Abstract
A clustered regularly interspaced short palindromic repeat (CRISPR) typing method has recently been developed and used for typing and subtyping of Salmonella spp., but it is complicated and labor intensive because it has to analyze all spacers in two CRISPR loci. Here, we developed a more convenient and efficient method, namely, CRISPR locus spacer pair typing (CLSPT), which only needs to analyze the two newly incorporated spacers adjoining the leader array in the two CRISPR loci. We analyzed a CRISPR array of 82 strains belonging to 21 Salmonella serovars isolated from humans in different areas of China by using this new method. We also retrieved the newly incorporated spacers in each CRISPR locus of 537 Salmonella isolates which have definite serotypes in the Pasteur Institute's CRISPR Database to evaluate this method. Our findings showed that this new CLSPT method presents a high level of consistency (kappa = 0.9872, Matthew's correlation coefficient = 0.9712) with the results of traditional serotyping, and thus, it can also be used to predict serotypes of Salmonella spp. Moreover, this new method has a considerable discriminatory power (discriminatory index [DI] = 0.8145), comparable to those of multilocus sequence typing (DI = 0.8088) and conventional CRISPR typing (DI = 0.8684). Because CLSPT only costs about $5 to $10 per isolate, it is a much cheaper and more attractive method for subtyping of Salmonella isolates. In conclusion, this new method will provide considerable advantages over other molecular subtyping methods, and it may become a valuable epidemiologic tool for the surveillance of Salmonella infections.
INTRODUCTION
Microbiologists have used serological and nutritional characteristics to subdivide pathogenic bacteria for nearly 100 years (1). Traditional serotyping according to the White-Kauffmann-Le Minor scheme, based on the agglutination of bacteria with specific sera, identifies somatic (O) and flagellar (H) antigens (2). However, although traditional serotyping has been widely used, it still has some drawbacks. First, traditional serotyping takes at least 3 days to complete (3) and requires the maintenance of more than 250 typing sera and 350 different antigens. Second, traditional serotyping does not provide a discriminatory level sufficient for the investigation of outbreaks of food-borne illness and cannot be used to infer phylogenetic relationships.
Differentiation between isolates within the most common serotypes requires the use of subtyping methods. DNA-based subtyping methods have been developed. These include multilocus sequence typing (MLST), which requires the sequencing of seven housekeeping genes, and pulsed-field gel electrophoresis (PFGE). The latter technique is based on analysis of the restriction pattern of high-molecular-weight DNA digested with a rare restriction enzyme (4). However, both methods have several limitations. MLST is highly expensive and has low throughput (5), and PFGE is a technically demanding and nonautomated method. Furthermore, the interpretation and comparison of banding profiles is not straightforward, even with standard protocols and BioNumerics specialized analysis software (Applied Maths).
The clustered regularly interspaced short palindromic repeat (CRISPR) that has been discovered in archaea and bacteria provides adaptive, heritable immunity against viruses, plasmids, and other mobile genetic elements (6). CRISPRs encode tandem sequences containing 21- to 47-bp direct repeats (DRs) and spacers of similar size. The spacers are short DNA sequences obtained from foreign nucleic acids, such as phage or plasmids, inserted into bacterial chromosomes to protect them from infection by homologous phage or plasmids (7). As such, different CRISPRs arise due to diverse phage and plasmid pools in an environment. Thus, CRISPRs differentiate outbreak strains/clones within epidemic clones (8). Since the middle of the 1990s, the CRISPR locus of Mycobacterium tuberculosis has been studied extensively, and the high degree of polymorphism of the spacer content has led to the development of a subtyping method known as spoligotyping (9). Several studies have reported the presence of two CRISPR loci in Salmonella (10, 11), and CRISPR polymorphisms are strongly correlated with serovars and subtypes (12). Recently, three studies suggested that CRISPR loci may provide information useful for typing and subtyping of Salmonella (8, 13, 14), and they presented a CRISPR typing method (hereinafter referred to as conventional CRISPR typing [CCT]) that is based on all spacers in both CRISPR loci. However, only a limited number of serotypes from a single geographic area were studied using the CCT method because it is complicated and labor intensive.
The functional studies of the CRISPR loci suggested that, upon infection with a foreign element, part of the Salmonella genome is typically incorporated into the leader end of the CRISPR array as a spacer and the repeat is duplicated (15). Therefore, these spacers are integrated into the CRISPR locus in a polarized manner and the newly incorporated spacers adjoin the leader array in the CRISPR locus (7). Therefore, we think the newly incorporated spacer adjoining the leader array may be an effective molecular marker for subtyping of Salmonella isolates. We evaluated these assumptions using CRISPR databases, and we propose a new method for Salmonella typing and subtyping, called CRISPR locus spacer pair typing (CLSPT), in which only the newly incorporated spacers in two CRISPR loci are analyzed.
MATERIALS AND METHODS
Bacterial isolates.
Eighty-two Salmonella isolates were separated from 4,901 fecal samples obtained during January 2009 to March 2011 from patients with diarrhea or dysentery at hospitals in 6 provinces within the following regions of China: Beijing, Jiangsu, Guangdong, Shandong, Liaoning, and Xinjiang provinces (Table 1). Immunological serotyping was completed using diagnostic sera (SSI Diagnostica, Hillerød, Denmark) for Salmonella according to the manufacturer's instructions. The collection of Salmonella enterica serotypes was comprised of 34 strains of S. Enteritidis, 9 strains of S. Infantis, 7 strains of S. Paratyphi B, 5 strains of S. Kentucky, 4 strains of S. Agona, 3 strains of S. Typhimurium, and other common epidemic serotypes (Table 1). All isolates were stored at −80°C in 20% glycerol. When necessary, some isolates were grown in Luria-Bertani broth at 37°C overnight. Total DNA was extracted using the TIANamp bacteria DNA kit (Tiangen Biotech, Beijing, China) according to the manufacturer's instructions and stored at −20°C before use.
TABLE 1.
