Abstract
The native plasmid of both Chlamydia muridarum and Chlamydia trachomatis has been shown to control virulence and infectivity in mice and in lower primates. We recently described the development of a plasmid-based genetic transformation protocol for Chlamydia trachomatis that for the first time provides a platform for the molecular dissection of the function of the chlamydial plasmid and its individual genes or coding sequences (CDS). In the present study, we transformed a plasmid-free lymphogranuloma venereum isolate of C. trachomatis, serovar L2, with either the original shuttle vector (pGFP::SW2) or a derivative of pGFP::SW2 carrying a deletion of the plasmid CDS5 gene (pCDS5KO). Female mice were inoculated with these strains either intravaginally or transcervically. We found that transformation of the plasmid-free isolate with the intact pGFP::SW2 vector significantly enhanced infectivity and induction of host inflammatory responses compared to the plasmid-free parental isolate. Transformation with pCDS5KO resulted in infection courses and inflammatory responses not significantly different from those observed in mice infected with the plasmid-free isolate. These results indicate a critical role of plasmid CDS5 in in vivo fitness and in induction of inflammatory responses. To our knowledge, these are the first in vivo observations ascribing infectivity and virulence to a specific plasmid gene.
INTRODUCTION
Plasmids of Chlamydia spp. are all small (∼7.5 kb), nonconjugative, nonintegrative, and highly conserved within species (1–3). The observation that naturally occurring plasmid-deficient Chlamydia trachomatis strains are exceedingly rare implies a significant but noncritical role for in vivo fitness and transmission. However, until the past few years, a role for the plasmid in C. trachomatis pathobiology remained to be fully explored.
In vivo studies have shown that the plasmid of C. trachomatis contributes significantly to fitness and induction of inflammatory responses (4–8). Cumulatively, these studies were each conducted using plasmid-deficient [plasmid(−)] or closely matched plasmid-bearing [plasmid(+)] isolates from different biovars: urogenital isolates, ocular (trachoma) isolates, and lymphogranuloma venereum (LGV) isolates. It is assuring that the results of these studies in mice and lower primates were consistent with regard to the observation that plasmid(−) isolates of all biovars exhibit reduced infectivity and virulence compared to plasmid(+) isolates. However, it should be noted that many observations using plasmid-deficient strains have been made in suboptimally controlled experimental conditions, since the comparator plasmid-containing strains were not always isogenic; thus, roles for any putative chromosomal variances could not be completely ruled out.
Results of the use of plasmid(+) and plasmid(−) isolates in the commonly utilized mouse model of chlamydial urogenital infection with the mouse pathogen Chlamydia muridarum have also been reported. Because a naturally occurring plasmid(−) isolate of C. muridarum has yet to be reported, the organism was treated with novobiocin to cure the plasmid (8, 9). Initial reports in this model indicated that plasmid deficiency caused no severe defect in infectivity or fitness but significantly reduced inflammatory responses and related sequelae of infection (8, 10). Recently, others have evaluated the role of the plasmid in this model and have reported somewhat disparate results, with the plasmid(−) C. muridarum showing reduced infectious burdens in vivo (11). The reason for these disparate results could be technical/procedural or may reside in chromosomal differences in the parental C. muridarum strains, as has been previously described (12). In summary, it is clear that the plasmid is, in some way, an important mediator of Chlamydia-host interactions.
While all of the aforementioned reports point to the chlamydial plasmid as a mediator of host interactions in some form, scant evidence is available to assign roles to individual plasmid genes in these interactions. We recently showed that a naturally occurring mutant in the C. trachomatis plasmid altered infectivity and virulence in a mouse model (7). However, isolating, identifying, and describing naturally occurring plasmid mutations will no doubt prove to be a significant bottleneck in discovery. Historically, a hindrance to the selective study of the role of chlamydial genes had been the refractory nature of chlamydial pathogens to commonly used molecular biological methods of selective and sustained genetic mutation and complementation (13).
Recently, we described the development of a transformation system for C. trachomatis that has been used by us, and subsequently by others, to modify the chlamydial plasmid and to begin to dissect the role of selected plasmid genes with regard to plasmid maintenance, host cell interactions, and in vitro phenotypic expression (e.g., inclusion morphology and glycogen accumulation) (14–16). In the present study, we further capitalize upon these findings and extend the transformation system to an analysis of in vivo plasmid-mediated pathobiology. We focused our experiments on the product of plasmid gene CDS5 (pgp3). We selected CDS5 (pgp3) because unlike the other 7 plasmid-encoded proteins, Pgp3 is secreted beyond the inclusion membrane and into the cytosol of the host cell (17), it has been found to be immunogenic in mice and women (18, 19), and DNA vaccination with CDS5 (pgp3) provides partial protection in a mouse model (20). Of the remaining plasmid genes, CDS1 appears to be noncritical for plasmid maintenance and in vitro phenotypic variation (2), and CDS2, -3, -4, and -8 (encoding proteins Pgp8, -1, -2, and -6, respectively) appear to be essential for plasmid maintenance and replication, whereas CDS6 and -7 (pgp4 and -5, respectively) may be involved in regulating expression of chromosomal genes (16, 21). Thus, we have hypothesized a role for plasmid CDS5 (pgp3) in directly affecting virulence, in vivo fitness, and induction of inflammatory responses. To test this hypothesis, we infected female mice in the urogenital tract or respiratory tract with either a naturally occurring plasmid(−) isolate; the same isolate transformed with a replication-competent vector containing plasmid CDSs (14), and the isolate transformed with the vector but with a knockout in CDS5 (pgp3) (15). Our results support the hypothesis that the product of the plasmid CDS5 (pgp3) gene plays a direct role in infectivity, in vivo fitness, and induction of host inflammatory responses.
MATERIALS AND METHODS
Chlamydial strains and vectors.
A clonal plasmid(−) isolate of the C. trachomatis LGV strain (serovar L2, strain 25667R [22]) was used as the transformation recipient of the vectors in this study. The genome of L2 strain P− 25667R has been sequenced and deposited in GenBank (reference sequence NC_020930) (23). The cloning vector pGFP::SW2 (14) and plasmid pCDS5KO (15) were described previously. pCDS5KO is a deletion version of plasmid pGFP::SW2, with the CDS5 promoter and approximately three-quarters of the CDS5 coding sequence deleted but carefully designed to retain the promoter of CDS6. The two plasmids were transformed into C. trachomatis L2P− 25667R (referred to here as L2P−) separately, and the transformants, L2P−.pGFP::SW2 and L2P−.pCDS5KO, were recovered under penicillin selection. The resulting transformed strains are described in Table 1. Quantitative PCR was conducted to confirm that copy number of the plasmid was comparable between the L2P−.pGFP::SW2 and L2P−.pCDS5KO transformants. It was found that both transformants had a plasmid-to-genome ratio of approximately 5 as determined by the method of Pickett et al. (24). In some cases, a wild-type plasmid(+) serovar L2 isolate, L2 434/Bu, was used (25, 26) (GenBank reference sequence NC_010287). It should be noted that while closely matched, this isolate is not fully isogenic with reference to the chromosome of L2P−. Thus, we did not believe this entirely suitable as an isogenic control, and this strain was used only as a serovar-matched comparator with a wild-type plasmid for the L2P−.pGFP::SW2 transformant.
