Abstract
Acinetobacter baumannii and Pseudomonas aeruginosa are nosocomial pathogens with overlapping sites of infection. This work reports that the two can coexist stably in mixed-culture biofilms. In a study intended to improve our understanding of the mechanism of their coexistence, it was found that pyocyanin, produced by P. aeruginosa that generally eliminates competition from other pathogens, led to the generation of reactive oxygen species (ROS) in A. baumannii cells, which in response showed a significant (P ≤ 0.05) increase in production of enzymes, specifically, catalase and superoxide dismutase (SOD). This work shows for the first time that the expression of catalase and SOD is under the control of a quorum-sensing system in A. baumannii. In support of this observation, a quorum-sensing mutant of A. baumannii (abaI::Km) was found to be sensitive to pyocyanin compared to its wild type and showed significantly (P ≤ 0.001) lower levels of the antioxidant enzymes, which increased on addition of 5 μM N-(3-hydroxydodecanoyl)-l-homoserine lactone. Likewise, in wild-type A. baumannii, there was a significant (P < 0.01) decrease in the level of anti-oxidant enzymes in the presence of salicylic acid, a known quencher of quorum sensing. In the presence of amikacin and carbenicillin, A. baumannii formed 0.07 and 0.02% persister cells, which increased 4- and 3-fold, respectively, in the presence of pyocyanin. These findings show that pyocyanin induces a protective mechanism in A. baumannii against oxidative stress and also increases its persistence against antibiotics which could be of clinical significance in the case of coinfections with A. baumannii and P. aeruginosa.
INTRODUCTION
Acinetobacter baumannii is an emerging opportunistic human pathogen that causes a plethora of nosocomial infections (1). The ability of this pathogen to sense and react to environmental and host stress signals allows it to persist in medical settings and in human hosts (2). A. baumannii is often associated with coinfection by other pathogens (3). Infection by A. baumannii and Pseudomonas aeruginosa in the upper respiratory tract and in lungs of patients with cystic fibrosis has been reported (4–6). P. aeruginosa competes with coinfecting organisms through the secretion of pyocyanin which is a redox-active phenazine compound, known to act through the generation of reactive oxygen species (ROS) (7). Pyocyanin was identified as the active component responsible for inhibition of Burkholderia cepacia in mixed-species biofilms with P. aeruginosa (8), while it did not have any such inhibitory effect on coexisting Klebsiella pneumoniae (9). A. baumannii has also been shown to coexist in vitro with P. aeruginosa, with evidence of interspecies interactions between them (10). Interspecies interactions may affect virulence and antibiotic sensitivities in the interacting pathogens (11).
ROS stress causes macromolecular damage, and bacteria respond to it by producing enzymatic scavengers, such as catalase and superoxide dismutase (SOD), the two conspicuous antioxidant enzymes widely distributed in nature (12). P. aeruginosa survives oxidative stress by raising cellular catalase and SOD enzyme activities, which have been demonstrated to be controlled by lasI-lasR- and rhlI-rhlR-mediated quorum-sensing systems (13). A. baumannii has only one quorum-sensing system, involving the transcriptional regulator AbaR, which forms a complex with N-(3-hydroxydodecanoyl)-l-homoserine lactone synthesized by the autoinducer synthase gene (abaI) (14). However, no link between quorum sensing and oxidative stress in A. baumannii has been reported.
Reports on multiple drug tolerance in A. baumannii are emerging in response to antibiotics (1). Generally, pathogens produce persister cells as phenotypic variants of the wild type that are tolerant to antibiotics. Persister cells can also form in response to starvation or oxidative stress (15). Very little is known about the phenotypic variant subpopulation of persister cells in A. baumannii. In view of coinfection by A. baumannii and P. aeruginosa in respiratory tract infections, especially in cystic fibrosis patients, where pyocyanin has been detected in the sputum and pulmonary secretions (16), it is pertinent to study the mechanism(s) by which A. baumannii survives oxidative stress caused by pyocyanin exposure and also its effect with regard to persistence. This work reports that A. baumannii and P. aeruginosa can coexist in mixed-species biofilms, as the oxidative stress caused by P. aeruginosa pyocyanin induced protective antioxidative enzymes in A. baumannii and enhanced its tolerance to antibiotics.
MATERIALS AND METHODS
Organisms and culture conditions.
Strains used in this study are listed in Table 1. For pyocyanin production, P. aeruginosa PAO1 was grown in peptone water (2% [wt/vol] peptone, pH 7.0), as it resulted in higher production of pyocyanin. All other strains were grown in Luria-Bertani broth at 37°C under shaking conditions.
TABLE 1.
A. baumannii and P. aeruginosa strains
| Strain | Characteristic(s) | Reference or source |
|---|---|---|
| A. baumannii | ||
| M2 | Clinical isolate | 14 |
| M2 (abaI::Km) | Autoinducer synthase (abaI) mutant of M2 strain; Kanr | 14 |
| M2, GFP tagged | pMU125 encoding GFP expressed under tet promoter; Ampr | This study |
| ATCC 17978 | Standard strain | American Type Culture Collection |
| ATCC 19606 | Standard strain | American Type Culture Collection |
| C1 to C4 | Clinical isolates from endoscopic tracheal secretions | Maintained in lab |
| P. aeruginosa | ||
| PAO1 | Wild type | 17 |
| PAO1, YFP tagged | Chromosomally encoded YFP | 18 |
Biofilm formation.
