ABSTRACT
HIV-1 modulates key host cellular pathways for successful replication and pathogenesis through viral proteins. By evaluating the hijacking of the host ubiquitination pathway by HIV-1 at the whole-cell level, we now show major perturbations in the ubiquitinated pool of the host proteins post-HIV-1 infection. Our overexpression- and infection-based studies of T cells with wild-type and mutant HIV-1 proviral constructs showed that Vpr is necessary and sufficient for reducing whole-cell ubiquitination. Mutagenic analysis revealed that the three leucine-rich helical regions of Vpr are critical for this novel function of Vpr, which was independent of its other known cellular functions. We also validated that this effect of Vpr was conserved among different subtypes (subtypes B and C) and circulating recombinants from Northern India. Finally, we establish that this phenomenon is involved in HIV-1-mediated diversion of host ubiquitination machinery specifically toward the degradation of various restriction factors during viral pathogenesis.
IMPORTANCE HIV-1 is known to rely heavily on modulation of the host ubiquitin pathway, particularly for counteraction of antiretroviral restriction factors, i.e., APOBEC3G, UNG2, and BST-2, etc.; viral assembly; and release. Reports to date have focused on the molecular hijacking of the ubiquitin machinery by HIV-1 at the level of E3 ligases. Interaction of a viral protein with an E3 ligase alters its specificity to bring about selective protein ubiquitination. However, in the case of infection, multiple viral proteins can interact with this multienzyme pathway at various levels, making it much more complicated. Here, we have addressed the manipulation of ubiquitination at the whole-cell level post-HIV-1 infection. Our results show that HIV-1 Vpr is necessary and sufficient to bring about the redirection of the host ubiquitin pathway toward HIV-1-specific outcomes. We also show that the three leucine-rich helical regions of Vpr are critical for this effect and that this ability of Vpr is conserved across circulating recombinants. Our work, the first of its kind, provides novel insight into the regulation of the ubiquitin system at the whole-cell level by HIV-1.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1), a primate lentivirus, infects primarily T cells, macrophages, and probably dendritic cells. This narrow tropism is determined by the cell surface receptors (CD4 and the coreceptor CXCR4/CCR5) required for HIV-1 to attach and gain entry (1). HIV-1 infection is characterized by a gradual deterioration in immune function because of a severe depletion of CD4-TH lymphocytes that ultimately causes AIDS in humans (2). A human cell harbors a number of host-encoded antiretroviral restriction factors to ensure protection from invading retroviruses. HIV-1, on the other hand, being a highly evolved retrovirus, has mechanisms to evade these restrictive host responses. A detailed understanding of how the virus establishes successful infection despite the presence of numerous antiretroviral factors is essential for identifying and developing effective therapeutics and vaccines.
The HIV-1 genome is unique compared to the genomes of other retroviruses because of the presence of highly evolved accessory genes (Vpr, Vif, Nef, and Vpu) (3). Most of these small open reading frame (ORF)-encoded proteins are involved in manipulating cellular physiology for immune evasion, replication, and transmission (4). In addition, these accessory proteins confer to HIV-1 an exceptional ability to overcome numerous cellular antiretroviral restriction factors (ARVs) (4, 5) present throughout the viral life cycle. Targeted degradation of specific restriction factors during infection is achieved by the diversion of the cellular ubiquitin (Ub) proteasomal pathway (UPP) by viral accessory proteins. The UPP involves a multienzyme cascade with three distinct enzymes, namely, E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase). Substrate specificity is mediated at the level of E3 ligases, which are further classified into three groups: the RING, HECT, and F-box-containing ligases (6). The sequential attachment of Ub to various cellular proteins is also regulated by deubiquitinases (cysteine proteases) that remove Ub from proteins (7). Attachment of ubiquitin is a reversible event that is induced by various stimuli that not only affects protein stability but also regulates functional interactions, thus controlling various cellular processes, such as localization, proliferation, and immune responses (8). The ability of ubiquitinated proteins to have myriad functions depends on the number of ubiquitin molecules attached and the type of linkage. Monoubiquitination regulates vesicular transport, DNA repair, and virus budding (9). Polyubiquitination at K48 mediates protein degradation and cell cycle arrest, and the same at K63 regulates the activation of protein kinases and DNA repair (9, 10). All these host cellular processes are of prime interest with respect to viral pathogenesis; hence, viruses have developed mechanisms to exploit the UPP to create a cellular state favorable to their replication and pathogenesis (8, 11).
