ABSTRACT
Human influenza cases caused by a novel avian H7N9 virus in China emphasize the zoonotic potential of that subtype. We compared the infectivity and pathogenicity of the novel H7N9 virus with those of a recent European avian H7N7 strain in chickens, pigeons, and ferrets. Neither virus induced signs of disease despite substantial replication in inoculated chickens and rapid transmission to contact chickens. Evidence of the replication of both viruses in pigeons, albeit at lower levels of RNA excretion, was also detected. No clear-cut differences between the two H7 isolates emerged regarding replication and antibody development in avian hosts. In ferrets, in contrast, greater replication of the avian H7N9 virus than of the H7N7 strain was observed with significant differences in viral presence, e.g., in nasal wash, lung, and cerebellum samples. Importantly, both viruses showed the potential to spread to the mammal brain. We conclude that efficient asymptomatic viral replication and shedding, as shown in chickens, facilitate the spread of H7 viruses that may harbor zoonotic potential. Biosafety measures are required for the handling of poultry infected with avian influenza viruses of the H7 subtype, independently of their pathogenicity for gallinaceous poultry.
IMPORTANCE This study is important to the field since it provides data about the behavior of the novel H7N9 avian influenza virus in chickens, pigeons, and ferrets in comparison with that of a recent low-pathogenicity H7N7 strain isolated from poultry. We clearly show that chickens, but not pigeons, are highly permissive hosts of both H7 viruses, allowing high-titer replication and virus shedding without any relevant clinical signs. In the ferret model, the potential of both viruses to infect mammals could be demonstrated, including infection of the brain. However, the replication efficiency of the H7N9 virus in ferrets was higher than that of the H7N7 strain. In conclusion, valuable data for the risk analysis of low-pathogenicity avian influenza viruses of the H7 subtype are provided that could also be used for the risk assessment of zoonotic potentials and necessary biosafety measures.
INTRODUCTION
In March 2013, a novel avian influenza A virus (AIV) strain of subtype H7N9 was found to infect humans in an outbreak in the People's Republic of China (1). The transmission of the China/2013 virus to humans probably occurred at live-bird markets and resulted in a high case fatality rate (2–4). Genetic analysis indicates that the virus represents a multiple reassortant with all of the gene segments being of complex avian ancestry. The hemagglutinin (HA) and neuraminidase (NA) genome segments probably descended from viruses of ducks and migratory birds, respectively, whereas the six “internal genes” might have originated from H9N2 viruses circulating in chickens in eastern China (1, 5, 6). The HA cleavage site of China/2013 contains a monobasic motif, indicating a low-pathogenicity phenotype in gallinaceous poultry (1, 2, 4–6). Moreover, sequence analysis revealed several genetic features probably associated with its ability to replicate in mammals, like alterations in the receptor binding site (H5 numbering: G195V, Q235L/I) and loss of a glycosylation site (T169A) within the HA protein, as well as either the E627K or the D701N substitution in PB2 of H7N9 China/2013 viruses isolated from humans (1, 5–9). It was shown that the novel avian H7N9 virus can bind to both avian-type (α2,3-linked sialic acid) and human-type (α2,6-linked sialic acid) receptors, that it can invade epithelial cells in the human lower respiratory tract and type II pneumocytes in alveoli, and that it replicates efficiently in human ex vivo lung and trachea explant cultures, as well as in several mammalian cell lines (9). Furthermore, the novel H7N9 virus exhibits a deletion of five amino acids at positions 69 to 73 within the NA stalk domain, which is supposed to be associated with the adaptation of AIVs to domestic, in particular gallinaceous, poultry (7, 10, 11) and increased virulence in mammals (12).
Outbreaks in poultry caused by low-pathogenicity AIVs (LPAIVs) or highly pathogenic AIVs (HPAIVs) of subtype H7 have occurred repeatedly during the last few years in Europe and North America (reviewed in reference 13). In addition, historic reports have described natural transmission of H7 viruses of avian origin to horses and seals (14, 15). Before 2013, human infections with LPAIV H7 strains (H7N7, H7N2, H7N3) were reported as well (16). However, these infections resulted in mild lower respiratory tract illness or conjunctivitis. Likewise, human infections with HPAIV H7 strains (H7N3 [Canada], H7N7 [The Netherlands, 2003; Italy, 2013]) resulted mainly in conjunctivitis and mild upper respiratory symptoms, with the exception of one death of a veterinarian in the Netherlands in 2003 (reviewed in reference 17). Studies with mammalian models of influenza A virus infection such as mice and ferrets indicated that H7 viruses, especially those of the Eurasian lineage, replicated efficiently in the respiratory tract without prior adaptation and spread systemically, including to the central nervous system (18, 19). In another study with selected H7 strains associated with human infections, it was demonstrated that although they display a low-pathogenicity phenotype, these viruses had a propensity to be transmitted between ferrets by direct contact but not by respiratory droplets (20). Recently, Lam and colleagues identified an H7N7 virus in chickens at live-poultry markets in eastern China that had the same genetic backbone as the avian H7N9 virus and that was able to infect mammals experimentally (21). Although no human infections with this virus have been reported yet, the authors concluded that H7 viruses in general may pose threats beyond the current outbreak of H7N9 in Southeast Asia.
In the present study, we characterized the infectivity and pathogenicity of a human isolate of the novel avian H7N9 virus, A/Anhui/1/13, in comparison with those of a recent LPAIV H7N7 strain from Germany, A/turkey/Germany/AR534/13. To this end, we infected chickens and pigeons; both species are involved as avian hosts in the current circulation of the H7N9 virus in China (4, 8). The ferret is considered a suitable mammalian model in which to study the pathogenicity (and transmissibility) of influenza viruses of various origins (22, 23) and was included here to comparatively investigate the pathogenicity to mammals of the novel avian H7N9 virus and a “standard” LP H7N7 virus. Neither the avian H7N9 virus nor the recent H7N7 strain from Germany induced influenza-like symptoms in either avian host species or the ferret model, even at the large infective dose of 106 50% tissue culture infective doses (TCID50) used in this study. Nevertheless, we observed greater replication of the novel avian H7N9 virus than of the H7N7 virus in the ferret model, in contrast to the low pathogenicity of both the H7N9 and H7N7 viruses in the avian host species.
MATERIALS AND METHODS
Ethics statement.
The animal experiments described here were evaluated by the responsible ethics committee of the State Office of Agriculture, Food Safety, and Fishery in Mecklenburg-Western Pomerania (LALLF M-V) and gained governmental approval under registration numbers LVL MV/TSD/7221.3-2.5-010/10, MV/TSD/7221.3-2.5-010/13, and MV/TSD/7221.3-2.1-052/09.
Viruses.
The novel avian H7N9 virus A/Anhui/1/13 was isolated from a 35-year-old woman from Anhui Province, China, who had visited a chicken market 1 week before the onset of symptoms and died of acute respiratory distress syndrome (1). The virus was kindly provided by the WHO Collaborating Centre, London, United Kingdom, under the auspices of the Pandemic Influenza Preparedness program. The low-pathogenicity H7N7 virus A/turkey/Germany/AR534/2013 represents an H7N7 virus recently detected in outbreaks among gallinaceous poultry in Germany. Both viruses were propagated in the allantoic cavities of 11-day-old embryonated chicken eggs and, after clarification of the harvested allantoic fluids by centrifugation, stored at −70°C until further use. The genome sequences of A/Anhui/1/13 (H7N9) and A/turkey/Germany/AR534/2013 (H7N7) [identical to A/turkey/Germany-NI/R534/2013 (H7N7), following the database nomenclature] are available at GISAID's EpiFlu database (www.gisaid.org) (accession numbers EPI439503 to EPI439510, EPI470366, EPI470367, and EPI490869 to EPI490874, respectively). All procedures involving H7N9 and H7N7 viruses were carried out in an approved, enhanced-biosafety level 3+ facility.
Experimental infection and sampling of chickens, pigeons, and ferrets.
Adult white Leghorn chickens (Gallus gallus domesticus) were purchased from Lohmann Animal Health, Cuxhaven, Germany, while adult racing pigeons (Columba species) were obtained from an AI-free racing pigeon loft. Four chickens and four pigeons per virus group were infected oculonasally (o.n.) with 106 TCID50 of A/Anhui/1/13 (H7N9) or A/turkey/Germany/AR534/13 (H7N7), respectively, and monitored daily for clinical signs for 14 days. They were classified as clinically healthy (score of 0), ill (exhibiting one or more of the following signs of disease: labored breathing, diarrhea, cyanosis, edema, and neurological symptoms) (score of 1), or dead (score of 2) as described previously (24). Oropharyngeal and cloacal swabs were taken at 2, 4, 7, 10, and 14 days postinfection (dpi) or as combined oropharyngeal-cloacal swabs prior to infection and stored in 1 ml medium. At 2 dpi, two further chickens were placed into direct contact with the H7N9- or H7N7-infected birds, serving as transmission controls. Blood sampling and a postmortem examination of one chicken and one pigeon per virus group were performed at 2 and 4 dpi, respectively, or at the end of the experiment for the remaining birds (two inoculated chickens, two contact chickens, two inoculated pigeons). Euthanized birds were dissected, and the lungs and intestines were removed and stored at −70°C until further use.