TABLE 1.
C. trachomatis isolates and transformants used in this study
| Strain nomenclature | Description |
|---|---|
| L2 P− 25667R | Naturally occurring plasmid-free isolate of C. trachomatis serovar L2 from a patient with proctocolitis (22); the genome has been sequenced and deposited at GenBank (reference sequence NC_020930) (23); for brevity, we use the designation L2P− here |
| L2 P−.pGFP::SW2 | L2P− 25667R transformed with a recombinant chlamydial plasmid (pGFP::SW2) derived from Swedish new variant C. trachomatis serovar E (14); this plasmid (pGFP::SW2) possesses an Escherichia coli origin of replication, bla, cat, and a red-shifted green fluorescent protein gene; the latter two are under the control of a Neisseria meningitidis promoter |
| L2 P−.pCDS5KO | L2P− 25667R transformed with pGFP::SW2 with a functional deletion in plasmid CDS5 which encodes the putative virulence effector pgp3; the creation of this recombinant plasmid was described in detail previously (15) |
| L2 434/Bu | Commonly referenced laboratory strain of C. trachomatis serovar L2 cultured from a bubo of a patient with LGV (25); the genome of this isolate has been sequenced and published elsewhere (26) (GenBank reference sequence NC_010287); in the present study, this isolate was used only as a comparator for L2P−.pGFP::SW2 |
Following transformation of L2P−, McCoy cells were used for the propagation and expansion of C. trachomatis transformants (Clarke lab; University of Southampton, Southampton, United Kingdom). Aliquots of the transformants L2P−.pGFP::SW2 and L2P−.pCDS5KO were provided at minimal passage number for in vivo applications with further expansion in HeLa 229 cells (Ramsey lab; Midwestern University, Downers Grove, IL, USA) as previously described (7, 12). L2P−.pCDS5KO stained for glycogen, but the mature inclusion phenotype was unaffected by the presence of the deletion compared with the plasmid-free parent. This was in contrast to our previous observations with the same plasmid in a urogenital tract background (27).
Mice, infections, and infection monitoring.
The use of animals in these studies was approved by the Midwestern University Institutional Animal Care and Use Committee. Outbred female Swiss Webster mice (Harlan Sprague Dawley, Indianapolis, IN) were purchased at 5 to 6 weeks of age and allowed to acclimate in the Animal Resources Facility at Midwestern University for a minimum of 10 days. Prior to urogenital infection, mice were treated with medroxyprogesterone acetate (Greenstone LLC, Peapack, NJ) at 2.5 mg subcutaneously as has been described previously (28). Seven days later, groups of 10 mice were infected with graded log10 doses of each strain either intravaginally (29) in 10 μl or in experiments using transcervical inoculation, i.e., a single dose of 105 inclusion-forming units (IFU) given transcervically in 4 μl using a nonsurgical embryo transfer (NSET) device (30). Following intravaginal inoculation, infection course and infectious burden were assessed by the collection of cervical-vaginal swabs at 4, 7, 10, 14, and 21 days postinfection. C. trachomatis was isolated from sucrose-potassium glutamate buffer (SPG, pH 7.2) swab fluid in HeLa 229 cell monolayers. Following transcervical inoculation, we confirmed infection by cervical-vaginal swabs at day 4 postinfection. In some experiments, swabs and upper genital tract tissues (UGT; uterine horns and oviducts, sans ovaries) were collected, and tissues were homogenized in 1 ml SPG as described previously (31). Inclusions isolated from swabs and from UGT were visualized by indirect immunofluorescence microscopy and quantitated as IFU as previously described (32).
For respiratory infection, mice were anesthetized and inoculated intranasally exactly as described previously (7, 12). We used inocula of either 104 or 105 IFU for each strain and transformant tested. These doses were chosen because they mirrored breakpoint doses for significant differences in weight change over time in previous studies (7, 12). Mice were weighed daily, and weight change from baseline was recorded as a measure of morbidity (7, 12).
Antibody responses.
Heparinized blood was collected at day 0, just prior to inoculation, and day 35 postinoculation. Chlamydia-specific plasma immunoglobulin G (IgG) antibody responses were assessed using enzyme-linked immunosorbent assay (ELISA) as described previously with minor modifications (33). Gradient-purified, UV light-inactivated elementary bodies (EBs) of the LGV isolate L2 434/Bu were used as the capture antigen. For quantitative comparison of antibody responses, titers were converted to log10, and the arithmetic mean of all seropositive animals was determined as described elsewhere (7, 12, 33).
Histopathological assessments.
Following transcervical inoculation, five excised genital tract samples from each experimental group on day 7 postinfection were preserved in buffered 10% formalin at neutral pH, then embedded in paraffin, and serially sectioned (5 μm) longitudinally. Attempts were made to include the cervix, both uterine horns, and both oviducts and to include the lumenal structures of each tissue in each section (AML Labs, Rosedale, MD). The tissues were stained with hematoxylin and eosin (H&E) (34). Each H&E-stained tissue section was assessed by a pathologist (B.C.J.) who was blinded to the animal numbers, treatments, and groupings in the experiment. Uninfected mouse urogenital tract tissues were included for reference to normal tissue. Histopathological assessment was scored in the cervix, uterus (considering both left and right uterine horns), and oviducts relative to normal, uninfected urogenital tract tissue. Parameters assessed were acute (neutrophil) and chronic (mononuclear cell) infiltrates. Each parameter was scored on a scale of 0 to 4 using a modification of the methods of Rank et al. (35) as follows: 0 = normal/no presence of parameter; 1 = rare or slight presence of parameter; 2 = scattered/mild or diffuse presence of parameter; 3 = consistent/numerous/frequent presence of parameter; 4 = confluent or severe presence of parameter. To control for variances in parameter foci according to cutting depth within the same tissue, we included 3 separate but roughly serial sections from each tissue in each assessment. Scoring was done on each section, but the mean score for each serial section was used for calculating means and for data reporting.
Statistics.
Infection course as assessed by shedding of viable organisms from the lower genital tract was compared at each inoculating dose by a two-factor analysis of variance (ANOVA) (plasmid type group and time) with repeated measures on the main effect of time and applying Tukey's post hoc analysis to compare plasmid type groups. Immunoglobulin G antibody titers and quantitative infection burdens in upper genital tract tissues were compared using a two-tailed t test. Histopathological scores were compared using a Kruskal-Wallis test. Statistical significance for all assessments was set at a P value of <0.05. The incidence of viable chlamydial shedding and seroconversion was used to determine the infectious dose that resulted in infection in 50% of mice (ID50), as estimated by the method of Reed and Muench (36).
