ABSTRACT
The N-terminal region of simian hemorrhagic fever virus (SHFV) nonstructural polyprotein 1a is predicted to encode three papain-like proteases (PLP1α, PLP1β, and PLP1γ). Catalytic residues and cleavage sites for each of the SHFV PLP1s were predicted by alignment of the SHFV PLP1 region sequences with each other as well as with those of other arteriviruses, and the predicted catalytic residues were shown to be proximal by homology modeling of the SHFV nsp1s on porcine respiratory and reproductive syndrome virus (PRRSV) nsp1 crystal structures. The functionality of the predicted catalytic Cys residues and cleavage sites was tested by analysis of the autoproteolytic products generated in in vitro transcription/translation reactions done with wild-type or mutant SHFV nsp1 constructs. Cleavage sites were also analyzed by mass spectroscopy analysis of selected immunoprecipitated cleavage products. The data showed that each of the three SHFV PLP1s is an active protease. Cys63 was identified as the catalytic Cys of SHFV PLP1α and is adjacent to an Ala instead of the canonical Tyr observed in other arterivirus PLP1s. SHFV PLP1γ is able to cleave at both downstream and upstream nsp1 junction sites. Although intermediate precursor polyproteins as well as alternative products generated by each of the SHFV PLP1s cleaving at sites within the N-terminal region of nsp1β were produced in the in vitro reactions, Western blotting of SHFV-infected, MA104 cell lysates with SHFV nsp1 protein-specific antibodies detected only the three mature nsp1 proteins.
IMPORTANCE SHFV is unique among arteriviruses in having three N-terminal papain-like protease 1 (PLP1) domains. Other arteriviruses encode one or two active PLP1s. This is the first functional study of the SHFV PLP1s. Analysis of the products of in vitro autoprocessing of an N-terminal SHFV nonstructural 1a polypeptide fragment showed that each of the three SHFV PLP1s is active, and the predicted catalytic Cys residues and cleavage sites for each PLP1 were confirmed by testing mutant constructs. Several unique features of the SHFV PLP1s were discovered. The SHFV PLP1α catalytic Cys63 is unique among arterivirus PLP1s in being adjacent to an Ala instead of a Trp. Other arterivirus PLP1s cleave only in cis at a single downstream site, but SHFV PLP1γ can cleave at both the downstream nsp1γ-nsp2 and upstream nsp1β-nsp1γ junctions. The three mature nsp1 proteins were produced both in the in vitro reactions and in infected cells.
INTRODUCTION
Simian hemorrhagic fever virus (SHFV) is a member of the family Arteriviridae. Other members include the prototype equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV), and porcine reproductive and respiratory syndrome virus (PRRSV). Based on similarities in genomic organization and replication strategy, the families Arteriviridae, Coronaviridae, and Roniviridae are classified in the order Nidovirales (1, 2). Arterivirus genomes are polycistronic, positive-sense, single-stranded RNAs with a 5′ type I cap and a 3′ poly(A) tract (3, 4). The ∼15.7-kb SHFV genome is the longest known arterivirus genome. The structural protein open reading frames (ORFs) are located at the 3′ end of the genome and expressed from a nested set of 5′ and 3′ coterminal subgenomic (sg) mRNAs (5–7). The nonstructural polyproteins 1a and 1ab are expressed from overlapping 5′ ORFs. Translation of polyprotein 1a terminates at the first in-frame UAA. Polyprotein 1ab is produced when a −1 ribosomal frameshift occurs on a slippery sequence located before the polyprotein 1a stop codon (2). The arterivirus ORF1a polyprotein contains a papain-like protease 1 (PLP1) domain in each of the three N-terminal nsp1s, a PLP2 domain in nsp2, and the main serine protease in nsp4 that cleaves at multiple sites in the 1a and 1ab polyproteins (2). All coronavirus PLPs (PLP1, PLP2, and PLpro) and the arterivirus PLP2 contain a catalytic Cys-His-Asp triad also found in other positive-sense RNA virus PLP sequences (8, 9). However, the active sites of arterivirus PLP1 proteases were predicted to consist of a Cys-His tandem and this was confirmed by the recent reports of the crystal structures of PRRSV nsp1α and nsp1β proteins (10, 11). The length of the sequence between the catalytic residues in arterivirus PLPs is half of that in coronavirus PLPs, making the arterivirus PLPs the smallest known.
All of the coronavirus PLPs as well as the arterivirus PLP2s also have deubiquitinating and deISGylating activities, but the arterivirus PLP1s do not (8). The coronavirus PLPs and arterivirus PLP2s both have cis and trans cleavage activities, while the arterivirus PLP1s have been reported to cleave exclusively in cis at a single site downstream of their catalytic domains (2, 8, 10–13). The crystal structures of type II PRRSV nsp1α and nsp1β suggest that these enzymes utilize an intramolecular cleavage mechanism (2, 10, 11). The N-terminal EAV PLP1 domain is inactive due to a Lys substitution of the catalytic Cys residue, but the downstream EAV PLP1 is active and produces an nsp1 fusion protein (14, 15). The LDV and PRRSV nonstructural polyproteins contain active PLP1α and PLP1β enzymes, each of which cleaves at a single downstream site in in vitro reactions (10, 11, 16). Although most coronavirus PLPs cleave at a canonical -Leu-X-Gly↓Gly site, the sequences of the cleavage sites used by arterivirus PLPs are not well conserved (8, 10, 11, 14, 16).
The EAV nsp1 and PRRSV nsp1α and nsp1β proteins function as interferon pathway antagonists (17–20). An N-terminal zinc finger domain in EAV nsp1 and PRRSV nsp1α regulates sg mRNA and genome transcription abundance (2, 21–23) and is also important for mediating the IFN-β and NF-κB immunosuppressive activities of PRRSV nsp1α (24).
In the present study, the catalytic residues and cleavage sites of the three SHFV PLP1 domains (PLP1α, PLP1β, and PLP1γ) were predicted by sequence alignment with each other as well as with other arterivirus PLP1s and the predicted catalytic residues were shown to be proximal by homology modeling on PRRSV nsp1α and nsp1β structures (10, 11). The results of analyses of the autocleavage products produced by wild-type and mutant in vitro-translated SHFV nsp1 region polyproteins indicated that each of the SHFV PLP1s is active and identified the catalytic Cys residues and cleavage sites for each SHFV PLP. In the in vitro reactions, expected products as well as precursor peptides and alternative products were detected and PLPγ was shown to be able to cleave at both downstream and upstream nsp1 junction sites. However, only mature nsp1α, nsp1β, and nsp1γ were detected in infected cells.
MATERIALS AND METHODS
Sequence and bioinformatics analyses.
Sequence analysis was performed on the ORF1a protein sequence that was conceptually translated from an updated SHFV genome sequence (GenBank accession no. AF180391.2). The SHFV PLP1α, PLP1β, and PLP1γ boundaries were estimated using the putative cleavage sites and catalytic domains of the PLP1s of LDV and PRRSV. The putative SHFV PLP1 sequences were aligned with each other and also with the PLP1s of other arteriviruses. All sequences were manipulated in Geneious software suite version 5.6.3 (BoiMatters, Ltd.), and multiple alignments of the PLP1 amino acid sequences were performed using ClustalX in Geneious, with similarity determined using the PAM250 substitution matrix. Once the putative boundaries were predicted, homology models were separately generated for SHFV nsp1α, SHFV nsp1β, and SHFV nsp1γ using the crystal structure of a PRRSV nsp1α protein (PDB code 3IFU) (10) and also the crystal structure of a PRRSV nsp1β protein (PDB code 3MTV) (11) as the template. Homology models were generated using MODELLER (25) (http://salilab.org/modeller/) as implemented in the Max Planck Institute's Bioinformatics Toolkit (http://toolkit.tuebingen.mpg.de/), and the structures were rendered and compared using MacPymol (Delano Scientific, LLC; http://www.pymol.org).
Cells and virus.
The MA104 cell line was a gift from O. Nainan, Centers for Disease Control and Prevention. These cells were grown in minimal essential medium supplemented with 10% fetal bovine serum and 10 μg/ml gentamicin at 37°C in a 5% CO2 atmosphere. An aliquot of SHFV, strain LVR 42-0/M6941, passage 2, was obtained from the American Type Culture Collection, sequentially plaque purified three times, and then amplified once on MA104 cell monolayers. To make experimental virus pools, confluent MA104 monolayers were infected with the stock virus at a multiplicity of infection (MOI) of 0.2 and culture media were harvested at 32 h after infection and clarified by centrifugation. Virus pools contained titers of ∼107 PFU/ml, and aliquots were stored at −80°C.
