Abstract
Decorin-binding protein A (DbpA) of Borrelia burgdorferi mediates bacterial adhesion to heparin and dermatan sulfate associated with decorin. Lysines K82, K163, and K170 of DbpA are known to be important for in vitro interaction with decorin, and the DbpA structure, initially solved by nuclear magnetic resonance (NMR) spectroscopy, suggests these lysine residues colocalize in a pocket near the C terminus of the protein. In the current study, we solved the structure of DbpA from B. burgdorferi strain 297 using X-ray crystallography and confirmed the existing NMR structural data. In vitro binding experiments confirmed that recombinant DbpA proteins with mutations in K82, K163, or K170 did not bind decorin, which was due to an inability to interact with dermatan sulfate. Most importantly, we determined that the in vitro binding defect observed upon mutation of K82, K163, or K170 in DbpA also led to a defect during infection. The infectivity of B. burgdorferi expressing individual dbpA lysine point mutants was assessed in mice challenged via needle inoculation. Murine infection studies showed that strains expressing dbpA with mutations in K82, K163, and K170 were significantly attenuated and could not be cultured from any tissue. Proper expression and cellular localization of the mutated DbpA proteins were examined, and NMR spectroscopy determined that the mutant DbpA proteins were structurally similar to wild-type DbpA. Taken together, these data showed that lysines K82, K163, and K170 potentiate the binding of DbpA to dermatan sulfate and that an interaction(s) mediated by these lysines is essential for B. burgdorferi murine infection.
INTRODUCTION
Lyme disease, caused by spirochetes in the Borrelia burgdorferi sensu lato complex, is the most prevalent vector-borne disease in the United States and Europe (1, 2). The disease develops after bacteria are introduced into a human via the bite of an infected Ixodes tick (3–5). Lyme disease initially presents as a localized skin infection at the bite site, which can develop into the pathognomonic erythema migrans rash and is typically accompanied by nondescript flu-like symptoms. If the early infection is not treated, bacteria can disseminate from the primary bite site through the circulatory and lymphatic systems and invade other tissues and organs, thereby causing the severe secondary multisystemic illnesses associated with Lyme disease (e.g., carditis, arthritis, and neuroborreliosis) (6–8). The ability of bacteria to disseminate, colonize, and persist within an infected host is a complex and multifactorial process (9). Surface-exposed bacterial proteins called adhesins promote interaction of the bacteria with a host cell or the extracellular matrix (ECM) and are recognized to be key mediators of bacterial colonization (10). A number of the host and bacterial factors that mediate B. burgdorferi attachment to mammalian and tick tissues have been identified (5, 11, 12). Specifically, B. burgdorferi adhesins bind host cell-associated integrins (13–16), as well as the mammalian ECM components fibronectin (17–23), type I collagen (23, 24), laminin (23, 25, 26), glycosaminoglycans (GAGs) (18, 27–29), and decorin (30–32). This capacity to interact with numerous host ligands is predicted to be responsible for the spirochete's ability to spread to and infect diverse host tissues, cause the various disease sequelae, and persist within the mammal and tick vector (5). Interestingly, strains of B. burgdorferi appear to differ in their abilities to cause specific systemic sequelae, and these differences have been linked in part to their capacities to bind/colonize certain host tissues (33–36).
Decorin, a small proteoglycan found in the ECM of numerous tissues (e.g., dermis and cartilage), is composed of a 36-kDa protein core covalently linked to a 40-kDa GAG chain of chondroitin sulfate or dermatan sulfate (37–39). The lp54 linear plasmid of B. burgdorferi sensu stricto carries a two-gene operon that encodes two surface lipoproteins, decorin-binding proteins A and B (DbpA and DbpB), which bind decorin (30–32). In vitro studies have shown that both adhesins mediate interaction with the GAGs heparin and dermatan sulfate (31), but only DbpB binds chondroitin sulfate (40). dbpA, and most likely dbpB, is upregulated by B. burgdorferi during tick feeding and expressed during mammalian infection (41–45). dbpA is presumed to remain highly expressed throughout infection based on the presence of high levels of reactive antibodies at 47 weeks postinoculation in macaques experimentally infected with B. burgdorferi (46). Considering that dbpA is expressed during mammalian infection and decorin/dermatan sulfate can be found associated with almost all mammalian tissues, DbpA and the capacity to bind decorin have long been hypothesized to be key determinants in B. burgdorferi colonization and dissemination within the host. In agreement with this, mutational studies have since confirmed that B. burgdorferi mutants in which dbpBA was disrupted have a diminished capacity for dissemination and infection in mice challenged via needle inoculation (47–50), but dbpA and dbpB are not required for colonization of Ixodes scapularis ticks (47). However, in contrast to the needle challenge studies, the dbpBA mutant was capable of being transmitted to and infecting mice via tick bite (47). A recent study also suggests that the attenuation observed in dbpBA mutants is limited to early stages of dissemination and that this defect is alleviated during chronic infection (51). Interestingly, this study also found that the dbpBA mutant was unable to migrate through the lymphatic system, suggesting that interaction of B. burgdorferi with decorin/GAGs might be important when utilizing this route of dissemination. A number of early biochemical and functional studies characterized the interaction between DbpA and decorin/GAGs (31, 52, 53). Comparison of DbpA sequences from 30 members of the B. burgdorferi sensu lato complex identified 11 lysine residues that were highly conserved among the different strains; five lysine residues were conserved in all sequences, and six lysine residues were found in >60% of the DbpA sequences (52). Analysis of recombinant DbpA proteins carrying mutations in these individual lysines showed that mutation of K82, K163, or K170 resulted in reduced decorin binding. A recent study also showed that the highly basic C terminus of DbpA was required for bacterial interaction with decorin, dermatan sulfate, and 293 (human kidney epithelial) cells (35). Although the in vitro data clearly implicate these residues in DbpA-decorin/GAG interaction, they did not address their contributions during mammalian infection.
Despite the abundance of functional and biochemical data available regarding DbpA, it was not until recently that the solution structure of DbpA from strain B31 was elucidated using nuclear magnetic resonance (NMR) spectroscopy (54). In the present study, we sought to complement the NMR studies by solving the structure of DbpA from B. burgdorferi strain 297 using X-ray crystallography, as well as to assess whether lysines K82, K163, and K170 are required during B. burgdorferi infection. Our crystal structure of a DbpA monomer (determined at a resolution of 1.60 Å) confirmed the prior NMR structure, as well as the localization of the lysines critical for the interaction of DbpA with decorin to a common basic patch near the C terminus of the protein. To test the contributions of lysines K82, K163, and K170 during mammalian infection, mice were challenged with B. burgdorferi dbpBA knockout strains expressing dbpA point mutants in which these lysine residues were changed to alanine. The dbpBA mutants complemented with K82, K163, and K170 alanine point mutants were unable to infect mice, thus correlating the physiological contributions of these residues to decorin/GAG binding and murine infection.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All the strains and plasmids used in this study are described in Table 1. Escherichia coli strain TOP10F′ (Invitrogen, Carlsbad, CA) was used for cloning. E. coli strains XL1-Blue (Stratagene, La Jolla, CA) and B834(DE3) (Novagen, Madison, WI) were used for protein expression. E. coli transformants were selected in Luria-Bertani (LB) medium supplemented with 100 μg/ml of ampicillin or 100 μg/ml of spectinomycin. Infectious, low-passage-number B. burgdorferi strain 297 (Bb297) and the previously characterized B. burgdorferi dbpBA mutant (BbKH500) were used for these studies (47, 55). B. burgdorferi was grown in Barbour-Stoenner-Kelley II (BSK-II) medium at 37°C and pH 7.5 with 3 to 5% CO2 and 150 μg/ml of streptomycin or 150 μg/ml of kanamycin (56).
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Descriptiona | Source |
|---|---|---|
| Plasmids | ||
| pGEM-T Easy | TA cloning vector; Ampr | Promega |
| pProEX-HTa | Expression construct; N-terminal and TEV-cleavable His6 tag; Ampr | Invitrogen |
| pProEX-DbpAWT | pProEX HTa::DbpA (aa 26–187); Ampr | This study |
| pProEX-DbpAΔC | pProEX HTa::DbpA (aa 26–173); Ampr | This study |
| pProEX-DbpAK51A | pProEX HTa::DbpAWT with K51A mutation; Ampr | This study |
| pProEX-DbpAK82A | pProEX HTa::DbpAWT with K82A mutation; Ampr | This study |
| pProEX-DbpAK124A | pProEX HTa::DbpAWT with K124A mutation; Ampr | This study |
| pProEX-DbpAK163A | pProEX HTa::DbpAWT with K163A mutation; Ampr | This study |
| pProEX-DbpAK170A | pProEX HTa::DbpAWT with K170A mutation; Ampr | This study |
| pProEX-DbpAK177A | pProEX HTa::DbpAWT with K177A mutation; Ampr | This study |
| pRevA | pQE30::RevA (aa 20–160); Ampr | 20 |
| pJD51 | B. burgdorferi and E. coli shuttle vector; Specr Strepr | 47 |
| pKH2000 | pJD51::PdbpBA-dbpA ORF complementation vector; Specr Strepr | 47 |
| pDbpAK51A | pKH2000 with DbpAK51A mutation; Specr Strepr | This study |
| pDbpAK82A | pKH2000 with DbpAK82A mutation; Spec/Strepr | This study |
| pDbpAK124A | pKH2000 with DbpAK124A mutation; Spec/Strepr | This study |
| pDbpAK163A | pKH2000 with DbpAK163A mutation; Spec/Strepr | This study |
| pDbpAK170A | pKH2000 with DbpAK170A mutation; Spec/Strepr | This study |
| pDbpAK177A | pKH2000 with DbpAK177A mutation; Spec/Strepr | This study |
| pJSB568 | pGEM-T Easy::flaB-actin qPCR standard; Ampr | This study |
| Strains | ||
| E. coli | ||
| TOP10F′ | F′ [lacIq Tn10 (Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 rec A1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIq ZΔM15::Tn10 (Tetr)] | Strategene |
| B834(DE3) | F− ompT hsdSB(rB− mB−) gal dcm met (DE3) | Novagen |
| B. burgdorferi | ||
| Bb297 | Strain 297; infectious, human spinal fluid isolate | 55 |
| BbKH500 | Strain 297; dbpBA::PflgB-Kan mutant; Kanr | 47 |
| BbKH501 | BbKH500 complemented with pKH2000; Kanr Strepr | 47 |
| BbDbpAK51A | BbKH500 complemented with pDbpAK51A; Kanr Strepr | This study |
| BbDbpAK82A | BbKH500 complemented with pDbpAK82A; Kanr Strepr | This study |
| BbDbpAK124A | BbKH500 complemented with pDbpAK124A; Kanr Strepr | This study |
| BbDbpAK163A | BbKH500 complemented with pDbpAK163A; Kanr Strepr | This study |
| BbDbpAK170A | BbKH500 complemented with pDbpAK170A; Kanr Strepr | This study |
| BbDbpAK177A | BbKH500 complemented with pDbpAK177A; Kanr Strepr | This study |
aa, amino acids; Ampr, ampicillin resistant; Kanr, kanamycin resistant; Specr, spectinomycin resistant; Strepr, streptomycin resistant.
