Abstract
The ESX-1 secreted virulence factor ESAT-6 is one of the major and most well-studied virulence factors of Mycobacterium tuberculosis, given that its inactivation severely attenuates virulent mycobacteria. In this work, we show that clinical isolates of M. tuberculosis produce and secrete larger amounts of ESAT-6 than the widely used M. tuberculosis H37Rv laboratory strain. A search for the genetic polymorphisms underlying this observation showed that whiB6 (rv3862c), a gene upstream of the ESX-1 genetic locus that has not previously been found to be implicated in the regulation of the ESX-1 secretory apparatus, presents a unique single nucleotide insertion in its promoter region in strains H37Rv and H37Ra. This polymorphism is not present in any of the other publicly available M. tuberculosis complex genomes or in any of the 76 clinical M. tuberculosis isolates analyzed in our laboratory. We demonstrate that in consequence, the virulence master regulator PhoP downregulates whiB6 expression in H37Rv, while it upregulates its expression in clinical strains. Importantly, reintroduction of the wild-type (WT) copy of whiB6 in H37Rv restored ESAT-6 production and secretion to the level of clinical strains. Hence, we provide clear evidence that in M. tuberculosis—with the exception of the H37Rv strain—ESX-1 expression is regulated by WhiB6 as part of the PhoP regulon, which adds another level of complexity to the regulation of ESAT-6 secretion with a potential role in virulence adaptation.
INTRODUCTION
Mycobacterium tuberculosis represents the paradigm of an intracellular pathogen, which has been suggested to have coevolved with human beings for a long time (1, 2). This pathogen is transmitted to new hosts by aerosol droplets and reaches alveoli, where it is engulfed by resident macrophages. Following phagocytosis, bacilli reside in phagosomes, exposing the bacilli to a harsh environment evolved to destroy invading agents (3). Many pathogenic bacteria have evolved mechanisms that interfere with the host-pathogen interplay in the phagosome, which largely depends on secretion of virulence effectors translocated through the bacterial cell wall by multiprotein complexes forming a secretory apparatus. Some relevant examples include the type III secretion systems of Salmonella spp., which allow pathogen invasion and replication (4), the Dot/Icm type IV secretion system of Legionella pneumophila, which translocates over 200 effectors to hijack various host cell processes (5), and secretion of listeriolysin O by Listeria monocytogenes, which mediates membrane lysis and cell-to-cell spread (6). M. tuberculosis also possesses a variety of mechanisms to efficiently survive within the phagosomal compartment (7) or escape from it (8, 9). Specifically, the type VII secretion system, namely, the ESX-1 system that allows specialized protein secretion of the 6-kDa early antigenic target (ESAT-6) and its partner protein, the 10-kDa culture filtrate protein CFP-10, through the mycobacterial cell envelope (10). The M. tuberculosis genome contains five ESX paralogs (11), and two of them, ESX-1 and ESX-5, are implicated in virulence (8, 12, 13). The ESX-1-encoding core region spans from espE (rv3864) to mycP1 (rv3883c) but also includes trans-acting elements outside the core region, namely, extended ESX-1 (i.e., espR [rv3849] and espACD [rv3616c to rv3614c]) (14, 15) (Fig. 1A).
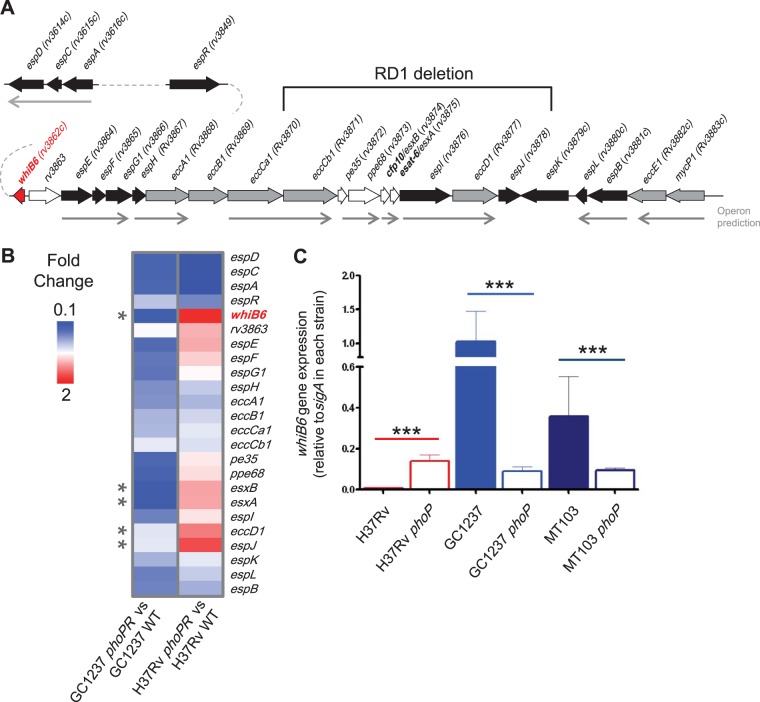
FIG 1.
The M. tuberculosis whiB6 gene is differentially regulated by PhoP, depending on the genetic background. (A) Schematic representation of ESX-1 and extended ESX-1 genes from M. tuberculosis. The diagram shows genetic regions from whiB6 (rv3862c) to mycP1 (rv3883c), espD (rv3614c) to espA (rv3616c), and espR (rv3849). Genes are represented by filled arrows and colored according to their putative function. Predicted operons are represented by thin arrows. The RD1 region deleted in M. bovis BCG is also indicated. (B) Heat map from microarray comparisons of H37Rv and GC1237 with their respective phoPR mutants. Only genes involved in ESAT-6 production and secretion are reported for clarity. The GC1237 phoPR mutant displays downregulation of most ESX-1 genes. In contrast, the H37Rv reference strain show divergent PhoP regulation of whiB6, esxB (rv3874), esxA (rv3875), eccD1 (rv3877), and espJ (rv3878) (indicated by asterisks) compared to strain GC1237. Note that whiB6 shows the most divergent PhoP regulation between both strains. (C) qRT-PCR analyses showing PhoP regulation over whiB6 in the H37Rv strain compared to those of MT103 and GC1237 clinical isolates. Note the divergent PhoP regulation in clinical strains compared to that of the reference standard H37Rv. Bars indicate fold changes in gene expression relative to the sigA gene used as endogenous control. Results represent the average of three independent experiments, and error bars indicate the standard deviation (SD) of the mean.
Two ESX-1-secreted proteins have attracted the most attention in recent years: ESAT-6 (EsxA [Rv3875]) and CFP-10 (EsxB [Rv3874]); both are translocated, forming a 1:1 heterodimer (16), and might dissociate under the acidic conditions found in the phagosome (17). Several roles have been attributed to ESAT-6, ranging from a secreted effector that induces macrophage apoptosis to a membrane lytic factor that enables phagosomal escape (9, 18–20). Overall, the ESX-1 region is considered a major pathogenic determinant of M. tuberculosis, and its absence, due to the deletion of the RD1 region (Fig. 1A), from the Mycbacterium bovis bacillus Calmette-Guérin (BCG) vaccine is thought to substantially contribute to the attenuation and safety of this strain (21). Although the core elements required for ESAT-6 secretion have been characterized (8, 22), other trans-acting proteins remain to be elucidated. Here we report that whiB6—a gene adjacent to the ESX-1 genetic locus—carries a point mutation in its promoter region that is exclusively found in H37R strain derivatives H37Rv and H37Ra. We provide evidence that this polymorphism leads to a differently regulated whiB6 expression by PhoP, which ultimately results in different ESAT-6 levels between H37Rv and clinical M. tuberculosis isolates.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The Mycobacterium tuberculosis H37Rv and GC1237 phoP mutants and wild-type (WT) strains were previously described (23). MT103, its isogenic phoP mutant (SO2), and the phoP complemented strain were also described elsewhere (24). Construction of the vaccine candidate MTBVAC by unmarked deletions of phoP and fadD26 genes was recently described (25). M. tuberculosis H37Rv::KIMt, H37Rv::KIRv, H37Rv phoP::KIMt, and H37Rv phoP::KIRv were constructed in this work. Mycobacterial strains were grown at 37°C in 7H9 medium (Difco) supplemented with 0.05% Tween 80 and 10% albumin-dextrose-catalase (ADC) (Middlebrook) or on 7H10 plates supplemented with 10% ADC. Escherichia coli DH5α used for cloning procedures was grown at 37°C in LB broth or on LB agar plates. Kanamycin (20 μg/ml) and hygromycin (20 μg/ml) were used as appropriate.