Construction of SHFV nsp1 clones.
A region consisting of the 5′ 1,934 nucleotides (nt) of the SHFV genome, which included the 5′ noncoding region (NCR) (209 nt) and the 5′ 1,725 nt of ORF1a, was amplified by reverse transcription-PCR (RT-PCR) from purified SHFV-LVR genomic RNA using the EcoRI- and TaqI-tailed forward primer 5′-AGgaattctcgaGATTAAAATAAAAGTGTGAAG-3′ (restriction sites are indicated by lowercase characters, and two 5′ nt were added at the end to enhance restriction enzyme cleavage) and the XbaI-tailed reverse primer (5′-GCtctagaACCGGCAGTACAGCATGGGT-3′). The cloned N-terminal region of ORF1a contained the entire nsp1 region as well as the first 273 nt of nsp2. An in-frame N-terminal Flag tag and an in-frame C-terminal c-Myc tag were added by subcloning nt 210 through 1934 of the SHFV genome into the pFlag/c-Myc expression vector (Sigma), generating pFlag-SHFV-C-Myc. To construct plasmid DNA for use as the template in coupled in vitro transcription/translation reactions, the Flag–SHFV–c-Myc region of the pFlag-SHFV-C-Myc clone was subcloned into the pTNT expression vector (Promega), generating pTNT-FSM-wt. Mutagenesis of the pTNT-FSM-wt construct was performed using a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. The primers used to change the putative PLP1 catalytic Cys residues to Ala or to replace the −2 and −1 residues of each predicted cleavage site with either Val or Ala are listed in Table 1. All wild-type and mutant sequences generated were confirmed by sequencing.
TABLE 1.
Primersa
| Construct | Mutation(s) | Primer direction | Primer sequence (5′–3′) |
|---|---|---|---|
| mα | Cys63 → Ala | F | GGGCGCTATgctGCTCTTGAGATGATA |
| R | TATCATCTCAAGAGCagcATAGCGCCC | ||
| mβ | Cys246 → Ala | F | CTTTGAGCATGGCCGCgcCTGGCTGAAGTTGTTC |
| R | GAACAACTTCAGCCAGgcGCGGCCATGCTCAAAG | ||
| mγ | Cys378 → Ala | F | CCTCACTGCTGGGTTCgcTTGGTTGCAGCTATTTCC |
| R | GGAAATAGCTGCAACCAAgcGAACCCAGCAGTGAGG | ||
| G164/G165 | Thr163 → Val/Gly164 → Val | F | CCATTCAACACGgtTGtAGGTGACGTTTATC |
| R | GATAAACGTCACCTaCAacCGTGTTGAATGG | ||
| G171/T172 | Leu170 → Ala/Gly171 → Ala | F | GGAGGTGACGTTTATCAGgcAGcAACCTGCACTATCGTTGAG |
| R | CTCAACGATAGTGCAGGTTgCTgcCTGATAAACGTCACCTCC | ||
| G271/V272 | Cys270 → Val/Gly271 → Val | F | CGGTTATCAATTGAACgtTGtCGTTCAAGGAAAATACATTGC |
| R | GCAATGTATTTTCCTTGAACGaCAacGTTCAATTGATAACCG | ||
| G350/G351 | Phe349 → Ala/Gly350 → Ala | F | TTAAGTTGCCAGGGAAGACTTATgcCGcTGGAAATGCCAGTTCGG |
| R | CCGAACTGGCATTTCCAgCGgcATAAGTCTTCCCTGGCAACTTAA | ||
| G484/G485 | Arg483 → Ala/Gly484 → Ala | F | CGTTACGGTCGCCGTgcgGcAGGCGGCAAGAAGTCAGGCCAATCG |
| R | CGATTGGCCTGACTTCTTGCCGCCTgCcgcACGGCGACCGTAACG | ||
| G485/G486 | Gly484 → Ala/Gly485 → Ala | F | CGTTACGGTCGCCGTCGAGcAGcCGGCAAGAAGTCAGGCCAATCG |
| R | CGATTGGCCTGACTTCTTGCCGgCTgCTCGACGGCGACCGTAACG |
F, forward; R, reverse. The substituted nucleotides in the primer sequences are indicated by lowercase letters.
In vitro, coupled transcription/translation reactions.
Wild-type and mutant pTNT-FSM cDNA constructs were used as the templates for in vitro coupled transcription/translation reactions performed using a TNT coupled wheat germ extract system (Promega) according to the manufacturer's protocol. Briefly, 1 μg of the plasmid DNA was mixed with 25 μl of wheat germ extract, 2 μl of TNT reaction buffer, 1 μl of amino acid mixture without Cys, 1 μl of TNT SP6 RNA polymerase, and 20 μCi of [35S]Cys (PerkinElmer) (1 mCi/mmol) in a total volume of 50 μl. Reaction mixtures were incubated at 30°C for 2 h prior to use for immunoprecipitation (IP).
Immunoprecipitation.
In vitro translation reaction mixtures (50 μl) were divided into four aliquots. One aliquot (∼5 μl) was stored at −20°C until use as the lysate control for gel electrophoresis. One 15-μl aliquot was incubated with control murine anti-IgG antibody and another with anti-c-Myc monoclonal antibody (Sigma) for 1 h with rotation at 4°C, after which protein G agarose (Roche) was added, and the samples were next rotated at 4°C overnight and then pelleted by centrifugation. The third aliquot (15 μl) was incubated with anti-Flag M2 affinity agarose beads (Sigma) with rotation at 4°C overnight. The beads were next washed 3 times in lysis buffer (1% Triton X-100, 0.1% SDS, 150 mM NaCl, and 50 mM Tris HCl, pH 7.4) containing Halt protease and phosphatase inhibitor cocktail (Pierce Scientific) and then pelleted by centrifugation. After addition of 30 μl of 2× sample loading buffer (20% SDS, 25% glycerol, 0.5 M Tris-HCl [pH 6.8], 0.5% bromophenol blue, and 5% β-mercaptoethanol) to the IP pellets and the lysate aliquot, the samples were boiled for 5 min. The peptides in half of each sample were separated by SDS-PAGE on a 13% polyacrylamide gel, and the remainder of the sample was frozen for use in a repeat analysis. The gels were fixed in 10% acetic acid and 30% methanol, incubated in Autofluor (National Diagnostics) and then in anticracking buffer (7% acetic acid, 7% methanol, and 1% glycerol), dried, and autoradiographed.
MS sequencing of autoprocessed peptides.
Large-volume (200-μl) in vitro translation reactions were performed as described above except that an amino acid mixture containing Cys was used. The wild-type nsp1 construct and the PLP1β catalytic Cys246Ala mutant construct were separately transcribed and translated and the peptides immunoprecipitated with anti-c-Myc monoclonal antibody or anti-Flag M2 affinity agarose beads. The precipitated peptides were separated on different lanes of a 12% NuPAGE bis-tris (2-[bisamino]-2–1,3-propanediol) gel using NuPAGE MOPS (morpholinepropanesulfonic acid) SDS buffer (Life Technology). Precise Plus protein standards (Bio-Rad) were run on a separate lane. The proteins were stained with a colloidal blue staining kit (Life Technology), and the gel was sent to the Wistar Institute proteomics facility, Philadelphia, PA. Selected bands were excised, and after trypsin digestion, the peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on an LTQ-Orbitrap XL mass spectrometer. The MS/MS spectra generated were searched against a custom database containing the SHFV nsp1-plus-N-terminal nsp2 (nsp1+N-terminal nsp2; amino acids [aa] 1 to 575) construct sequence, the Triticum proteome, and common contaminants using SEQUEST.
Western blotting.