Viruses may encode either ubiquitin, E3 ligases, or deubiquitinases in their genome. In addition, viral proteins often act as adaptors, altering the specificity of E3 ligases to bring about specific protein ubiquitination, thereby hijacking the cellular Ub ligase complex (11). During HIV-1 infection, degradation of the antiretroviral factor APOBEC3G requires the association of the Vif protein with the cullin-5 ElonginB-ElonginC complex (12–17). Vpr-mediated G2 arrest involves the DDB1-CUL4A (VPRBP) E3 ubiquitin ligase (18–20), which is essential for viral replication. Furthermore, degradation of interferon-induced BST-2/Tetherin and CD4 by the Vpu protein depends on the ability of the Vpu protein to bind the β-TrCP subunit of SCF (Skp1–Cullin–F-box)–β-TrCP ubiquitin ligase complex (21–23). In addition, HIV-1 Nef is multiply ubiquitinated, which is critical for CD4 downregulation (24), and Gag is ubiquitinated, which is essential for virus budding (25). The diversion of E3 substrate specificity by viral proteins in certain cases can be so drastic that the natural function of the Ub ligase complex is inhibited or compromised. Previous reports supported this notion and showed that the interaction of Vpu with the SCF–β-TrCP E3 complex results in the accumulation of many of its natural substrates (β-catenin, ATF4, IκB, and p53), contributing to viral pathogenesis (26–29). The UPP is also critical for transactivation (30), the NF-κB pathway (31), as well as the assembly and release of virions from infected cells (32). Hence, exploitation of the UPP via extensive interplay between multiple viral proteins and different cellular Ub ligase complexes seems to play a major role in driving HIV-1 pathogenesis. The broad cellular consequences resulting from these changes within infected cells, however, remain unidentified.
In this report, we show that whole-cell ubiquitination is reduced within HIV-1-infected cells. The ubiquitination of known ARVs, however, was found to be protected from this inhibition. Interestingly, Vpr contributes to this major perturbation in the pool of ubiquitinated proteins during infection as well as in overexpression assays. The structural integrity of the three helical domains of Vpr was found to be critically important for this newly identified function that is conserved across various subtypes and primary isolates of the virus. In agreement with the previously demonstrated ubiquitination of ARV factors by numerous viral proteins, this inhibitory role of Vpr in host-specific cellular ubiquitination possibly suggests a redirection of the host ubiquitin pathway toward specific outcomes important for HIV-1 infection, i.e., degradation of antiviral factors. This study is first of its kind, where the effect of a viral infection on whole-cell ubiquitination has been pursued from a host perspective.
MATERIALS AND METHODS
Plasmid constructs and proviral DNAs.
Vpr, Tat, Rev, Nef, Vpu, and Vif from subtype B (pNL4-3) and Vpr from subtype C (Indian isolate 93IN905) HIV-1 isolates (obtained from the National Institute of Allergy and Infectious Diseases [NIAID], National Institutes of Health [NIH]) were amplified by PCR and cloned into the mammalian expression vector pCMV-Myc (Clontech) to generate Myc-VprB, Myc-TatB, Myc-NefB, Myc-VpuB, Myc-VifB, and Myc-VprC constructs. The Myc-VprB L22A, L64A, L64P, L67A, R62P, R80A, Δ17-33, Δ38-48, and Δ53-77 mutants were generated by site-directed mutagenesis using Kappa Hi-Fi DNA polymerase (Kappa Biosystems). Hemagglutinin (HA)-APOBEC3G was cloned by using cDNA from TZM-bl cells. LTR-luc (which has a firefly luciferase gene under the control of the HIV-1 LTR promoter) was obtained from NIAID, NIH. The pNL4-3 HIV-1 clone as well as pNL4-3Δvpr mutant (having a deletion in Vpr initiation codon) variants were kind gifts from K. Strebel (NIH) (26). The pNL4-3ΔvprΔvif mutant was a kind gift from Kathleen Boris-Lawrie (33). UNG2-HA was a kind gift from Serge Benichou (34).