Plasmid construction.
Sequences of whiB6 and its own promoter were amplified from H37Rv (KIRv) and MT103 (KIMt) genomic DNA using the primers whiB6-KI fw and whiB6-KI rv (Table 1). PCR products were digested with NheI and inserted into the unique NheI site of the integrative plasmid pMV361 (26). Plasmids were analyzed by sequencing, confirming the polymorphisms representative of each strain in the final construction.
TABLE 1.
Primers used in this work
| Primera | Use | Sequence |
|---|---|---|
| RT whiB6 fw | qRT-PCR | CGCGGCAGAGGCTACAAC |
| RT whiB6 rv | qRT-PCR | GGCGGTTACTGTCATGTCTACGT |
| whiB6seq fw | Sequencing | ATCCCTTATTCGCGGGTACT |
| whiB6seq rv | Sequencing | GGGCGGTTACTGTCATGTCT |
| RT whiB6 prom fw | ChIP qRT-PCR | GCGCACCGCCGAGTAC |
| RT whiB6 prom rw | ChIP qRT-PCR | CGGCGAGGCCAGATACAC |
| RT cfp10 fw | qRT-PCR | GCAGGAGGCAGGTAATTTCG |
| RT cfp10 rv | qRT-PCR | CCTGGTCGATCTGGGTTTTC |
| RT esat6 fw | qRT-PCR | AGGGTGTCCAGCAAAAATGG |
| RT esat6 rv | qRT-PCR | CTGCAGCGCGTTGTTCAG |
| RT espA fw | qRT-PCR | GGCACCCTCGGAGAAGTGT |
| RT espA rv | qRT-PCR | AGCTCTTTCAGGCCGTTGAG |
| RT espC fw | qRT-PCR | TGTACTTGACTGCCCACAATGC |
| RT espC rv | qRT-PCR | TCGACACCGGCCGTATG |
| RT espD fw | qRT-PCR | CAACAGGTCGATGCAGATGAA |
| RT espD rv | qRT-PCR | TCGCCCACGGTCTTACGTA |
| whiB6-KI fw | Knock-in mutant | CAGGCTAGCGCCTGCAAAACGTCAATCTC |
| whiB6-KI rv | Knock-in mutant | CGCGCTAGCCACCGTCCATCATCGGATAC |
All primers were designed in this study. fw, forward; rv, reverse.
Sequence analysis of clinical isolates and spoligotyping.
H37Rv sequence (GenBank accession no. AL123456.3) was compared, in the whiB6 promoter region, to available sequenced genomes of the Mycobacterium complex (ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/) using NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Clinical isolate genomic DNAs from different lineages were sequenced using the primers whiB6seq fw and whiB6seq rv. Spoligotyping was performed as described in reference 27, and the results were compared with those in the SpolDB4 database (28). The site http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE is a proprietary database maintained at the Pasteur Institute of Guadeloupe, which contains both spoligotype and mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) patterns of M. tuberculosis. At the time of this comparison (March 2013), the database contained data on about 70,000 strains from 160 countries of origin.
ChIP experiments.
Chromatin immunoprecipitation (ChIP) experiments were performed as previously described (29) with MT103, MTBVAC, and H37Rv and its phoP mutant, with the following modifications. Briefly, M. tuberculosis cultures were grown to the exponential phase (optical density at 600 nm [OD600] of 0.4) and cross-linked with 1% formaldehyde for 10 min at 37°C. Cross-linking was quenched by addition of glycine (125 mM). Cells were then washed twice with Tris-buffered saline (TBS) (20 mM Tris-HCl [pH 7.5], 150 mM NaCl), resuspended in 4 ml immunoprecipitation (IP) buffer (50 mM HEPES-KOH [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail from Roche), and sonicated to shear DNA with a Bioruptor (Diagenode). Cell debris was removed by centrifugation, and the supernatant was used in IP experiments. Nucleoprotein extracts were incubated with 50 μl of rabbit polyclonal anti-PhoP antibodies at 4°C for 2 days on a rotating wheel. Complexes were subsequently precipitated with Dynal Dynabeads (anti-rabbit; Invitrogen) for 3 h at 4°C. Beads were washed twice with IP buffer, once with IP buffer plus 500 mM NaCl, once with buffer III (10 mM Tris-HCl [pH 8], 250 mM LiCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate), and once with Tris-EDTA buffer (pH 7.5). Elution was performed in 50 mM Tris-HCl (pH 7.5)–10 mM EDTA–1% SDS for 40 min at 65°C. Samples were finally treated with RNase A for 1 h at 37°C, and cross-links were reversed by incubation for 2 h at 50°C and for 8 h at 65°C in 0.5× elution buffer with 50 μg proteinase K (Eurogentec). DNA was purified by phenol-chloroform extraction and quantified by Nanodrop and Qubit fluorometer according to the manufacturer's recommendations (Invitrogen).
RNA extraction and qRT-PCR experiments.
Cultures of MT103, MTBVAC, GC1237 and its phoP mutant, H37Rv and its phoP mutant and their knock-in (KI) derivatives were grown to exponential phase (OD600 of 0.5 to 0.6) and pelleted by centrifugation. To minimize RNA degradation, bacteria were resuspended in 1 ml RNAprotect bacterial reagent (Qiagen), incubated for 5 min at room temperature, and then centrifuged. Bacterial pellets were resuspended in 0.4 ml lysis buffer (0.5% SDS, 20 mM NaAc, 0.1 mM EDTA) and 1 ml 1:1 phenol-chloroform (pH 4.5). Suspensions were transferred to tubes containing glass beads (Qbiogene) and lysed using a ribolyser (Fast-prep instrument) with a three-cycle program (15 s at speed 6.5 m/s) including cooling the samples on ice for 5 min between pulses. Samples were then centrifuged, and the homogenate was removed from the beads and transferred to a tube containing 24:1 chloroform-isoamyl alcohol. Tubes were inverted carefully before centrifugation, and the upper (aqueous) phase was then transferred to a fresh tube containing 0.3 M Na-acetate (pH 5.5) and isopropanol. Precipitated nucleic acids were collected by centrifugation. The pellets were rinsed with 70% ethanol and air dried before being redissolved in RNase-free water. DNA was removed from RNA samples with Turbo DNA free (Ambion) by incubation at 37°C for 1 h. RNA integrity was assessed by agarose gel electrophoresis, and absence of contaminating DNA was checked by lack of amplification products after 30 PCR cycles. One microgram of RNA was converted to cDNA using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's recommendations. All PCR primers were designed using Primer Express software (Applied Biosystems). The PCR mixture consisted of 1× SYBR green PCR master mix (Applied Biosystems), 0.25 μM each primer, and 1 μl of 1:10 diluted cDNA or IP DNA from immunoprecipitation reactions (total volume, 10 μl). Reactions were carried out in triplicate in an Applied Biosystems StepOnePlus sequence detection system (Applied Biosystems) according to the manufacturer's instructions. Melting curves were constructed to ensure that only one amplification product was obtained. In the case of quantitative reverse transcription-PCR (qRT-PCR) for microarray data confirmation, normalization was obtained to the number of sigA molecules in each sample. Regarding the qPCR for ChIP-seq data validation, the number of target molecules was normalized to the mutant (control) sample after subtraction of the background represented by the mock IP (no-antibody control). Primers used in these experiments are listed in Table 1.