Infected MA104 monolayers were lysed by addition of radioimmunoprecipitation assay (RIPA) buffer (1× phosphate-buffered saline, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) containing Halt protease inhibitor cocktail (Thermo Scientific). Following separation by SDS-PAGE, cell proteins were electrophoretically transferred to a nitrocellulose membrane. Membranes were blocked with 1× Tris-buffered saline (TBS; 0.01 M Tris and 0.15 M NaCl, pH 8) containing 5% nonfat dry milk and 0.1% Tween 20 before incubation in the presence of blocking buffer with one of the SHFV nsp1 monospecific, polyclonal antibodies made by Abgent. The peptides used to make these antibodies were as follows: for nsp1α, GDLTRPEETPLPGGC; for the nsp1β N terminus, FAQKVITAFPEGVLC; for the nsp1β C terminus, DESVPPDCQIIARF; and for nsp1γ, FPPLSRKSEAQRAIL. Actin was used as a loading control and was detected with antibody C-11 (Santa Cruz Biotechnology). Blots were washed with 1× TBS and then incubated with either anti-rabbit horseradish peroxidase (Santa Cruz Biotechnology) or anti-mouse horseradish peroxidase (Santa Cruz Biotechnology). After washes, the blots were processed for chemiluminescence using a Super-Signal West Pico detection kit (Pierce Scientific) according to the manufacturer's protocol.
RESULTS
Analysis of SHFV nsp1s by sequence alignment and homology modeling.
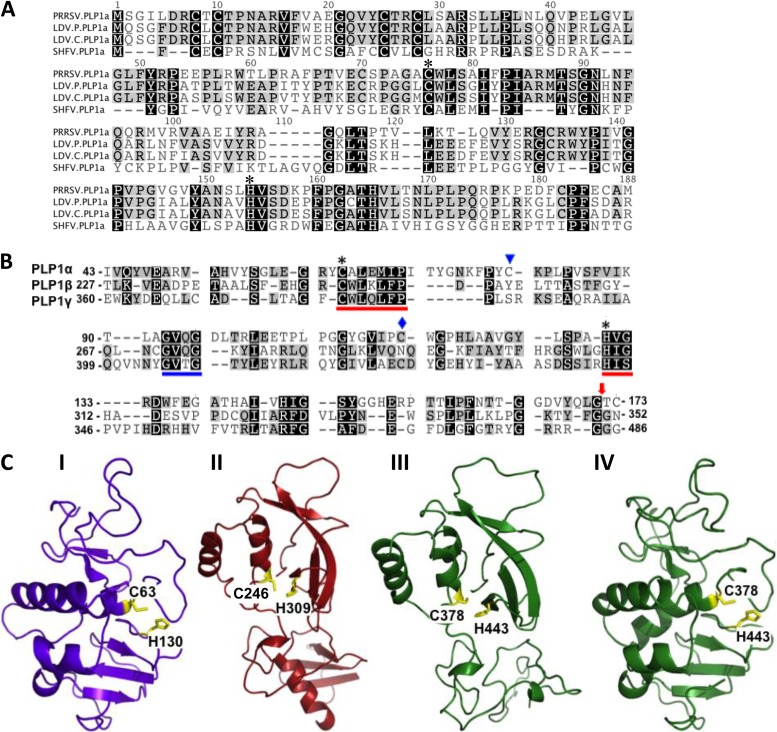
In all previously studied active arterivirus PLP1s, a Trp follows the catalytic Cys (10, 11, 16), but this is not the case for all of the coronavirus PLP1s or for arterivirus PLP2s (8, 13). An initial inspection of the sequences of the three SHFV PLP1 catalytic domains revealed a single Cys-Trp upstream of a His for each PLP1: Cys115/Trp116/His130 (indicated by a blue diamond in Fig. 1B) in PLPα, Cys246/Trp247/His309 in PLP1β, and Cys378/Trp379/His443 in PLP1γ (indicated by an asterisk in Fig. 1B). However, the PLP1α Cys115/Trp116 was located only 14 aa upstream from the catalytic His130 residue (indicated by an asterisk in Fig. 1B) whereas the Cys-Trp of PLP1β and that of PLP1γ were located 62 and 64 aa, respectively, upstream of their His residues (indicated by an asterisk in Fig. 1B). A previous study predicted that the SHFV PLP1α catalytic Cys would be Cys79 (indicated by a blue triangle in Fig. 1B) based on an alignment of the nsp1α sequences of PRRSV, EAV, LDV, and SHFV (10). The sequence context of this SHFV Cys is -X-Cys-Lys-Pro- instead of the conserved -X-Cys-Trp-Lys- context of the PRRSV and LDV nsp1α proteins (Fig. 1A). However, the catalytic function of SHFV Cys79 was not functionally tested in the previous study. In the present study, the putative SHFV PLP1α domain boundaries were first estimated by comparison with the previously reported cleavage sites and catalytic residues of the LDV and PRRSV PLP1α domains (Fig. 1A). The EAV PLP1α was not included in the alignment since it is inactive and shorter than the others. This alignment predicted Cys63 as the SHFV PLP1α catalytic Cys. An additional alignment of the three SHFV PLP1 regions with each other identified a conserved GV[Q/T]G motif approximately halfway between the catalytic Cys and His residues in the putative SHFV PLP1β and PLP1γ domains (blue underline, Fig. 1B). This GV[X]G motif was also conserved in the PLP1β regions of PRRSV and LDV (11). The position of this motif in SHFV PLP1α (aa positions 93 to 96) was then used as an anchor to align the three SHFV PLP1s with each other. Cys63 aligned with the predicted catalytic Cys residues of both SHFV PLP1β and PLP1γ and was located 67 aa from the PLP1α catalytic His130 (indicated by an asterisk in Fig. 1A and B). However, Cys63 is followed by an Ala instead of by the Trp present in all of the other active arterivirus PLP1s (Fig. 1A) (10, 11).
FIG 1.
Prediction of the three SHFV PLP domains. (A) Alignment of SHFV PLP1α with LDV and PRRSV PLP1α domains. (B) Alignment of the three SHFV PLP1 (α, β, and γ) domains. Asterisks indicate the predicted catalytic residues of each protease. A blue diamond indicates the only Cys adjacent to a Trp in SHFV PLP1α. A blue triangle indicates the Cys previously predicted to be the SHFV PLP1α catalytic residue. A red arrow indicates the position of the predicted SHFV PLP1 cleavage sites. Red lines indicate the conserved catalytic residue motifs. A blue line indicates a conserved motif used to anchor the alignment. (C) Homology models of the three SHFV PLP1 proteins generated using the crystal structure of the PRRSV nsp1α protein for PLP1α (I) and the PRRSV nsp1β structure for SHFV PLP1β (II) and PLP1γ (III). For comparison, SHFV nsp1γ was also modeled on the PRRSV nsp1α structure (IV). The predicted catalytic Cys and His residues for each SHFV PLP1 are indicated in yellow.
Homology models of the three SHFV nsp1 proteins were next generated as described in Materials and Methods to determine whether the predicted catalytic residues were proximal to one another in a manner that would facilitate activity. The homology model of SHFV nsp1α generated using the crystal structure of the type II PRRSV (strain XH-GD) nsp1α (10) and the homology model of SHFV nsp1β generated using the crystal structure of the type II PRRSV (strain XH-GD) nsp1β are shown in Fig. 1C. The homology models suggested that the sequence and structure of SHFV nsp1α were more similar to those of PRRSV nsp1α whereas the sequence and structure of SHFV nsp1β were more similar to those of PRRSV nsp1β. Because the homology modeling did not clearly suggest which of the PRRSV nsp1 structures was the most similar to that of SHFV nsp1γ, the SHFV nsp1γ homology models generated on the basis of the structures of both PRRSV nsp1α and PRRSV nsp1β are shown (Fig. 1C). The SHFV nsp1γ homology model made on the basis of the PRRSV nsp1β structure appeared to be less structured in the lower half than the model made on the basis of the PRRSV nsp1α structure. However, homology modeling is limited in that it models a nonidentical but similar amino acid sequence on the basis of a fixed structural template. The actual structures of the SHFV nsp1 proteins will not be known until crystal structures for each have been solved. In the modeled structure of SHFV nsp1α, Cys63 was located close to His130 in positions spatially similar to those of the catalytic Cys and His residues of PRRSV nsp1α. The alignment and modeling data supported the hypothesis that the SHFV PLPα catalytic residues are Cys63 and His130.
Most coronavirus PLPs recognize and cleave canonical -Lys-X-Gly↓Gly- sites, but some can cleave at -Lys-X-Gly↓Ala- sites (8). In contrast, the arterivirus PLP1 cleavage site sequences are not well conserved. Type II PRRSV nsp1α cleaves at -Cys-Ala-Met↓Ala-Asp- and type II PRRSV nsp1β at -Trp-Tyr-Gly↓Ala-Gly- (10, 11). Both a previous alignment (10) and the new arterivirus PLP1α alignment shown in Fig. 1A predicted the SHFV nsp1α cleavage site to be -Thr-Thr-Gly↓Gly-Asp-. However, the alignment of the three SHFV PLP1s predicted that cleavage would occur at a downstream -Gln-Leu-Gly↓Thr-Cys- site (Fig. 1B, red arrow). Both the previous alignment (10) and the three SHFV PLP1 alignments predicted that SHFV nsp1β cleavage would occur at a -Thr-Phe-Gly↓Gly- site, and the alignment of the three SHFV PLP1s predicted that SHFV nsp1γ cleavage would occur at a -Arg-Arg-Gly↓Gly-Gly- site (Fig. 1B, red arrow).
