Abstract
We evaluated the use of the molecular bacterial load (MBL) assay, for measuring viable Mycobacterium tuberculosis in sputum, in comparison with solid agar and liquid culture. The MBL assay provides early information on the rate of decline in bacterial load and has technical advantages over culture in either form.
TEXT
Assessment of Mycobacterium tuberculosis sputum bacterial load has routinely been performed using solid culture (1). Automated liquid culture has been proposed as an alternative (2), but all culture-based methods are hampered by contamination with other microorganisms (3), protracted time to obtain results (4), and populations of viable, nonculturable bacteria (5, 6). We recently described the molecular bacterial load (MBL) assay, based on 16S rRNA, observing that MBL declined biphasically in response to treatment (7). In the present study, we directly compared the MBL assay with solid and liquid culture on 148 sputum samples (collected overnight from 1600 h until 0800 h the following morning) from 20 patients enrolled and hospitalized at the EBA Unit, Tanzanian National Institute for Medical Research-Mbeya Medical Research Centre, Tanzania. Samples were collected pretreatment (2 overnight samples) and longitudinally (days 2, 4, 5, 7, 10, and 14) during standard WHO treatment for drug-sensitive tuberculosis. The OEBA (Observation of Early Bactericidal Activity) study was a capacity building project of the PanACEA consortium. Before patient enrolment, the study was approved by the site's local ethics board (reference MRH/T.30/44/2) and national ethics board (reference NIMR/HQ/R.8a/Vol.IX/1169) and the sponsor (University of Munich) ethics board. The study was conducted in compliance with the declaration of Helsinki. All patients provided written informed consent to study participation, including use of their samples for evaluation of novel molecular assays. The study was registered in the Pan African Clinical Trials Registry (pactr.org) under PACTR201209000394102.
Sputum was homogenized, and one half was decontaminated with N-acetyl-l-cysteine-sodium hydroxide (1% final NaOH concentration) prior to inoculation into mycobacterial growth indicator tubes (BBL MGIT; Becton, Dickinson and Company, MD, USA), according to the manufacturer's instructions, in order to determine time to positivity (TTP) in the Bactec MGIT 960 (Becton, Dickinson and Company, MD, USA). The second portion was mixed with an equal volume of Sputasol (Oxoid, Limited, United Kingdom), and quadruplicate 100-μl volumes of 10-fold dilutions were inoculated onto 7H11 agar containing Mycobacteria Selectatab Kirchner supplement (Mast Group, Limited, United Kingdom). Nine hundred fifty microliters of sputum/Sputasol was preserved in guanidine thiocyanate containing 1% β-mercaptoethanol, and the MBL assay was performed as detailed in the work of Honeyborne et al. (7). MBL value was assigned based on 16S rRNA cycle threshold (CT) using the normalized 16S rRNA CT in the following equation: bacterial load (log10) = (normalized 16S CT − 31.76)/−3.171 (7).
Artificial sputum (see Text S1 in the supplemental material) developed as a matrix for high assay standards was found to be a good substitute for pooled human sputum. High standards had mean 7.46 log10 bacilli (standard error of the mean [SEM], 0.08) and 7.49 log10 bacilli (SEM, 0.07) (P = 0.80) for pooled human sputum and artificial sputum, respectively (see Fig. S1 in the supplemental material). The low standard mean of 3.70 log10 bacilli (SEM, 0.13) for pooled human sputum was statistically different from that of artificial sputum (mean, 3.38 log10 bacilli; SEM, 0.13; P = 0.01) (see Fig. S1). However, the SEMs are identical for the two matrices, and therefore, the critical factor, reproducibility of extraction of an assigned bacterial number, was met.
Spearman rank correlations for 148 sputum samples revealed a high degree of correlation between the assays: log10 MBL compared to solid agar log10 CFU (r = +0.84; 95% confidence interval [CI], 0.78, 0.88), log10 MBL compared to solid agar log10 TTP (r = −0.81; 95% CI, −0.86, −0.74), and log10 TTP compared to solid agar log10 CFU (r = −0.78; 95% CI, −0.84, −0.71; for all, P < 0.0001) (see Fig. S2 and Text S2 in the supplemental material). Correlation between log10 CFU and log10 TTP for our study was comparable to that in a study of >2,000 sputum samples tested in South Africa (r = −0.72) (2), although TTP was not log-transformed in this study.
