Abstract
Changes in periodontal status are associated with shifts in the composition of the bacterial community in the periodontal pocket. The relative abundances of several newly recognized microbial species, including Filifactor alocis, as-yet-unculturable organisms, and other fastidious organisms have raised questions on their impact on disease development. We have previously reported that the virulence attributes of F. alocis are enhanced in coculture with Porphyromonas gingivalis. We have evaluated the proteome of host cells and F. alocis during a polymicrobial infection. Coinfection of epithelial cells with F. alocis and P. gingivalis strains showed approximately 20% to 30% more proteins than a monoinfection. Unlike F. alocis ATCC 35896, the D-62D strain expressed more proteins during coculture with P. gingivalis W83 than with P. gingivalis 33277. Proteins designated microbial surface component-recognizing adhesion matrix molecules (MSCRAMMs) and cell wall anchor proteins were highly upregulated during the polymicrobial infection. Ultrastructural analysis of the epithelial cells showed formation of membrane microdomains only during coinfection. The proteome profile of epithelial cells showed proteins related to cytoskeletal organization and gene expression and epigenetic modification to be in high abundance. Modulation of proteins involved in apoptotic and cell signaling pathways was noted during coinfection. The enhanced virulence potential of F. alocis may be related to the differential expression levels of several putative virulence factors and their effects on specific host cell pathways.
INTRODUCTION
While recent attention has focused on the study of the composition of the human microbiome, the inherent mechanisms underlying the complex interpathogen and host-pathogen interactions leading to polymicrobial infectious diseases of an inflammatory nature are still poorly defined. One such inflammatory disease, periodontitis, has a multifactorial etiology which is influenced by host genetics and several environmental factors. Further, there is evidence that this inflammatory disease affecting the periodontium represents an increased risk for several systemic diseases, including atherosclerosis (1), diabetes (2), and rheumatoid arthritis (3, 4). Historically, periodontal disease is associated with several pathogens contributing to a complex microbial milieu which can initiate or directly contribute to host tissue destruction (5). Bacteria such as Porphyromonas gingivalis, Prevotella intermedia, Aggregatibacter (Actinobacillus) actinomycetemcomitans, Tannerella forsythia, and Treponema denticola have previously been demonstrated to be major pathogens associated with periodontal diseases (6–8). A comparative oral microbiome analysis of the healthy and diseased states has indicated diversity in the microbial communities (9, 10). Collectively, these studies have demonstrated that changes in the periodontal status are associated with shifts in the composition of the bacterial community in the periodontal pocket (11, 12). The relative abundances of several newly recognized microbial species, as-yet-unculturable organisms, and other fastidious organisms (9, 13, 14) have raised questions on their impact on disease development.
Filifactor alocis, a Gram-positive, asaccharolytic, obligate anaerobic rod, based on the emerging microbiome data, is one of the marker organisms associated with periodontal inflammation and is suggested to be an important organism for periodontal disease (15–19). Further, in comparison to the other traditional periodontal pathogens, the high incidence of F. alocis in the periodontal pocket compared to its absence in healthy or periodontitis-resistant patients could support the idea of its importance in the infectious state of the disease (16, 17, 20). This organism, first isolated in 1985 from the gingival sulcus in gingivitis and periodontics patients, was originally classified as Fusobacterium alocis (21). However, based on phylogenetic analysis using 16S rRNA sequences, it was reclassified in 1999 into the genus Filifactor (22).
We have earlier demonstrated that F. alocis has virulence properties that may enhance its ability to survive and persist in the periodontal pocket (23). For example, it was relatively resistant to oxidative stress and its stimulated growth under those conditions could be an important attribute (23). As reported elsewhere, others have shown that F. alocis can induce secretion of proinflammatory cytokines, including interleukin-1β (IL-1β), IL-6, and tumor necrosis factor alpha (TNF-α), from gingival epithelial cells and can trigger apoptosis of these cells (24). Colonization and survival of F. alocis in a mouse model showed proapoptotic local infection that was rapidly resolved by host neutrophil influx (25). A comparative analysis of several F. alocis isolates showed heterogeneity in their levels of virulence potential (23). F. alocis can interact with other important periodontal pathogens such as P. gingivalis (26). Further, in coculture with P. gingivalis, these F. alocis strains showed variations in their capacity for invasion of epithelial cells (23) While synergistic interactions during polymicrobial infections have resulted in enhanced pathogenesis of periodontopathogens such as P. gingivalis (27), whether there is a similar mechanism(s) for F. alocis is unclear. It is likely that surface and secretory proteins from F. alocis play a role in this process.
Host-pathogen interactions are known to induce significant changes in the transcriptional program of the host cells resulting in the mobilization of genes involved in key processes that mediate the appropriate response. Some of these changes may lead to epigenetic modifications that are associated with a variety of biological processes, including cell differentiation, proliferation, and immunity (28, 29). Successful pathogens have developed novel strategies, including bacterially induced epigenetic deregulation that may affect host cell function to facilitate their survival and persistence. Proteomics analyses have significantly contributed toward a deeper understanding of the molecular mechanisms utilized by several oral pathogens such as Streptococcus mutans (30), Streptococcus oralis (31), Fusobacterium nucleatum (32), and P. gingivalis (33–35) during their interaction with the host. In a previous host-pathogen interaction study performed with epithelial cells, we showed proteome variation in F. alocis with upregulation of many important bacterial proteins (36) that could potentially trigger direct or indirect epigenetic modifications in the host. Because virulence heterogeneity has been observed in F. alocis (23), it is unclear which key host pathways were modulated in this interaction that may lead to the differential host response during the infectious process. In this study, we used shotgun proteomics-based differential protein expression analysis and relative quantification of both F. alocis and host proteins to study pathogen-dependent host modulations. We have also used metabolomics to evaluate the changes associated with metabolic pathways and networks that could influence the variation in cell responses during the infectious process.
MATERIALS AND METHODS
Bioinformatics analysis.
The DNA and amino acid sequences were aligned using Bioedit (http://www.mbio.ncsu.edu/bioedit/bioedit.html). The phylogenetic relationship between the sequences of these oral pathogens was analyzed using MEGA version 4.0 (37). The signal peptide and potential cleavage sites were predicted using both neural network and hidden Markov model methods (38). Metabolic pathway analysis was carried out using the Kyoto Encyclopedia of Genes and Genomes (KEGG) (www.genome.jp/kegg/) (39).
Bacterial strains and growth conditions.
F. alocis ATCC 35896 was purchased from the American Type Culture Collection (Rockville, MD). F. alocis D-62D was a gift from Floyd Dewhirst, the custodian of the Moores' anaerobic microbial collection (The Forsyth Institute, Boston, MA). The identity of the F. alocis D-62D strain was confirmed by 16S rRNA gene sequencing (D-62D; GenBank accession no. GU968904). F. alocis strains were grown initially in Robertson's bullock heart medium followed by adaptation to brain heart infusion (BHI) broth supplemented with hemin (5 μg/ml), vitamin K (0.5 μg/ml), cysteine (1 μg/ml), and arginine (17.42 μg/ml). P. gingivalis strains were grown in BHI broth (Difco) supplemented with hemin (5 μg/ml), vitamin K (0.5 μg/ml), and cysteine (0.1%). Blood agar medium was prepared by the addition of sheep blood (5%) and agar (2%). The bacterial cultures were incubated at 37°C in an anaerobic chamber (Coy Manufacturing) in an atmosphere that included 10% H2, 10% CO2, and 80% N2. Growth rates were determined spectrophotometrically (optical density at 600 nm [OD600]).
Epithelial cell culture.
HeLa cells were grown and maintained at a density of 2 × 105 cells/ml in a humid incubator with 5% CO2 at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (100 IU/ml), streptomycin (100 IU/ml), and amphotericin B (2.5 mg/ml) (Invitrogen, Carlsbad, CA). The volume of cells was split into two halves, in fresh prewarmed medium. Confluent stock cultures were trypsinized, adjusted to approximately 5 × 103 cells/ml, seeded (1 ml per well) into 12-well plates (Nunc, Rochester, NY), and further incubated for 48 h to reach semiconfluence (105 cells per well).
Coculture of F. alocis and P. gingivalis and standard antibiotic protection assay.
Invasion of epithelial cells was quantified using the standard antibiotic protection assay (40). Briefly, an isolated bacterial colony harvested from a solid agar plate was grown to the exponential phase in BHI broth. The bacterial cells were then centrifuged, washed three times in phosphate-buffered saline (PBS), and adjusted to 107 CFU/ml of bacteria in DMEM. The epithelial cell monolayer was washed three times with PBS, infected with bacteria at a multiplicity of infection (MOI) of 1:100 (105 epithelial cells), and then incubated at 37°C for 30 and 45 min under 5% CO2. Nonadherent bacteria were removed by washing with PBS, while cell surface-bound bacteria were killed with metronidazole (200 μg/ml, 60 min). F. alocis is sensitive to 100 μg/ml of metronidazole. After removal of the antibiotic, the internalized bacteria were released by osmotic lysis of the epithelial cells in sterile distilled water. Lysates were serially diluted, plated (in duplicate) on BHI agar, and incubated for 6 to 10 days. The number of bacterial cells recovered was expressed as a percentage of the original inoculum. The number of adherent bacteria was obtained by subtracting the number of intracellular bacteria from the total number of bacteria obtained in the absence of metronidazole (41). Coinfection was performed as described previously (7). F. alocis and P. gingivalis inocula were prepared by mixing equal volumes (1 × 107 cells/well) of bacterial suspension which was then incubated for 5 min in the anaerobic chamber prior to infection. The serially diluted lysate was plated on BHI blood agar and incubated for 6 to 10 days. The bacterial colonies were phenotypically identified.