Analysis of the peptides produced in vitro by autoprocessing of wild-type and mutant SHFV N-terminal ORF1a polyprotein fragments.
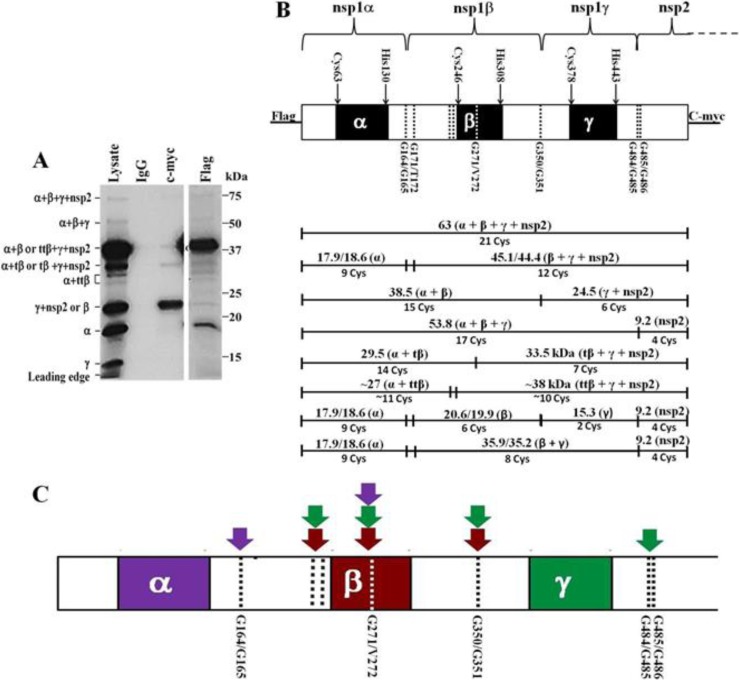
An expression plasmid containing the N-terminal region of ORF1a (genome nt 210 through 1934) fused to an N-terminal Flag tag and a C-terminal c-Myc tag was constructed as described in Materials and Methods. The expressed polyprotein included the complete SHFV nsp1 (nsp1α, nsp1β, and nsp1γ) region plus the N-terminal (91-aa) region of nsp2. Wild-type cDNA was first used as the template in a coupled in vitro transcription/translation reaction done in the presence of [35S]Cys. The peptides produced by PLP1 autoprocessing were immunoprecipitated (IP) with anti-Flag, anti-c-Myc, or a control IgG antibody. The peptides in the IP samples and also an aliquot of the unprecipitated reaction (lysate) were separated by SDS-PAGE (Fig. 2A). The wild-type construct produced 9 bands (∼63, ∼54, ∼39, ∼30, ∼28, ∼27, ∼23, ∼18, and ∼15 kDa), and these peptides were identified by their predicted sizes and by the presence or absence of terminal tags (Fig. 2B). The full-length peptide (63 kDa), the intermediate precursors nsp1α plus nsp1β plus nsp1γ (here designated nsp1α+β+γ) (54 kDa) and nsp1α +β (39 kDa), and nsp1α (18 kDa) were pulled down by anti-Flag antibody, while the full-length peptide (63 kDa) and the intermediate precursor nsp1γ +nsp2 (23 kDa) were pulled down by the anti-c-Myc antibody (Fig. 2A). The 15-kDa band present only in the lysate lane was most likely nsp1γ since neither the anti-Flag nor the anti-c-Myc antibody pulled down this peptide (see Fig. 2B). The nsp1γ band was not as efficiently detected as the other peptides due to the presence of fewer Cys residues in this peptide (Fig. 2B). The 22.1-kDa nsp1β peptide would be expected to migrate to a position on the gel similar to that of the 24.5-kDa nsp1γ+nsp2 precursor. Because nsp1β is an internal peptide, it would not be tagged and would be detected only in the lysate lane whereas the c-Myc-tagged nsp1γ+nsp2 band would be detected in both the lysate and c-Myc IP lanes. The detection of both nsp1α and nsp1γ indicated that efficient cleavage occurred at both the nsp1α-nsp1β and the nsp1β-nsp1γ junctions.
FIG 2.
SHFV nsp1 polyprotein cleavage map. The N-terminal 1,725 nt of SHFV ORF1a were cloned into the pTNT vector. The polyprotein expressed had an N-terminal Flag tag and a C-terminal c-Myc tag. (A) Products of the wild-type SHFV nsp1 polyprotein autoprocessing. The wild-type cDNA was in vitro transcribed and translated in a coupled TNT reaction (Promega) as described in Materials and Methods. Protein products labeled by incorporation of [35S]Cys were immunoprecipitated with murine IgG, anti-c-Myc, or anti-Flag antibody prior to separation by 13% SDS-PAGE. The positions of protein standards are indicated on the right, and the identities of the SHFV peptides generated are indicated on the left. (B) Diagram showing the relative locations of the predicted PLP1 domains and cleavage sites in the SHFV nsp1 polyprotein and the predicted cleavage products. (Top panel) Black boxes indicate the predicted PLP1 domains, and dotted lines indicate the cleavage sites. (Bottom panel) The sizes and identities of the predicted cleavage products are indicated above the lines, and the number of Cys residues in each peptide is indicated below the lines. (C) The sites shown by the data to be cleaved by each SHFV PLP1 in in vitro autoprocessing reactions are indicated by color-coordinated arrows.
In addition to the expected precursor and product bands, an ∼30-kDa band was present in the lysate, Flag, and c-Myc IP lanes but not in the IgG IP lane, suggesting that this lysate band contained both N-terminal and C-terminal peptides (Fig. 2A). Consistent with this hypothesis, the intensities of the 30-kDa bands in each of the IP lanes were lower than that of the band in the lysate lane. It is likely that the 30-kDa lysate band contained two precursor peptides, one consisting of nsp1α plus an N-terminal portion of nsp1β and a second consisting of a C-terminal portion of nsp1 plus nsp2. The N- and C-terminal 30-kDa bands were thus designated nsp1α+tβ and tβ+nsp1γ+nsp2, respectively. Three additional, unexpected bands of 23, 27, and 28 kDa were present in both the lysate and the Flag IP lanes (Fig. 2A). Based on the presence of the Flag tag and the estimated sizes of these peptides, they likely represented nsp1α plus various lengths of the N-terminal region of nsp1β (see Fig. 2B). These bands were designated nsp1α+ttβ due to the shorter nsp1β sequences they contained. An ∼39-kDa band was also detected in the c-Myc IP lane that likely represents ttβ+nsp1γ+nsp2. The detection of bands containing the C-terminal portions of each of the alternative N-terminal bands indicated that these peptides were generated by a single cleavage of the polyprotein and not by a mechanism such as premature translation termination.
Functional analysis of the predicted PLP1 catalytic Cys residues.