Outbred ferrets (Mustela putorius furo) were reared at the Friedrich-Loeffler-Institut and housed in cages in groups of one or three animals per control group or virus group, respectively, during the experiment. The cages were separated by stainless steel grids to prevent direct contact. Six 8-week-old healthy and influenza-negative ferrets in each virus group were infected intranasally (i.n.) under anesthesia by inhalation of 5% isoflurane in O2 with 106 TCID50 of A/Anhui/1/13 (H7N9) or A/turkey/Germany/AR534/2013 (H7N7), respectively, and monitored daily for clinical signs (nasal discharge, labored breathing, reduced activity, fever, body weight loss, or neurological symptoms). Two additional ferrets, which were kept in the same room but remained uninfected, served as control animals. Animals were weighed every second day, and body temperatures were recorded with a temperature-logging device (Plexx, Elst, The Netherlands) implanted subcutaneously. Blood samples were drawn from the vena saphena under anesthesia with isoflurane prior to infection or on the day of euthanasia, respectively. Nasal wash samples were collected from all of the ferrets under anesthesia by applying 0.7 ml phosphate-buffered saline (PBS) to each nostril prior to infection and at 2, 4, 6, 8, 10, 12, and 17 dpi. Euthanasia of four animals per virus group and one control ferret at 4 dpi and of the remaining animals at 17 dpi was performed under general anesthesia with isoflurane by means of exsanguination. Euthanized ferrets were dissected, and their spleens, tracheas, lungs (divided into left and right parts), conchae, cerebella, and cerebra were removed and stored at −70°C until further use.
Organ homogenization.
Tissue samples of individual animals were suspended in 1 ml of medium supplemented with 5% fetal bovine serum plus antibiotics. A single stainless steel bead (diameter, 5 mm) per organ sample was added, and samples were homogenized in 2-ml collection tubes for 2 min in a TissueLyser (Qiagen, Hilden, Germany). After subsequent centrifugation at 13,000 rpm for 2 min to remove cellular debris, the supernatant was used to extract viral RNA.
RNA isolation.
Viral RNA was extracted from 200 μl of the supernatant of organ homogenates or swab samples or nasal wash samples with the MagAttract Virus Mini M48 kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions on a Biosprint 96 platform (Qiagen).
Real-time RT-qPCR.
The primer and probe set of the recently described FLI-H7generic-2 assay (25) was used to determine the quantification cycle (Cq) in swabs, nasal wash samples, and organs with the AgPath-ID One-Step RT-PCR kit (Applied Biosystems, Foster City, CA). The quantitative reverse transcription-PCR (RT-qPCR) assay was optimized by using a total volume of 12.5 μl. Briefly, for a single well, 2.25 μl RNase-free water, 6.25 μl 2× RT-PCR buffer, 0.5 μl 25× RT-PCR Enzyme Mix, and 1.0 μl primer-probe mixture were pooled as a master mixture. Finally, 2.5 μl RNA template was added and the reaction was run on an ABI cycler 7500 machine.
Internal extraction control.
For IAV screening investigation, the FLI-H7generic-2 assay was combined with an internal control system in a duplex assay as described earlier (26).
Correlation of TCID50 and viral genome load.
For both viruses (H7N9, H7N7), a log10 dilution series of infectious virus was analyzed by RT-qPCR and by virus titration with Madin-Darby canine kidney (MDCK) cells. Both experiments were performed in triplicate. Regression curves for the H7N9 virus (y = −3.5289x + 39.719; R2 = 0.9997) and for the H7N7 virus (y = −3.7378x + 38.194; R2 = 0.9996) were established and used to calculate TCID50 ml−1 equivalents (TCID50-eq) from Cq values measured by RT-qPCR in swab and organ samples.
Histopathological and immunohistochemical analyses.
Tissues from infected and control animals were placed into 4% phosphate-buffered neutral formaldehyde and processed for paraffin embedding. Paraffin wax sections (2 μm) were dewaxed and stained with hematoxylin and eosin. To investigate the presence of influenza A antigen, and sections were stained as described previously (27).
Serology.
Serum samples from all animals were heat inactivated at 56°C for 30 min and analyzed by means of a commercial enzyme-linked immunosorbent assay (ELISA) for the presence or absence of antibodies against IAV nucleoprotein (NP) (ID Screen Influenza A Antibody Competition ELISA kit; ID-vet, Montpellier, France) and H7 (ID Screen Influenza H7 Antibody Competition ELISA kit; ID-vet, Montpellier, France) according to the manufacturer's instructions. HI assays against A/Anhui/1/13 (H7N9) and A/turkey/Germany/AR534/13 (H7N7) were performed according to standard protocols (2006/437/EC). A virus neutralization assay was performed as described previously (28) but with a few modifications. Threefold serial dilutions were prepared in 50-μl volumes of medium in 96-well plates. Diluted serum samples were mixed with an equal volume of medium containing either the H7N9 or the H7N7 virus at a concentration of 102 TCID50/well. After incubation (1 h, 37°C, 5% CO2), 100 μl of MDCK cells at 1 × 105 cells/liter was added to each well. The plates were incubated for 3 days (37°C, 5% CO2), and viral replication was assessed by visual scoring of the cytopathic effect without staining. Each assay was validated by comparison with positive- and negative-control sera and by titration of the virus dilutions used.
Statistics.
We used the Wilcoxon rank sum test to evaluate the statistical significance of data measured by qRT-PCR. Because of the small group sizes, only ferret nasal wash samples were suitable for analysis. A P value of <0.05 was considered significant.
RESULTS
To compare the infectivity, virulence, and organ distribution of the novel H7N9 virus from China with those of an LPAIV H7N7 virus recently circulating in Europe in avian host species, four chickens and four pigeons per virus were inoculated o.n. with 106 TCID50 of A/Anhui/1/13 (H7N9) or A/turkey/Germany/AR534/13 (H7N7); two chickens and two pigeons, respectively, were sampled and monitored for clinical signs for 14 days, and one bird per virus and species group was euthanized at 2 and 4 dpi for assessment of the viral RNA loads in different organs. To evaluate the transmission of the H7N9 and H7N7 viruses by infected birds, two additional chickens per virus were placed into direct contact at 2 dpi. For experiments with a mammalian model, six ferrets per virus were inoculated i.n. with 106 TCID50; two animals were sampled and monitored for symptoms for 14 days, and four ferrets were euthanized at 4 dpi to investigate viral dissemination.
Virulence of H7N9 and H7N7 viruses.
After o.n. infection with A/Anhui/1/13 (H7N9) or A/turkey/Germany/AR534/13 (H7N7), none of the chickens showed any signs of disease or died during the 14-day observation period. The contact chickens remained healthy, too. Therefore, the clinical score for both viruses was 0.0, confirming that the novel avian H7N9 virus and the recent H7N7 virus are apathogenic in chickens.
For comparison of the pathogenicity of avian H7N9 virus and the H7N7 virus in the other avian host species, four pigeons per virus group were infected o.n. and observed daily for symptoms. However, infection with A/Anhui/1/13 (H7N9) or A/turkey/Germany/AR534/13 (H7N7) did not result in any clinical signs, proving that both viruses are apathogenic in pigeons as well.
To examine the pathogenicity and zoonotic potential of the novel avian H7N9 virus in comparison with those of a recent H7N7 virus, we infected two ferrets per virus group i.n. and included two uninfected control ferrets. Infection of ferrets with the H7N7 virus caused fever from 2 to 6 dpi, whereas the H7N9 virus caused slightly elevated body temperatures from 2 to 4 dpi, followed by higher temperatures from 6 to 8 dpi (Fig. 1A). Of note, a minor body temperature elevation was also seen in the control animal, indicating that the infection procedure itself contributed partially to these observations. The ferrets inoculated with A/Anhui/1/13 (H7N9) or A/turkey/Germany/AR534/13 (H7N7) showed a slight delay in body weight gain from 2 to 4 dpi but developed well after that time point (Fig. 1B). The control ferret was much heavier at the beginning of the experiment and therefore had a more constant body weight throughout the study. Neither the H7N9- or H7N7-infected ferrets nor the mock-infected animals developed clinical signs like sneezing, nasal discharge, coughing, diarrhea, or lethargy as described previously for ferrets infected with an HPAIV of subtype H7N7 (18).
FIG 1.