EM.
Transmission electron microscopy (TEM) was performed using an FEI G2 TEM per the method of Harris (42). The processed grids were placed in stain solution containing neutral 1% aqueous phosphotungstic acid for 30 s. After being blotted dry, the slides were examined using an FEI G2 TEM.
Ultrathin sections were made per the method described by Massey (43). The trypsinized HeLa cell monolayers were pelleted and postfixed in 1% OsO4–0.1 M sodium cacodylate for 1 h, and the ultrathin sections were contrasted using lead citrate and uranyl acetate before examination using an FEI-Technai G2 transmission electron microscope.
Scanning electron microscopy was performed per the method described in reference 44 using a Philips XL30 FEG (FEI). The trypsinized culture of HeLa epithelial cells (5 × 103 cells/ml) was grown on coverslips in 4-well BD Falcon culture slides (BD Falcon, Bedford, MA) in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (100 IU/ml), streptomycin (100 IU/ml), and amphotericin B (2.5 mg/ml) (Invitrogen, Carlsbad, CA), at 37°C under 5% CO2. The infected epithelial cells were later grown in Eagle's DMEM without serum several times to ensure complete transition from growth media to fixative, and fixation was carried for 30 min at room temperature. The processed coverslips were subjected to carbon coating using a vacuum evaporator and sputter coated. The slides were then viewed using a Philips XL30 FEG.
Sample preparation and labeling for MS.
The infected epithelial cells were trypsinized followed by centrifugation at 5,000 × g, and the pellet was freeze-thawed twice. Approximately 300 μg of protein sample in 100 mM Tris-HCl (pH 8.6)–0.1% SDS buffer solution was reduced by incubation in 10 mM dithiothreitol (DTT) at 50°C for 1 h followed by carboxymethylation with 25 mM iodoacetamide in the dark for 2 h. Proteins were precipitated using cold acetone at −20°C overnight. The protein pellet was obtained by centrifugation at 15,000 rpm for 10 min, and the supernatant was removed using a glass Pasteur pipette. The protein pellet was dissolved in 80 mM triethyl ammonium bicarbonate buffer. Proteins were subsequently digested using trypsin at a protein/enzyme ratio of 40:1 (by mass), and the samples were incubated at 37°C overnight. A tandem mass tag (TMT) isobaric mass tagging kit (Thermo Fisher Scientific) was used for labeling the samples following the manufacturer's recommended conditions. The U937 control protein digests were labeled with TMT labels 128 and 130, whereas the phorbol myristate acetate (PMA)-treated protein digests were labeled with TMT labels 127 and 129. Equal amounts of the labeled control and PMA-treated protein digests were combined for mass spectrometry (MS) analysis.
MS and data analysis.
Protein samples were analyzed using a Thermo Scientific LTQ Orbitrap Velos mass spectrometer and the data processed as previously reported (45). The four-part protocol used for the MS and tandem MS (MS/MS) analyses was carried out, and the data collection was achieved using Xcalibur software (Thermo Electron) followed by screening performed with Bioworks 3.1.
Data processing and functional analysis.
The data from the Orbitrap were processed and searched using Thermo Scientific Proteome Discoverer software suite 1.1. The MASCOT software was used for each analysis to produce unfiltered data and output files. Statistical validation of peptide and protein findings was achieved using X TANDEM (www.thegpm.org) and SCAFFOLD 2 meta-analysis software. The presence of two different peptides at a probability of at least 95% was required for consideration of a result as representing positive identification. General protein database searches were conducted using the UniProtKb protein knowledge base database (http://www.uniprot.org/uniprot). The F. alocis open reading frame (ORF) database is based on the latest release of the Filifactor alocis genome at the NCBI genome project (http://www.ncbi.nlm.nih.gov/nuccore/CP002390.1). Human protein identifications were performed using the Human IPI database (ftp://ftp.ebi.ac.uk/pub/databases/IPI) (46). A precursor ion mass tolerance of 10 ppm and fragment ion tolerance of 0.01 Da and MUDPIT scoring were applied with a peptide cutoff score of 10 and a peptide relevance score of 1. Peptides were filtered based on false-discovery-rate cutoff values of 1% (strict) and 5% (relaxed). Using MASCOT searching, 98% of the total peptides detected were quantified. Protein ratios were reported as the mean values for the observed peptides.
Functional analysis.
The NCBI RefSeq symbols of the modulated genes were mapped to their corresponding gene names in Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems). The corresponding lists were processed using the IPA software to arrive at the canonical pathways, biological functions, and networks significantly associated with the gene lists. Ingenuity Pathways Analysis is a knowledge database and Web-based analysis system that permits classification of the molecular networks and biological function and metabolic canonical pathways that are most significantly represented in the set of genes of interest. The P value associated with biological process or pathway annotation was calculated according to the right-tailed Fisher exact test. This statistical test assesses the null hypothesis that the proportion of genes that map to a particular function or pathway in the sample is similar to the proportion that map in the entire population (IPA reference set). Only overrepresented functions or pathways that are more abundant than expected by chance are reported as significant and given the respective color codes for up- and downregulation. The gene ontology classification was used for referencing the eukaryotic proteome through manual curation and also using QuickGo (www.ebi.ac.uk/quickgo/).
RESULTS
Several proteins from F. alocis D-62D were increased in abundance during coinfection of epithelial cells with P. gingivalis W83.
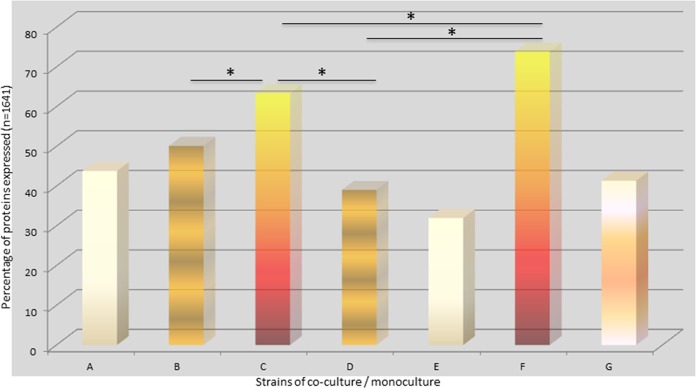
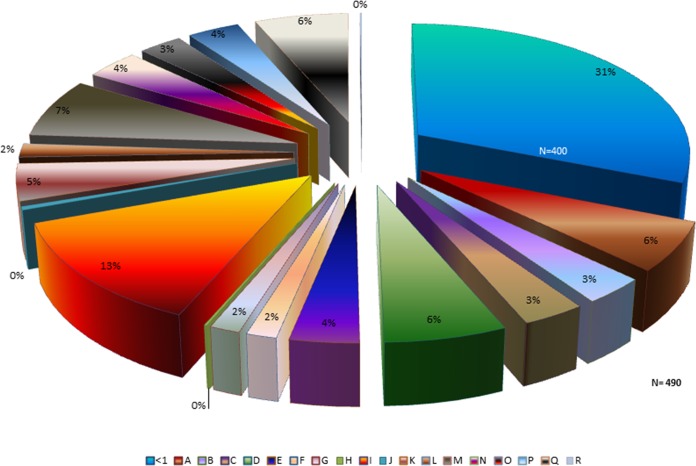
Because the variations in the pathogenic potential of the F. alocis strains in coculture with P. gingivalis may be related to the relative abundances of specific bacterial protein factors, we examined the proteome of F. alocis during coinfection of epithelial cells with P. gingivalis. As shown in Fig. 1, approximately 20% to 30% (comparing bars C and F with bar G) more proteins were observed in F. alocis during coinfection with P. gingivalis (P < 0.01). Unlike F. alocis ATCC 35896, the D-62D strain expressed more proteins interacting with P. gingivalis W83 than with P. gingivalis 33277. The proteome modulation of F. alocis during coinfection with P. gingivalis W83 showed a total of 490 proteins with a change in expression of >1-fold in contrast to 400 proteins that had a change in expression of 0.5-fold to 1.0-fold. The proteins that were most highly upregulated were classified as hypothetical (13%), regulatory (7%), and transport and binding (6%) proteins and related to cellular processes (6%) and amino acid biosynthesis (6%) (Fig. 2). Several surface adhesion proteins that were upregulated during coinfection include collagen adhesion protein, fibronectin binding protein, calcium binding acid repeat proteins, and hemolysin III calcium binding protein. Furthermore, many hypothetical proteins with cell wall anchor motifs (HMPREF0389_1719, HMPREF0389_00599, HMPREF0389_00019, HMPREF0389_00672, HMPREF0389_1172, and HMPREF0389_1476) were also found in abundance. The hypothetical protein HMPREF0389_00967 was highly (6.4 times) upregulated in coinfection of F. alocis clinical strain D-62D with P. gingivalis W83 (Table 1).
FIG 1.
