ABSTRACT
The myxovirus resistance 2 (MX2) protein of humans has been identified recently as an interferon (IFN)-inducible inhibitor of human immunodeficiency virus type 1 (HIV-1) that acts at a late postentry step of infection to prevent the nuclear accumulation of viral cDNA (C. Goujon et al., Nature 502:559–562, 2013, http://dx.doi.org/10.1038/nature12542; M. Kane et al., Nature 502:563–566, 2013, http://dx.doi.org/10.1038/nature12653; Z. Liu et al., Cell Host Microbe 14:398–410, 2013, http://dx.doi.org/10.1016/j.chom.2013.08.015). In contrast, the closely related human MX1 protein, which suppresses infection by a range of RNA and DNA viruses (such as influenza A virus [FluAV]), is ineffective against HIV-1. Using a panel of engineered chimeric MX1/2 proteins, we demonstrate that the amino-terminal 91-amino-acid domain of MX2 confers full anti-HIV-1 function when transferred to the amino terminus of MX1, and that this fusion protein retains full anti-FluAV activity. Confocal microscopy experiments further show that this MX1/2 fusion, similar to MX2 but not MX1, can localize to the nuclear envelope (NE), linking HIV-1 inhibition with MX accumulation at the NE. MX proteins are dynamin-like GTPases, and while MX1 antiviral function requires GTPase activity, neither MX2 nor MX1/2 chimeras require this attribute to inhibit HIV-1. This key discrepancy between the characteristics of MX1- and MX2-mediated viral resistance, together with previous observations showing that the L4 loop of the stalk domain of MX1 is a critical determinant of viral substrate specificity, presumably reflect fundamental differences in the mechanisms of antiviral suppression. Accordingly, we propose that further comparative studies of MX proteins will help illuminate the molecular basis and subcellular localization requirements for implementing the noted diversity of virus inhibition by MX proteins.
IMPORTANCE Interferon (IFN) elicits an antiviral state in cells through the induction of hundreds of IFN-stimulated genes (ISGs). The human MX2 protein has been identified as a key effector in the suppression of HIV-1 infection by IFN. Here, we describe a molecular genetic approach, using a collection of chimeric MX proteins, to identify protein domains of MX2 that specify HIV-1 inhibition. The amino-terminal 91-amino-acid domain of human MX2 confers HIV-1 suppressor capabilities upon human and mouse MX proteins and also promotes protein accumulation at the nuclear envelope. Therefore, these studies correlate the cellular location of MX proteins with anti-HIV-1 function and help establish a framework for future mechanistic analyses of MX-mediated virus control.
INTRODUCTION
Virus infections elicit the production of interferons (IFNs), a family of immunomodulatory cytokines that promote both innate and adaptive immunity (1–3). During acute viral infections, type-1 IFNs (α, β, and ω) are rapidly expressed in response to the interactions of pathogen-associated molecular patterns with cell-encoded pattern recognition receptors. Following the engagement of type-1 IFN with its receptor, cascades of host-encoded IFN-stimulated genes (ISGs) are induced which collectively establish an antiviral state through myriad mechanisms (2, 4). It has long been recognized that type-1 IFN suppresses the replication of human and simian immunodeficiency viruses (HIVs and SIVs) in cultured cells (5–10), and it has been shown more recently that serum IFN-α levels during acute experimental SIV infection of rhesus macaques inversely correlate with viral loads (11), supporting the conclusion that type 1 IFN plays an important role in viral control in acute infection in vivo.
The human myxovirus resistance 2 (MX2) protein recently has been established as a novel ISG with potent HIV type 1 (HIV-1) inhibitory properties (12–14). More specifically, human MX2 efficiently inhibits infection by diverse HIV-1 strains, is typically less active against HIV type 2 and SIVs derived from various nonhuman primates, and does not inhibit other retroviruses, such as feline immunodeficiency virus (FIV) or murine leukemia virus (MLV) (12). MX2 blocks HIV-1 infection at a relatively late postentry phase, which is manifested as the failure of newly synthesized viral cDNAs to accumulate in the nucleus (12, 13), and viral specificity is determined by the capsid (CA) region of Gag in that polymorphisms in CA that are known to impact interactions with cellular factors (15–22) can confer insensitivity to MX2-mediated suppression (12–14).
MX2 is a member of the dynamin-like IFN-inducible guanosine triphosphatase (GTPase) superfamily (23, 24). The most closely related family member is MX1 (denoted Mx1 in mice), a well-known inhibitor of a broad range of RNA and DNA viruses that includes influenza A virus (FluAV), La Crosse encephalitis virus, and hepatitis B virus but not retroviruses such as HIV-1 (12, 25–28). While the antiviral capability of MX2 has only just been recognized, a considerable body of elegant work focusing on the biology of MX1 already exists. MX1 is a mechanoenzyme, and structural studies have demonstrated that it is organized into an amino-terminal GTPase domain and a carboxy-terminal stalk domain that are connected by a bundle signaling element (BSE) formed from three noncontiguous helical regions (Fig. 1) (29). GTPase function typically is considered to be required for antiviral function (30, 31), and the stalk (or effector) domain, together with the hinge that links the BSE to the stalk, mediates MX1 oligomerization and, presumably, assembly into the higher-order ring-like structures that can be visualized with recombinant protein in vitro (29, 32, 33). In some cases, antiviral function involves interactions between components of virus replication complexes, for instance, the nucleoproteins (NP) of the orthomyxoviruses FluAV and Thogoto virus (THOV), and a disordered surface-exposed region of the MX1 stalk domain, termed loop 4 (L4) (34–39). Current mechanistic models suggest that substrate engagement promotes higher-order MX1 oligomerization leading to GTPase activation and nucleotide hydrolysis, coupled with conformational changes and the enforced disintegration/perturbation of viral nucleoprotein replication complexes (27, 29). However, many important questions remain. For instance, how does MX1 recognize its diversity of viral substrates, particularly in light of the positive selection pressures that act upon L4 (35), defining the precise stage(s) of replication at which MX1 exerts its effects on different viruses and elucidating how GTPase activity translates to viral suppression?
FIG 1.
Human MX1 and MX2 sequence alignment. The different domains of the proteins, namely, the N-terminal region (N-term), the bundle signaling element (BSE), the GTPase domain (G domain), the stalk domain (Stalk), and L4 loop (L4), are indicated by colored boxes above (for MX1) or underneath (for MX2) the sequences. Identical amino acids are highlighted in black, and those with similar properties are in gray.
Here, we have exploited the dichotomy in HIV-1 inhibitory function between the human MX proteins to identify determinants of antiviral activity. By analyzing a panel of chimeric MX1/MX2 proteins, we demonstrate that the amino-terminal 91 residues of MX2 are essential for anti-HIV-1 function, and that appending this domain to the amino terminus of MX1 is sufficient to confer a potent HIV-1 suppression phenotype and to promote targeting to the nuclear envelope (NE).
MATERIALS AND METHODS
Cell culture and plasmids.
Human 293T, HeLa, and U87-MG/CD4/CXCR4 (12) cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum plus l-glutamine and penicillin-streptomycin. The HIV-1IIIB provirus, HIV-1/Nef-internal ribosome entry signal (IRES)-Renilla reporter virus, the HIV-1-, FIV-, equine infectious anemia virus (EIAV)-, and MLV-derived lentiviral and retroviral vectors (LVs and RVs, respectively) and the human MX1 and MX2 expressing pEasiLV-based vectors have been described (12, 40–44), as have the HIV-1 P90A capsid mutant (19) and the HIV-1/SIVMAC capsid chimera (45). Murine Mx1 and Mx2 cDNAs (provided by Georg Kochs) were amplified by PCR and cloned into pEasiLV-MCS using the BamHI and XhoI restriction sites. cDNAs encoding the MX1(L4MX2), MX1(stalkMX2), MX1(GMX2), MX1(G stalkMX2), MX1(NMX2), and reciprocal MX2-based chimeras, as well as the murine Mx1- and Mx2-based chimeras and the GTPase domain mutants, were obtained by overlapping PCRs and cloned into pEasiLV-MCS using the BamHI and XhoI restriction sites (see Fig. S1 in the supplemental material for the amino acid sequences of all chimeras). cDNA fragments for certain MX/Mx proteins, chimeras, and GTPase mutants were subcloned from pEasiLV into pCAGGS (Addgene) using the BclI and XhoI restriction sites. All chimeras and wild-type MX proteins harbor a Flag tag at either the C terminus (HIV-1 and microscopy experiments) or the N terminus (FluAV experiments). The cDNA for turbo-red fluorescent protein (tbRFP; Evrogen) fused to the C terminus of residues 1 to 91 of human MX2 (NMX2-tbRFP) was obtained by overlapping PCRs and cloned, along with wild-type tbRFP, into pCAGGS as a BamHI-XhoI fragment.