List of 82 Salmonella isolates that were analyzed in this study
| Strain | Serovar | Origin | Source | Yr of isolation | Serotype (O:H1:H2) |
|---|---|---|---|---|---|
| BJ0047 | Agona | Beijing | Human | 2009 | 1,4,12:f,g,s:− |
| BJ0048 | Agona | Beijing | Human | 2009 | 1,4,13:f,g,s:− |
| NJ92234 | Agona | Nanjing | Human | 2011 | 1,4,15:f,g,s:− |
| SY628 | Agona | Shenyang | Human | 2010 | 1,4,14:f,g,s:− |
| B3V3-2 | Albany | Xinjiang | Human | 2009 | 8,20:Z4,Z24:− |
| NJ81658-1 | Choleraesuis | Nanjing | Human | 2011 | 6,7:C:1,5 |
| NJ81658-2 | Choleraesuis | Nanjing | Human | 2011 | 6,7:C:1,5 |
| BJ0053 | Derby | Beijing | Human | 2009 | 1,4,5,12:f,g:1,2 |
| SY454 | Derby | Shenyang | Human | 2010 | 1,4,5,12:f,g:1,2 |
| B43V2 | Enteritidis | Xinjiang | Human | 2009 | 1,9,12:g,m:1,7 |
| B63V1 | Enteritidis | Xinjiang | Human | 2009 | 1,9,12:g,m:1,7 |
| BJ0046 | Enteritidis | Beijing | Human | 2009 | 1,9,12:g,m:1,7 |
| BJ0049 | Enteritidis | Beijing | Human | 2009 | 1,9,12:g,m:1,7 |
| BJ0051 | Enteritidis | Beijing | Human | 2009 | 1,9,12:g,m:1,7 |
| BJ0065 | Enteritidis | Beijing | Human | 2009 | 1,9,12:g,m:1,7 |
| BJ0068 | Enteritidis | Beijing | Human | 2009 | 1,9,12:g,m:1,7 |
| BJ0069 | Enteritidis | Beijing | Human | 2009 | 1,9,12:g,m:1,7 |
| C3V1 | Enteritidis | Xinjiang | Human | 2009 | 1,9,12:g,m:1,7 |
| E19V2 | Enteritidis | Xinjiang | Human | 2009 | 1,9,12:g,m:1,7 |
| GZ0010 | Enteritidis | Guangzhou | Human | 2009 | 1,9,12:g,m:1,7 |
| GZ0013 | Enteritidis | Guangzhou | Human | 2009 | 1,9,12:g,m:1,7 |
| GZ0015 | Enteritidis | Guangzhou | Human | 2009 | 1,9,12:g,m:1,7 |
| GZ0020 | Enteritidis | Guangzhou | Human | 2009 | 1,9,12:g,m:1,7 |
| GZ0023 | Enteritidis | Guangzhou | Human | 2009 | 1,9,12:g,m:1,7 |
| GZ0027 | Enteritidis | Guangzhou | Human | 2009 | 1,9,12:g,m:1,7 |
| GZ0042 | Enteritidis | Guangzhou | Human | 2009 | 1,9,12:g,m:1,7 |
| NJ81625 | Enteritidis | Nanjing | Human | 2011 | 1,9,12:g,m:1,7 |
| NJ82854 | Enteritidis | Nanjing | Human | 2011 | 1,9,12:g,m:1,7 |
| NJ92143 | Enteritidis | Nanjing | Human | 2011 | 1,9,12:g,m:1,7 |
| NJ92341-1 | Enteritidis | Nanjing | Human | 2011 | 1,9,12:g,m:1,7 |
| NJ92341-2 | Enteritidis | Nanjing | Human | 2011 | 1,9,12:g,m:1,7 |
| NJ92374 | Enteritidis | Nanjing | Human | 2011 | 1,9,12:g,m:1,7 |
| NJ92432 | Enteritidis | Nanjing | Human | 2011 | 1,9,12:g,m:1,7 |
| NJ92663 | Enteritidis | Nanjing | Human | 2011 | 1,9,12:g,m:1,7 |
| SY020 | Enteritidis | Shenyang | Human | 2009 | 1,9,12:g,m:1,7 |
| SY070 | Enteritidis | Shenyang | Human | 2009 | 1,9,12:g,m:1,7 |
| SY204 | Enteritidis | Shenyang | Human | 2010 | 1,9,12:g,m:1,7 |
| SY330 | Enteritidis | Shenyang | Human | 2010 | 1,9,12:g,m:1,7 |
| SY609 | Enteritidis | Shenyang | Human | 2010 | 1,9,12:g,m:1,7 |
| SY637 | Enteritidis | Shenyang | Human | 2010 | 1,9,12:g,m:1,7 |
| XJ001 | Enteritidis | Xinjiang | Human | 2009 | 1,9,12:g,m:1,7 |
| XJ002 | Enteritidis | Xinjiang | Human | 2009 | 1,9,12:g,m:1,7 |
| XJ003 | Enteritidis | Xinjiang | Human | 2009 | 1,9,12:g,m:1,7 |
| NJ92713 | Heidelberg | Nanjing | Human | 2011 | 1,4,15,12:r:1,2 |
| SY588-3 | Indiana | Shenyang | Human | 2010 | 1,4,12:Z:1,7 |
| JN0005 | Infantis | Jinan | Human | 2009 | 6,7:r:1,5 |
| JN0011 | Infantis | Jinan | Human | 2009 | 6,7:r:1,5 |
| JN0014 | Infantis | Jinan | Human | 2009 | 6,7:r:1,5 |
| JN0015 | Infantis | Jinan | Human | 2009 | 6,7:r:1,5 |
| JN0016 | Infantis | Jinan | Human | 2009 | 6,7:r:1,5 |
| JN0025 | Infantis | Jinan | Human | 2009 | 6,7:r:1,5 |
| JN008 | Infantis | Jinan | Human | 2009 | 6,7:r:1,5 |
| JN009 | Infantis | Jinan | Human | 2009 | 6,7:r:1,5 |
| JN0096 | Infantis | Jinan | Human | 2009 | 6,7:r:1,5 |
