler WL. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086. 10.1016/S0140-6736(05)71144-2 [DOI] [PubMed] [Google Scholar]
- 6.Karmali MA, Gannon V, Sargeant JM. 2010. Verocytotoxin-producing Escherichia coli (VTEC). Vet. Microbiol. 140:360–370. 10.1016/j.vetmic.2009.04.011 [DOI] [PubMed] [Google Scholar]
- 7.Pennington H. 2010. Escherichia coli O157. Lancet 376:1428–1435. 10.1016/S0140-6736(10)60963-4 [DOI] [PubMed] [Google Scholar]
- 8.Pennington H. 2011. Escherichia coli O104, Germany 2011. Lancet Infect. Dis. 11:652–653. 10.1016/S1473-3099(11)70166-9 [DOI] [PubMed] [Google Scholar]
- 9.Buchholz U, Bernard H, Werber D, Böhmer MM, Remschmidt C, Wilking H, Deleré Y, an der Heiden M, Adlhoch C, Dreesman J, Ehlers J, Ethelberg S, Faber M, Frank C, Fricke G, Greiner M, Höhle M, Ivarsson S, Jark U, Kirchner M, Koch J, Krause G, Luber P, Rosner B, Stark K, Kühne M. 2011. German outbreak of Escherichia coli O104:H4 associated with sprouts. N. Engl. J. Med. 365:1763–1770. 10.1056/NEJMoa1106482 [DOI] [PubMed] [Google Scholar]
- 10.Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, Bernard H, Fruth A, Prager R, Spode A, Wadl M, Zoufaly A, Jordan S, Kemper MJ, Follin P, Müller L, King LA, Rosner B, Buchholz U, Stark K, Krause G, HUS Investigation Team 2011. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 365:1771–1780. 10.1056/NEJMoa1106483 [DOI] [PubMed] [Google Scholar]
- 11.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. 10.1371/journal.pone.0022751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monecke S, Mariani-Kurkdjian P, Bingen E, Weill FX, Balière C, Slickers P, Ehricht R. 2011. Presence of enterohemorrhagic Escherichia coli ST678/O104:H4 in France prior to 2011. Appl. Environ. Microbiol. 77:8784–8786. 10.1128/AEM.06524-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robert Koch Institut. 2011. Final presentation and evaluation of the epidemiological findings in the EHEC O104:H4 outbreak, Germany 2011. Robert-Koch Institut, Berlin, Germany: http://www.rki.de/EN/Home/EHEC_final_report.pdf;jsessionid=34C1A193129444B79D5164ECD0589B98.2_cid372?__blob=publicationFile [Google Scholar]
- 14.Betz J, Bauwens A, Kunsmann L, Bielaszewska M, Mormann M, Humpf HU, Karch H, Friedrich AW, Müthing J. 2012. Uncommon membrane distribution of Shiga toxin glycosphingolipid receptors in toxin-sensitive human glomerular microvascular endothelial cells. Biol. Chem. 393:133–147. 10.1515/hsz-2011-0288 [DOI] [PubMed] [Google Scholar]
- 15.Sandvig K, Bergan J, Dyve AB, Skotland T, Torgersen ML. 2010. Endocytosis and retrograde transport of Shiga toxin. Toxicon 56:1181–1185. 10.1016/j.toxicon.2009.11.021 [DOI] [PubMed] [Google Scholar]
- 16.Bielaszewska M, Friedrich AW, Aldick T, Schürk-Bulgrin R, Karch H. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43:1160–1167. 10.1086/508195 [DOI] [PubMed] [Google Scholar]
- 17.Friedrich AW, Bielaszewska M, Zhang WL, Pulz M, Kuczius T, Ammon A, Karch H. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74–84. 10.1086/338115 [DOI] [PubMed] [Google Scholar]
- 18.Persson S, Olsen KEP, Ethelberg S, Scheutz F. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45:2020–2024. 10.1128/JCM.02591-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheutz F, Teel LD, Beutin L, Piérard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O'Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 50:2951–2963. 10.1128/JCM.00860-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monecke S, Ehricht R. 2005. Rapid genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolates using miniaturised oligonucleotide arrays. Clin. Microbiol. Infect. 11:825–833. 10.1111/j.1469-0691.2005.01243.x [DOI] [PubMed] [Google Scholar]
- 21.Monecke S, Slickers P, Ehricht R. 2008. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol. Med. Microbiol. 53:237–251. 10.1111/j.1574-695X.2008.00426.x [DOI] [PubMed] [Google Scholar]
- 22.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O'Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan HL, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. 10.1371/journal.pone.0017936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stieber B, Monecke S, Müller E, Baier V, W Coombs G, Ehricht R. 2013. Development and usage of protein microarrays for the quantitative measurement of Panton-Valentine leukocidin. Mol. Cell. Probes 28:123–132. 10.1016/j.mcp.2013.11.003 [DOI] [PubMed] [Google Scholar]
- 24.SantaLucia J., Jr 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U. S. A. 95:1460–1465. 10.1073/pnas.95.4.1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geue L, Monecke S, Engelmann I, Braun S, Slickers P, Ehricht R. 2014. Rapid microarray-based DNA genoserotyping of Escherichia coli. Microbiol. Immunol. 58:77–86. 10.1111/1348-0421.12120 [DOI] [PubMed] [Google Scholar]
- 26.Schilling AK, Hotzel H, Methner U, Sprague LD, Schmoock G, El-Adawy H, Ehricht R, Wöhr AC, Erhard M, Geue L. 2012. Zoonotic agents in small ruminants kept on city farms in southern Germany. Appl. Environ. Microbiol. 78:3785–3793. 10.1128/AEM.07802-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geue L, Schares S, Mintel B, Conraths FJ, Müller E, Ehricht R. 2010. Rapid microarray-based genotyping of enterohemorrhagic Escherichia coli serotype O156:H25/H-/Hnt isolates from cattle and clonal relationship analysis. Appl. Environ. Microbiol. 76:5510–5519. 10.1128/AEM.00743-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.