Percentage of protein expressed during coculture or monoculture using various strains of P. gingivalis and F. alocis. HeLa cells were infected with F. alocis ATCC 35896 and D-62D strains (MOI of 1:100 [105 epithelial cells]) in monoculture or coculture with P. gingivalis W83 as previously reported (6). Tandem isobaric mass tagging analysis of cocultures and monocultures was carried out using Orbitrap. (*, P < 0.01.) Bar A, P. gingivalis (33277) plus F. alocis (D-62D) versus P. gingivalis (33277) plus F. alocis (ATCC); bar B, P. gingivalis (33277) plus F. alocis (D-62D) versus P. gingivalis (W83) plus F. alocis (D-62D); bar C, P. gingivalis (33277) plus F. alocis (D-62D) versus F. alocis (D-62D); bar D, P. gingivalis (33277) plus F. alocis (ATCC) versus P. gingivalis (W83) plus F. alocis (ATCC); bar E, P. gingivalis (W83) plus F. alocis (D-62D) versus P. gingivalis (W83) plus F. alocis (ATCC); bar F, P. gingivalis (W83) plus F. alocis (D-62D) versus F. alocis (D-62D); bar G, F. alocis (D-62D).
FIG 2.
Upregulation in proteome profile showing various protein classes noted during coculture infection with P. gingivalis and Filifactor alocis in epithelial cells. HeLa cells were infected with F. alocis ATCC 35896 and D-62D strains (MOI of 1:100 [105 epithelial cells]) in monoculture or coculture with P. gingivalis W83 as previously reported (6). Tandem isobaric mass tagging analysis of cocultures and monocultures was carried out using Orbitrap. The F. alocis proteins were analyzed using MASCOT, and functional analysis was carried out using the UNIPROT proteome database. A, amino acid biosynthesis; B, biosynthesis of cofactors, prosthetic groups, and carriers; C, cell envelope; D, cellular processes; E, central intermediary metabolism; F, DNA metabolism; G, energy metabolism; H, fatty acid and phospholipid metabolism; I, hypothetical/unassigned/uncategorized/unknown functions; J, mobile and extrachromosomal element functions; K, protein fate; L, purines, pyrimidines, nucleosides, and nucleotides; M, regulatory functions; N, replication; O, transcription; P, translation; Q, transport and binding; R, transposon functions. Protein expression change > 1-fold, n = 490; protein expression change < 1-fold, n = 400.
TABLE 1.
F. alocis coinfection with P. gingivalis: relative abundance of proteins with cell wall motif
| Annotation | Name |
|---|---|
| HMPREF0389_01580 | Leukotoxin translocation ATP binding protein |
| HMPREF0389_00575 | Fibronectin binding protein |
| HMPREF0389_00426 | Type IV pilus assembly protein |
| HMPREF0389_00816 | Signal recognition particle protein |
| HMPREF0389_01719 | Hypothetical protein |
| HMPREF0389_00599 | Hypothetical protein |
| HMPREF0389_00019 | Hypothetical protein |
| HMPREF0389_01532 | Calcium binding acid repeat protein |
| HMPREF0389_01139 | S-layer Y domain-containing protein |
| HMPREF0389_00672 | Hypothetical protein |
| HMPREF0389_01172 | Hypothetical protein |
| HMPREF0389_00415 | Fimbrial assembly protein |
| HMPREF0389_01657 | Membrane protein |
| HMPREF0389_01476 | Hypothetical protein |
| HMPREF0389_01477 | Hemolysin III type calcium binding protein |
| HMPREF0389_01478 | Protein export membrane protein |
| HMPREF0389_01006 | Collagen adhesion protein |
Upregulation of F. alocis proteins involved in host cell signaling.
F. alocis strains ATCC-35896 and D-62D cocultured with P. gingivalis showed upregulation of bacterial proteins that are reported to be involved in eukaryotic transcription (Table 2). Coinfection also showed relative abundances of noncoding RNAs such as clustered regularly interspaced short palindromic repeat (CRISPR) RNA and toxin-antitoxin system proteins. Peptidyl prolyl cis-trans isomerase, an enzyme involved in the histone-modifying pathway, was also upregulated. Relevant gene network data based on host proteome data mining suggest upregulation of host cell signal transduction processes; however, analysis also showed downregulation of kinase and protein binding activities. Many vital cellular processes such as phosphorylation, apoptosis, gene expression, cell proliferation, and cell growth were modulated (see Fig. S1 and S2 in the supplemental material).
TABLE 2.
F. alocis proteins found in relative abundance during coinfection
| Gene ID | Annotation | Fold change |
|
|---|---|---|---|
| D-62D | ATCC | ||
| HMPREF0389_ 00905 | Sodium neurotransmitter symporter family protein | 3.64 | 2.76 |
| HMPREF0389_ 00296 | PP loop family protein | 6.38 | 1.15 |
| HMPREF0389_ 01047 | TetR family transcriptional regulator | 1.75 | 1.04 |
| HMPREF0389_ 01180 | Anti-anti-sigma factor RsbV | 2.92 | 1.09 |
| HMPREF0389_ 00038 | Hypoxanthine phosphoribosyl transferase | 1.70 | 2.09 |
| HMPREF0389_ 01084 | Hypothetical protein (containing −7TM receptors with diverse intracellular signaling molecules) | 1.63 | 1.20 |
| HMPREF0389_ 01060 | GMP synthase | 1.63 | 1.10 |
| HMPREF0389_ 01081 | Caax aminoprotease family protein | 1.68 | 1.75 |
| HMPREF0389_ 00290 | Translational regulator LacI family | 2.38 | 1.20 |
| HMPREF0389_ 01109 | FeS assembly ATPase SufC | 2.08 | 1.50 |
| HMPREF0389_ 01107 | Iron-regulated ABC-type transporter | 1.51 | 2.5 |
| HMPREF0389_ 00519 | Hypothetical protein (containing putative zinc ribbon domain) | 2.18 | 3.3 |
| HMPREF0389_ 00052 | CDP-diacyl glycerol-glycerol 3 phosphate 3 phosphatidyl transferase | 2.06 | 1.90 |
| HMPREF0389_ 00948 | Phosphoglycerate dehydrogenase | 1.95 | 1.52 |
| HMPREF0389_ 01590 | Transcriptional regulator AraC family | 1.55 | 1.03 |
| HMPREF0389_ 01398 | Oxygen-independent coproporphyrinogen III oxidase | 1.83 | 1.08 |
| HMPREF0389_ 01164 | Hypothetical protein | 2.28 | 1.31 |
| HMPREF0389_ 00472 | Gamma glutamyl ligase family | 1.92 | 1.40 |
| HMPREF0389_ 00387 | Pyruvate kinase | 2.47 | 0.74 |
| HMPREF0389_ 00382 | Hypothetical protein (VCBS domain protein) | 4.11 | 1.13 |
| HMPREF0389_ 00294 | Ribose ABC transporter permease protein | 1.74 | 2.4 |
| HMPREF0389_ 00212 | CRISPR-associated protein 2 Cas2 | 2.12 | 1.4 |
| HMPREF0389_ 00211 | CRISPR-associated protein 1 Cas1 | 1.4 | 0.70 |
| HMPREF0389_ 00210 | CRISPR-associated protein Csn1 | 1.046 | 1.00 |
| HMPREF0389_ 01169 | CRISPR-associated protein Csd1 | 0.960 | 0.95 |
| HMPREF0389_ 01170 | CRISPR-associated protein Cas5 | 0.748 | 0.78 |
| HMPREF0389_ 01165 | CRISPR-associated protein Cas2 | 1.30 | 1.20 |
| HMPREF0389_ 01167 | CRISPR-associated protein Cas4 | 1.01 | 1.00 |
| HMPREF0389_ 00275 | Peptidyl prolyl isomerase | 1.243 | 1.0 |
| HMPREF0389_ 01236 | 30S ribosomal protein S18 | 3.9 | 1.5 |
| HMPREF0389_ 00068 | Hypothetical protein | 1.8 | 1.1 |
| HMPREF0389_ 00044 | Glutamate synthase, RluA family | 2.08 | 0.8 |
| HMPREF0389_ 00019 | Outer membrane protein | 2.34 | 1.8 |
| HMPREF0389_ 00569 | tRNA delta(2) isopentenyl pyrophosphate/glucosamine 1 phosphate N acetyltransferase | 1.83 | 1.1 |
| HMPREF0389_ 00756 | Cytidylate kinase | 1.70 | 2.3 |
| HMPREF0389_ 00967 | Hypothetical protein (CHASE 3 domain) | 3.10 | 6.4 |
| HMPREF0389_ 00243 | Toxin-antitoxin component ribbon-helix-helix fold protein | 0.821 | 0.88 |
| HMPREF0389_ 00201 | Hypothetical protein (HTH3 domain protein) | 1.38 | 1.36 |
| HMPREF0389_ 00275 | Peptidyl prolyl isomerase | 1.24 | 0.99 |
| HMPREF0389_ 00359 | Peptidyl prolyl cis-trans isomerase | 1.24 | 1.10 |
Proteome profile of epithelial cells coinfected with F. alocis and P. gingivalis.