Transcriptome analysis.
Transcriptome analysis was performed following a previously used protocol (30). In brief, RNA quality was monitored with a Bioanalyzer RNA Nano assay (Bio-Rad), and appropriate preparations were used for further hybridization experiments. Two biological replicates were done for each RNA sample. The design of oligonucleotides covering all protein coding sequences was done using OligoArray version 2.1 (31) on the basis of the 3,924 predicted coding sequences composing the entire M. tuberculosis genome (32). We used Agilent-manufactured customized microarrays. The reverse transcription and indirect labeling with Cy5 or Cy3 (GE Healthcare Life Sciences) were performed with the SuperScript indirect cDNA labeling system (Invitrogen), and cDNA hybridization was performed as described by the manufacturer (Agilent). Expression of all genes on the array was simultaneously analyzed through competitive hybridization of the probes and scanning. Signal quantification for each spot was performed with the image analysis GenePix Pro 6.0 software (Axon Instruments). All data generated were then imported and statistically analyzed by using the statistical programming language R (R 2.0.1 at http://www.R-project.org) as described previously (30).
Protein extraction and Western blot procedures.
In order to avoid albumin contamination in the secreted protein fraction, cultures of MT103, SO2, H37Rv and its phoP mutant, H37Rv::KIMt, H37Rv::KIRv, H37Rv phoP::KIMt, and H37Rv phoP::KIRv were grown in 7H9 (Difco)–0.05% Tween 80 supplemented with 0.2% dextrose–0.085% NaCl. After 2 to 3 weeks of incubation at 37°C, cultures were pelleted by centrifugation. The supernatant containing secreted proteins was incubated with 10% trichloroacetic acid (TCA) for 1 h in ice and then centrifuged at 4°C for 30 min. Pelleted proteins were rinsed with cold acetone and then resuspended in 150 mM Tris-HCl (pH 8). Protein integrity and absence of albumin contamination were checked by SDS-PAGE and Coomassie staining. The pelleted fraction of bacterial cultures was used for extraction of cell proteins. The pellet was resuspended in phosphate-buffered saline (PBS) containing 1% Triton X-100 and a cocktail of protease inhibitors (Roche) and sonicated for 30 min at 4°C using a Bioruptor (Diagenode). Samples were then centrifuged, and the upper phase containing cell lysate was used in downstream experiments.
Protein samples were quantified using the RC DC protein assay (Bio-Rad), and equal amounts of protein preparations were loaded per well. Proteins were separated on 12 to 15% SDS-PAGE gels and transferred onto polyvinylidene difluoride (PVDF) membranes using a semidry electrophoresis transfer apparatus (Bio-Rad). Membranes were incubated in TBS-T blocking buffer (25 mM Tris [pH 7.5], 150 mM NaCl, 0.05% Tween 20) with 5% (wt/vol) skimmed milk powder for 30 min prior to overnight incubation with primary antibodies at the dilution indicated below. Membranes were washed in TBS-T three times and then incubated with secondary antibodies for 1 h before washing. Antibodies were used at the following dilutions: 1:1,000 for anti-ESAT-6 (abcam ab 26246; HYB 076-08), 1:500 for anti-GroEL2 (abcam ab69618; 3F7), and 1:1,000 for anti-SigA (kindly provided by Ida Rosenkrands). Horseradish peroxidase (HRP)-conjugated IgG secondary antibodies (Sigma-Aldrich) were used at a 1:20,000 dilution. Signals were detected using chemiluminescent substrates (GE Healthcare).
RESULTS
The M. tuberculosis whiB6 gene is differentially regulated between clinical isolates and the H37Rv laboratory strain.
PhoP is a virulence regulator required for ESAT-6 secretion in M. tuberculosis (33). Previous transcriptome analyses of the PhoP regulon using M. tuberculosis phoP mutants from different genetic backgrounds, such as the MT103 clinical isolate (34) or the reference strain H37Rv (35), revealed a largely overlapping gene set but also showed strain-specific differences. Although some of these divergences might have been due to the relatively low sensitivity of previously employed microarray techniques, particularly for genes whose expression levels were barely detectable, other variations were obviously caused by the different genetic strain background. To get deeper insights into the molecular mechanisms responsible for these differences, we constructed isogenic phoP mutants of strain H37Rv and a clinical isolate from the Beijing family (GC1237) and subjected them to transcriptomic analyses, which showed that the expression of several genes in the ESX-1 locus was differentially regulated by PhoP in strain GC1237 compared to strain H37Rv (Fig. 1B). Since the whiB6 gene exhibited one of the most divergent regulation profiles by PhoP between H37Rv and GC1237 (Fig. 1B), we focused on this gene as it seems to represent a novel ESX-1-associated regulator. Analysis by qRT-PCR confirmed that whiB6 was higher expressed in the H37Rv phoP mutant compared to WT H37Rv. On the contrary, whiB6 mRNA was barely detectable in the phoP mutants of GC1237 and MT103 clinical isolates (Fig. 1C). We extended our qRT-PCR analysis to genes coding for the ESAT-6/CFP-10 pair (esxA and esxB) and genes from the extended ESX-1 (espACD) previously reported to be regulated by PhoP (33). Although espACD showed an unambiguous PhoP-dependent regulation in the H37Rv and GC1237 strains, the expression pattern of ESAT-6/CFP-10-encoding genes correlated with that of whiB6 and showed divergent PhoP regulation between the clinical isolate and the laboratory strain (Fig. 2).
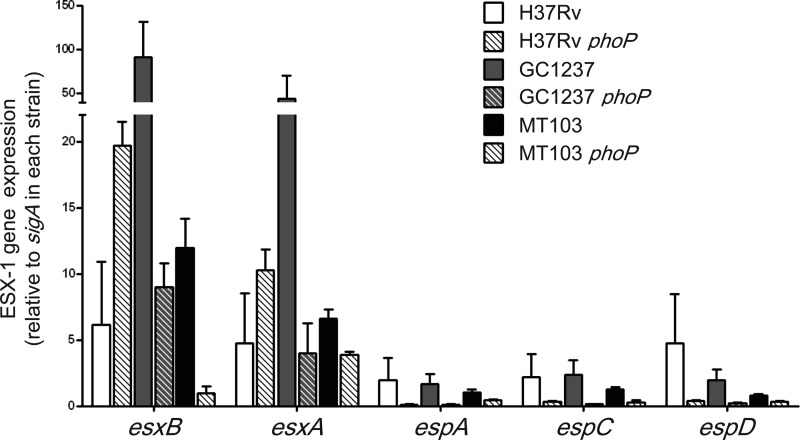
FIG 2.
Expression profiles of ESX-1 genes show differential PhoP regulation in clinical and laboratory strains of M. tuberculosis. qRT-PCR measurements of representative genes from ESX-1 and extended ESX-1 regions involved in ESAT-6 secretion, showing PhoP-dependent expression in the H37Rv, MT103, and GC1237 strains. Expression of esxA and esxB genes correlates with that of the whiB6 gene in Fig. 1C, showing a divergent PhoP regulation in H37Rv compared to those of the MT103 and GC1237 clinical strains. In contrast, PhoP regulation of the espA, espC, and espD genes is independent of the genetic background. Bars indicate fold change in gene expression relative to the sigA gene used as an endogenous control. Results represent the average of three independent experiments, and error bars indicate the SD of the mean.