| BJ0055 | Kentucky | Beijing | Human | 2009 | 8,20:i:Z6 |
| BJ0058 | Kentucky | Beijing | Human | 2009 | 8,20:i:Z6 |
| BJ0059 | Kentucky | Beijing | Human | 2009 | 8,20:i:Z6 |
| BJ0060 | Kentucky | Beijing | Human | 2009 | 8,20:i:Z6 |
| BJ0061 | Kentucky | Beijing | Human | 2009 | 8,20:i:Z6 |
| BJ0063 | Lexington | Beijing | Human | 2009 | 6,7:f,g:− |
| GZ001 | Litchfield | Guangzhou | Human | 2009 | 6,8:1,v:1,2 |
| NJ85022 | Litchfield | Nanjing | Human | 2011 | 6,8:1,v:1,2 |
| NJ91889 | Montevideo | Nanjing | Human | 2011 | 6,7:g,m,p,s:1,2,7 |
| JN0010 | Newport | Jinan | Human | 2009 | 6,8:e,h:1,2 |
| BJ0054 | Newwington | Beijing | Human | 2009 | 3,15:e,h:1,6 |
| BJ0062 | Newwington | Beijing | Human | 2009 | 3,15:e,h:1,6 |
| NJ92291 | Paratyphi a | Nanjing | Human | 2011 | 1,21,2:a:1,5 |
| BJ0064 | Paratyphi b | Beijing | Human | 2009 | 1,4,5,12:b:1,2 |
| BJ0066 | Paratyphi b | Beijing | Human | 2009 | 1,4,5,12:b:1,2 |
| BJ0067 | Paratyphi b | Beijing | Human | 2009 | 1,4,5,12:b:1,2 |
| GZ0012 | Paratyphi b | Guangzhou | Human | 2009 | 1,4,5,12:b:1,2 |
| GZ0036 | Paratyphi b | Guangzhou | Human | 2009 | 1,4,5,12:b:1,2 |
| GZ0041 | Paratyphi b | Guangzhou | Human | 2009 | 1,4,5,12:b:1,2 |
| XJ004 | Paratyphi b | Xinjiang | Human | 2009 | 1,4,5,12:b:1,2 |
| BJ0052 | Senftenberg | Beijing | Human | 2009 | 1,3,19:g,s,t:− |
| NJ92176 | Senftenberg | Nanjing | Human | 2011 | 1,3,19:g,s,t:− |
| GZ0045 | Stanley | Guangzhou | Human | 2009 | 1,4,5,12,27:d:1,2 |
| NJ92671 | Thompson | Nanjing | Human | 2011 | 6,7,k,1,5 |
| XJ005 | Typhi | Xinjiang | Human | 2009 | 9,12,Vi:d:− |
| BJ302 | Typhimurium | Beijing | Human | 2010 | 1,4,5,12:i:1,2 |
| NJ81674 | Typhimurium | Nanjing | Human | 2011 | 1,4,5,12:i:1,2 |
| NJ85592 | Typhimurium | Nanjing | Human | 2011 | 1,4,5,12:i:1,2 |
To evaluate our method in silico, we obtained CRISPR arrays from 537 Salmonella isolates in the Pasteur Institute's CRISPR Database (http://www.pasteur.fr/recherche/genopole/PF8/crispr/CRISPRDB.html) which comprised 131 strains of S. Enteritidis, 102 strains of S. Typhimurium, 28 strains of S. Paratyphi B, 15 strains of S. Newport, 15 strains of S. Typhi, 13 strains of S. Kentucky, 11 strains of S. Agona, and other common epidemic serotypes. These serotypes belonged to the two species of the Salmonella genus, S. enterica and S. bongori, and the six S. enterica subspecies, S. enterica subsp. enterica, salamae, arizonae, diarizonae, indica, and houtenae.
PCR amplification and sequencing of the CRISPR loci.
We amplified the CRISPR 1 locus with forward primer A1 (5′-GTRGTRCGGATAATGCTGCC-3′) and reverse primer A2 (5′-CGTATTCCGGTAGATBTDGATGG-3′). To amplify the CRISPR 2 locus, we used forward primer B1 (5′-GAGCAATACYYTRATCGTTAACGCC-3′) and reverse primer B2 (5′-GTTGCDATAKGTYGRTRGRATGTRG-3′). All the primers were designed by L. Fabre (14) and synthesized by Sangon Biotech (Shanghai, China).
A 50-μl PCR mixture volume contained 0.25 μl TaKaRa Ex Taq DNA polymerase, 5 μl 10× Ex Taq buffer (Mg2+ plus), 4 μl deoxynucleoside triphosphate (dNTP) mix (2.5 mM each), 5 μl DNA template, 2 μl each forward primer and reverse primer (final concentration, 0.2 μM), and 31.75 μl sterile double-distilled water. The cycling conditions were as follows: 10 min at 94°C for denaturation (1 cycle), followed by 35 cycles of 1 min at 94°C for denaturation, 1 min at 55°C for annealing, and 1 min at 72°C for polymerization, followed by an additional 10 min at 72°C for extension. The PCR products were sequenced with a BigDye Terminator kit, version 3.1 (Sangon Biotech, Shanghai, China), using an ABI 3730XL apparatus.
CLSPT.