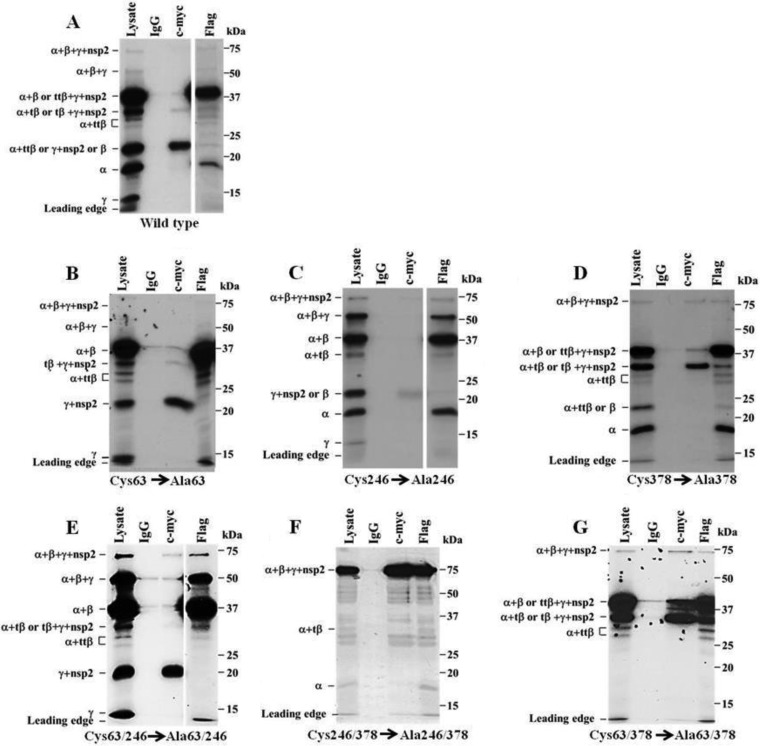
To determine whether the predicted nsp1α catalytic Cys residue was functional, Cys63 was substituted with Ala. The resulting construct was transcribed and translated in vitro, and the products were analyzed either directly or after IP with anti-Flag or anti-c-Myc antibody (Fig. 3B). The nsp1α (18-kDa) band produced by autoprocessing of the wild-type polyprotein was absent from both the lysate and Flag IP lanes, indicating that Cys63 is required for PLP1α cleavage of nsp1α from the polyprotein. Interestingly, the nsp1α+ttβ (27- and 28-kDa) bands were present in both the lysate and Flag lanes and the intensities of these bands were increased compared to those produced by the wild-type construct, suggesting that cleavage at sites within the N-terminal region of nsp1β increased when the nsp1α PLP1 was not active. A construct with the predicted nsp1β catalytic Cys246 substituted with Ala produced full-length polyprotein (63 kDa) and nsp1α+β+γ polyprotein precursor (54-kDa) bands with increased intensity compared to the wild-type construct, indicating that processing at the nsp1β-nsp1γ junction was decreased when the predicted nsp1β catalytic Cys was mutated (Fig. 3C). However, detection of nsp1γ (15 kDa) in the lysate lane and nsp1γ+nsp2 (23 kDa) in the lysate and c-Myc lanes indicated that cleavage at the nsp1β-nsp1γ junction still occurred when PLPβ was inactive and suggested that cleavage at this junction was not exclusively dependent on PLPβ. For the construct with the predicted nsp1γ catalytic Cys378 substituted with an Ala, the absence of nsp1γ (15 kDa) in the lysate lane and also the absence of nsp1α+β+γ (54 kDa) in the lysate and Flag lanes indicated that cleavage at the nsp1γ-nsp2 junction did not occur (Fig. 3D). The detection of the ttβ/tβ+nsp1γ+nsp2 peptides (39- and 30-kDa bands in the lysate and c-Myc lanes) provided additional evidence that nsp1γ was not cleaved from nsp2 when the PLPγ catalytic Cys was mutated. Cleavage occurred efficiently at both the nsp1α-nsp1β and nsp1β-nsp1γ junctions when the predicted nsp1γ catalytic Cys378 was substituted.
FIG 3.
Cleavage products produced by nsp1 polyproteins containing mutations in one or two catalytic Cys residues. (A) Wild-type polyprotein. (B) PLP1α Cys63 mutant. (C) PLP1β Cys246 mutant. (D) PLP1γ Cys378 mutant. (E) PLP1α Cys63 and PLP1β Cys246 mutant. (F) PLP1β Cys246 and PLP1γ Cys378 mutant. (G) PLP1α Cys63 and PLP1γ Cys378 mutant. Wild-type and mutant cDNAs were in vitro transcribed and translated and the cleavage products immunoprecipitated and analyzed as described in the legend of Fig. 2A. A shorter exposure is shown for the Flag IP lanes in panels A, C, and E.
Mutation of PLP1α Cys63 inhibited cleavage at the nsp1α-nsp1β junction, and mutation of PLP1γ Cys378 inhibited cleavage at the nsp1γ-nsp2 junction. However, cleavage at the nsp1β-nsp1γ junction was still observed when PLP1β Cys246 was mutated, suggesting that either PLP1α or PLP1γ can also cleave at the nsp1β-nsp1γ junction. To analyze the activity of each PLP1 individually, double-mutant constructs that produced polyproteins with only one active PLP were made. The construct with both PLP1α Cys63 and PLP1β Cys246 substituted with Ala produced an increased amount of nsp1α+β+γ (54 kDa) in the Flag IP lane compared to the wild-type construct, indicating that the nsp1γ-nsp2 junction was efficiently cleaved (Fig. 3E). The detection of nsp1γ (15 kDa) in the lysate lane and nsp1α+β (39 kDa) in the Flag IP lane indicated that the nsp1β-nsp1γ junction was also cleaved when only PLPγ was active. Interestingly, the nsp1α+tβ and nsp1α+ttβ (30-, 28-, and 27-kDa) bands were also detected in the Flag IP lanes, suggesting that PLP1γ can cleave at multiple sites within nsp1β. The absence of an nsp1α (18-kDa) band in both the lysate and Flag IP lanes as well as the observed increase in the intensity of the nsp1α+β (39-kDa) band in the lysate and Flag IP lanes compared to the bands produced by the wild-type construct indicated that the nsp1α-nsp1β junction was not cleaved by PLPγ.
A double mutant with PLP1β Cys246 and PLPγ Cys378 substituted with Ala produced a high-intensity full-length polypeptide (63-kDa) band in the lysate, c-Myc IP, and Flag IP lanes as well as nsp1α+tβ (30 kDa) and nsp1α (18-kDa) bands in the lysate and Flag IP lanes (Fig. 3F). Although an nsp1α band was detected when only PLPα was active, the efficiency of cleavage at the nsp1α-nsp1β junction was decreased compared to that of the wild-type polyprotein. The detection of the 30-kDa band in both the lysate and Flag IP lanes indicated that PLP1α is also able to cleave within the catalytic region of nsp1β, but this cleavage was also inefficient when PLP1α was the only protease. The lack of detection of nsp1γ+nsp2 (23 kDa) in the lysate and c-Myc lanes and of nsp1α+β (39 kDa) in the lysate and Flag lanes indicated that cleavage did not occur at the nsp1β-nsp1γ junction. Likewise, the lack of detection of nsp1γ (15 kDa) in the lysate lane and of nsp1α+β+γ (54 kDa) in the lysate and Flag lanes indicated that cleavage also did not occur at the nsp1γ-nsp2 junction. Multiple weak background bands were detected in the lysate, Flag IP, and c-Myc IP lanes, suggesting that either some premature termination or polyprotein breakdown had occurred.
A mutant construct with both PLP1α Cys63 and PLP1γ Cys378 substituted with Ala was next tested (Fig. 3G). Detection of nsp1α+β (39 kDa) in the Flag lane indicated that cleavage occurred at the nsp1β-nsp1γ junction. The detection of nsp1α+ttβ/tβ (30, 28 and 27 kDa) and ttβ/tβ+nsp1γ+nsp2 (39 and 30 kDa) unexpectedly indicated that PLP1β can also cleave within its own catalytic region as well as within the N-terminal region of nsp1β. The lack of detection of nsp1γ (15 kDa) in the lysate lane and of nsp1α (18 kDa) in the lysate and Flag lanes indicated that PLP1β is not able to cleave at either the nsp1α-nsp1β or nsp1γ-nsp2 junctions. Because self-cleavage by a protease within its own catalytic region seemed unlikely, additional bioinformatics analyses were done to determine the possible existence of an additional protease with a catalytic domain located between Cys115/Trp116 and His181 or His185. However, neither sequence alignment nor homology modeling supported the existence of an additional protease between aa 100 and 203 (data not shown).
Functional analysis of the cleavage sites utilized by the SHFV PLP1s.
