ABSTRACT
Approximately 8% of the human genome is made up of endogenous retroviral sequences. As the HIV-1 Tat protein activates the overall expression of the human endogenous retrovirus type K (HERV-K) (HML-2), we used next-generation sequencing to determine which of the 91 currently annotated HERV-K (HML-2) proviruses are regulated by Tat. Transcriptome sequencing of total RNA isolated from Tat- and vehicle-treated peripheral blood lymphocytes from a healthy donor showed that Tat significantly activates expression of 26 unique HERV-K (HML-2) proviruses, silences 12, and does not significantly alter the expression of the remaining proviruses. Quantitative reverse transcription-PCR validation of the sequencing data was performed on Tat-treated PBLs of seven donors using provirus-specific primers and corroborated the results with a substantial degree of quantitative similarity.
IMPORTANCE The expression of HERV-K (HML-2) is tightly regulated but becomes markedly increased following infection with HIV-1, in part due to the HIV-1 Tat protein. The findings reported here demonstrate the complexity of the genome-wide regulation of HERV-K (HML-2) expression by Tat. This work also demonstrates that although HERV-K (HML-2) proviruses in the human genome are highly similar in terms of DNA sequence, modulation of the expression of specific proviruses in a given biological situation can be ascertained using next-generation sequencing and bioinformatics analysis.
INTRODUCTION
Human endogenous retroviruses (HERVs), which make up 8% of the human genome, are the result of the rare and successful individual infections of germ line cells that have occurred over millions of years of human evolution (1–5). In the genome, the proviruses are made up of the basic retroviral genes (gag, prt, pol, and env) flanked by two long terminal repeats (LTRs). Due to host selection events, however, most of these proviral sequences appear to be nonfunctional (3, 4, 6). Interestingly, a limited number are still active at both the transcriptional and translational levels, with HERV-K (HML-2) being the prime example [HERV-K (HML-2) is a subgroup of HERV-K] (4, 7–9). This group of retroviruses is one of the most recent entrants into the human genome, with some proviruses having integrated either by reinfection or by intracellular transposition within the last 200,000 years, and they are the only HERVs with conserved and potentially functional open reading frames (ORFs) for all viral proteins (6, 9–12). Thousands of HERV-K (HML-2) solo LTRs are found in the human genome, along with approximately 91 proviruses (4, 9), some of which may encode the basic retroviral proteins and the accessory, putative oncogenes np9 and rec (4, 13–16). The accessory genes np9 and rec are expressed by the two types of HERV-K (HML-2), type 1 and type 2 (9, 17), respectively, which differ by only a 292-bp deletion in the env gene of type 1 viruses. This deletion causes a new splice site that results in the synthesis of Np9 in type 1 viruses.
The basal transcriptional activity of human endogenous retroviruses varies in different human cell and tissue types, both for those viruses that have intact genes and for those with mutated cistrons (18–23). It appears that expression depends upon individual host factors, like ethnicity (10, 21, 22, 24), cell populations (18, 25, 26), and tissue types (27–29). Additionally, external stimuli, such as infections, viral transactivators, chemical exposures, cytokines, hormones, and inflammation, can induce expression of these proviruses (30–46). We and others have shown that HIV-1 infection is associated with increased HERV-K (HML-2) expression in vitro and in vivo (33, 45–53). Compared to healthy individuals, HIV-1-infected individuals show high levels of HERV-K (HML-2) RNA, DNA, protein, and viral particles in their plasma (45, 47, 48, 50). This HIV-1-mediated activation of HERV-K (HML-2) is partially due to the HIV-1 Tat protein activating the NF-κB and NF-AT transcription factors, which then interact with the HERV-K (HML-2) LTR promoter to stimulate transcription (46).
As expression of HERV-K (HML-2) transcripts and proteins has been associated with several cancers as well as autoimmune, inflammatory, and neurological conditions, the possibility of it participating in the development of HIV-1-associated disease or malignancies cannot be ruled out (54–61). In fact, Np9 and Rec have been shown to be highly expressed in transformed tissues (Np9), to cause carcinoma in situ in mice (Rec), and to be associated with the Notch pathway and pathways affecting c-myc expression (Np9 and both, respectively) (13, 15, 16, 62–65). Our laboratory has noted high expression levels of both proteins during HIV-1-associated lymphoma (unpublished observations). However, although all HERV-K (HML-2) type 1 and type 2 proviruses have the potential to be expressed in both health and disease, it is currently not known which are transcribed or if expression of only certain proviruses predominates under particular circumstances. Such information ultimately could help us assess whether these proviruses are important to disease development.
The main purpose of this study was to implement RNA sequencing (RNA-Seq) coupled with quantitative reverse transcription-PCR (qRT-PCR) validation to analyze the HERV-K (HML-2) transcriptome, as seen in peripheral blood lymphocytes (PBLs) responding to the HIV-1 Tat protein. Our additional goal was to provide insight into HIV-1 pathogenesis, i.e., the modulation of endogenous retroviruses that could affect host cellular function and/or immunity. We show which of the 91 annotated HERV-K (HML-2) proviruses are modulated by treatment with Tat, whether the two types of HERV-K (HML-2) proviruses (type 1 and type 2) are differentially affected, their ORF status, and whether the viruses targeted by Tat are the same as those expressed in patients during HIV-1 infection. We discovered that while many HERV-K (HML-2) proviruses are activated by Tat, expression of multiple others is repressed. In addition, the mechanisms by which Tat regulates HERV-K (HML-2) expression as detected in this genome-wide screen are surprisingly complex. Although beyond the scope of the analyses presented here, we hypothesize that the endogenous retroviral gene modulation induced by Tat during HIV-1 infection influences the development of HIV-1-associated disease, as these HERV sequences are associated with many pathological conditions, as noted above. These results also demonstrate that, in spite of the marked sequence similarity between the individual HERV-K (HML-2) proviruses, deep sequencing technologies enable the detection of transcription (or lack thereof) of any specific endogenous proviruses that might have a role in health or disease.
MATERIALS AND METHODS
Isolation and culture of primary cells.
Peripheral blood mononuclear cells (PBMCs) were obtained by venipuncture from healthy donors, and monocytes were separated from peripheral blood lymphocytes (PBLs) by differential adhesion to plates as previously described (66). PBLs were washed three times with phosphate-buffered saline (PBS) and stimulated with 5 μg/ml phytohemagglutinin (PHA-P; Sigma-Aldrich, St. Louis, MO) for 3 days in RPMI 1640 complete media containing 10% heat-inactivated fetal bovine serum (FBS) and 10 U/ml of interleukin-2 (IL-2; Sigma-Aldrich, St. Louis, MO).
Addition of exogenous Tat protein.
The purified recombinant 86-amino-acid form of the HIV-1 Tat protein was obtained from the NIH AIDS Research and Reference Reagent Program (the late John Brady and DAIDS, NIAID [67]) or from ProSpec Protein Specialists (catalog no. HIV-129; ProSpec-Tany TechnoGene Ltd., East Brunswick, NJ). The protein was resuspended in sterile PBS (Gibco/Invitrogen, Carlsbad, CA) containing 1 mg/ml bovine serum albumin (BSA) and 0.1 mM dithiothreitol (DTT) (both from Sigma-Aldrich, St. Louis, MO), deaerated, and protected from light. Tat protein was added at a concentration of 50 ng/ml for 12 h.
RNA extraction.
Total cellular RNA was isolated from cells using the RNeasy minikit (Qiagen, Valencia, CA) and subjected to on-column RNase-free DNase treatment (Qiagen, Valencia, CA) for 15 min at room temperature. After elution, another round of DNase treatment was performed using the DNA-free DNase removal kit (Ambion, Austin, TX) and by following the manufacturer's protocol. RNA concentration and purity were measured using a spectrophotometer, calculating the 260-nm/280-nm ratio. RNA integrity (as well as the absence of DNA contamination) was confirmed by one-step reverse transcription-PCR (RT-PCR) using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) amplification with primers that can bind both genomic DNA and cDNA, employing the PCR conditions described below, as well as no-RT controls. If DNA contamination was detected, another round of DNase treatment was performed.
cDNA library preparation and RNA-Seq.
Total RNA was isolated from PBLs of a healthy Caucasian female volunteer that had been treated with Tat for 12 h or with vehicle only, purified, and DNase treated. RNA integrity was verified using the Agilent RNA 6000 Nano kit (Agilent, Santa Clara, CA), and a NanoDrop was used for measuring quantity (Thermo Scientific, Rockford, IL). cDNA was prepared for the RNA-Seq library using the NuGen Encore complete library preparation kit (Invitrogen, Carlsbad, CA), and quality was controlled using a Bioanalyzer DNA 1000 (Agilent, Santa Clara, CA). Quantitative PCR (qPCR) for quantification of the library was performed with the KAPA kit (Kapa Biosystems, Woburn, MA). Samples were sequenced on a HiSeq2000 (Illumina, Madison, WI), one sample per lane, in a paired-end 100-cycle run, using TruSeq SBS v3 reagents (Illumina, Madison, WI).
In silico sequence analysis and differential expression determination: RNA-Seq.