Cell culture, transfections, and immunoblot analysis.
HEK 293T (human embryonic kidney 293T) and TZM-bl (HIV indicator HeLa-derived cells, acquired from the NIH AIDS Research and Reagent Program) cells were maintained in Dulbecco's modified Eagle's medium (DMEM; HiMedia) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37°C with 5% CO2. Jurkat E6.1 T cells (leukemic T cell lymphoblasts) were maintained in RPMI medium (HiMedia) supplemented with glutamine, 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37°C with 5% CO2. Plasmid transfections were performed by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Viral stocks of pNL4-3, pNL4-3Δvpr, pNL4-3Δvif, and pNL4-3ΔvprΔvif were prepared by cotransfecting different proviral DNAs and a plasmid encoding vesicular stomatitis virus glycoprotein (VSV-G) into HEK 293T cells, followed by collection of viral particles from the culture supernatant at 48 and 72 h. The levels of different cellular proteins were compared by immunoblot analysis. Cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Cell Signaling Technology), and protein estimation was done by using a BCA Protein Estimation kit (Pierce Biotechnology, Inc.). The primary antibodies used were anti-Myc, anti-HA (Clontech), anti-Ub, anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Cell Signaling Technology), anti-His (Sigma-Aldrich), anti-p24 (catalog no. 6457; NIH) and anti-tubulin (Santa Cruz Biotechnology, Inc.). The secondary antibodies used were anti-rabbit/mouse-horseradish peroxidase (HRP)-conjugated (Jackson Immuno Research) and anti-mouse-fluorescein isothiocyanate (FITC)-conjugated (BD Biosciences) antibodies. The proteins of interest were detected with EZ Western horseradish peroxidase substrate (Biological Industries, Israel). GAPDH was used as a loading control.
In vivo ubiquitination assay.
For detection of whole-cell ubiquitination, HEK 293T cells were grown in 35-mm dishes and transfected with 1 μg of a 6×His-ubiquitin expression plasmid (35) along with equal amounts of the indicated plasmids. After 36 h of transfection, 25 μM MG132 (Sigma-Aldrich) was added, and cells were further incubated for 8 h. Thereafter, cells were collected in phosphate-buffered saline (PBS), resuspended in 1 ml of lysis buffer (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris [pH 8.0], 10 mM imidazole, and 10 mM β-mercaptoethanol), sonicated, and centrifuged. Protein estimation was done using a BCA protein estimation kit (Pierce Biotechnology, Inc.). To lysates containing equal amounts of whole-cell protein, 50 μl of Ni-nitrilotriacetic acid (NTA) beads were added, and the mixture was incubated at room temperature for 4 h with rotation. Subsequently, the beads were washed for 5 min at room temperature with 750 μl of each of the following buffers: lysis buffer, buffer A (1.5 M guanidinium-HCl, 0.025 M Na2HPO4/NaH2PO4, 0.01 M Tris [pH 8.0], 10 mM β-mercaptoethanol), and buffer B (0.025 M Tris [pH 6.8], 20 mM imidazole, 0.2% Triton X-100). Ubiquitinated proteins were eluted by incubating the beads in 75 μl of buffer containing 200 mM imidazole, 5% SDS, 0.15 M Tris (pH 6.7), 30% glycerol, and 0.72 M β-mercaptoethanol for 20 min at room temperature. The eluates were mixed in a 1:1 ratio with 2× Laemmli buffer and resolved by SDS-PAGE followed by immunoblotting with the indicated antibodies.
Infection by HIV-1 pNL4-3 or HIV-1 mutants.
Jurkat E6.1 T, HEK 293T, and TZM-bl cells were infected with pNL4-3/pNL4-3Δvpr/pNL4-3Δvif/pNL4-3ΔvifΔvpr viral stocks. Infection was accomplished by incubating cells for 4 h with equal amounts of infectious virus, as assessed by β-galactosidase staining using HIV-1 indicator TZM-bl cells (36). The infected cells were harvested 48 h after infection and divided into two halves. One half was subjected to immunoblotting with the indicated antibodies. The other half was evaluated for the extent of HIV-1 infection by intracellular p24 staining with primary p24 mouse antibody (catalog no. 6457; NIH) followed by secondary anti-mouse antibody (FITC conjugated). The intracellular p24 levels of infected cells were assessed by flow cytometry.