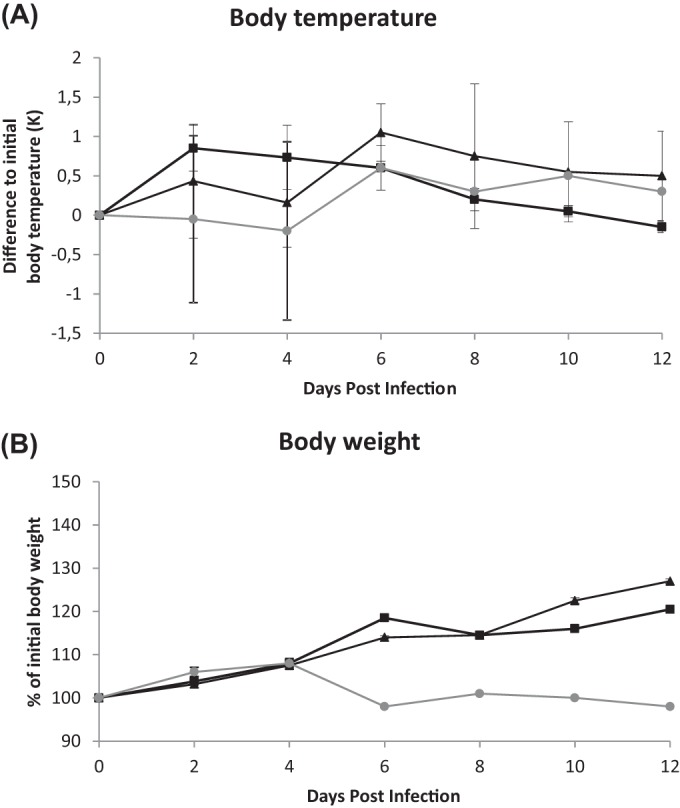
Body temperatures (A) and weights (B) of ferrets infected with avian H7N9 (squares) or H7N7 (triangles) virus and uninfected controls (gray line). Measurements were made at the time points indicated while the ferrets were under anesthesia. Body temperature is given as the mean value and standard deviation of the difference from the temperature prior to infection. Body weight is given as the mean value and standard deviation compared to the weight prior to infection.
Taken together, exposure to H7N9 and H7N7 showed comparable clinically inconspicuous profiles in chickens, pigeons, and ferrets.
Viral replication and shedding of H7N9 and H7N7 viruses.
In order to compare the replication of the avian H7N9 and H7N7 viruses in chickens and pigeons and shedding via the respiratory or digestive tract, viral RNA loads in oropharyngeal and cloacal swabs were determined at different time points during the observation period. Transmission of the H7N9 virus and the H7N7 strain by the birds was examined by investigating swab and organ samples from contact chickens. Moreover, the extent of viral replication in the upper respiratory tracts of ferrets was determined by measuring the viral RNA loads in nasal wash samples.
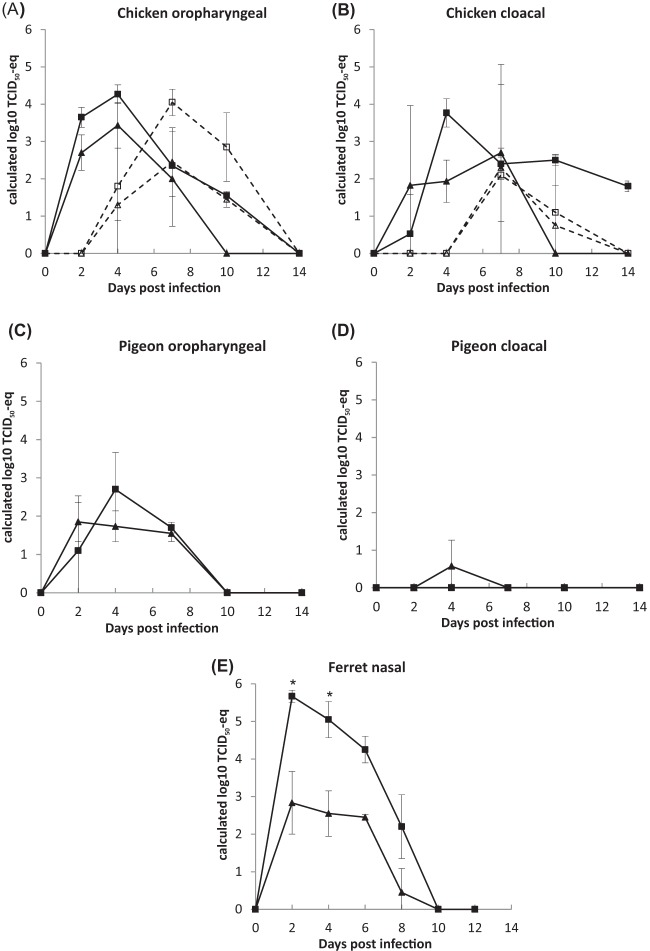
Chickens infected with A/Anhui/1/13 (H7N9) (C1, C2, C5, and C6) shed large viral RNA loads corresponding to substantial calculated TCID50-eqs from 2 to 10 dpi via the respiratory route (Fig. 2A; Table 1), whereas chickens infected with A/turkey/Germany/AR534/13 (H7N7) (C7, C8, C11, and C12) shed virus at similar levels but only until 7 dpi. Chickens placed into direct contact with the H7N9-infected birds (C3 and C4) shed virus on days 2 to 8 postexposure (p.e.) via the respiratory route, indicating successful transmission of the virus. The chickens with contact with H7N7-infected birds (C9 and C10) shed virus via the respiratory tract as well but with smaller RNA loads. Therefore, the transmission of both viruses could be demonstrated. Remarkably, the avian H7N9 virus could be detected in considerable amounts in cloacal swabs until 14 dpi (Fig. 2B), indicating efficient and prolonged replication in the digestive tract. In contrast, the recent H7N7 virus was only detected until 7 dpi in cloacal swabs. Chickens with direct contact with H7N9- or H7N7-infected birds cloacally shed virus from day 5 p.e. to day 8 p.e., with slightly reduced amounts shed by chickens placed into direct contact with H7N7-infected birds. Serum antibodies reacting to the NP and H7 proteins were detected in all chicken samples by 14 dpi (Table 2), with the exception of one chicken (C9) that was in direct contact with H7N7-infected chickens and developed antibodies against H7 but tested negative for the presence of antibodies against NP. Virus neutralization and HI assays revealed a subtype-specific cross-reactivity of the antibodies directed against the H7 protein by 14 dpi.
FIG 2.
Viral loads in oropharyngeal (A, C) or cloacal (B, D) swabs obtained from chickens (A, B) or pigeons (C, D) and in nasal wash samples from ferrets (E) infected (solid lines) with avian H7N9 (squares) or H7N7 (triangles) virus and in contact chickens (dotted lines). Swab samples or nasal wash samples taken at the time points indicated were analyzed for the presence of H7 gene-specific RNA by RT-qPCR. Shown are the mean values and standard deviations of the calculated TCID50-eqs. Significant differences (P value = 0.002165; exact Wilcoxon rank sum test) between H7N9- and H7N7-infected ferrets are indicated (*).
TABLE 1.
Viral loads in oropharyngeal and cloacal swabs of chickens infected with avian H7N9 or H7N7
| Virus, chicken no., and swab typea |
Cq value (calculated log10 TCID50-eq) at: |
|||||
|---|---|---|---|---|---|---|
| 0c dpi | 2 dpi | 4 dpi | 7 dpi | 10 dpi | 14 dpi | |
| A/Anhui/1/13 (H7N9) | ||||||
| C1 | ||||||
| OP | Noneb | 26.1 (3.9) | 23.9 (4.5) | 31.7 (2.3) | 34.1 (1.6) | Noned |
| C | None | None | 27.2 (3.5) | 32.3 (2.1) | 31.4 (2.4) | 32.9 (1.9)d |
| C2 | ||||||
| OP | None | 28.1 (3.3) | 25.5 (4.0) | 31.4 (2.4) | 34.5 (1.5) | Noned |
| C | None | 32.3 (2.1) | 24.8 (4.2) | 30.3 (2.7) | 30.7 (2.6) | 33.8 (1.7)d |
| C3e | ||||||
| OP | None | None | None | 26.2 (3.8) | 27.4 (3.5) | Noned |
| C | None | None | None | None | 32.0 (2.2) | Noned |
| C4e | ||||||
| OP | None | None | 26.9 (3.6) | 24.5 (4.3) | 30.3 (2.7) | Noned |
| C | None | None | None | 24.8 (4.2) | None | Noned |
| C5 | ||||||
| OP | None | 26.9 (3.6)d | ||||
| C | None | Noned | ||||
| C6 | ||||||
| OP | None | 26.4 (3.8) | 24.7 (4.3)d | |||
| C | None | None | 27.1 (3.6)d | |||
| A/turkey/Germany/AR534/13 (H7N7) | ||||||
| C7 | ||||||
| OP | None | 29.2 (2.4) | 25.9 (3.3) | 27.2 (2.9) | None | Noned |
| C | None | None | 33.3 (1.3) | 23.4 (4.0) | None | Noned |
| C8 | ||||||
| OP | None | 27.0 (3.0) | 27.3 (2.9) | 34.0 (1.1) | None | Noned |
| C | None | 26.1 (3.2) | 30.4 (2.1) | 33.0 (1.4) | None | Noned |
| C9e | ||||||
| OP | None | None | 32.3 (1.6) | 31.4 (1.8) | 32.4 (1.6) | Noned |
| C | None | None | None | 30.5 (2.1) | None | Noned |
| C10e | ||||||
| OP | None | None | 34.3 (1.0) | 26.6 (3.1) | 33.5 (1.3) | Noned |
| C | None | None | None | 29.0 (2.5) | 32.6 (1.5) | Noned |
| C11 | ||||||
| OP | None | 26.4 (3.2)d | ||||
| C | None | Noned | ||||
| C12 | ||||||
| OP | None | 29.8 (2.2) | 22.8 (4.1)d | |||
| C | None | 22.7 (4.1) | 29.2 (2.4)d | |||
OP, oropharyngeal, C, cloacal.