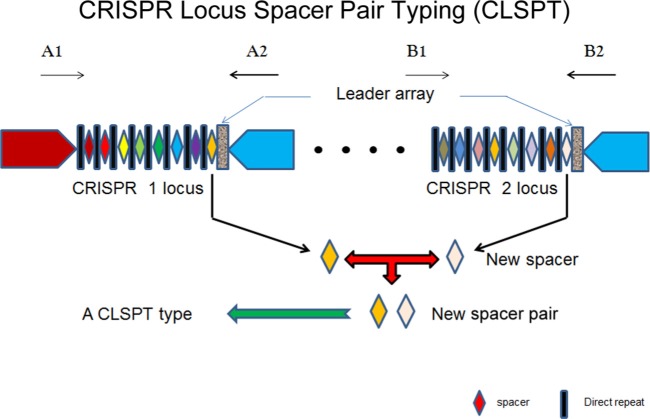
In CLSPT, we performed sequencing with the corresponding reverse primer and identified the newly incorporated spacers, which are located between the first and second direct repeats (DRs) adjoining the leader array. The CLSPT profiles are the sequences which are composed of the two newly incorporated spacers from the CRISPR 1 and CRISPR 2 loci. In order to predict their serovars, we also assigned each CLSPT profile a unique type composed of the two newly incorporated spacers' names (Fig. 1). The CLSPT profiles were clustered with the BioNumerics software (Applied Maths, Austin, TX, USA) using a categorical coefficient and a graphing method called the minimum spanning tree.
FIG 1.
The new method is based only on the newly incorporated spacers adjoining the leader array in both CRISPR loci. We used the spacer adjoining the leader array in each CRISPR locus to form a spacer pair to represent each isolate. Spacers and direct repeats were visualized as described by L. Fabre (14) et al.
CCT.
The sequences of direct repeats and spacers in CRISPR 1 and CRISPR 2 were identified by using CRISPRfinder (http://crispr.u-psud.fr/Server/). All sequences from this study were submitted as a batch to a private database in CRISPRdb (http://crispr.u-psud.fr/CRISPRcompar/private/PrivateDatabase.php) under accession numbers 403_15795 to 403_15895. The analyses of the spacer arrangements were performed using CRISPRcompar (6). Different allelic types (ATs; sequences with at least a 1-nucleotide difference, or a 1-spacer difference in the case of CRISPRs) were assigned arbitrary numbers. The combination of 2 alleles (CRISPR 1 and CRISPR 2) determined the allelic profile, and each unique allelic profile was designated a unique CCT type.
MLST.
MLST was carried out using the protocols described on the MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Senterica/documents/primersEnterica_html). The PCR conditions were as follows: 94°C for 5 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and 72°C for 5 min. PCR amplicons were sequenced at Sangon Biotech (Shanghai Biotech, China). Sequences were assembled and analyzed using Lasergene 7.1 software (DNAStar). Sequence type (ST) numbers were assigned by submitting the sequences to the Salmonella MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Senterica).
PFGE.
Restriction endonuclease digestion was carried out using XbaI (TaKaRa, Dalian, China) at 37°C for 3 h. DNA macrorestriction fragments were resolved on 1% SeaKem Gold agarose (Lonza, Rockland, ME, USA) using a CHEF Mapper PFGE system for over 19 h. S. enterica serovar Braenderup strain H9812 was used as the reference strain (16). BioNumerics version 6.0 was used to analyze the PFGE patterns. Similarity analysis was performed using the Dice coefficient, and clustering was performed using the unweighted-pair group method by arithmetic mean with a 1.5% tolerance limit.
RESULTS
CLSPT.
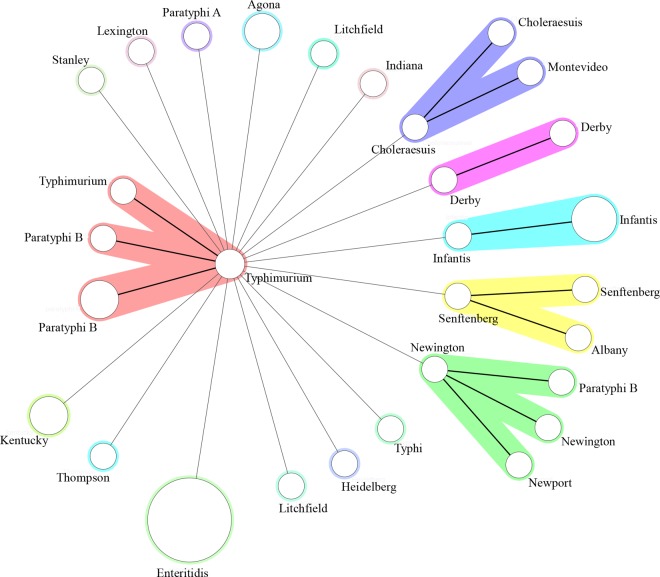
For 82 Salmonella isolates, both CRISPR 1 and CRISPR 2 loci were identified. In total, there are 2,998 spacers in both CRISPR loci. The newly incorporated spacer adjoining the leader array in the CRISPR 1 locus has 23 different alleles, by which 77 of the 82 (93%) isolates' serotypes could be correctly predicted. The isolates that could not be predicted included S. Paratyphi B BJ0066, S. Typhimurium NJ85436, S. Newport JN0010, S. Newington BJ0062, and S. Paratyphi B BJ0064. In addition, there were 25 different newly incorporated spacer alleles in the CRISPR 2 locus, by which 78 of the 82 (95%) isolates' serotypes were correctly predicted. The isolates that could not be predicted included S. Choleraesuis NJ81658-1, S. Montevideo NJ91889, S. Albany XJB3V3-2, and S. Senftenberg NJ92176. We obtained 30 different alleles and predicted 82 of 82 (100%) serotypes correctly when the combination of the two newly incorporated spacers was considered (Table 2). Therefore, 82 Salmonella enterica strains were subtyped into 30 different CRISPR locus spacer pair types (CLSPTs) and all of their serotypes were clearly separated. In addition, the minimum spanning tree constructed based on CLSPTs in BioNumerics (Fig. 2) also supported the fact that the CLSPTs are highly correlated with the serotypes.
TABLE 2.