RESULTS
A deletion mutation in the plasmid gene CDS5 (pgp3) results in reduced infectivity and attenuated fitness following intravaginal inoculation.
The establishment of an ID50 is commonly used to estimate infectivity in models of infectious diseases. To establish an ID50 with the subject chlamydial strains, we inoculated groups of 10 mice intravaginally with log10 graded doses of each of the isolates in Table 1. Subsequent to inoculation, and to verify infection, we collected cervical-vaginal swabs at 4, 7, 10, 14, and 21 days postinfection. In addition, we have found that seroconversion postinfection can detect infection at levels below the sensitivity of our culture methods that are sometimes observed with low-dose inocula (7, 12). Thus, at day 35 postinoculation, we assessed plasma IgG antibody responses in an ELISA that uses gradient-purified UV-inactivated L2 434/Bu as the capture antigen. The results of these assessments are shown in Table 2. By both culture and serology, comparable ID50s were obtained for the L2P− isolate, with estimates of 3.2 × 105 IFU and 2.5 × 105 IFU, respectively. This is comparable to the results reported by Olivares-Zavaleta et al. for this isolate (5). When the strain was transformed with the recombinant vector pGFP::SW2, the infectivity of the resultant transformant was greatly enhanced, with L2P−.pGFP::SW2 yielding an ID50 approximately 3 log10 lower at 103 IFU by culture. By serology, the ID50 for L2P−.pGFP::SW2 could not be determined but was less than the lowest dose of 103 IFU. The results of inoculation with the L2P−.pGFP::SW2 transformant were comparable to those obtained with a nonisogenic but serovar-matched isolate, C. trachomatis L2 434/Bu, containing a wild-type plasmid (ID50 estimated at 3.1 × 103 for culture and 5.6 × 103 for seroconversion; data not shown). Interestingly, transformation with the pCDS5KO vector yielded an estimated ID50 approximately 1 log10 unit lower than that of the plasmid-deficient isolate L2P− and 1.4 log10 units higher than that of the L2P−.pGFP::SW2 transformant. These results were consistent regardless of whether culture or seroconversion was used to determine ID50. From these observations, we conclude that plasmid deficiency represents a significant infectivity defect in the mouse urogenital infection model and that transformation with a vector containing a full complement of chlamydial plasmid genes restores infectivity. Second, we conclude that deficiency in that CDS5 (pgp3) similarly and significantly degrades infectivity.
TABLE 2.
ID50 determination for strains in urogenital tract infections
| Strain and method | No. of positive mice (n = 10) inoculated with dose (IFU) ofa: |
ID50 (IFU)b | |||
|---|---|---|---|---|---|
| 103 | 104 | 105 | 106 | ||
| L2P− | |||||
| Culture | 1 | 2 | 3 | 7 | 316,227 |
| Seroconversion | 2 | 4 | 7 | 9 | 251,116 |
| L2P−.pGFP::SW2 | |||||
| Culture | 5 | 9 | 10 | 10 | 1,000 |
| Seroconversion | 7 | 9 | 10 | 10 | <1,000 |
| L2P−.pCDS5KO | |||||
| Culture | 1 | 2 | 5 | 9 | 26,827 |
| Seroconversion | 3 | 4 | 5 | 7 | 21,258 |
For culture, data are numbers of mice that were culture positive at any time point postinfection. Culture detection of infection was attempted on swabs collected at 4, 7, 10, 14, and 21 days postinfection (Fig. 1). For seroconversion, data are numbers of mice displaying seroconversion for plasma IgG antibody (day 35 postinfection; positive seroconversion was empirically set at a titer of 20).
Calculated dose that will achieve a 50% infection rate. Data were calculated by the method of Reed and Muench (36).
To determine a role for the plasmid and for CDS5 (pgp3) with regard to in vivo fitness, we measured infectious burden over time at each of the inoculating doses. Figure 1 depicts the results of the infection course over time in mice inoculated with the plasmid(−) L2P− isolate and the transformants L2P−.pGFP::SW2 and L2P−.pCDS5KO. Because evidence of infection by either culture or by seroconversion was seen only in very low numbers of mice inoculated with the L2P− isolate and the L2P−.pCDS5KO transformant at the 103 and 104 inoculating doses (1 or 2 mice culture positive and 2 to 4 seropositive), valid statistical comparisons could not be garnered from these groups. However, a distinct trend toward abbreviated infection courses and lower infectious burden can be observed at these doses compared to the results for the L2P−.pGFP::SW2-inoculated mice. Nonetheless, at doses of 105 and 106 IFU, mice inoculated with the L2P−.pGFP::SW2 transformant displayed significantly greater infectious burdens and more protracted infection courses than those inoculated with L2P− and the L2P−.pCDS5KO transformant. The latter (pCDS5KO transformant and host) were not significantly different from each other at any dose tested. Similarly, when the infection course in mice inoculated with L2P−.pGFP::SW2 was compared to that seen with the nonisogenic L2 434/Bu isolate (with wild-type plasmid), there were no statistical differences at any dose (data not shown).
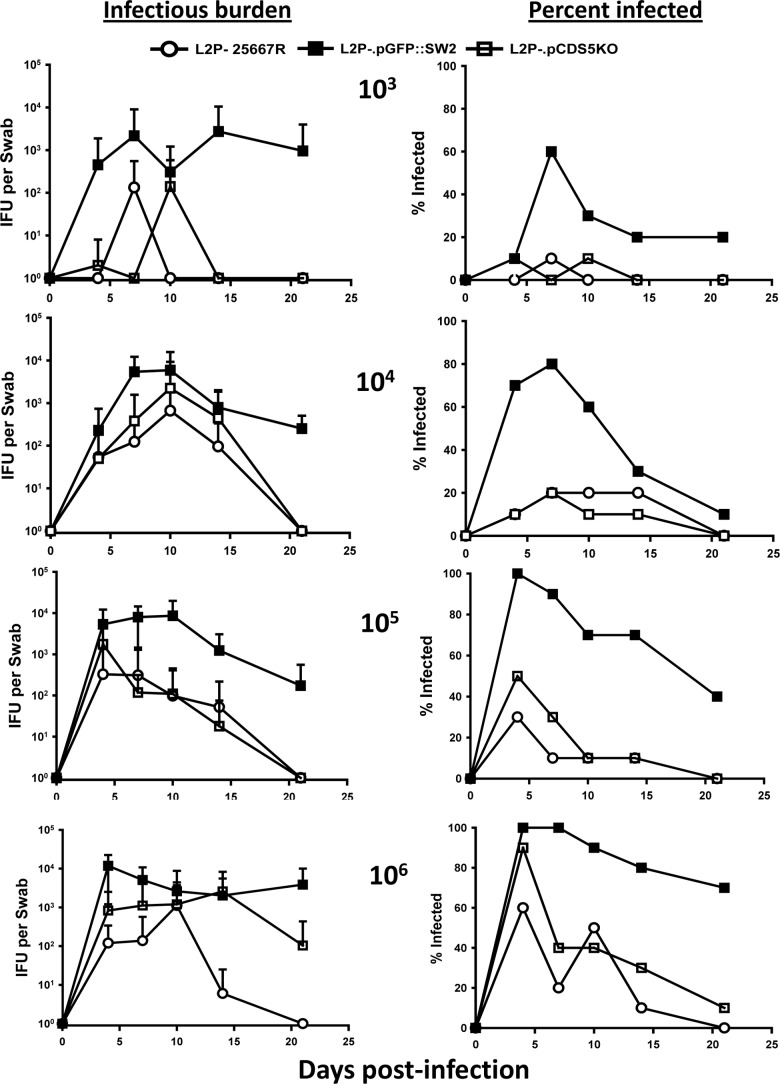
FIG 1.