None, >35.
Combined pharyngeal and cloacal swabs (only at 0 dpi).
Euthanasia and dissection.
Placed into direct contact with infected chicken at 2 dpi.
TABLE 2.
Serum antibody titers of chickens infected with avian H7N9 or H7N7 determined by ELISA, virus neutralization assay, and HI assay
| Virus and chicken no. (dpi) | ELISA |
Neutralization of: |
HI of: |
|||
|---|---|---|---|---|---|---|
| Anti-NPa | Anti-H7b | H7N9 | H7N7 | H7N9 | H7N7 | |
| A/Anhui/1/13 (H7N9) | ||||||
| C1 (14) | 6.3 | 18.8 | 160 | 202 | 128 | 32 |
| C2 (14) | 7.7 | 12.0 | 127 | 63.5 | 256 | 64 |
| C3c (14) | 6.6 | 22.5 | 640 | 127 | 64 | 64 |
| C4c (14) | 11.4 | 18.9 | 80 | 127 | 128 | 32 |
| C5 (2) | 93.3 | 113.6 | <8 | <8 | <2 | <2 |
| C6 (4) | 96.9 | 117.8 | <8 | <8 | <2 | <2 |
| A/turkey/Germany/AR534/13 (H7N7) | ||||||
| C7 (14) | 8.3 | 13.8 | 127 | 403 | 128 | 128 |
| C8 (14) | 8.6 | 27.1 | 25.2 | 202 | 64 | 128 |
| C9c (14) | 101.8 | 24.2 | 101 | 320 | 64 | 128 |
| C10c (14) | 7.6 | 27.6 | 63.5 | 101 | 32 | 128 |
| C11 (2) | 104.1 | 119.0 | <8 | <8 | <2 | <2 |
| C12 (4) | 74.3 | 115.4 | <8 | <8 | <2 | <2 |
Anti-NP ELISA: positive (bold), <55; negative, >65; questionable, 55 to 65.
Anti-H7 ELISA: positive (bold), <50; negative, >60; questionable, 50 to 60.
Placed into direct contact with infected chickens at 2 dpi.
In addition, we determined the viral replication and shedding patterns of the avian H7N9 virus and the recent H7N7 virus in infected pigeons. After infection with A/Anhui/1/13 (H7N9), the pigeons (P1 to P4) shed virus via the respiratory tract from 2 dpi (P2 and P3) to 4 dpi (P1 and P4) (Fig. 2C; Table 3), although their RNA loads were far smaller than those of infected chickens. In oropharyngeal swabs, viral RNA was detectable until 7 dpi. Pigeons infected with A/turkey/Germany/AR534/13 (H7N7) (P5 to P8) shed similarly small RNA loads via respiratory secretions from 2 to 7 dpi. No cloacal shedding could be detected in pigeons infected with the avian H7N9 virus (Fig. 2D; Table 3). In contrast, two of the H7N7-infected pigeons (P5 and P8) shed viral RNA in cloacal swabs at 4 dpi, indicating replication of the virus in the intestines. Antibodies against NP were detected by ELISA in the serum of every individually infected pigeon by 14 dpi, but all of the pigeons tested were negative by HI for the presence of antibodies against H7 (Table 4). The results indicate that both viruses are capable of an initial infection and, to a certain extent, of replication in pigeons but that the induction of an H7-specific antibody-mediated immune reaction in that bird species is weak.
TABLE 3.
Viral loads in oropharyngeal and cloacal swabs from pigeons infected with avian H7N9 or H7N7
| Virus, pigeon no., and swab typea |
Cq value (calculated log10 TCID50-eq) at: |
|||||
|---|---|---|---|---|---|---|
| 0c dpi | 2 dpi | 4 dpi | 7 dpi | 10 dpi | 14 dpi | |
| A/Anhui/1/13 (H7N9) | ||||||
| P1 | ||||||
| OP | Noneb | None | 31.5 (2.3) | 34.2 (1.6) | None | Noned |
| C | None | None | None | None | None | Noned |
| P2 | ||||||
| OP | None | 29.3 (3.0) | 26.2 (3.8) | 33.3 (1.8) | None | Noned |
| C | None | None | None | None | None | Noned |
| P3 | ||||||
| OP | None | 34.7 (1.4)d | ||||
| C | None | Noned | ||||
| P4 | ||||||
| OP | None | None | 32.6 (2.0)d | |||
| C | None | None | Noned | |||
| A/turkey/Germany/AR534/13 (H7N7) | ||||||
| P5 | ||||||
| OP | None | 30.7 (2.0) | 31.6 (1.8) | 31.8 (1.7) | None | Noned |
| C | None | None | 34.7 (0.9) | None | None | Noned |
| P6 | ||||||
| OP | None | 30.0 (2.0) | 33.5 (1.3) | 32.8 (1.4) | None | Noned |
| C | None | None | None | None | None | Noned |
| P7 | ||||||
| OP | None | 33.9 (1.1)d | ||||
| C | None | Noned | ||||
| P8 | ||||||
| OP | None | 30.5 (2.1) | 30.5 (2.1)d | |||
| C | None | None | 33.1 (1.4)d | |||
OP, oropharyngeal, C, cloacal.
None, >35.
Combined pharyngeal and cloacal swabs (only at 0 dpi).
Euthanasia and dissection.
TABLE 4.
Serum antibody titers of pigeons infected with avian H7N9 or H7N7 determined by ELISA, virus neutralization assay, and HI assay
| Virus and pigeon no. (dpi) | ELISA |
Neutralization of: |
HI of: |
|||
|---|---|---|---|---|---|---|
| Anti-NPa | Anti-H7b | H7N9 | H7N7 | H7N9 | H7N7 | |
| A/Anhui/1/13 (H7N9) | ||||||
| P1 (14) | 19.5 | 104.1 | <10 | <10 | <2 | <2 |
| P2 (14) | 29.9 | 94.3 | <10 | <10 | 2 | <2 |
| P3 (2) | 100.0 | 111.9 | <8 | <8 | 4 | <2 |
| P4 (4) | 95.3 | 115.5 | <8 | <8 | <2 | <2 |
| A/turkey/Germany/AR534/13 (H7N7) | ||||||
| P5 (14) | 44.7 | 93.7 | <10 | <10 | <2 | <2 |
| P6 (14) | 47.0 | 111.6 | <10 | <10 | <2 | <2 |
| P7 (2) | 92.9 | 112.6 | <8 | <8 | <2 | <2 |
| P8 (4) | 98.1 | 132.4 | <8 | <8 | <2 | <2 |
Anti-NP ELISA: positive (bold), <55; negative, >65; questionable, 55 to 65.
Anti-H7 ELISA: positive (bold), <50; negative, >60; questionable, 50 to 60.
For the detection of viral RNA in nasal wash samples from ferrets, samples were collected every 2 days from all of the animals. All of the ferrets infected with A/Anhui/1/13 (H7N9) (F1 to F6) or A/turkey/Germany/AR534/13 (H7N7) (F7 to F12) shed large viral genome loads into nasal wash samples as early as 2 dpi and continued viral shedding until 8 dpi (Fig. 2E; Table 5), but the H7N9 virus replicated to significantly larger amounts, as detected in nasal secretions than the H7N7 strain (P value = 0.002165). No viral RNA was detected in the control animals (F13 and F14) except for in one animal on 4 dpi (euthanasia and dissection), with a very low viral-RNA load. All of the ferrets tested negative for influenza A virus NP-specific antibodies prior to infection (data not shown). Seroconversion of all of the infected ferrets was confirmed by the detection of antibodies against NP and H7 by 17 dpi (F1, F2, F7, and F8) (Table 6). The sera of H7N9-infected ferrets were able to neutralize the homologous H7N9 virus in the neutralization assay; however, neutralization of the H7N7 strain was weak. Antibodies induced after infection with the recent H7N7 virus displayed weak neutralizing activity against both the H7N9 and H7N7 viruses. Similar results were obtained with the HI assay.