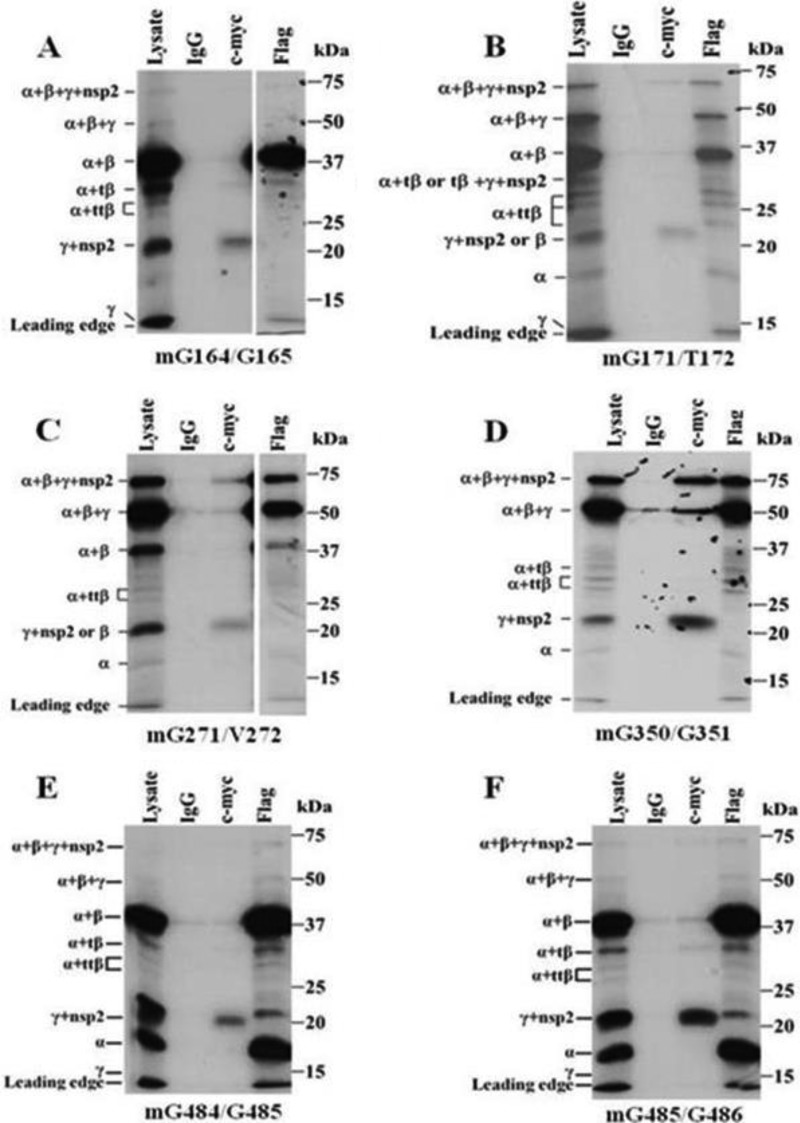
Previous studies showed that substitutions of the −1 and −2 amino acids of PLP cleavage sites prevented their recognition and cleavage (26, 27). The −1 and −2 positions of each of the predicted SHFV PLP1 cleavage sites were substituted with Ala or Val (Table 1). The PLP1α cleavage site, -Thr-Thr-Gly↓Gly- (aa 162 to 165), was predicted by the multiple arterivirus PLP1α alignment shown in Fig. 1A and also by a previously published alignment (10). When the peptides produced by a construct with the −1 and −2 positions of this site mutated were analyzed (Fig. 4A), the nsp1α (18-kDa) band was not detected in the lysate or Flag IP lanes consistent with this being the preferred nsp1α cleavage site. When the PLP1α cleavage site, -Gln-Leu-Gly↓Thr- (aa 169 to 172), predicted by the alignment of the three SHFV PLP1s and homology modeling (Fig. 1B and C) was mutated, the nsp1α (18-kDa) band was still detected in the lysate and Flag IP lanes but the intensity of these bands was lower than in the wild-type reactions (Fig. 4B). The intensities of the full-length (63-kDa) and nsp1α+β+γ (54-kDa) precursor protein bands were increased relative to the intensities of these bands in wild-type autoprocessing reactions, suggesting that the overall cleavage efficiency of this mutant polypeptide was reduced. Both the alignment of the three SHFV PLP1s (Fig. 1B) and a previous arterivirus PLP1β alignment predicted -Tyr-Phe-Gly↓Gly- (aa 348 to 351) to be the nsp1β cleavage site. A construct with the −2 and −1 aa residues of this site substituted produced nsp1α+β+γ (54-kDa) bands with increased intensity and nsp1α+β (39-kDa) bands with decreased intensity in the lysate and Flag IP lanes (Fig. 4D), indicating that the -Tyr-Phe-Gly↓Gly- (aa 348 to 351) site is the main cleavage site at the nsp1β-nsp1γ junction. The alignment of the SHFV PLP1s predicted -Arg-Arg-Gly↓Gly-Gly- (aa 482 to 486) to be the PLPγ cleavage site (Fig. 1B). To determine whether cleavage occurs between G484 and G485, the −2 and −1 aa residues of this site were substituted. This construct produced an nsp1γ+nsp2 (23-kDa) band that was detected in the c-Myc lane, but no nsp1γ (15-kDa) band was detected in the lysate lane, indicating that cleavage did not occur at the nsp1γ-nsp2 junction (Fig. 4E). To determine whether cleavage can also occur between G485 and G486, the −2 and −1 aa residues of this site were substituted. The bands produced by this construct were the same as those produced by the G484/G485 mutant construct (Fig. 4F). Because G484 was mutated in both constructs (−2 aa and −1 aa), it was not possible to distinguish experimentally which of these sites was used.
FIG 4.
Cleavage products produced by nsp1 polyproteins with a single predicted PLP cleavage site mutated. mG164/G165 (A), mG171/T172 (B), mG271/V272 (C), mG350/G351 (D), mG484/G485 (E), and mG485/G486 (F) cDNAs were in vitro transcribed and translated in a coupled TNT reaction in the presence of [35S]Cys, immunoprecipitated, and analyzed as described in the legend of Fig. 2A. A shorter exposure was used for the Flag IP lane in panels A and C.
Production of the 30-kDa nsp1α+tβ and tβ+nsp1γ+nsp2 bands by cleavage within the catalytic region of PLP1β was observed with the wild-type and many of the mutant polyproteins. Each of the SHFV PLP1s was predicted to cleave between two Gly residues at each of the nsp1 junctions. Analysis of the PLP1β sequence did not reveal any -Gly-Gly- tandems. However, an -Asp-Cys-Gly-Val- (269- to 272-aa) sequence was found within the predicted nsp1β catalytic region (Fig. 1B). A construct with the −2 and −1 aa residues of this site substituted with Val produced no 30-kDa band in the lysate, Flag IP, or c-Myc IP lanes, suggesting that the N- and C-terminal 30-kDa bands were produced by cleavage at the Asp-Cys-Gly↓Val- (269- to 272-aa) site (Fig. 4C). The increased intensity of the full-length polyprotein (63-kDa) and nsp1α+β+γ (54-kDa) bands indicated that mutation of this site decreased the overall cleavage efficiency of the polyprotein.
The wild-type construct produced two other unexpected nsp1α+ttβ (28- and 27-kDa) bands that were precipitated by anti-Flag but not by anti-c-Myc antibody, indicating that these two proteins contained nsp1α and part of nsp1β. Based on their sizes, these two proteins are likely to be produced by cleavages within the N-terminal region of nsp1β. However, the sites at which these cleavages occur are not currently known.
MS sequencing of selected in vitro autoprocessing cleavage products.
As an additional means of identifying the cleavage sites for each SHFV nsp1, selected autoprocessing products were subjected to sequence analysis by MS. The positions of the SHFV peptide bands were estimated by comparison to the positions of the protein standards and also to the positions of radiolabeled SHFV peptide bands on gels obtained in previous experiments because the small amounts of immunoprecipitated peptides could not be detected by protein staining. The estimated nsp1α (18-kDa) and nsp1α+β (39-kDa) precursor regions were separately excised from the Flag IP lane of a wild-type construct autoprocessing reaction, while the nsp1α+β+γ (54-kDa) region was excised from the Flag IP lane of a Cys246Ala mutant construct autoprocessing reaction. Trypsin peptides aligning to the expected regions in each product were detected (Fig. 5). For nsp1α, 10 trypsin-generated peptides ending at aa 164 were detected, indicating that the -Thr-Thr-Gly↓Gly- (aa 162 to 165) site was the preferred site for PLP1α cleavage (Fig. 5A). One peptide ending at aa 171 was also detected, suggesting that this alternative site can also be used by PLP1α in vitro. No peptides matching the sequence downstream of the alternative cleavage site were detected.
FIG 5.