Direct comparison of solid culture and the MBL assay for individual patients revealed that the declines in bacterial load closely matched during the first 14 days of therapy (Fig. 1). rRNA is downregulated in bacteria that are entering dormancy (8), and so one might expect that the MBL assay is an underestimate of bacterial load. However, the baseline median bacterial load was 0.43 log10 (95% CI, 0.12, 0.73; P = 0.008) higher when measured using the MBL assay than when measured by the solid agar CFU method (paired t test, P = 0.008). This may be explained by the selective antibiotics in the solid agar method killing a proportion of the bacteria present. We observed a similar effect for six clinical isolates (M. tuberculosis [n = 5] and Mycobacterium bovis [n = 1]) isolated from decontaminated sputum on Lowenstein-Jensen (LJ) slopes (Southern Group Laboratories, Limited, United Kingdom). Determination of bacterial number for these was done using the Miles-Misra method (9) on 7H10 agar with 10% oleic acid-albumin-dextrose-catalase (OADC) supplement (Becton, Dickinson and Company, MD, USA) with and without the addition of antibiotics using bacteria subcultured from the LJ slope into 7H9 medium containing 0.2% Tween 80 and 10% ADC supplement (Becton, Dickinson and Company, MD, USA). Addition of antibiotics (Mycobacteria Selectatabs Kirchner; Mast Group, Limited, United Kingdom) reduced the bacterial count by 0.47 log10 (95% CI, 0.23, 0.71; paired t test, P = 0.004). This may explain the increased MBL readout observed here for patient sputum in comparison to that for solid culture despite comparable rates of decline.
FIG 1.
Individual patient plots comparing bacterial load measured using solid agar and MBL during the first 14 days of treatment for 20 individuals.
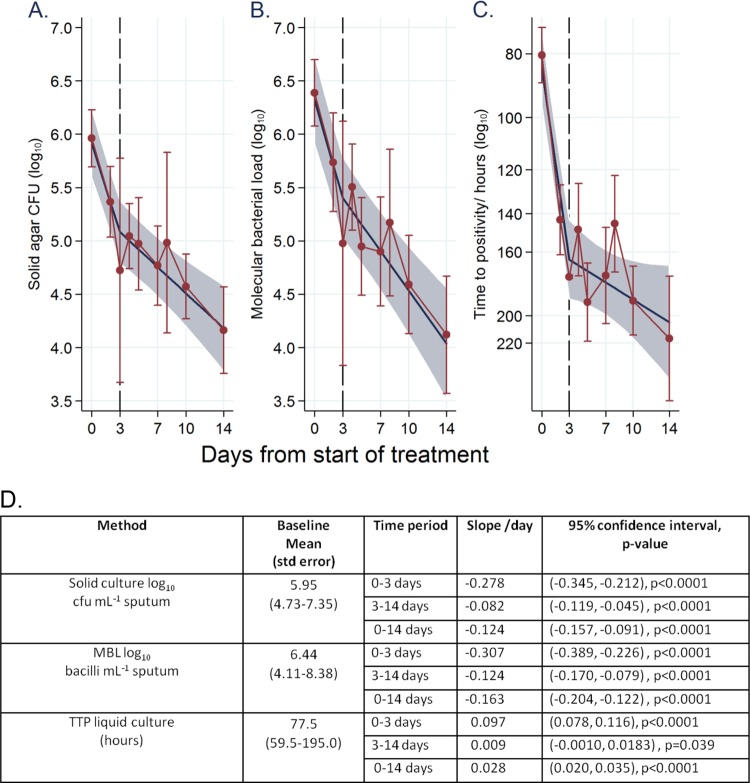
A biphasic decline in bacterial load was observed for liquid and solid culture and the MBL assay with a node at day 3 (Fig. 2). Full details of statistical analysis are given in Text S2 in the supplemental material. The decline rate for days 0 to 3 for solid agar was −0.278 log10 day−1 (95% confidence interval, −0.345, −0.212), and that for MBL was −0.307 log10 day−1 (−0.389, −0.226). MBL decline was therefore slightly quicker over the first 3 days, although the confidence intervals overlap. The use of mRNA for the isocitrate lyase gene and the use of noncoding ribosomal promoter region mRNA as amplification targets for bacterial quantification have previously been found to respond comparably to culture during days 2 to 7 of therapy but not during the first 