WhiB6 divergent regulation correlates to ESAT-6 production and secretion in clinical and laboratory strains.
WhiB6 has a phylogenetic profile identical to espG (rv3866) (36), a gene contained in the genomic proximity of whiB6 that belongs to the ESX-1 locus and is thought to act as a specific type VII secretion pathway chaperon (37) involved in M. tuberculosis virulence (38). Moreover, it has been suggested that WhiB6 might bind to the upstream regions of espA and eccD1 (rv3877) (39). EspA is essential for ESAT-6 secretion (40, 41), and EccD1 is predicted to be part of the translocon of the ESX-1 type VII secretion system (22, 42). To experimentally evaluate the role of WhiB6 in ESX-1 regulation that was also suggested by bioinformatic predictions (36), we sought to determine ESAT-6 production and secretion in the WT strain and phoP mutants of different genetic backgrounds by Western blotting. As a first striking result, we found that the amount of ESAT-6 produced by the WT H37Rv reference strain was far less than that produced by WT clinical strains MT103 and GC1237 (Fig. 3A). For phoP mutants, we observed that mutants constructed in the MT103 and GC1237 strains produced less ESAT-6 than their respective WT strains (Fig. 3A). In contrast, the H37Rv phoP mutant produced equivalent amounts of ESAT-6 to H37Rv WT (Fig. 3A). We next investigated whether protein production was correlated with protein secretion and observed that the H37Rv strain secreted less ESAT-6 than the MT103 and GC1237 clinical isolates (Fig. 3B), whereas all three M. tuberculosis phoP mutants failed to secrete ESAT-6 due to EspACD downregulation (Fig. 2) (data not shown), confirming previous reports (33). Finally, we also examined whether this defect in ESAT-6 expression is exclusive of the H37Rv strain relative to other commonly used laboratory strains. Interestingly, we found that M. tuberculosis CDC1551 and Erdman reference strains produce and secrete more ESAT-6 than H37Rv (Fig. 3C), thus resembling the phenotype previously observed with clinical isolates.
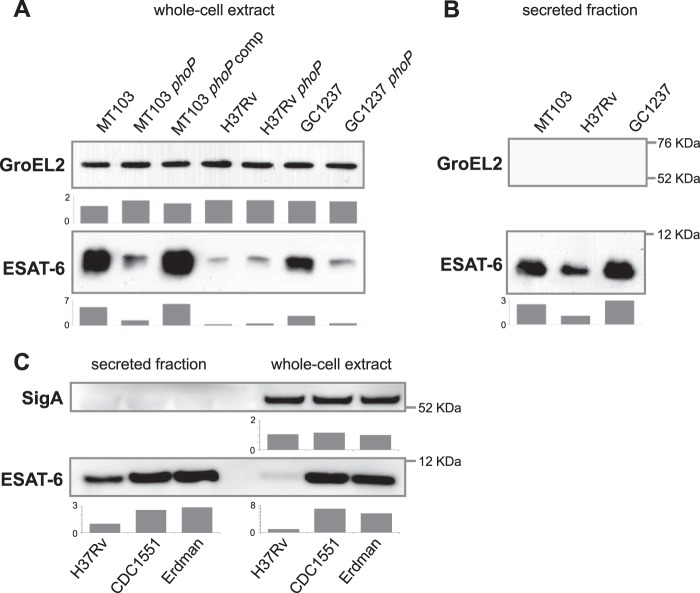
FIG 3.
Production and secretion of ESAT-6 vary between clinical isolates and the H37Rv reference strain. (A) Immunoblots against ESAT-6 and GroEL2 in whole-cell extracts of MT103, H37Rv and GC1237 parent strains and their respective phoP mutants. MT103 and GC1237 phoP mutants show decreased production of ESAT-6 with respect to their WT strains. The H37Rv WT strain produces a smaller amount of ESAT-6 than the MT103 and GC1237 clinical strains. GroEL2 was used as a control of protein loading. (B) Immunoblots against ESAT-6 and GroEL2 in the secreted fraction of the MT103, H37Rv, and GC1237 WT strains. Note the decreased secretion of ESAT-6 in H37Rv compared to that in the clinical isolates. GroEL2 was used as a control for absence of cell lysis. (C) Immunoblots against ESAT-6 and SigA in the whole-cell lysate and secreted fraction of M. tuberculosis H37Rv, CDC1551, and Erdman strains, grown in Sauton medium for 7 days. Note the decreased amount of ESAT-6 in the whole-cell lysate and secreted fraction of H37Rv compared to those in the two other reference strains. Bars represent densitometric analysis of band intensity and are referred to the H37Rv strain. Results are representative of three independent experiments.
The promoter region of whiB6 carries a point mutation in the PhoP binding site.
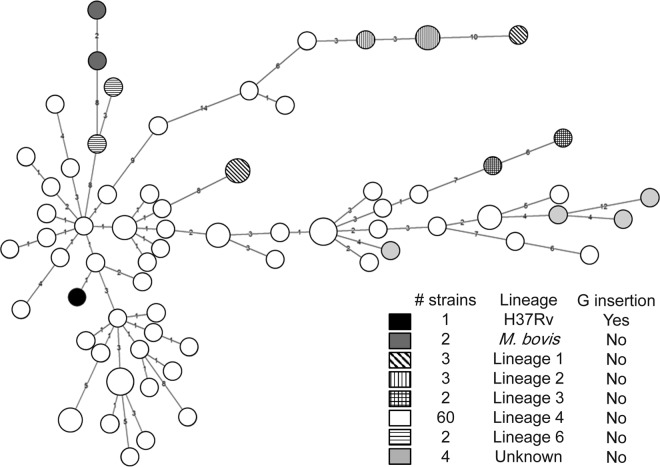
In a search for the genetic determinants responsible for the observed divergent whiB6 regulation in strain H37Rv, we focused on the sequence of the whiB6 gene and its flanking region. First, this analysis showed that whiB6 is present in all of the different members of the M. tuberculosis complex, including the more distantly related Mycobacterium canettii strains (2), as well as in many other mycobacterial species that harbor an orthologous ESX-1 system, including Mycobacterium smegmatis. When we analyzed the intergenic region between whiB6 and the adjacent gene rv3863, we identified two unique mutations exclusively present in strain H37Rv/Ra. The first mutation corresponds to a G insertion at the −74 position upstream of the whiB6 start codon (Fig. 4A), while the second was a G-to-A mutation found 117 nucleotides upstream of the start codon of gene rv3863. Our transcriptome analysis demonstrated that whiB6, but not rv3863, was subjected to PhoP regulation (data not shown), and we decided to focus on the G insertion upstream of whiB6. This region corresponds to a confirmed binding site of PhoP as determined by chromatin immunoprecipitation (ChIP) experiments using anti-PhoP antibodies, which showed an enrichment of this region in the immunoprecipitated fraction of WT strains relative to phoP mutants (Fig. 4B). Surprisingly, this G insertion generates an inverted repeat in the whiB6 promoter from H37Rv/Ra, which might result in the formation of a stem-loop structure (Fig. 4C). We confirmed that this mutation is absent from all members of the M. tuberculosis complex (sequences taken from 100 available mycobacterial genome sequences in the NCBI's ftp database) other than H37Rv/Ra strains (data not shown), including 76 M. tuberculosis clinical isolates screened in our laboratory, belonging to different M. tuberculosis lineages (Fig. 5; see Fig. S1 in the supplemental material).