CRISPR locus spacer pair types, conventional CRISPR types, multilocus sequence types, and pulsed-field gel electrophoresis types of the 82 Salmonella isolatesa
| Strain | Salmonella serotype | ST | PT | CLSPTb | CCT |
|---|---|---|---|---|---|
| NJ92234 | Agona | 13 | PT36 | Ago13 AgoB8 | 1 |
| BJ0047 | Agona | 13 | PT59 | Ago13 AgoB8 | 1 |
| BJ0048 | Agona | 13 | PT59 | Ago13 AgoB8 | 1 |
| SY628 | Agona | 13 | PT59 | Ago13 AgoB8 | 1 |
| B3V3-2 | Albany | 292 | PT65 | Abl4* AlbB17 | 2 |
| NJ81658-1 | Choleraesuis | 145 | PT33 | Chol3 CholB6 | 3 |
| NJ81658-2 | Choleraesuis | 145 | PT58 | Chol3 MonB54 | 4 |
| SY454 | Derby | 40 | PT76 | Der21 DerB11 | 6 |
| BJ0053 | Derby | 40 | PT77 | Der21 DerB12 | 5 |
| SY609 | Enteritidis | 11 | PT37 | Ent8 EntB9 | 8 |
| NJ82854 | Enteritidis | 11 | PT82 | Ent8 EntB9 | 7 |
| BJ0049 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 7 |
| BJ0051 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 7 |
| BJ0069 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 7 |
| E19V2 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 7 |
| GZ0010 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 7 |
| GZ0015 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 7 |
| GZ0042 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 7 |
| NJ92143 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 7 |
| NJ92341-1 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 7 |
| NJ92341-2 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 7 |
| SY020 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 7 |
| SY070 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 7 |
| SY204 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 7 |
| SY330 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 8 |
| SY637 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 8 |
| XJ001 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 9 |
| XJ002 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 10 |
| XJ003 | Enteritidis | 11 | PT84 | Ent8 EntB9 | 10 |
| B63V1 | Enteritidis | 11 | PT85 | Ent8 EntB9 | 7 |
| C3V1 | Enteritidis | 11 | PT85 | Ent8 EntB9 | 7 |
| NJ92374 | Enteritidis | 11 | PT87 | Ent8 EntB9 | 7 |
| NJ92663 | Enteritidis | 11 | PT87 | Ent8 EntB9 | 7 |
| BJ0065 | Enteritidis | 11 | PT88 | Ent8 EntB9 | 7 |
| GZ0020 | Enteritidis | 11 | PT88 | Ent8 EntB9 | 7 |
| GZ0023 | Enteritidis | 11 | PT88 | Ent8 EntB9 | 7 |
| BJ0046 | Enteritidis | 11 | PT91 | Ent8 EntB9 | 7 |
| BJ0068 | Enteritidis | 11 | PT92 | Ent8 EntB9 | 7 |
| B43V2 | Enteritidis | 11 | PT93 | Ent8 EntB9 | 7 |
| GZ0013 | Enteritidis | 11 | PT93 | Ent8 EntB9 | 7 |
| NJ81625 | Enteritidis | 11 | PT95 | Ent8 EntB9 | 7 |
| NJ92432 | Enteritidis | 11 | PT97 | Ent8 EntB9 | 7 |
| GZ0027 | Enteritidis | 11 | PT98 | Ent8 EntB9 | 7 |
| NJ92713 | Heidelberg | 15 | PT45 | Heid14* STMB24 | 11 |
| SY588-3 | Indiana | 17 | PT31 | Ind13 IndB13 | 12 |
| JN0005 | Infantis | 32 | PT05 | Inf34* InfB26 | 13 |
| JN0011 | Infantis | 32 | PT05 | Inf31 InfB26 | 14 |
| JN0014 | Infantis | 32 | PT05 | Inf31 InfB26 | 14 |
| JN0015 | Infantis | 32 | PT05 | Inf31 InfB26 | 14 |
| JN0016 | Infantis | 32 | PT05 | Inf31 InfB26 | 14 |
| JN0025 | Infantis | 32 | PT05 | Inf31 InfB26 | 14 |
| JN008 | Infantis | 32 | PT09 | Inf31 InfB26 | 14 |
| JN009 | Infantis | 32 | PT09 | Inf31 InfB26 | 14 |
| JN0096 | Infantis | 32 | PT09 | Inf31 InfB26 | 14 |
| BJ0055 | Kentucky | 198 | PT78 | Ken26 KenB41 | 15 |
| BJ0058 | Kentucky | 198 | PT78 | Ken26 KenB41 | 15 |
| BJ0059 | Kentucky | 198 | PT78 | Ken26 KenB41 | 15 |
| BJ0060 | Kentucky | 198 | PT78 | Ken26 KenB41 | 15 |
| BJ0061 | Kentucky | 198 | PT78 | Ken26 KenB41 | 15 |
| BJ0063 | Lexington | 247 | PT74 | Lex16 LexB6 | 16 |
| GZ001 | Litchfield | 1499 | PT24 | Bovis13 BovB10var1 | 17 |
| NJ85022 | Litchfield | 214 | PT28 | Lit1 LitB1 | 18 |
| NJ91889 | Montevideo | 4 | PT14 | Mon43 CholB6 | 19 |
| BJ0054 | Newington | 516 | PT71 | Der26 NewB1* | 21 |
| BJ0062 | Newington | 516 | PT71 | Inf34* NewB1* | 22 |
| JN0010 | Newport | 33 | PT11 | Inf34* HadB30 | 20 |
| NJ92291 | Paratyphi A | 85 | PT75 | ParA5 ParAB2 | 23 |
| BJ0064 | Paratyphi B | 34 | PT39 | Inf34* STMB33 | 24 |
| BJ0066 | Paratyphi B | 34 | PT39 | STM6 STM31 | 25 |
| BJ0067 | Paratyphi B | 34 | PT39 | STM6 STM31 | 25 |
| GZ0041 | Paratyphi B | 34 | PT39 | STM6 STM31 | 25 |
| GZ0012 | Paratyphi B | 34 | PT41 | STM6 STM31 | 25 |
| XJ004 | Paratyphi B | 328 | PT42 | STM6 STM24 | 26 |
| GZ0036 | Paratyphi B | 34 | PT43 | STM6 STM31 | 25 |
| NJ92176 | Senftenberg | 14 | PT01 | Sen16 AlbB17 | 28 |
| BJ0052 | Senftenberg | 14 | PT13 | Sen16 SenB25 | 27 |
| GZ0045 | Stanley | 29 | PT17 | Sta3 StaB5 | 29 |
| NJ92671 | Thompson | 26 | PT23 | Tho15 ThoB7 | 30 |
| XJ005 | Typhi | 2 | PT62 | Typhi6 AnaB14 | 31 |
| BJ302 | Typhimurium | 19 | PT47 | STM24 STMB31 | 32 |
| NJ81674 | Typhimurium | 19 | PT52 | STM6 STMB31 | 33 |
| NJ85592 | Typhimurium | 19 | PT54 | STM6 STMB31 | 33 |
CLSPT, CRISPR locus spacer pair type; CCT, conventional CRISPR type; ST, multilocus sequence type; PT, PFGE pulsotype.