TABLE 5.
Viral loads in nasal wash samples from ferrets infected with avian H7N9 or H7N7
| Virus and ferret no. |
Cq value (calculated log10 TCID50-eq) at: |
||||||
|---|---|---|---|---|---|---|---|
| 0 dpi | 2 dpi | 4 dpi | 6 dpi | 8 dpi | 10 dpi | 12 dpi | |
| A/Anhui/1/13 (H7N9) | |||||||
| F1 | Nonea | 19.5 (5.7) | 21.4 (5.2) | 23.7 (4.5) | 34.1 (1.6) | None | Noneb |
| F2 | None | 19.5 (5.7) | 23.6 (4.6) | 25.6 (4.0) | 29.8 (2.8) | None | Noneb |
| F3 | None | 18.9 (5.9) | 22.1 (5.0)b | ||||
| F4 | None | 19.6 (5.7) | 24.1 (4.4)b | ||||
| F5 | None | 20.0 (5.6) | 19.8 (5.6)b | ||||
| F6 | None | 20.7 (5.4) | 20.2 (5.5)b | ||||
| A/turkey/Germany/AR534/13 (H7N7) | |||||||
| F7 | None | 28.8 (2.5) | 27.9 (2.8) | 29.3 (2.4) | None | None | None |
| F8 | None | 23.3 (4.0) | 26.3 (3.2) | 28.8 (2.5) | 34.8 (0.9) | None | None |
| F9 | None | 30.3 (2.1) | 27.7 (2.8)b | ||||
| F10 | None | 28.2 (2.7) | 32.6 (1.5)b | ||||
| F11 | None | 30.8 (2.0) | 29.9 (2.2)b | ||||
| F12 | None | 24.4 (3.7) | 27.8 (2.8)b | ||||
| Controlc | |||||||
| F13 | None | None | None | None | None | None | None |
| F14 | None | None | 33.1 (1.4)b | ||||
None, >35.
Euthanasia and dissection.
PBS-inoculated animals.
TABLE 6.
Serum antibody titers of ferrets infected with avian H7N9 or H7N7 determined by ELISA, virus neutralization assay, and HI assay
| Virus and ferret no. (dpi) | ELISA |
Neutralization of: |
HI of: |
|||
|---|---|---|---|---|---|---|
| Anti-NPa | Anti-H7b | H7N9 | H7N7 | H7N9 | H7N7 | |
| A/Anhui/1/13 (H7N9) | ||||||
| F1 (17) | 7.5 | 24.1 | 202 | 101 | 256 | 8 |
| F2 (17) | 12.4 | 40.5 | 403 | 25.2 | 256 | 16 |
| F3 (4) | 96.0 | 102.7 | <8 | <8 | 8 | <2 |
| F4 (4) | 83.8 | 96.4 | <8 | <8 | NDd | ND |
| F5 (4) | 80.8 | 97.6 | <8 | <8 | ND | ND |
| F6 (4) | 62.7 | 97.0 | <8 | <8 | ND | ND |
| A/turkey/Germany/AR534/13 (H7N7) | ||||||
| F7 (17) | 13.6 | 33.8 | 40 | 33.8 | 32 | 4 |
| F8 (17) | 16.0 | 38.3 | 40 | 38.3 | 64 | 16 |
| F9 (4) | 109.6 | 98.0 | <8 | <8 | 8 | <2 |
| F10 (4) | 84.6 | 99.8 | <8 | <8 | ND | ND |
| F11 (4) | 73.7 | 96.1 | <8 | <8 | ND | ND |
| F12 (4) | 73.9 | 103.0 | <8 | <8 | ND | ND |
| Controlc | ||||||
| F13 (17) | 103.4 | 97.2 | <8 | <8 | <2 | <2 |
| F14 (4) | 76.7 | 95.4 | <8 | <8 | ND | ND |
Anti-NP ELISA: positive (bold), <55; negative, >65; questionable: 55 to 65.
Anti-H7 ELISA: positive (bold), <50; negative, >60; questionable: 50 to 60.
PBS-inoculated animals.
ND, not determined.
Overall, the kinetics of H7N9 and H7N7 shedding were comparable in the avian and mammalian species investigated, with indications of enhanced replication of the H7N9 strain in ferrets.
Organ distribution of H7N9 and H7N7 viral RNA loads.
In order to compare the distribution of the novel avian H7N9 virus and that of the recent H7N7 virus in the respiratory or digestive tract, one infected chicken and pigeon in each virus group was euthanized at 2 and 4 dpi, respectively, and their lungs and intestines were removed for viral RNA detection. Furthermore, a panel of organs was taken at 4 dpi from four additionally inoculated ferrets per virus group and one uninfected control animal. Immunohistochemical analysis was performed to detect viral antigen in the organs of infected animals.
Viral RNA of the H7N9 virus or the H7N7 strain was not detected in the lungs of infected chickens at any time point (Fig. 3A; Table 7). At 4 dpi, the replication of both A/Anhui/1/13 (H7N9) (C6) and A/turkey/Germany/AR534/13 (H7N7) (C12) could be detected in the intestines of infected chickens, with a larger load of the genome of the H7N9 virus. Interestingly, at 14 dpi, the H7N9 virus (C1 and C2) could still be detected in the intestine. No viral RNA was detected in the organs of contact chicken on day 12 p.e. Furthermore, no viral antigen could be detected by immunostaining of organ (lung, intestine) samples from infected chickens (data not shown).
FIG 3.
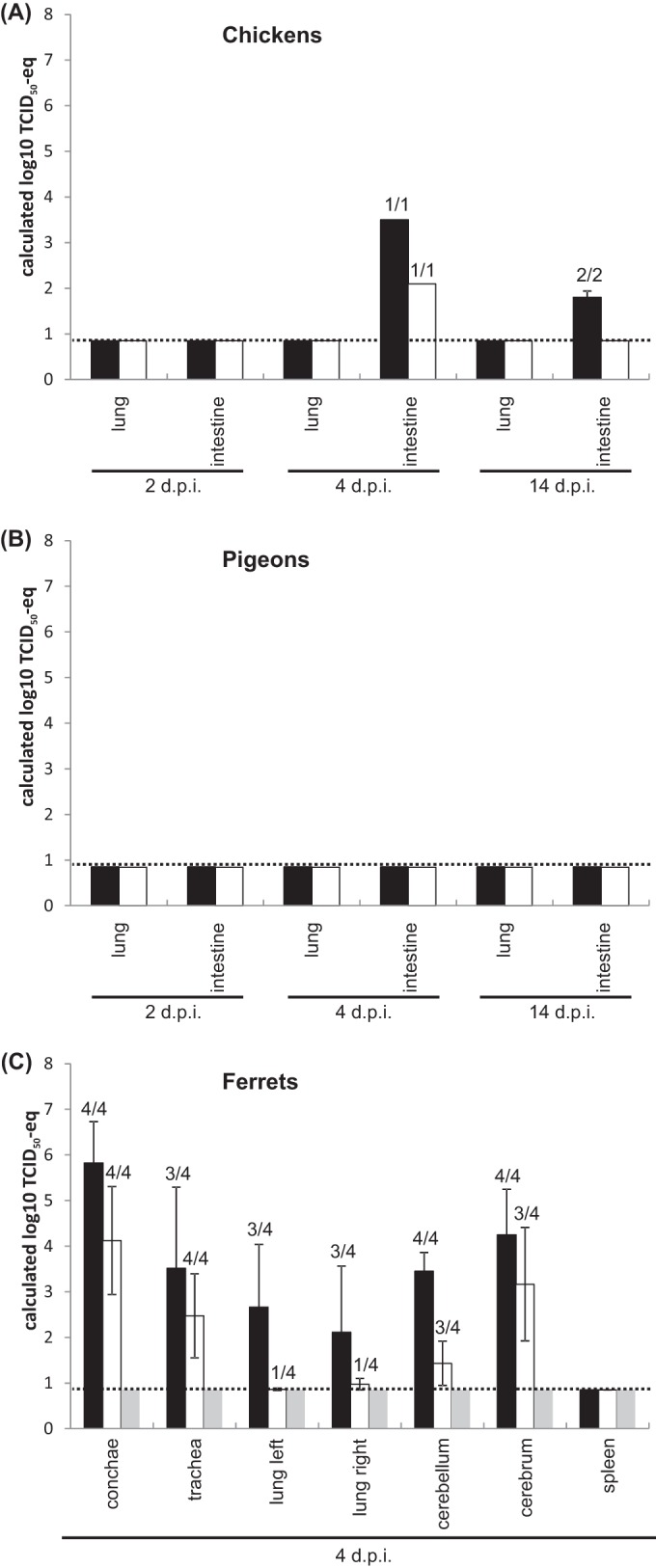
Viral loads in organs of chickens (A), pigeons (B), or ferrets (C) infected with avian H7N9 (black bars) or H7N7 virus (shaded bars) and an uninfected ferret (gray bars). Organ samples taken at the time points indicated were analyzed for the presence of H7 gene-specific RNA by RT-qPCRs. Shown are the mean values and standard deviations of the calculated TCID50-eqs. The number of positive samples/total number of samples is shown above each bar.