Cloning wild-type, carboxyl-terminally truncated, and lysine mutant recombinant DbpA proteins.
The first 25 amino acids of DbpA from Bb297 contain the putative signal peptide terminated by a potential consensus sequence for lipoprotein modification (31, 32). To facilitate expression of the nonlipidated wild-type (WT) protein in E. coli, a DNA fragment encoding amino acids 26 to 187 of DbpA was amplified by PCR using Bb297 genomic DNA as the template and the oligonucleotides 5′-DbpA(26) and 3′-DbpA(187) (Table 2); these primers contained BamHI and HindIII restriction sites, respectively. The 3′-DbpA(187) primer also contained tandem stop codons. A variant of DbpA with a C-terminal truncation was also generated by PCR amplification with the oligonucleotide pair 5′-DbpA(26) and 3′-DbpA(173) (Table 2); 3′-DbpA(173) introduces a HindIII site and two tandem stop codons on the 3′ end of the amplified open reading frame (ORF). PCR amplification was performed using Ex-Taq polymerase (TaKaRa, Madison, WI). The amplified fragments were then digested with BamHI and HindIII and ligated into pProEX-HTa digested with the same enzymes. The ligation was transformed into E. coli, and the resulting clones were verified by DNA sequencing. Confirmed clones were designated pProEX-DbpAWT and pProEX-DbpAΔC (Table 1).
TABLE 2.
Oligonucleotide primers used in this study

Relevant restriction sites are italicized. Stop codons are underlined. Nucleotides changed to generate mutant codons are in boldface.
The construct for the production of the His6-tagged RevA (recombinant RevA [rRevA]) control protein was previously described (20). Expression constructs for rDbpA variants carrying the lysine-to-alanine point mutation K51A, K82A, K124A, K163A, K170A, or K177A were generated by PCR amplifying a DNA fragment corresponding to amino acids 26 to 187 of DbpA using oligonucleotides 5′-DbpA(26) and 3′-DbpA(187). The pJD51 lysine point mutant shuttle vectors, the derivation of which is discussed below, were used as the template in these PCRs. Cloning of the DbpA ORFs into pProEX-HTa was carried out as described above.
Expression and purification of rDbpA.
Soluble His6-tagged rRevA was purified as previously described (35). Expression of rDbpA from pProEX-HTa generates recombinant protein with an N-terminal His6 tag followed by a tobacco etch virus (TEV) protease cleavage site. For expression, cultures of XL1-Blue transformed with pProEX-DbpAWT or pProEX-DbpAΔC were grown in LB medium containing ampicillin. When the cell density reached an optical density at 600 nm (OD600) of 0.6, the culture was induced for 3 h with 400 μM isopropyl-1-thio-β-d-galactopyranoside (IPTG). Cells were collected by centrifugation; suspended in 20 mM HEPES (pH 7.5), 20 mM NaCl; and lysed by extrusion with an EmulsiFlex (Avestin, Ontario, Canada). To pellet cell debris, the lysate was centrifuged at 27,000 × g for 15 min. rDbpA was purified from the supernatant on Ni2+ affinity resin (Qiagen Inc., Valencia, CA) and eluted with 20 mM HEPES (pH 7.5), 300 mM NaCl, 500 mM imidazole. The elution buffer was exchanged with 20 mM HEPES (pH 7.5), 50 mM NaCl, 15 mM β-mercaptoethanol (buffer A) using an Amicon Ultra-15 10,000-molecular-weight cutoff (MWCO) centrifugal-filtration unit (Millipore, Billerica, MA). The His6 tag was cleaved from rDbpA by adding 1 mg of His6-TEV protease per 30 mg of recombinant protein. Proteolysis reaction mixtures were incubated overnight at room temperature and stopped using an Ni2+ affinity column to remove the cleaved His6 tag and His6-TEV protease. The rDbpA protein was then concentrated to a final volume of 1 ml and resolved on a HiLoad 16/60 Superdex 75 gel filtration column using an Äkta fast-performance liquid chromatography (FPLC) system (GE Healthcare Biosciences, Piscataway, NJ). The protein was eluted with buffer A at a flow rate of 1 ml/min, and samples of the fractions containing protein were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to assess purity. SDS-PAGE indicated that the protein was pure to apparent homogeneity (i.e., >95%). Fractions containing purified DbpA were pooled and concentrated to 18 to 40 mg/ml in buffer A for crystallization and binding experiments.
For the production of selenomethionine (SeMet)-labeled protein, pProEX-DbpAWT or pProEX-DbpAΔC was transformed into the E. coli methionine auxotroph B834(DE3). Cells were cultured in 5% LB-95% M9 minimal medium containing 125 mg/liter each of adenine, uracil, thymine, and guanosine; 2.5 mg/liter thiamine; 4 mg/liter d-biotin; 20 mM glucose; 2 mM MgSO4; 50 mg/liter each of 19 l-amino acids (with the exception of methionine); and 50 mg/liter l-selenomethionine. The cultures were induced, and the protein was purified as described above.
Crystallization, data collection, and structure determination.
Crystals of a proteolytic fragment of wild-type rDbpA (rDbpAWT) were grown at 20°C using the vapor diffusion method in hanging-drop mode by mixing 2 μl protein (25 mg/ml) in 20 mM HEPES, pH 7.5, 50 mM NaCl, 15 mM β-mercaptoethanol with 2 μl reservoir solution composed of 30 to 35% (wt/vol) polyethylene glycol (PEG) 3000, 100 mM 2-(N-cyclohexylamino)ethanesulfonic acid (CHES), pH 9.5, 50 mM NaCl and equilibrating against 1 ml of reservoir solution. The crystals exhibit the symmetry of space group P43 and were cryoprotected in reservoir solution supplemented with 10% (vol/vol) ethylene glycol and then flash cooled in liquid nitrogen.
Crystals of selenomethionyl-derivatized rDbpAΔC were grown at 20°C using the vapor diffusion method in hanging-drop mode by mixing 1 μl protein (25 mg/ml) in 20 mM HEPES, pH 7.5, 50 mM NaCl, 15 mM β-mercaptoethanol with 1 μl reservoir solution, 30 to 35% (wt/vol) PEG 3000, 0.1 M CHES, pH 9.5, 50 mM NaCl and equilibrating against 1 ml of reservoir solution. The crystals exhibit the symmetry of space group P212121 and were cryoprotected in reservoir solution supplemented with 10% (vol/vol) ethylene glycol and then flash cooled in liquid nitrogen.
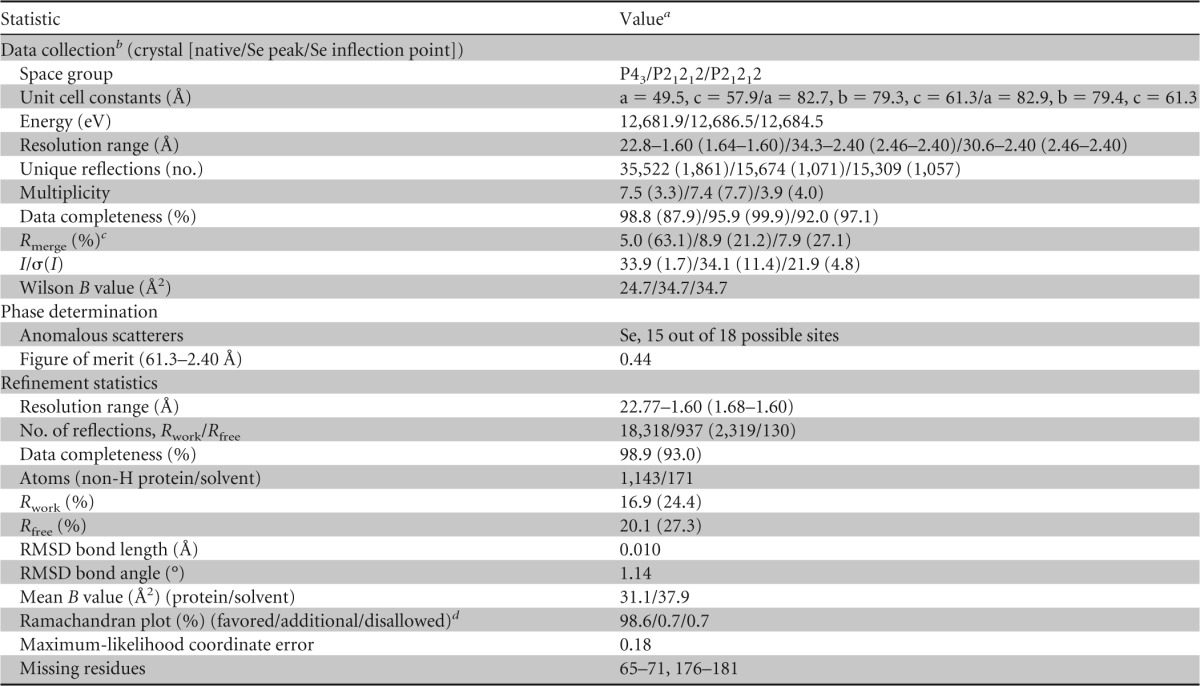
Phases obtained from a two-wavelength selenium (Se) anomalous-dispersion experiment were refined with the program MLPHARE (57), resulting in an overall figure of merit of 0.44 for data between 61.3 and 2.40 Å. The phases were further improved by density modification and 3-fold averaging with the program DM (58), resulting in a figure of merit of 0.63. An initial model containing 89% of all rDbpA residues was automatically generated by alternating cycles of the programs Resolve (59, 60) and ARP/wARP (61). Phases for the proteolytic fragment of rDbpA in the P43 cell were obtained via molecular replacement in the program PHASER (62), using the coordinates of DbpA from the orthorhombic cell as a search model. Additional residues for rDbpA were manually modeled in the program O (63). Refinement was performed with the data collected on the proteolytic fragment of rDbpAWT to a resolution of 1.60 Å using the program PHENIX (64), with a random 5% of all data set aside for free refinement (R) factor (Rfree) calculation. Phasing and model refinement statistics are provided in Table 3.
TABLE 3.
Data collection, phasing, and refinement statistics for DbpA structure
Data for the outermost shell are given in parentheses.
For SE, Bijvoet pairs were kept separate for data processing.
Rmerge = 100 ΣhΣi|Ih,i − 〈Ih〉|/ΣhΣiIh,i, where the outer sum (h) is over the unique reflections and the inner sum (i) is over the set of independent observations of each unique reflection.
As defined by the validation suite MolProbity (81).
Site-directed mutagenesis of DbpA.