2 days of therapy (10). In contrast, MBL robustly matched solid and liquid culture over the early phase of treatment. Extended early bactericidal activities (EBAs) from days 3 to 14 were also comparable, with −0.082 log10 (−0.119, −0.045) and −0.124 log10 (−0.170, −0.079) for CFU on solid medium and MBL, respectively. These data match other EBA studies measuring treatment response with solid culture (11–16). The close comparability between MBL and culture contrasts with data for treatment monitoring using the GeneXpert MTB/RIF assay (Xpert; Cepheid, CA, USA). GeneXpert MTB/RIF did not respond to changes in bacterial load as rapidly as did culture (17). Within-subject variation of Xpert was high, at 56.7%. In contrast, MBL had a 9.6% variance between 2 baseline samples for 16 subjects (18), suggesting that MBL is more reproducible. To corroborate this, within-patient variabilities for solid (17.9%) and liquid (21.6%) culture in our study were compared to those in the study by Kayigire et al., with 16.5% and 22%, respectively (18). Prior to exclusion of outliers, solid culture had invalid results in 11% of samples (18 of 169), whereas MBL had only 1.2% of samples (2 of 169) unreadable.
FIG 2.
Bacterial load during the first 14 days of treatment: linear mixed-effects modeling using splines (splines allow for estimating different slopes before and after the specified node). (A) CFU on solid culture. (B) MBL. (C) Liquid culture (the scale on the vertical axis for panel C is reversed to aid comparison with panels A and B). (D) Declines of bacteria per day for the 3 methods. The red line shows the raw mean with 95% confidence interval, and the thicker blue line shows the fitted biphasic slope with 95% confidence region.
In summary, our data show that the MBL assay is at least as good as culture for measuring EBA during standard tuberculosis therapy to day 14, with higher precision, fewer missing data, and a shorter time to result (24 h compared to weeks). This assay shows promise as a replacement for culture in future EBA trials testing new drugs.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Denis Ngatemelela and Chacha Mangu for clinical support.
The work was supported by the following grants European Metrology Research Programme (EMRP) EURAMET grant HLT-08 INFECT-MET. The EMRP is jointly funded by the EMRP participating countries within EURAMET and the European Union. PanACEA is funded by the European and Developing Countries Clinical Trials Partnership, grant IP.2007.32011.013, and the German Ministry for Education and Research, grant 01KA0901.