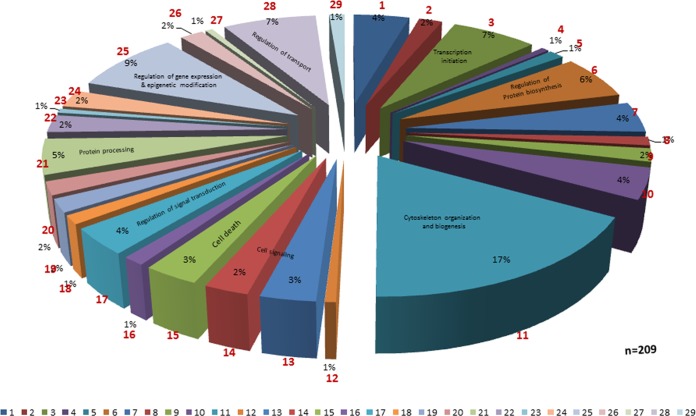
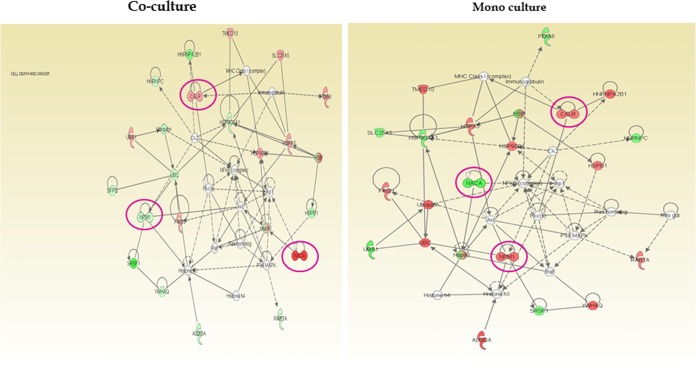
Epithelial cells coinfected with F. alocis and P. gingivalis strains showed activation of several eukaryotic proteins involved in the inflammatory response, cell signaling, and cell death (Fig. 3). The global proteome analysis of the host showed modulation in expression of 209 proteins. Among them, proteins involved in cytoskeleton organization and biogenesis were affected most (17%) followed by proteins involved in the regulation of gene expression and epigenetic modification (9%), regulation of protein transport (7%), transcription initiation (7%), regulation of protein biosynthesis (6%), protein processing (5%), regulation of signal transduction (4%), and cell death-apoptosis (3%). Highly downregulated proteins during coinfection include eukaryotic translation initiation factor, splicing factors, histone protein clusters, and other signaling proteins such as vimentin, prohibitin, and redox proteins. Highly upregulated proteins include the RAS oncogene family proteins, proteins involved in granzyme signaling, and cytoskeletal matrix proteins (Table 3). An Ingenuity pathway analysis showed increased expression of eukaryotic genes involved in antigen presentation, cellular movement, the hematological system, cell trafficking, and inflammatory response (see Fig. S3 in the supplemental material).
FIG 3.
Eukaryotic proteome profile showing various protein classes modulated during coculture infection with P. gingivalis and Filifactor alocis. Section 1, GO:0005975 (carbohydrate metabolism); section 2, GO:0006260 (DNA replication); section 3, GO:0006352 (transcription initiation); section 4, GO:0006353 (transcription termination); section 5, GO:0006360 (transcription from RNA polymerase I promoter); section 6, GO:0006417 (regulation of protein biosynthesis); section 7, GO:0006457 (protein folding); section 8, GO:0006512 (ubiquitin cycle); section 9, GO:0006839 (mitochondrial transport); section 10, GO:0007005 (mitochondrion organization and biogenesis); section 11, GO:0007010 (cytoskeleton organization and biogenesis); section 12, GO:0007047 (cell wall organization and biogenesis); section 13, GO:0007166 (cell surface receptor-linked signal transduction); section 14, GO:0007264 (small-GTPase-mediated signal transduction); section 15, GO:0008219 (cell death); section 16, GO:0009890 (negative regulation of biosynthesis); section 17, GO:0009966 (regulation of signal transduction); section 18, GO:0015931 (vesicle organization and biogenesis);section 19, GO:0016071 (mRNA metabolism); section 20, GO:0016481 (negative regulation of transcription); section 21, GO:0016485 (protein processing); section 22, GO:0018193 (peptidyl amino acid modification); section 23, GO:0019932 (secondary messenger-mediated signaling); section 24, GO:0030705 (cytoskeleton-dependent intracellular transport); section 25, GO:0040029 (regulation of gene expression, epigenetics); section 26, GO:0045333 (cellular respiration); section 27, GO:0045454 (cell redox homeostasis); section 28, GO:0051049 (regulation of transport); section 29, GO:0051052 (regulation of DNA metabolism).
TABLE 3.
Modulated host proteins during coinfection with F. alocis and P. gingivalis
| Gene | Fold change | Annotation |
|---|---|---|
| Downregulated proteins | ||
| EIF4B | −3.675 | Eukaryotic translation initiation factor 4B |
| HIST1H1C | −2.742 | Histone cluster 1, H1c |
| HIST1H1E | −2.643 | Histone cluster 1, H1e |
| HIST1H2BL | −2.790 | Histone cluster 1, H2bl |
| PPIA | −3.307 | Peptidyl prolyl isomerase A (cyclophilin A) |
| VIM | −2.158 | Vimentin |
| SRSF1 | 4.049 | Serine/arginine-rich splicing factor 1 |
| PHB | Prohibin | |
| TRAP 1 | −1.035 | TNF receptor-associated protein 1 |
| PRDX1 | −1.219 | Peroxiredoxin 1 and 5 |
| PRDX5 | −1.201 | |
| Upregulated proteins | ||
| NACA | 2.310 | Nascent polypeptide-associated complex alpha subunit |
| IMPDH2 | 2.472 | Inosine-5′-monophosphate dehydrogenase 2 |
| AGF3 | 1.774 | AFG3-like protein 2 |
| RAB 10 | 1.128 | Member of RAS oncogene family |
| RAB 7A | 1.028 | Member of RAS oncogene family |
| ITGB1 | 1.356 | Integrin |
| PHB | 1.260 | Prohibitin |
| VCL | 2.189 | Vinculin |
| IGAAD | Granzyme A signaling proteins | |
| HSP10 | 2.160 | 10-kDa heat shock protein (mitochondrial) |
| LDHA | 1.516 | Isoform 1 of l-lactate dehydrogenase A chain |
| 1.512 | 46-kDa protein | |
| 1.907 | 26-kDa protein | |
| NACA | 2.310 | Nascent polypeptide-associated complex |
| KRT1 | 1.461 | Keratin type II cytoskeletal I |
| HNRPR | 1.728 | Heterogenous nuclear ribonucleoprotein R |
Many host cell regulatory proteins that are involved in cytoskeleton integrity were modulated. The integrin beta-1 (ITGB1) genes are downregulated in coculture. The valosin-containing protein (VCP) gene involved in ubiquitin-dependent protein degradation is downregulated in coculture. In both coculture and monoculture of F. alocis, there was upregulation of the poly-β-hydroxybutyrate (PHB) gene involved in negative regulation of cell proliferation (see Fig. S4 in the supplemental material). The vinculin (VCL) gene coding for the membrane cytoskeleton protein vinculin and the voltage-dependent anion channel (VDAC1) gene were upregulated during coinfection. Proteins involved in important cell regulatory networks such as serine/arginine-rich splicing factor 1 (SRSF1), annexin-2 (ANXA2), heat shock proteins (HSPA8, HSP9, and HSPE1), synaptotagmin binding protein (SYNCRIP), eukaryotic initiation factor 4A-1, 19-kDa protein-SRP-dependent translational protein, protein S100A11, and other cytoskeletal proteins such as the lamin A/C proteins were also found to be modulated (see Table S2). There was a generalized downregulation of the actin pathway (see Fig. S5).
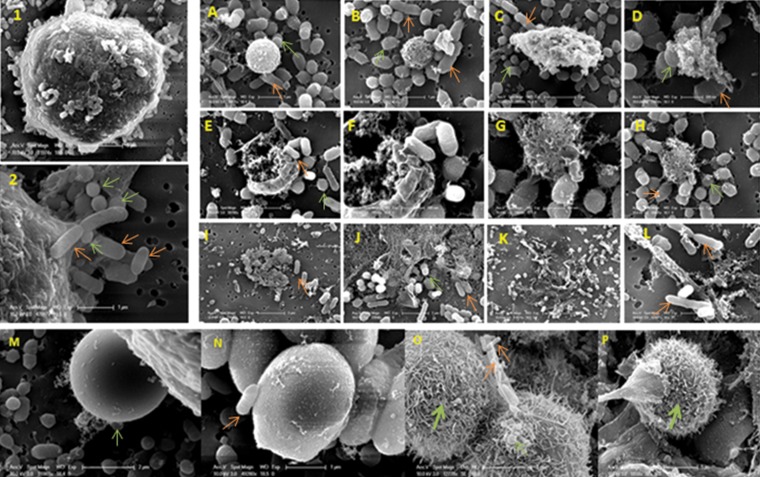
In order to identify variations in host cell surface morphology and cell death between coinfection and monoinfection, epithelial cells coinfected with F. alocis and P. gingivalis were subjected to electron microscopy study. Wide morphological variation of the host cell was noted after coinfection compared to monoinfection with either F. alocis or P. gingivalis. The infected epithelial cells show modification of cell surface filopodial projections that were used by both F. alocis and P. gingivalis to adhere to the cell surface and formation of membrane microdomains such as the lipid rafts (Fig. 4, panels 1 and 2). Infected cells showed early apoptosis during coinfection compared to monoinfection (Fig. 4A to L). Surface modifications of the coinfected epithelial cells were noted in both F. alocis ATCC 35896 and the clinical strain D-62D; however, the level of filopodial projections was greater in the F. alocis D-62D strain than in the F. alocis ATCC 35896 strain (Fig. 4O and P). Such morphological variations were not noted in monoinfected epithelial cells (Fig. 4M and N).