Mutation of selected lysine residues in DbpA was performed by overlap extension PCR. Primers 5′ DbpA prom and 3′ DbpA ORF represent the 5′ and 3′ outermost primers, respectively, used for all amplifications. The internal primers used to mutate selected lysine amino acids to alanine (e.g., K51A, K82A, K124A, K163A, K170A, and K177A) are indicated with their intended mutation and denoted as sense or antisense (Table 2). The numbering of the mutated lysine residues is based on the full-length dbpA ORF from Bb297 (31). Oligonucleotides 5′ DbpA prom and 3′ DbpA ORF were paired with the mutational antisense and sense oligonucleotides, respectively. DNA fragments were amplified with Ex-Taq polymerase using pKH2000 as the template. pKH2000 (47), which encodes the native promoter for the dbpBA operon fused directly to the dbpA ORF, is a borrelial shuttle vector used in prior studies for genetic complementation of dbpA expression in the B. burgdorferi dbpBA mutant (BbKH500). Amplicons from the individual reactions were gel purified and combined to serve as the DNA template in a second PCR using the outermost oligonucleotides, 5′ DbpA prom and 3′ DbpA ORF. The resulting amplicons for the point mutants were cloned in pGEM-T Easy and confirmed by DNA sequencing with vector-specific primers and the 5′ DbpA internal seq oligonucleotide. The primers 5′ DbpA prom and 3′ DbpA ORF incorporated BglII and SphI restriction sites, respectively, to facilitate cloning of the mutated dbpA alleles into the borrelial shuttle vector pJD51 (Table 1).
Quantitative ELISA to measure decorin and dermatan sulfate binding.
Quantitative enzyme-linked immunosorbent assay (ELISA) for decorin and dermatan sulfate binding by rDbpA proteins was performed similarly to that previously described (65). Briefly, wells of 96-well microtiter plates were coated with 1 μg of decorin, dermatan sulfate, or bovine serum albumin (BSA); 100 μl of increasing concentrations (0.007, 0.015, 0.03125, 0.0625, 0.125, 0.25, 0.5, and 1 μM for decorin binding; 0.03125, 0.0625, 0.125, 0.25, 0.5, 1, and 2 μM for dermatan sulfate binding) of His6-tagged rRevA protein (20), rDbpAWT, or the lysine rDbpA mutant proteins was then added to the wells. To detect the binding of recombinant proteins, a 1:200 dilution of mouse anti-His6 antibody (Sigma, St. Louis, MO) and a 1:1,000 dilution of horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Invitrogen) were used as primary and secondary antibodies, respectively. The plates were washed three times with PBST (0.05% Tween 20 in phosphate-buffered saline [PBS]), and 100 μl of tetramethylbenzidine (TMB) solution (KPL, Gaithersburg, MD) was added to each well and incubated for 5 min. The reaction was stopped by adding 100 μl of 0.5% hydrosulfuric acid to each well. The absorbance was then read at 405 nm using an ELISA plate reader (SpectraMAX 250; Molecular Devices, Sunnyvale, CA). Statistical significance was determined using the Student t test; P values of ≤0.05 were considered significant. To determine the dissociation constant (Kd), the data were fitted by the following equation using KaleidaGraph software (version 2.1.3; Abekbecj software, Reading, PA) where OD450(MAX) denotes the maximum absorbance at an optical density (OD) of 450 nm: OD450 = OD450(MAX) × [DbpA protein]/Kd + [DbpA protein].
SPR.
Surface plasmon resonance (SPR) was measured using a Biacore 3000 (GE Healthcare). Dermatan sulfate (EMD Millipore, Billerica, MA) and decorin (Sigma) were biotinylated using EZ-Link Biocytin Hydrazide (Pierce, Rockford, IL) and then dialyzed in PBS using a Slide-A-Lyzer cassette (Pierce) with a 10,000-kDa-molecular-mass cutoff. Ten micrograms of biotinylated dermatan sulfate or decorin was conjugated on a streptavidin SA sensor chip (GE Healthcare). A control flow cell was injected with PBS buffer lacking dermatan sulfate or decorin. For quantitative SPR experiments to measure decorin binding, 20 μl of increasing concentrations (0, 0.03125, 0.0625, 0.125, 0.25, 0.5, and 1 μM) of rDbpAWT or the rDbpA lysine mutants was injected into the control cell and decorin-bound flow cells. To measure dermatan sulfate binding, 20 μl of various concentrations (0, 0.09375, 0.1875, 0.375, 0.75, 1.5, and 3 μM) of recombinant protein was added to flow cells containing immobilized dermatan sulfate. The proteins were loaded into the cells at a flow rate of 10 μl/min, and binding was carried out at 25°C. All sensogram data for the dermatan sulfate- and decorin-immobilized cells were subtracted from the negative-control flow cell. To obtain the kinetic parameters of the interaction, the data from the sensograms were fitted by BIAevaluation software version 3.0 using the one-step biomolecular association reaction model (1:1 Langmuir model), which resulted in optimum mathematical fits using the lowest χ2 values.
NMR spectroscopy.
To express rDbpAWT for NMR characterization, bacteria were grown in M9 minimal medium supplemented with 1 g/liter 15NH4Cl as the sole nitrogen source. The 15N-labeled recombinant protein was purified, and the His6 tag was cleaved with TEV protease as described above. The rDbpA proteins were then concentrated to a final volume of 2 ml and resolved on a HiLoad 16/60 Superdex 75 gel filtration column by FPLC. The protein was eluted with 50 mM sodium phosphate, 50 mM NaCl (pH 6.5), and samples of the fractions containing protein were analyzed by SDS-PAGE to assess purity. Data were collected on 500 μM protein samples in 50 mM sodium phosphate, 50 mM NaCl (pH 6.5) at 25°C. 1H, 15N heteronuclear single quantum coherence (HSQC) spectra of labeled protein were acquired by using a Varian Inova 500-MHz spectrometer and utilizing NMRPipe (66) and NMRView (67) for data processing and analysis, respectively.
Transformation of B. burgdorferi.
Electroporation of the B. burgdorferi BbKH500 dbpBA mutant with the pJD51 shuttle vectors carrying the dbpA lysine point mutants was carried out as previously described (68). Streptomycin-resistant (Strepr) clones were confirmed by DNA sequence analysis of the transformed shuttle vector and PCR-based profiling to assess the endogenous borrelial plasmid content (47). Confirmed clones were designated according to the DbpA lysine mutation that each transformant carried; BbDbpAK51A, BbDbpAK82A, BbDbpAK124A, BbDbpAK163A, BbDbpAK170A, or BbDbpAK177A.
Mouse infection experiments.
The University of Arkansas for Medical Sciences (UAMS) is accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC), and all animal experiments were approved by the UAMS Institutional Animal Care and Use Committee (IACUC). The infectivity of BbKH500 and dbpA-complemented strains was assessed using the murine needle challenge model of Lyme borreliosis (32, 47). Clones were grown to the mid-log phase with antibiotic selection, at which time the bacterial density in each culture was enumerated using dark-field microscopy. Three- to 4-week-old C3H/HeN (Harlan, Indianapolis, IN) mice were infected with 1.5 × 105 bacteria via intradermal injection. At 2, 6, and 10 weeks postinfection, ear punch, lymph node (brachial and inguinal), tarsal, and heart tissue samples were collected from the mice and placed in BSK-II medium containing borrelia antibiotic cocktail (Monserate Biotechology Group, San Diego, CA). Skin, tibiotarsal, and heart samples were collected, stored in RNAlater Stabilization Solution (Life Technologies, Carlsbad, CA), and frozen at −80°C until analyzed by quantitative PCR (qPCR). Dark-field microscopy was used to assess the growth of spirochetes in each of these cultures 1 to 2 weeks postinoculation. Aliquots of growth-positive cultures were transferred to fresh medium with antibiotic selection for genotypic confirmation of infectious clones (data not shown).
qPCR analysis of the spirochete tissue burden.
To generate a copy number standard for qPCR measurement of bacterial tissue burdens, a fragment of the murine β-actin gene was amplified using primers ActF-STD and ActR-STD with C3H/HeN DNA (Table 2) (69). These primers introduced a BamHI restriction site at one end. The amplicon was cloned into pGEM-T Easy. A portion of flaB was then amplified using primers flaBF-STD and flaBR-STD and Bb297 genomic DNA (gDNA) (Table 2), which introduced BamHI restriction sites on both ends of the fragment. The flaB amplicon was ligated into pGEM-T Easy containing the actin fragment via the BamHI sites. The pGEM-T Easy::β-actin/flaB construct was designated pJSB568.
For qPCR, DNA was extracted from skin, tibiotarsal, and heart samples from infected mice as previously described by Maruskova et al. (69). A High Pure PCR template preparation kit (Roche Applied Sciences, Indianapolis, IN) was used to extract DNA according to the manufacturer's instructions, except that 200 μg of collagenase (Sigma) was added during lysis. Purified DNA was analyzed via qPCR on a StepOnePlus Real Time PCR System using TaqMan Fast Advanced master mix (Life Technologies). The primer set and probe used to detect flaB were FlaB-ABI-F, FlaB-ABI-R, and FlaB-ABI-Probe (Table 2). The primer set and probe used to detect the β-actin gene were Act-ABI-F, Act-ABI-R, and Actin-ABI-Probe (Table 2). The flaB and β-actin probes were labeled on the 5′ end with 6-carboxyfluorescein (FAM) and MAX Freedom dyes, respectively. Both probes were 3′ labeled with Iowa Black FQ and internally quenched with ZEN (Integrated DNA Technologies, Coralville, IA). A copy number standard curve using the pJSB568 flaB/β-actin vector as the standard was used to measure the number of flaB copies and to quantify spirochetal burdens. Bacterial burdens were reported as the number of flaB copies per 106 copies of mouse β-actin. Analysis of variance (ANOVA) models were used to compare qPCR measurements between strains, and Tukey's procedure was used to perform pairwise comparisons (Prism v6.0c; GraphPad software).
In vitro growth analyses.
B. burgdorferi cultures were inoculated to a starting density of 103 spirochetes/ml and grown for 7 days at 37°C and pH 7.5. Beginning at 3 days postinoculation, spirochetes were counted daily using dark-field microscopy to determine culture densities. For each culture and time point, bacteria were enumerated in 20 individual microscope fields. To assess RpoS-dependent gene activation in B. burgdorferi, bacteria were grown in BSK-II at pH 6.8 until they reached the stationary growth phase (>108 bacteria/ml) (70). Bacteria were collected and prepared for SDS-PAGE and immunoblot analysis as described below.
Proteinase K accessibility assay.
Borrelia cultures were grown to the late exponential growth phase. Cells were collected by centrifugation, washed twice with PBS, and suspended to a density of 2 × 109 cells/ml in PBS. Spirochetes (1 × 109) were treated with 200 μg of proteinase K (10 mg/ml; Fisher Scientific, Pittsburgh, PA) or sham treated for 60 min at room temperature. Proteinase K digestion was stopped by adding 15 μl of phenylmethylsulfonyl fluoride (PMSF) (50 mg/ml in isopropanol; Sigma), and samples were prepared for SDS-PAGE and immunoblotting.