The Pan African Consortium for the Evaluation of Anti-tuberculosis Antibiotics (PanACEA) comprises the following institutions and individuals: Medical Centre of the University of Munich, Munich, Germany (Sonja Henne, Anna Maria Mekota, Norbert Heinrich, Anna Framhein, Andrea Rachow, Anke Kohlenberg, Elmar Saathoff, and Michael Hoelscher); University of St. Andrews, St Andrews, United Kingdom (Stephen Gillespie); Radboud University Nijmegen Medical Centre. Nijmegen, the Netherlands (Georgette Plemper van Balen, Marloes Weijers, Rob Aarnoutse, and Martin Boeree); University College of London, London, United Kingdom (Anna Bateson, Timothy McHugh, Saraswathi Murthy, Robert Hunt, and Alimuddin Zumla); MRC Clinical Trials Unit, London, United Kingdom (Andrew Nunn, Patrick Phillips, and Sunita Rehal); University of Cape Town, Cape Town, South Africa (Rodney Dawson and Kim Narunsky); University of Stellenbosch, Cape Town, South Africa (Andreas Diacon, Jeannine du Bois, Amour Venter, and Sven Friedrich); University of the Witswatersrand, Johannesburg, South Africa (Ian Sanne, Karla Mellet, and Eefje de Jong); The Aurum Institute, Johannesburg, South Africa (Gavin Churchyard and Salome Charalambous); University of Zambia, Lusaka, Zambia (Peter Mwaba); NIMR-Mbeya Medical Research Centre, Mbeya, Tanzania (Nyanda Elias, Chacha Mangu, Gabriel Rojas-Ponce, Bariki Mtafya, and Leonard Maboko); Ifakara Health Institute-Bagamoyo Research and Training Centre, Bagamoyo, Tanzania (Lilian Tina Minja and Mohamed Sasamalo); Swiss Tropical and Public Health Institute, Basel, Switzerland (Klaus Reither and Levan Jugheli); Kilimanjaro Clinical Research Institute, Moshi, Tanzania (Noel Sam, Gibson Kibiki, Hadija Semvua, Norah Ndusilo, and Stellah Mpagama); Medical Research Unit-Albert Schweitzer Hospital, Lambarene, Gabon (Abraham Alabi and Ayola Akim Adegnika); Kenya Medical Research Institute, Nairobi, Kenya (Evans Amukoye); Makerere University, Kampala, Uganda (Alphonse Okwera).
Footnotes
Published ahead of print 28 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01128-14.
REFERENCES
- 1.Donald PR, Sirgel FA, Venter A, Parkin DP, Seifart HI, van de Wal BW, Maritz JS, Fourie PB. 2003. Early bactericidal activity of antituberculosis agents. Expert Rev. Anti Infect. Ther. 1:141–155. 10.1586/14787210.1.1.141 [DOI] [PubMed] [Google Scholar]
- 2.Diacon AH, Maritz JS, Venter A, van Helden PD, Dawson R, Donald PR. 18 August 2011. Time to liquid culture positivity can substitute for colony counting on agar plates in early bactericidal activity studies of antituberculosis agents. Clin. Microbiol. Infect. 10.1111/j.1469-0691.2011.03626.x [DOI] [PubMed] [Google Scholar]
- 3.Cruciani M, Scarparo C, Malena M, Bosco O, Serpelloni G, Mengoli C. 2004. Meta-analysis of BACTEC MGIT 960 and BACTEC 460 TB, with or without solid media, for detection of mycobacteria. J. Clin. Microbiol. 42:2321–2325. 10.1128/JCM.42.5.2321-2325.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyrrell FC, Budnick GE, Elliott T, Gillim-Ross L, Hildred MV, Mahlmeister P, Parrish N, Pentella M, Vanneste J, Wang YF, Starks AM. 2012. Probability of negative Mycobacterium tuberculosis complex cultures based on time to detection of positive cultures: a multicenter evaluation of commercial-broth-based culture systems. J. Clin. Microbiol. 50:3275–3282. 10.1128/JCM.01225-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukamolova GV, Turapov O, Malkin J, Woltmann G, Barer MR. 2010. Resuscitation-promoting factors reveal an occult population of tubercle bacilli in sputum. Am. J. Respir. Crit. Care Med. 181:174–180. 10.1164/rccm.200905-0661OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shleeva MO, Bagramyan K, Telkov MV, Mukamolova GV, Young M, Kell DB, Kaprelyants AS. 2002. Formation and resuscitation of “non-culturable” cells of Rhodococcus rhodochrous and Mycobacterium tuberculosis in prolonged stationary phase. Microbiology 148:1581–1591 [DOI] [PubMed] [Google Scholar]