FIG 4.
Coinfection of P. gingivalis and Filifactor alocis, showing morphological variations during adherence leading to apoptosis of the epithelial cells. Scanning electron microscopy images show modification of the cell surface with filapodial projections in infected epithelial cells that were used by both F. alocis and P. gingivalis to adhere to the cell surface (panels 1 and 2). Infected cells showed early apoptosis during coinfection compared to monoinfection (A to L). Surface morphological variations were not noted in monoinfected epithelial cells (M and N). Filopodial projections were more increased in the F. alocis D-62D strain (O and P). Orange arrows, F. alocis; light-green arrows, P. gingivalis; dark-green arrows, filapodial projections.
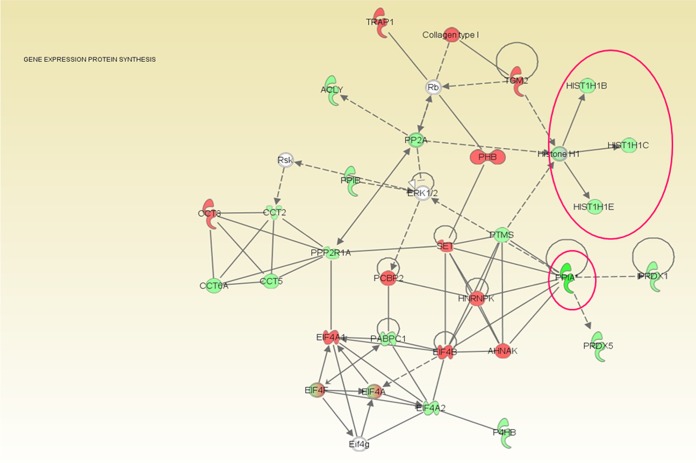
Many host proteins involved in chromatin function and remodeling were downregulated during F. alocis coinfection with P. gingivalis. An overall downregulation of histone cluster proteins and PPIA-peptidyl prolyl isomerase was noted (Fig. 5) (Table 2). The relative expression levels of such proteins were the lowest in F. alocis clinical strain coinfection compared to the type strain coinfection (data not shown). Heterogeneous nuclear ribonuclear protein A2/B1 was found in least abundance during coinfection. Proteins involved in transport and secretory pathways such as the transferrin receptor protein 1 (involved in iron transport), transmembrane emp24 domain-containing protein 10 and dynein (involved in vesicular protein trafficking), surfeit 4, and solute carrier family proteins were also less expressed during coinfection.
FIG 5.
Coinfection of Filifactor alocis with P. gingivalis showing downregulation of proteins involved in gene expression and protein synthesis pathways. HeLa cells were infected with the F. alocis ATCC 35896 and D-62D strains (MOI of 1:100 [105 epithelial cells]) in monoculture or coculture with P. gingivalis strains as previously reported (6). Tandem isobaric mass tagging analysis of cocultures and monocultures was carried out using Orbitrap. The eukaryotic proteins were analyzed using MASCOT, and functional analysis was carried out using Ingenuity pathway analysis software. The gene ontology classification was used for referencing the proteome. F. alocis coinfection with P. gingivalis showed overall downregulation of histone cluster proteins (histone [Hist] H1, Hist 1H1B, Hist 1H1C, and Hist 1H1E) (shown within the circle), peptidyl prolyl isomerase (PPIA and PPIB), and antioxidant enzymes (PRDX1 and PRDX5). Green, downregulation; red, upregulation; solid lines, direct interaction; dotted lines, indirect interaction.
Proteins that are known to activate oncogenes directly or indirectly were expressed in high abundance during F. alocis coculture. The major proteins that were present in high abundance during coinfection were RAB 7A, RAB10 proteins belonging to the RAS oncogene family, poly(rC) binding protein 2 (PCBP2), voltage-dependent anion channel protein 1, copine-1 (calcium-dependent membrane binding protein), and collagen alpha-2 (V) chain precursor (Table 3).
Many host cell proteins that are involved in cell adhesion and cytoskeletal interactions were upregulated during coinfection of F. alocis with P. gingivalis. Host cytoskeletal proteins such as vimentin, actin, plectin, vinculin, profilin, and transgelin and chaperone proteins such as HSP90 and endoplasmin and proteins such as filamin B and filamin C involved in cell communication were modulated. Signal transduction proteins galectin, proteasome subunit alpha type 6, and 14-3-3 protein theta were relatively less abundant. Inosine 5′ monophosphate dehydrogenase (IMPDH), receptor kinectin, and peroxiredoxins were found to be highly expressed during coinfection (see Table S2).
Oribtrap analysis of the whole proteome of F. alocis coculture with P. gingivalis revealed many upregulated host cell proteins that are involved in apoptosis, cell regulation, and differentiation pathways. Proteins such as prohibitins and Ras-related protein Rab 10 and Ras-related protein Rab 7 SET translocation proteins that are involved in apoptosis and histone binding were upregulated. Also, SRSF1 protein encoded by a proto-oncogene, nascent-polypeptide-associated complex alpha (NACA) protein (transcriptional coactivator), NPM1 (nucleophosmin) involved in apoptosis and tumorigenesis, and CALR (calreticulin) involved in calcium binding and storage were highly upregulated (Fig. 6). Further, our analysis showed upregulation of proteins involved in the ubiquitin proteasome pathway and the granzyme-mediated apoptotic signaling pathway (see Fig. S6 in the supplemental material).
FIG 6.
Coinfection of Filifactor alocis with P. gingivalis showing upregulation of proteins involved in cancer and cell death pathways. HeLa cells were infected with F. alocis ATCC 35896 and D-62D strains (MOI of 1:100 [105 epithelial cells]) in monoculture or coculture with P. gingivalis strains as previously reported (6). Tandem isobaric mass tagging analysis of cocultures and monocultures was carried out using Orbitrap. The eukaryotic proteins were analyzed using MASCOT, and functional analysis was carried out using Ingenuity pathway analysis software. The gene ontology classification was used for referencing the proteome. Proteins such as prohibitins, Ras-related protein Rab 10, and the Ras-related protein Rab 7 SET translocation protein that are involved in apoptosis and histone binding were upregulated. Also, the SRSF1 protein encoded by a proto-oncogene, NACA protein (transcriptional coactivator), NPM1 (nucleophosmin; involved in apoptosis and tumorigenesis), and CALR (calreticulin; involved in calcium binding and storage) were highly upregulated. Green, downregulation; red, upregulation.
The host metabolic pathways modulated during coinfection are given in Table 4. Pathways relating to ammonia synthesis, urate biosynthesis, amino acid degradation (valine and aspartate), lipid synthesis, and palmitate and fatty acid synthesis were highly upregulated. There was downregulation of amino acid excretory pathways and transport pathways such as the glutamyl/arginine exchange and tryptophan pathway. Coinfection of F. alocis was shown to affect basic energy pathways such as glycolysis and acetyl coenzyme A (CoA) biosynthesis.
TABLE 4.
Metabolome variation in host protein during F. alocis coinfection
| Expression category and pathway |
|---|
| Upregulated |
| Ammonia production |
| Urate biosynthesis through inosine 5′phosphate degradation |
| Valine degradation |
| Aspartate degradation |
| l-Asparagine synthesis |
| Glutaraldehyde CoA degradation |
| Fatty acid biosynthesis |
| Pentose phosphate pathway |
| TCA cyclea |
| Pyruvate fermentation to lactate |
| Thioredoxin pathway |
| Palmitate biosynthesis |
| Purine de novo synthesis |
| Downregulated |
| Sodium-independent glutamyl/arginine exchange |
| l-Tryptophan transport |
| Sucrose degradation |
| Peptidyl proline synthesis |
| Phosphoprotein synthesis |
| Acetyl CoA biosynthesis from citrate |
| Glycolysis |
| NADH repair |
aTCA, tricarboxylic acid.
DISCUSSION
F. alocis and P. gingivalis are important members of a complex multispecies biofilm that occupies the gingival crevice. Multiple interbacterial interactions are required for developing and maintaining the subgingival microbial community (47, 48). The impact of these interspecies interactions on the host is significant for their survival and their ability to cause disease. In contrast to previous approaches that have used purified proteins and isogenic mutants to identify specific molecular pathways responsible for many of the complex cellular responses involving host-microbe interactions, this study has used a comprehensive proteomic assessment that simultaneously evaluated the modulation of proteins in key pathways in both the microbe and the host. We previously reported variations in the pathogenic potential of F. alocis strains which may be partly related to the differential expression of several putative virulence factors, including several proteases, neutrophil activating protein A, and calcium binding acid repeat protein (36). Expression of these factors was increased during the invasion of HeLa cells (23).