SDS-PAGE and immunoblot analysis.
SDS-PAGE and immunoblotting were performed as previously described (47). A volume of cell lysate equivalent to 2 × 107 bacteria was loaded in each gel lane. The molecular-weight standard used for all immunoblots was All Blue Precision Plus standard (Bio-Rad, Hercules, CA). For colorimetric detection of DbpA, FlaB, and OspC, 4-chloro-1-naphthol was used as the substrate. Antibodies used to detect DbpA, FlaB, and OspC were described in prior studies (47, 71).
Protein structure accession number.
The coordinates and structure factors for DbpA have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB) under the PDB ID 4ONR.
RESULTS AND DISCUSSION
rDbpA adopts a four-helical-bundle fold.
In the present study, we sought to expand our knowledge of DbpA by solving the structure of DbpA from B. burgdorferi strain 297 using X-ray crystallography. A nonlipidated form of Bb297 rDbpA was expressed in E. coli, and crystals of a proteolytic fragment of rDbpA were grown using the vapor diffusion method in hanging-drop mode. The structure of the proteolytic fragment of rDbpA that encompasses residues 26 to 175 was determined at a resolution of 1.60 Å via a combination of single-wavelength anomalous dispersion and molecular replacement (Table 3). Analogous to the structure determined via NMR methods (54), rDbpA is a monomer and contains an up-down four-alpha-helical bundle (Fig. 1A), with a fifth helix packed against helices 2, 3, and 4. Residues 65 to 71 of the flexible loop between helices 1 and 2 were disordered in the electron density map and were not modeled. An alignment of the X-ray coordinates via DaliLite (72) with the deposited NMR coordinates (PDB ID 2LQU) yielded a root mean square deviation (RMSD) of 2.2 Å for 143 equivalent C-alpha atoms and a Z-score of 15.4. Mapping of the refined atomic displacement parameters (B factors) on the X-ray structure show that the largest values occur at the ends of helices 1, 2, and 5 and along the distal surface of helix 1 (Fig. 1B). This is in contrast to the published NMR study by Wang (54), which indicated that helix 4 may undergo more internal motion than the other helices. The larger values for atomic displacement parameters found in helix 1 do not appear to be an artifact of crystalline lattice packing, as the solvent content of the lattice is quite low (approximately 40%) and the largest solvent channel is in the vicinity of the disordered loop.
FIG 1.
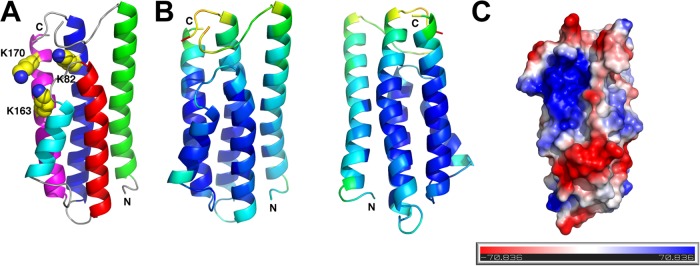
Crystal structure of rDbpA. (A) Helix 1 is colored green, helix 2 is blue, helix 3 is red, helix 4 is cyan, and helix 5 is magenta. The lysine residues known to be involved in GAG binding (K82, K163, and K170) are shown as yellow spheres, and the N and C termini are labeled. (B) Refined atomic displacement parameters (ADP) from the X-ray structure mapped onto the structure of rDbpA. ADP values from low to high are colored from dark blue to red. The view on the left is in the same orientation as in panel A, and the view on the right is rotated by 180°. (C) Electrostatic potential surface map of rDbpA revealing a positively charged cleft in the vicinity of the GAG binding lysines. This view has been rotated slightly from the view in panel A for clarity.
The three lysine residues (K82, K163, and K170) identified as essential for decorin binding in vitro interaction studies (52) are mapped to a common positively charged cleft near the disordered helix 1-helix 2 loop (Fig. 1C). Additional lysines mutated in this study either are solvent exposed on the outer surface of the protein distal to the disordered loop (e.g., K51 and K124) or were not located in the electron density at the C terminus of the molecule but would be too distant from the positively charged cleft to contribute to substrate binding (K177). The proximity of the flexible lysine side chains in the positively charged cleft provides a useful motif for interaction with the GAG of decorin, and the high mobility of the helix 1-helix 2 loop allows easy accessibility of the substrate to the cleft.
Contribution of conserved lysine residues in DbpA to dermatan sulfate binding.
In vitro binding studies identified several lysine residues in DbpA that were involved in mediating adherence to decorin. Specifically, lysines K82, K163, and K170 are absolutely essential for decorin binding (52), and the structural data (Fig. 1) (54) confirmed that these critical lysines are located on proximal unique alpha helices that form a basic pocket in which GAGs are speculated to bind. To confirm that these lysine mutations had the intended effect on decorin binding, we first generated recombinant DbpA protein for wild-type DbpA and each of the lysine mutants. In addition to K82, K163, and K170, several conserved lysine residues in DbpA that have not been implicated in decorin binding were also targeted (52). Overlap extension PCR was used to mutate individual lysine codons in the dbpA ORF to alanine, which were then expressed and purified as N-terminally His6-tagged recombinant proteins: rDbpAK51A, rDbpAK82A, rDbpAK124A, rDbpAK163A, rDbpAK170A, and rDbpAK177A. Decorin-binding activities of the rDbpA derivatives were quantitatively measured by both ELISA and quantitative SPR (Table 4; see Fig. S1 and S2 in the supplemental material). As shown in Table 4, rDbpAK51A, rDbpAK124A, and rDbpAK177A bound decorin at levels comparable to rDbpAWT. In contrast, rDbpAK82A, rDbpAK163A, and rDbpAK170A exhibited no decorin binding, confirming that K82, K163, and K170 of DbpA are essential for interaction with decorin. The reduction in binding with rDbpAK82A, rDbpAK163A, and rDbpAK170A at 0.25, 0.5, and 1 μM decorin was statistically significant relative to rDbpAWT (see Fig. S1 in the supplemental material). Since the GAGs associated with the protein core mediate the interaction between decorin and DbpA, we also wanted to assess the interaction of each of these mutants with dermatan sulfate. In agreement with the decorin-binding data, rDbpAK51A, rDbpAK124A, and rDbpAK177A bound dermatan sulfate at levels equivalent to rDbpAWT, while dermatan sulfate binding was completely abolished with rDbpAK82A, rDbpAK163A, and rDbpAK170A. The reduced binding at 0.5, 1, and 2 μM dermatan sulfate with rDbpAK82A, rDbpAK163A, and rDbpAK170A was statistically significant relative to rDbpAWT (see Fig. S1 in the supplemental material). Taken together, these results agree with those of previous decorin-binding studies and indicate that K82, K163, and K170 of DbpA are essential for interaction with the dermatan sulfate moiety of decorin (52).
TABLE 4.
Kinetic data and dissociation constants for the interaction of rDbpA proteins with decorin or dermatan sulfate determined by SPR and quantitative ELISA
| Protein | Ligand | Kd (μM)a | Kd (μM)b | kon (104 s−1 M−1)b | koff (s−1)b |
|---|---|---|---|---|---|
| rDbpA | Decorin | 0.32 ± 0.05 | 0.29 ± 0.06 | 35.05 ± 6.15 | 0.099 ± 0.003 |
| Dermatan sulfate | 0.47 ± 0.05 | 0.46 ± 0.08 | 5.21 ± 0.16 | 0.024 ± 0.004 | |
| rDbpAK51A | Decorin | 0.74 ± 0.05 | 0.79 ± 0.03 | 12.6 ± 1.70 | 0.099 ± 0.018 |
| Dermatan sulfate | 1.45 ± 0.37 | 1.17 ± 0.28 | 2.57 ± 0.32 | 0.030 ± 0.003 | |
| rDbpAK82A | Decorin | NBc | NB | NB | NB |
| Dermatan sulfate | NB | NB | NB | NB | |
| rDbpAK124A | Decorin | 0.28 ± 0.04 | 0.28 ± 0.14 | 29.25 ± 7.50 | 0.084 ± 0.01 |
| Dermatan sulfate | 0.47 ± 0.02 | 0.54 ± 0.11 | 5.92 ± 0.53 | 0.032 ± 0.009 | |
| rDbpAK163A | Decorin | NB | NB | NB | NB |
| Dermatan sulfate | NB | NB | NB | NB | |
| rDbpAK170A | Decorin | NB | NB | NB | NB |
| Dermatan sulfate | NB | NB | NB | NB | |
| rDbpAK177A | Decorin | 0.30 ± 0.03 | 0.20 ± 0.06 | 35.70 ± 1.10 | 0.071 ± 0.004 |
| Dermatan sulfate | 0.48 ± 0.04 | 0.53 ± 0.09 | 3.34 ± 0.18 | 0.018 ± 0.004 |
Calculated from the ELISA experiments shown in Fig. S1 in the supplemental material. The Kd values are the means ± standard deviations (SD) from three independent experiments.
Calculated from the SPR experiments shown in Fig. S2 in the supplemental material. kon, association rate constant; koff, dissociation rate constant. The Kd, kon, and koff values are the means ± SD from three independent experiments.
NB, no binding. rDbpAK82A, rDbpAK163A, and rDbpAK170A bound decorin and dermatan sulfate so poorly that curve-fitting software could not accurately calculate the Kd, kon, and koff values. Since these values were not available, it was not possible to do a statistical analysis of decorin or dermatan sulfate binding between these mutants and WT DbpA.
While the previous data suggest that these three lysine residues contribute to decorin and dermatan sulfate binding, it is possible that these substitutions could have altered binding activity by triggering structural changes that destabilized the DbpA protein. To confirm that the lysine mutations did not significantly alter the structure of DbpA, rDbpAWT, rDbpAK82A, rDbpAK163A, and rDbpAK170A were expressed as 15N-labeled recombinant proteins, purified, and analyzed by NMR (Fig. 2). The 1H-15N HSQC spectra for rDbpAK82A, rDbpAK163A, and rDbpAK170A were then superimposed on that of rDbpAWT to assess gross structural variations between the individual lysine mutants and wild-type DbpA. Comparisons revealed that DbpAK82A, DbpAK163A, and DbpAK170A were correctly folded, suggesting that these lysine mutations had little or no impact on the overall protein conformation and that K82, K163, and K170 are directly involved in DbpA binding to dermatan sulfate.
FIG 2.