TABLE 7.
Viral loads in organs of chickens infected with avian H7N9 or H7N7
| Virus and chicken no. (dpi)a |
Cq value (calculated log10 TCID50-eq) in: |
|
|---|---|---|
| Lung | Intestine | |
| A/Anhui/1/13 (H7N9) | ||
| C1 (14) | Noneb | 32.9 (1.9) |
| C2 (14) | None | 33.8 (1.7) |
| C3c (14) | None | None |
| C4c (14) | None | None |
| C5 (2) | None | None |
| C6 (4) | None | 27.3 (3.5) |
| A/turkey/Germany/AR534/13 (H7N7) | ||
| C7 (14) | None | None |
| C8 (14) | None | None |
| C9c (14) | None | None |
| C10c (14) | None | None |
| C11 (2) | None | None |
| C12 (4) | None | 30.2 (2.1) |
Day postinfection on which the organ was taken.
None, >35.
Placed into direct contact with infected chicken at 2 dpi.
The viral RNA loads in the lungs and intestines of pigeons after infection with the avian H7N9 virus or the recent H7N7 virus were determined at 2 and 4 dpi as well, but no A/Anhui/1/13 (H7N9) or A/turkey/Germany/AR534/13 (H7N7) viral RNA could be detected at these time points or at the end of the experiment (14 dpi) (Fig. 3B). By immunohistochemical analysis, no viral antigen could be detected in organ (lung, intestine) samples from infected pigeons (data not shown).
In order to characterize the virus distribution in organs of ferrets infected with the avian H7N9 virus or the recent H7N7 strain, four animals per group and one uninfected control animal were euthanized at 4 dpi and their spleens, tracheas, left and right lungs, conchae, cerebella, and cerebra were taken. The RNAs of both the A/Anhui/1/13 (H7N9) (F3 to F6) and A/turkey/Germany/AR534/13 (H7N7) (F9 to F12) viruses were detected in the upper respiratory tract (conchae, trachea) (Fig. 3C; Table 8), whereas the RNA loads of both viruses were smaller in the lower respiratory tract and several samples, especially those from the H7N7-infected ferrets, were even negative. Interestingly, RNA of the H7N9 virus could be detected in the brains of all four ferrets and three of the four H7N7-infected animals. In all of the organs tested, the genome loads of the H7N9 virus were larger than those of the H7N7 strain, indicating better replication of A/Anhui/1/13 in that mammalian animal model. No virus was detected in the spleens of inoculated ferrets, excluding systemic infection/viremia. No viral RNA was detectable in any organ at 17 dpi, indicating that the ferrets were able to clear the viruses (data not shown). Immunohistological investigation of lung samples of infected ferrets at 4 dpi revealed an interstitial bronchopneumonia affecting single endothelial cells, pneumocytes, and bronchial epithelial cells in two of four H7N9-inoculated ferrets examined (F3 and F4) but in only one of four H7N7-infected animals (F11) (data not shown). Viral antigen could not be detected in the ferret brain samples examined by immunohistological techniques.
TABLE 8.
Viral loads in organs of ferrets infected with avian H7N9 or H7N7
| Virus and ferret no. (dpi)a |
Cq value (calculated log10 TCID50-eq) in: |
||||||
|---|---|---|---|---|---|---|---|
| Conchae | Trachea | Left lung | Right lung | Cerebellum | Cerebrum | Spleen | |
| A/Anhui/1/13 (H7N9) | |||||||
| F1 (17) | Noneb | None | None | None | None | None | None |
| F2 (17) | None | None | None | None | None | None | None |
| F3 (4) | 24.2 (4.4) | 24.5 (4.3) | 24.8 (4.2) | 24.9 (4.2) | 28.9 (3.1) | 29.6 (2.9) | None |
| F4 (4) | 17.4 (6.3) | 23.6 (4.6) | 29.5 (2.9) | 33.9 (1.6) | 28.9 (3.1) | 24.5 (4.3) | None |
| F5 (4) | 16.9 (6.5) | None | None | None | 26.0 (3.9) | 21.1 (5.3) | None |
| F6 (4) | 18.4 (6.0) | 24.4 (4.3) | 30.2 (2.7) | 33.3 (1.8) | 26.7 (3.7) | 23.8 (4.5) | None |
| A/turkey/Germany/AR534/13 (H7N7) | |||||||
| F7 (17) | 34.4 (1.0) | None | None | None | None | None | None |
| F8 (17) | None | None | None | None | None | None | None |
| F9 (4) | 29.1 (2.4) | 26.9 (3.0) | 34.9 (0.9) | 34.0 (1.1) | None | None | None |
| F10 (4) | 20.0 (4.9) | 34.0 (0.9) | None | None | 32.3 (1.6) | 26.1 (3.2) | None |
| F11 (4) | 22.2 (4.3) | 27.9 (2.8) | None | None | 34.8 (0.9) | 28.6 (2.6) | None |
| F12 (4) | 19.8 (4.9) | 26.9 (3.0) | None | None | 31.5 (1.8) | 24.5 (3.7) | None |
| Control | |||||||
| F13 (17) | None | None | None | None | None | None | None |
| F14 (4) | None | None | None | None | None | None | None |
Day postinfection on which the organ was taken.
None, >35.
In summary, the H7N9 and H7N7 viruses were comparably disseminated in the organs of infected chickens and pigeons; however, RNA of both viruses could be detected in the respiratory organs of ferrets, with the potential to disseminate into the brain and with lower Cq values for the H7N9 virus.
DISCUSSION
In this study, we compared the infectivity and virulence of the novel avian H7N9 virus A/Anhui/1/13 with those of H7N7 LPAIV A/turkey/Germany/AR534/13, a representative of the H7N7 viruses recently circulating in poultry farms in Germany, in natural avian hosts (chickens, pigeons) and in a model mammalian host species (ferrets). Neither of the two strains induced any clinical signs in either inoculated or contact control chickens or in inoculated pigeons. Comparable patterns of oropharyngeal and cloacal shedding were observed in chickens, and both viruses were transmitted to contact chickens, which shed slightly smaller amounts of the H7N7 isolate than of the H7N9 virus. Viral RNA was detected in the intestines of infected chickens at 4 dpi with larger RNA loads of the H7N9 virus than of the H7N7 strain, indicating a replication advantage in this avian host species. Neither the H7N9 virus nor the H7N7 strain was detected in the lungs of infected chickens. Obviously, lung tissue is not the primary location of viral replication, but rather tissues of the upper respiratory tract or the air sac epithelia. The lack of detection of viral antigen in lung tissues despite the presence of infectious virus in oropharyngeal swabs is in accordance to experimental data from Pantin-Jackwood and colleagues (29). Therefore, oropharyngeal swab samples are more adequate than lung tissue for testing for the presence of the virus (or the viral genome). Antibodies directed against the H7 protein in chicken serum revealed only partial subtype-specific cross-reactivity, indicating that the overall antigenic difference between the HAs of the novel avian H7N9 virus and the H7N7 viruses currently circulating in Europe is low. Thus, reference antigens of contemporary European H7 viruses are suitable for the detection of H7N9/China-specific antibodies.
Respiratory shedding of both viruses by pigeons was detected, although only in very small amounts compared to those shed by infected chickens. Cloacal shedding by pigeons could be observed only for the H7N7 virus. Neither A/Anhui/1/13 (H7N9) nor A/turkey/Germany/AR534/13 (H7N7) could be detected in the organs of infected pigeons. Interestingly, both H7 viruses induced antibodies against NP in pigeons, but the infected pigeons failed to develop antibodies detectable by HI or ELISA against the H7 HA protein. Most probably, these data reflect the different sensitivities of the serological test systems used.
Zhang and colleagues have analyzed different strains of the novel avian H7N9 viruses isolated from different bird species from different habitats and from humans in different provinces of China for pathogenicity and transmission in chickens, mice, and ferrets (8). Determining virulence in chickens, they obtained an intravenous pathogenicity index of 0, which is consistent with our result, although we chose the o.n. and therefore more natural application route. In contrast to our data, Ku and colleagues detected virus shedding by inoculated chickens for no longer than 2 days (30). Likewise transmission between inoculated and naive chickens could not be demonstrated by the experimental setup of Ku and colleagues. Reasonable explanations for these differences from our data might be based on the different application routes (oculo-oro-nasal versus intranasal/intratracheal) and different methods of measurement of viral excretion. Further data from experimental infections of chickens, as well as genuine samples (swabs sampled from different poultry species), from the affected region rather support our findings of substantial replication in and shedding by chickens (29, 31, 32).