An average of 88 million paired-end reads per sample passed the Casava 1.8 filters. A subset of the RNA-Seq data in FASTQ format representing the sequence and quality scores for all reads that map to all known HERV-K (HML-2) genomes is available at the Markovitz laboratory website (http://markovitzlab.com/?page_id=507). The data set can be treated as plain text files and opened with any word-processing software. All hits were deduplicated (Samtools rmdup) to remove PCR artifacts, yielding an average of 62 million paired-end reads per sample. Tophat v2.0.4 (Bowtie2) (68, 69) was used to map the reads to the human genome (hg19), and Cufflinks and Cuffdiff v2.0.2 (69) were used to determine transcripts and isoforms, as well as for differential expression analysis. The genomic locations of the 91 currently known HERV-K (HML-2) genomes were organized in a BED-formatted file, and BEDTools (70) was used to intersect the HERV-K (HML-2) locations with the aligned cDNA reads. Results were analyzed as single reads. A customized Perl script was written to count the reads, which were aligned to those regions. Tophat software output, in SAM/BAM format, provided a mapping quality (MQ) field reported as bins for unique mapping and multimapping. For each read, the MQ score reflects a bin into which the read could map: 0, maps to more than 10 locations; 1, 4 to 9 locations; 2, maps to 3 locations; 3, maps to 2 locations; 255, unique mapping. We counted total reads for each HERV-K (HML-2) proviral locus (all reads, regardless of MQ), and we counted unique reads (MQ = 255). Read counts were converted to reads per kilobase of transcript per million mapped reads (RPKM) by normalizing for the potential length of the provirus (in kilobases) at specific loci and normalizing the read counts to 1 million reads for each treatment (see Table 2 for these values). Quartiles and percentiles then were calculated for all RPKM values from Tat-treated cells, including those differentially expressed and those not tested or for which no reads were found (as truer measurements of overall gene expression or repression). An RPKM of 0.02 (our cutoff determinant) means we obtained about 0.02 reads aligned across the whole of a 1,000-bp length (reads per kilobase of transcript). Since there were approximately 120 million reads per sample, we have an average of approximately 0.4× coverage of the unique segments of the proviral transcripts. As a separate analysis, without regard to unique mapping status, we assessed what proportion of the total reads per sample mapped to a composite of all HERV-K proviral sequences. Approximately 1.6% of the total reads aligned to these endogenous retroviruses. For differential expression determination, P values were calculated from a proportions test (using R software's function prop.test) based on both the total read counts and the unique read counts for each provirus. To calculate conservative estimates, the maximum P value between those two tests was corrected for multiple testing using the false-discovery-rate (FDR) method, which then was used to calculate a Q value. Significance then was determined to be a Q value of 0.05 or less.
TABLE 2.
All known HERV-K (HML-2) proviruses and their RPKM values as detected in PBLs after HIV-1 Tat or vehicle-alone treatment using RNA-Seqa
| Chromosome | Length (kb) | Locus | Name and alternative name(s) | Unique RPKM value by treatment |
|
|---|---|---|---|---|---|
| Tat | Vehicle | ||||
| 1 | 6,104 | 1p36.21a | 1p36.21a | 0.0000 | 0.0000 |
| 1 | 9,521 | 1p36.21b | K(OLDAL023753), K6, K76 | 0.0000 | 0.0000 |
| 1 | 9,392 | 1p36.21c | K6, K76 | 0.0000 | 0.0000 |
| 1 | 1,238 | 1p34.3 | 1p34.3 | 0.0000 | 0.0000 |
| 1 | 6,372 | 1p31.1 | K4, K116, ERVK-1 | 0.2416 | 0.1461 |
| 1 | 3,077 | 1q21.3 | 1q21.3 | 3.0075 | 0.8425 |
| 1 | 9,179 | 1q22 | K102, K(C1b), K50a, ERVK-7 | 0.7907 | 0.2809 |
| 1 | 9,231 | 1q23.3 | K110, K18, K(C1a), ERVK-18 | 0.4454 | 0.2110 |
| 1 | 5,655 | 1q24.1 | K12 | 0.0060 | 0.0000 |
| 1 | 4,179 | 1q32.2 | 1q32.2 | 0.0000 | 0.0514 |
| 1 | 2,178 | 1q43 | 1q43 | 0.0000 | 0.0000 |
| 2 | 2,671 | 2q21.1 | 2q21.1 | 0.0000 | 0.0590 |
| 3 | 6,890 | 3p25.3 | K11, ERVK-2 | 0.0123 | 0.0270 |
| 3 | 8,685 | 3p12.3 | 3p12.3 | 0.1286 | 0.0016 |
| 3 | 9,122 | 3q12.3 | K(II), ERVK-5 | 1.8324 | 0.7050 |
| 3 | 8,803 | 3q13.2 | K106, K(C3), K68, ERVK-3 | 0.0058 | 0.0049 |
| 3 | 9,114 | 3q21.2 | K(I), ERVK-4 | 0.0835 | 0.0566 |
| 3 | 3,919 | 3q24 | ERVK-13 | 0.0000 | 0.0000 |
| 3 | 9,179 | 3q27.2 | K50b, K117, ERVK-11 | 0.0424 | 0.0328 |
| 4 | 4,470 | 4p16.3a | 4p16.3a | 0.0114 | 0.0384 |
| 4 | 8,562 | 4p16.3b | K77 | 0.0553 | 0.0736 |
| 4 | 9,560 | 4p16.1a | K17b | 0.0637 | 0.0839 |
| 4 | 9,062 | 4p16.1b | K50c | 0.5227 | 0.0980 |
| 4 | 5,514 | 4q13.2 | 4q13.2 | 0.0000 | 0.0000 |
| 4 | 2,422 | 4q32.1 | 4q32.1 | 0.1746 | 0.0000 |
| 4 | 7,228 | 4q32.3 | K5, ERVK-12 | 0.0000 | 0.0119 |
| 4 | 7,287 | 4q35.2 | 4q35.2 | 0.0720 | 0.0609 |
| 5 | 9,091 | 5p13.3 | K104, K50d | 0.0037 | 0.0016 |
| 5 | 9,843 | 5p12 | K8 | 0.0017 | 0.0422 |
| 5 | 7,712 | 5q33.2 | K18b | 0.0088 | 0.0316 |
| 5 | 9,179 | 5q33.3 | K107/K10, K(C5), ERVK-10 | 0.0608 | 0.0312 |
| 6 | 10,368 | 6p22.1 | K(OLDAL121932), K69, K20 | 0.0000 | 0.0000 |
| 6 | 9,958 | 6p21.1 | K(OLDAL035587), KOLD35587 | 0.0374 | 0.0302 |
| 6 | 4,808 | 6p11.2 | K23 | 0.0035 | 0.0923 |
| 6 | 9,064 | 6q14.1 | K109, K(C6), ERVK-9 | 0.0000 | 0.0032 |
| 6 | 2,825 | 6q25.1 | 6q25.1 | 0.0180 | 0.0862 |
| 7 | 9,471 | 7p22.1a | K108L, K(HML.2-HOM), K(C7), ERVK-6 | 0.0000 | 0.0030 |
| 7 | 9,470 | 7p22.1b | K108R, ERVK-6, HERV-K (HML-2).HOM | 0.0518 | 0.0212 |
| 7 | 2,695 | 7q11.21 | 7q11.21 | 0.0000 | 0.0000 |
| 7 | 4,897 | 7q22.2 | ERVK-14 | 0.1244 | 0.0000 |
| 7 | 4,977 | 7q34 | K(OLDAC004979), ERVK-15 | 1.7302 | 0.3914 |
| 8 | 9,462 | 8p23.1a | K115, ERVK-8 | 0.1037 | 0.0378 |
| 8 | 1,025 | 8p23.1b | K27 | 0.0000 | 0.0000 |
| 8 | 9,527 | 8p23.1c | 8p23.1c | 0.0586 | 0.0060 |
| 8 | 9,515 | 8p23.1d | KOLD130352 | 0.0658 | 0.0030 |
| 8 | 8,738 | 8p22 | 8p22 | 0.0174 | 0.0344 |
| 8 | 8,011 | 8q11.1 | K70, K43 | 0.0000 | 0.0536 |
| 8 | 3,087 | 8q24.3a | 8q24.3a | 0.0000 | 0.0000 |
| 8 | 7,563 | 8q24.3b | K29 | 0.1633 | 0.0170 |
| 9 | 7,221 | 9q34.11 | K31 | 0.3889 | 0.1686 |
| 9 | 9,462 | 9q34.3 | K30 | 0.0054 | 0.0212 |
| 10 | 7,526 | 10p14 | K(C11a), K33, ERVK-16 | 0.0090 | 0.0837 |
| 10 | 981 | 10p12.1 | K103, K(C10) | 0.0172 | 0.0000 |
| 10 | 7,147 | 10q24.2 | ERVK-17, c10_B | 0.0071 | 0.0040 |
| 11 | 9,553 | 11p15.4 | K7 | 0.3507 | 0.0825 |
| 11 | 5,748 | 11q12.1 | 11q12.1 | 0.0000 | 0.0050 |
| 11 | 14,600 | 11q12.3 | K(OLDAC004127) | 0.1506 | 0.0275 |
| 11 | 9,465 | 11q22.1 | K(C11c), K36, K118, ERVK-25 | 0.0018 | 0.0076 |
| 11 | 9,159 | 11q23.3 | K(C11b), K37, ERVK-20 | 0.0092 | 0.0407 |
| 12 | 9,662 | 12p11.1 | K50e | 0.0000 | 0.0163 |
| 12 | 968 | 12q13.2 | 12q13.2 | 0.0000 | 0.0000 |
| 12 | 9,456 | 12q14.1 | K(C12), K41, K119, ERVK-21 | 0.0000 | 0.0121 |
| 12 | 1,482 | 12q24.11 | 12q24.11 | 0.0114 | 0.0000 |
| 12 | 6,012 | 12q24.33 | K42 | 0.0338 | 0.0048 |
| 14 | 3,496 | 14q11.2 | K(OLDAL136419), K71 | 0.5904 | 0.2048 |
| 14 | 2,881 | 14q32.33 | 14q32.33 | 0.0000 | 0.0000 |
| 15 | 3,344 | 15q25.2 | 15q25.2 | 0.0000 | 0.0000 |
| 16 | 1,501 | 16p13.3 | K(OLDAC004034) | 0.3156 | 0.0286 |
| 16 | 2,668 | 16p11.2 | 16p11.2 | 0.0254 | 0.0000 |
| 17 | 6,862 | 17p13.1 | 17p13.1 | 0.0099 | 0.0605 |
| 19 | 2,542 | 19p13.3 | ERVK-22 | 0.0000 | 0.0056 |
| 19 | 10,112 | 19p12a | K52 | 0.0033 | 0.0269 |
| 19 | 6 | 19p12b | K113 (potential insertion site, 6 bp long) | 0.0000 | 0.0000 |
| 19 | 6,737 | 19p12c | K51 | 0.0075 | 0.0000 |
| 19 | 8,863 | 19q11 | K(C19), ERVK-19 | 0.0630 | 0.0259 |
| 19 | 4,227 | 19q13.12a | 19q13.12a | 0.1201 | 0.0000 |
| 19 | 9,517 | 19q13.12b | K50F | 0.9475 | 0.5252 |
| 19 | 4,317 | 19q13.41 | 19q13.41 | 0.1254 | 0.0597 |
| 19 | 5,696 | 19q13.42 | LTR13 | 0.0149 | 0.0503 |
| 20 | 9,634 | 20q11.22 | K(OLDAL136419), K59 | 0.0386 | 0.0253 |
| 21 | 8,046 | 21q21.1 | K60, ERVK-23 | 0.0000 | 0.0285 |
| 22 | 9,120 | 22q11.21 | K101, K(C22), ERVK-24 | 0.0519 | 0.0031 |
| 22 | 8,880 | 22q11.23 | K(OLDAP000345), KOLD345 | 0.0038 | 0.0500 |
| X | 2,505 | Xq11.1 | Xq11.1 | 0.0135 | 0.1029 |
| X | 2,052 | Xq12 | Xq12 | 0.0000 | 0.0000 |
| X | 2,399 | Xq28a | K63 | 0.0000 | 0.0000 |
| X | 7,340 | Xq28b | K63 | 0.0000 | 0.0020 |
| Y | 6,943 | Yp11.2 | Yp11.2 | 0.0000 | 0.0000 |
| Y | 3,198 | Yq11.23a | Yq11.23a | 0.0000 | 0.0000 |
| Y | 3,199 | Yq11.23b | Yq11.23b | 0.0000 | 0.0000 |
The different names shown represent alternative ways to refer to the same virus based on the literature. Tat or vehicle unique RPKM values are the reads that had a single unique genomic alignment, mapping to a particular provirus, not shared with any other known provirus.