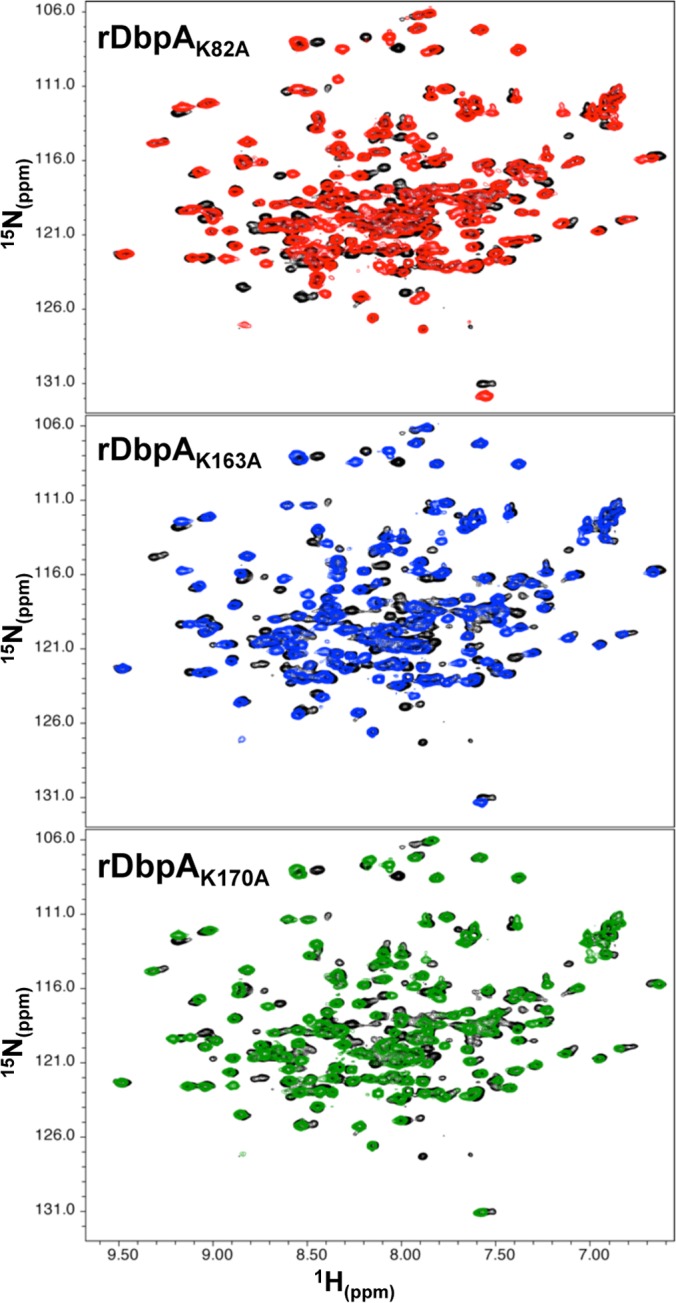
Superposition of 1H-15N HSQC spectra of rDbpAWT and rDbpA lysine point mutants. The NMR spectra of 15N-labeled rDbpAWT (black), rDbpAK82A (red), rDbpAK163A (blue), and rDbpAK170A (green) were determined, and each of the lysine mutant spectra was overlaid relative to rDbpAWT to assess changes in protein structure. The data were collected from a single analysis of each recombinant protein.
Contributions of conserved lysine residues in DbpA during infection.
While the contributions of lysines K82, K163, and K170 to the binding of decorin and dermatan sulfate have been demonstrated in vitro (52), it has not been assessed whether the impaired in vitro binding observed with these mutants correlates with a defect in B. burgdorferi infectivity. To test this, overlap extension PCR was used to mutate individual lysine codons to alanine (e.g., K51, K82, K124, K163, K170, and K177) in the dbpA ORF of pKH2000, a borrelial shuttle vector that encodes the native promoter for the dbpBA operon fused directly to the dbpA ORF. Prior studies confirmed that transformation of the B. burgdorferi dbpBA mutant, BbKH500, with pKH2000 is sufficient to restore dbpA expression and infectivity to the mutant (47). BbKH500 was transformed with each of the shuttle vectors carrying the lysine variants to generate BbDbpAK51A, BbDbpAK82A, BbDbpAK124A, BbDbpAK163A, BbDbpAK170A, and BbDbpAK177A.
To assess the impacts of these DbpA lysine mutations on infectivity, groups of five C3H/HeN mice were needle challenged with one of the six clones expressing the individual dbpA lysine mutants. As controls, groups of mice were also infected with the BbHK500 dbpBA mutant and BbKH500 complemented with the shuttle vector carrying the wild-type dbpA (BbKH501) (47). At 2 weeks postinfection, animals were sacrificed, and ear punch, lymph node, tibiotarsal, and heart tissue samples were collected for culture. The infection results are summarized in Table 5. As seen in previous studies (47), spirochetes could not be cultured from mice infected with BbKH500, and complementation of BbKH500 with pKH2000 expressing wild-type dbpA (BbKH501) restored the infectivity of this mutant. Similar to BbKH500, spirochetes were not recovered from mice infected with the lysine mutant strain BbDbpAK82A, BbDbpAK163A, or BbDbpAK170A. This noninfectious phenotype was specific for these three lysine mutants because infection rates with BbDbpAK51A, BbDbpAK124A, and BbDbpAK177A were equivalent to that observed with BbKH501. To further evaluate the infectivity of the DbpA lysine mutants, spirochete burdens in skin, tibotarsal, and heart tissue samples collected at 14 days postinfection were measured via qPCR from mice infected with BbKH500, BbKH501, BbDbpAK51A, BbDbpAK82A, BbDbpAK124A, BbDbpAK163A, BbDbpAK170A, or BbDbpAK177A (Fig. 3). qPCR results demonstrated that bacterial burdens were undetectable in tissues isolated from BbKH500, BbDbpAK82A, BbDbpAK163A, and BbDbpAK170A. In contrast, spirochete burdens in analyzed tissues from BbDbpAK51A, BbDbpAK124A, and BbDbpAK177A were comparable to those fromBbKH501.
TABLE 5.
Infectivity of B. burgdorferi dbpA lysine point mutants in C3H/HeN mice during early infection
| Strain | No. of positive cultures/total |
No. of positive mice/total | ||||
|---|---|---|---|---|---|---|
| Ear punch | Lymph nodesa | Heart | Tibiotarsal | All sites | ||
| BbKH500b | 0/5 | 0/5 | 0/5 | 0/5 | 0/20 | 0/5 |
| BbKH501 | 5/5 | 5/5 | 5/5 | 4/5 | 19/20 | 5/5 |
| BbDbpAK51A | 5/5 | 5/5 | 5/5 | 4/5 | 19/20 | 5/5 |
| BbDbpAK82Ab | 0/5 | 0/5 | 0/5 | 0/5 | 0/20 | 0/5 |
| BbDbpAK124A | 5/5 | 5/5 | 5/5 | 4/5 | 19/20 | 5/5 |
| BbDbpAK163Ab | 0/5 | 0/5 | 0/5 | 0/5 | 0/20 | 0/5 |
| BbDbpAK170Ab | 0/5 | 0/5 | 0/5 | 0/5 | 0/20 | 0/5 |
| BbDbpAK177A | 5/5 | 4/5 | 4/5 | 4/5 | 17/20 | 5/5 |
A single brachial and inguinal lymph node were collected from each mouse and cultured together.
The reduction in infection rates observed in individual tissues, at all sites, and the overall number of positive mice were statistically significantly different from BbKH501 (P ≤ 0.05 based on Fisher's exact test).
FIG 3.
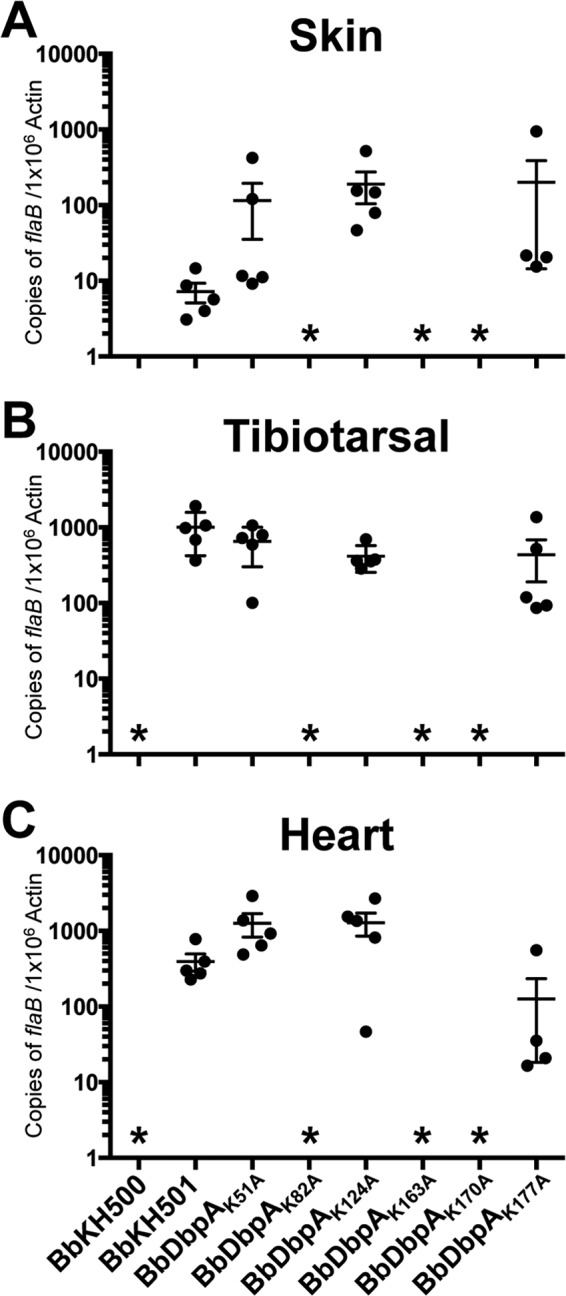
Spirochete burdens in tissues from mice infected with B. burgdorferi expressing lysine point mutants. Tissues were collected at 2 weeks postinfection from mice needle inoculated with BbKH500, BbKH501, BbDbpAK51A, BbDbpAK82A, BbDbpAK124A, BbDbpAK163A, BbDbpAK170A, or BbDbpAK177A at a dose of 105 spirochetes. DNAs from tissue samples were analyzed by qPCR to measure spirochete burdens and are represented as copies of flaB/106 copies of mouse β-actin. The results from skin (A), tibiotarsal (B), and heart (C) tissues are derived from a single infection experiment (n = 5 mice for each strain). The results represent the mean values from two independent assays, with each sample measured in triplicate during each assay. The error bars represent standard errors of the mean (SEM). Statistical significance was determined by ANOVA and Tukey's procedure. The asterisks indicate a statistically significant difference relative to BbH501 within a given tissue set (P ≤ 0.05).