Cell cycle staining.
HEK 293T cells were collected at 48 h posttransfection. The cells were fixed in 70% alcohol and kept at 4°C overnight. The cells were then washed twice with 1× PBS and stained with 10 μg/ml of propidium iodide. The cells were then analyzed on a BD FACSVerse instrument.
Luciferase assay.
HEK 293T cells were transfected with wild-type (wt) or mutant green fluorescent protein (GFP)/LTR-luc/Myc-Tat/Vpr. Cells were lysed in 1× passive lysis buffer (Promega). Luciferase activity was measured by using a luminometer after the addition of the substrate. GFP was used as a transfection control, and relative luciferase units (RLU) were calculated.
Sequence and phylogenetic analyses.
Vpr DNA sequences of different subtypes of HIV-1 (HIV Sequence Database [http://clustalw.ddbj.nig.ac.jp/]) and of natural variants were aligned by using ClustalW-DDBJ (http://www.hiv.lanl.gov/content/index). The Guide tree file was downloaded and was analyzed by using MEGA4 software. The protein sequence alignment was done by using ClustalW-EBI (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The alignment file was analyzed by using CLC sequence viewer.
Nucleotide sequence accession numbers.
The GenBank accession numbers for Indian vpr sequences reported in this study are KF724967 to KF724972.
RESULTS
Whole-cell ubiquitination is reduced during HIV-1 infection.
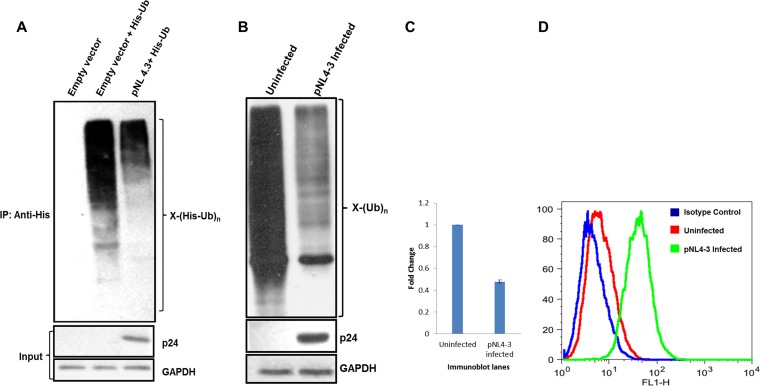
In order to determine the effect of HIV-1 infection on whole-cell ubiquitination of host cells, a molecular infectious clone, pNL4-3, was used. For probing of the whole-cell ubiquitination profile, HEK 293T cells were cotransfected with a His-Ub plasmid and/or an empty vector/pNL4-3, followed by enrichment of ubiquitinated proteins using Ni-NTA beads. Immunoblot analysis using anti-His antibody showed that the expression of the full-length HIV-1 clone reduces ubiquitination at the whole-cell level (Fig. 1A). To validate these results in an infection-based scenario in a physiologically relevant T cell line, VSV-G-pseudotyped pNL4-3 virus particles were used to infect Jurkat E6.1 cells. The infected cells were divided into two parts. One half was used to probe endogenous whole-cell ubiquitination by using an anti-Ub antibody. It was observed that HIV-1 infection reduces whole-cell ubiquitination compared to the control (Fig. 1B). Figure 1C shows the quantitation of ubiquitination for the blot shown in Fig. 1B. The other half was used to determine the extent of infection by intracellular p24 staining (Fig. 1D). Seventy-eight percent of Jurkat cells were infected.