Virus production.
To produce EasiLV particles, 293T cells were cotransfected with pEasiLV, p8.91, pptTRKrab (46), and pMD.G at a ratio of 1:1:0.5:0.25, respectively, with TransIT-2020 reagent (Mirus Bio). The medium was replaced after overnight incubation and viral particles were harvested 36 h later, filtered, and used directly to transduce target cells. After a few hours, the medium was replaced and doxycycline added (0.05 to 0.5 μg/ml, depending on the transgene; Sigma-Aldrich) to induce transgene expression. The percentage of E2-Crimson-positive cells was scored by flow cytometry (FACSCalibur; BD Biosciences) after 48 to 72 h and typically was >85%.
HIV-1IIIB particles were produced by standard polyethylenimine (PEI) transfection of 293T monolayers. The culture medium was changed ∼6 h later, and virus-containing supernatant was harvested at ∼36 h. LV and RV stocks were obtained by PEI-mediated triple transfection of 293T cells with vectors expressing Gag-Pol, vectors expressing miniviral genomes bearing a cytomegalovirus-enhanced green fluorescent protein (CMV-EGFP) cassette, and pMD.G at a ratio of 1:1:0.5, respectively.
Viral particles were filtered, and, when required (i.e., for HIV-1IIIB infections that were followed by real-time quantitative PCR [qPCR] analysis of viral cDNA), the virus-containing supernatants were treated with RQ1 DNase (Promega) for 3 h, purified by ultracentrifugation through a sucrose cushion (20%, wt/vol; 75 min; 4°C at 28,000 rpm using a Sorvall Surespin630 rotor), resuspended in RPMI 1640 medium without serum, and stored in aliquots at −80°C.
Viral particles were normalized according to HIV-1 p24Gag enzyme-linked immunosorbent assay (ELISA) (PerkinElmer) and/or by determining their infectious titers on U87-MG cells. The multiplicity of infection (MOI) for LV and RV stocks was determined by infecting a known number of U87-MG cells with standardized amounts of viral particles and evaluating, by flow cytometry, the percentage of infected cells 2 to 3 days later. For instance, an MOI of 0.25 equates to the volume of virus necessary to obtain 25% GFP-expressing U87-MG cells.
Retroviral infection.
All infections were initiated using standard conditions (12). Briefly, for infection with NL4-3/Nef-IRES-Renilla or GFP-expressing LVs and RVs, U87-MG/CD4/CXCR4 cells were plated at ∼2.5 × 104 to 5 × 104 per well in 96-well plates. When HIV-1 infection was followed by DNA extraction, U87-MG/CD4/CXCR4 cells were seeded at ∼2 × 105 cells per well in 24-well plates. The efficiency of productive infection was analyzed after ∼48 h by evaluating the percentage of GFP-expressing cells using flow cytometry or by measuring renilla luciferase activity (Promega).
Influenza A virus infection.
pCAGGS vectors expressing N-terminally Flag-tagged MX1, MX2, and MX1(NMX2), as well as IFITM3 or the empty control, were transfected into 293T cells (0.3 μg) in 48-well plates using Lipofectamine 2000 (Invitrogen) simultaneously with a firefly luciferase minigenome-expressing plasmid (pHSPOM1-Firefly; 0.1 μg) (47) and a renilla luciferase expression plasmid (T7-Renilla; 0.03 μg). At 24 h, cells were infected with FluAV A/Victoria/3/75 (H3N2) at an MOI of 2 or were mock infected. Activities of firefly and renilla luciferases were measured 18 h postinfection, and firefly luciferase signals were normalized to renilla luciferase signals from mock-infected cells.
Immunoblot analysis.
Cell pellets were lysed in sample buffer (200 mM Tris-HCl, pH 6.8, 5.2% SDS, 20% glycerol, 0.1% bromphenol blue, 5% β-mercaptoethanol), resolved by SDS-PAGE, and analyzed by immunoblotting using primary antibodies specific for Flag (mouse monoclonal M2; Sigma-Aldrich), Hsp90 (rabbit; Santa Cruz Biotechnology), and tubulin (mouse monoclonal DM1A; Sigma-Aldrich), followed by secondary horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin antibody and chemiluminescence (ECL Western blotting substrate; Pierce).
GTPase enzymatic activity.
Subconfluent 293T cell monolayers in 10-cm dishes were transfected with 15 μg of the pCAGGS-based Flag-tagged MX expression constructs using PEI. At 48 h posttransfection, the cells were washed twice with cold phosphate-buffered saline and lysed for 10 min on ice with lysis buffer {10 mM 3-([3-cholamidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate (CHAPSO), 50 mM Tris, pH 8.0, 150 mM NaCl, 0.5% Triton X-100, 0.1% sodium deoxycholate} in the presence of complete protease inhibitor cocktail (Roche). The lysates were sonicated (3 times for 10 s each), clarified by centrifugation for 10 min at 1,000 × g, and incubated with anti-Flag M2 magnetic beads (Sigma-Aldrich) for 2 h at 4°C on a rotational tumbler. The beads were washed 5 times with lysis buffer lacking Triton X-100, and the Flag-tagged MX proteins were eluted by incubation in GTPase assay buffer (50 mM Tris, pH 8.0, 5 mM MgCl2, 10% glycerol, 50 μM GTP, 2 mM DTT) in the presence of 150 μg/ml of the 3× Flag peptide (Sigma-Aldrich) for 2 h on ice. The MX protein levels were evaluated by SDS-PAGE followed by silver staining, and normalized amounts were incubated in 50 μl GTPase assay buffer in the presence of 13 nM [α-32P]GTP for 1 h at 37°C before stopping the reactions with 2 mM EDTA, 0.5% SDS. The reaction products were resolved by thin-layer chromatography (TLC; Merck) using TLC buffer (1 M LiCl, 1 M acetic acid) and detected using phosphor screen autoradiography (GE Amersham Typhoon).
qPCR analysis of HIV-1 reverse transcription products.
A total of 2 × 105 U87-MG/CD4/CXCR4 cells per well in 24-well plates were transduced with CD8-, MX2-, or MX1(NMX2)-expressing EasiLV. The cells then were challenged with HIV-1IIIB (10 ng p24Gag) for 4 h. The cells then were washed twice, incubated in complete medium (supplemented with 0.5 or 0.05 μg/ml doxycycline and/or reverse transcription inhibitors as required), and harvested at 4, 24, or 48 h postinfection. Total DNA was prepared using the DNeasy kit (Qiagen), and 30 ng of each sample was used for qPCR analysis. First (minus)-strand transfer cDNA products were detected using primers that amplify the region between nucleotides 53 and 175 of pIIIB in U3 (U3for, 5′-TCTACCACACACAAGGCTAC; U3rev, 5′-CTTCTAACTTCTCTGGCTCAAC) and with U3probe (5′-6-carboxyfluorescein [FAM]-CAGAACTACACACCAGGACCAGGGATCA-6-carboxytetramethylrhodamine [TAMRA]); 2-long-terminal-repeat (2-LTR) circular forms were detected using primers that amplify the region between nucleotides 585 of pIIIB (3′ terminus of U5) and 102 of pIIIB (5′ terminus of U3) (2LTRfor, 5′-GTAACTAGAGATCCCTCAGACC; 2LTRrev, 5′-TCCTGGTGTGTAGTTCTGCC) with 2LTRprobe (5′-FAM-CTACCACACACAAGGCTACTTCCCTGAT-TAMRA). qPCRs were performed in triplicate in TaqMan Universal PCR master mix using 900 nM each primer and 250 nM probe. After 10 min at 95°C, reactions were cycled through 15 s at 95°C followed by 1 min at 60°C for 40 repeats. pIIIB or pTOPO-2LTR (containing a 2-LTR circle junction amplified from HIV-1IIIB-infected CEM-SS cell DNA using primers oHC64 [5′-TAACTAGGGAACCCACTGC] and U3rev and cloned into pPCR-Blunt II-TOPO) was diluted in 20 ng/ml salmon sperm DNA to create dilution standards that were used to calculate relative cDNA copy numbers and confirm the linearity of all assays.