While the colonization data from 14 days postinfection suggested that BbDbpAK82A, BbDbpAK163A, and BbDbpAK170A are noninfectious, prior studies have shown that the dissemination defect observed in mice infected with a dbpBA mutant was not evident if the infection was allowed to progress to the later stages (50, 51). To evaluate the impact of these DbpA lysine mutations during chronic infection, groups of 10 C3H/HeN mice were needle challenged with BbKH500, BbKH501, BbDbpAK82A, BbDbpAK163A, or BbDbpAK170A at a dose of 105 bacteria. Ear punch tissues were collected from all animals at 2 weeks postinfection to assess early infection (Table 6). In agreement with Table 5, spirochetes were not recovered from mice infected with BbKH500, BbDbpAK82A, BbDbpAK163A, or BbDbpAK170A at 2 weeks postinfection. At six and 10 weeks postinfection, ear punch, lymph node, tibiotarsal, and heart samples were collected from five mice and cultured (Table 6). In contrast to the results of Imai et al. (51), no spirochetes were recovered from mice infected with the BbKH500 dbpBA mutant at either time point. Additionally, no spirochetes were recovered from mice infected with BbDbpAK82A, BbDbpAK163A, or BbDbpAK170A at 6 or 10 weeks postinfection. Infectivity was restored in mice infected with BbKH501 at 2, 6, and 10 weeks postinfection. Imai et al. also described a specific defect in the ability of the dbpBA mutant to disseminate through the lymphatic system (51). Our findings demonstrated a similar defect, since lymph nodes were taken from both a proximal (brachial lymph node) and a distal (inguinal lymph node) site in relation to the site of inoculation, and no spirochetes were recovered in these cultures. Taken together, these data suggest that the loss of binding observed in vitro upon mutation of K82, K160, and K163 correlates with significant attenuation of the lysine mutants during both early and late stages of mammalian infection, and unlike the results observed with the strain B31 dbpBA mutant, this defect was not overcome during chronic infection with the strain 297 dbpBA mutant.
TABLE 6.
Infectivity of B. burgdorferi dbpA lysine point mutants in C3H/HeN mice during chronic infection
| Strain | No. of days postinfection | No. of positive cultures/total |
No. of positive mice/total | ||||
|---|---|---|---|---|---|---|---|
| Ear punch | Lymph nodesa | Heart | Tibiotarsal | All sites | |||
| BbKH500 | 14 | 0/10b | NDc | ND | ND | 0/10b | 0/10b |
| 42 | 0/5 | 0/5b | 0/5b | 0/5 | 0/20b | 0/5b | |
| 70 | 0/5b | 0/5b | 0/5b | 0/5b | 0/20b | 0/5b | |
| BbKH501 | 14 | 10/10 | ND | ND | ND | 10/10b | 10/10b |
| 42 | 3/5 | 4/5 | 5/5 | 3/5 | 15/20b | 5/5b | |
| 70 | 5/5 | 5/5 | 5/5 | 5/5 | 20/20b | 5/5b | |
| BbDbpAK82A | 14 | 0/10b | ND | ND | ND | 0/10b | 0/10b |
| 42 | 0/5 | 0/5b | 0/5b | 0/5 | 0/20b | 0/5b | |
| 70 | 0/5b | 0/5b | 0/5b | 0/5b | 0/20b | 0/5b | |
| BbDbpAK163A | 14 | 0/10b | ND | ND | ND | 0/10b | 0/10b |
| 42 | 0/5 | 0/5b | 0/5b | 0/5 | 0/20b | 0/5b | |
| 70 | 0/5b | 0/5b | 0/5b | 0/5b | 0/20b | 0/5b | |
| BbDbpAK170A | 14 | 0/10b | ND | ND | ND | 0/10b | 0/10b |
| 42 | 0/5 | 0/5b | 0/5b | 0/5 | 0/20b | 0/5b | |
| 70 | 0/5b | 0/5b | 0/5b | 0/5b | 0/20b | 0/5b | |
A single brachial and inguinal lymph node were collected from each mouse and cultured together.
The reductions in infection rates were statistically significantly different from BbKH501 (P ≤ 0.05 based on Fisher's exact test).
ND, not determined.
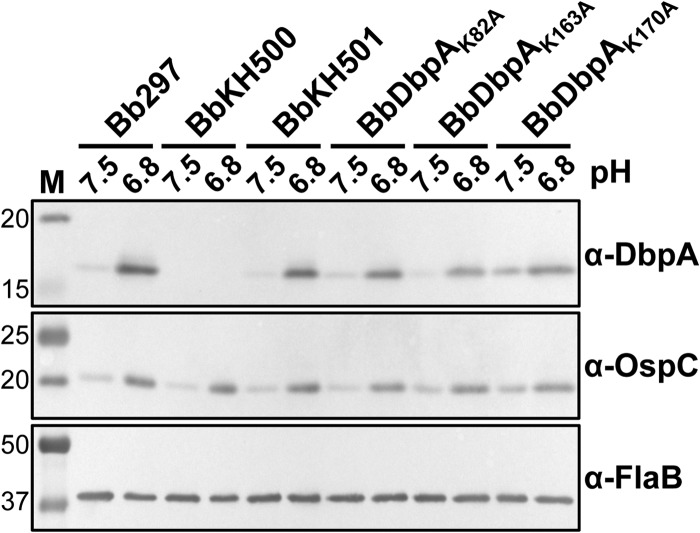
B. burgdorferi strains in which dbpA has been inactivated are unable to infect mice (47–50); therefore, to confirm that loss of infectivity with strain BbDbpAK82A, BbDbpAK163A, or BbDbpAK170A was not due to aberrant expression or localization of the mutated DbpA proteins, a number of in vitro studies were performed. In vitro growth curve analyses comparing Bb297 and BbKH501 to each of the attenuated lysine mutant clones determined that their growth rates were similar; therefore, the loss of infectivity observed with BbDbpAK82A, BbDbpAK163A, or BbDbpAK170A was not due to a general growth defect in the point mutant strains (see Fig. S3 in the supplemental material). The attenuated phenotypes of BbDbpAK82A, BbDbpAK163A, and BbDbpAK170A could also be due to these mutated dbpA alleles not being properly expressed during infection. While it is difficult to directly assess dbpA expression in vivo, it is possible to grow B. burgdorferi in vitro under conditions known to induce rpoS-dependent expression of dbpBA (e.g., elevated temperature and reduced pH [41, 70, 73]). Cultures of BbDbpAK82A, BbDbpAK163A, or BbDbpAK170A were grown at pH 7.5 and 6.8, and induction of DbpA production was evaluated using immunoblotting (Fig. 4). The production of OspC, an outer surface lipoprotein of B. burgdorferi known to be activated by rpoS (70, 73), was assessed as a positive control. When cultured at pH 6.8, all of the clones carrying the DbpA lysine point mutants showed elevated OspC and DbpA at levels comparable to that observed with BbKH501. The noninfectious phenotype of BbDbpAK82A, BbDbpAK163A, or BbDbpAK170A could also be attributed to the mutated DbpA proteins not being properly localized to the bacterial surface, where they can interact with host GAGs. To assess surface localization, bacteria were subjected to proteinase K digestion and then immunoblotted for DbpA. In all strains expressing dbpA, levels of DbpA decreased significantly when the cells were treated with proteinase K, suggesting that DbpA was appropriately localized on the cell surface (Fig. 5). Immunoblot detection of FlaB was included as a control to confirm that the outer membranes of the cells remained intact and only outer surface proteins were digested. Taken together, these data support the conclusion that loss of infectivity in BbDbpAK82A, BbDbpAK163A, or BbDbpAK170A was due to the impact of the lysine mutations on decorin/dermatan sulfate binding and was not due to an unknown secondary effect of these lysine residues on overall dbpA expression or adhesin localization.
FIG 4.
Assessment of DbpA expression in B. burgdorferi expressing lysine point mutants. Shown is immunoblot analysis of DbpA, OspC, and FlaB in Bb297, BbKH500, BbKH501, BbDbpAK82A, BbDbpAK163A, and BbDbpAK170A. The strains were cultivated in vitro at pH 7.5 or 6.8 to induce production of DbpA. Antibodies used to detect the respective proteins are indicated on the right. The values on the left denote relevant molecular masses (kDa) of the standard (lane M). Samples from three independent biological replicates were analyzed and shown to yield similar results. Depicted is an immunoblot from one of these biological replicates.
FIG 5.
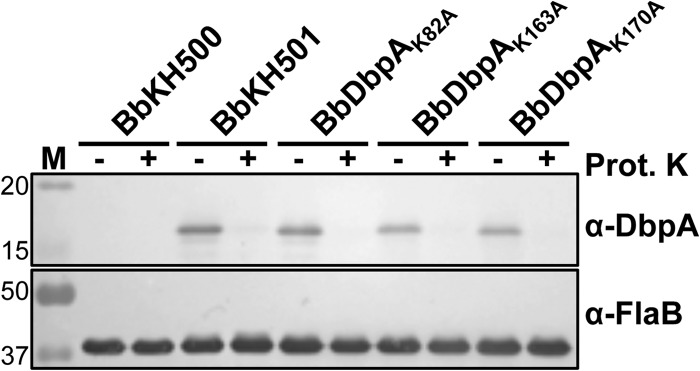
Confirmation of surface localization of DbpA in B. burgdorferi expressing lysine point mutants. BbKH500, BbKH501, BbDbpAK82A, BbDbpAK163A, and BbDbpAK170A were grown in vitro at pH 7.5 to the late exponential growth phase. Whole-cell lysates from intact bacteria subjected to proteinase K digestion (+) or mock treated (−) were analyzed by immunoblotting. Antibodies used to detect the respective proteins are indicated on the right. The values on the left denote relevant molecular masses (kDa) of the standard (lane M). Samples from three independent biological replicates were analyzed and shown to yield similar results. Depicted is an immunoblot from one of these biological replicates.
Summary and conclusions.
There is a growing consensus that NMR and crystallographic methods of structural determination are complementary and should be pursued in tandem to fully elucidate the structures of proteins of interest (74, 75). Therefore, we utilized X-ray crystallography to confirm the previously published NMR structure for DbpA (54). Although the flexible loop between helices 1 and 2 that contains a BXBB (where B is a basic amino acid) motif recently shown to contribute to GAG binding (76) could not be modeled in the X-ray structure, the two structures were very similar and confirmed the clustering of lysine residues on helices 2 (e.g., K82) and 5 (e.g., K163 and K170) in an exposed binding cleft near the C terminus of the adhesin. In addition, recombinant DbpA proteins containing mutations in K82, K163, or K170 demonstrated a complete loss of decorin and dermatan sulfate binding during in vitro binding experiments (52), and this binding defect correlated with significant attenuation observed in B. burgdorferi strains expressing dbpAK82A, dbpAK163A, and dbpAK170A during murine infection. Considering that a number of B. burgdorferi surface proteins have been shown to interact with multiple host ligands (e.g., DbpA [35, 40, 76], OspC [77, 78], Bgp [18], and CRASP-1 [23, 79]), it remains possible that K82, K163, or K170 mediates interaction of DbpA with additional unidentified host ligands. However, these data support early studies showing the importance of decorin binding during mammalian infection (80). The findings are also in agreement with studies showing that a B. burgdorferi strain expressing a dbpA allele lacking the 11 C-terminal amino acids, which is unable to bind decorin or dermatan sulfate (35), is also noninfectious in mice (J.M. Leong, unpublished results).