FIG 1.
pNL4-3 transfection or infection reduces whole-cell ubiquitination. (A) HEK 293T cells were cotransfected with plasmids encoding His-Ub and pCMV-Myc or pNL4-3. After 36 h, cells were treated with MG132 for 8 h, followed by Ni-NTA pulldown. Immunoblot analysis was done, and whole-cell ubiquitination was probed by using an anti-His antibody. p24 was indicative of infection. GAPDH was used as a loading control (input). IP, immunoprecipitation. (B) Jurkat cells were infected with pNL4-3 viral supernatants for 4 h, and after 36 h, they were treated with MG132 for 8 h. The lysate was prepared in RIPA buffer containing 1× protease inhibitor and 5 mM N-ethylmaleimide. Immunoblot analysis was done, and whole-cell ubiquitination was probed by using an anti-Ub antibody. p24 was indicative of infection. GAPDH was used as a loading control. (C) Quantitation of ubiquitination for the immunoblot shown in panel B. (D) Jurkat cells were evaluated for the extent of HIV-1 infection by intracellular p24 staining with primary p24 mouse antibody (NIH) followed by secondary anti-mouse antibody (FITC conjugated). FITC staining in flow cytometer channel 1 (FL1) was analyzed; H indicates height.
Vpr is sufficient for reducing whole-cell ubiquitination in host cells.
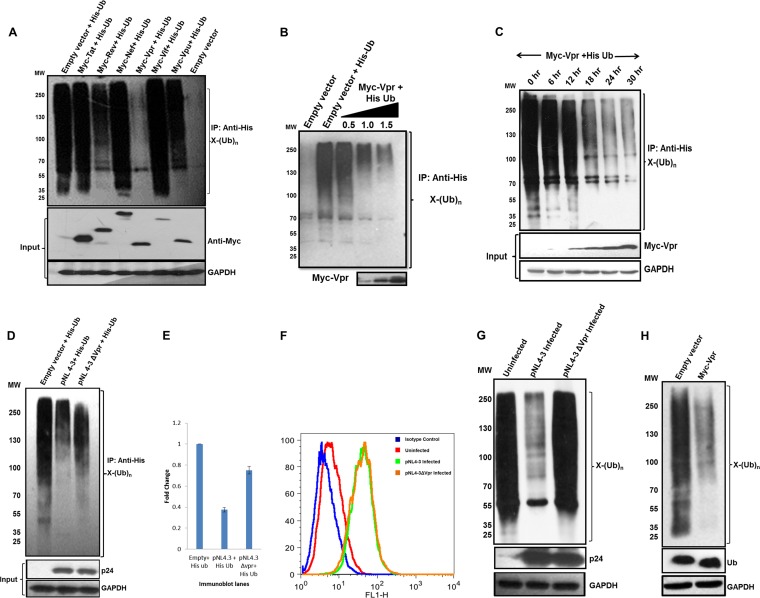
To identify the viral gene responsible for this perturbation, each of the auxiliary viral genes (Myc tagged) were independently cotransfected with the His-Ub plasmid. Ubiquitinated proteins were enriched by using Ni-NTA beads (as described in Materials and Methods), and immunoblot analysis was done. Of the six regulatory/accessory genes (tat, rev, nef, vpr, vif, and vpu), vpr drastically decreased whole-cell ubiquitination compared to the control and other genes (Fig. 2A). We further investigated if this phenomenon was dose dependent. Three different doses (0.5, 1.0, and 1.5 μg) of Myc-VprB were cotransfected with the His-Ub plasmid, and ubiquitinated proteins were enriched. The results showed that the reduction in whole-cell ubiquitination was dose dependent (Fig. 2B). In order to assess if this function of Vpr is upstream or downstream of its other known functions, a time course analysis was done. Expression of Vpr was detected as soon as 12 h following transfection, and there was a progressive decrease in the whole-cell ubiquitination profile as levels of Vpr increased (Fig. 2C). To confirm this finding in a viral scenario, pNL4-3 and pNL4-3Δvpr were cotransfected with the His-Ub plasmid. Immunoblot analysis, after pulldown using Ni-NTA beads, showed that the reduction that was observed with pNL4-3 was reverted after vpr deletion from the backbone (Fig. 2D). Figure 2E shows the quantitation of ubiquitination for the blot shown in Fig. 2D. In order to reproduce the results in an infection-based scenario, VSV-G-pseudotyped viral particles (pNL4-3 and pNL4-3Δvpr) were used to infect Jurkat E6.1 cells. One half of the cells was used to determine the extent of infection by intracellular p24 staining. Flow cytometric analysis revealed that the extent of infection was equal for pNL4-3- and pNL4-3Δvpr-infected samples (Fig. 2F). pNL4-3 infection reduced whole-cell ubiquitination, but pNL4-3Δvpr infection showed ubiquitination comparable to that of the control (Fig. 2G). A similar experiment was done with the monocytic cell line Thp1. These cells were differentiated into macrophages by using phorbol myristate acetate (PMA) and were infected with the pNL4-3 and pNL4-3Δvpr viruses. Whole-cell ubiquitination was again reduced in the presence of Vpr (see Fig. S1 in the supplemental material). In the endogenous scenario (where Ub is not overexpressed), expression of Vpr showed a similar effect with respect to the reduction of whole-cell ubiquitination when probed by using an anti-Ub antibody (Fig. 2H). Also, we observed no change in the levels of free ubiquitin upon Vpr expression. To rule out the possibility that the effect on whole-cell ubiquitination is due to the overexpression of Vpr above physiological levels, we showed that expression levels of Vpr were in a similar range in infected and transfected samples (see Fig. S2 in the supplemental material). We thus conclude that Vpr is sufficient for bringing about a reduction in the whole-cell ubiquitination of host cells.