Urogenital infection course in female mice inoculated intravaginally with the plasmid(−), plasmid(+), and plasmid CDS5 deletion strains of C. trachomatis serovar L2. The graphs on the left depict the mean infectious burden over time in mice inoculated with L2P− (open circles), L2P−.pGFP::SW2 (filled squares), and the derivative of L2P−.pGFP::SW2 but with a deletion mutation in CDS5 (pgp3), L2P−.pCDS5KO (open squares). The graphs on the right depict the percentage of mice that were culture positive at the same time points. Results are for doses of 103, 104, 105, and 106 IFU per mouse (top to bottom; n = 10 mice inoculated per group). There were significant differences between the infectious burden (IFU) over time at the 105 dose in the L2P−. pGFP::SW2 group and that in the L2P− or the L2P−.pCDS5KO group (P < 0.001 and P = 0.004 by two-way ANOVA), but there was no difference between groups receiving L2P− and L2P−.pCDS5KO (P = 0.663). Similarly, there were significant differences between the infection courses in the 106 dose group receiving L2P−. pGFP::SW2 and the group receiving L2P− or L2P−.pCDS5KO (P < 0.001 for both groups), and there was no difference between the L2P− and L2P−.pCDS5KO groups at this dose (P = 0.956). Statistical analyses were not conducted on the 103 and 104 IFU doses due to low numbers of mice sustaining infection, as assessed either by culture or by seroconversion, in the groups inoculated with L2P− and L2P−.pCDS5KO.
Lastly, we inoculated groups of 10 mice intranasally with each of the subject strains at doses of 104 and 105 IFU, and the weight change from baseline was monitored daily for 21 days as previously described (7). No differences were seen between the groups and doses, with the exception of a trend toward more weight gain over time in the group inoculated with L2P− that did not prove to be significantly different from the gain in the L2P−.pGFP::SW2-inoculated group (data not shown.).
In summary, a deletion in CDS5 (pgp3) resulted in reduced infectivity and fitness in vivo following urogenital inoculation and thereby establishes a role for a specific plasmid-based virulence effector gene in this model.
A deletion mutation in the plasmid gene CDS5 (pgp3) results in reduced antibody responses following intravaginal inoculation.
Plasma samples collected at day 35 postinoculation in the aforementioned experiments (Table 2 and Fig. 1) and plasma IgG antibody responses in mice that seroconverted were quantitated by ELISA (12, 33). Figure 2 shows that for each of the inoculating doses, mice infected with the plasmid(+) transformant L2P−.pGFP::SW2 induced a significantly more robust antibody response than mice infected with either the plasmid(−) L2P− isolate or the transformant L2P−.pCDS5KO. Thus, in light of the data in Table 2, the data in Fig. 1 further data support and serve to confirm the assertion that the infectivity defect is conveyed by the absence of a functional CDS5 gene and additionally shows that the plasmid(−) isolate and the CDS5 deletion mutant may have comparable defects in the induction of adaptive immunity. An alternative explanation could be that the lower infectious burden resulted in lower antibody responses vis-à-vis a lower antigenic burden. However, a comparison of the lowest dose (103 IFU) of L2P−.pGFP::SW2 and the highest doses (105 to 106 IFU) of plasmid(−) L2P− isolate or the transformant L2P−.pCDS5KO does not seem to support this interpretation, since the higher doses did not result in higher antibody responses (Fig. 2) despite higher infectious burdens (Fig. 1).
FIG 2.
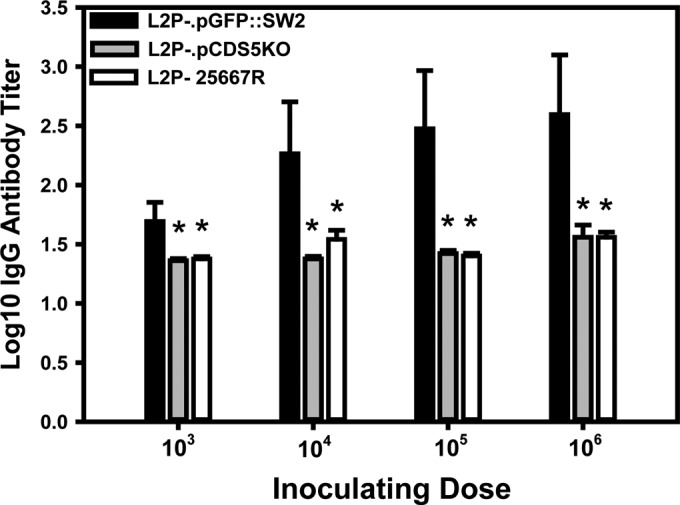
Immunoglobulin G antibody responses in mice inoculated intravaginally with plasmid(−), plasmid(+), and plasmid CDS5 deletion mutant of C. trachomatis serovar L2. Plasma collected at day 35 after intravaginal inoculation was assessed by ELISA as previously described (33) but using gradient purified UV-inactivated EBs of strain L2 434/Bu as the capture antigen. The arithmetic mean of the log10 antibody responses in mice that seroconverted (titer ≥ 20) is shown (error bars show standard deviations). Numbers of mice seroconverting are shown in Table 2. Significant differences (asterisks) between L2P−.pGFP::SW2-infected mice and L2P−- and L2P−.pCDS5KO-infected mice were observed at each inoculating dose: 103 IFU, P < 0.04 and P < 0.04, respectively; 104 IFU, P < 0.002 and P < 0.004, respectively; 105 IFU, P < 0.0008 and P < 0.0009, respectively; and 106 IFU, P < 0.002 and P < 0.0001, respectively. The antibody responses were not significantly different at any inoculating dose for mice inoculated with L2P− compared to mice inoculated L2P−.pCDS5KO.
Deletion mutation in the plasmid gene CDS5 (pgp3) reduces infectivity in upper genital tract tissues following transcervical inoculation.