For mixed-culture biofilms, A. baumannii C4, a clinical isolate from tracheal secretion, was grown with P. aeruginosa PAO1, as C4 is resistant to amikacin and could be differentially separated from sensitive P. aeruginosa on LB agar plates containing amikacin. Pure-culture biofilms were formed by inoculating LB containing glycerol (1% vol/vol) with A. baumannii C4 or P. aeruginosa PAO1 (108 CFU/ml) in polystyrene tubes at 37°C under batch conditions for a period of 48 h. A binary-culture biofilm was formed by cocultivation of A. baumannii C4 and P. aeruginosa PAO1 (108 CFU/ml each) under similar conditions. The loosely adherent planktonic cells in the biofilm were removed by three washings with 10 mM phosphate-buffered saline (PBS, pH 7.0) while the tightly adherent cells were harvested by three consecutive cycles of alternate vortexing (15 s) and sonication (35% amplitude, 4 cycles of 30 s on and 30 s off) with the sonicator probe outside the tube in the water bath (19). Viable-cell counting of recovered cells was done to determine the growth of individual species in coculture using LB agar plates containing amikacin (100 μg/ml) for A. baumannii C4 and Pseudomonas agar with 0.03% (wt/vol) cetrimide for P. aeruginosa PAO1.
Microscopy of biofilms.
Electrocompetent A. baumannii M2 cells (20) were electroporated with pMU125 (21), an Escherichia coli-Acinetobacter shuttle vector expressing green fluorescent protein (GFP), using a Bio-Rad Gene Pulser (1.8 kV, 200 W, 25 μF) in a sterile prechilled electroporation cuvette (1 mm), and the transformants were selected on LB agar containing ampicillin (100 μg/ml). GFP was induced by tetracycline (100 ng/ml), and tagging of A. baumannii M2 was confirmed by fluorescence microscopy (excitation, 488 nm; emission, 530 nm) of the transformed culture.
The distribution of GFP-tagged A. baumannii M2 cocultured with yellow fluorescent protein (YFP)-tagged P. aeruginosa PAO1 in the mixed biofilm was studied by confocal laser scanning microscopy (CLSM) (Nikon). A one-day-old static biofilm was developed on the surface of a glass coverslip by immersing it in LB containing A. baumannii M2 and P. aeruginosa PAO1 (108 CFU/ml each) at 37°C. The coverslip was washed three times with 10 mM PBS (pH 7.0) to remove the weakly adherent cells. GFP fluorescence was recorded at 517 nm after excitation at 488 nm, while YFP fluorescence was captured at 527 nm after excitation at 514 nm. The spatial arrangement of both GFP-tagged A. baumannii and YFP-tagged P. aeruginosa under coculture conditions was observed at a magnification of ×40. Images from three independent biofilms were analyzed using NIS Elements AR 4.12.01 software, and the coefficient of variation (CoV) was calculated as the standard deviation divided by the mean fluorescence intensity (22).
Extraction of pyocyanin from P. aeruginosa PAO1.
P. aeruginosa PAO1 was inoculated in peptone water (2% [wt/vol], pH 7.0) and incubated at 37°C and 180 rpm for 24 h. The soluble pyocyanin pigment was extracted with chloroform, followed by re-extraction in 0.5 N HCl, and quantitated by measuring the absorbance at 520 nm (23). The aqueous fraction was lyophilized and dissolved in dimethyl sulfoxide (DMSO) to make a stock solution of 500 mM. The presence of pyocyanin in the extract was confirmed by liquid chromatography-mass spectrometry (LC-MS) (m/z as 210).
Effect of pyocyanin on A. baumannii growth.
A. baumannii was grown in the presence (250 μM) and absence of pyocyanin at 37°C for 24 h at 180 rpm. Viable cells which escaped pyocyanin stress converted triphenyl tetrazolium chloride (TTC) dye (0.005% [wt/vol]) to red formazan, which was quantitated at an optical density at 560 nm (OD560) (24). A culture grown without pyocyanin was treated as the control, and the percent survival was calculated as the OD560 of pyocyanin-treated cells divided by the OD560 of untreated cells.
Detection of pyocyanin-stimulated reactive oxygen species (ROS) in A. baumannii.
A. baumannii M2 cells were grown in the presence of pyocyanin (250 μM) as described earlier. Pyocyanin was removed by centrifugation at 8,000 × g for 10 min at 4°C. The cell pellet was suspended in PBS (pH 7.2) containing a 5 μM concentration of 5 (and 6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA; Invitrogen), and incubated at 37°C for 30 min in the dark (25). Stained cells were then pelleted and resuspended in PBS to obtain a dilute cell suspension (105 CFU/ml), a wet mount of which was imaged on an Olympus fluorescence microscope (excitation/emission, 495 nm/510 nm). Quantitation of fluorescence of the stained bacterial suspension was done on a plate reader fluorimeter (Spectramax) by taking the ratio of the OD510 to the OD600. A. baumannii M2 cells without exposure to pyocyanin were stained in a similar way to be used as controls.
Effect of pyocyanin on catalase and superoxide dismutase activities of A. baumannii. (i) Enzyme preparation.
Ten milliliters of LB was inoculated with 1% (vol/vol) of overnight grown culture of A. baumannii and grown in the presence (250 μM) and absence of pyocyanin at 37°C and 180 rpm for 16 h. The culture was centrifuged at 10,000 × g for 15 min at 4°C. The cell pellet was washed twice and resuspended in 2 ml of 50 mM phosphate buffer (pH 7.0) before disruption by sonication (five cycles of a pulse, 30 s on and 30 s off, 21% amplitude). The cell lysate was cleared of cell debris by centrifugation at 10,000 × g and 4°C for 15 min. The protein was quantitated at 280 nm with a Nanodrop spectrophotometer and used as crude enzyme (26).
(ii) Catalase assay.
H2O2 (500 μl; 5 mM) and phosphate buffer (400 μl; 50 mM, pH 7.0) were mixed with 100 μl of crude enzyme. The mixture was scanned at 240 nm every 10 s for 5 min. The enzyme solution containing H2O2-free phosphate buffer served as control. One unit of catalase activity was the amount of enzyme that decomposed 1 mM H2O2 per min at room temperature. The catalase activity was expressed as units per mg of protein (13).
(iii) Superoxide dismutase (SOD) assay.