Microscopy.
HeLa cells were seeded on coverslips at ∼105 cells per well in 6-well plates 24 h prior to transfection with 500 ng of the pCAGGS-based expression constructs using TransIT-LT1 reagent (Mirus Bio). Sixteen h posttransfection, the cells were washed twice with phosphate-buffered saline and fixed in 3% paraformaldehyde (EM Sciences) for 15 min, permeabilized with 0.2% Triton X-100 for 12 min, and blocked/quenched in buffer NGB (50 mM NH4Cl, 1% goat serum, 1% bovine serum albumin) for 30 min. MX proteins and the NE were detected using Flag- or NUP358-specific antibodies, respectively (mouse M2 anti-Flag and rabbit anti-RANBP2; Abcam) and secondary anti-mouse antibody conjugated to Alexa Fluor 488 and anti-rabbit antibody conjugated to Alexa 594 (or Alexa 488 for cells expressing tbRFP). Cells were visualized using a Nikon A1 point-scanning laser confocal microscope (Nikon Instruments). The images correspond to single optical sections of 0.53 μm.
RESULTS
The amino-terminal domain of MX2 specifies HIV-1 suppression.
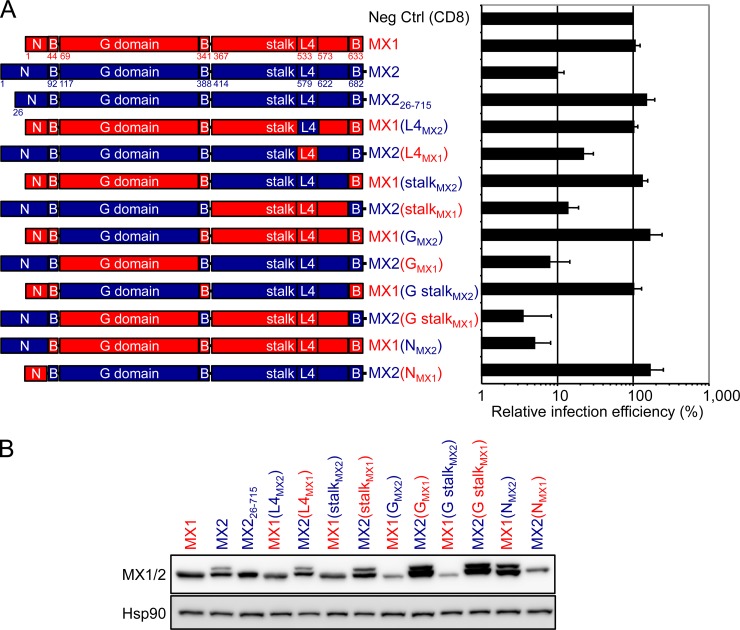
The human MX1 and MX2 proteins share an overall amino acid identity of 63%: an alignment of the two sequences is displayed in Fig. 1, with domains delineated in accordance with the resolved structure of MX1 (29). There is evident homology throughout most of the sequence, though the predicted isoelectric points are different (5.6 for MX1 and 9.1 for MX2; calculated using the Geneious software). The feature that obviously distinguishes the two proteins is the length and sequence of the extension that lies amino-terminal to the first helix of the BSE: 43 amino acids for MX1 compared with 91 residues for MX2. To identify peptide segments that endow MX2, but not MX1, with the capacity to inhibit HIV-1 infection, we constructed a series of Flag-tagged chimeric MX genes in which the GTPase domain, L4 loop, stalk, and amino-terminal extensions were systematically substituted back and forth (Fig. 2A; MX1 sequences are shown in red, MX2 sequences are in blue).
FIG 2.
N-terminal domain of MX2 confers potent anti-HIV-1 activity to human MX1. (A, left) Schematic representation illustrating the MX1 (red) and MX2 (blue) origins of the different domains comprising the chimeric MX1/MX2 proteins. (Right) U87-MG/CD4/CXCR4 cells were transduced with EasiLV expressing CD8 (negative control [Neg Ctrl]), MX1, MX2, or chimeric MX1/MX2 cDNAs and treated with doxycycline [0.5 μg/ml except for MX1(NMX2), which received 0.05 μg/ml] for 48 h prior to infection with 25 ng p24Gag of NL4-3/Nef-IRES-Renilla. Infection efficiency relative to the Neg Ctrl was monitored at 48 h by measuring renilla activity, and mean relative infection efficiencies with standard deviations from four independent experiments are shown. (B) Immunoblot analysis of parallel samples from panel A. Protein levels of wild-type MX1 and MX2 proteins and the MX1/MX2 chimeras were determined using a Flag-specific antibody, and staining for Hsp90 served as a loading control.
All gene chimeras, as well as CD8 (negative control), were inserted into the pEasiLV-MCS doxycycline-inducible lentiviral vector (12), where levels of transgene expression are regulated by doxycycline addition and the efficiency of transduction is monitored by visualizing E2-Crimson fluorescent protein expression. Vector stocks were prepared in 293T cells and used to transduce U87-MG/CD4/CXCR4 monolayers. Following doxycycline induction, all chimeric proteins were readily detected in whole-cell lysates (Fig. 2B), and at least 85% of the cells reliably expressed E2-Crimson (data not shown). The cultures then were challenged with an HIV-1 reporter virus, NL4-3/Nef-IRES-Renilla, and productive infection measured at 48 h by quantitating renilla luciferase expression (Fig. 2A; infection levels were normalized to that seen in the presence of the CD8 negative control). As previously demonstrated (12), MX2 inhibited single-cycle HIV-1 infection by ∼90%, whereas MX1 had no effect. Examination of the data in toto revealed immediately that the sole MX2-derived determinant that is necessary for specifying anti-HIV-1 function is its amino-terminal domain. In other words, provided that a GTPase domain, stalk, and BSE are present, a human MX1/2 protein can inhibit HIV-1 infection whenever it contains the amino-terminal 91 amino acids of MX2. This conclusion is most succinctly illustrated by the MX1(NMX2) chimera, which displays potency similar to that of wild-type MX2. Notably, these findings were unexpected, since it is the L4 region of MX1 that previously had been identified as a determinant of virus substrate selection, at least for FluAV and THOV. Accordingly, this analysis also specifically tested the contribution of L4 through the reciprocal exchange of this region between MX1 and MX2 [MX1(L4MX2) and MX2(L4MX1)]; however, neither substitution substantially altered anti-HIV-1 activity relative to the wild-type proteins, supporting the notion that L4 does not determine MX-mediated HIV-1 inhibition.