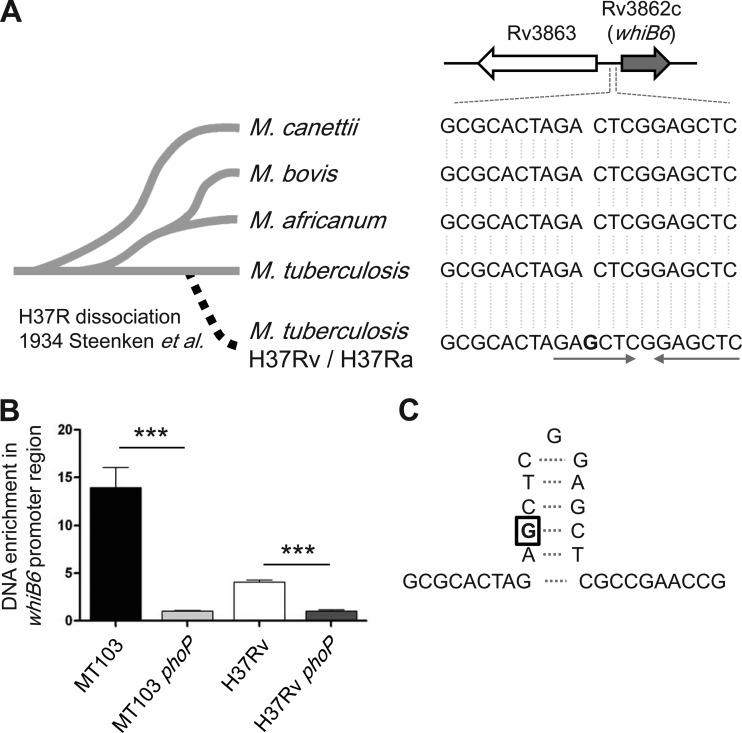
FIG 4.
Study of M. tuberculosis complex strains reveals a point mutation in the whiB6 promoter exclusive of the H37Rv strain. (A) The whiB6 gene and flanking sequences were aligned with 100 available mycobacterial genomes. The image shows a single nucleotide insertion (labeled as a G in boldface) upstream of the whiB6 gene exclusively present in H37Rv/Ra, being absent in other M. tuberculosis complex strains. Note the presence of inverted repeats (depicted by arrows) that originated as a consequence of the guanine insertion in H37Rv. (B) Immunoprecipitation of M. tuberculosis DNA with anti-PhoP antibodies and subsequent qRT-PCR quantification of the region adjacent to the G-insertion in the whiB6 promoter. The results show an enrichment of immunoprecipitated DNA in the MT103 and H37Rv WT strains relative to their phoP mutants, indicative of PhoP binding to the whiB6 promoter. (C) Inverted repeats in panel A probably result in formation of the indicated stem-loop structure.
FIG 5.
Extensive sequence analysis of M. tuberculosis clinical isolates reveals that whiB6 mutation is exclusive of H37Rv and independent of the M. tuberculosis lineage. The whiB6 promoter region of 76 clinical isolates from our laboratory collection was sequenced. The dendrogram based on spoligotyping of the clinical isolates reveals that the G insertion in H37Rv is independent of the lineage. Spoligotyping is further detailed in Fig. S1 in the supplemental material.
Reintroduction of the WT copy of whiB6 in H37Rv causes an increase in ESAT-6 production and secretion.
WhiB6 is divergently regulated by PhoP in H37Rv and clinical strains (Fig. 1), and the same trend is also seen for ESAT-6 production and secretion in these strains (Fig. 3): together, these results suggest that WhiB6 might be regulating ESAT-6 expression via transcriptional regulation of ESX-1 genes. To test this hypothesis, we constructed a knock-in (KI) strain in which the WT copy of whiB6 present in clinical isolates was restored in the H37Rv genetic background (H37Rv::KIMt). We also constructed a KI strain of the mutated version of the whiB6 mutant (H37Rv::KIRv) to control for undesirable effects due to the different genetic dosage of the whiB6 gene. The results showed that restoration of the whiB6 WT allele is able to boost ESAT-6 production in H37Rv to the levels of the clinical strains (Fig. 6A), which was also followed by increased secretion of ESAT-6 to the levels of the clinical isolates (Fig. 6B). In contrast, doubling the genetic dosage of the “mutant” whiB6 allele in the H37Rv::KIRv strain did not impact ESAT-6 production or secretion (Fig. 6A and B), linking the effect to the specific single nucleotide polymorphism (SNP) in the WhiB6 promoter. As expected, expression of the WT or mutant variants of whiB6 in an H37Rv phoP mutant did not change the absence of ESAT-6 secretion, due to independent EspACD downregulation (Fig. 6B). We also used these KIMt and KIRv strains to confirm that some ESX-1 genes could be regulated by PhoP in a WhiB6-dependent fashion. Interestingly, we observed that the esxA and esxB genes require both PhoP and a WT whiB6 gene in order to achieve gene expression levels comparable to those of clinical isolates (Fig. 6C).
FIG 6.

Reintroduction of a WT whiB6 allele in H37Rv restores ESAT-6 production and secretion to the levels of clinical strains. (A) Immunoblots against ESAT-6 and GroEL2 in whole-cell extracts of H37Rv-derived strains containing either a WT (KIMt) or a mutated (KIRv) whiB6 allele. MT103 and its phoP mutant were used as controls for ESAT-6 phenotypes in clinical strains. Introduction of a WT copy of whiB6 in H37Rv restores ESAT-6 production to levels comparable to those in the MT103 clinical isolate. Introduction of either a WT or mutated whiB6 allele in H37Rv phoP mutant does not significantly impact ESAT-6 production. GroEL2 was used as a control of protein loading. (B) Immunoblots against ESAT-6 and GroEL2 in the secreted fraction from strains used in panel A. The whiB6 WT allele is able to enhance ESAT-6 secretion in H37Rv to the levels of the MT103 clinical strain. The phoP mutants show absence of ESAT-6 secretion, independent of whiB6 expression. GroEL2 was used as a control for absence of cell lysis. Note that introduction of an additional “mutated” allele into H37Rv (H37Rv::KIRv) has no effect over ESAT-6 production or secretion. Bars represent densitometric analysis of band intensity and are referred to the H37Rv strain. (C) qRT-PCR measurements of the esxA, esxB, and espJ genes in KIMt and KIRv derivatives of H37Rv and its phoP mutant. Note that esxA, esxB, and, to a lesser extent, espJ require both WT alleles of phoP and whiB6 in order to reach expression levels comparable to those of the clinical isolates in Fig. 2. Bars indicate fold changes in gene expression relative to the sigA gene used as an endogenous control. Results are representative of three independent experiments, and error bars indicate the SD of the mean.