The assay mixture contained 140 μl of 25 mM phosphate buffer (pH 8.3), 10 μl of 186 μM PMS (phenazine methosulfate), 30 μl of 300 μM NBT (nitroblue tetrazolium), and 100 μl of the enzyme preparation in a total volume of 280 μl. The reaction was initiated by the addition of 20 μl of 780 μM NADH, the mixture was incubated at 30°C for 90 s, and the reaction was stalled by the addition of 100 μl of glacial acetic acid. The reaction mixture was shaken with 400 μl of butanol, allowed to stand for 10 min, and centrifuged. The intensity of the chromogen in the butanol layer was measured at 560 nm. One unit of enzyme activity was the amount of enzyme that resulted in 50% inhibition of NBT reduction (27). The SOD activity was expressed as units per mg of protein.
Quorum sensing and oxidative stress in A. baumannii.
A 10-ml culture of A. baumannii M2 and its quorum-sensing mutant M2 (abaI::Km) were grown in the presence (0.5 and 5 μM) and absence of synthetic 3-OH-C12-HSL (Cayman Chemicals; 10 mM stock in dimethyl formamide), the quorum-sensing signal molecule of A. baumannii, at 37°C and 180 rpm for 16 h. Both the cultures were also grown in the absence and presence of 20 and 40 μM salicylic acid (50 mM stock in DMSO), a known quencher of quorum sensing, at 37°C and 180 rpm for 16 h. Enzyme assays for catalase and SOD were performed as described earlier. Overnight-grown cultures of A. baumannii M2 and M2 (abaI::Km) were inoculated (10% [vol/vol]) into 10 ml fresh LB broth. A 200-μl portion of each freshly inoculated culture was exposed to 100 mM H2O2 and incubated at 37°C and 180 rpm for 30 min. The reaction was stopped with 5% (wt/vol) sodium thiosulfate. Viable cells which escaped H2O2 stress were quantitated at OD560 with TTC dye (0.005% [wt/vol]) (24). The surviving cells were also enumerated as CFU by spreading appropriate dilutions of H2O2-treated culture on LB agar. Percent survival was calculated as the CFU in the H2O2-treated culture divided by the CFU in the untreated culture.
Determination of persister cell populations.
The MICs of carbenicillin and amikacin against A. baumannii M2 were determined (28). The persister cell population in A. baumannii M2 was determined in a time-dependent manner by growing cells to log phase followed by antibiotic exposure (MIC) at 37°C. Samples were withdrawn at various intervals (0, 3, 5, and 24 h), washed once to remove antibiotic completely, serially diluted, and spread on LB plates for a persister cell count (29). To determine dose-dependent persister cell formation in A. baumannii, the log-phase culture was exposed to high concentrations of antibiotic (3× MIC and 5× MIC) for 3 h followed by a wash with LB to remove antibiotic. The culture was then serially diluted and plated for enumeration of persister cells (30).
The effect of pyocyanin on persister cell formation by A. baumannii was determined by growing the cells in the presence (250 μM) of pyocyanin followed by challenge with antibiotic under conditions as described above. Following the challenge, the cells were washed, and an appropriate dilution was spread on LB agar for determination of surviving persister cells.
The nonheritable nature of the persister cells was determined by inoculating the persister cells surviving the antibiotic into LB without antibiotic and allowing it to grow for 24 h at 37°C. These cells were further reinoculated into LB with antibiotic (amikacin or carbenicillin at MIC). This cycle was repeated three times, and samples from each set were withdrawn from antibiotic medium at various times (0, 3, 5 and 24 h), washed once to remove antibiotic, and then plated on LB agar to determine persister cells.
Statistical analysis.
Data were expressed as means ± standard deviations (SD), and all experiments were repeated at least three times in triplicate. Student's t test was used to determine the significance. A P value of ≤0.05 was considered significant.
RESULTS
Biofilm formation by A. baumannii and P. aeruginosa.
Both A. baumannii C4 and P. aeruginosa PAO1 adhered well to polystyrene tubes used for the formation of biofilm under static conditions. After 4 h of incubation, 6.2 ± 1.6 and 6.5 ± 1.0 log CFU/ml of A. baumannii C4 and P. aeruginosa PAO1, respectively, were recovered from their pure-culture biofilms (Fig. 1a), which increased to 8.1 ± 0.81 and 7.2 ± 0.23 log CFU/ml, respectively, after 24 h. A. baumannii C4 counts decreased to 5.6 ± 0.81 log CFU/ml, compared to 7.5 ± 0.34 log CFU/ml for P. aeruginosa PAO1 after 48 h. When the coculture of biofilms composed of A. baumannii C4 and P. aeruginosa PAO1 was assessed, it was found that both showed growth patterns similar to those of the pure-culture biofilms (Fig. 1b). After 4 h of incubation, 2 ± 0.6 and 5.7 ± 1.4 log CFU/ml of A. baumannii C4 and P. aeruginosa PAO1 were recovered from mixed-culture biofilm, and the counts steadily increased to 4.7 ± 0.8 log CFU/ml and 7.5 ± 0.3 log CFU/ml, respectively, after 24 h, clearly indicating that the two organisms coexisted in the mixed-species biofilm. From 48-h-old cocultured biofilm, 4.8 ± 0.8 and 7.3 ± 0.3 log CFU/ml A. baumannii C4 and P. aeruginosa PAO1, respectively, were recovered. The decrease in A. baumannii C4 counts in mixed biofilm was similar to the trend seen in its pure-culture biofilm.
FIG 1.