The analysis of MX protein expression shows that any construct containing the 5′ terminus of MX2 yields two isoforms (Fig. 2B). We wished to determine which of these isoforms is antiviral and confirm that the methionine at position 26 serves as an alternative translation start site that generates the shorter isoform (48). Two additional MX2 cassettes were constructed: MX2Kozak contains optimal sequences for translation 5′ of the ATG for the methionine at position 1 (gccgccaccATG), and MX226-715 lacks the amino-terminal 25 codons. MX2Kozak expressed the longer isoform, which displayed robust anti-HIV-1 activity, whereas MX226-715 yielded the shorter isoform and was inactive (see Fig. S2 in the supplemental material). Therefore, HIV-1 infection is inhibited exclusively by the 715-amino-acid full-length form of MX2, consistent with previous results (13).
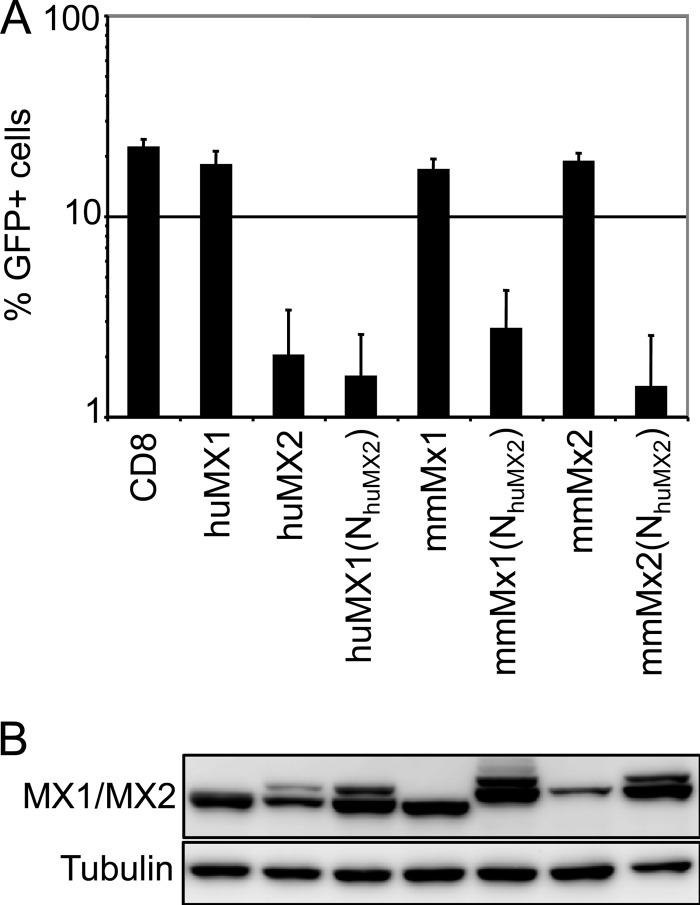
Like humans, mice contain two Mx genes, although both are more closely related to human MX1 than to human MX2 on the basis of primary sequence (49), and neither carries the amino-terminal extension that distinguishes human MX2. Therefore, we examined the HIV-1 inhibitory capabilities of both wild-type murine Mx proteins as well as chimeras containing the amino-terminal 91 amino acids of human MX2 (Fig. 3). Both murine proteins were inactive, but each acquired marked anti-HIV-1 function upon addition of the MX2 amino-terminal region, revealing that this sequence can confer HIV-1 suppressor function when appended to multiple MX proteins.
FIG 3.
N-terminal region of MX2 confers anti-HIV-1 activity to mouse Mx1 and Mx2. (A) U87-MG/CD4/CXCR4 cells were transduced with EasiLV expressing CD8, human MX1 (huMX1) or MX2 (huMX2), mouse Mx1 (mmMx1) or Mx2 (mmMX2), or huMX1/mmMx1/mmMx2 containing the N-terminal domain of human MX2 [huMX1(NhuMX2), mmMx1(NhuMX2), and mmMx2(NhuMX2), respectively]. The cells were treated with doxycycline for 48 to 72 h and challenged with an HIV-1-based lentiviral vector expressing GFP at an MOI of 0.25. The percentage of GFP-expressing cells was evaluated by flow cytometry 2 days later. Mean percentages of transduced cells from three independent experiments are shown. (B) Immunoblot analysis of parallel samples from panel A. Protein levels of Flag-tagged MX1/MX2 proteins were determined, and tubulin staining served as a loading control.
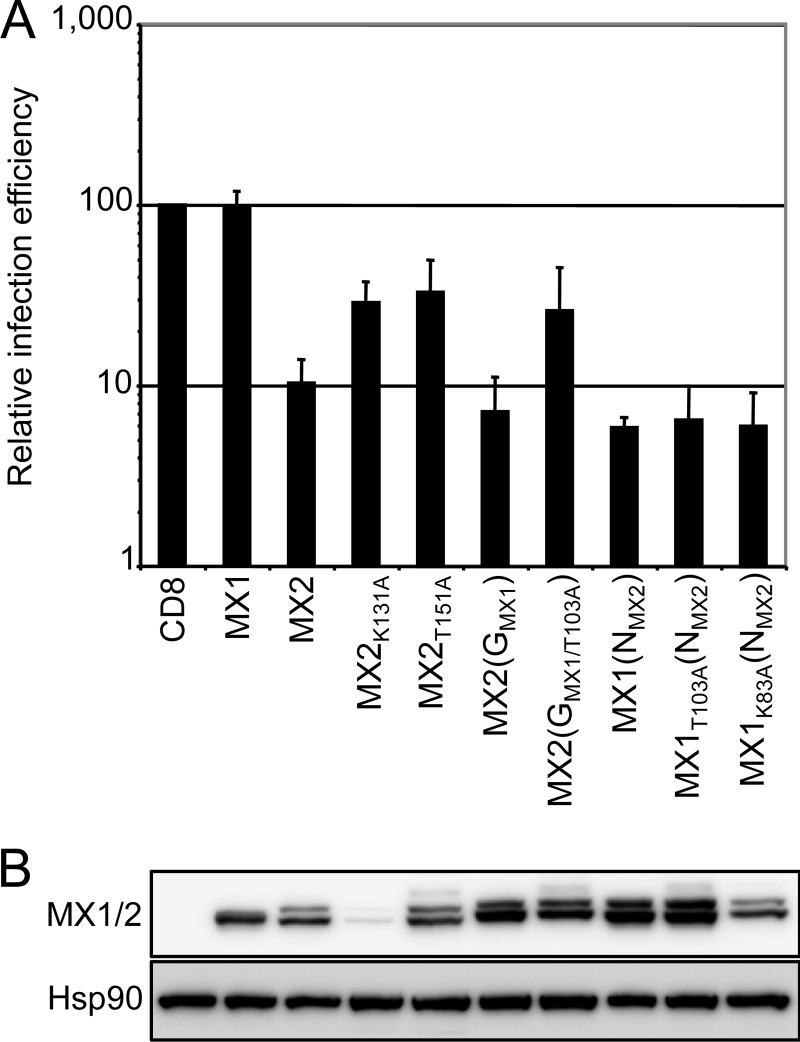
GTPase function is dispensable for MX-mediated inhibition of HIV-1 infection.
Previous work has established that an enzymatically active GTPase domain generally is required for MX1-mediated antiviral function (30, 31), but that GTP binding and hydrolysis are not prerequisites for moderate inhibition of HIV-1 by MX2 (12, 13). We next evaluated if this trend was maintained in MX1/2 chimeras containing the amino-terminal region of MX2 and the GTPase domain of MX1 (Fig. 4). MX2(GMX1), which contains the GTPase domain of MX1 in an otherwise MX2 background, displayed strong antiviral activity, and this was partially retained by the GTP-binding mutant MX2(GMX1/T103A), findings that mirror what is seen for MX2 GTPase domain mutants K131A and T151A. More extremely, the GTP-binding and GTPase-defective derivatives of the MX1(NMX2) chimera, MX1K83A(NMX2) and MX1T103A(NMX2), both exhibited anti-HIV-1 potencies equivalent to those of MX1(NMX2) as well as wild-type human MX2 itself.
FIG 4.