*, novel spacer (not found in the Pasteur Institute's CRISPR database) identified in the current study. We used the spacer nomenclature described by L. Fabre (14). The three- or four-letter prefix in the spacer name indicates the serotype from which the spacer was first extracted, the suffix B indicates that the spacer was found in the CRISPR 2 locus, and spacers were numbered consecutively in order of discovery. Abl4*, CACATGTCAGATGTTATTTCCAAGGCGGAGC; Heid14*, ACCGTTACGCGCATCTTGAAAATCAGCTTCGA; Inf34*, GTTCCGTACTGCAGTGGTGAATTATCAGTAAT; NewB1*, TTAGTATTTGTAGCGGTTTTCCGGAGAAAACG.
FIG 2.
A minimum spanning tree has been constructed based on CLSPTs using the strains listed in Table 2. In the tree, the corresponding serotypes are circled. CLSPT types are represented by circles, and the size of a circle indicates the number of strains with this particular type. There is no ambiguous result such as one CLSPT type corresponding to two or more serotypes. The halos surrounding the various types denote the groupings obtained by Bionumerics analysis, which indicate that they may be separated from the related phage/plasmid pool. A minimum neighbor difference of 1 was used for the creation of groups.
High consistency of typing results between CLSPT and traditional serotyping.
To evaluate the consistency between the results of CLSPT and those of the traditional serotyping method, 537 Salmonella strains belonging to 101 serovars in the Pasteur Institute's CRISPR Database were selected. Ninety-three different alleles were identified using CLSPT. Then, we established a CLSPT/serotype dictionary (see Table S1 in the supplemental material) by which the serovars of 514 of 537 (95%) strains were correctly correlated to the CLSPTs. In general, one or several CLSPTs are specific to one serovar. For example, the CLSPT of 13/15 S. Typhi strains is Typhi6 EntB0var1, while 2/15 are Typhi3 EntB0var1. However, nine CLSPTs corresponded to several serotypes, indicating that isolates of the different serotypes might have been separated from the same phage/plasmid pool, infected with the same phage, or had the same evolutionary origin. In general, a majority of the CLSPTs are specific to corresponding serotypes, and the addition of an extra spacer in the CRISPR locus is an alternative way for the ambiguous results to occur.
In order to check the consistency of the results of CLSPT and serotyping, we proposed to use the most likely serovar to resolve these ambiguous results. For example, 121 isolates had the same CLSPT, Ent8 EntB9, including 115 isolates of S. Enteritidis, 3 isolates of S. enterica Nitra, 2 of S. enterica Rosenberg, and 1 of S. enterica Blegdam. Thus, S. Enteritidis was the most likely predicted serotype. The kappa coefficient demonstrated a kappa of 0.9872, the 95% confidence interval (CI) was 0.9771 to 0.9974, and the Matthew's correlation coefficient (MCC) was 0.9712.
Comparison of discriminatory power between CLSPT, CCT, MLST, and PFGE.
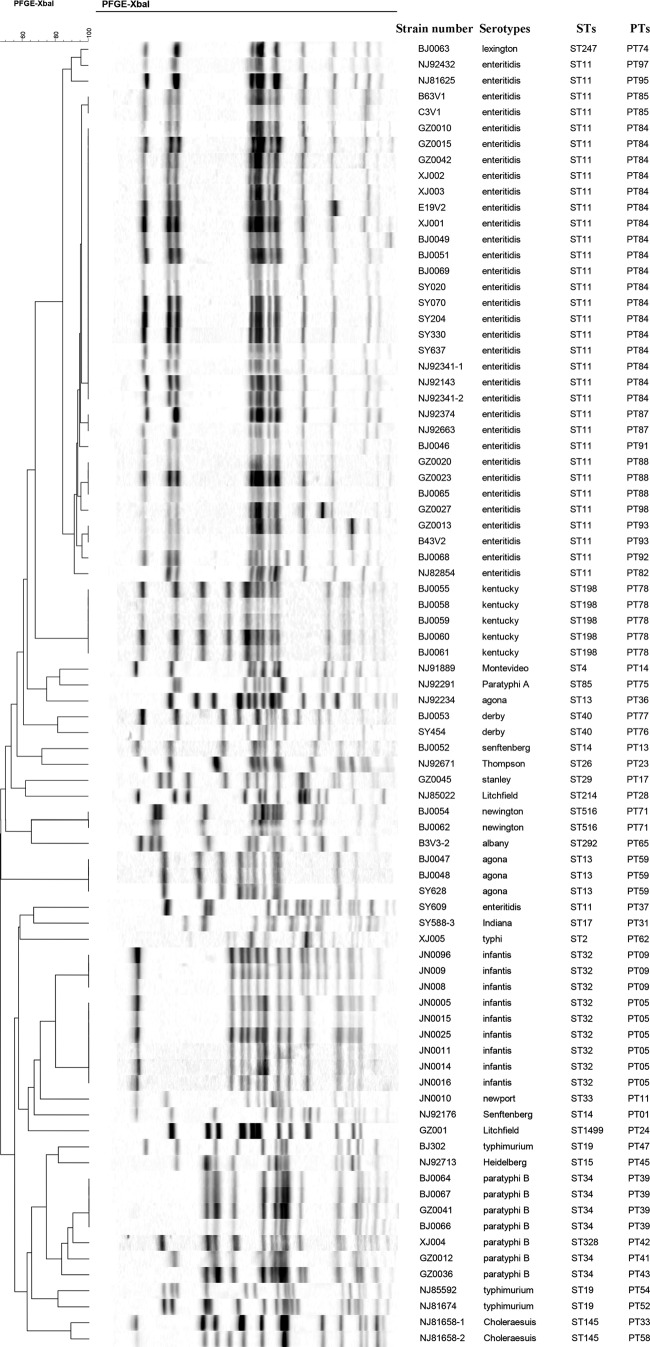
By MLST, we divided the 82 Salmonella isolates belonging to 21 serovars into 23 STs (see Table S2 in the supplemental material). Most STs (n = 21, 91.3%) were completely consistent with their serovars. Two of the S. Litchfield isolates were divided into 2 of the STs (ST214 and ST1499), and the S. Paratyphi B isolates were divided into 2 of the STs, ST34 (n = 6; 85.7%) and ST328 (n = 1; 14.3%). By CLSPT, we subtyped the 82 Salmonella isolates into 30 CLSPTs. The most frequent CLSPT type was Ent8 EntB9 (n = 34, 41.5%), and we were able to further subtype isolates of ST145, ST40, ST32, ST516, ST34, ST14, and ST19 into 14 different CLSPT types (Table 2), while by using conventional CRISPR typing, we subtyped the 82 Salmonella isolates into 33 CCTs by analyzing all the spacers contained in two CRISPR loci. The most frequent CCT was CCT7 (n = 28, 34.1%), and we subtyped isolates ST145, ST11, ST40, ST32, ST516, ST34, ST14, and