Similar to the results of Imai et al. (51), BbKH500, BbDbpAK82A, BbDbpAK163A, and BbDbpAK170A could not be recovered from the brachial (proximal) or inguinal (distal) lymph nodes, which is supportive of their hypothesis that DbpA is important for dissemination via the lymphatic system. Recent studies have shown that the infectivity defect observed with dbpA knockouts in the strain B31 background could be overcome during later stages of infection (50, 51), which is presumably due to compensation by another bacterial adhesin(s) or alternate mechanisms of dissemination. It should be noted that our results demonstrated that BbDbpAK82A, BbDbpAK163A, and BbDbpAK170A were noninfectious during both the early and chronic stages of B. burgdorferi infection. Considering that DbpA is a prominent antigen (41–46), one possible explanation is that BbDbpAK82A, BbDbpAK163A, and BbDbpAK170A express a nonfunctional adhesin that affords no dissemination benefit and the bacteria would be recognized and cleared by the humoral response. However, the fact that the BbKH500 dbpBA mutant also could not be recovered from mice at 10 weeks postinfection puts this conclusion into question. At this time, we are unable to definitively reconcile the disparate phenotypes observed during persistent-infection studies. Because the same challenge dose was employed in each of these studies (105 bacteria), it does not appear to be a result of an inoculum difference. One possible explanation for these variations could be the different parental strain used in our study (e.g., Bb297) versus clonal derivatives of strain B31 (e.g., B31-A3 or ML23), so that the compensatory bacterial adhesin(s) or alternative dissemination mechanisms in strain B31 are more effective than those in strain 297. Future analysis of the function of DbpA in B. burgdorferi should include a comprehensive and comparative analysis of these different dbpBA mutants in a single challenge study to assess whether strain-dependent differences exist regarding the relative contribution of dbpBA during chronic infection. Because our prior studies, albeit a relatively limited assessment of systemic colonization, have shown that BbKH500 can infect mice when challenged via tick feeding, future dbpBA mutant comparisons in mice should also employ the tick colonization and feeding model. Despite the discrepancy regarding the phenotype of the dbpBA mutant during chronic infection, these data clearly demonstrate that lysines K82, K163, and K170 are required for binding of DbpA to dermatan sulfate and that the interaction(s) mediated by these lysines is critical during B. burgdorferi murine infection.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by funding through the Arkansas Biosciences Institute (a major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000), NIH/NIAID R01-AI087678, the Translational Research Institute (UL1-TR000039; NIH National Center for Research Resources and National Center for Advancing Translational Sciences), the UAMS Center for Microbial Pathogenesis and Host Inflammatory Responses (COBRE grant P20-GM103625), and the UAMS IMSD program (NIH/NIGMS R25-GM083247). The work was also supported by NIH R01-AI1093104 to John M. Leong and American Heart Association Postdoctoral Fellowship 12POST11660031 to Yi-Pin Lin. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357.
We thank Michael Norgard for his helpful discussions and critical evaluation of the manuscript. We also thank Allen Gies in the UAMS Sequencing Core Facility for his technical assistance and the staff at the Advanced Photon Source for synchrotron data collection support.
Footnotes
Published ahead of print 19 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02036-14.
REFERENCES
- 1.Adams DA, Gallagher KM, Jajosky RA, Kriseman J, Sharp P, Anderson WJ, Aranas AE, Mayes M, Wodajo MS, Onweh DH, Abellera JP. 2013. Summary of notifiable diseases—United States, 2011. MMWR Morb. Mortal. Wkly. Rep. 60:1–117. [PubMed] [Google Scholar]
- 2.Rizzoli A, Hauffe H, Carpi G, Vourc HG, Neteler M, Rosa R. 2011. Lyme borreliosis in Europe. Euro Surveill. 16:19906. [PubMed] [Google Scholar]
- 3.Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317–1319. 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 4.Steere AC, Grodzicki RL, Kornblatt AN, Craft JE, Barbour AG, Burgdorfer W, Schmid GP, Johnson E, Malawista SE. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308:733–740. 10.1056/NEJM198303313081301. [DOI] [PubMed] [Google Scholar]
- 5.Radolf JD, Caimano MJ, Stevenson B, Hu LT. 2012. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat. Rev. Microbiol. 10:87–99. 10.1038/nrmicro2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanek G, Wormser GP, Gray J, Strle F. 2012. Lyme borreliosis. Lancet 379:461–473. 10.1016/S0140-6736(11)60103-7. [DOI] [PubMed] [Google Scholar]
- 7.Steere AC. 2001. Lyme disease. N. Engl. J. Med. 345:115–125. 10.1056/NEJM200107123450207. [DOI] [PubMed] [Google Scholar]
- 8.Steere AC, Coburn J, Glickstein L. 2004. The emergence of Lyme disease. J. Clin. Invest. 113:1093–1101. 10.1172/JCI200421681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finlay BB, Falkow S. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linke D, Goldman A. 2011. Bacterial adhesion: chemisty, biology, and physics, vol 715 Springer Scientific, New York, NY. [Google Scholar]
- 11.Coburn J, Leong J, Chaconas G. 2013. Illuminating the roles of the Borrelia burgdorferi adhesins. Trends Microbiol. 21:372–379. 10.1016/j.tim.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabello FC, Godfrey HP, Newman SA. 2007. Hidden in plain sight: Borrelia burgdorferi and the extracellular matrix. Trends Microbiol. 15:350–354. 10.1016/j.tim.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Coburn J, Leong JM, Erban JK. 1993. Integrin αIIbβ3 mediates binding of the Lyme disease agent Borrelia burgdorferi to human platelets. Proc. Natl. Acad. Sci. U. S. A. 90:7059–7063. 10.1073/pnas.90.15.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coburn J, Magoun L, Bodary SC, Leong JM. 1998. Integrins αVβ3 and αVβ1 mediate attachment of Lyme disease spirochetes to human cells. Infect. Immun. 66:1946–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coburn J, Chege W, Magoun L, Bodary SC, Leong JM. 1999. Characterization of a candidate Borrelia burgdorferi β3-chain integrin ligand identified using a phage display library. Mol. Microbiol. 34:926–940. 10.1046/j.1365-2958.1999.01654.x. [DOI] [PubMed] [Google Scholar]
- 16.Behera AK, Durand E, Cugini C, Antonara S, Bourassa L, Hildebrand E, Hu LT, Coburn J. 2008. Borrelia burgdorferi BBB07 interaction with integrin α3β1 stimulates production of pro-inflammatory mediators in primary human chondrocytes. Cell Microbiol. 10:320–331. 10.1111/j.1462-5822.2007.01043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Probert WS, Johnson BJ. 1998. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol. Microbiol. 30:1003–1015. 10.1046/j.1365-2958.1998.01127.x. [DOI] [PubMed] [Google Scholar]
- 18.Fischer JR, LeBlanc KT, Leong JM. 2006. Fibronectin binding protein BBK32 of the Lyme disease spirochete promotes bacterial attachment to glycosaminoglycans. Infect. Immun. 74:435–441. 10.1128/IAI.74.1.435-441.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Probert WS, Kim JH, Hook M, Johnson BJ. 2001. Mapping the ligand-binding region of Borrelia burgdorferi fibronectin-binding protein BBK32. Infect. Immun. 69:4129–4133. 10.1128/IAI.69.6.4129-4133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moriarty TJ, Shi M, Lin YP, Ebady R, Zhou H, Odisho T, Hardy PO, Salman-Dilgimen A, Wu J, Weening EH, Skare JT, Kubes P, Leong J, Chaconas G. 2012. Vascular binding of a pathogen under shear force through mechanistically distinct sequential interactions with host macromolecules. Mol. Microbiol. 86:1116–1131. 10.1111/mmi.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brissette CA, Bykowski T, Cooley AE, Bowman A, Stevenson B. 2009. Borrelia burgdorferi RevA antigen binds host fibronectin. Infect. Immun. 77:2802–2812. 10.1128/IAI.00227-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaultney RA, Gonzalez T, Floden AM, Brissette CA. 2013. BB0347, from the Lyme disease spirochete Borrelia burgdorferi, is surface exposed and interacts with the CS1 heparin-binding domain of human fibronectin. PLoS One 8:e75643. 10.1371/journal.pone.0075643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hallstrom T, Haupt K, Kraiczy P, Hortschansky P, Wallich R, Skerka C, Zipfel PF. 2010. Complement regulator-acquiring surface protein 1 of Borrelia burgdorferi binds to human bone morphogenic protein 2, several extracellular matrix proteins, and plasminogen. J. Infect. Dis. 202:490–498. 10.1086/653825. [DOI] [PubMed] [Google Scholar]
- 24.Zambrano MC, Beklemisheva AA, Bryksin AV, Newman SA, Cabello FC. 2004. Borrelia burgdorferi binds to, invades, and colonizes native type I collagen lattices. Infect. Immun. 72:3138–3146. 10.1128/IAI.72.6.3138-3146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verma A, Brissette CA, Bowman A, Stevenson B. 2009. Borrelia burgdorferi BmpA is a laminin-binding protein. Infect. Immun. 77:4940–4946. 10.1128/IAI.01420-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brissette CA, Verma A, Bowman A, Cooley AE, Stevenson B. 2009. The Borrelia burgdorferi outer-surface protein ErpX binds mammalian laminin. Microbiology 155:863–872. 10.1099/mic.0.024604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell TM, Johnson BJ. 2013. Lyme disease spirochaetes possess an aggrecan-binding protease with aggrecanase activity. Mol. Microbiol. 90:228–240. 10.1111/mmi.12276. [DOI] [PubMed] [Google Scholar]
- 28.Parveen N, Leong JM. 2000. Identification of a candidate glycosaminoglycan-binding adhesin of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:1220–1234. 10.1046/j.1365-2958.2000.01792.x. [DOI] [PubMed] [Google Scholar]
- 29.Leong JM, Morrissey PE, Ortega-Barria E, Pereira ME, Coburn J. 1995. Hemagglutination and proteoglycan binding by the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo BP, Norris SJ, Rosenberg LC, Hook M. 1995. Adherence of Borrelia burgdorferi to the proteoglycan decorin. Infect. Immun. 63:3467–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo BP, Brown EL, Dorward DW, Rosenberg LC, Hook M. 1998. Decorin-binding adhesins from Borrelia burgdorferi. Mol. Microbiol. 30:711–723. 10.1046/j.1365-2958.1998.01103.x. [DOI] [PubMed] [Google Scholar]
- 32.Hagman KE, Lahdenne P, Popova TG, Porcella SF, Akins DR, Radolf JD, Norgard MV. 1998. Decorin-binding protein of Borrelia burgdorferi is encoded within a two-gene operon and is protective in the murine model of Lyme borreliosis. Infect. Immun. 66:2674–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang G, Ojaimi C, Iyer R, Saksenberg V, McClain SA, Wormser GP, Schwartz I. 2001. Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect. Immun. 69:4303–4312. 10.1128/IAI.69.7.4303-4312.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wormser GP, Liveris D, Nowakowski J, Nadelman RB, Cavaliere LF, McKenna D, Holmgren D, Schwartz I. 1999. Association of specific subtypes of Borrelia burgdorferi with hematogenous dissemination in early Lyme disease. J. Infect. Dis. 180:720–725. 10.1086/314922. [DOI] [PubMed] [Google Scholar]