Heat map generation.
Log2 RPKM values of uniquely mapped reads were calculated and clustered using average linkage hierarchical clustering with the Euclidean distance matrix. Proviruses with zero reads in both conditions were removed before clustering.
qRT-PCR.
To validate the RNA-Seq data, we performed two-step quantitative RT-PCR (qRT-PCR) on cDNA synthesized from RNA isolated from Tat-treated PBLs from healthy volunteers using the Bio-Rad Supermix iScript SYBR green kit (Bio-Rad, Hercules, CA). Briefly, 500 ng of total cellular RNA was reverse transcribed for 30 min at 50°C using the iScript reverse transcription supermix for qRT-PCR (Bio-Rad, Hercules, CA) and diluted to 100 μl. This kit contains a mix of oligo(dT) primers and random primers for cDNA synthesis. A 20-μl qPCR was made using 5 μl of cDNA and 0.6 μM specific primers designed and verified by BLAST analysis to amplify 9 specific HERV-K (HML-2) proviruses. The efficiency of these primers was verified by comparing the linear slopes of their standard curve after amplification using reference DNA to the linear slope of a standard curve generated by amplification of the reference gene used for normalization, GAPDH. The slopes were comparable, reaching values of −3.3 ± 0.12, with primer efficiencies close to 99 to 100%. The PCR for amplification of specific proviruses in the samples consisted of an initial denaturing step of 15 min at 95°C, followed by 40 cycles with the following optimal conditions: 94°C for 15 s, 57°C for 30 s, and 72°C for 10 s. This was followed by a melting curve with fluorescence captured at 78°C or 81°C, which determined that the product amplified did not have any signal generated by primer dimers. Data were collected and recorded by an ABI StepOne plus (Applied Biosystems). GAPDH amplification was used to normalize samples to an endogenous reference gene using the 2−ΔΔCT method, as stated in the figure legends.
List of primers used.
The amplification product was designed to contain one primer or both primers binding to each provirus's unique regions. Sequences were verified through BLAST analysis so that any other possible proviruses or cellular genes that could be amplified with the primer pairs had 75% or less sequence identity. Sequences can be found in Table 1.
TABLE 1.
Primer sequences for specific HERV-K (HML-2) proviruses
| Primer | Sequence |
|
|---|---|---|
| Forward | Reverse | |
| 7q34 KOLD/ERVK-15 | CCAACCCTGTGCTCGTAGAAACAA | GCACATCCTACATAGCCCTAAATCC |
| 8q11.1 K70/K43 | GTGAAGAAGAGGCAGGAAGAGAG | TTGCTTTGACTGAGCCACTACGGA |
| 5p12 | GGTCGAGCTCTTCAACCAGTGAGTTT | CAGATGCTATTGCCAGTCCTGCAT |
| 6p11.2 K23 | GGTATGGTATGGAATGATTGGGCCA | GGGCATCAGACACTGAAACACT |
| 3p12.3 | CTGTTATAACTGTGGTCAAATCGGTC | AGGCCAGGTGCCTCCTTT |
| 4q32.1 | GCTCGGAAGAAGCTAGGGTGATAA | TGGTTCTTCTGTTTGAAATGGCTTG |
| 11q12.3 (KOLD) | ACAGATGATCGTTGCCCTGCCAAA | AACATCCTGGCGCTAAACATCCTG |
| 16p13.3 (KOLD) | CTCTAAAGAGCCCTACCCTGACTT | TGGCCAATTGACATTCCG |
Analysis of the LTRs of HERV-K (HML-2) proviruses.
Analysis of the HERV-K (HML-2) long terminal repeats (LTRs) for mutations and deletions was performed using published sequences (9, 45). Potential transcription factor binding sites were analyzed using the online prediction software tools ALGGEN PROMO and version 8.3 of TRANSFAC (BioBase Co., Beverly, MA) (71, 72).
Statistical analysis.
The mean numbers of HERV-K (HML-2) cDNA relative fold expression units between Tat and control treatments were compared using a Student's t test for samples exhibiting normally distributed values. Two-tailed P values were considered significant at P < 0.05.
RESULTS
HIV-1 Tat both activates and silences specific HERV-K (HML-2) proviruses in normal PBLs.
In order to examine the HERV-K (HML-2) transcriptome in response to HIV-1 Tat, normal PBLs isolated from the blood of a healthy volunteer were cultured for 3 days under PHA and IL-2 stimulation. No specific cell selection methods were performed, and total PBLs were subjected to treatment with physiologically relevant levels of recombinant Tat protein or vehicle buffer alone for 12 h. The amounts of Tat used have been shown to activate the HIV-1 LTR promoter (46). Paired-end RNA sequencing was performed, with over 100 million high-quality reads obtained per sample, and results were aligned to the human genome and transcriptome (hg19). As a separate analysis, we assessed what proportion of the total reads per sample mapped to a composite of all HERV-K (HML-2) proviral sequences and found that approximately 1.6% of the total reads aligned with these endogenous retroviruses. This means that at least 1.6% of the expressed transcripts are of endogenous retrovirus origin. We next examined the reads aligned with the human genome and transcriptome for expression of each of the 91 known HERV-K (HML-2) proviruses. In view of the high degree of similarity between proviruses and multiple potential locations for mapping, we identified unique portions of each provirus (see Data S1 in the supplemental material) and used Tophat to map them to their unique genomic location (score of 255). Transcript levels were determined by the calculation of RPKM (73), since the sensitivity of RNA-Seq is a function of both sequencing depth and transcript length. This provides us a transparent comparison of transcript levels both within and between treatments. Reads mapping uniquely to transcripts were counted. Resulting RPKM values are reported in Table 2 as unique RPKM, the resulting reads that had a single unique genomic alignment and that do not match any provirus other than the one stated. Table 2 denotes all HERV-K (HML-2) proviruses (with any alternative names), their lengths, and chromosomal locations as detected (or not) in each treatment. As can be seen, most HERV-K (HML-2) proviruses are already expressed at what we deemed a basal level (vehicle-treatment columns). Treatment with HIV-1 Tat protein caused modifications in these expression levels, yet RPKM values were never higher than approximately 3. A comparison was made between expression levels of HERV-K (HML-2) proviruses and other genes in order to define a threshold value above which we can have the highest confidence in the validity of expression. We compiled RPKM values for several genes, ranging from almost undetectable (e.g., CD27) to over 17,000 (e.g., ribosomal protein S12). A summary of these results are shown in Table 3. We then designated an RPKM cutoff value of ≥0.02, after determining the false-discovery rate, as the values at which to allocate significant expression of HERV-K (HML-2). This represented an optimal compromise between false-positive and false-negative values, taking into account that we had a sample number of 1 (n = 1; see Materials and Methods). To put this number in simpler terms, an RPKM of 0.02 roughly corresponds to about a 40% read coverage of each expressed transcript (see Materials and Methods). To place this gene expression in context, we calculated percentile ranges for all of the RPKM values of Tat-induced gene expression (summarized in Table 4) and observed where most of the HERV-K (HML-2) RPKMs fall. As such, 78 proviruses appear to fit within the 30th and 40th percentiles, while the rest fit at or above the median (50th and 75th percentiles). As an example, the lowest HERV-K (HML-2) RPKM value (besides 0) is 0.0017 for HERV-K (HML-2) 5p12, falling in the 30th percentile, and the highest RPKM value is 3.0075 for HERV-K (HML-2) 1q21.3, above the 75th percentile. This means that unique HERV-K (HML-2) provirus gene expression in response to Tat is, in the vast majority of cases, 30% to 50% higher than that of all other annotated genes. Thus, the expression of HERV-K (HML-2) proviruses is within the range of that of most other cellular genes. Of all annotated HERV-K (HML-2) sequences, 43 (47%) had an RPKM of >0.02 in the control sample, while Tat treatment caused 35 (38%) unique proviruses to have an RPKM of >0.02. All of these proviruses are above the 30th percentile bin of RPKM values for all annotated genes. The 43 unique proviruses expressed in the vehicle treatment mostly map to chromosomes 1, 3, 4, 5, 6, 11, and 19. Thirty-five percent of hits fall between the 30th and 40th percentile, ∼63% between the 40th and the median, and ∼2% are above the 60th percentile of the RPKMs of all cellular genes. Of the 35 Tat-treated, expressed HERV-K (HML-2) proviruses, 20% had an RPKM at or above the median, 11% were above the 60th percentile, and 3% were above the 75th percentile. Most of these proviruses are located in chromosomes 1, 3, 4, 7, 8, 16, and 19.