It is a common observation that, unlike with C. muridarum, intravaginal inoculation of human C. trachomatis strains into mice yields minimal upper genital tract infection and thus induces scant pathological changes. We confirmed this observation for all of the isolates used in this study (data not shown). Thus, to induce a significant upper genital tract infection, we employed a method of transcervical intrauterine inoculation using an NSET device as described by Gondek et al. (30). A preliminary experiment was then conducted by using plasmid(+) L2 434/Bu and assessing infectious burden in UGT homogenates at 4, 7, 10, and 14 days after transcervical inoculation. It was determined in this experiment that day 7 postinfection represents the optimal time for assessment in this model, a time frame roughly mirroring that reported by Gondek et al. using the same strain (30). We then applied the same technique but used the subject plasmid variants described in Table 1. Figure 3 shows that transcervical inoculation with L2P−.pGFP::SW2 induces a greater infectious burden in the upper genital tract than does inoculation with L2P− or with L2P−.pCDS5KO. Though there was a trend toward higher IFU counts in UGT tissues in the mice inoculated with L2P−.pCDS5KO than in those given the plasmidless parental isolate, L2P−, the difference did not prove to be significantly different.
FIG 3.
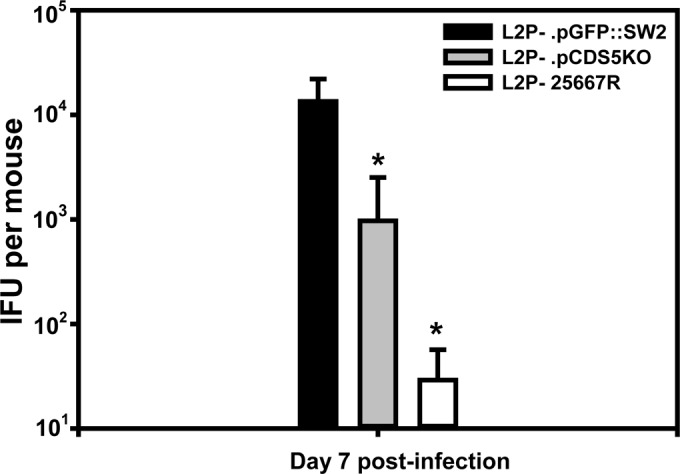
Infectious burden in mice following transcervical inoculation. At day 7 after transcervical inoculation, mice were euthanized, and UGT tissues (uterus and oviduct) were extracted for chlamydial culture. Data are the mean IFU counts for mice (n = 8) from each group, and error bars show standard deviations. Asterisks indicated significant differences compared to L2P−.pGFP::SW2 as assessed by a two-tailed t test (P < 0.005 for L2P−.pCDS5KO, and P < 0.004 for L2P−). The L2P−.pCDS5KO and L2P− groups were not significantly different (P = 0.12).
In summary, and in consideration of the results of intravaginal ID50 determination, infection course, quantitative antibody responses, and direct transcervical inoculation into the upper genital tract, we conclude that the plasmid is a critical mediator of in vivo fitness and infectivity and that pCDS5 (pgp3) plays a necessary role in this regard.
A deletion mutation in plasmid gene CDS5 (pgp3) reduces inflammatory infiltrates in upper genital tract tissues.
In the experiment whose results are presented in Fig. 3, we harvested UGT tissues from parallel groups of mice following transcervical inoculation and assessed H&E-stained sections of these tissues for pathological changes. As before, we chose the day 7 postinoculation time point as optimal for these assessments, as framed by results of preliminary experiments. The main features assessed and scored were acute (mostly neutrophil) and chronic (monocytic and lymphocytic) infiltrates. We also made note of tissue changes potentially attributable to the inflammatory response, such as edema and mucosal erosion. Figure 4A and B show examples of the observations noted, whereas Fig. 4C and D graphically depict the semiquantitative pathological scores assessed by a pathologist (B.C.J.) who was blinded to the study groups and experimental design.
FIG 4.
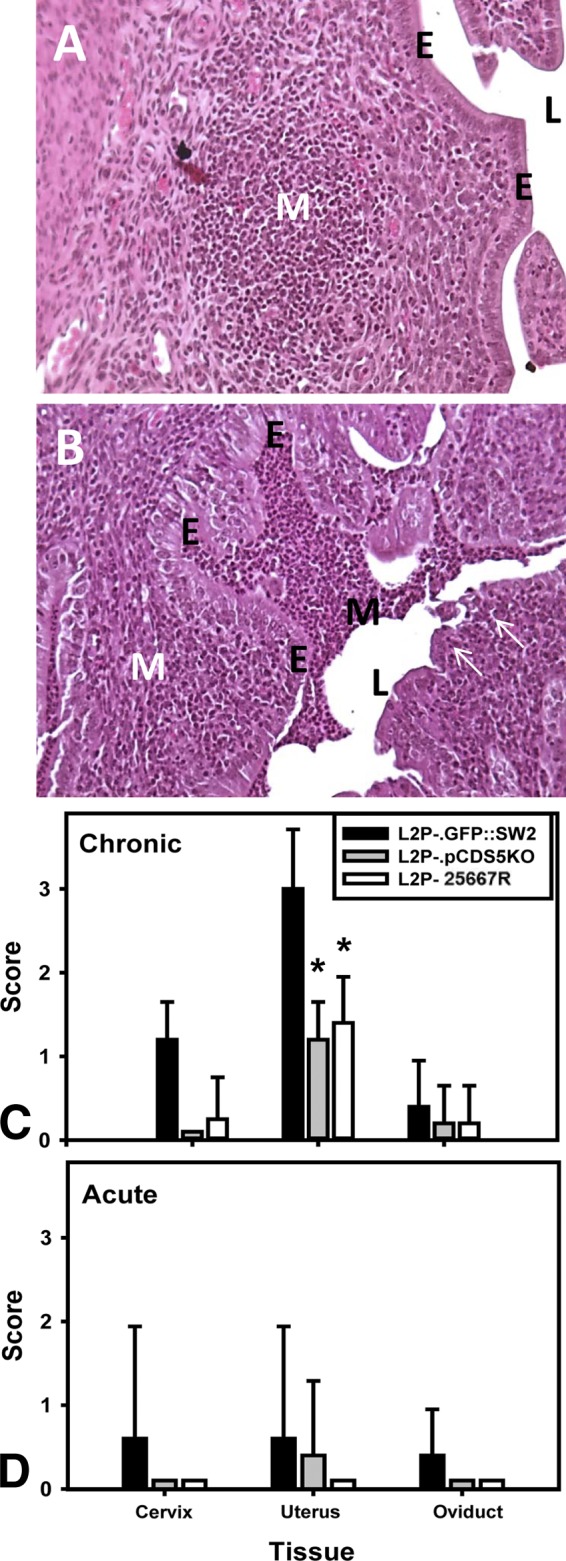
Histopathology in mice following transcervical inoculation. (A) Typical H&E-stained tissues from mice inoculated with either L2P− or L2P−.pCDS5KO. Scattered subepithelial mononuclear aggregates were observed (white M) in the uterine horns. (B) Typical results from a mouse inoculated with L2P−.pGFP::SW2. While substantial amounts of subepithelial mononuclear cells (white M) were also observed, they tended not to be found in aggregates but were dispersed throughout the endometrium. Also, in several mice, mononuclear inflammatory cells could be seen filling or nearly filling the lumen (black M). Variable amounts of acute infiltrates could also be seen in these tissues. Arrows point to areas of mucosal erosion. E, epithelium; L, lumen of the uterus. (C and D) Mean scores for the parameters of chronic infiltrates (monocytes and lymphocytes) (C) and acute infiltrates (mostly polymorphonuclear leukocytes) (D) in the cervix, uterus, and oviduct. Asterisks represent significant differences compared to mice inoculated with L2P−.pGFP::SW2 (P < 0.008; Kruskal-Wallis ANOVA on ranks). No significant difference was observed between the mice infected with L2P− and L2P−.pCDS5KO.