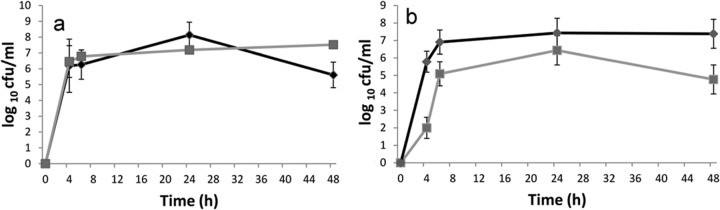
Viable-cell counts of A. baumannii C4 (■) and P. aeruginosa PAO1 (◆) from (a) pure cultures and (b) mixed-culture biofilms. A. baumannii C4 and P. aeruginosa PAO1 were cocultivated in LB to form batch biofilms at 37°C. The cells were harvested from the biofilm in a water bath sonicator at the indicated times. Viable-cell counts were obtained by spread plating appropriate dilutions on LB plates containing amikacin (100 μg/ml) and Pseudomonas agar plates (containing 0.03% cetrimide [wt/vol]), respectively. Data are means from three experiments.
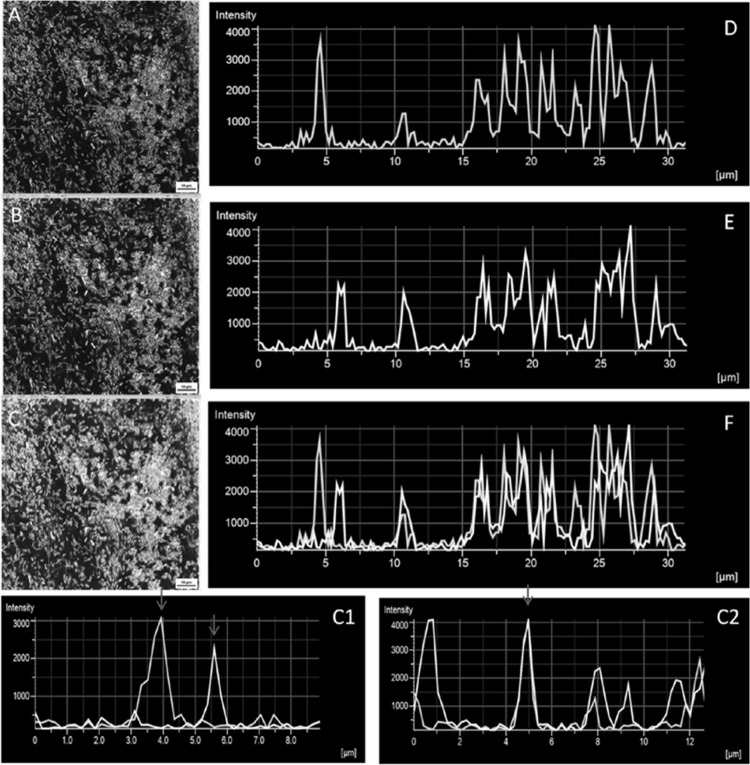
To study the spatial distribution of A. baumannii M2 and P. aeruginosa PAO1 in mixed-culture biofilm, GFP- and YFP-tagged cultures were used. Confocal microscopy confirmed that both A. baumannii M2 and P. aeruginosa PAO1 were able to coexist in mixed-species biofilms and formed mat-like structures that covered extensive surface areas of the glass coverslip (Fig. 2A, B, and C). P. aeruginosa PAO1 appeared to colonize the substratum before A. baumannii, as only yellow signal could be captured at 4 h of incubation (data not shown). Both green and yellow fluorescence signals of A. baumannii M2 and P. aeruginosa PAO1, respectively, were captured from the 6-h-old mixed biofilm specimen, indicating the coexistence of the two species. Intensity graphs of fluorescence generated from a 1-day-old mixed-species biofilm showed close proximity of the two signals, indicating cell-to-cell contact and coaggregation (Fig. 2C1). At certain places in the micrograph, a complete overlap was also observed (Fig. 2C2). Quantification of the variable distribution of A. baumannii M2 and P. aeruginosa PAO1 in 1-day-old biofilm was done by averaging the signal intensity of green and yellow fluorescence captured from three randomly selected areas of three different biofilms. The coefficients of variation of the averaged intensities of green and yellow fluorescence in the selected area were 0.22 and 0.32, respectively, indicating the coexistence of A. baumannii M2 and P. aeruginosa PAO1.
FIG 2.
(A to C) CLSM images of biofilms (cultured under batch conditions), formed on glass coverslips by (A) a GFP-tagged culture of A. baumannii M2, (B) a YFP-tagged culture of P. aeruginosa PAO1, and (C) cocultured A. baumannii M2 and P. aeruginosa PAO1. (D to F) Intensity graphs of fluorescence quantitated along the line indicated by arrows in the boxes in panels A, B, and C using NIS Elements viewer software. Highlighted area 1 in image C shows the two species in close proximity to each other (corresponding to the intensity graph in panel C1), and highlighted area 2 shows overlapping A. baumannii M2 and P. aeruginosa PAO1 cells (intensity graph in panel C2). Red arrows in panels C1 and C2 indicate intensity peaks. Magnification, ×63.
Response of A. baumannii to pyocyanin.
P. aeruginosa produces pyocyanin, a phenazine with redox activity, as one of the virulence factors which have been detected in lungs and sputum of cystic fibrosis patients. It has biocidal activity against coinfecting organisms. Since A. baumannii M2 and P. aeruginosa PAO1 were found to coexist, the effect of pyocyanin on the growth of A. baumannii M2 was checked, and the MIC was found to be >18 mM (data not shown). A. baumannii isolates grown in the presence of exogenous pyocyanin (250 μM) showed growth inhibition ranging from 0.8 to 15% only, indicating that A. baumannii isolates could survive pyocyanin-related stress (Fig. 3).
FIG 3.