The infected ferrets did not show any symptoms, and both of the viruses tested are therefore considered to be of low virulence in this mammalian model. Nevertheless, both the H7N9 virus and the H7N7 isolate replicated efficiently in the upper respiratory tract and were detected in nasal wash samples, with significantly larger loads of the H7N9 virus than of the H7N7 strain. Replication of both viruses in the lower respiratory tracts of ferrets was detectable, but the RNA loads were smaller than those in their noses and tracheas. Importantly, both viruses were able to spread into the brains of infected ferrets according to positive RT-qPCR results. It remains to be determined whether this occurred hematogenically as a result of systemic replication or by cranial nerve pathways, as has previously been shown for highly pathogenic H5N1 viruses in the ferret model (33). Since no infection was detected in the spleen, systemic infection and hence hematogenic spread appear unlikely. However, the absence of neurological symptoms and a lack of detection of influenza viral antigen by immunohistopathology indicate that there is no massive virus replication activity in brain tissues. But it is also important to take into consideration the sensitivity limitations of direct antigen detection by staining, e.g., due to missing amplification in comparison with the real-time PCR. This is also reflected by our experience with HPAIV H5N1, where cycle threshold (CT) values of <20 are regularly connected with positive antigen staining, while higher CT values, as observed here, often do not allow direct antigen detection.
Interestingly, the results of a mouse infection study indicate that the human H7N9 isolates replicated more efficiently and were more lethal than the H7N9 viruses of direct avian origin (8). They also tested two H7N9 viruses isolated from birds and three H7N9 strains isolated from humans in the ferret model and showed that all of the viruses replicated to similar levels in the nasal turbinates but that the viral loads in the tonsils, tracheas, and lungs were larger in ferrets infected with the human isolates, which were also able to replicate in the brain. Importantly, that group also reported respiratory droplet transmission between ferrets infected with H7N9 viruses isolated from humans, indicating the potential for human-to-human transmission of these viruses (8). In contrast, Belser and coworkers found no evidence of efficient droplet-mediated transmission between ferrets (34), and therefore, experimental conditions or strain variations might also influence transmission. In another study, Zhu and coworkers used the ferret model to evaluate the infectivity and transmissibility of another avian H7N9 virus isolated from a human, A/Shanghai/2/13, and found that the virus replicated in the upper and lower respiratory tracts and was shed at high titers for 6 to 7 days (35), correlating with the data of our study. But the ferrets infected with A/Shanghai/2/13 displayed mild clinical signs (sneezing, nasal discharge, coughing, mild lethargy, brief fever) and transmitted the virus to naive ferrets via direct contact but less efficiently so by air. Viral RNA was detected in their nasal turbinates, tracheas, lungs, and hilar lymph nodes, as well as in their brains, which is therefore consistent with our findings (35). However, RNA loads of the H7N7 virus could also be detected in the brains of ferrets. In a study by Lam and colleagues, an H7N7 strain of chicken origin cocirculating and sharing the same internal genes with the avian H7N9 virus caused a significant infection in ferrets under experimental conditions, although viral shedding was lower than that of the H7N9 virus (21). This suggests, together with our results, that also avian H7N7 viruses of low pathogenicity might bear zoonotic potential and that heightened awareness extending beyond the avian H7N9 virus is indicated.
Both the H7N9 virus and the H7N7 strain possess a monobasic motif at the cleavage site of the HA and the expected low-pathogenicity phenotype in chickens and pigeons was confirmed in our experiments.
The low-pathogenicity phenotype of the novel H7N9 viruses, leading to efficient yet asymptomatic shedding in avian species, serves as a stealth mechanism that facilitates their silent spread among poultry. However, our data clearly suggest a minor role for pigeons. The transmission to and replication in humans raise its chances to accumulate adaptive mutations, rendering it more transmissible among humans. Although several signature mutations signaling increased zoonotic potential of avian influenza virus isolates have been described, their predictive value for newly discovered field viruses remains low. Therefore, well-characterized animal model species such as the ferret constitute the most important tool for evaluation of the zoonotic potential of these viruses. Confirming and extending previous data on the replication of H7N9/China in ferrets, we show here that other Eurasian H7 viruses of low pathogenicity currently circulating in Europe harbor some potential to infect and replicate in mammals and possibly in humans as well. Handling of poultry infected with such viruses therefore requires a risk assessment and adequate levels of personal protection.
ACKNOWLEDGMENTS
We thank Bärbel Hammerschmidt of the Department of Experimental Animal Facilities and Biorisk Management, Friedrich Loeffler Institute, for breeding the ferrets. We thank Mareen Lange, Diana Wessler, and Conny Illing for excellent technical assistance; Mario Ziller for his help with the statistical analysis; and the animal caretakers. We are thankful to the WHO Collaborating Centre (John McCauley and Vicky Gregory) at the MRC National Institute for Medical Research, Mill Hill, London, United Kingdom, for providing the H7N9 virus. We gratefully acknowledge L. Yang and the WHO Chinese National Influenza Center as the originating and submitting laboratory of the sequence of A/Anhui/1/13 (H7N9) from GISAID's EpiFlu Database.
This work was supported by the European Union FP7 project European Management Platform for Emerging and Re-emerging Infectious Disease Entities (EMPERIE; no. 223498).
Footnotes
Published ahead of print 4 June 2014
REFERENCES
- 1.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368:1888–1897. 10.1056/NEJMoa1304459 [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Liang W, Yang S, Wu N, Gao H, Sheng J, Yao H, Wo J, Fang Q, Cui D, Li Y, Yao X, Zhang Y, Wu H, Zheng S, Diao H, Xia S, Zhang Y, Chan KH, Tsoi HW, Teng JL, Song W, Wang P, Lau SY, Zheng M, Chan JF, To KK, Chen H, Li L, Yuen KY. 2013. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 381:1916–1925. 10.1016/S0140-6736(13)60903-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, Xiang N, Chen E, Tang F, Wang D, Meng L, Hong Z, Tu W, Cao Y, Li L, Ding F, Liu B, Wang M, Xie R, Gao R, Li X, Bai T, Zou S, He J, Hu J, Xu Y, Chai C, Wang S, Gao Y, Jin L, Zhang Y, Luo H, Yu H, Gao L, Pang X, Liu G, Shu Y, Yang W, Uyeki TM, Wang Y, Wu F, Feng Z. 24 April 2013. Preliminary report: epidemiology of the avian influenza A (H7N9) outbreak in China. N. Engl. J. Med. 10.1056/NEJMoa1304617 [DOI] [Google Scholar]
- 4.Shi J, Deng G, Liu P, Zhou J, Guan L, Li W, Li X, Guo J, Wang G, Fan J, Wang J, Li Y, Jiang Y, Liu L, Tian G, Li C, Chen H. 2013. Isolation and characterization of H7N9 viruses from live poultry markets—implication of the source of current H7N9 infection in humans. Chin. Sci. Bull. 58:7. 10.1007/s11434-012-5577-1 [DOI] [Google Scholar]
- 5.Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. 2013. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 18:20459 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20453 [PMC free article] [PubMed] [Google Scholar]
- 6.Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, Liu W, Zhao G, Yang W, Wang Y, Ma J, Shu Y, Lei F, Gao GF. 2013. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 381:1926–1932. 10.1016/S0140-6736(13)60938-1 [DOI] [PubMed] [Google Scholar]