TABLE 3.
Representative RPKM values of cellular genes
| Gene | RPKM value by treatment type |
|
|---|---|---|
| Tat | Vehicle only | |
| CD27 | 0 | 0.17 |
| IL-2Rα | 0.7 | 0.5 |
| IL-2Rβ | 47.3 | 58.9 |
| IL-2Rγ | 290.6 | 245.8 |
| IFN-γa | 47.1 | 39.8 |
| β-Actin | 1,365 | 812 |
| GAPDH | 42.2 | 42.9 |
| RPS12 | 17,070 | 17,125 |
IFN-γ, gamma interferon.
TABLE 4.
Percentile ranges of RPKM values
| RPKM | Percentile |
|---|---|
| 0 | 30 |
| 0.03542 | 40 |
| 0.207821 | 50 |
| 2.659523 | 75 |
| 9.985367 | 90 |
| 19.73389 | 95 |
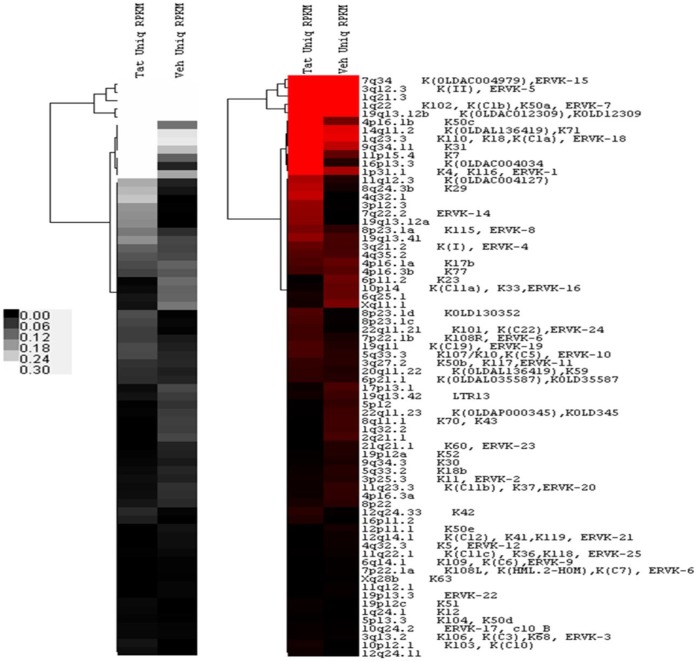
In order to determine which proviruses are significantly different between Tat and vehicle treatments, we first performed a proportion test based on total and unique read counts using a chi-squared statistic (74). For each HERV-K provirus, this statistic tests whether the proportion of total reads mapping to the provirus (considered the binomial success rate) is significantly different between the two samples, assuming the proportion of reads approximately follows a binomial distribution. We then used the Benjamini-Hochberg false-discovery-rate approach to adjust the P values for multiple tests (75) and expressed the adjusted P values as Q values for those proviruses that had RPKM values above 0.02. The false-discovery-rate control is a statistical method used to determine if a result would be statistically significant when using multiple-hypothesis testing or different testing methods to compare results. In findings where a null hypothesis is rejected, for example, the false discovery rate controls the expected proportion of incorrectly rejected null hypotheses (false discoveries). As such, it controls the number of false discoveries (or false positives) in those tests that result in a significant result. A false-discovery-rate-adjusted P value (or Q value) of 0.05 implies that 5% of significant tests will result in false positives. Removal of proviruses with zero as the RPKM value resulted in 70 candidates that were differentially modified by Tat (Fig. 1). Using a Q value cutoff of ≤0.05, we determined that the expression levels of 38 proviruses were significantly different between Tat and vehicle treatments (Table 5). Tat treatment induced the expression of 26 unique read-mapping proviruses and caused the silencing of 12 unique proviruses compared to vehicle treatment (Fig. 1). These results suggest that Tat treatment, in addition to activating, also represses HERV-K (HML-2) expression. Interestingly, of these activated proviruses, K108 and K115 (considered among the most likely candidates to remain replication competent in modern humans) (76–79) are significantly activated by Tat treatment. K109 and K113, also likely candidates for replication competence (76, 77, 79, 80), had RPKM values below our limit of detection or were not able to be analyzed in this study, respectively. K113 is the most recent germ line integration known (10, 76), with an allele frequency of ∼16% in the population (10, 76); thus, it is not yet fixed in the human population. As such, it is possible that K113 is not present in the genome of the PBL donor.
FIG 1.
Heat map of the unique sequence HERV-K (HML-2) expression levels in Tat- and vehicle-treated samples (Tat Uniq RPKM and Veh Uniq RPKM), respectively. Uniquely mapped log2 RPKM values were clustered using average linkage hierarchical clustering with the Euclidean distance matrix, as shown in the hierarchical tree. Proviruses with zero reads under both conditions were removed before clustering, resulting in 70 proviruses. The proviruses with the highest levels of expression are in white or red.
TABLE 5.
Statistical significance of differential expression (either up or down) of proviruses between Tat treatment and vehicle treatmenta
| Chromosome | Locus | Name and alternate name(s) |
P value |
Significance rank | Q value | |
|---|---|---|---|---|---|---|
| Unique | Max | |||||
| 7 | 7q34 | K(OLDAC004979), ERVK-15 | 1.8742E−63 | 1.8742E−63 | 1 | 1.6868E−61 |
| 3 | 3q12.3 | K(II), ERVK-5 | 7.9669E−68 | 7.9669E−68 | 2 | 3.5851E−66 |
| 1 | 1q21.3 | 1q21.3 | 2.1190E−56 | 2.1190E−56 | 3 | 6.3569E−55 |
| 4 | 4p16.1b | K50c | 1.8560E−40 | 1.8560E−40 | 4 | 4.1759E−39 |
| 1 | 1q22 | K102, K(C1b), K50a, ERVK-7 | 6.3468E−34 | 6.3468E−34 | 5 | 1.1424E−32 |
| 11 | 11p15.4 | K7 | 8.2763E−25 | 8.2763E−25 | 6 | 1.2414E−23 |
| 11 | 11q12.3 | K(OLDAC004127) | 8.1670E−20 | 8.1670E−20 | 7 | 1.0500E−18 |
| 3 | 3p12.3 | 3p12.3 | 1.4842E−17 | 1.4842E−17 | 8 | 1.6697E−16 |
| 19 | 19q13.12b | K50F | 4.7480E−18 | 4.7480E−18 | 9 | 4.7480E−17 |
| 8 | 8q24.3b | K29 | 1.0238E−14 | 1.0238E−14 | 10 | 9.2144E−14 |
| 1 | 1q23.3 | K110, K18, K(C1a), ERVK-18 | 1.3727E−12 | 1.3727E−12 | 11 | 1.1231E−11 |
| 14 | 14q11.2 | K(OLDAL136419), K71 | 6.7613E−11 | 6.7613E−11 | 12 | 5.0710E−10 |
| 9 | 9q34.11 | K31 | 1.6819E−10 | 1.6819E−10 | 13 | 1.1644E−09 |
| 7 | 7q22.2 | ERVK-14 | 2.1008E−10 | 2.1008E−10 | 14 | 1.3505E−09 |
| 8 | 8p23.1d | KOLD130352 | 2.1788E−09 | 2.1788E−09 | 15 | 1.3073E−08 |
| 19 | 19q13.12a | 19q13.12a | 7.9436E−09 | 7.9436E−09 | 16 | 4.4683E−08 |
| 4 | 4q32.1 | 4q32.1 | 1.6633E−07 | 1.6633E−07 | 17 | 8.8058E−07 |
| 8 | 8p23.1c | 8p23.1c | 2.9556E−07 | 2.9556E−07 | 18 | 1.4778E−06 |
| 22 | 22q11.21 | K101, K(C22), ERVK-24 | 4.7333E−07 | 4.7333E−07 | 19 | 2.2421E−06 |
| 8 | 8q11.1 | K70, K43 | 1.1955E−06 | 1.1955E−06 | 20 | 5.3795E−06 |
| 16 | 16p13.3 | K(OLDAC004034) | 1.6706E−06 | 1.6706E−06 | 21 | 7.1599E−06 |
| 6 | 6p11.2 | K23 | 2.9726E−06 | 2.9726E−06 | 22 | 1.2161E−05 |
| 5 | 5p12 | K8 | 7.1232E−06 | 7.1232E−06 | 23 | 2.7873E−05 |
| 22 | 22q11.23 | K(OLDAP000345), KOLD345 | 1.0234E−05 | 1.0234E−05 | 24 | 3.8378E−05 |
| 10 | 10p14 | K(C11a), K33, ERVK-16 | 3.9553E−07 | 3.9553E−07 | 25 | 1.4239E−06 |
| 8 | 8p23.1a | K115, ERVK-8 | 1.8342E−05 | 1.8342E−05 | 26 | 6.3492E−05 |
| 17 | 17p13.1 | 17p13.1 | 2.0446E−04 | 2.0446E−04 | 27 | 6.8154E−04 |
| 21 | 21q21.1 | K60, ERVK-23 | 6.0462E−04 | 6.0462E−04 | 28 | 1.9434E−03 |
| 19 | 19p12a | K52 | 1.7990E−03 | 1.7990E−03 | 29 | 5.5831E−03 |
| 1 | 1p31.1 | K4, K116, ERVK-1 | 2.2897E−03 | 2.2897E−03 | 30 | 6.8692E−03 |
| X | Xq11.1 | Xq11.1 | 2.7634E−03 | 2.7634E−03 | 31 | 8.0228E−03 |
| 19 | 19q11 | K(C19), ERVK-19 | 4.0119E−03 | 4.0119E−03 | 32 | 1.1283E−02 |
| 12 | 12q24.33 | K42 | 6.4178E−03 | 6.4178E−03 | 33 | 1.7503E−02 |
| 7 | 7p22.1b | K108R, ERVK-6, HERV-K (HML-2).HOM | 7.1651E−03 | 7.1651E−03 | 34 | 1.8966E−02 |
| 6 | 6q25.1 | 6q25.1 | 1.0955E−02 | 1.0955E−02 | 35 | 2.8170E−02 |
| 19 | 19q13.41 | 19q13.41 | 1.4918E−02 | 1.4918E−02 | 36 | 3.7295E−02 |
| 2 | 2q21.1 | 2q21.1 | 5.9765E−03 | 5.9765E−03 | 37 | 1.4537E−02 |
| 19 | 19q13.42 | LTR13 | 1.6717E−02 | 1.6717E−02 | 38 | 3.9593E−02 |
The different names shown represent alternative ways to refer to the same virus based on the literature. Tat or vehicle unique RPKM values are the reads that had a single unique genomic alignment, mapping to a particular provirus, not shared with any other known provirus. P values were calculated after a proportions test (R function), taking total and unique reads for each treatment. The maximum P value between those tests was selected and corrected for multiple testing using the false discovery rate, generating Q values. Shown are proviruses with significant differential expression among treatments (Q < 0.05), ordered according to significance rank.