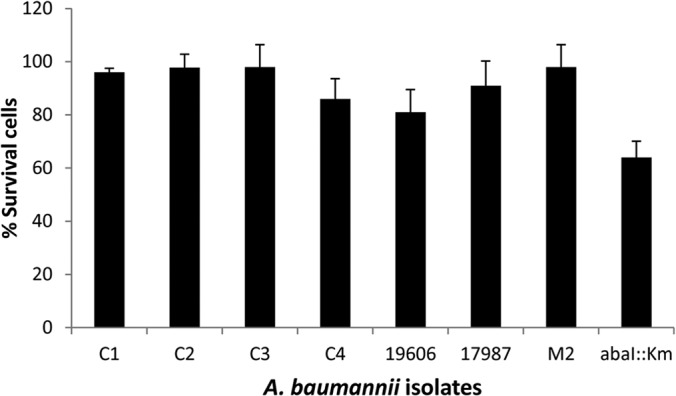
Survival profile of A. baumannii isolates (C1 to C4, M2 [clinical isolates], ATCC 19606, ATCC 17978, and the abaI::Km quorum-sensing mutant of M2) in the presence of pyocyanin (250 μM). Data are means from three experiments. The level corresponding to 100% survival in the absence of pyocyanin was 2 to 6 × 1012 CFU/ml in different isolates.
A. baumannii M2 cells grown in the presence of pyocyanin showed the generation of ROS (Fig. 4A and B), which led to 1.8-fold-higher (P < 0.001) fluorescence in pyocyanin-treated cells compared to the untreated cells stained with CM-H2DCFDA dye (Fig. 4C).
FIG 4.
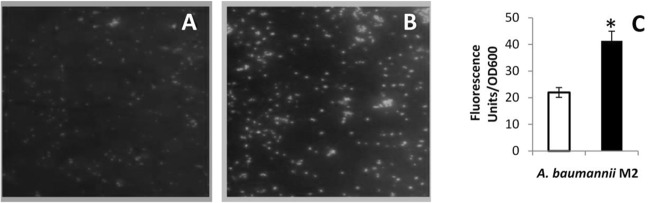
(A and B) Representative fluorescence microscope images of CM-H2DCFDA-stained A. baumannii M2 grown in the absence (A) and presence of pyocyanin (B). Magnification, ×40. (C) Quantitation of fluorescence in A. baumannii M2 grown in the absence (□) and presence (■) of pyocyanin. Data are means from three experiments. *, P ≤ 0.001.
In order to study the protective mechanism(s) against ROS in A. baumannii generated by pyocyanin, the levels of antioxidant enzymes were evaluated. Significant increases in catalase (1.3- to 6.9-fold) and SOD (1.2- to 6.1-fold) activities in the clinical isolates of A. baumannii were observed (Fig. 5a and b). In A. baumannii ATCC 17978, catalase and SOD activities increased 3.3 (P ≤ 0.01)- and 1.1-fold, respectively, whereas A. baumannii ATCC 19606 showed 2.3-fold (P ≤ 0.05) less catalase activity 1.6-fold (P ≤ 0.05) more SOD activity (Fig. 5b), which alone may not have been sufficient to protect it against pyocyanin stress. This was reflected as maximum (15%) growth inhibition of this strain in the presence of pyocyanin (Fig. 3).
FIG 5.
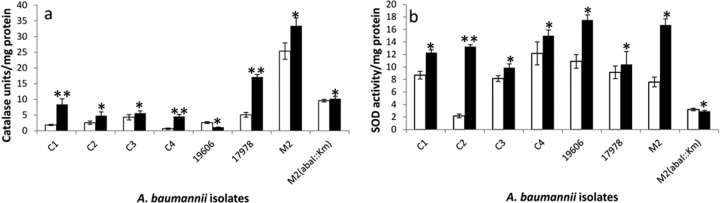
Catalase (a) and SOD (b) activities in A. baumannii isolates (C1 to C4, M2 [clinical isolates], ATCC 19606, ATCC 17978, and the abaI::Km quorum-sensing mutant of M2) grown in the absence (□) and presence (■) of pyocyanin. Data are means from three experiments. *, P ≤ 0.05; **, P ≤ 0.01.
Quorum sensing and oxidative stress management in A. baumannii.
A. baumannii M2 (abaI::Km), the quorum-sensing mutant, showed 30% growth inhibition, compared to only a 4% reduction in growth of the wild type, in response to pyocyanin (Fig. 3). Exposure of A. baumannii M2 to H2O2 (100 mM) also resulted in >2-fold-higher (P < 0.001) survival in the wild type than the quorum-sensing mutant of M2, as determined by TTC dye reduction assay. This was also confirmed from viable-cell counts of M2 (6.6 ± 0.6 log CFU/ml) and M2 (abaI::Km) (6.3 ± 0.6 log CFU/ml) after exposure to H2O2, which showed 55 and 35% survival compared to the unexposed cells (7 ± 0.3 log CFU/ml was taken as 100% survival).
The catalase and SOD activities in the mutant were 3-fold (P ≤ 0.001) and 2.6-fold (P ≤ 0.001) lower than those in its wild-type strain, which could be the reason for its increased sensitivity to pyocyanin compared to the wild type (Fig. 6a and b). Catalase and SOD activities of M2 (abaI::Km) increased 2-fold (P ≤ 0.001) and 1.7-fold (P ≤ 0.001) in response to pyocyanin in the presence of exogenously added 5 μM 3-OH-C12 homoserine lactone (HSL), the quorum-sensing signal of A. baumannii (Fig. 6a and b), although a lower concentration (0.5 μM) was not effective. The wild-type strain did not show any appreciable change in the catalase and SOD activities on addition of 3-OH-C12-HSL, as it has a quorum sensing system that produces 3-OH-C12-HSL. Likewise, salicylic acid, a known quencher of quorum sensing, showed a significant (P < 0.01) decrease in catalase and SOD activities of A. baumannii M2 in a concentration-dependent manner (20 and 40 μM), while there was no effect on the quorum-sensing mutant strain, indicating that the expression of catalase and SOD activities is under the control of the quorum-sensing system in A. baumannii (Fig. 7a and b).
FIG 6.