- 7.Honeyborne I, McHugh TD, Phillips PP, Bannoo S, Bateson A, Carroll N, Perrin FM, Ronacher K, Wright L, van Helden PD, Walzl G, Gillespie SH. 2011. Molecular bacterial load assay, a culture-free biomarker for rapid and accurate quantification of sputum Mycobacterium tuberculosis bacillary load during treatment. J. Clin. Microbiol. 49:3905–3911. 10.1128/JCM.00547-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717–731. 10.1046/j.1365-2958.2002.02779.x [DOI] [PubMed] [Google Scholar]
- 9.Miles AA, Misra SS, Irwin JO. 1938. The estimation of the bactericidal power of the blood. J. Hyg. (Lond.) 38:732–749. 10.1017/S002217240001158X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Mahan CS, Palaci M, Horter L, Loeffelholz L, Johnson JL, Dietze R, Debanne SM, Joloba ML, Okwera A, Boom WH, Eisenach KD. 2010. Sputum Mycobacterium tuberculosis mRNA as a marker of bacteriologic clearance in response to antituberculosis therapy. J. Clin. Microbiol. 48:46–51. 10.1128/JCM.01526-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies GR, Brindle R, Khoo SH, Aarons LJ. 2006. Use of nonlinear mixed-effects analysis for improved precision of early pharmacodynamic measures in tuberculosis treatment. Antimicrob. Agents Chemother. 50:3154–3156. 10.1128/AAC.00774-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brindle R, Odhiambo J, Mitchison D. 2001. Serial counts of Mycobacterium tuberculosis in sputum as surrogate markers of the sterilising activity of rifampicin and pyrazinamide in treating pulmonary tuberculosis. BMC Pulm. Med. 1:2. 10.1186/1471-2466-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rustomjee R, Lienhardt C, Kanyok T, Davies GR, Levin J, Mthiyane T, Reddy C, Sturm AW, Sirgel FA, Allen J, Coleman DJ, Fourie B, Mitchison DA. 2008. A phase II study of the sterilising activities of ofloxacin, gatifloxacin and moxifloxacin in pulmonary tuberculosis. Int. J. Tuberc. Lung Dis. 12:128–138 [PubMed] [Google Scholar]
- 14.Gillespie SH, Gosling RD, Charalambous BM. 2002. A reiterative method for calculating the early bactericidal activity of antituberculosis drugs. Am. J. Respir. Crit. Care Med. 166:31–35. 10.1164/rccm.2112077 [DOI] [PubMed] [Google Scholar]
- 15.Jindani A, Dore CJ, Mitchison DA. 2003. Bactericidal and sterilizing activities of antituberculosis drugs during the first 14 days. Am. J. Respir. Crit. Care Med. 167:1348–1354. 10.1164/rccm.200210-1125OC [DOI] [PubMed] [Google Scholar]
- 16.Diacon AH, Dawson R, von Groote-Bidlingmaier F, Symons G, Venter A, Donald PR, van Niekerk C, Everitt D, Winter H, Becker P, Mendel CM, Spigelman MK. 2012. 14-day bactericidal activity of PA-824, bedaquiline, pyrazinamide, and moxifloxacin combinations: a randomised trial. Lancet 380:986–993. 10.1016/S0140-6736(12)61080-0 [DOI] [PubMed] [Google Scholar]
- 17.Friedrich SO, Rachow A, Saathoff E, Singh K, Mangu CD, Dawson R, Phillips PP, Venter A, Bateson A, Boehme CC, Heinrich N, Hunt RD, Boeree MJ, Zumla A, McHugh TD, Gillespie SH, Diacon AH, Hoelscher M, Pan African Consortium for the Evaluation of Anti-tuberculosis Antibiotics (PanACEA) 2013. Assessment of the sensitivity and specificity of Xpert MTB/RIF assay as an early sputum biomarker of response to tuberculosis treatment. Lancet Respir. Med. 1:462–470. 10.1016/S2213-2600(13)70119-X [DOI] [PubMed] [Google Scholar]
- 18.Kayigire XA, Friedrich SO, Venter A, Dawson R, Gillespie SH, Boeree MJ, Heinrich N, Hoelscher M, Diacon AH, Pan African Consortium for the Evaluation of Anti-tuberculosis Antibiotics 2013. Direct comparison of Xpert MTB/RIF assay with liquid and solid mycobacterial culture for quantification of early bactericidal activity. J. Clin. Microbiol. 51:1894–1898. 10.1128/JCM.03290-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.