qRT-PCR validation of specific HERV-K (HML-2) provirus activation and silencing by HIV-1 Tat corroborates RNA-Seq data analysis.
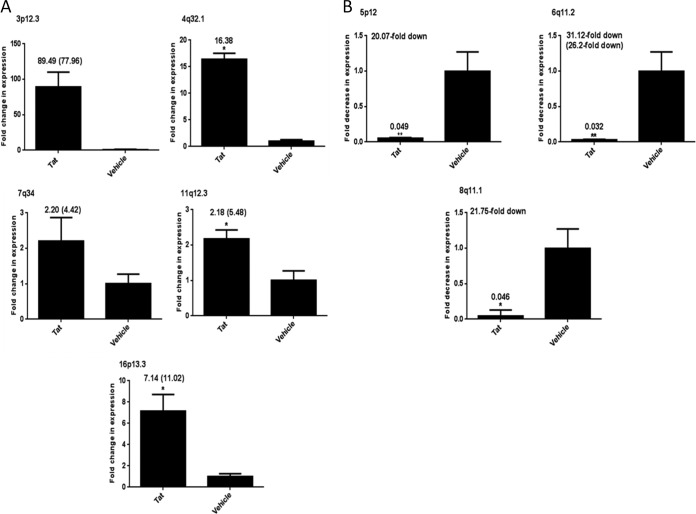
RNA-Seq results were validated via two-step qRT-PCR, using PBLs from six additional donors. First, we looked for regions in the HERV-K (HML-2) proviruses with an RPKM of ≥0.02 in the Tat treatment that had sufficient sequence diversity to allow for the design of qRT-PCR primers specific to a given provirus. Only 8 of the 35 proviruses fit this description: 3p12.3, 4q32.1, 5p12, 6p11.2 (K23), 7q34 (KOLD/ERVK-15), 8q11.1 (K70/K43), 11q12.3 (KOLD), and 16p13.3 (KOLD). Among these proviruses, five (3p12.3, 4q32.1, 7q34, 11q12.3, and 16p13.3) were upregulated by Tat treatment as measured by RNA-Seq, while three (5p12, 6p11.2, and 8q11.1) were downregulated or appeared to be silenced by said treatment with Tat. We designed specific primers to target these eight proviruses, validated their efficiency for amplification through comparison of equivalency with reference gene primers (data not shown), and validated the provirus-specific PCR amplification by direct sequencing. The quantitative expression of these proviruses by two-step qRT-PCR from the RNA (cDNA) used for RNA-Seq, as well as from the PBLs of six additional healthy volunteers, showed similar patterns of expression overall: the HERV-K (HML-2) proviruses that were upregulated by RNA-Seq also were upregulated as analyzed by qRT-PCR and vice versa (Fig. 2). In fact, fold increases were very similar to those obtained by RNA-Seq (where it was able to be calculated; fold values in parentheses in Fig. 2A correspond to fold values obtained from RNA-Seq). The same held true for those proviruses that were downregulated by Tat (Fig. 2B). PCR products were sent for sequence verification and confirmed as the single provirus product the primers were targeting. It is interesting that the RNA sample from the PBLs used for the RNA-Seq experiment showed almost the same fold activation when determined by two-step qRT-PCR as that calculated from RNA-Seq differential expression determination, and the proviruses 5p12, 6p11.2, and 8q11.1 were undetectable (data not shown). Additional healthy volunteer PBL RNA samples showed that proviruses 5p12, 6p11.2, and 8q11.1 could indeed be detected by two-step qRT-PCR, although they were clearly downregulated (Fig. 2B). As a control for Tat function, we also measured genes whose RNA expression has been shown to be downregulated by Tat, class I major histocompatibility (MHC) molecules. We observed that in the PBLs from approximately 50% of the donors, Tat treatment decreased expression of HLA-A by about 1.5-fold, as measured by qPCR analysis (data not shown). In PBLs from the other individuals, no significant difference was seen. The data obtained from RNA-Seq results from the one subject also show diminished expression of HLA genes (1.20- to 2.16-fold decreased after Tat treatment). In order to best determine whether the two transcript analysis methods (i.e., qRT-PCR and RNA-Seq) are actually correlated, we performed a statistical analysis. The Wilcoxon matched-pairs signed-rank test was not sufficiently powered to be used accurately to compare the fold values for each specific provirus tested in the PBL sample that was originally used for RNA-Seq, as two of the proviruses measured have no RPKM values in either the Tat or vehicle treatment (8q11.1 and 4q32.1, respectively). However, using the Spearman's rank correlation test, which assesses how well the relationship between two variables can be described, we obtained a correlation coefficient of 1 (n = 6). This means that we have a positive monotonic relationship between the qRT-PCR data and the RNA-Seq data where, as the value of one variable increases, so does the value of the other variable. As such, qRT-PCR and the RNA-Seq data are positively correlated, and this correlation is significantly different from zero (P < 0.0001).
FIG 2.
Validation of RNA-Seq data for HERV-K(HML-2)-specific proviruses and HLA-A. Total cellular RNA was isolated from primary cells (PBLs) that were treated with recombinant Tat protein or vehicle alone for 12 h. RNA was DNase treated, and cDNA was synthesized and amplified by SYBR green two-step qRT-PCR. Data are expressed as fold increase in RNA (Tat treatment over vehicle alone) (A) or fold decrease in RNA (Tat treatment over vehicle alone) (B), with the fold written on top of the bar. Numbers in parentheses indicate the fold activation as measured by RNA-Seq for ease of comparison for those proviruses whose RPKM was not zero in either treatment. (C) Expression of HLA-A RNA in PBLs treated with Tat or vehicle using two groups of PBLs (3 replicates each). PBLs 1 to 3 had a 1.5-fold decrease in RNA expression (2.16-fold decreased by RNA-Seq), and PBLs 4 to 6 had a 0.28-fold increase in HLA-A RNA expression. All qRT-PCR results were normalized to those for the GAPDH reference gene after analysis using the 2−ΔΔCT method, and relative expression is plotted. Error bars indicate standard errors of the means for results from the PBLs of seven individual volunteers (except for panel C, which represents three individual samples each). Significance was calculated by comparing Tat treatment to vehicle treatment using a Student's t test. Significant results are indicated (*, P < 0.05).
While these results point to the fact that donor responsiveness or sensitivity to Tat treatment might play a role with regard to which proviruses or additional genes are activated or silenced after Tat treatment, overall the qRT-PCR data again show strong agreement with the data obtained by RNA-Seq.