MassSpec analyses of selected polyprotein cleavage products. (A) nsp1α. (B) nsp1α+β. (C) nsp1α+β+γ. Products from in vitro transcription/translation reactions were separated on a 12% NuPAGE bis-tris gel and stained with colloidal blue. The nsp1α (18-kDa) band and the nsp1α+β (39-kDa) precursor band were separately excised from the Flag IP lane of a wild-type construct reaction, while the nsp1α+β+γ (54-kDa) band was excised from the Flag IP lane of a Cys246Ala mutant construct reaction. Proteins in the excised bands were subjected to in-gel trypsin digestion and the peptides analyzed by LC-MS/MS on an LTQ-Orbitrap XL mass spectrometer. The MS/MS spectra generated were searched against a custom database containing the SHFV nsp1+N-terminal nsp2 construct sequence, the Triticum proteome, and common contaminants using SEQUEST. Regions of the nsp1 sequences for which peptides were detected are highlighted in yellow. Oxidized amino acids are indicated in green, and methylated amino acids are indicated in red. The predicted PLP1 cleavage sites are indicated with red arrows. The N-terminal Flag tag is boxed in red, and the first amino acid of the nsp1 polyprotein is indicated by a red asterisk. The entire nsp1α sequence is shown. For nsp1α+β and nsp1α+β+γ, only the C-terminal portion of the peptide is shown.
Peptide coverage for the nsp1α+β (39-kDa) precursor was lower than that for nsp1α (Fig. 5B). Seven peptides were detected that extended to aa 342. Trypsin primarily cleaves on the C-terminal side of Lys or Arg residues except when a Pro follows one of these amino acids. Two Lys residues were located on the N-terminal side of the predicted -Tyr-Phe-Gly↓Gly- (aa 348 to 351) nsp1β cleavage site (−5 and −9 positions), which resulted in peptide coverage ceasing just before the predicted cleavage site because peptides shorter than 6 aa were not aligned. The positions of upstream trypsin cleavage sites and the lack of detection of peptides aligning to downstream sequences indicated that cleavage occurred at or very near the predicted nsp1β cleavage site. Three Arg residues are located immediately upstream of the predicted nsp1γ cleavage site -Arg-Arg-Arg-Gly↓Gly-Gly- (aa 481 to 486). The detection of 14 peptides ending at Arg478 and no peptides aligning to downstream sequences strongly suggested that cleavage occurred at the predicted nsp1γ cleavage site (Fig. 5C).
Analysis of nsp1 processing in infected cells.
Nsp1α, nsp1β, and nsp1γ processing was next analyzed in infected cells. MA104 cells were infected with SHFV-LVR at an MOI of 1. Cell lysates were collected at various times after infection and analyzed by Western blotting with anti-nsp1α, anti-nsp1β, or anti-nsp1γ antibody. The anti-nsp1α antibody detected a single band at 22 kDa (Fig. 6A). A single band at 22 kDa was detected with an anti-N-terminal nsp1β antibody (Fig. 6B) and also with an anti-C-terminal nsp1β antibody (data not shown). Anti-nsp1γ antibody detected a single 18-kDa band (Fig. 6C). The estimated sizes for both nsp1α (22 versus 18 kDa) and nsp1γ (18 versus 15 kDa) were higher in cell lysates than in the in vitro translation assays, suggesting that these proteins are posttranslationally modified in infected cells. The lack of detection of any precursors suggested that rapid processing of the nsp1 proteins from the polyprotein likely occurs in infected cells. Also, no truncated forms of nsp1β or longer forms of nsp1α were detected in cell lysates during the course of the infection, suggesting that cleavage at sites within nsp1β may not occur in infected cells or that these alternative proteins are in low abundance or unstable and rapidly degraded.
FIG 6.
Autoprocessing of SHFV nsp1 proteins in infected cells. MA104 monolayers were infected with SHFV, strain LVR, at an MOI of 1. Cell lysates were harvested in RIPA buffer at the indicated times after infection, and the individual nsp1 viral proteins were detected by Western blotting using anti-SHFV nsp1α (A)-, nsp1β (B)-, or nsp1γ (C)-specific antibody. Anti-actin antibody was used to detect actin on stripped blots. The positions of the protein markers are indicated on the left, and those of the viral protein bands are indicated on the right.
DISCUSSION
Each of the EAV, LDV, and PRRSV genomes encodes two PLP1 domains that rapidly cleave one (the EAV PLP1α is inactive) or two nsp1 proteins from the N terminus of the 1a/1ab nonstructural polyproteins (2). The SHFV genome is unique in that it encodes the three PLP1 domains PLP1α, PLP1β, and PLP1γ (14). The present study showed that each of the SHFV PLP1s is an active protease and that three mature nsp1 proteins are produced in in vitro reactions and in infected cells. In the PLP1 domains of other arteriviruses, the catalytic Cys is adjacent to an aromatic Trp. Consistent with this conservation, the identified catalytic Cys of SHFV PLP1β (Cys246) and that of SHFV PLP1γ (Cys378) are both adjacent to a Trp residue. The only Cys within the SHFV nsp1α region with an adjacent Trp was located only 14 aa from the predicted catalytic His. However, Cys63, which is adjacent to the nonaromatic residue Ala, was predicted by both alignment and homology modeling to be the nsp1α catalytic Cys, and substitution of Cys63 resulted in the loss of the nsp1α cleavage product, confirming that Cys63 is the PLP1α catalytic Cys. While the distances between the catalytic Cys and His residues of SHFV PLP1β and SHFV PLP1γ are 62 and 65 aa, respectively, similar to the distances in other arterivirus PLP1s, Cys63 is 67 aa upstream of its catalytic His residue. The crystal structures obtained for PRRSV nsp1α and nsp1β show that these proteins have similar but distinct structures (10, 11). Both of the PRRSV nsp1 structures were used as templates for modeling each SHFV nsp1. The sequence analysis and homology modeling data suggested that SHFV nsp1α is more similar to PRRSV nsp1α, SHFV nsp1β is more similar to PRRSV nsp1β, and SHFV nsp1γ is more similar to PRRSV nsp1α than to PRRSV nsp1β. However, the actual structures of the SHFV nsp1 proteins will not be known until they are solved by X-ray crystallography.
Although arterivirus PLP1α cleavage site sequences are not conserved, arterivirus PLP1β cleavage sites show some conservation (-Trp/Tyr/Asn-Tyr/Phe-Gly↓Gly-) (10, 11). Each of the three SHFV PLP1s cleaves preferentially between two Gly residues, but the amino acids upstream of these cleavage sites are not conserved. Inefficient PLP1α cleavage was also observed at an alternative downstream -Gln-Leu-Gly↓Thr- (aa 169 to 172) site in vitro. Only cis cleavage at a single site downstream of the catalytic His residue was observed in in vitro autocleavage reactions previously done with EAV, PRRSV, and LDV nsp1 polyprotein fragments (14, 16). Because cleavage of the single expected EAV nsp1 protein occurs rapidly in in vitro polyprotein autoprocessing reactions, it was concluded that cleavage occurred cotranslationally as the growing peptide folded into the appropriate conformation with the cleavage site located in the active site binding pocket (11, 14). The detection of “free” SHFV nsp1γ when the catalytic Cys residues of both PLP1α and PLP1β were substituted indicated that nsp1γ can cleave at both downstream and upstream junction sites. The PLP1s encoded by the coronaviruses murine hepatitis virus A59 and human coronavirus 229E were previously reported to cis cleave at a downstream site and trans cleave at a site upstream of the catalytic Cys residue in in vitro cleavage assays (28, 29). The arterivirus PLP2 in nsp2 was also shown to possess trans cleavage activity (12, 13).
The 575-aa SHFV N-terminal polyprotein (599 aa with the tags) expressed in the in vitro transcription/translation assays in the present study is the longest arterivirus nsp1 polyprotein to have been tested to date and contains three instead of two tandem PLPs. The sites cleaved by each of the SHFV PLP1s in the in vitro autocleavage reactions are summarized in Fig. 2C. PLP1α cleaves preferentially at Gly164↓Gly165. PLP1α cleavage at Gly271↓Val272 located within the catalytic region of PLP1β may occur when the polyprotein is misfolded. The abundance of the 30-kDa bands generated by cleavage at the Gly271↓Val272 site was typically low, suggesting that only a small proportion of the in vitro-synthesized polyproteins were misfolded. However, the intensity of the 30-kDa bands increased when PLP1α was mutated, suggesting that retention of nsp1α may facilitate misfolding of nsp1β. The detection of cleavage at multiple sites within nsp1β when only PLP1β was active was surprising. Misfolding of this region may expose alternative sites for PLP1β cis or trans cleavage. PLP1γ cleaves at the downstream nsp1γ-nsp2 junction (Gly484/Gly485/Gly486) but also at the upstream nsp1β-nsp1γ junction (Gly350/Gly351) and at sites within nsp1β. Cleavage by PLP1γ at the upstream sites may occur in trans similarly to upstream cleavage by coronavirus PLPs (8).