FIG 2.
Vpr is sufficient to reduce whole-cell ubiquitination. (A) HEK 293T cells were cotransfected with His-Ub and pCMV-Myc or each viral accessory/regulatory gene (Myc-TatB/RevB/NefB/VprB/VifB/VpuB). After 36 h, cells were treated with MG132 for 8 h, followed by Ni-NTA pulldown. Immunoblot analysis was done, and whole-cell ubiquitination was probed by using an anti-His antibody. MW, molecular weight marker (in thousands). (B) Increasing doses of Myc-VprB were cotransfected with His-Ub, and immunoblotting was done as described above for panel A. The level of VprB is shown as the input. (C) A time course assay was done. After 12 h of transfection, Vpr was detected in lysates, and whole-cell ubiquitination was probed at the same time intervals. (D) HEK 293T cells were cotransfected with His-Ub and pNL4-3/pNL4-3Δvpr, and whole-cell ubiquitination was probed as described above for panel A. (E) Quantitation of ubiquitination for the immunoblot shown in panel D. (F and G) Jurkat cells were infected with pNL4-3/pNL4-3Δvpr viral supernatants for 4 h, and after 36 h, they were treated with MG132 for 8 h. Cells were divided; one set was used for intracellular p24 staining (as described in the legend of Fig. 1D), the other set was lysed, and whole-cell ubiquitination was probed by using an anti-Ub antibody. GAPDH was used as a loading control. (H) Endogenous ubiquitination was probed by using an anti-Ub antibody after overexpression of Myc-VprB in HEK 293T cells. Levels of free ubiquitin are also shown. GAPDH was used as a loading control.
Analysis of Vpr determinants critical for reducing whole-cell ubiquitination.
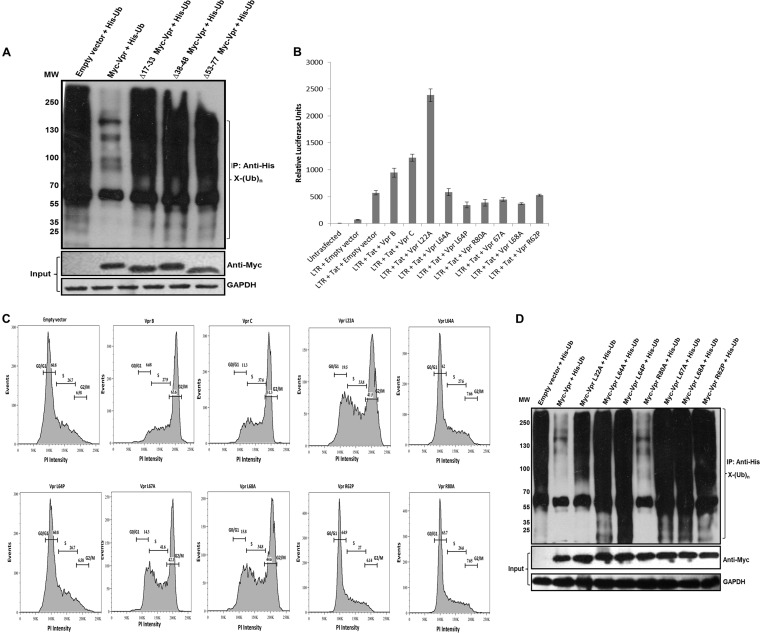
Structure analysis by nuclear magnetic resonance (NMR) and circular dichroism (CD) spectroscopy showed that Vpr contains three helical domains with a basic-amino-acid-enriched C terminus. These helical domains are rich in leucine residues with classical steroid signature motifs (LxxLL) in the helical domains (37). The three helical regions (residues 17 to 33, 38 to 48, and 53 to 77) were independently deleted, and it was shown that each mutant was able to revert the reduction in the ubiquitination profile compared to the wild-type protein (Fig. 3A), which suggests that the determinants lie in all three helices. Next, we investigated whether this reduction of whole-cell ubiquitination correlates with other known functions of Vpr, i.e., induction of G2 cell cycle arrest (18, 38) and transcriptional coactivation of viral and host genes (39). To accomplish this, functional point mutants of Vpr, such as L22A, L64A, L64P, L67A, L68A, R62P, and R80A, were cloned with a Myc backbone. The ability of these mutants to transactivate the long terminal repeat (LTR) was determined. Wild-type and mutant Vpr constructs were cotransfected with GFP, LTR-luc, and Myc-Tat. Luciferase activity was normalized by using GFP as a transfection