MX GTPase activity is not necessary for HIV-1 inhibition. (A) U87-MG/CD4/CXCR4 cells were transduced with EasiLV expressing CD8, MX1, MX2, MX2(GMX1), MX1(NMX2), or the GTPase-deficient mutants MX2K131A, MX2T151A, MX2(GMX1/T103A), MX1K83A(NMX2), and MX1T103A(NMX2) and treated with doxycycline [0.5 μg/ml for CD8, MX1, MX2, MX2(GMX1), MX2K131A, MX2(GMX1/T103A), and MX1K83A(NMX2) or 0.05 μg/ml for MX2T151A, MX1(NMX2), and MX1T103A(NMX2)]. The cells were infected with 25 ng p24Gag of NL4-3/Nef-IRES-Renilla, and infection efficiency was monitored at 48 h by measuring renilla activity. Mean relative infection efficiencies from three independent experiments are shown. (B) Immunoblot analysis of parallel samples from panel A. Protein levels of Flag-tagged MX1/MX2 proteins were determined, and Hsp90 served as a loading control.
To confirm that these mutations in the MX1 GTPase domain prevent enzymatic activity in the context of MX1(NMX2) chimeric proteins, we assessed GTP hydrolysis using proteins immunoprecipitated from transfected 293T cells using a Flag-specific antibody (see Fig. S3 in the supplemental material). As observed by the conversion of GTP to GDP, MX1(NMX2) exhibited activity similar to that of human MX1, whereas the MX1K83A(NMX2)- and MX1T103A(NMX2)-containing samples displayed the same background (or null) activity as the well-established inactive T103A mutant version of MX1. Taken together, these observations further strengthen the argument that GTPase activity is not required for MX-mediated suppression of HIV-1 infection.
The MX1(NMX2) chimera recapitulates the mechanism and specificity of MX2-mediated viral inhibition.
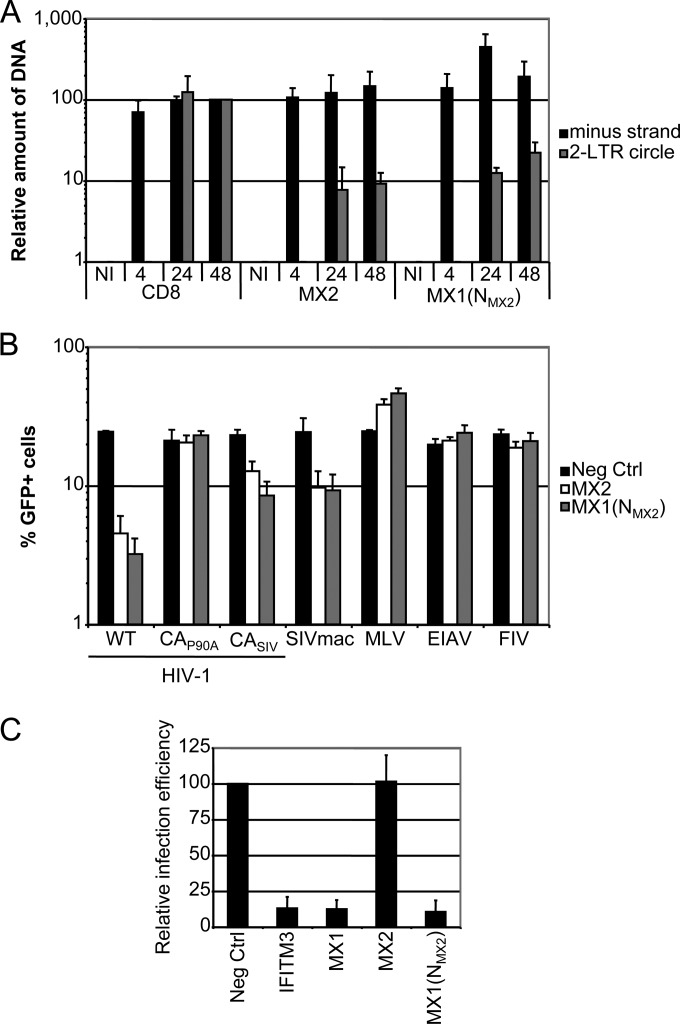
As summarized earlier, the current weight of evidence indicates that MX2 inhibits the nuclear entry of nascent HIV-1 reverse transcripts rather than their initial synthesis. To examine whether MX1(NMX2) also acts in this manner, U87-MG/CD4/CXCR4 cells expressing CD8, human MX2, or the MX1(NMX2) chimera were challenged with wild-type HIV-1 or mock infected (Fig. 5A). Total DNA was extracted after 4, 24, and 48 h, and the generation of viral DNA replication intermediates was measured using quantitative PCR. As has been shown previously for MX2, the synthesis of viral minus (first)-strand cDNA was not repressed by MX1(NMX2), whereas the accumulation of 2-LTR circles, a marker for the nuclear uptake of viral cDNA, was inhibited by ∼90% by this fusion protein.
FIG 5.
MX1(NMX2) inhibits HIV-1 nuclear import with the same specificity as MX2 and retains antiviral activity against influenza A virus. (A) U87-MG/CD4/CXCR4 cells were transduced with EasiLV expressing CD8, MX2, or MX1(NMX2) and treated with doxycycline for 48 to 72 h prior to infection. The cells either were not infected (NI) or were challenged with 10 ng p24Gag HIV-1IIIB and harvested at 4, 24, or 48 h postinfection for DNA extraction and qPCR analysis of minus-strand and 2-LTR circle DNAs. Mean values of relative amounts of DNA (normalized to the CD8 control at 48 h) from three independent experiments are shown. (B) U87-MG cells were transduced with EasiLV expressing CD8, MX2, or MX1(NMX2) and treated with doxycycline for 48 to 72 h. Cells were challenged with HIV-1-based lentiviral vectors containing either wild-type capsid (WT), the P90A capsid mutant (CAP90A), capsid from SIVmac (CASIV), or SIVmac-, EIAV-, FIV-, or MLV-based retroviral vector expressing GFP at an MOI of 0.25. The percentage of GFP-expressing cells was evaluated by flow cytometry 2 days later. Mean percentages of transduced cells from three independent experiments are shown. (C) 293T cells were cotransfected with expression plasmids for IFITM3, Flag-tagged MX1, MX2, or MX1(NMX2), or empty plasmid (Neg Ctrl), along with a FluAV firefly luciferase minigenome plasmid and a renilla luciferase expression plasmid. At 24 h, cells were infected with FluAV A/Victoria/3/75 (H3N2) at an MOI of 2, and firefly and renilla luciferase activities were measured 18 h postinfection. Mean relative infection efficiencies from three independent experiments are shown.
We next evaluated the viral substrate specificity of MX1(NMX2) compared to that of MX2 using a panel of green fluorescent protein (GFP)-expressing viral vectors derived from HIV-1, MLV, FIV, EIAV, or SIV from rhesus macaque (SIVmac), as well as HIV-1 vectors carrying the capsid (CA) region of SIVmac or the proline-to-alanine mutation at position 90 of CA (Fig. 5B). The inhibitory characteristics of MX1(NMX2) closely matched those of MX2, with viruses containing SIVmac CA displaying reduced sensitivity and the P90A mutation conferring resistance (12). We conclude that transfer of the amino-terminal domain of MX2 is sufficient to confer fully the antiretroviral properties and specificity of MX2 upon MX1.
The finding that the amino-terminal region of human MX2 bestows full anti-HIV-1 activity on MX1 is surprising in light of data demonstrating that the L4 region of MX1, a completely distinct element, also dictates the specificity of viral inhibition. Therefore, it was of interest to assess the FluAV inhibitory phenotype of the MX1(NMX2) chimera, in particular asking whether it would be compromised once anti-HIV-1 activity is acquired. FluAV infectivity was measured by viral challenge of 293T cells previously transfected with a vector encoding a firefly luciferase-containing FluAV minigenome and expressing MX1, MX2, MX1(NMX2), IFITM3 (positive control), or an empty vector control, followed by luciferase quantification at 18 h (Fig. 3C). Consistent with previous reports, MX1 inhibited FluAV infection by >80%, whereas MX2 had no effect; importantly, MX1(NMX2) maintained full FluAV suppressor function, indicating that the determinants of HIV-1 and FluAV inhibition are discrete from each other and are not mutually exclusive from a functional standpoint in that both can be operative in the context of a single engineered MX protein.