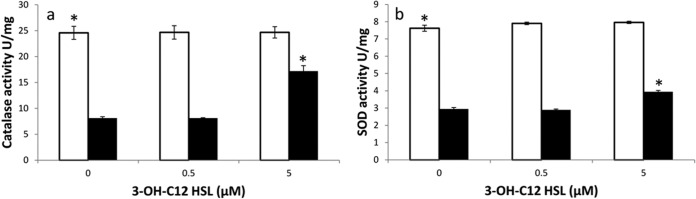
Catalase (a) and SOD (b) activities in A. baumannii M2 (□) and its autoinducer synthase mutant A. baumannii M2 (abaI::Km) (■) grown in the presence of 3-OH-C12-HSL. Data are means from three experiments. *, P ≤ 0.001.
FIG 7.
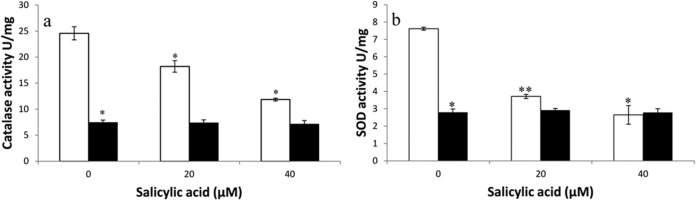
Catalase (a) and SOD (b) activities in A. baumannii M2 (□) and its autoinducer synthase mutant A. baumannii M2 (abaI::Km) (■) grown in the presence of salicylic acid. Data are means from three experiments. *, P ≤ 0.01; **, P ≤ 0.001.
Exogenous pyocyanin increases the persister cell population of A. baumannii.
Persister cells are a bacterial subpopulation that is tolerant to killing by antibiotics, while the majority of the sensitive cells are killed. Exposure of a logarithmic-phase culture of A. baumannii M2 to the MICs of amikacin (4 μg/ml) and carbenicillin (50 μg/ml) in a time-dependent assay showed biphasic killing. A rapid decline in the antibiotic-sensitive population was followed by a plateau formed by antibiotic-tolerant persister cells. Amikacin and carbenicillin exposure led to the generation of 0.07 and 0.02% persister cells, which significantly (P ≤ 0.05) increased 4- and 3-fold, respectively, in the presence of pyocyanin (Fig. 8a and b). A similar biphasic killing pattern was observed when A. baumannii M2 was exposed to amikacin and carbenicillin at concentrations well above their MICs (3× and 5×), and once again, pyocyanin increased the number of persister cells significantly (data not shown). Since multidrug tolerance is an epigenetic, nonheritable change, the persister cells formed in response to amikacin and carbenicillin were shown to retain their sensitivity to these antibiotics when regrown in LB. This was confirmed by performing the assay for three consecutive cycles (Fig. 9a and b).
FIG 8.
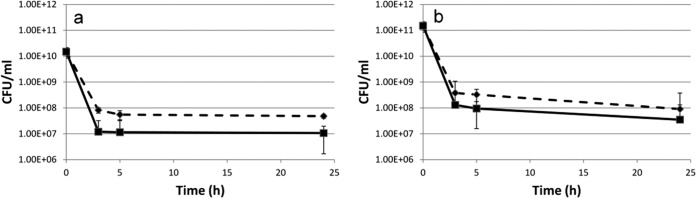
Effect of pyocyanin on the A. baumannii M2 persister cell population. The culture was grown to mid-log phase at 37°C with aeration and challenged with (a) amikacin and (b) carbenicillin at MIC in the presence (◆) and absence (■) of pyocyanin.
FIG 9.
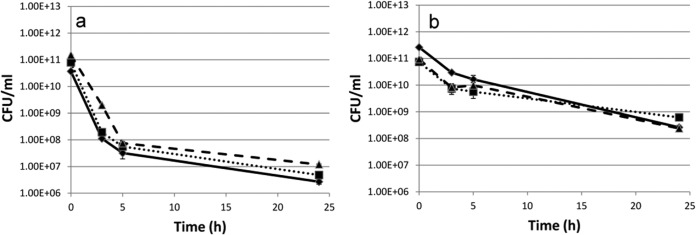
Nonheritability of the A. baumannii M2 persister phenotype on exposure to (a) 4 μg/ml of amikacin or (b) 50 μg/ml of carbenicillin. An overnight culture of A. baumannii M2 was diluted 1:20 with LB and exposed to antibiotics for 24 h. The surviving cells were then regrown in fresh antibiotic-free LB until stationary phase and exposed to antibiotic again for 24 h. The procedure was repeated three times. ◆, passage 1; ■, passage 2; ▲, passage 3.
DISCUSSION
Mixed-species biofilms are ubiquitous and are not uncommon in hospital environments. P. aeruginosa is a major cause of chronic lung infection in cystic fibrosis patients and may form mixed-species biofilms with coinfecting pathogens. A. baumannii is an emerging human pathogen having overlapping sites of infection with P. aeruginosa and has been shown previously in our lab to stably coexist with it under planktonic growth conditions (10). The present study shows a stable in vitro association of the two in a static mixed-species biofilm using fluorescent-dye-tagged A. baumannii and P. aeruginosa cultures.
P. aeruginosa secretes pyocyanin, a redox-active compound which generates ROS. Pyocyanin in its reduced form is highly unstable and reacts with molecular O2 to form superoxide (O2−), hydrogen peroxide (H2O2), or hydroxyl radical (OH−), all of which affect the growth and viability of a number of organisms (31). This benefits P. aeruginosa by reducing competition from coinfecting pathogens during infection. However, pyocyanin showed no growth inhibition of A. baumannii (MIC > 18 mM), as it counteracted the pyocyanin-mediated oxidative stress by enhanced production of catalase and SOD. A. baumannii has been reported to combat H2O2-generated oxidative stress by increasing its catalase activity (32).