In this study, F. alocis in coculture with P. gingivalis showed enhanced adhesion to epithelial cells, altering the cell morphology and inducing cell death. This was in contrast to monoinfections with either F. alocis or P. gingivalis, which did not trigger the same morphological alteration, although the monoinfections were still able to induce cell death over a longer time period. These observations are consistent with previous reports which showed that the virulence potential of F. alocis is enhanced by its coinfection with P. gingivalis (23, 36). While there was uniform expression of several membrane proteins that might play a role in attachment and virulence modulation (23), proteomic analysis of F. alocis during coinfection of epithelial cells with P. gingivalis revealed upregulation of several membrane adhesion proteins. This suggests that the interaction of F. alocis and P. gingivalis may result in the upregulation of a specific factor(s) that may enhance its virulence potential. Furthermore, several of these proteins are structurally related to other microbial surface component-recognizing adhesion matrix molecules (MSCRAMMs) that are known to play an important role in Gram-positive bacterial virulence by mediating adherence to and colonization of host tissues, which are early steps in clinical infection (49). Given the relative abundances of the many collagen binding MSCRAMMs, this may suggest that collagen is a likely target for F. alocis. Several unique hypothetical proteins with transmembrane domains were also observed to be upregulated. Proteins with these domains show interaction with extracellular matrix components which in turn can act as proinflammatory signal molecules (50). It is also noteworthy that there are no homologues of these proteins identified in P. gingivalis. Their functional role in F. alocis pathogenesis is under further investigation in the laboratory.
There is evidence that extracellular matrix adhesion proteins can be regulated by quorum sensing (51), which implies that environmental signals can modulate their expression and hence adhesion and colonization. Putative F. alocis proteins that could be involved in quorum sensing and signal transduction pathways were upregulated during coinfection. One of the highly upregulated hypothetical proteins, HMPREF0389_00967, contains a CHASE3 extracellular sensory domain. This domain, which is commonly found in histidine kinase, adenylate cyclases, and chemotaxis proteins (52), is involved in signal transduction pathways in bacteria (53). It is tempting to speculate that this protein plays a role in cell signaling and mediation of quorum sensing. Some of the other major proteins upregulated during coinfection include the noncoding RNA, CRISPR RNA, and toxin-antitoxin system proteins. CRISPR regulation of gene expression (54) is implicated in biofilm formation (55) and horizontal gene transfer (56, 57) in other oral bacteria. These proteins have also been implicated in stress response and chaperone function, mediating important signal transduction events (58). It was also noted that HMPREF0389_00382, a hypothetical protein that contains two VCBS domains (repeat domains in Vibrio, Colwellia, Bradyrhizobium, and Shewanella), was highly upregulated. These VCBS domain-containing proteins are involved in bacterial adhesion and play a role in virulence in other pathogenic bacteria (TIGRfam1965).
Our coinfection study showed relative abundances of many bacterial methyltransferases which could imply that they may play a role in targeting host DNA hypermethylation and chromatin modification. Similar variations in protein expression were also noted in a previous study performed with gingival epithelial cells coinfected with P. gingivalis and F. nucleatum (59). Note that histone modification through the mitogen-activated protein kinase (MAPK) pathway was found to be mediated by peptidyl prolyl cis-trans isomerase in other pathogenic bacteria (60). Both P. gingivalis and F. alcois possess the gene coding for peptidyl prolyl cis-trans isomerase (36). Our study showed increases in the abundance of these proteins during coinfection. Further, our study also showed variations in the host proteins that are involved indirectly in chromatin modification during F. alocis coinfection with P. gingivalis. Coinfection showed relative abundances of proteins such as parathymosin, prothymosin a14, prothymosine alpha (PTMA), SET translocation protein, and zinc finger BED domain-containing protein 1 that affect histone binding to nucleosomes causing histone binding (61) and chromatin remodeling (62). These proteins also regulate histone acetylation (63). Bacterially induced DNA methylation was shown earlier to affect the host cell proliferation (64). While host-pathogen methyltransferase similarities among many virulent strains of bacteria were noticed earlier (65), chromatin modification through bacterial proteins can regulate expression of host genes and enzymes such as histone deacetylase (HDAC) (66). Such DNA methylation and histone acetylation are commonly associated with cancer and tumor growth (59). The F. alocis genome is annotated with 18 methyltransferase genes; their role in host chromatin modification and epigenetic changes awaits further confirmation. Our study showed upregulation of many methyltransferase genes during coinfection.
Coinfection of F. alocis with P. gingivalis showed modulation of host proteins involved in signaling, cell-cell interaction, and chaperone function. Several proteins involved in maintaining cell shape and the integrity of the cytoplasm and in stabilizing cytoskeletal interactions and cellular integrity were downregulated. This is correlated with the results of the electron microscopic study, which revealed variations in cell surface morphology in epithelial cells coinfected with F. alocis and P. gingivalis. Formations of lipid rafts due to the host cell plasma membrane response to pathogens were demonstrated in several invasive pathogens (67–70). Such modifications of the host cell could be used as a protective mechanism for both F. alocis and P. gingivalis to evade the degradative lysosomal pathway (69). The electron microscopy findings could be corroborated with the proteome data showing modulation of actin and other proteins involved in cytoskeletal modification. Such characteristic cytoskeletal remodeling and transcellular processes mediated by F. alocis could help in cointernalization of F. alocis and P. gingivalis. The increase in adhesion of F. alocis observed in this study would likely be due to expression of host adhesion proteins such as vinculin and VDAC1 protein that could favor pathogen adherence (71). Compared to monoinfection, coinfection with P. gingivalis showed an overall dysfunction of protein function and transport due to a generalized downregulation of many host chaperone proteins such as Hsp90 (heat shock protein 90), protein disulfide isomerase, and endoplasmins. Note that proteins involved in cell growth and proliferation were also affected during coinfection.
Coinfection of F. alocis with P. gingivalis showed regulation of many proteins that are involved in the host regulatory network. Among them, serine/arginine-rich splicing factor 1 (SRSF1) protein was highly downregulated. This protein imparts genomic stability and prevents slicing variants and is also implicated in many critical functions such as cell viability and programmed cell death (72). Annexin was found to be downregulated in coculture compared to monoculture. Annexin is believed to be involved in membrane-related functions of the cell and in the endocytic pathway, regulating the onset of cell degradation (73). Other major variations during coinfection include downregulation of lectin galactose binding soluble protein 1 (LGALS1), annexin 2 (ANXA2), heat shock proteins (HSPA8, HSP9, and HSPE1), and synaptotagmin binding protein (SYNCRIP). Eukaryotic initiation factor 4A-1 is involved in the cytokine-mediated signaling pathway and in host-pathogen interaction. The relative abundances of lamin A/C proteins were much lower during coinfection. They are essential proteins that make up the nuclear matrix and are involved in nuclear stability, chromatin structure, and gene expression and in lamin-associated signaling pathways. Additionally, we have noted downregulation of host cell nuclear ribonuclear proteins. These nuclear envelope (NE) proteins act as regulators of MAPK, Wnt–β-catenin, transforming growth factor β (TGF-β), and Notch signaling cascades (74). It is noteworthy that protein S100A11, implicated as a potential biomarker of infective endocarditis, was more abundant during F. alocis monoinfection (75). S100A11 proteins are involved in endocytosis and exocytosis (76), regulation of enzyme activity, cell growth regulation, apoptosis, and inflammation (77). Periodontitis is an inflammatory disease; however, its role in other systemic inflammatory diseases is unknown.
Metabolic host responses to bacterial infections favor survival and are important in the pathogenic process (78). The relative abundances of proteins involved in arginine metabolism and citrulline synthesis, namely, arginine deiminase (HMPREF0389_01584), acetyl ornithine transferase (HMPREF0389_01570) (36), aminotransferase (HMPREF0389_01352 and HMPREF0389_01353), amidotransferase family protein (HMPREF 0389_00349), arginine-tRNA ligase (HMPREF0389_00390), and arginine decarboxylase (HMPREF0389_00102), indicate that the nutritional needs of the bacteria are met during infection. Since F. alocis and P. gingivalis are asaccharolytic in nature and resort to protein breakdown for energy and survival, this process could lead to a heavy production of ammonia. A well-developed arginine cycle noted in F. alocis could help the coinfected pathogen partners to survive well in periodontal pockets by utilizing the toxic metabolites of the host cell. This could be one of the many processes that lead to pathogen synergy. F. alocis genome annotation also showed F. alocis to possess a well-developed citrulline synthesis mechanism from arginine. Citrullination of proteins has already been shown to be an important posttranslational modification. Upregulation of peptidyl arginine deiminase (PAD) expression and an associated increase in the levels of citrullinated proteins are found in the synovium of patients with rheumatoid arthritis (79). Bioinformatic analysis has shown that P. gingivalis possesses a form of PAD that shares major sequence and structural homology with the F. alocis arginine deiminase enzyme (our unpublished data). Arginine deiminase of the pathogens was shown to possess multiple regulatory roles and was also shown to possess PAD function (80).