ST19 into 18 different CCTs (Table 2). Using PFGE, we identified 43 profiles among the 82 isolates (Fig. 3), and the most frequent PFGE pulsotype (PT) was PT84 (n = 17, 21%). In addition, we subtyped 10 STs (ST145, ST11, ST13, ST198, ST40, ST32, ST516, ST34, ST14, and ST19) into 30 different PTs. In the 82 Salmonella isolates, the discriminatory power (discriminatory index [DI]) of CLSPT was 0.8145. This means that there should be an 81% probability that two unrelated isolates can be separated using the CLSPT scheme. The discriminatory powers of MLST, CCT, and PFGE were 0.8088, 0.8684, and 0.9455, respectively, among these isolates.
FIG 3.
PFGE dendrogram of 82 Salmonella strains, with strain number, serotype, ST, and PT for each strain.
DISCUSSION
Several molecular subtyping methods have been developed for studying the epidemiology of Salmonella, including PFGE and MLST (17). MLST has commonly been used in the subtyping of bacteria (18), and PFGE is currently the gold standard method used by public health surveillance laboratories for tracking food-borne pathogens (17), but both of them still have some disadvantages. Two studies recently suggested that CRISPR loci might provide information useful for typing (8, 13). With the use of the CRISPR typing method in Salmonella, more strategies have emerged. For example, (i) variations in the number and type of spacers can be used to track strains (14), (ii) CRISPOL, for CRISPR polymorphism, a bead-based liquid hybridization assay, is a high-throughput method for subtyping a serotype or a monophasic variant in real time (14), and (iii) a novel MLST scheme, CRISPR-multi-virulence-locus sequence typing (MVLST), using the virulence genes sseL and fimH and CRISPRs, is even better than PFGE in discrimination (14).
In this study, we proposed a new method, CRISPR locus spacer pair typing (CLSPT), to type and subtype Salmonella isolates. This new method only needs three steps. First, PCR is used to amplify both CRISPR loci. Second, the newly incorporated spacer in both CRISPR loci is investigated by reverse sequencing of the PCR products using reverse primers. Third, the sequenced newly incorporated spacers in CRISPR 1 and CRISPR 2 are used to form a pair to represent a new CRISPR type, and the explicit serovar is identified with the CLSPT/serotype dictionary (see Table S1 in the supplemental material). Thus, the CLSPT method makes the CRISPR typing method for Salmonella simpler and CRISPOL (14) more realizable in all Salmonella bacteria. Meanwhile, our study indicated that (i) CLSPT has a considerable discriminatory power (DI = 0.8145) and may provide an ideal balance between a high discriminatory power and a convenient process, (ii) CLSPT results have a high level of consistency (kappa = 0.9872, MCC = 0.9712) with the results of traditional serotyping, such that CLSPT may become a new procedure in Salmonella serovar prediction, and (iii) CLSPT may be the least expensive method for typing and subtyping of Salmonella. Traditional serotyping costs $35 to $185 per isolate (19), and the cost of 7-gene MLST is about $35 per isolate (20), whereas CLSPT was predicted to only cost about $5 to $10 per isolate. Besides the excellent time savings, low cost, high throughput, and considerable discriminatory power, CLSPT as a typing and subtyping method in Salmonella also contains geographic information, which other methods do not. A previous study demonstrated that bacteria from distant geographic locations had strikingly different spacer arrangements, possibly due to the existence of unique phage/plasmid pools in these different geographic locations (21). However, we speculate that newly incorporated spacers may represent unique ecotypes that are distinct from STs and serovars. Of note, an acquisition of a newly incorporated spacer in response to phage and/or plasmids has not yet been reported for Salmonella.