- 7.Liu Q, Lu L, Sun Z, Chen GW, Wen Y, Jiang S. 2013. Genomic signature and protein sequence analysis of a novel influenza A (H7N9) virus that causes an outbreak in humans in China. Microbes Infect. 15:432–439. 10.1016/j.micinf.2013.04.004 [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, Wang G, Chen Y, Liu L, Liang L, Li Y, Fan J, Wang J, Li W, Guan L, Li Q, Yang H, Chen P, Jiang L, Guan Y, Xin X, Jiang Y, Tian G, Wang X, Qiao C, Li C, Bu Z, Chen H. 2013. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 341:410–414. 10.1126/science.1240532 [DOI] [PubMed] [Google Scholar]
- 9.Zhou J, Wang D, Gao R, Zhao B, Song J, Qi X, Zhang Y, Shi Y, Yang L, Zhu W, Bai T, Qin K, Lan Y, Zou S, Guo J, Dong J, Dong L, Zhang Y, Wei H, Li X, Lu J, Liu L, Zhao X, Li X, Huang W, Wen L, Bo H, Xin L, Chen Y, Xu C, Pei Y, Yang Y, Zhang X, Wang S, Feng Z, Han J, Yang W, Gao GF, Wu G, Li D, Wang Y, Shu Y. 2013. Biological features of novel avian influenza A (H7N9) virus. Nature 499:500–503. 10.1038/nature12379 [DOI] [PubMed] [Google Scholar]
- 10.Castrucci MR, Kawaoka Y. 1993. Biologic importance of neuraminidase stalk length in influenza A virus. J. Virol. 67:759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munier S, Larcher T, Cormier-Aline F, Soubieux D, Su B, Guigand L, Labrosse B, Cherel Y, Quere P, Marc D, Naffakh N. 2010. A genetically engineered waterfowl influenza virus with a deletion in the stalk of the neuraminidase has increased virulence for chickens. J. Virol. 84:940–952. 10.1128/JVI.01581-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuoka Y, Swayne DE, Thomas C, Rameix-Welti MA, Naffakh N, Warnes C, Altholtz M, Donis R, Subbarao K. 2009. Neuraminidase stalk length and additional glycosylation of the hemagglutinin influence the virulence of influenza H5N1 viruses for mice. J. Virol. 83:4704–4708. 10.1128/JVI.01987-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capua I, Alexander DJ. 2004. Avian influenza: recent developments. Avian Pathol. 33:393–404. 10.1080/03079450410001724085 [DOI] [PubMed] [Google Scholar]
- 14.Röhm C, Horimoto T, Kawaoka Y, Suss J, Webster RG. 1995. Do hemagglutinin genes of highly pathogenic avian influenza viruses constitute unique phylogenetic lineages? Virology 209:664–670. 10.1006/viro.1995.1301 [DOI] [PubMed] [Google Scholar]
- 15.Webster RG, Hinshaw VS, Bean WJ, Van Wyke KL, Geraci JR, St Aubin DJ, Petursson G. 1981. Characterization of an influenza A virus from seals. Virology 113:712–724. 10.1016/0042-6822(81)90200-2 [DOI] [PubMed] [Google Scholar]
- 16.Kalthoff D, Globig A, Beer M. 2010. (Highly pathogenic) avian influenza as a zoonotic agent. Vet. Microbiol. 140:237–245. 10.1016/j.vetmic.2009.08.022 [DOI] [PubMed] [Google Scholar]
- 17.Belser JA, Bridges CB, Katz JM, Tumpey TM. 2009. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg. Infect. Dis. 15:859–865. 10.3201/eid1506.090072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belser JA, Lu X, Maines TR, Smith C, Li Y, Donis RO, Katz JM, Tumpey TM. 2007. Pathogenesis of avian influenza (H7) virus infection in mice and ferrets: enhanced virulence of Eurasian H7N7 viruses isolated from humans. J. Virol. 81:11139–11147. 10.1128/JVI.01235-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Wit E, Munster VJ, Spronken MI, Bestebroer TM, Baas C, Beyer WE, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2005. Protection of mice against lethal infection with highly pathogenic H7N7 influenza A virus by using a recombinant low-pathogenicity vaccine strain. J. Virol. 79:12401–12407. 10.1128/JVI.79.19.12401-12407.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belser JA, Blixt O, Chen LM, Pappas C, Maines TR, Van Hoeven N, Donis R, Busch J, McBride R, Paulson JC, Katz JM, Tumpey TM. 2008. Contemporary North American influenza H7 viruses possess human receptor specificity: implications for virus transmissibility. Proc. Natl. Acad. Sci. U. S. A. 105:7558–7563. 10.1073/pnas.0801259105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X, Li L, Hong W, Chai Y, Zhou L, Liang H, Ou Z, Liu Y, Farooqui A, Kelvin DJ, Poon LL, Smith DK, Pybus OG, Leung GM, Shu Y, Webster RG, Webby RJ, Peiris JS, Rambaut A, Zhu H, Guan Y. 2013. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502:241–244. 10.1038/nature12515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belser JA, Katz JM, Tumpey TM. 2011. The ferret as a model organism to study influenza A virus infection. Dis. Model. Mech. 4:575–579. 10.1242/dmm.007823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuoka Y, Lamirande EW, Subbarao K. 2009. The ferret model for influenza. Curr. Protoc. Microbiol. Chapter 15:Unit 15G.2 [DOI] [PubMed] [Google Scholar]
- 24.Alexander DJ. 2008. Avian influenza, p 465–481 In Linnane S. (ed), Manual of diagnostic tests and vaccines for terrestrial animals (mammals, birds and bees), 6th ed, vol 1 World Organisation for Animal Health, Paris, France [Google Scholar]
- 25.Kalthoff D, Bogs J, Harder T, Grund C, Pohlmann A, Beer M, Hoffmann B. 2014. Nucleic acid-based detection of influenza A virus subtypes H7 and N9 with a special emphasis on the avian H7N9 virus. Euro Surveill. 19:20731 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20731 [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann B, Depner K, Schirrmeier H, Beer M. 2006. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J. Virol. Methods 136:200–209. 10.1016/j.jviromet.2006.05.020 [DOI] [PubMed] [Google Scholar]
- 27.Breithaupt A, Kalthoff D, Dale J, Bairlein F, Beer M, Teifke JP. 2011. Neurotropism in blackcaps (Sylvia atricapilla) and red-billed queleas (Quelea quelea) after highly pathogenic avian influenza virus H5N1 infection. Vet. Pathol. 48:924–932. 10.1177/0300985810386467 [DOI] [PubMed] [Google Scholar]
- 28.Rowe T, Abernathy RA, Hu-Primmer J, Thompson WW, Lu X, Lim W, Fukuda K, Cox NJ, Katz JM. 1999. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 37:937–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pantin-Jackwood MJ, Miller PJ, Spackman E, Swayne DE, Susta L, Costa-Hurtado M, Suarez DL. 2014. Role of poultry in spread of novel H7N9 influenza virus in China. J. Virol. 88:5381–5390. 10.1128/JVI.03689-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ku KB, Park EH, Yum J, Kim HM, Kang YM, Kim JC, Kim JA, Kim HS, Seo SH. 2014. Transmissibility of novel H7N9 and H9N2 avian influenza viruses between chickens and ferrets. Virology 450–451:316–323. 10.1016/j.virol.2013.12.022 [DOI] [PubMed] [Google Scholar]
- 31.Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, Takashita E, McBride R, Noda T, Hatta M, Imai H, Zhao D, Kishida N, Shirakura M, de Vries RP, Shichinohe S, Okamatsu M, Tamura T, Tomita Y, Fujimoto N, Goto K, Katsura H, Kawakami E, Ishikawa I, Watanabe S, Ito M, Sakai-Tagawa Y, Sugita Y, Uraki R, Yamaji R, Eisfeld AJ, Zhong G, Fan S, Ping J, Maher EA, Hanson A, Uchida Y, Saito T, Ozawa M, Neumann G, Kida H, Odagiri T, Paulson JC, Hasegawa H, Tashiro M, Kawaoka Y. 2013. Characterization of H7N9 influenza A viruses isolated from humans. Nature 501:551–555. 10.1038/nature12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu X, Jin T, Cui Y, Pu X, Li J, Xu J, Liu G, Jia H, Liu D, Song S, Yu Y, Xie L, Huang R, Ding H, Kou Y, Zhou Y, Wang Y, Xu X, Yin Y, Wang J, Guo C, Yang X, Hu L, Wu X, Wang H, Liu J, Zhao G, Zhou J, Pan J, Gao GF, Yang R. 2014. Influenza H7N9 and H9N2 viruses: coexistence in poultry linked to human H7N9 infection and genome characteristics. J. Virol. 88:3423–3431. 10.1128/JVI.02059-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamada M, Bingham J, Payne J, Rookes J, Lowther S, Haining J, Robinson R, Johnson D, Middleton D. 2012. Multiple routes of invasion of wild-type clade 1 highly pathogenic avian influenza H5N1 virus into the central nervous system (CNS) after intranasal exposure in ferrets. Acta Neuropathol. 124:505–516. 10.1007/s00401-012-1010-8 [DOI] [PubMed] [Google Scholar]
- 34.Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, Katz JM, Tumpey TM. 2013. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501:556–559. 10.1038/nature12391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu H, Wang D, Kelvin DJ, Li L, Zheng Z, Yoon SW, Wong SS, Farooqui A, Wang J, Banner D, Chen R, Zheng R, Zhou J, Zhang Y, Hong W, Dong W, Cai Q, Roehrl MH, Huang SS, Kelvin AA, Yao T, Zhou B, Chen X, Leung GM, Poon LL, Webster RG, Webby RJ, Peiris JS, Guan Y, Shu Y. 2013. Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science 341:183–186. 10.1126/science.1239844 [DOI] [PubMed] [Google Scholar]