As expected, of the tissues assessed, the uterus (site of NSET device inoculation) was the tissue most affected. Endometritis was noted in most mice that had been inoculated. The predominant infiltrate in all infected tissues was mononuclear. Mice infected with L2P− or L2P−.pCDS5KO displayed somewhat sparsely distributed subepithelial aggregates of lymphoid and mononuclear uterine infiltrates (a typical aggregate is pictured in Fig. 4A). Infiltrates were rarely seen in the lumen of the uterus in these mice. The mucosae remained intact, and no erosion or epithelial sloughing was observed. In contrast, in mice infected with L2P−.pGFP::SW2, the mononuclear infiltrate tended to be less aggregated but was distributed throughout large sections of the subepithelial layers. The related pathological scores for chronic infiltrates were blunted in mice infected with L2P− and L2P−.pCDS5KO compared to mice infected with L2P−.pGFP::SW2. Mucosal erosion with lumenal infiltrates was commonly observed in mice inoculated with L2P−.pGFP::SW2 (a typical section is shown in Fig. 4B). In addition, we observed a trend toward the presence of more acute infiltrates in tissues from L2P−.pGFP::SW2 infected mice, but the trend did not prove significant. Also of note was the observation that some mice in all of the groups exhibited modest infiltrates in the submucosae of the oviduct but intraluminal mononuclear infiltrates were observed in one oviduct in the group infected with L2P−.pGFP::SW2. These data indicate that deletion of CDS5 (pgp3) results in less severe histopathological changes and reduced inflammatory infiltration in the UGT in this model.
DISCUSSION
For years it has been known that many chlamydial pathogens, including all biovars of C. trachomatis, routinely carry a highly conserved plasmid. However, the exact function of the plasmid was unknown. It was assumed that it conveyed some criticality of function, since it is exceedingly rare for live, naturally occurring plasmid(−) isolates of C. trachomatis to be found. Some roles for the plasmid have been identified in vitro, such as controlling the expression of certain chromosomal genes and inducing glycogen accumulation within inclusions, while other reports indicated that the plasmid variously contributes to infectivity and pathogenicity in selected hosts in vivo (5, 6, 8, 10, 37, 38). However, no critical in vivo role has been assigned to the plasmid in Chlamydia psittaci intraperitoneal infections of mice (38) or Chlamydia caviae urogenital infections of guinea pigs (37). Thus, one can reasonably conclude that whatever role the plasmid plays, it is likely specific to the chlamydial species, the host, and possibly the anatomical site of infection.
The molecular basis of plasmid-mediated in vivo fitness and pathogenicity is still not understood but has been the impetus behind the growing interest in understanding plasmid function in chlamydial pathobiology. The recent elaboration of a system of transformation via the plasmid and the fact that selected plasmid genes can undergo deletion mutation has allowed investigators to begin to dissect in vitro roles for each gene (14–16, 21, 27). For example, it is accepted that plasmid effectors enhance expression of selected chromosomal genes which also likely affect inclusion morphology and glycogen accumulation and thereby mimic the phenotype of naturally occurring plasmid(−) isolates (10, 16, 27).
In the present study, we further capitalized upon this transformation system to explore the role of a specific plasmid gene, CDS5 (pgp3) in vivo, using a specific deletion mutation designed to inactivate this gene alone, although deletion of CDS5 might have impacts that are not solely related to the deletion. We focused on CDS5 because this gene and its protein product, Pgp3, are associated with intriguing properties that imply a role in virulence. For example, Pgp3, unlike other plasmid gene products, appears to be secreted beyond the inclusion into the host cell cytosol (17). The cytosolic nature of this protein makes it a likely target for endogenous antigen processing, but so far, this has not been explored. However, Pgp3 is immunogenic in that antibodies are routinely made against it in the context of infection in humans and animals (18, 39) and DNA immunization provides at least partial protection (20). All of these qualities led us to hypothesize that this gene encodes an in vivo virulence effector.
We used a genomically sequenced naturally occurring plasmid(−) isolate of C. trachomatis, L2P−, as the transformation recipient of the intact vector pGFP::SW2, as well as a derivative of the vector with a deletion mutation in CDS5 (pgp3), to infect female mice in the urogenital tract. This system is ideal from the standpoint of starting experimentation with a known transformation recipient that will be isogenic to all subsequent transformants. We observed that transformation with C. trachomatis carrying the intact vector greatly enhanced infectivity by decreasing the ID50 by approximately 3 log10 IFU, which results in an infectivity within the range of, if not greater than, the infectivity of a wild-type plasmid(+) isolate. By ID50 estimation, the transformant with the vector containing a deletion mutation in CDS5 incrementally but substantially reduced infectivity compared to the L2P−.pGFP::SW2 transformant with the intact vector (Table 2). Several additional observations led us to conclude that, in this model, deletion of CDS5 (pgp3) is tantamount, or nearly so, to absolute plasmid deficiency. These observations include comparable infection courses at all inoculating doses (Fig. 1), comparable induction of adaptive immune responses (Fig. 2), and similar patterns of histopathological changes (Fig. 4). All of these parameters were indistinguishable between mice inoculated with plasmid(−) L2P− and the L2P−.pCDS5KO and were significantly attenuated compared to infection in mice with the L2P−.pGFP::SW2 transformants containing the intact vector. In summary, it is reasonable to conclude that CDS5 (pgp3) contributes to infectivity, heightens in vivo fitness, and enhances virulence in this model. The data support our hypothesis that plasmid CDS5 indeed encodes a virulence effector (Pgp3) and validates the use of chlamydial transformants in vivo to assess chlamydial plasmid-based pathobiology.
It is also interesting that we did not find a strict plasmid tropism in C. trachomatis species between biovars, since the plasmid backbone for these experiments was derived from a disparate biovar (serovar E) isolate but was readily accepted by the serovar L2 host. This has proved to ably restore all in vitro and in vivo characteristics associated with wild-type nonrecombinant plasmid(+) C. trachomatis isolates (14). Considering the homogeneous nature of the plasmid and our present results, C. trachomatis plasmids appear to be functionally interchangeable within the species. However, there may be subtle differences that need to be explored more in depth (40).