DISCUSSION
The deep impact of mutations in transcriptional regulators on the outcome of infection is well known. Some relevant examples include a K220T substitution in the pleiotropic PrfA regulator with profound effects on Listeria monocytogenes virulence (43), point mutations in the streptococcal regulator of virulence (Srv) that alter protein-DNA binding (44), or the S219L substitution in PhoP, which is a major cause of M. tuberculosis H37Ra attenuation (45, 46). However, polymorphisms in noncoding regions have been less explored. In this work, we highlight a single nucleotide insertion within the whiB6 promoter region that influences the PhoP regulatory mechanism. The presence of a sole G insertion at the −74 position relative to whiB6 start codon is able to alter the PhoP binding affinity to this region (Fig. 4B). It could be possible that PhoP binds as a multimer in order to activate transcription, and the presence of point mutations within the binding site results in transcriptional deregulation. Our sequence analyses indicate the exclusive presence of this point mutation in H37Rv and H37Ra. In 1934, Steenken and colleagues reported that after successive cultures, the parental H37R strain dissociated into an avirulent form (H37Ra) and a virulent form (H37Rv) (47). The results presented here support the hypothesis that in vitro passages of the H37R parent strain led to this single nucleotide insertion. Furthermore, since H37Rv and H37Ra share the same polymorphism, this mutation likely occurred prior to segregation of both strains. If mutation in the whiB6 promoter plausibly occurred in in vitro cultures, we could also hypothesize that a proper expression of this gene in clinical isolates might increase fitness in the host. Supporting this observation, we demonstrate that WhiB6 modulates ESAT-6 secretion, a phenotype dispensable for growth in vitro. However, the mutation in the whiB6 promoter retains a virulent phenotype in H37Rv, probably because this strain is still able to secrete ESAT-6 in vitro, albeit at lower levels than other reference strains and clinical isolates (Fig. 3). Subsequent culture passages probably resulted in the deleterious S219L mutation in PhoP from H37Ra (45, 48), which impaired ESAT-6 secretion and consequently strongly contributes to the attenuation of H37Ra (33, 46). In parallel, the H37Rv strain retained a WT phoP gene, allowing it to secrete at least some ESAT-6 and consequently maintaining virulence (49), which might, however, mainly reflect virulence in the mouse infection model, because since the isolation of the H37 strain from a tuberculosis patient more than 100 years ago, the strain was passaged in axenic media and in mice. However, it might also be possible that regulation of ESAT-6 expression or secretion during cellular or animal infections differs from that observed in vitro. This hypothesis would explain why different M. tuberculosis strains (H37Rv, CDC1551, and Erdman) show different levels of ESAT-6 expression under laboratory culture conditions, albeit they exhibit similar levels of virulence in the mouse infection model (49–51). It would also explain the absence of differences between H37Rv and clinical isolates from the Beijing family in induction of apoptosis, recently reported to be an ESAT-6-mediated phenotype (20). On the other hand, the possibility remains that mouse infection models are not sensitive enough to detect virulence changes due to ESAT-6 fluctuations. Indeed, in the sensitive guinea pig infection model, the H37Rv strain was shown to be less virulent than the Erdman, CDC1551, and HN878 strains (52), which produce more ESAT-6 than the H37Rv strain.
WhiB proteins are bifunctional regulators, acting either as transcription factors or by modulating the redox state of target proteins (53–55). Whereas WhiB1, WhiB2, WhiB3, and WhiB7 have been extensively studied, little is known about WhiB4, WhiB5, and WhiB6, apart from their expression profiles and disulfide reductase activity (53, 54, 56, 57). WhiB-like proteins exert their regulatory activity depending on the redox state, which is linked to the presence of environmental metals. This fact is especially relevant in the context of the phagosomal presence of M. tuberculosis since this phagovacuole is rich in metals and oxidative or nitrosative species that may restrict growth or survival of pathogens (7, 58). As an example, WhiB1 is an essential protein able to react with nitric oxide (NO) and thereby can shift its DNA binding capacity (59). This ability suggests a possible role for WhiB1 in reprograming M. tuberculosis gene expression in response to NO generated by macrophages. M. tuberculosis resists the acidic and metabolic stresses in the hostile environment of the macrophage in multiple ways (60–62). One of the recently demonstrated mechanisms is that the bacterium escapes from the microbicidal phagosome by inducing pore formation and disruption of the phagosomal membrane, a phenotype dependent on the function of the ESX-1 secretion system (8, 9, 19). It is not yet clear, however, which signal(s) might be regulating ESX-1 expression. The ability of the pleotropic two-component regulator PhoPR to sense the acidic phagosomal pH (63) could be involved in this process in two independent ways. First, it is known that inactivation of PhoP leads to loss of ESAT-6 secretion due to interference with the EspACD/EspR regulation cascade (15, 33). In parallel, here we show that PhoPR may also interfere with WhiB6 regulation, which seems to act as an alternative regulation loop to respond to nitrosative/oxidative stresses encountered within the phagosome, thereby increasing the expression and production of ESAT-6. In support of this hypothesis, it has been shown that whiB6 expression in in vitro-grown M. tuberculosis cultures increases upon treatment with NO (56, 57), and whiB6 gene expression gets upregulated when M. tuberculosis infects macrophages (55). This model orchestrates a regulatory network involving sensor proteins (PhoR and WhiB6), which might modulate ESX-1 expression and ESAT-6 secretion in favor of M. tuberculosis disrupting the phagosomal membrane and getting access to the cytosol. From recent literature, it is well documented that cytosolic access triggers a wide range of cellular changes that are specific for virulent tubercle bacilli, as they are not induced in ESX-1-deleted, attenuated strains, such as the BCG vaccine strains (8, 9, 20). It seems that PhoP-WhiB6-ESX-1-associated functions play an important role in these processes.
Aside from PhoP implications in ESAT-6 secretion, a recent work uses whole-sequencing approaches to demonstrate that PhoP is also involved in secretion of TAT-dependent proteins, which include members of the Ag85 complex (64).
Finally, apart from the new insights into the various regulation cascades described above, our data also highlight a generally applicable point about the impact of genetic polymorphisms potentially present in often passaged and widely used reference strains. Our findings argue against the exclusive use of laboratory strains as universal comparators for scientific experiments and suggest that parallel validation with clinical strains should be undertaken prior to universalizing results to the whole M. tuberculosis lineages.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the European Community's Framework Programme 7 grants NEWTBVAC 241745 and MM4TB 260872, the Spanish Ministry of Economy and Competitiveness grants BIO2011-23555 and FIS12/1970, the Fondation pour la Recherche Médicale FRM no. DEQ20130326471, and the Pyrenees biomedical network Refbio grant SecRegulTBC. Jesús Gonzalo-Asensio was supported by the Juan de la Cierva Programme (JCI-2009-03799) from the Spanish Ministry of Science and Innovation. Luis Solans was supported by grant BES-2009-013037.
We thank Marie-Agnès Dillies and Jean-Yves Coppée from the Transcriptome Platform PF2 at the Institut Pasteur for advice and Ida Rosenkrands from Statens Serum Institute for kindly providing the polyclonal anti-SigA antibodies, and we also gratefully acknowledge Alberto Cebollada for help with the sequence analysis of M. tuberculosis strains.
Footnotes
Published ahead of print 2 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01824-14.