DISCUSSION
Next-generation sequencing technology has allowed us to analyze the transcriptome of HERV-K (HML-2) proviruses in response to HIV-1 Tat in peripheral blood lymphocytes. Here, we show a new strategy for analyzing transcript expression of specific HERV-K (HML-2) proviruses through RNA-Seq, using a uniquely compiled list of specific sequences for each provirus. This was performed by normalizing the number of reads obtained to the size of the transcript fragment (RPKM method) and then determining how many of the obtained reads aligned to a unique sequence in a single provirus at its specific chromosomal location (and not to any other provirus in the genome). This removes the potential biases that can be obtained by both the size of the transcripts and the high degree of similarity that all HERV-K (HML-2) proviruses inherently have. Additionally, we validated the RNA-Seq findings, which were based on a single donor, with two-step qRT-PCR technology on several independent donors (seven samples total, including the one used for RNA-Seq) by designing unique sets of primers to quantitate the expression of particular proviruses. The pairing of these two techniques allowed us to better understand which proviruses become activated by HIV-1 Tat treatment, as well as the magnitude of activation. Of the 91 annotated HERV-K (HML-2) proviruses, we found that Tat treatment significantly activates 26 and silences 12. Importantly, the RNA expression levels detected by RNA-Seq were validated with high correlation in fold changes by amplification through two-step qRT-PCR of RNA isolated from 6 additional healthy volunteers, as well as from the same sample used for RNA-Seq analysis (7 samples total). Our analysis is consistent with the dual nature of the effect of HIV-1 Tat on cellular genes, as it can be both an activator and a suppressor protein (81–84). However, it should be noted that although the expression of a number of proviruses was downregulated by Tat treatment, the magnitude of the effect was not as marked as that seen with activated proviruses, consistent with our earlier observations that Tat treatment increases HERV-K (HML-2) expression on a global level (46). Additionally, it is worth noting that we did not look in detail for genetic variations, such as SNPs (single-nucleotide polymorphisms) or any other transcript polymorphisms, in the RNA-Seq data. If present in splice sites, for example, these modifications to a transcript sequence could affect the expression level of a transcript and, as such, the RPKM values a standard transcript sequence receives. As our use of a Tophat MQ score of 255 implied only unique mapping and not perfect mapping, it is possible that our expression results are both higher or lower than presented here if we were to finely analyze the parameters of inclusion of polymorphism in the data.
As we know from our previous observations about Tat treatment of PBLs (46), it is important to note that during HIV-1 infection, Tat may not be the sole contributor to HERV-K (HML-2) activation, as Vif and potential immune responses to the infection may also induce provirus expression. Tat effects most likely are mediated by cellular transcription factors, such as NF-κB and NF-AT, and alterations in chromatin structure (46, 85).
Interestingly, upon examination of proviruses that are activated by Tat treatment, we observed that some do not contain viable 5′-LTR promoter regions (i.e., the promoters have large regions missing or are truncated or mutated). It is known that proviruses and solo LTRs can be transcribed as part of other cellular transcription units if they are integrated into them (52, 86); thus, HERV sequences may arise from the sense strand, the antisense strand, or both depending on where active promoters are present. However, while cellular promoters close to sequences upstream of these proviruses might mediate their activation, we do not have any evidence for that, and further analyses to test this are being assessed. Meanwhile, several possibilities should be considered explanations for the activation of those proviruses lacking a promoter. First, as stated above, cellular gene promoters (proximal or distal) could be mediating proviral gene activation, as the ENCODE gene mapping results have shown for multiple genes (87, 88). Second, some activation may be due to nearby HERV-K (HML-2) proviruses. This could be the case for K52 (19q13.12a), for example. This provirus lacks both 5′- and 3′-LTRs, yet its expression is detected during HIV-1 Tat treatment. As K52 is flanked by K(C19) (19q11, which has an intact 3′-LTR) and KOLD12309 (19q13.12b, which has both LTRs), activation could be mediated by these proviruses in cis. Lastly, expression could be mediated by intact 3′-LTRs, by LTRs from retrotransposons, or by solo LTRs.
Our data, originating from analyses of the HERV-K (HML-2) Tat-induced transcriptome, make it clear that there is a broad and complex array of mechanisms at play in the regulation of HERV-K (HML-2) expression by HIV-1. We demonstrate that in spite of the extraordinary degree of sequence similarity between different HERV-K (HML-2) proviruses, we can accurately use next-generation sequencing to determine which specific proviruses are modulated under particular biological conditions. This should prove helpful for future investigations of the roles of HERV-K (HML-2) in health and disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeffery de Wet, Nirit Mor-Vaknin, Maureen Legendre, Seagal Teitz-Tennenbaum, and Lili Zhao for their help in experimental design and data acquisition, analysis, and interpretation and for their thoughtful comments. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: clade B recombinant HIV-1 Tat86 protein.
M.J.G.-H. was supported by a Rackham Merit Fellowship, the Molecular Mechanisms of Microbial Pathogenesis Training Grant from the University of Michigan (5T32AI007528), and by the NIH Ruth L. Kirschstein NRSA Individual Predoctoral Fellowship to Promote Diversity in Health-Related Research (1F31CA150523). R.C.-G. was supported by a Research Supplement to Promote Diversity in Health-Related Research (3R01CA144043-03S1) from the National Institutes of Health. D.D. was supported by the Molecular Mechanisms of Microbial Pathogenesis Training Grant from the University of Michigan (5T32AI007528). A.K.S. was supported through the University of Michigan Medical Scientist Training Program. J.D.C., F.M., M.D., and M.A.S. were supported by NIH grant RO1CA144043 to D.M.M. G.S.O. acknowledges support from NIH grants RM-08-029, P30U54ES017885, U54DA021519, and UL1RR24986. S.D.G. was supported by a grant from the Veterans Education and Research Association of Michigan. This research was additionally supported by a generous grant from the Concerned Parents for AIDS Research (05-5089) to M.H.K. and by grants RO1AI062248 and RO1CA144043 to D.M.M. from the National Institutes of Health.
Footnotes
Published ahead of print 28 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00556-14.
REFERENCES
- 1.Gifford R, Tristem M. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26:291–315. 10.1023/A:1024455415443 [DOI] [PubMed] [Google Scholar]
- 2.Feschotte C, Gilbert C. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 13:283–296. 10.1038/nrg3199 [DOI] [PubMed] [Google Scholar]
- 3.Jern P, Coffin JM. 2008. Effects of retroviruses on host genome function. Annu. Rev. Genet. 42:709–732. 10.1146/annurev.genet.42.110807.091501 [DOI] [PubMed] [Google Scholar]
- 4.Bannert N, Kurth R. 2006. The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genomics Hum. Genet. 7:149–173. 10.1146/annurev.genom.7.080505.115700 [DOI] [PubMed] [Google Scholar]
- 5.Belshaw R, Pereira V, Katzourakis A, Talbot G, Paces J, Burt A, Tristem M. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. U. S. A. 101:4894–4899. 10.1073/pnas.0307800101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannert N, Kurth R. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. U. S. A. 101(Suppl 2):S14572–S14579. 10.1073/pnas.0404838101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flockerzi A, Ruggieri A, Frank O, Sauter M, Maldener E, Kopper B, Wullich B, Seifarth W, Muller-Lantzsch N, Leib-Mosch C, Meese E, Mayer J. 2008. Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV transcriptome project. BMC Genomics 9:354. 10.1186/1471-2164-9-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkhout B, Jebbink M, Zsiros J. 1999. Identification of an active reverse transcriptase enzyme encoded by a human endogenous HERV-K retrovirus. J. Virol. 73:2365–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. 2011. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8:90. 10.1186/1742-4690-8-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531–1535. 10.1016/S0960-9822(01)00455-9 [DOI] [PubMed] [Google Scholar]
- 11.Barbulescu M, Turner G, Seaman MI, Deinard AS, Kidd KK, Lenz J. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861–868. 10.1016/S0960-9822(99)80390-X [DOI] [PubMed] [Google Scholar]
- 12.Mayer J, Sauter M, Racz A, Scherer D, Mueller-Lantzsch N, Meese E. 1999. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat. Genet. 21:257–258. 10.1038/6766 [DOI] [PubMed] [Google Scholar]
- 13.Armbruester V, Sauter M, Krautkraermer E, Meese E, Kleiman A, Best B, Roemer K, Mueller-Lantzsch N. 2002. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin. Cancer Res. 8:1800–1807 [PubMed] [Google Scholar]