These findings should be carried forward to determine if the same or similar observations can be garnered in other mouse models (e.g., C. muridarum) and anatomical sites of inoculation and to assess other putative plasmid-based virulence effectors such as CDS6 (pgp4) and CDS7 (pgp5) (16, 41). The former appears to regulate the expression of chromosomal genes and, interestingly, the CDS5 (pgp3) virulence effector we have identified in the present study. In this regard, we would hypothesize that the presence of CDS6 (pgp4) would be necessary for virulence in this model through regulating the expression of CDS5 (pgp3). One could also envision a scenario in which the accumulation of glycogen, while associated with virulence phenotype, is merely an association and thus a red herring or, at best, a surrogate marker of pathogenicity. This assertion is supported by the observation that although L2P−.pCDS5KO has no such defect in glycogen accumulation (but does have a subtle change in mature inclusion morphology), it was certainly attenuated for infectivity and virulence in vivo.
As with most technological breakthroughs, the advent of the chlamydial transformation system has opened a window of opportunity and simultaneously generated many more questions than it has answered. It will be only through carefully planned and executed experimentation that a detailed dissection of the roles of plasmid genes can be conducted.
ACKNOWLEDGMENTS
The work conducted at Midwestern University was supported by intramural funds from the Office of Research and Sponsored Programs and by funds supplied the Microbiology and Immunology Department. B.J.S. was a recipient of a Chicago College of Osteopathic Medicine Summer Research Fellowship. The construction of the plasmid transformants and mutants was funded by Wellcome Trust grant no 091296.
We thank Lesley T. Cutcliffe for her excellent technical assistance.
Footnotes
Published ahead of print 27 May 2014
REFERENCES
- 1.Thomas NS, Lusher M, Storey CC, Clarke IN. 1997. Plasmid diversity in Chlamydia. Microbiology 143:1847–1854. 10.1099/00221287-143-6-1847. [DOI] [PubMed] [Google Scholar]
- 2.Seth-Smith HM, Harris SR, Persson K, Marsh P, Barron A, Bignell A, Bjartling C, Clark L, Cutcliffe LT, Lambden PR, Lennard N, Lockey SJ, Quail MA, Salim O, Skilton RJ, Wang Y, Holland MJ, Parkhill J, Thomson NR, Clarke IN. 2009. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics 10:239. 10.1186/1471-2164-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer L, Falkow S. 1986. A common plasmid of Chlamydia trachomatis. Plasmid 16:52–62. 10.1016/0147-619X(86)90079-X. [DOI] [PubMed] [Google Scholar]
- 4.Carlson JH, Whitmire WM, Crane DD, Wicke L, Virtaneva K, Sturdevant DE, Kupko JJ, III, Porcella SF, Martinez-Orengo N, Heinzen RA, Kari L, Caldwell HD. 2008. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect. Immun. 76:2273–2283. 10.1128/IAI.00102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olivares-Zavaleta N, Whitmire W, Gardner D, Caldwell HD. 2010. Immunization with the attenuated plasmidless Chlamydia trachomatis L2(25667R) strain provides partial protection in a murine model of female genitourinary tract infection. Vaccine 28:1454–1462. 10.1016/j.vaccine.2009.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kari L, Whitmire WM, Olivares-Zavaleta N, Goheen MM, Taylor LD, Carlson JH, Sturdevant GL, Lu C, Bakios LE, Randall LB, Parnell MJ, Zhong G, Caldwell HD. 2011. A live-attenuated chlamydial vaccine protects against trachoma in nonhuman primates. J. Exp. Med. 208:2217–2223. 10.1084/jem.20111266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigar IM, Schripsema JH, Wang Y, Clarke IN, Cutcliffe LT, Seth-Smith HM, Thomson NR, Bjartling C, Unemo M, Persson K, Ramsey KH. 2014. Plasmid deficiency in urogenital isolates of Chlamydia trachomatis reduces infectivity and virulence in a mouse model. Pathog. Dis. 70:61–69. 10.1111/2049-632X.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Connell CM, Ingalls RR, Andrews CW, Jr, Scurlock AM, Darville T. 2007. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J. Immunol. 179:4027–4034. 10.4049/jimmunol.179.6.4027. [DOI] [PubMed] [Google Scholar]
- 9.O'Connell CM, Nicks KM. 2006. A plasmid-cured Chlamydia muridarum strain displays altered plaque morphology and reduced infectivity in cell culture. Microbiology 152:1601–1607. 10.1099/mic.0.28658-0. [DOI] [PubMed] [Google Scholar]
- 10.O'Connell CM, Abdelrahman YM, Green E, Darville HK, Saira K, Smith B, Darville T, Scurlock AM, Meyer CR, Belland RJ. 2011. Toll-like receptor 2 activation by Chlamydia trachomatis is plasmid dependent, and plasmid-responsive chromosomal loci are coordinately regulated in response to glucose limitation by C. trachomatis but not by C. muridarum. Infect. Immun. 79:1044–1056. 10.1128/IAI.01118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lei L, Chen J, Hou S, Ding Y, Yang Z, Zeng H, Baseman J, Zhong G. 2014. Reduced live organism recovery and lack of hydrosalpinx in mice infected with plasmid-free Chlamydia muridarum. Infect. Immun. 82:983–992. 10.1128/IAI.01543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramsey KH, Sigar IM, Schripsema JH, Denman CJ, Bowlin AK, Myers GA, Rank RG. 2009. Strain and virulence diversity in the mouse pathogen Chlamydia muridarum. Infect. Immun. 77:3284–3293. 10.1128/IAI.00147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyrick PB. 2002. C. trachomatis: infection strategies of the ultimate intracellular pathogen. ASM News 68:70–76. [Google Scholar]
- 14.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 7:e1002258. 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Persson K, Bjartling C, Clarke IN. 2013. Genetic transformation of a clinical (genital tract), plasmid-free isolate of Chlamydia trachomatis: engineering the plasmid as a cloning vector. PLoS One 8:e59195. 10.1371/journal.pone.0059195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, Watkins H, Zhou B, Sturdevant GL, Porcella SF, McClarty G, Caldwell HD. 2013. Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect. Immun. 81:636–644. 10.1128/IAI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Chen D, Zhong Y, Wang S, Zhong G. 2008. The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect. Immun. 76:3415–3428. 10.1128/IAI.01377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D, Lei L, Lu C, Galaleldeen A, Hart PJ, Zhong G. 2010. Characterization of Pgp3, a Chlamydia trachomatis plasmid-encoded immunodominant antigen. J. Bacteriol. 192:6017–6024. 10.1128/JB.00847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Lu C, Peng B, Zeng H, Zhou Z, Wu Y, Zhong G. 2012. Induction of protective immunity against Chlamydia muridarum intravaginal infection with a chlamydial glycogen phosphorylase. PLoS One 7:e32997. 10.1371/journal.pone.0032997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Wang S, Wu Y, Zhong G, Chen D. 2008. Immunization with chlamydial plasmid protein pORF5 DNA vaccine induces protective immunity against genital chlamydial infection in mice. Sci. China C Life Sci. 51:973–980. 10.1007/s11427-008-0130-9. [DOI] [PubMed] [Google Scholar]