The amino-terminal domain of MX2 is a transferable nuclear envelope targeting signal.
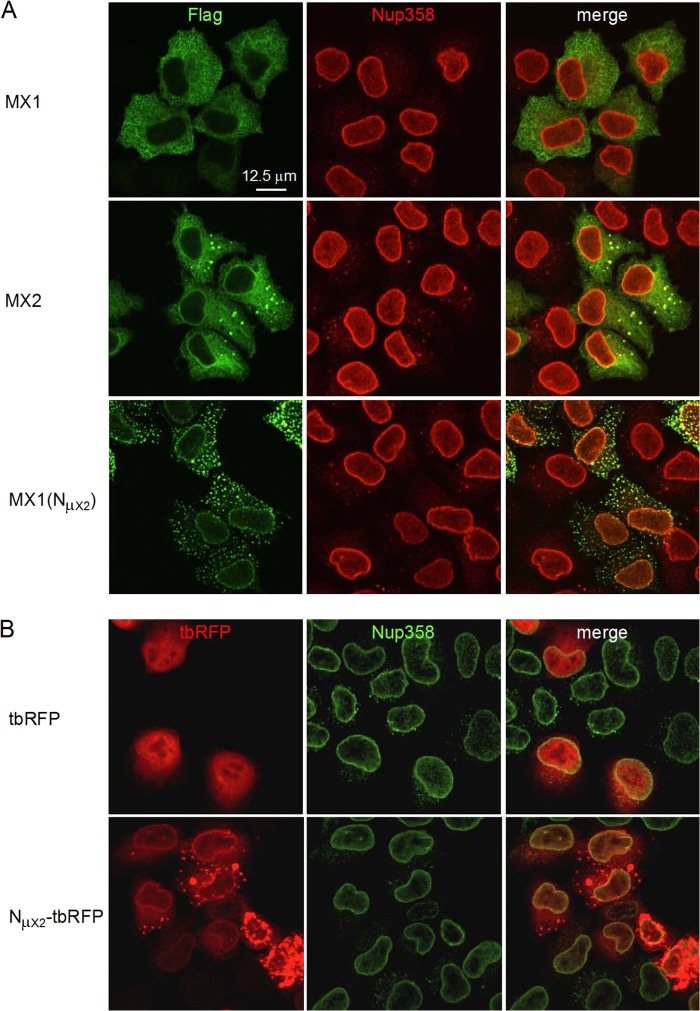
Previous studies are somewhat inconsistent in their descriptions of the subcellular localization of human MX2, with accumulation in the nucleus and toward the inner face of the NE, at the NE, particularly at its cytoplasmic side, and in cytoplasmic granules having been reported (13, 48, 50). Human MX1, in contrast, localizes throughout the cytoplasm (28, 51). Therefore, we used indirect immunofluorescence and confocal microscopy to examine the localization of Flag-tagged MX1, MX2, and MX1(NMX2) in transiently transfected HeLa cell monolayers. The samples were doubly stained with an antibody specific for the nucleoporin NUP358 to identify the NE (Fig. 6A).
FIG 6.
Amino-terminal domain of human MX2 is a transferable nuclear envelope targeting domain. HeLa cells were seeded on glass coverslips and transfected with Flag-tagged MX1, MX2, or MX1(NMX2) (A) or with turboRFP (tbRFP) or NMX2-tbRFP (B) expression constructs and fixed 16 h posttransfection. MX proteins and the NE were visualized by indirect immunofluorescence using Flag- or NUP358-specific antibodies, respectively, and confocal microscopy. Scale bar, 12.5 μm.
In keeping with previous reports, human MX1 localized to the cytoplasm with a filigree-like staining pattern. MX2 accumulated at the NE, throughout the cytoplasm, and in large cytoplasmic bodies/granules/puncta, although we note that cells expressing lower levels of MX2 tended to display reduced granule staining, indicating that this characteristic reflects higher expression levels. Importantly, the localization of the MX1(NMX2) chimera changed dramatically compared to that of MX1, with a marked acquisition of accumulation at both the NE and in cytoplasmic puncta: the latter appeared different from those seen with MX2, being both smaller and more numerous.
In contrast to human MX1, murine Mx1 localizes to the nucleus, with both nucleoplasmic and punctate staining patterns having been described (52, 53); murine Mx2, on the other hand, is cytoplasmic and appears rather similar to human MX1 (54). Therefore, we proceeded to compare the localization phenotypes of these proteins with and without the swapping in of the amino-terminal 91 residues of human MX2 (see Fig. S4 in the supplemental material). In each case, and irrespective of the localization of the parental Mx protein, there was a profound retargeting of the chimeric protein to the NE.
Lastly, to assess whether the amino-terminal domain of human MX2 can dictate the localization of a heterologous and unrelated substrate, it was fused to the amino terminus of turbo-red fluorescent protein (tbRFP) and expressed in HeLa cells (Fig. 6B). While untagged tbRFP was distributed through the nucleus and cytoplasm, the NMX2-tbRFP fusion accumulated at the NE and in large cytoplasmic granules in cells that expressed higher protein levels. Therefore, we conclude that the amino-terminal 91-amino-acid region of MX2 is both necessary and sufficient for localization to the NE.
DISCUSSION
Here, we have exploited chimeric MX proteins to define the amino-terminal 91 amino acids of human MX2 as a critical determinant of this protein's capacity to inhibit HIV-1 infection. Transfer of this region onto human MX1 as well as murine Mx1 or Mx2 confers potent HIV-1 inhibitory functions (Fig. 2 and 3). Subcellular localization studies further revealed that the amino-terminal region of human MX2 is sufficient to redirect a substantial fraction of human MX1, murine Mx1 and Mx2, and tbRFP to the NE (Fig. 6; also see Fig. S4 in the supplemental material). Accordingly, we have identified a strong correspondence between the MX-mediated suppression of HIV-1 and localization at the NE. Previous work, confirmed by the experiments discussed here, has shown that MX2 inhibits HIV-1 infection at a relatively late postentry stage, following substantial cDNA synthesis but prior to the accumulation of nascent DNAs in the nucleus (Fig. 5A) (12, 13). Taken together, these findings suggest that MX2 inhibits the nuclear import of HIV-1 replication complexes and that positioning at the NE is an attribute that facilitates such an activity.
The precise mechanism(s) of viral inhibition remains to be determined and presumably will require identification of MX2 interacting partners, particularly those that bind to the amino-terminal 91 amino acids. Based on the evidence that this region is an NE targeting signal (Fig. 6), one model is that MX2 interacts with a component of the NE, possibly the nuclear pore complexes (13, 50). Inhibition then could be achieved by occluding key interactions between viral replication complexes and nuclear docking and/or import pathways. On the other hand, mutations in CA are sufficient to allow effective escape from MX2 and MX1(NMX2) inhibition (Fig. 5B) (12–14), perhaps arguing that the amino terminus of MX2 interacts with viral replication complexes in the vicinity of the NE. Since earlier work has shown that polymorphisms in CA can reroute HIV-1 replication complexes to different or alternative nuclear import pathways (17, 19), it is also plausible that escape occurs because such pathways are insensitive to interference by human MX2. Whichever of these (or other) models turns out to have merit, we also cannot exclude the possibility that NE localization is merely a surrogate for critical interactions that occur elsewhere in the cell, although we currently view the late postentry nature of MX-mediated inhibition as arguing against this possibility.