- 14.Buscher K, Hahn S, Hofmann M, Trefzer U, Ozel M, Sterry W, Lower J, Lower R, Kurth R, Denner J. 2006. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec and Np9 proteins in melanomas and melanoma cell lines. Melanoma Res. 16:223–234. 10.1097/01.cmr.0000215031.07941.ca [DOI] [PubMed] [Google Scholar]
- 15.Boese A, Sauter M, Galli U, Best B, Herbst H, Mayer J, Kremmer E, Roemer K, Mueller-Lantzsch N. 2000. Human endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene 19:4328–4336. 10.1038/sj.onc.1203794 [DOI] [PubMed] [Google Scholar]
- 16.Galli UM, Sauter M, Lecher B, Maurer S, Herbst H, Roemer K, Mueller-Lantzsch N. 2005. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene 24:3223–3228. 10.1038/sj.onc.1208543 [DOI] [PubMed] [Google Scholar]
- 17.Buscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. 2005. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 65:4172–4180. 10.1158/0008-5472.CAN-04-2983 [DOI] [PubMed] [Google Scholar]
- 18.Krieg AM, Gourley MF, Klinman DM, Perl A, Steinberg AD. 1992. Heterogeneous expression and coordinate regulation of endogenous retroviral sequences in human peripheral blood mononuclear cells. AIDS Res. Hum. Retrovir. 8:1991–1998. 10.1089/aid.1992.8.1991 [DOI] [PubMed] [Google Scholar]
- 19.Medstrand P, Lindeskog M, Blomberg J. 1992. Expression of human endogenous retroviral sequences in peripheral blood mononuclear cells of healthy individuals. J. Gen. Virol. 73(Part 9):2463–2466. 10.1099/0022-1317-73-9-2463 [DOI] [PubMed] [Google Scholar]
- 20.Brodsky I, Foley B, Haines D, Johnston J, Cuddy K, Gillespie D. 1993. Expression of HERV-K proviruses in human leukocytes. Blood 81:2369–2374 [PubMed] [Google Scholar]
- 21.Tonjes RR, Lower R, Boller K, Denner J, Hasenmaier B, Kirsch H, Konig H, Korbmacher C, Limbach C, Lugert R, Phelps RC, Scherer J, Thelen K, Lower J, Kurth R. 1996. HERV-K: the biologically most active human endogenous retrovirus family. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13(Suppl 1):S261–S267. 10.1097/00042560-199600001-00039 [DOI] [PubMed] [Google Scholar]
- 22.Blomberg JMP, Yin H, Andersson ML, Lindeskog M, Borg A, Olsson H. 1995. Expression of human endogenous retroviral sequences: differences between individuals and cell types. Increased expression in a human breast cancer. J. Cancer Res. Clin. Oncol. 121:3 [Google Scholar]
- 23.Paces J, Huang YT, Paces V, Ridl J, Chang CM. 2012. New insight into transcription of human endogenous retroviral elements. N. Biotechnol. 30:314–318. 10.1016/j.nbt.2012.11.009 [DOI] [PubMed] [Google Scholar]
- 24.Moyes DL, Martin A, Sawcer S, Temperton N, Worthington J, Griffiths DJ, Venables PJ. 2005. The distribution of the endogenous retroviruses HERV-K113 and HERV-K115 in health and disease. Genomics 86:337–341. 10.1016/j.ygeno.2005.06.004 [DOI] [PubMed] [Google Scholar]
- 25.Gourley MF, Kisch WJ, Mojcik CF, King LB, Krieg AM, Steinberg AD. 1994. Role of endogenous retroviruses in autoimmune diseases. Tohoku J. Exp. Med. 173:105–114. 10.1620/tjem.173.105 [DOI] [PubMed] [Google Scholar]
- 26.Bieda K, Hoffmann A, Boller K. 2001. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J. Gen. Virol. 82:591–596 [DOI] [PubMed] [Google Scholar]
- 27.Seifarth W, Frank O, Zeilfelder U, Spiess B, Greenwood AD, Hehlmann R, Leib-Mosch C. 2005. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 79:341–352. 10.1128/JVI.79.1.341-352.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okahara G, Matsubara S, Oda T, Sugimoto J, Jinno Y, Kanaya F. 2004. Expression analyses of human endogenous retroviruses (HERVs): tissue-specific and developmental stage-dependent expression of HERVs. Genomics 84:982–990. 10.1016/j.ygeno.2004.09.004 [DOI] [PubMed] [Google Scholar]
- 29.La Mantia G, Maglione D, Pengue G, Di Cristofano A, Simeone A, Lanfrancone L, Lania L. 1991. Identification and characterization of novel human endogenous retroviral sequences preferentially expressed in undifferentiated embryonal carcinoma cells. Nucleic Acids Res. 19:1513–1520. 10.1093/nar/19.7.1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsiao FC, Lin M, Tai A, Chen G, Huber BT. 2006. Cutting edge: Epstein-Barr virus transactivates the HERV-K18 superantigen by docking to the human complement receptor 2 (CD21) on primary B cells. J. Immunol. 177:2056–2060. 10.4049/jimmunol.177.4.2056 [DOI] [PubMed] [Google Scholar]
- 31.Tai AK, Luka J, Ablashi D, Huber BT. 2009. HHV-6A infection induces expression of HERV-K18-encoded superantigen. J. Clin. Virol. 46:47–48. 10.1016/j.jcv.2009.05.019 [DOI] [PubMed] [Google Scholar]
- 32.Turcanova VL, Bundgaard B, Hollsberg P. 2009. Human herpesvirus-6B induces expression of the human endogenous retrovirus K18-encoded superantigen. J. Clin. Virol. 46:15–19. 10.1016/j.jcv.2009.05.015 [DOI] [PubMed] [Google Scholar]
- 33.Contreras-Galindo R, Lopez P, Velez R, Yamamura Y. 2007. HIV-1 infection increases the expression of human endogenous retroviruses type K (HERV-K) in vitro. AIDS Res. Hum. Retrovir. 23:116–122. 10.1089/aid.2006.0117 [DOI] [PubMed] [Google Scholar]
- 34.Kwun HJ, Han HJ, Lee WJ, Kim HS, Jang KL. 2002. Transactivation of the human endogenous retrovirus K long terminal repeat by herpes simplex virus type 1 immediate early protein 0. Virus Res. 86:93–100. 10.1016/S0168-1702(02)00058-8 [DOI] [PubMed] [Google Scholar]
- 35.Chu WM, Ballard R, Carpick BW, Williams BR, Schmid CW. 1998. Potential Alu function: regulation of the activity of double-stranded RNA-activated kinase PKR. Mol. Cell. Biol. 18:58–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang KL, Collins MK, Latchman DS. 1992. The human immunodeficiency virus tat protein increases the transcription of human Alu repeated sequences by increasing the activity of the cellular transcription factor TFIIIC. J. Acquir. Immune Defic. Syndr. 5:1142–1147 [PubMed] [Google Scholar]
- 37.Katsumata K, Ikeda H, Sato M, Ishizu A, Kawarada Y, Kato H, Wakisaka A, Koike T, Yoshiki T. 1999. Cytokine regulation of env gene expression of human endogenous retrovirus-R in human vascular endothelial cells. Clin. Immunol. 93:75–80. 10.1006/clim.1999.4762 [DOI] [PubMed] [Google Scholar]
- 38.Ono M, Kawakami M, Ushikubo H. 1987. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J. Virol. 61:2059–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hohenadl C, Germaier H, Walchner M, Hagenhofer M, Herrmann M, Sturzl M, Kind P, Hehlmann R, Erfle V, Leib-Mosch C. 1999. Transcriptional activation of endogenous retroviral sequences in human epidermal keratinocytes by UVB irradiation. J. Investig. Dermatol. 113:587–594. 10.1046/j.1523-1747.1999.00728.x [DOI] [PubMed] [Google Scholar]
- 40.Khan AS, Muller J, Sears JF. 2001. Early detection of endogenous retroviruses in chemically induced mouse cells. Virus Res. 79:39–45. 10.1016/S0168-1702(01)00280-5 [DOI] [PubMed] [Google Scholar]
- 41.Reiche J, Pauli G, Ellerbrok H. 2010. Differential expression of human endogenous retrovirus K transcripts in primary human melanocytes and melanoma cell lines after UV irradiation. Melanoma Res. 20:435–440. 10.1097/CMR.0b013e32833c1b5d [DOI] [PubMed] [Google Scholar]
- 42.Evans LH, Alamgir AS, Owens N, Weber N, Virtaneva K, Barbian K, Babar A, Malik F, Rosenke K. 2009. Mobilization of endogenous retroviruses in mice after infection with an exogenous retrovirus. J. Virol. 83:2429–2435. 10.1128/JVI.01926-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeda S, Sugimoto K, Otsuki H, Hirochika H. 1998. Transcriptional activation of the tobacco retrotransposon Tto1 by wounding and methyl jasmonate. Plant Mol. Biol. 36:365–376. 10.1023/A:1005911413528 [DOI] [PubMed] [Google Scholar]
- 44.Toufaily C, Landry S, Leib-Mosch C, Rassart E, Barbeau B. 2011. Activation of LTRs from different human endogenous retrovirus (HERV) families by the HTLV-1 tax protein and T-cell activators. Viruses 3:2146–2159. 10.3390/v3112146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Contreras-Galindo R, Kaplan MH, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Ferlenghi I, Giusti F, Lorenzo E, Gitlin SD, Dosik MH, Yamamura Y, Markovitz DM. 2012. Characterization of human endogenous retroviral elements in the blood of HIV-1-infected individuals. J. Virol. 86:262–276. 10.1128/JVI.00602-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalez-Hernandez MJ, Swanson MD, Contreras-Galindo R, Cookinham S, King SR, Noel RJ, Jr, Kaplan MH, Markovitz DM. 2012. Expression of human endogenous retrovirus type K (HML-2) is activated by the Tat protein of HIV-1. J. Virol. 86:7790–7805. 10.1128/JVI.07215-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Contreras-Galindo R, Almodovar-Camacho S, Gonzalez-Ramirez S, Lorenzo E, Yamamura Y. 2007. Comparative longitudinal studies of HERV-K and HIV-1 RNA titers in HIV-1-infected patients receiving successful versus unsuccessful highly active antiretroviral therapy. AIDS Res. Hum. Retrovir. 23:1083–1086. 10.1089/aid.2007.0054 [DOI] [PubMed] [Google Scholar]
- 48.Contreras-Galindo R, Gonzalez M, Almodovar-Camacho S, Gonzalez-Ramirez S, Lorenzo E, Yamamura Y. 2006. A new real-time-RT-PCR for quantitation of human endogenous retroviruses type K (HERV-K) RNA load in plasma samples: increased HERV-K RNA titers in HIV-1 patients with HAART non-suppressive regimens. J. Virol. Methods 136:51–57. 10.1016/j.jviromet.2006.03.029 [DOI] [PubMed] [Google Scholar]
- 49.Contreras-Galindo R, Kaplan MH, Leissner P, Verjat T, Ferlenghi I, Bagnoli F, Giusti F, Dosik MH, Hayes DF, Gitlin SD, Markovitz DM. 2008. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J. Virol. 82:9329–9336. 10.1128/JVI.00646-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Contreras-Galindo R, Kaplan MH, Markovitz DM, Lorenzo E, Yamamura Y. 2006. Detection of HERV-K(HML-2) viral RNA in plasma of HIV type 1-infected individuals. AIDS Res. Hum. Retrovir. 22:979–984. 10.1089/aid.2006.22.979 [DOI] [PubMed] [Google Scholar]