The results in this study are very preliminary, and further study is necessary to enlarge the relatively small number of S. Enteritidis isolates causing infection and to test isolates that are not S. enterica subsp. enterica. Second, it is imperative that we enlarge our CLSPT/serotype dictionary, since some common serovars, such as S. Newington, have not yet been recorded in this dictionary. It would be ideal for this dictionary to include a majority of the >2,600 Salmonella serovars or at least a majority of the serovars that are typically encountered. As studies about CRISPRs move along, we believe that the typing and subtyping methods based on CRISPRs will become more critical for Salmonella characterization and that this new CLSPT method will provide considerable advantages over other molecular serotyping methods. Particularly because this new method is simple and rapid and its results have high accordance with serotyping, it can become a valuable epidemiologic tool and may be widely used in laboratory surveillance of Salmonella infections. For example, it may be especially useful and time saving to predict the serovars of unknown isolates using CLSPT before doing the traditional serotyping. Also, the method is feasible for more laboratories or even primary units of disease control without well trained laboratory staff or sophisticated typing equipment, such as PFGE systems. Thus, the CLSPT method could greatly improve the efficiency and scope of epidemiological investigations.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Major Scientific and Technological Special Projects for Infectious Diseases during the Twelfth Five-year Plan Period (grants no. 2013ZX10004607, 2012ZX10004215, and 2013ZX10004218), the National Nature Science Foundation of China (grants no. 81371854, 81373053, 81202252, and 31200942), and the Beijing Science and Technology Nova program (grant no. xx2013061).
Footnotes
Published ahead of print 4 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00696-14.
REFERENCES
- 1.Popoff M, Le Minor L. 2001. Antigenic formulas of the Salmonella serovars, 8th ed. WHO Collaborating Centre for Reference and Research on Salmonella, World Health Organization, Geneva, Switzerland [Google Scholar]
- 2.Grimont PA, Weill F-X. 2007. Antigenic formulae of the Salmonella serovars, 9th ed. WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, France: http://www.pasteur.fr/ip/portal/action/WebdriveActionEvent/oid/01s-000036-089 [Google Scholar]
- 3.Ranieri ML, Shi C, Moreno Switt AI, den Bakker HC, Wiedmann M. 2013. Comparison of typing methods with a new procedure based on sequence characterization for Salmonella serovar prediction. J. Clin. Microbiol. 51:1786–1797. 10.1128/JCM.03201-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsen J, Skov MN, Threlfall E, Brown D. 1994. Clonal lines of Salmonella enterica serotype Enteritidis documented by IS200-, ribo-, pulsed-field gel electrophoresis and RFLP typing. J. Med. Microbiol. 40:15–22. 10.1099/00222615-40-1-15 [DOI] [PubMed] [Google Scholar]
- 5.Wattiau P, Boland C, Bertrand S. 2011. Methodologies for Salmonella enterica subsp. enterica subtyping: gold standards and alternatives. Appl. Environ. Microbiol. 77:7877–7885. 10.1128/AEM.05527-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grissa I, Vergnaud G, Pourcel C. 2008. CRISPRcompar: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 36:W145–W148. 10.1093/nar/gkn228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 8.Liu F, Kariyawasam S, Jayarao BM, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. 2011. Subtyping Salmonella enterica serovar enteritidis isolates from different sources by using sequence typing based on virulence genes and clustered regularly interspaced short palindromic repeats (CRISPRs). Appl. Environ. Microbiol. 77:4520–4526. 10.1128/AEM.00468-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamerbeek J, Schouls L, Kolk A, Van Agterveld M, Van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fricke WF, Mammel MK, McDermott PF, Tartera C, White DG, LeClerc JE, Ravel J, Cebula TA. 2011. Comparative genomics of 28 Salmonella enterica isolates: evidence for CRISPR-mediated adaptive sublineage evolution. J. Bacteriol. 193:3556–3568. 10.1128/JB.00297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Touchon M, Rocha EP. 2010. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One 5:e11126. 10.1371/journal.pone.0011126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weill F-X, Fabre-Berland L, Guibert V, Diancourt L, Brisse S. 2009. Molecular typing and subtyping of Salmonella by identification of the variable nucleotide sequences of the crispr loci. WO patent 2009115861
- 13.Liu F, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. 2011. Novel virulence gene and clustered regularly interspaced short palindromic repeat (CRISPR) multilocus sequence typing scheme for subtyping of the major serovars of Salmonella enterica subsp. enterica. Appl. Environ. Microbiol. 77:1946–1956. 10.1128/AEM.02625-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabre L, Zhang J, Guigon G, Le Hello S, Guibert V, Accou-Demartin M, de Romans S, Lim C, Roux C, Passet V, Diancourt L, Guibourdenche M, Issenhuth-Jeanjean S, Achtman M, Brisse S, Sola C, Weill FX. 2012. CRISPR typing and subtyping for improved laboratory surveillance of Salmonella infections. PLoS One 7:e36995. 10.1371/journal.pone.0036995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richter C, Chang JT, Fineran PC. 2012. Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated (Cas) systems. Viruses 4:2291–2311. 10.3390/v4102291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, Wrigley D, Barrett T, Ribot E. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045–1050. 10.1128/JCM.43.3.1045-1050.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerner-Smidt P, Hise K, Kincaid J, Hunter S, Rolando S, Hyytiä-Trees E, Ribot E, Swaminathan B. 2006. PulseNet USA: a five-year update. Foodborne Pathog. Dis. 3:9–19. 10.1089/fpd.2006.3.9 [DOI] [PubMed] [Google Scholar]
- 18.Cho S, Boxrud DJ, Bartkus JM, Whittam TS, Saeed M. 2007. Multiple-locus variable-number tandem repeat analysis of Salmonella Enteritidis isolates from human and non-human sources using a single multiplex PCR. FEMS Microbiol. Lett. 275:16–23. 10.1111/j.1574-6968.2007.00875.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guard J, Sanchez-Ingunza R, Morales C, Stewart T, Liljebjelke K, Kessel J, Ingram K, Jones D, Jackson C, Fedorka-Cray P. 2012. Comparison of dkgB-linked intergenic sequence ribotyping to DNA microarray hybridization for assigning serotype to Salmonella enterica. FEMS Microbiol. Lett. 337:61–72. 10.1111/1574-6968.12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Achtman M, Wain J, Weill F-X, Nair S, Zhou Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uesbeck A. 2012. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 8:e1002776. 10.1371/journal.ppat.1002776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunin V, He S, Warnecke F, Peterson SB, Martin HG, Haynes M, Ivanova N, Blackall LL, Breitbart M, Rohwer F. 2008. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 18:293–297. 10.1101/gr.6835308 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.