The analyses of viral reverse transcripts (Fig. 5A) shows that the levels of total cDNA are somewhat higher in the context of MX1(NMX2)-mediated inhibition, most clearly at 24 h postinfection. One explanation for this finding is that MX1(NMX2) affords HIV-1 replication complexes with a degree of stability (or delayed uncoating) that may help protect their associated cDNA from cellular nucleases, such as three prime repair exonuclease 1 (TREX1) or the structure-specific endonuclease (SSE) complex that can act to degrade them (55, 56). Such an effect could be manifested whether the amino terminus of MX2 interacts with viral complexes or nuclear import pathways, with both models being consistent with the notion that HIV-1 uncoating and nuclear import are linked processes.
In light of what is known about MX1 and the role of L4 in dictating the specificity of FluAV inhibition, it was unexpected to find that the amino-terminal 91 residues of MX2 also functions as a primary determinant of viral substrate specificity, particularly given the wide separation of these protein elements (Fig. 1) (29). Coupled with the observation that HIV-1 inhibition evidently does not require MX GTPase activity (Fig. 4; also see Fig. S3 in the supplemental material) (12, 13) in the same way that FluAV does, we conclude that there are fundamental differences between the modes of MX1/FluAV and MX2/HIV-1 inhibition. However, there may still be aspects of commonality, because, at least in the case of human MX1 protein function, there is strong evidence that the nuclear import of FluAV and THOV ribonucleoprotein complexes is inhibited, which, for these RNA viruses, leads to the suppression of primary viral transcription (57–59). Interestingly, such MX1-mediated inhibition of virus infection can be accomplished irrespective of whether the protein is distributed throughout the cytoplasm or is concentrated at the NE (Fig. 5C and 6A). In sum, much remains to be discovered regarding the subcellular positioning, intermolecular interactions, and molecular processes that underpin the MX-mediated inhibition of viral infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stéphane Hué, Georg Kochs, and Darja Pollpeter for the generous provision of reagents and for helpful discussions and John Harris from the King's College London Nikon Imaging Centre for advice with confocal microscopy and image acquisition.
This work was supported by the U.K. Medical Research Council, the Wellcome Trust (to W.S.B.), the National Institutes of Health (DA033773), the European Commission's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. PIEF-GA-2009-237501 (to C.G.), a Wellcome Trust Research Training Fellowship (to T.D.), and the Department of Health via a National Institute for Health Research comprehensive Biomedical Research Centre award to Guy's and St. Thomas' NHS Foundation Trust in partnership with King's College London and King's College Hospital NHS Foundation Trust.
Footnotes
Published ahead of print 4 Jun 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01269-14.
REFERENCES
- 1.Ivashkiv LB, Donlin LT. 2014. Regulation of type I interferon responses. Nat. Rev. Immunol. 14:36–49. 10.1038/nri3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47. 10.1099/vir.0.83391-0 [DOI] [PubMed] [Google Scholar]
- 3.Samuel CE. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778–809. 10.1128/CMR.14.4.778-809.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schoggins JW. 2014. Interferon-stimulated genes: roles in viral pathogenesis. Curr. Opin. Virol. 6:40–46. 10.1016/j.coviro.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho DD, Hartshorn KL, Rota TR, Andrews CA, Kaplan JC, Schooley RT, Hirsch MS. 1985. Recombinant human interferon alfa-A suppresses HTLV-III replication in vitro. Lancet i:602–604 [DOI] [PubMed] [Google Scholar]
- 6.Poli G, Orenstein JM, Kinter A, Folks TM, Fauci AS. 1989. Interferon-alpha but not AZT suppresses HIV expression in chronically infected cell lines. Science 244:575–577. 10.1126/science.2470148 [DOI] [PubMed] [Google Scholar]
- 7.Gendelman HE, Baca LM, Turpin J, Kalter DC, Hansen B, Orenstein JM, Dieffenbach CW, Friedman RM, Meltzer MS. 1990. Regulation of HIV replication in infected monocytes by IFN-alpha. Mechanisms for viral restriction. J. Immunol. 145:2669–2676 [PubMed] [Google Scholar]
- 8.Meylan PR, Guatelli JC, Munis JR, Richman DD, Kornbluth RS. 1993. Mechanisms for the inhibition of HIV replication by interferons-alpha, -beta, and -gamma in primary human macrophages. Virology 193:138–148. 10.1006/viro.1993.1110 [DOI] [PubMed] [Google Scholar]
- 9.Goujon C, Malim MH. 2010. Characterization of the alpha interferon-induced postentry block to HIV-1 infection in primary human macrophages and T cells. J. Virol. 84:9254–9266. 10.1128/JVI.00854-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheney KM, McKnight A. 2010. Interferon-alpha mediates restriction of human immunodeficiency virus type-1 replication in primary human macrophages at an early stage of replication. PLoS One 5:e13521. 10.1371/journal.pone.0013521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraietta JA, Mueller YM, Yang G, Boesteanu AC, Gracias DT, Do DH, Hope JL, Kathuria N, McGettigan SE, Lewis MG, Giavedoni LD, Jacobson JM, Katsikis PD. 2013. Type I interferon upregulates Bak and contributes to T cell loss during human immunodeficiency virus (HIV) infection. PLoS Pathog. 9:e1003658. 10.1371/journal.ppat.1003658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goujon C, Moncorgé O, Bauby H, Doyle T, Ward CC, Schaller T, Hué S, Barclay WS, Schulz R, Malim MH. 2013. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502:559–562. 10.1038/nature12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, Bieniasz PD. 2013. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502:563–566. 10.1038/nature12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, Cen S, Guo F, Liang C. 2013. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14:398–410. 10.1016/j.chom.2013.08.015 [DOI] [PubMed] [Google Scholar]
- 15.Braaten D, Aberham C, Franke EK, Yin L, Phares W, Luban J. 1996. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J. Virol. 70:5170–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koh Y, Wu X, Ferris AL, Matreyek KA, Smith SJ, Lee K, KewalRamani VN, Hughes SH, Engelman A. 2013. Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. J. Virol. 87:648–658. 10.1128/JVI.01148-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee K, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, Vandegraaff N, Baumann JG, Wang R, Yuen W, Takemura T, Shelton K, Taniuchi I, Li Y, Sodroski J, Littman DR, Coffin JM, Hughes SH, Unutmaz D, Engelman A, KewalRamani VN. 2010. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7:221–233. 10.1016/j.chom.2010.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matreyek KA, Yucel SS, Li X, Engelman A. 2013. Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog. 9:e1003693. 10.1371/journal.ppat.1003693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hue S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. 2011. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 7:e1002439. 10.1371/journal.ppat.1002439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah VB, Shi J, Hout DR, Oztop I, Krishnan L, Ahn J, Shotwell MS, Engelman A, Aiken C. 2013. The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. J. Virol. 87:422–432. 10.1128/JVI.07177-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. 10.1038/nature02343 [DOI] [PubMed] [Google Scholar]
- 22.Matreyek KA, Engelman A. 2013. Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes. Viruses 5:2483–2511. 10.3390/v5102483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haller O, Kochs G. 2011. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J. Interferon Cytokine Res. 31:79–87. 10.1089/jir.2010.0076 [DOI] [PubMed] [Google Scholar]