- 51.van der Kuyl AC. 2012. HIV infection and HERV expression: a review. Retrovirology 9:6. 10.1186/1742-4690-9-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones RB, Garrison KE, Mujib S, Mihajlovic V, Aidarus N, Hunter DV, Martin E, John VM, Zhan W, Faruk NF, Gyenes G, Sheppard NC, Priumboom-Brees IM, Goodwin DA, Chen L, Rieger M, Muscat-King S, Loudon PT, Stanley C, Holditch SJ, Wong JC, Clayton K, Duan E, Song H, Xu Y, Sengupta D, Tandon R, Sacha JB, Brockman MA, Benko E, Kovacs C, Nixon DF, Ostrowski MA. 2012. HERV-K-specific T cells eliminate diverse HIV-1/2 and SIV primary isolates. J. Clin. Investig. 122:4473–4489. 10.1172/JCI64560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones RB, John VM, Hunter DV, Martin E, Mujib S, Mihajlovic V, Burgers PC, Luider TM, Gyenes G, Sheppard NC, Sengupta D, Tandon R, Yue FY, Benko E, Kovacs C, Nixon DF, Ostrowski MA. 2012. Human endogenous retrovirus K(HML-2) Gag- and Env-specific T-cell responses are infrequently detected in HIV-1-infected subjects using standard peptide matrix-based screening. Clin. Vaccine Immunol. 19:288–292. 10.1128/CVI.05583-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freimanis G, Hooley P, Ejtehadi HD, Ali HA, Veitch A, Rylance PB, Alawi A, Axford J, Nevill A, Murray PG, Nelson PN. 2010. A role for human endogenous retrovirus-K (HML-2) in rheumatoid arthritis: investigating mechanisms of pathogenesis. Clin. Exp. Immunol. 160:340–347. 10.1111/j.1365-2249.2010.04110.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Antony JM, Deslauriers AM, Bhat RK, Ellestad KK, Power C. 2011. Human endogenous retroviruses and multiple sclerosis: innocent bystanders or disease determinants? Biochim. Biophys. Acta 1812:162–176. 10.1016/j.bbadis.2010.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nelson PN, Carnegie PR, Martin J, Davari Ejtehadi H, Hooley P, Roden D, Rowland-Jones S, Warren P, Astley J, Murray PG. 2003. Demystified. Human endogenous retroviruses. Mol. Pathol. 56:11–18. 10.1136/mp.56.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ehlhardt S, Seifert M, Schneider J, Ojak A, Zang KD, Mehraein Y. 2006. Human endogenous retrovirus HERV-K(HML-2) Rec expression and transcriptional activities in normal and rheumatoid arthritis synovia. J. Rheumatol. 33:16–23 [PubMed] [Google Scholar]
- 58.Sekigawa I, Ogasawara H, Kaneko H, Hishikawa T, Hashimoto H. 2001. Retroviruses and autoimmunity. Intern. Med. 40:80–86. 10.2169/internalmedicine.40.80 [DOI] [PubMed] [Google Scholar]
- 59.Herrmann M, Neidhart M, Gay S, Hagenhofer M, Kalden JR. 1998. Retrovirus-associated rheumatic syndromes. Curr. Opin. Rheumatol. 10:347–354. 10.1097/00002281-199807000-00012 [DOI] [PubMed] [Google Scholar]
- 60.Löwer R, Löwer J, Kurth R. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. U. S. A. 93:5177–5184. 10.1073/pnas.93.11.5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh S, Kaye S, Francis N, Peston D, Gore M, McClure M, Bunker C. 2013. Human endogenous retrovirus K (HERV-K) rec mRNA is expressed in primary melanoma but not in benign naevi or normal skin. Pigment Cell Melanoma Res. 26:426–428. 10.1111/pcmr.12066 [DOI] [PubMed] [Google Scholar]
- 62.Armbruester V, Sauter M, Roemer K, Best B, Hahn S, Nty A, Schmid A, Philipp S, Mueller A, Mueller-Lantzsch N. 2004. Np9 protein of human endogenous retrovirus K interacts with ligand of numb protein X. J. Virol. 78:10310–10319. 10.1128/JVI.78.19.10310-10319.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Denne M, Sauter M, Armbruester V, Licht JD, Roemer K, Mueller-Lantzsch N. 2007. Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. J. Virol. 81:5607–5616. 10.1128/JVI.02771-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaufmann S, Sauter M, Schmitt M, Baumert B, Best B, Boese A, Roemer K, Mueller-Lantzsch N. 2010. Human endogenous retrovirus protein Rec interacts with the testicular zinc-finger protein and androgen receptor. J. Gen. Virol. 91:1494–1502. 10.1099/vir.0.014241-0 [DOI] [PubMed] [Google Scholar]
- 65.Chen T, Meng Z, Gan Y, Wang X, Gu Y, Xu X, Tang J, Zhou H, Zhang X, Gan X, Van Ness C, Xu F, Xu G, Huang L, Fang Y, Wu J, Zheng S, Jin J, Huang W, Xu R. 2013. The viral oncogene Np9 acts as a critical molecular switch for co-activating beta-catenin, ERK, Akt and Notch1 and promoting the growth of human leukemia stem/progenitor cells. Leukemia 27:1469–1478. 10.1038/leu.2013.8 [DOI] [PubMed] [Google Scholar]
- 66.Swanson MD, Winter HC, Goldstein IJ, Markovitz DM. 2010. A lectin isolated from bananas is a potent inhibitor of HIV replication. J. Biol. Chem. 285:8646–8655. 10.1074/jbc.M109.034926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bohan CA, Kashanchi F, Ensoli B, Buonaguro L, Boris-Lawrie KA, Brady JN. 1992. Analysis of Tat transactivation of human immunodeficiency virus transcription in vitro. Gene Expr. 2:391–407 [PMC free article] [PubMed] [Google Scholar]
- 68.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7:562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, Messeguer X. 2003. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 31:3651–3653. 10.1093/nar/gkg605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. 2002. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18:333–334. 10.1093/bioinformatics/18.2.333 [DOI] [PubMed] [Google Scholar]
- 73.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5:621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 74.Newcombe RG. 1998. Interval estimation for the difference between independent proportions: comparison of eleven methods. Stat. Med. 17:873–890. [DOI] [PubMed] [Google Scholar]
- 75.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. 57:289–300 [Google Scholar]
- 76.Jha AR, Pillai SK, York VA, Sharp ER, Storm EC, Wachter DJ, Martin JN, Deeks SG, Rosenberg MG, Nixon DF, Garrison KE. 2009. Cross-sectional dating of novel haplotypes of HERV-K 113 and HERV-K 115 indicate these proviruses originated in Africa before Homo sapiens. Mol. Biol. Evol. 26:2617–2626. 10.1093/molbev/msp180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dewannieux M, Blaise S, Heidmann T. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J. Virol. 79:15573–15577. 10.1128/JVI.79.24.15573-15577.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mayer J, Stuhr T, Reus K, Maldener E, Kitova M, Asmus F, Meese E. 2005. Haplotype analysis of the human endogenous retrovirus locus HERV-K(HML-2.HOM) and its evolutionary implications. J. Mol. Evol. 61:706–715. 10.1007/s00239-005-0066-7 [DOI] [PubMed] [Google Scholar]
- 79.Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 16:1548–1556. 10.1101/gr.5565706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boller K, Schonfeld K, Lischer S, Fischer N, Hoffmann A, Kurth R, Tonjes RR. 2008. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J. Gen. Virol. 89:567–572. 10.1099/vir.0.83534-0 [DOI] [PubMed] [Google Scholar]
- 81.Coiras M, Camafeita E, Urena T, Lopez JA, Caballero F, Fernandez B, Lopez-Huertas MR, Perez-Olmeda M, Alcami J. 2006. Modifications in the human T cell proteome induced by intracellular HIV-1 Tat protein expression. Proteomics 6(Suppl 1):S63–S73. 10.1002/pmic.200500437 [DOI] [PubMed] [Google Scholar]
- 82.Pocernich CB, Poon HF, Boyd-Kimball D, Lynn BC, Nath A, Klein JB, Butterfield DA. 2005. Proteomic analysis of oxidatively modified proteins induced by the mitochondrial toxin 3-nitropropionic acid in human astrocytes expressing the HIV protein tat. Brain Res. Mol. Brain Res. 133:299–306. 10.1016/j.molbrainres.2004.10.024 [DOI] [PubMed] [Google Scholar]
- 83.Poggi A, Carosio R, Spaggiari GM, Fortis C, Tambussi G, Dell'Antonio G, Dal Cin E, Rubartelli A, Zocchi MR. 2002. NK cell activation by dendritic cells is dependent on LFA-1-mediated induction of calcium-calmodulin kinase II: inhibition by HIV-1 Tat C-terminal domain. J. Immunol. 168:95–101. 10.4049/jimmunol.168.1.95 [DOI] [PubMed] [Google Scholar]
- 84.Rautonen N, Rautonen J, Martin NL, Wara DW. 1994. HIV-1 Tat induces cytokine synthesis by uninfected mononuclear cells. AIDS 8:1504–1506. 10.1097/00002030-199410000-00023 [DOI] [PubMed] [Google Scholar]
- 85.Contreras-Galindo R, Kaplan MH, He S, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Kappes F, Dube D, Chan SM, Robinson D, Meng F, Dai M, Gitlin SD, Chinnaiyan AM, Omenn GS, Markovitz DM. 2013. HIV infection reveals widespread expansion of novel centromeric human endogenous retroviruses. Genome Res. 23:1505–1513. 10.1101/gr.144303.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Illarionova AE, Vinogradova TV, Sverdlov ED. 2007. Only those genes of the KIAA1245 gene subfamily that contain HERV(K) LTRs in their introns are transcriptionally active. Virology 358:39–47. 10.1016/j.virol.2006.06.027 [DOI] [PubMed] [Google Scholar]
- 87.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Ernst J, Furey TS, Gerstein M, Giardine B, Greven M, Hardison RC, Harris RS, Herrero J, Hoffman MM, Iyer S, Kelllis M, Kheradpour P, Lassmann T, Li Q, Lin X, Marinov GK, Merkel A, Mortazavi A, Parker SC, Reddy TE, Rozowsky J, Schlesinger F, Thurman RE, Wang J, et al. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sanyal A, Lajoie BR, Jain G, Dekker J. 2012. The long-range interaction landscape of gene promoters. Nature 489:109–113. 10.1038/nature11279 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.