control. Figure 3B shows relative luciferase units (RLU) for wild-type and mutant Vpr constructs. The L22A mutant was observed to drastically increase luciferase activity, while the other mutants failed (RLU values were compared to those of the control). These mutants were then checked for G2/M arrest by propidium iodide staining (40). Expression of wt Vpr and the L22A, L67A, and L68A Vpr mutants showed G2/M arrest, while the L64A, L64P, R62P, and R80A mutants failed to show any arrest (Fig. 3C). Furthermore, each of these mutants was coexpressed with a His-Ub plasmid, and ubiquitinated proteins were enriched by using Ni-NTA beads. We observed that all point mutants except the R80A mutant reverted the reduction in global ubiquitination compared to wild-type Vpr (Fig. 3D). The functionality of these mutants is summarized in Table 1. The data on transactivation and cell cycle arrest were consistent with data from previous reports (37, 41–44). This led us to conclude that the integrity of the three helices is critical for this effect and is independent of other Vpr functions.
FIG 3.
The three helical regions of Vpr critical for reducing whole-cell ubiquitination. (A) The three helices were independently deleted (Δ17-33, Δ38-48, and Δ53-77 Myc-tagged Vpr deletions). Expression was checked in HEK 293T cells, and each deletion construct was then cotransfected with His-Ub. After 36 h, MG132 treatment was given for 8 h, and ubiquitinated proteins were enriched by using Ni-NTA beads (as described in Materials and Methods). Immunoblotting was done by using an anti-His antibody to probe whole-cell ubiquitination. (B) wt Vpr and point mutants were checked for their abilities to transactivate the LTR by using the LTR-luc construct (as described in Materials and Methods). The results are representative of three independent experiments. (C) wt Vpr and point mutants were checked for their abilities to cause G2/M arrest. HEK 293T cells were collected at 48 h posttransfection and were stained with propidium iodide as described in Materials and Methods. (D) wt Vpr and point mutants were then cotransfected with His-Ub in HEK 293T cells. After 36 h, MG132 treatment was given for 8 h, and ubiquitinated proteins were enriched by using Ni-NTA beads (as described in Materials and Methods). Immunoblotting was done by using an anti-His antibody to probe whole-cell ubiquitination. GAPDH was used as a loading control.
TABLE 1.
Functional comparison of Vpr mutantsa
| Vpr protein | Cell cycle arrest | LTR transactivation | Reduction of whole-cell ubiquitination |
|---|---|---|---|
| VprB | + | + | + |
| VprC | + | + | + |
| L22A | + | ++ | − |
| L64A | − | − | − |
| L64P | − | − | − |
| L67A | + | − | − |
| L68A | + | − | − |
| R62P | − | ND | − |
| R80A | − | − | + |
Comparison of Vpr proteins from HIV-1 subtypes B and C and natural variants with respect to their effects on whole-cell ubiquitination.