- 24.Kim BH, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. 2012. IFN-inducible GTPases in host cell defense. Cell Host Microbe 12:432–444. 10.1016/j.chom.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frese M, Kochs G, Feldmann H, Hertkorn C, Haller O. 1996. Inhibition of bunyaviruses, phleboviruses, and hantaviruses by human MxA protein. J. Virol. 70:915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordien E, Rosmorduc O, Peltekian C, Garreau F, Brechot C, Kremsdorf D. 2001. Inhibition of hepatitis B virus replication by the interferon-inducible MxA protein. J. Virol. 75:2684–2691. 10.1128/JVI.75.6.2684-2691.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kochs G, Janzen C, Hohenberg H, Haller O. 2002. Antivirally active MxA protein sequesters La Crosse virus nucleocapsid protein into perinuclear complexes. Proc. Natl. Acad. Sci. U. S. A. 99:3153–3158. 10.1073/pnas.052430399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavlovic J, Zürcher T, Haller O, Staeheli P. 1990. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol. 64:3370–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao S, von der Malsburg A, Dick A, Faelber K, Schroder GF, Haller O, Kochs G, Daumke O. 2011. Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity 35:514–525. 10.1016/j.immuni.2011.07.012 [DOI] [PubMed] [Google Scholar]
- 30.Pitossi F, Blank A, Schroder A, Schwarz A, Hussi P, Schwemmle M, Pavlovic J, Staeheli P. 1993. A functional GTP-binding motif is necessary for antiviral activity of Mx proteins. J. Virol. 67:6726–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponten A, Sick C, Weeber M, Haller O, Kochs G. 1997. Dominant-negative mutants of human MxA protein: domains in the carboxy-terminal moiety are important for oligomerization and antiviral activity. J. Virol. 71:2591–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kochs G, Haener M, Aebi U, Haller O. 2002. Self-assembly of human MxA GTPase into highly ordered dynamin-like oligomers. J. Biol. Chem. 277:14172–14176. 10.1074/jbc.M200244200 [DOI] [PubMed] [Google Scholar]
- 33.von der Malsburg A, Abutbul-Ionita I, Haller O, Kochs G, Danino D. 2011. Stalk domain of the dynamin-like MxA GTPase protein mediates membrane binding and liposome tubulation via the unstructured L4 loop. J. Biol. Chem. 286:37858–37865. 10.1074/jbc.M111.249037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mänz B, Dornfeld D, Götz V, Zell R, Zimmerman P, Haller O, Kochs G, Schwemmle M. 2013. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog. 9:e1003279. 10.1371/journal.ppat.1003279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell PS, Patzina C, Emerman M, Haller O, Malik HS, Kochs G. 2012. Evolution-guided identification of antiviral specificity determinants in the broadly acting interferon-induced innate immunity factor MxA. Cell Host Microbe 12:598–604. 10.1016/j.chom.2012.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmermann P, Mänz B, Haller O, Schwemmle M, Kochs G. 2011. The viral nucleoprotein determines Mx sensitivity of influenza A viruses. J. Virol. 85:8133–8140. 10.1128/JVI.00712-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turan K, Mibayashi M, Sugiyama K, Saito S, Numajiri A, Nagata K. 2004. Nuclear MxA proteins form a complex with influenza virus NP and inhibit the transcription of the engineered influenza virus genome. Nucleic Acids Res. 32:643–652. 10.1093/nar/gkh192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kochs G, Haller O. 1999. GTP-bound human MxA protein interacts with the nucleocapsids of Thogoto virus (Orthomyxoviridae). J. Biol. Chem. 274:4370–4376. 10.1074/jbc.274.7.4370 [DOI] [PubMed] [Google Scholar]
- 39.Patzina C, Haller O, Kochs G. 2014. Structural requirements for the antiviral activity of the human MxA protein against Thogoto and influenza A virus. J. Biol. Chem. 289:6020–6027. 10.1074/jbc.M113.543892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267. 10.1126/science.272.5259.263 [DOI] [PubMed] [Google Scholar]
- 41.Saenz DT, Teo W, Olsen JC, Poeschla EM. 2005. Restriction of feline immunodeficiency virus by Ref1, Lv1, and primate TRIM5alpha proteins. J. Virol. 79:15175–15188. 10.1128/JVI.79.24.15175-15188.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Rourke JP, Newbound GC, Kohn DB, Olsen JC, Bunnell BA. 2002. Comparison of gene transfer efficiencies and gene expression levels achieved with equine infectious anemia virus- and human immunodeficiency virus type 1-derived lentivirus vectors. J. Virol. 76:1510–1515. 10.1128/JVI.76.3.1510-1515.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarrosson-Wuilleme L, Goujon C, Bernaud J, Rigal D, Darlix JL, Cimarelli A. 2006. Transduction of nondividing human macrophages with gammaretrovirus-derived vectors. J. Virol. 80:1152–1159. 10.1128/JVI.80.3.1152-1159.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon JH, Southerling TE, Peterson JC, Meyer BE, Malim MH. 1995. Complementation of vif-defective human immunodeficiency virus type 1 by primate, but not nonprimate, lentivirus vif genes. J. Virol. 69:4166–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cordeil S, Nguyen XN, Berger G, Durand S, Ainouze M, Cimarelli A. 2013. Evidence for a different susceptibility of primate lentiviruses to type I interferons. J. Virol. 87:2587–2596. 10.1128/JVI.02553-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mangeot PE, Dollet S, Girard M, Ciancia C, Joly S, Peschanski M, Lotteau V. 2011. Protein transfer into human cells by VSV-G-induced nanovesicles. Mol. Ther. 19:1656–1666. 10.1038/mt.2011.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cauldwell AV, Moncorge O, Barclay WS. 2013. Unstable polymerase-nucleoprotein interaction is not responsible for avian influenza virus polymerase restriction in human cells. J. Virol. 87:1278–1284. 10.1128/JVI.02597-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melén K, Keskinen P, Ronni T, Sareneva T, Lounatmaa K, Julkunen I. 1996. Human MxB protein, an interferon-alpha-inducible GTPase, contains a nuclear targeting signal and is localized in the heterochromatin region beneath the nuclear envelope. J. Biol. Chem. 271:23478–23486. 10.1074/jbc.271.38.23478 [DOI] [PubMed] [Google Scholar]
- 49.Aebi M, Fah J, Hurt N, Samuel CE, Thomis D, Bazzigher L, Pavlovic J, Haller O, Staeheli P. 1989. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol. Cell. Biol. 9:5062–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.King MC, Raposo G, Lemmon MA. 2004. Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proc. Natl. Acad. Sci. U. S. A. 101:8957–8962. 10.1073/pnas.0403167101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zürcher T, Pavlovic J, Staeheli P. 1992. Mechanism of human MxA protein action: variants with changed antiviral properties. EMBO J. 11:1657–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dreiding P, Staeheli P, Haller O. 1985. Interferon-induced protein Mx accumulates in nuclei of mouse cells expressing resistance to influenza viruses. Virology 140:192–196. 10.1016/0042-6822(85)90460-X [DOI] [PubMed] [Google Scholar]
- 53.Engelhardt OG, Ullrich E, Kochs G, Haller O. 2001. Interferon-induced antiviral Mx1 GTPase is associated with components of the SUMO-1 system and promyelocytic leukemia protein nuclear bodies. Exp. Cell Res. 271:286–295. 10.1006/excr.2001.5380 [DOI] [PubMed] [Google Scholar]
- 54.Zürcher T, Pavlovic J, Staeheli P. 1992. Mouse Mx2 protein inhibits vesicular stomatitis virus but not influenza virus. Virology 187:796–800. 10.1016/0042-6822(92)90481-4 [DOI] [PubMed] [Google Scholar]
- 55.Laguette N, Bregnard C, Hue P, Basbous J, Yatim A, Larroque M, Kirchhoff F, Constantinou A, Sobhian B, Benkirane M. 2014. Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell 156:134–145. 10.1016/j.cell.2013.12.011 [DOI] [PubMed] [Google Scholar]
- 56.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. 2010. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 11:1005–1013. 10.1038/ni.1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao H, Killip MJ, Staeheli P, Randall RE, Jackson D. 2013. The human interferon-induced MxA protein inhibits early stages of influenza A virus infection by retaining the incoming viral genome in the cytoplasm. J. Virol. 87:13053–13058. 10.1128/JVI.02220-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kochs G, Haller O. 1999. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc. Natl. Acad. Sci. U. S. A. 96:2082–2086. 10.1073/pnas.96.5.2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matzinger SR, Carroll TD, Dutra JC, Ma ZM, Miller CJ. 2013. Myxovirus resistance gene A (MxA) expression suppresses influenza A virus replication in alpha interferon-treated primate cells. J. Virol. 87:1150–1158. 10.1128/JVI.02271-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.