REFERENCES
- 1.Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, Parkhill J, Malla B, Berg S, Thwaites G, Yeboah-Manu D, Bothamley G, Mei J, Wei L, Bentley S, Harris SR, Niemann S, Diel R, Aseffa A, Gao Q, Young D, Gagneux S. 2013. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 45:1176–1182. 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, Majlessi L, Criscuolo A, Tap J, Pawlik A, Fiette L, Orgeur M, Fabre M, Parmentier C, Frigui W, Simeone R, Boritsch EC, Debrie AS, Willery E, Walker D, Quail MA, Ma L, Bouchier C, Salvignol G, Sayes F, Cascioferro A, Seemann T, Barbe V, Locht C, Gutierrez MC, Leclerc C, Bentley SD, Stinear TP, Brisse S, Medigue C, Parkhill J, Cruveiller S, Brosch R. 2013. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat. Genet. 45:172–179. 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flannagan RS, Cosio G, Grinstein S. 2009. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7:355–366. 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- 4.Steele-Mortimer O. 2008. The Salmonella-containing vacuole: moving with the times. Curr. Opin. Microbiol. 11:38–45. 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo ZQ. 2012. Legionella secreted effectors and innate immune responses. Cell. Microbiol. 14:19–27. 10.1111/j.1462-5822.2011.01713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnupf P, Portnoy DA. 2007. Listeriolysin O: a phagosome-specific lysin. Microbes Infect. 9:1176–1187. 10.1016/j.micinf.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Ehrt S, Schnappinger D. 2009. Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell. Microbiol. 11:1170–1178. 10.1111/j.1462-5822.2009.01335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houben D, Demangel C, van Ingen J, Perez J, Baldeon L, Abdallah AM, Caleechurn L, Bottai D, van Zon M, de Punder K, van der Laan T, Kant A, Bossers-de Vries R, Willemsen P, Bitter W, van Soolingen D, Brosch R, van der Wel N, Peters PJ. 2012. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell. Microbiol. 14:1287–1298. 10.1111/j.1462-5822.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- 9.Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, Enninga J. 2012. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 8:e1002507. 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. 2007. Type VII secretion—mycobacteria show the way. Nat. Rev. Microbiol. 5:883–891. 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 11.Bitter W, Houben EN, Bottai D, Brodin P, Brown EJ, Cox JS, Derbyshire K, Fortune SM, Gao LY, Liu J, Gey van Pittius NC, Pym AS, Rubin EJ, Sherman DR, Cole ST, Brosch R. 2009. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 5:e1000507. 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdallah AM, Bestebroer J, Savage ND, de Punder K, van Zon M, Wilson L, Korbee CJ, van der Sar AM, Ottenhoff TH, van der Wel NN, Bitter W, Peters PJ. 2011. Mycobacterial secretion systems ESX-1 and ESX-5 play distinct roles in host cell death and inflammasome activation. J. Immunol. 187:4744–4753. 10.4049/jimmunol.1101457. [DOI] [PubMed] [Google Scholar]
- 13.Bottai D, Di Luca M, Majlessi L, Frigui W, Simeone R, Sayes F, Bitter W, Brennan MJ, Leclerc C, Batoni G, Campa M, Brosch R, Esin S. 2012. Disruption of the ESX-5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Mol. Microbiol. 83:1195–1209. 10.1111/j.1365-2958.2012.08001.x. [DOI] [PubMed] [Google Scholar]
- 14.Raghavan S, Manzanillo P, Chan K, Dovey C, Cox JS. 2008. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature 454:717–721. 10.1038/nature07219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blasco B, Chen JM, Hartkoorn R, Sala C, Uplekar S, Rougemont J, Pojer F, Cole ST. 2012. Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog. 8:e1002621. 10.1371/journal.ppat.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renshaw PS, Lightbody KL, Veverka V, Muskett FW, Kelly G, Frenkiel TA, Gordon SV, Hewinson RG, Burke B, Norman J, Williamson RA, Carr MD. 2005. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J. 24:2491–2498. 10.1038/sj.emboj.7600732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jonge MI, Pehau-Arnaudet G, Fretz MM, Romain F, Bottai D, Brodin P, Honore N, Marchal G, Jiskoot W, England P, Cole ST, Brosch R. 2007. ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J. Bacteriol. 189:6028–6034. 10.1128/JB.00469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derrick SC, Morris SL. 2007. The ESAT6 protein of Mycobacterium tuberculosis induces apoptosis of macrophages by activating caspase expression. Cell. Microbiol. 9:1547–1555. 10.1111/j.1462-5822.2007.00892.x. [DOI] [PubMed] [Google Scholar]
- 19.van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. 2007. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 129:1287–1298. 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 20.Aguilo JI, Alonso H, Uranga S, Marinova D, Arbues A, de Martino A, Anel A, Monzon M, Badiola J, Pardo J, Brosch R, Martin C. 2013. ESX-1-induced apoptosis is involved in cell-to-cell spread of Mycobacterium tuberculosis. Cell. Microbiol. 15:1994–2005. 10.1111/cmi.12169. [DOI] [PubMed] [Google Scholar]
- 21.Hsu T. 2003. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. U. S. A. 100:12420–12425. 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brodin P, Majlessi L, Marsollier L, de Jonge MI, Bottai D, Demangel C, Hinds J, Neyrolles O, Butcher PD, Leclerc C, Cole ST, Brosch R. 2006. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect. Immun. 74:88–98. 10.1128/IAI.74.1.88-98.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalo Asensio J, Maia C, Ferrer NL, Barilone N, Laval F, Soto CY, Winter N, Daffe M, Gicquel B, Martin C, Jackson M. 2006. The virulence-associated two-component PhoP-PhoR system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. J. Biol. Chem. 281:1313–1316. 10.1074/jbc.C500388200. [DOI] [PubMed] [Google Scholar]
- 24.Perez E, Samper S, Bordas Y, Guilhot C, Gicquel B, Martin C. 2001. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol. Microbiol. 41:179–187. 10.1046/j.1365-2958.2001.02500.x. [DOI] [PubMed] [Google Scholar]
- 25.Arbues A, Aguilo JI, Gonzalo-Asensio J, Marinova D, Uranga S, Puentes E, Fernandez C, Parra A, Cardona PJ, Vilaplana C, Ausina V, Williams A, Clark S, Malaga W, Guilhot C, Gicquel B, Martin C. 2013. Construction, characterization and preclinical evaluation of MTBVAC, the first live-attenuated-based vaccine to enter clinical trials. Vaccine 31:4867–4873. 10.1016/j.vaccine.2013.07.051. [DOI] [PubMed] [Google Scholar]
- 26.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460. 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 27.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demay C, Liens B, Burguiere T, Hill V, Couvin D, Millet J, Mokrousov I, Sola C, Zozio T, Rastogi N. 2012. SITVITWEB—a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect. Genet. Evol. 12:755–766. 10.1016/j.meegid.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Sala C, Haouz A, Saul FA, Miras I, Rosenkrands I, Alzari PM, Cole ST. 2009. Genome-wide regulon and crystal structure of BlaI (Rv1846c) from Mycobacterium tuberculosis. Mol. Microbiol. 71:1102–1116. 10.1111/j.1365-2958.2008.06583.x. [DOI] [PubMed] [Google Scholar]
- 30.Pawlik A, Garnier G, Orgeur M, Tong P, Lohan A, Le Chevalier F, Sapriel G, Roux AL, Conlon K, Honore N, Dillies MA, Ma L, Bouchier C, Coppee JY, Gaillard JL, Gordon SV, Loftus B, Brosch R, Herrmann JL. 2013. Identification and characterization of the genetic changes responsible for the characteristic smooth-to-rough morphotype alterations of clinically persistent Mycobacterium abscessus. Mol. Microbiol. 90:612–629. 10.1111/mmi.12387. [DOI] [PubMed] [Google Scholar]