- 21.Gong S, Yang Z, Lei L, Shen L, Zhong G. 2013. Characterization of Chlamydia trachomatis plasmid-encoded open reading frames. J. Bacteriol. 195:3819–3826. 10.1128/JB.00511-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson EM, Markoff BA, Schachter J, De La Maza LM. 1990. The 7.5-kb plasmid present in Chlamydia trachomatis is not essential for the growth of this microorganism. Plasmid 23:144–148. 10.1016/0147-619X(90)90033-9. [DOI] [PubMed] [Google Scholar]
- 23.Harris SR, Clarke IN, Seth-Smith HM, Solomon AW, Cutcliffe LT, Marsh P, Skilton RJ, Holland MJ, Mabey D, Peeling RW, Lewis DA, Spratt BG, Unemo M, Persson K, Bjartling C, Brunham R, de Vries HJ, Morre SA, Speksnijder A, Bebear CM, Clerc M, de Barbeyrac B, Parkhill J, Thomson NR. 2012. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat. Genet. 44:413–419, S1. 10.1038/ng.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickett MA, Everson JS, Pead PJ, Clarke IN. 2005. The plasmids of Chlamydia trachomatis and Chlamydophila pneumoniae (N16): accurate determination of copy number and the paradoxical effect of plasmid-curing agents. Microbiology 151:893–903. 10.1099/mic.0.27625-0. [DOI] [PubMed] [Google Scholar]
- 25.Schachter J, Meyer KF. 1969. Lymphogranuloma venereum. II. Characterization of some recently isolated strains. J. Bacteriol. 99:636–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomson NR, Holden MT, Carder C, Lennard N, Lockey SJ, Marsh P, Skipp P, O'Connor CD, Goodhead I, Norbertzcak H, Harris B, Ormond D, Rance R, Quail MA, Parkhill J, Stephens RS, Clarke IN. 2008. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 18:161–171. 10.1101/gr.7020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Cutcliffe LT, Skilton RJ, Persson K, Bjartling C, Clarke IN. 2013. Transformation of a plasmid-free, genital tract isolate of Chlamydia trachomatis with a plasmid vector carrying a deletion in CDS6 revealed that this gene regulates inclusion phenotype. Pathog. Dis. 67:100–103. 10.1111/2049-632X.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cotter TW, Meng Q, Shen Z, Zhang Y, Su H, Caldwell HD. 1995. Protective efficacy of major outer membrane protein specific immunoglobulin A (IgA) and IgG monoclonal antibodies in a murine model of Chlamydia trachomatis genital tract infection. Infect. Immun. 63:4704–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cotter TW, Miranpuri GS, Ramsey KH, Poulsen CE, Byrne GI. 1997. Reactivation of chlamydial genital tract infection in mice. Infect. Immun. 65:2067–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gondek DC, Olive AJ, Stary G, Starnbach MN. 2012. CD4+ T cells are necessary and sufficient to confer protection against Chlamydia trachomatis infection in the murine upper genital tract. J. Immunol. 189:2441–2449. 10.4049/jimmunol.1103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HY, Schripsema JH, Sigar IM, Lacy SR, Kasimos JN, Murray CM, Ramsey KH. 2010. A role for CXC chemokine receptor-2 in the pathogenesis of urogenital Chlamydia muridarum infection in mice. FEMS Immunol. Med. Microbiol. 60:49–56. 10.1111/j.1574-695X.2010.00715.x. [DOI] [PubMed] [Google Scholar]
- 32.Ramsey KH, Cotter TW, Salyer RD, Miranpuri GS, Yanez MA, Poulsen CE, DeWolfe JL, Byrne GI. 1999. Prior genital tract infection with a murine or human biovar of Chlamydia trachomatis protects mice against heterotypic challenge infection. Infect. Immun. 67:3019–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramsey KH, Newhall WJ, Rank RG. 1989. Humoral immune response to chlamydial genital infection of mice with the agent of mouse pneumonitis. Infect. Immun. 57:2441–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah AA, Schripsema JH, Imtiaz MT, Sigar IM, Kasimos J, Matos PG, Inouye S, Ramsey KH. 2005. Histopathologic changes related to fibrotic oviduct occlusion after genital tract infection of mice with Chlamydia muridarum. Sex. Transm. Dis. 32:49–56. 10.1097/01.olq.0000148299.14513.11. [DOI] [PubMed] [Google Scholar]
- 35.Rank RG, Sanders MM, Patton DL. 1995. Increased incidence of oviduct pathology in the guinea pig after repeat vaginal inoculation with the chlamydial agent of guinea pig inclusion conjunctivitis. Sex Transm. Dis. 22:48–54. 10.1097/00007435-199501000-00008. [DOI] [PubMed] [Google Scholar]
- 36.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 27:493–497. [Google Scholar]
- 37.Frazer LC, Darville T, Chandra-Kuntal K, Andrews CW, Jr, Zurenski M, Mintus M, Abdelrahman YM, Belland RJ, Ingalls RR, O'Connell CM. 2012. Plasmid-cured Chlamydia caviae activates TLR2-dependent signaling and retains virulence in the guinea pig model of genital tract infection. PLoS One 7:e30747. 10.1371/journal.pone.0030747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyairi I, Laxton JD, Wang X, Obert CA, Arva Tatireddigari VR, van Rooijen N, Hatch TP, Byrne GI. 2011. Chlamydia psittaci genetic variants differ in virulence by modulation of host immunity. J. Infect. Dis. 204:654–663. 10.1093/infdis/jir333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Zhong Y, Lei L, Wu Y, Wang S, Zhong G. 2008. Antibodies from women urogenitally infected with C. trachomatis predominantly recognized the plasmid protein pgp3 in a conformation-dependent manner. BMC Microbiol. 8:90. 10.1186/1471-2180-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song L, Carlson JH, Zhou B, Virtaneva K, Whitmire WM, Sturdevant GL, Porcella SF, McClarty G, Caldwell HD. 2014. Plasmid-mediated transformation tropism of chlamydial biovars. Pathog. Dis. 70:189–193. 10.1111/2049-632X.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Chen C, Gong S, Hou S, Qi M, Liu Q, Baseman J, Zhong G. 2014. Transformation of Chlamydia muridarum reveals a role for Pgp5 in suppression of plasmid-dependent gene expression. J. Bacteriol. 196:989–998. 10.1128/JB.01161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]