- 35.Benoit VM, Fischer JR, Lin YP, Parveen N, Leong JM. 2011. Allelic variation of the Lyme disease spirochete adhesin DbpA influences spirochetal binding to decorin, dermatan sulfate, and mammalian cells. Infect. Immun. 79:3501–3509. 10.1128/IAI.00163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang G, van Dam AP, Schwartz I, Dankert J. 1999. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin. Microbiol. Rev. 12:633–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher LW, Termine JD, Young MF. 1989. Deduced protein sequence of bone small proteoglycan I (biglycan) shows homology with proteoglycan II (decorin) and several nonconnective tissue proteins in a variety of species. J. Biol. Chem. 264:4571–4576. [PubMed] [Google Scholar]
- 38.Poole AR, Webber C, Pidoux I, Choi H, Rosenberg LC. 1986. Localization of a dermatan sulfate proteoglycan (DS-PGII) in cartilage and the presence of an immunologically related species in other tissues. J. Histochem. Cytochem. 34:619–625. 10.1177/34.5.3701029. [DOI] [PubMed] [Google Scholar]
- 39.Scott JE, Orford CR. 1981. Dermatan sulphate-rich proteoglycan associates with rat tail-tendon collagen at the d band in the gap region. Biochem. J. 197:213–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer JR, Parveen N, Magoun L, Leong JM. 2003. Decorin-binding proteins A and B confer distinct mammalian cell type-specific attachment by Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. U. S. A. 100:7307–7312. 10.1073/pnas.1231043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagman KE, Yang X, Wikel SK, Schoeler GB, Caimano MJ, Radolf JD, Norgard MV. 2000. Decorin-binding protein A (DbpA) of Borrelia burgdorferi is not protective when immunized mice are challenged via tick infestation and correlates with the lack of DbpA expression by B. burgdorferi in ticks. Infect. Immun. 68:4759–4764. 10.1128/IAI.68.8.4759-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hodzic E, Feng S, Freet KJ, Borjesson DL, Barthold SW. 2002. Borrelia burgdorferi population kinetics and selected gene expression at the host-vector interface. Infect. Immun. 70:3382–3388. 10.1128/IAI.70.7.3382-3388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barthold SW, Hodzic E, Tunev S, Feng S. 2006. Antibody-mediated disease remission in the mouse model of Lyme borreliosis. Infect. Immun. 74:4817–4825. 10.1128/IAI.00469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanson MS, Cassatt DR, Guo BP, Patel NK, McCarthy MP, Dorward DW, Hook M. 1998. Active and passive immunity against Borrelia burgdorferi decorin binding protein A (DbpA) protects against infection. Infect. Immun. 66:2143–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cassatt DR, Patel NK, Ulbrandt ND, Hanson MS. 1998. DbpA, but not OspA, is expressed by Borrelia burgdorferi during spirochetemia and is a target for protective antibodies. Infect. Immun. 66:5379–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Embers ME, Hasenkampf NR, Jacobs MB, Philipp MT. 2012. Dynamic longitudinal antibody responses during Borrelia burgdorferi infection and antibiotic treatment of rhesus macaques. Clin. Vaccine Immunol. 19:1218–1226. 10.1128/CVI.00228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blevins JS, Hagman KE, Norgard MV. 2008. Assessment of decorin-binding protein A to the infectivity of Borrelia burgdorferi in the murine models of needle and tick infection. BMC Microbiol. 8:82. 10.1186/1471-2180-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Y, Xu Q, McShan K, Liang FT. 2008. Both decorin-binding proteins A and B are critical for the overall virulence of Borrelia burgdorferi. Infect. Immun. 76:1239–1246. 10.1128/IAI.00897-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi Y, Xu Q, Seemanaplli SV, McShan K, Liang FT. 2008. Common and unique contributions of decorin-binding proteins A and B to the overall virulence of Borrelia burgdorferi. PLoS One 3:e3340. 10.1371/journal.pone.0003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weening EH, Parveen N, Trzeciakowski JP, Leong JM, Hook M, Skare JT. 2008. Borrelia burgdorferi lacking DbpBA exhibits an early survival defect during experimental infection. Infect. Immun. 76:5694–5705. 10.1128/IAI.00690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Imai DM, Samuels DS, Feng S, Hodzic E, Olsen K, Barthold SW. 2013. The early dissemination defect attributed to disruption of decorin-binding proteins is abolished in chronic murine Lyme borreliosis. Infect. Immun. 81:1663–1673. 10.1128/IAI.01359-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown EL, Guo BP, O'Neal P, Hook M. 1999. Adherence of Borrelia burgdorferi. Identification of critical lysine residues in DbpA required for decorin binding. J. Biol. Chem. 274:26272–26278. [DOI] [PubMed] [Google Scholar]
- 53.Pikas DS, Brown EL, Gurusiddappa S, Lee LY, Xu Y, Hook M. 2003. Decorin-binding sites in the adhesin DbpA from Borrelia burgdorferi: a synthetic peptide approach. J. Biol. Chem. 278:30920–30926. 10.1074/jbc.M303979200. [DOI] [PubMed] [Google Scholar]
- 54.Wang X. 2012. Solution structure of decorin-binding protein A from Borrelia burgdorferi. Biochemistry 51:8353–8362. 10.1021/bi3007093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hughes CA, Kodner CB, Johnson RC. 1992. DNA analysis of Borrelia burgdorferi NCH-1, the first northcentral U.S. human Lyme disease isolate. J. Clin. Microbiol. 30:698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pollack RJ, Telford SR, III, Spielman A. 1993. Standardization of medium for culturing Lyme disease spirochetes. J. Clin. Microbiol. 31:1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763. 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 58.Cowtan K, Main P. 1998. Miscellaneous algorithms for density modification. Acta Crystallogr. D Biol. Crystallogr. 54:487–493. 10.1107/S0907444997011980. [DOI] [PubMed] [Google Scholar]
- 59.Terwilliger TC. 2002. Rapid automatic NCS identification using heavy-atom substructures. Acta Crystallogr. D Biol. Crystallogr. 58:2213–2215. 10.1107/S0907444902016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Terwilliger TC. 2003. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 59:38–44. 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langer G, Cohen SX, Lamzin VS, Perrakis A. 2008. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3:1171–1179. 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Read RJ. 2001. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 57:1373–1382. 10.1107/S0907444901012471. [DOI] [PubMed] [Google Scholar]
- 63.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110–119. 10.1107/S0108767390010224. [DOI] [PubMed] [Google Scholar]
- 64.Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC. 2004. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11:53–55. 10.1107/S0909049503024130. [DOI] [PubMed] [Google Scholar]
- 65.Lin YP, Lee DW, McDonough SP, Nicholson LK, Sharma Y, Chang YF. 2009. Repeated domains of leptospira immunoglobulin-like proteins interact with elastin and tropoelastin. J. Biol. Chem. 284:19380–19391. 10.1074/jbc.M109.004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293. [DOI] [PubMed] [Google Scholar]
- 67.Johnson BA, Blevins RA. 1994. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4:603–614. 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 68.Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 199:641–648. 10.1084/jem.20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maruskova M, Esteve-Gassent MD, Sexton VL, Seshu J. 2008. Role of the BBA64 locus of Borrelia burgdorferi in early stages of infectivity in a murine model of Lyme disease. Infect. Immun. 76:391–402. 10.1128/IAI.01118-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang X, Goldberg MS, Popova TG, Schoeler GB, Wikel SK, Hagman KE, Norgard MV. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 37:1470–1479. 10.1046/j.1365-2958.2000.02104.x. [DOI] [PubMed] [Google Scholar]
- 71.Smith AH, Blevins JS, Bachlani GN, Yang XF, Norgard MV. 2007. Evidence that RpoS (σS) in Borrelia burgdorferi is controlled directly by RpoN (σ54/σN). J. Bacteriol. 189:2139–2144. 10.1128/JB.01653-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holm L, Park J. 2000. DaliLite workbench for protein structure comparison. Bioinformatics 16:566–567. 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 73.Hubner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. U. S. A. 98:12724–12729. 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Snyder DA, Chen Y, Denissova NG, Acton T, Aramini JM, Ciano M, Karlin R, Liu J, Manor P, Rajan PA, Rossi P, Swapna GV, Xiao R, Rost B, Hunt J, Montelione GT. 2005. Comparisons of NMR spectral quality and success in crystallization demonstrate that NMR and X-ray crystallography are complementary methods for small protein structure determination. J. Am. Chem. Soc. 127:16505–16511. 10.1021/ja053564h. [DOI] [PubMed] [Google Scholar]
- 75.Yee AA, Savchenko A, Ignachenko A, Lukin J, Xu X, Skarina T, Evdokimova E, Liu CS, Semesi A, Guido V, Edwards AM, Arrowsmith CH. 2005. NMR and X-ray crystallography, complementary tools in structural proteomics of small proteins. J. Am. Chem. Soc. 127:16512–16517. 10.1021/ja053565+. [DOI] [PubMed] [Google Scholar]
- 76.Morgan A, Wang X. 2013. The novel heparin-binding motif in decorin-binding protein A from strain B31 of Borrelia burgdorferi explains the higher binding affinity. Biochemistry 52:8237–8245. 10.1021/bi401376u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Onder O, Humphrey PT, McOmber B, Korobova F, Francella N, Greenbaum DC, Brisson D. 2012. OspC is potent plasminogen receptor on surface of Borrelia burgdorferi. J. Biol. Chem. 287:16860–16868. 10.1074/jbc.M111.290775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramamoorthi N, Narasimhan S, Pal U, Bao F, Yang XF, Fish D, Anguita J, Norgard MV, Kantor FS, Anderson JF, Koski RA, Fikrig E. 2005. The Lyme disease agent exploits a tick protein to infect the mammalian host. Nature 436:573–577. 10.1038/nature03812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kraiczy P, Hellwage J, Skerka C, Becker H, Kirschfink M, Simon MM, Brade V, Zipfel PF, Wallich R. 2004. Complement resistance of Borrelia burgdorferi correlates with the expression of BbCRASP-1, a novel linear plasmid-encoded surface protein that interacts with human factor H and FHL-1 and is unrelated to Erp proteins. J. Biol. Chem. 279:2421–2429. 10.1074/jbc.M308343200. [DOI] [PubMed] [Google Scholar]
- 80.Brown EL, Wooten RM, Johnson BJ, Iozzo RV, Smith A, Dolan MC, Guo BP, Weis JJ, Hook M. 2001. Resistance to Lyme disease in decorin-deficient mice. J. Clin. Invest. 107:845–852. 10.1172/JCI11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375–W383. 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.