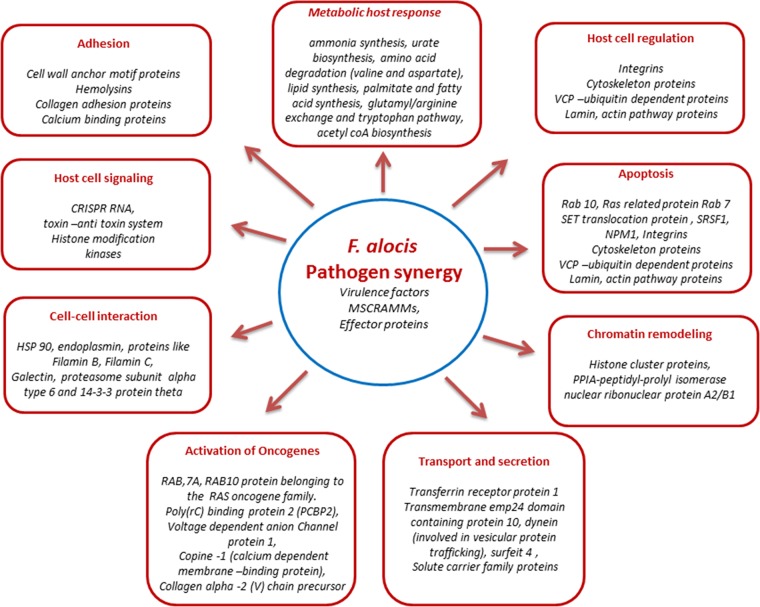
Our present study has shown the putative ability of the specific factors from F. alocis to modulate multiple changes in the host cell proteome (Fig. 7). It is likely that such variations at the molecular level are responsible for the functional changes required to mediate the pathogenic process. The relative resistance of F. alocis to oxidative stress (23) and its enhanced virulence potential in association with P. gingivalis collectively suggest its importance as a periodontal pathogen. The relative significance of specific F. alocis putative virulence factors that may trigger the key host response and hence the pathology awaits further clarification in the laboratory.
FIG 7.
Summary overview showing the major role of F. alocis pathogen synergy in the host cell response. The major modulated proteins are shown under each category heading.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Loma Linda University and Public Health Services grants DE13664, DE019730, DE019730 04S1, DE022508, and DE022724 from NIDCR (to H.F.).
Footnotes
Published ahead of print 27 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01727-14.
REFERENCES
- 1.Genco RJ, Van Dyke TE. 2010. Prevention: reducing the risk of CVD in patients with periodontitis. Nat. Rev. Cardiol. 7:479–480. 10.1038/nrcardio.2010.120. [DOI] [PubMed] [Google Scholar]
- 2.Astolphi RD, Curbete MM, Colombo NH, Shirakashi DJ, Chiba FY, Prieto AK, Cintra LT, Bomfim SR, Ervolino E, Sumida DH. 2013. Periapical lesions decrease insulin signal and cause insulin resistance. J. Endod. 39:648–652. 10.1016/j.joen.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 3.Kaur S, White S, Bartold PM. 2013. Periodontal disease and rheumatoid arthritis: a systematic review. J. Dent. Res. 92:399–408. 10.1177/0022034513483142. [DOI] [PubMed] [Google Scholar]
- 4.Bingham CO, Moni M. 2013. Periodontal disease and rheumatoid arthritis: the evidence accumulates for complex pathobiologic interactions. Curr. Opin. Rheumatol. 25:345–353. 10.1097/BOR.0b013e32835fb8ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamont RJ, Jenkinson HF. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudney JD, Chen R, Sedgewick GJ. 2001. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect. Immun. 69:2700–2707. 10.1128/IAI.69.4.2700-2707.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito A, Inagaki S, Kimizuka R, Okuda K, Hosaka Y, Nakagawa T, Ishihara K. 2008. Fusobacterium nucleatum enhances invasion of human gingival epithelial and aortic endothelial cells by Porphyromonas gingivalis. FEMS Immunol. Med. Microbiol. 54:349–355. 10.1111/j.1574-695X.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- 8.Saito A, Inagaki S, Ishihara K. 2009. Differential ability of periodontopathic bacteria to modulate invasion of human gingival epithelial cells by Porphyromonas gingivalis. Microb. Pathog. 47:329–333. 10.1016/j.micpath.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J. Bacteriol. 192:5002–5017. 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, Gibbons TR, Treangen TJ, Chang YC, Li S, Stine OC, Hasturk H, Kasif S, Segre D, Pop M, Amar S. 2012. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One 7:e37919. 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berezow AB, Darveau RP. 2011. Microbial shift and periodontitis. Periodontol. 2000 55:36–47. 10.1111/j.1600-0757.2010.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6:1176–1185. 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA, Asnani K, Griffen AL. 2010. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J. Clin. Microbiol. 48:4121–4128. 10.1128/JCM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770–3783. 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahlén G, Leonhardt A. 2006. A new checkerboard panel for testing bacterial markers in periodontal disease. Oral Microbiol. Immunol. 21:6–11. 10.1111/j.1399-302X.2005.00243.x. [DOI] [PubMed] [Google Scholar]
- 16.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82:338–344. 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 17.Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. 2006. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J. Clin. Microbiol. 44:3665–3673. 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonhardt A, Carlen A, Bengtsson L, Dahlen G. 2011. Detection of periodontal markers in chronic periodontitis. Open Dent. J. 5:110–115. 10.2174/1874210601105010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlafer S, Riep B, Griffen AL, Petrich A, Hübner J, Berning M, Friedmann A, Göbel UB, Moter A. 2010. Filifactor alocis—involvement in periodontal biofilms. BMC Microbiol. 10:66. 10.1186/1471-2180-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wade WG. 2011. Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J. Clin. Periodontol. 38(Suppl 11):7–16. 10.1111/j.1600-051X.2010.01679.x. [DOI] [PubMed] [Google Scholar]
- 21.Cato EP, Moore LVH, Moore WEC. 1985. Fusobacterium alocis sp. nov. and Fusobacterium sulci sp. nov. from the human gingival sulcus. Int. J. Syst. Bacteriol. 35:475–477. 10.1099/00207713-35-4-475. [DOI] [Google Scholar]
- 22.Jalava J, Eerola E. 1999. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int. J. Syst. Bacteriol. 49(Pt 4):1375–1379. 10.1099/00207713-49-4-1375. [DOI] [PubMed] [Google Scholar]
- 23.Aruni AW, Roy F, Fletcher HM. 2011. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect. Immun. 79:3872–3886. 10.1128/IAI.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moffatt CE, Whitmore SE, Griffen AL, Leys EJ, Lamont RJ. 2011. Filifactor alocis interactions with gingivalis epitheial cells. Mol. Oral Microbiol. 26:365–373. 10.1111/j.2041-1014.2011.00624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Jotwani R, Le J, Krauss JL, Potempa J, Coventry SC, Uriarte SM, Lamont RJ. 30 December 2013. Filifactor alocis infection and inflammatory responses in the mouse subcutaneous chamber model. Infect. Immun. 10.1128/IAI.01434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q, Wright CJ, Dingming H, Uriarte SM, Lamont RJ. 2013. Oral community interactions of Filifactor alocis in vitro. PLoS One 8:e76271. 10.1371/journal.pone.0076271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki N, Yoneda M, Hirofuji T. 6 March 2013. Mixed red-complex bacterial infection in periodontitis. Int. J. Dent. 2013:587279. 10.1155/2013/587279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma S, Kelly TK, Jones PA. 2010. Epigenetics in cancer. Carcinogenesis 31:27–36. 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivenbark AG, Strahl BD. 2007. Molecular biology. Unlocking cell fate. Science 318:403–404. 10.1126/science.1150321. [DOI] [PubMed] [Google Scholar]
- 30.Guo LH, Wang HL, Liu XD, Duan J. 2008. Identification of protein differences between two clinical isolates of Streptococcus mutans by proteomic analysis. Oral Microbiol. Immunol. 23:105–111. 10.1111/j.1399-302X.2007.00394.x. [DOI] [PubMed] [Google Scholar]
- 31.Wilkins JC, Homer KA, Beighton D. 2001. Altered protein expression of Streptococcus oralis cultured at low pH revealed by two-dimensional gel electrophoresis. Appl. Environ. Microbiol. 67:3396–3405. 10.1128/AEM.67.8.3396-3405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Haroni M, Skaug N, Bakken V, Cash P. 2008. Proteomic analysis of ampicillin-resistant oral Fusobacterium nucleatum. Oral Microbiol. Immunol. 23:36–42. 10.1111/j.1399-302X.2007.00387.x. [DOI] [PubMed] [Google Scholar]
- 33.Xia Q, Wang T, Park Y, Lamont RJ, Hackett M. 2007. Differential quantitative proteomics of Porphyromonas gingivalis by linear ion trap mass spectrometry: non-label methods comparison, q-values and LOWESS curve fitting. Int. J. Mass Spectrom. 259:105–116. 10.1016/j.ijms.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia Q, Wang T, Taub F, Park Y, Capestany CA, Lamont RJ, Hackett M. 2007. Quantitative proteomics of intracellular Porphyromonas gingivalis. Proteomics 7:4323–4337. 10.1002/pmic.200700543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamont RJ, Meila M, Xia Q, Hackett M. 2006. Mass spectrometry-based proteomics and its application to studies of Porphyromonas gingivalis invasion and pathogenicity. Infect. Disord. Drug Targets 6:311–325. 10.2174/187152606778249935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aruni AW, Roy F, Sandberg L, Fletcher HM. 2012. Proteome variation among Filifactor alocis strains. Proteomics 12:3343–3364. 10.1002/pmic.201200211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599. 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 38.Johnson LS, Eddy SR, Portugaly E. 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11:431. 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. 2010. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38:D355–D360. 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yilmaz O, Young PA, Lamont RJ, Kenny GE. 2003. Gingival epithelial cell signalling and cytoskeletal responses to Porphyromonas gingivalis invasion. Microbiology 149:2417–2426. 10.1099/mic.0.26483-0. [DOI] [PubMed] [Google Scholar]