Precursor proteins as well as alternative peptides were produced in the SHFV in vitro autoprocessing reactions but were not detected in infected cells. Each of the SHFV nsp1 proteins is predicted to contain multiple posttranslational modification motifs, including myristoylation, phosphorylation, and glycosylation sites (data not shown). That modifications at some of these predicted sites occur in vivo was suggested by the observed increase in the sizes of the nsp1α and nsp1γ proteins produced in infected cells. These posttranslational modifications may facilitate the folding of the SHFV polyprotein into the optimal conformation for autoprocessing in vivo. The failure to visualize the 30-kDa proteins in infected cells could be due to low abundance or the inability of the individual anti-nsp1 antibodies to detect these proteins. However, the possibility of the rapid degradation of uncleaved intermediate precursor peptides and alternative cleavage products in infected cells cannot be ruled out. The additional nsp1γ region in the SHFV genome is expected to have been generated by a recombination event that resulted in gene duplication. The sequence analysis and homology modeling data suggested that nsp1γ is more similar to nsp1α than to nsp1β. Only nsp1γ was shown to be able to cleave in vitro at both downstream and upstream junction sites. However, there was no junction site upstream of PLP1α. Preliminary data suggest that the individual SHFV nsp1 proteins may differ in their cellular localizations and functions.
ACKNOWLEDGMENTS
This work was supported by a Public Health Service research grant (AI073824) to M.A.B. from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. H.A.V. and H.D. were supported by Molecular Basis of Disease Fellowships from Georgia State University.
Footnotes
Published ahead of print 4 June 2014
REFERENCES
- 1.deVries AA, Horzinek MC, Rottier PJ, Groot RJd. 1997. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin. Virol. 8:33–47. 10.1006/smvy.1997.0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snijder EJ, Kikkert M. 2013. Arteriviruses, p 859–879 In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. (ed), Fields virology, 6th ed, vol 1 Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 3.Sagripanti JL. 1985. Polyadenylic acid sequences in the genomic RNA of the togavirus of simian hemorrhagic fever. Virology 145:350–355. 10.1016/0042-6822(85)90171-0 [DOI] [PubMed] [Google Scholar]
- 4.Sagripanti JL, Zandomeni RO, Weinmann R. 1986. The cap structure of simian hemorrhagic fever virion RNA. Virology 151:146–150. 10.1016/0042-6822(86)90113-3 [DOI] [PubMed] [Google Scholar]
- 5.Godeny EK, deVries AAF, Wang XC, Smith SL, deGroot RJ. 1998. Identification of the leader-body junctions for the viral subgenomic mRNAs and organization of the simian hemorrhagic fever virus genome: evidence for gene duplication during arterivirus evolution. J. Virol. 72:862–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godeny EK, Zeng L, Smith SL, Brinton MA. 1995. Molecular characterization of the 3′ terminus of the simian hemorrhagic fever virus genome. J. Virol. 69:2679–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng L, Godeny EK, Methven SL, Brinton MA. 1995. Analysis of simian hemorrhagic fever virus (SHFV) subgenomic RNAs, junction sequences, and 5′ leader. Virology 207:543–548. 10.1006/viro.1995.1114 [DOI] [PubMed] [Google Scholar]
- 8.Mielech AM, Chen Y, Mesecar AD, Baker SC. 7 February 2014. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 10.1016/j.virusres.2014.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Kasteren PB, Bailey-Elkin BA, James TW, Ninaber DK, Beugeling C, Khajehpour M, Snijder EJ, Mark BL, Kikkert M. 2013. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc. Natl. Acad. Sci. U. S. A. 110:E838–E847. 10.1073/pnas.1218464110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y, Xue F, Guo Y, Ma M, Hao N, Zhang XC, Lou Z, Li X, Rao Z. 2009. Crystal structure of porcine reproductive and respiratory syndrome virus leader protease Nsp1alpha. J. Virol. 83:10931–10940. 10.1128/JVI.02579-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue F, Sun Y, Yan L, Zhao C, Chen J, Bartlam M, Li X, Lou Z, Rao Z. 2010. The crystal structure of porcine reproductive and respiratory syndrome virus nonstructural protein Nsp1beta reveals a novel metal-dependent nuclease. J. Virol. 84:6461–6471. 10.1128/JVI.00301-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han J, Rutherford MS, Faaberg KS. 2009. The porcine reproductive and respiratory syndrome virus nsp2 cysteine protease domain possesses both trans- and cis-cleavage activities. J. Virol. 83:9449–9463. 10.1128/JVI.00834-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snijder EJ, Wassenaar AL, Spaan WJ, Gorbalenya AE. 1995. The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J. Biol. Chem. 270:16671–16676 [DOI] [PubMed] [Google Scholar]
- 14.Snijder EJ, Wassenaar AL, Spaan WJ. 1992. The 5′ end of the equine arteritis virus replicase gene encodes a papainlike cysteine protease. J. Virol. 66:7040–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ziebuhr J, Snijder EJ, Gorbalenya AE. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853–879 [DOI] [PubMed] [Google Scholar]
- 16.den Boon JA, Faaberg KS, Meulenberg JJ, Wassenaar AL, Plagemann PG, Gorbalenya AE, Snijder EJ. 1995. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papainlike cysteine proteases. J. Virol. 69:4500–4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song C, Krell P, Yoo D. 2010. Nonstructural protein 1alpha subunit-based inhibition of NF-kappaB activation and suppression of interferon-beta production by porcine reproductive and respiratory syndrome virus. Virology 407:268–280. 10.1016/j.virol.2010.08.025 [DOI] [PubMed] [Google Scholar]
- 18.Kim O, Sun Y, Lai FW, Song C, Yoo D. 2010. Modulation of type I interferon induction by porcine reproductive and respiratory syndrome virus and degradation of CREB-binding protein by non-structural protein 1 in MARC-145 and HeLa cells. Virology 402:315–326. 10.1016/j.virol.2010.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beura LK, Sarkar SN, Kwon B, Subramaniam S, Jones C, Pattnaik AK, Osorio FA. 2010. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J. Virol. 84:1574–1584. 10.1128/JVI.01326-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beura LK, Subramaniam S, Vu HL, Kwon B, Pattnaik AK, Osorio FA. 2012. Identification of amino acid residues important for anti-IFN activity of porcine reproductive and respiratory syndrome virus non-structural protein 1. Virology 433:431–439. 10.1016/j.virol.2012.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroese MV, Zevenhoven-Dobbe JC, Bos-de Ruijter JN, Peeters BP, Meulenberg JJ, Cornelissen LA, Snijder EJ. 2008. The nsp1alpha and nsp1 papain-like autoproteinases are essential for porcine reproductive and respiratory syndrome virus RNA synthesis. J. Gen. Virol. 89:494–499. 10.1099/vir.0.83253-0 [DOI] [PubMed] [Google Scholar]
- 22.Nedialkova DD, Gorbalenya AE, Snijder EJ. 2010. Arterivirus Nsp1 modulates the accumulation of minus-strand templates to control the relative abundance of viral mRNAs. PLoS Pathog. 6:e1000772. 10.1371/journal.ppat.1000772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Dinten LC, den Boon JA, Wassenaar AL, Spaan WJ, Snijder EJ. 1997. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. U. S. A. 94:991–996. 10.1073/pnas.94.3.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang Y, Snijder EJ. 2010. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 154:61–76. 10.1016/j.virusres.2010.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815. 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- 26.Bonilla PJ, Pinon JL, Hughes S, Weiss SR. 1995. Characterization of the leader papain-like protease of MHV-A59. Adv. Exp. Med. Biol. 380:423–430. 10.1007/978-1-4615-1899-0_68 [DOI] [PubMed] [Google Scholar]
- 27.van Dinten LC, Wassenaar AL, Gorbalenya AE, Spaan WJ, Snijder EJ. 1996. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J. Virol. 70:6625–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teng H, Pinon JD, Weiss SR. 1999. Expression of murine coronavirus recombinant papain-like proteinase: efficient cleavage is dependent on the lengths of both the substrate and the proteinase polypeptides. J. Virol. 73:2658–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herold J, Gorbalenya AE, Thiel V, Schelle B, Siddell SG. 1998. Proteolytic processing at the amino terminus of human coronavirus 229E gene 1-encoded polyproteins: identification of a papain-like proteinase and its substrate. J. Virol. 72:910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]