Pyocyanin exerts oxidative stress on P. aeruginosa cells also, but it survives because of limited redox cycling of pyocyanin and increased levels of catalase and SOD (7). The production of antioxidant enzymes is controlled by the quorum-sensing system in P. aeruginosa (13), but no such relationship has been reported for A. baumannii. In the present study, we report for the first time that catalase and SOD are under the control of a quorum-sensing system in A. baumannii. A quorum-sensing mutant of A. baumannii M2, M2 (abaI::Km), lacking the autoinducer synthase gene, had significantly low levels of catalase and SOD enzymes even on exposure to pyocyanin, compared to its wild type. Supplementation of the mutant with 3-OH-C12-HSL, the quorum-sensing signal molecule in A. baumannii, increased catalase and SOD enzyme activities significantly. Likewise, interfering with quorum sensing in A. baumannii M2 by salicylic acid, a known quencher of quorum sensing (33), reduced the catalase and SOD levels. Greater resistance of A. baumannii M2 to H2O2 than its quorum-sensing mutant further supports the idea that quorum sensing is involved in tolerance of oxidative stress. Hence, a combination of a quorum-sensing quencher and ROS-generating agents like H2O2 would be more effective in disinfection of fomites to control A. baumannii, which can persist in the hospital environment for a long time.
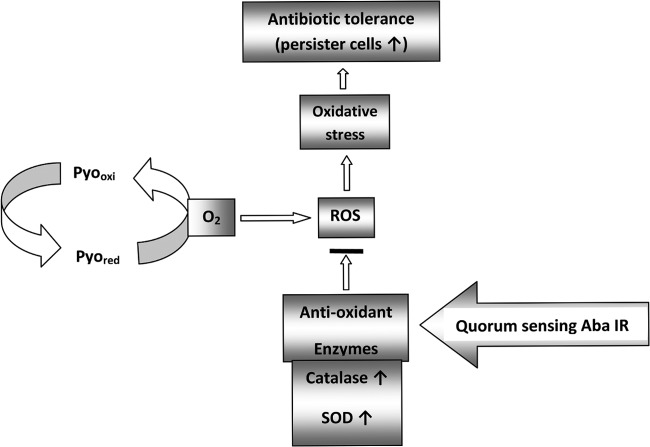
Bacterial tolerance to antibiotics has been linked with the emergence of persister cells, the phenotypic variants that are multidrug tolerant and are responsible for chronicity of infections. Persistence against antibiotics is genus or even strain specific, as P. aeruginosa has been reported to produce significantly fewer persisters than E. coli and Staphylococcus aureus under standard laboratory conditions (30), and persister cell formation in A. baumannii showed a highly heterogeneous pattern, ranging from 0.0007 to 10.1% (polymyxin B) and 0.0003 to 11.84% (tobramycin) in different clinical strains of A. baumannii (1). The present study shows persister cell formation in A. baumannii in response to carbenicillin (an extended-spectrum β-lactam) and amikacin (an aminoglycoside). These studies indicate that the persistence phenomenon is widely distributed among clinical A. baumannii strains, probably due to the antibiotic stress prevalent in hospital settings that may also be responsible for the selection of genetic determinants at different intensities in different strains. However, the genes involved in the persistence phenotype in A. baumannii are yet to be decoded. Since carbenicillin kills dividing cells only and amikacin kills both dividing and nondividing cells, it seems that for every antibiotic, specific sets of genes are induced for persistence. Many antibiotics have been shown to induce respiratory stress by generating ROS for killing the cells. Increased levels of catalase and SOD in A. baumannii in response, as shown in this work for pyocyanin-induced stress as well, may protect the cells which have developed persistence as a mechanism to survive (Fig. 10). Pyocyanin significantly (P ≤ 0.05) enhanced the persister cell formation induced by amikacin and carbenicillin. A 90-fold increase in the fraction of persister cells in P. aeruginosa in response to pyocyanin or 3-oxo-C12 HSL (its quorum-sensing signal molecule) has been reported (30). These observations have clinical relevance, as pyocyanin has been detected in the sputum and pulmonary secretions of cystic fibrosis patients with P. aeruginosa infections (16). These studies warrant further investigations, as coinfection with A. baumannii and P. aeruginosa could have serious implications for disease management.
FIG 10.
Proposed mechanism of A. baumannii survival of pyocyanin-induced oxidative stress.
ACKNOWLEDGMENTS
We are grateful to P. N. Rather, B. H. Iglewski, S. Molin, and L. A. Actis for providing the strains used in the study.
Nidhi Bhargava acknowledges Council of Scientific and Industrial Research, New Delhi, India for Senior Research Fellowship.
We declare that no competing interests exist.