- 31.Rouillard JM, Zuker M, Gulari E. 2003. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 31:3057–3062. 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 33.Frigui W, Bottai D, Majlessi L, Monot M, Josselin E, Brodin P, Garnier T, Gicquel B, Martin C, Leclerc C, Cole ST, Brosch R. 2008. Control of M. tuberculosis ESAT-6 secretion and specific T cell recognition by PhoP. PLoS Pathog. 4:e33. 10.1371/journal.ppat.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalo-Asensio J, Mostowy S, Harders-Westerveen J, Huygen K, Hernandez-Pando R, Thole J, Behr M, Gicquel B, Martin C. 2008. PhoP: a missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS One 3:e3496. 10.1371/journal.pone.0003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walters SB, Dubnau E, Kolesnikova I, Laval F, Daffe M, Smith I. 2006. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 60:312–330. 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 36.Das C, Ghosh TS, Mande SS. 2011. Computational analysis of the ESX-1 region of Mycobacterium tuberculosis: insights into the mechanism of type VII secretion system. PLoS One 6:e27980. 10.1371/journal.pone.0027980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daleke MH, van der Woude AD, Parret AH, Ummels R, de Groot AM, Watson D, Piersma SR, Jimenez CR, Luirink J, Bitter W, Houben EN. 2012. Specific chaperones for the type VII protein secretion pathway. J. Biol. Chem. 287:31939–31947. 10.1074/jbc.M112.397596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bottai D, Majlessi L, Simeone R, Frigui W, Laurent C, Lenormand P, Chen J, Rosenkrands I, Huerre M, Leclerc C, Cole ST, Brosch R. 2011. ESAT-6 secretion-independent impact of ESX-1 genes espF and espG1 on virulence of Mycobacterium tuberculosis. J. Infect. Dis. 203:1155–1164. 10.1093/infdis/jiq089. [DOI] [PubMed] [Google Scholar]
- 39.Reddy TB, Riley R, Wymore F, Montgomery P, DeCaprio D, Engels R, Gellesch M, Hubble J, Jen D, Jin H, Koehrsen M, Larson L, Mao M, Nitzberg M, Sisk P, Stolte C, Weiner B, White J, Zachariah ZK, Sherlock G, Galagan JE, Ball CA, Schoolnik GK. 2009. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 37:D499–D508. 10.1093/nar/gkn652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fortune SM, Jaeger A, Sarracino DA, Chase MR, Sassetti CM, Sherman DR, Bloom BR, Rubin EJ. 2005. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc. Natl. Acad. Sci. U. S. A. 102:10676–10681. 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacGurn JA, Raghavan S, Stanley SA, Cox JS. 2005. A non-RD1 gene cluster is required for Snm secretion in Mycobacterium tuberculosis. Mol. Microbiol. 57:1653–1663. 10.1111/j.1365-2958.2005.04800.x. [DOI] [PubMed] [Google Scholar]
- 42.Houben EN, Bestebroer J, Ummels R, Wilson L, Piersma SR, Jimenez CR, Ottenhoff TH, Luirink J, Bitter W. 2012. Composition of the type VII secretion system membrane complex. Mol. Microbiol. 86:472–484. 10.1111/j.1365-2958.2012.08206.x. [DOI] [PubMed] [Google Scholar]
- 43.Velge P, Herler M, Johansson J, Roche SM, Temoin S, Fedorov AA, Gracieux P, Almo SC, Goebel W, Cossart P. 2007. A naturally occurring mutation K220T in the pleiotropic activator PrfA of Listeria monocytogenes results in a loss of virulence due to decreasing DNA-binding affinity. Microbiology 153:995–1005. 10.1099/mic.0.2006/002238-0. [DOI] [PubMed] [Google Scholar]
- 44.Doern CD, Holder RC, Reid SD. 2008. Point mutations within the streptococcal regulator of virulence (Srv) alter protein-DNA interactions and Srv function. Microbiology 154:1998–2007. 10.1099/mic.0.2007/013466-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chesne-Seck ML, Barilone N, Boudou F, Asensio JG, Kolattukudy PE, Martin C, Cole ST, Gicquel B, Gopaul DN, Jackson M. 2008. A point mutation in the two-component regulator PhoP-PhoR accounts for the absence of polyketide-derived acyltrehaloses but not that of phthiocerol dimycocerosates in Mycobacterium tuberculosis H37Ra. J. Bacteriol. 190:1329–1334. 10.1128/JB.01465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JS, Krause R, Schreiber J, Mollenkopf HJ, Kowall J, Stein R, Jeon BY, Kwak JY, Song MK, Patron JP, Jorg S, Roh K, Cho SN, Kaufmann SH. 2008. Mutation in the transcriptional regulator PhoP contributes to avirulence of Mycobacterium tuberculosis H37Ra strain. Cell Host Microbe 3:97–103. 10.1016/j.chom.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Steenken W, Oatway WH, Petroff SA. 1934. Biological studies of the tubercle bacillus. iii. Dissociation and pathogenicity of the R and S variants of the human tubercle bacillus (H(37)). J. Exp. Med. 60:515–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalo-Asensio J, Soto CY, Arbues A, Sancho J, del Carmen Menendez M, Garcia MJ, Gicquel B, Martin C. 2008. The Mycobacterium tuberculosis phoPR operon is positively autoregulated in the virulent strain H37Rv. J. Bacteriol. 190:7068–7078. 10.1128/JB.00712-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manca C, Tsenova L, Barry CE, III, Bergtold A, Freeman S, Haslett PA, Musser JM, Freedman VH, Kaplan G. 1999. Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J. Immunol. 162:6740–6746. [PubMed] [Google Scholar]
- 50.Kelley CL, Collins FM. 1999. Growth of a highly virulent strain of Mycobacterium tuberculosis in mice of differing susceptibility to tuberculous challenge. Tuber. Lung Dis. 79:367–370. 10.1054/tuld.1999.0214. [DOI] [PubMed] [Google Scholar]
- 51.Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, Musser JM, Barry CE, III, Freedman VH, Kaplan G. 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc. Natl. Acad. Sci. U. S. A. 98:5752–5757. 10.1073/pnas.091096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palanisamy GS, Smith EE, Shanley CA, Ordway DJ, Orme IM, Basaraba RJ. 2008. Disseminated disease severity as a measure of virulence of Mycobacterium tuberculosis in the guinea pig model. Tuberculosis (Edinb.) 88:295–306. 10.1016/j.tube.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garg S, Alam MS, Bajpai R, Kishan KVR, Agrawal P. 2009. Redox biology of Mycobacterium tuberculosis H37Rv: protein-protein interaction between GlgB and WhiB1 involves exchange of thiol-disulfide. BMC Biochem. 10:1. 10.1186/1471-2091-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alam MS, Garg SK, Agrawal P. 2009. Studies on structural and functional divergence among seven WhiB proteins of Mycobacterium tuberculosis H37Rv. FEBS J. 276:76–93. 10.1111/j.1742-4658.2008.06755.x. [DOI] [PubMed] [Google Scholar]
- 55.Saini V, Farhana A, Glasgow JN, Steyn AJ. 2012. Iron sulfur cluster proteins and microbial regulation: implications for understanding tuberculosis. Curr. Opin. Chem. Biol. 16:45–53. 10.1016/j.cbpa.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geiman DE, Raghunand TR, Agarwal N, Bishai WR. 2006. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 50:2836–2841. 10.1128/AAC.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larsson C, Luna B, Ammerman NC, Maiga M, Agarwal N, Bishai WR. 2012. Gene expression of Mycobacterium tuberculosis putative transcription factors whiB1-7 in redox environments. PLoS One 7:e37516. 10.1371/journal.pone.0037516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soldati T, Neyrolles O. 2012. Mycobacteria and the intraphagosomal environment: take it with a pinch of salt(s)! Traffic 13:1042–1052. 10.1111/j.1600-0854.2012.01358.x. [DOI] [PubMed] [Google Scholar]
- 59.Smith LJ, Stapleton MR, Fullstone GJ, Crack JC, Thomson AJ, Le Brun NE, Hunt DM, Harvey E, Adinolfi S, Buxton RS, Green J. 2010. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochem. J. 432:417–427. 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan S, Sukumar N, Abramovitch RB, Parish T, Russell DG. 2013. Mycobacterium tuberculosis responds to chloride and pH as synergistic cues to the immune status of its host cell. PLoS Pathog. 9:e1003282. 10.1371/journal.ppat.1003282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gouzy A, Larrouy-Maumus G, Bottai D, Levillain F, Dumas A, Wallach JB, Caire-Brandli I, de Chastellier C, Wu TD, Poincloux R, Brosch R, Guerquin-Kern JL, Schnappinger D, Sorio de Carvalho LP, Poquet Y, Neyrolles O. 2014. Mycobacterium tuberculosis exploits asparagine to assimilate nitrogen and resist acid stress during infection. PLoS Pathog. 10:e1003928. 10.1371/journal.ppat.1003928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ. 2013. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 155:1296–1308. 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abramovitch RB, Rohde KH, Hsu FF, Russell DG. 2011. aprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol. Microbiol. 80:678–694. 10.1111/j.1365-2958.2011.07601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Solans L, Gonzalo-Asensio J, Sala C, Benjak A, Uplekar S, Rougemont J, Guilhot C, Malaga W, Martin C, Cole ST. 2014. The PhoP-dependent ncRNA Mcr7 modulates the TAT secretion system in Mycobacterium tuberculosis. PLoS Pathog. 10:e1004183. 10.1371/journal.ppat.1004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.