- 41.Castañeda-Roldán EI, Avelino-Flores F, Dall'Agnol M, Freer E, Cedillo L, Dornand J, Girón JA. 2004. Adherence of Brucella to human epithelial cells and macrophages is mediated by sialic acid residues. Cell. Microbiol. 6:435–445. 10.1111/j.1462-5822.2004.00372.x. [DOI] [PubMed] [Google Scholar]
- 42.Harris JR. 2007. Negative staining of thinly spread biological samples. Methods Mol. Biol. 369:107–142. 10.1007/978-1-59745-294-6_7. [DOI] [PubMed] [Google Scholar]
- 43.Massey BW. 1953. Ultra-thin sectioning for electron microscopy. Stain Technol. 28:19–26. [DOI] [PubMed] [Google Scholar]
- 44.Wyffels JT. 2001. Principles and techniques of electron microscopy: biological applications, 4th edition, by M. A. Hayat. Microsc. Microanal. 7:66. [DOI] [PubMed] [Google Scholar]
- 45.Xiong L, Darwanto A, Sharma S, Herring J, Hu S, Filippova M, Filippov V, Wang Y, Chen CS, Duerksen-Hughes PJ, Sowers LC, Zhang K. 2011. Mass spectrometric studies on epigenetic interaction networks in cell differentiation. J. Biol. Chem. 286:13657–13668. 10.1074/jbc.M110.204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. 2004. The International Protein Index: an integrated database for proteomics experiments. Proteomics 4:1985–1988. 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 47.Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ., Jr 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486–505. 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolenbrander PE, Palmer RJ, Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000 42:47–79. 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 49.Patti JM, Allen BL, McGavin MJ, Hook M. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585–617. 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 50.Fenno JC. 21 February 2012. Treponema denticola interactions with host proteins. J. Oral Microbiol. 10.3402/jom.v4i0.9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinkston KL, Gao P, Diaz-Garcia D, Sillanpaa J, Nallapareddy SR, Murray BE, Harvey BR. 2011. The Fsr quorum-sensing system of Enterococcus faecalis modulates surface display of the collagen-binding MSCRAMM Ace through regulation of gelE. J. Bacteriol. 193:4317–4325. 10.1128/JB.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhulin IB, Nikolskaya AN, Galperin MY. 2003. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J. Bacteriol. 185:285–294. 10.1128/JB.185.1.285-294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mougel C, Zhulin IB. 2001. CHASE: an extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem. Sci. 26:582–584. 10.1016/S0968-0004(01)01969-7. [DOI] [PubMed] [Google Scholar]
- 54.Jorth P, Whiteley M. 2012. An evolutionary link between natural transformation and CRISPR adaptive immunity. mBio 3:e00309-12. 10.1128/mBio.00309-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cady KC, O'Toole GA. 2011. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J. Bacteriol. 193:3433–3445. 10.1128/JB.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845. 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Díez-Villaseñor C, Almendros C, García-Martínez J, Mojica FJ. 2010. Diversity of CRISPR loci in Escherichia coli. Microbiology 156(Pt 5):1351–1361. 10.1099/mic.0.036046-0. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe T, Nozawa T, Aikawa C, Amano A, Maruyama F, Nakagawa I. 2013. CRISPR regulation of intraspecies diversification by limiting IS transposition and intercellular recombination. Genome Biol. Evol. 5:1099–1114. 10.1093/gbe/evt075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin L, Chung WO. 2011. Epigenetic regulation of human beta-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria. Mucosal Immunol. 4:409–419. 10.1038/mi.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pathak SK, Basu S, Bhattacharyya A, Pathak S, Banerjee A, Basu J, Kundu M. 2006. TLR4-dependent NF-kappaB activation and mitogen- and stress-activated protein kinase 1-triggered phosphorylation events are central to Helicobacter pylori peptidyl prolyl cis-, trans-isomerase (HP0175)-mediated induction of IL-6 release from macrophages. J. Immunol. 177:7950–7958. 10.4049/jimmunol.177.11.7950. [DOI] [PubMed] [Google Scholar]
- 61.Díaz-Jullien C, Pérez-Estévez A, Covelo G, Freire M. 1996. Prothymosin alpha binds histones in vitro and shows activity in nucleosome assembly assay. Biochim. Biophys. Acta 1296:219–227. 10.1016/0167-4838(96)00072-6. [DOI] [PubMed] [Google Scholar]
- 62.Martic G, Karetsou Z, Kefala K, Politou AS, Clapier CR, Straub T, Papamarcaki T. 2005. Parathymosin affects the binding of linker histone H1 to nucleosomes and remodels chromatin structure. J. Biol. Chem. 280:16143–16150. 10.1074/jbc.M410175200. [DOI] [PubMed] [Google Scholar]
- 63.Gomez-Marquez J, Rodriguez P. 1998. Prothymosin alpha is a chromatin-remodelling protein in mammalian cells. Biochem. J. 333(Pt 1):1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ushijima T, Hattori N. 2012. Molecular pathways: involvement of Helicobacter pylori-triggered inflammation in the formation of an epigenetic field defect, and its usefulness as cancer risk and exposure markers. Clin. Cancer Res. 18:923–929. 10.1158/1078-0432.CCR-11-2011. [DOI] [PubMed] [Google Scholar]
- 65.Champion MD. 2011. Host-pathogen o-methyltransferase similarity and its specific presence in highly virulent strains of Francisella tularensis suggests molecular mimicry. PLoS One 6:e20295. 10.1371/journal.pone.0020295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, Curry HM, Zwilling BS, Lafuse WP. 2005. Mycobacteria inhibition of IFN-gamma induced HLA-DR gene expression by up-regulating histone deacetylation at the promoter region in human THP-1 monocytic cells. J. Immunol. 174:5687–5694. 10.4049/jimmunol.174.9.5687. [DOI] [PubMed] [Google Scholar]
- 67.Seveau S, Bierne H, Giroux S, Prevost MC, Cossart P. 2004. Role of lipid rafts in E-cadherin– and HGF-R/Met–mediated entry of Listeria monocytogenes into host cells. J. Cell Biol. 166:743–753. 10.1083/jcb.200406078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lafont F, Abrami L, van der Goot FG. 2004. Bacterial subversion of lipid rafts. Curr. Opin. Microbiol. 7:4–10. 10.1016/j.mib.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 69.Wang M, Hajishengallis G. 2008. Lipid raft-dependent uptake, signalling and intracellular fate of Porphyromonas gingivalis in mouse macrophages. Cell Microbiol. 10:2029–2042. 10.1111/j.1462-5822.2008.01185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Knodler LA, Vallance BA, Hensel M, Jackel D, Finlay BB, Steele-Mortimer O. 2003. Salmonella type III effectors PipB and PipB2 are targeted to detergent-resistant microdomains on internal host cell membranes. Mol. Microbiol. 49:685–704. 10.1046/j.1365-2958.2003.03598.x. [DOI] [PubMed] [Google Scholar]
- 71.Ezzell RM, Goldmann WH, Wang N, Parashurama N, Ingber DE. 1997. Vinculin promotes cell spreading by mechanically coupling integrins to the cytoskeleton. Exp. Cell Res. 231:14–26. 10.1006/excr.1996.3451. [DOI] [PubMed] [Google Scholar]
- 72.Gautrey HL, Tyson-Capper AJ. 2012. Regulation of Mcl-1 by SRSF1 and SRSF5 in cancer cells. PLoS One 7:e51497. 10.1371/journal.pone.0051497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mayran N, Parton RG, Gruenberg J. 2003. Annexin II regulates multivesicular endosome biogenesis in the degradation pathway of animal cells. EMBO J. 22:3242–3253. 10.1093/emboj/cdg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andrés V, González JM. 2009. Role of A-type lamins in signaling, transcription, and chromatin organization. J. Cell Biol. 187:945–957. 10.1083/jcb.200904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thuny F, Textoris J, Amara AB, Filali AE, Capo C, Habib G, Raoult D, Mege JL. 2012. The gene expression analysis of blood reveals S100A11 and AQP9 as potential biomarkers of infective endocarditis. PLoS One 7:e31490. 10.1371/journal.pone.0031490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seemann J, Weber K, Gerke V. 1997. Annexin I targets S100C to early endosomes. FEBS Lett. 413:185–190. 10.1016/S0014-5793(97)00911-3. [DOI] [PubMed] [Google Scholar]
- 77.He H, Li J, Weng S, Li M, Yu Y. 2009. S100A11: diverse function and pathology corresponding to different target proteins. Cell Biochem. Biophys. 55:117–126. 10.1007/s12013-009-9061-8. [DOI] [PubMed] [Google Scholar]
- 78.Eisenreich W, Heesemann J, Rudel T, Goebel W. 2013. Metabolic host responses to infection by intracellular bacterial pathogens. Front. Cell Infect. Microbiol. 3:24. 10.3389/fcimb.2013.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Foulquier C, Sebbag M, Clavel C, Chapuy-Regaud S, Al Badine R, Méchin MC, Vincent C, Nachat R, Yamada M, Takahara H, Simon M, Guerrin M, Serre G. 2007. Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum. 56:3541–3553. 10.1002/art.22983. [DOI] [PubMed] [Google Scholar]
- 80.Touz MC, Ropolo AS, Rivero MR, Vranych CV, Conrad JT, Svard SG, Nash TE. 2008. Arginine deiminase has multiple regulatory roles in the biology of Giardia lamblia. J. Cell Sci. 121:2930–2938. 10.1242/jcs.026963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.