Footnotes
Published ahead of print 2 June 2014
REFERENCES
- 1.Barth VC, Jr, Rodrigues BA, Bonatto GD, Gallo SW, Pagnussatti VE, Ferreira CAS, Oliveira DS. 2013. Heterogeneous persister cells formation in Acinetobacter baumannii. PLoS One 8:e84361. 10.1371/journal.pone.0084361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiester SE, Actis LA. 2013. Stress responses in the opportunistic pathogen Acinetobacter baumannii. Future Microbiol. 8:353–365. 10.2217/fmb.12.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dent LL, Marshall DR, Pratap S, Hulette RB. 2010. Multidrug resistant Acinetobacter baumannii: a descriptive study in a city hospital. BMC Infect. Dis. 10:196–202. 10.1186/1471-2334-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riedel K, Hentzer M, Geisenberger O, Huber B, Steidle A, Wu H, Hoiby N, Givskov M, Molin S, Eberl L. 2001. N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147:3249–3262. [DOI] [PubMed] [Google Scholar]
- 5.Wagner VE, Iglewski BH. 2008. P. aeruginosa biofilms in CF infections. Clin. Rev. Allergy Immunol. 35:124–134. 10.1007/s12016-008-8079-9. [DOI] [PubMed] [Google Scholar]
- 6.Rogers GB, Carroll MP, Hoffman LR, Walker AW, Fine DA, Bruce KD. 2010. Comparing the microbiota of the cystic fibrosis lung and human gut. Gut Microbes 1:85–93. 10.4161/gmic.1.2.11350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassett DJ, Charniga L, Bean K, Ohman DE, Cohen MS. 1992. Response of Pseudomonas aeruginosa to pyocyanin: mechanisms of resistance, antioxidant defences, and demonstration of a manganese-cofactored superoxide dismutase. Infect. Immun. 60:328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomlin KL, Coll OP, Ceri H. 2001. Interspecies biofilms of Pseudomonas aeruginosa and Burkholderia cepacia. Can. J. Microbiol. 47:949–954. [PubMed] [Google Scholar]
- 9.Stewart PS, Camper AK, Handran SD, Huang CT, Warnecke M. 1997. Spatial distribution and co-existence of Klebsiella pneumonia and Pseudomonas aeruginosa in biofilms. Microb. Ecol. 33:2–10. 10.1007/s002489900002. [DOI] [PubMed] [Google Scholar]
- 10.Bhargava N, Sharma P, Capalash N. 2012. N-acyl homoserine lactone mediated interspecies interactions between A. baumannii and P. aeruginosa. Biofouling 28:813–822. 10.1080/08927014.2012.714372. [DOI] [PubMed] [Google Scholar]
- 11.Hobley L, King JR, Sockett E. 2006. Bdellovibrio predation in the presence of decoys: three-way bacterial interactions revealed by mathematical and experimental analyses. Appl. Environ. Microbiol. 72:6757–6765. 10.1128/AEM.00844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sies H. 1997. Oxidative stress: oxidants and antioxidants. Exp. Physiol. 82:291–295. [DOI] [PubMed] [Google Scholar]
- 13.Hassett DJ, Ma J-F, Elkins JG, McDermott TR, Ochsner UA, West SEA, Huang C-T, Fredericks J, Burnett S, Stewart PS, McFeters G, Passador L, Iglewski BH. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34:1082–1093. 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 14.Niu C, Clemmer KM, Bonomo RA, Rather PN. 2008. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 190:3386–3392. 10.1128/JB.01929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Wood TK. 2011. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 77:5577–5583. 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caldwell CC, Chen Y, Goetzmann HS, Hao Y, Borchers MT, Hassett DJ, Young LR, Mavrodi D, Thomashow L, Lau GW. 2009. Pseudomonas aeruginosa exotoxin pyocyanin causes cystic fibrosis airway pathogenesis. Am. J. Pathol. 175:2473–2488. 10.2353/ajpath.2009.090166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iglewski BH, Pesci EC, Pearson JP, Seed PC. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haagensen JAS, Klausen M, Ernst RK, Miller SI, Folkesson A, Tolker-Nielsen T, Molin S. 2007. Differentiation and distribution of colistin- and sodium dodecyl sulfate-tolerant cells in Pseudomonas aeruginosa biofilms. J. Bacteriol. 189:28–37. 10.1128/JB.00720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraigsley AM, Finkel SE. 2009. Adaptive evolution in single species bacterial biofilms. FEMS Microbiol. Lett. 293:135–140. 10.1111/j.1574-6968.2009.01526.x. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 21.Dorsey CW, Tomaras AP, Actis LA. 2002. Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl. Environ. Microbiol. 68:6353–6360. 10.1128/AEM.68.12.6353-6360.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart PS, Camper AK, Handran SD, Huang C-T, Warnecke M. 1997. Spatial distribution and coexistence of Klebsiella pneumonia and Pseudomonas aeruginosa in biofilms. Microb. Ecol. 33:2–10. 10.1007/s002489900002. [DOI] [PubMed] [Google Scholar]
- 23.Essar DW, Eberly L, Hadero A, Crawford IP. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S, Kim MJ, Kang HY, Seol SY, Cho DT, Kim J. 2010. A simple colorimetric method for testing antimicrobial susceptibility of biofilmed bacteria. J. Microbiol. 48:709–711. 10.1007/s12275-010-0299-z. [DOI] [PubMed] [Google Scholar]
- 25.Hillegass JM, Shukla A, MacPherson MB, Lathrop SA, Alexeeva V, Perkins T, Vliet AVA, Vacek PM, Gunter ME, Brooke T. 2010. Mechanisms of oxidative stress and alterations in gene expression by Libby six-mix in human mesothelial cells. Part. Fibre Toxicol. 7:26–40. 10.1186/1743-8977-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desjardins P, Hansen JB, Allen M. 2009. Microvolume protein concentration determination using the NanoDrop 2000c spectrophotometer. J. Vis. Exp. 2009(33):e1610 http://www.jove.com/details.php?id=1610. 10.3791/1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kakkar P, Das B, Viswanathan PN. 1984. A modified spectrophotometric assay of superoxide dismutase. Indian J. Biochem. Biophys. 21:130–132. [PubMed] [Google Scholar]
- 28.Andrews JM. 2001. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48:5–16. 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 29.Leung V, Levesque CM. 2012. A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J. Bacteriol. 194:2265–2274. 10.1128/JB.06707-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moker N, Dean CR, Tao J. 2010. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum sensing signaling molecules. J. Bacteriol. 192:1946–1955. 10.1128/JB.01231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price-Whelan A, Dietrich LE, Newman DK. 2006. Rethinking ‘secondary' metabolism: physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2:71–78. 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 32.Di Capua C, Bortolotti A, Farias ME, Cortez N. 2011. UV-resistant Acinetobacter sp. isolates from Andean wetlands display high catalase activity. FEMS Microbiol. Lett. 317:181–189. 10.1111/j.1574-6968.2011.02231.x. [DOI] [PubMed] [Google Scholar]
- 33.Rasmussen TB, Givskov M. 2006. Quorum sensing inhibitors; a bargain of effects. Microbiol. 152:895–904. 10.1099/mic.0.28601-0. [DOI] [PubMed] [Google Scholar]