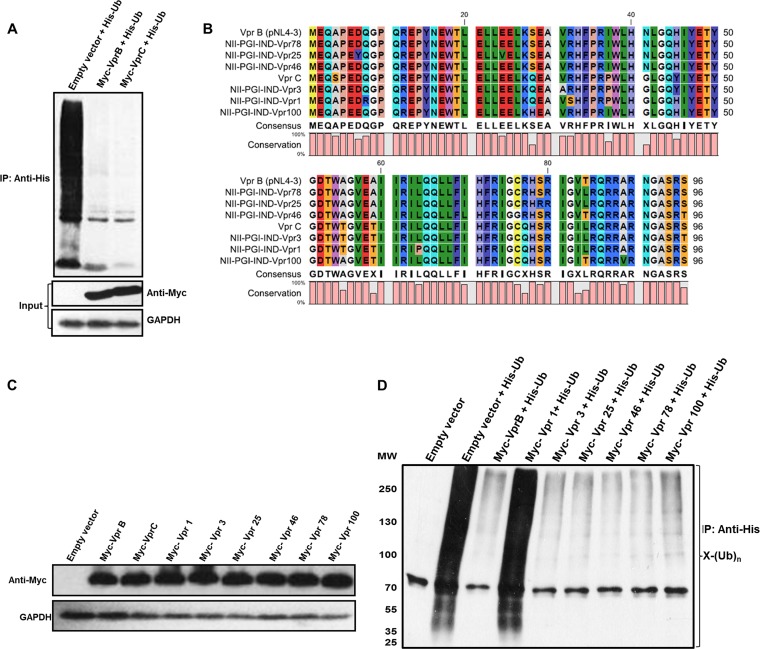
Of the nine pure subtypes that HIV-1 group M (main) viruses are divided into, subtype C is responsible for the majority of global HIV-1 infections (45). We went on to compare Vpr proteins from subtype B (laboratory-adapted strain pNL4-3) (46) and subtype C for their abilities to inhibit whole-cell ubiquitination. Myc-VprB/VprC was cotransfected with the His-Ub plasmid, and enrichment of ubiquitinated proteins was done by using Ni-NTA beads. Both VprB and VprC were found to be equally potent in reducing whole-cell ubiquitination (Fig. 4A). HIV-1 has a remarkable ability to exhibit genetic diversity, and in an infected individual, there is continuous generation of genetic variants (47). To explore if this novel function of Vpr is conserved across its natural variants in Northern India, Vpr from genomic DNA of 6 HIV-1-infected patients was amplified, cloned into the pCMV-Myc vector, and sequenced. Amino acid sequence alignment was carried out, and sequences were compared to consensus subtype B and C sequences (Fig. 4B). Phylogenetic analysis using MEGA4 software revealed that the natural variants are closely related to either subtype B or C (data not shown). The expression levels of the variants were normalized (Fig. 4C), and each variant was cotransfected with His-Ub to check their ability to affect whole-cell ubiquitination. Immunoblot analysis was done after enrichment of ubiquitinated proteins. As shown in Fig. 4D, the reduction in global ubiquitination by Vpr is largely conserved across natural variants, except for one mutant (Vpr-1) that harbors the L64P mutation, which disrupted the three helical regions and hence reverted the reduction of global ubiquitination, consistent with data from mutagenesis studies.
FIG 4.
Comparison of HIV-1 subtypes B and C and natural variants. (A) HEK 293T cells were cotransfected with Myc-VprB/VprC and His-Ub. After 36 h, MG132 treatment was given for 8 h, and ubiquitinated proteins were enriched by using Ni-NTA beads (as described in Materials and Methods). Immunoblotting was done by using an anti-His antibody to probe whole-cell ubiquitination. Levels of Myc-VprB and subtype C are shown as the input. (B) Protein sequence alignment of HIV-1 VprB, VprC, and 6 samples. (C) The expression levels of 6 variant samples (Vpr-1, -3, -25, -46, -78, and -100), Myc-VprB, and Myc-VprC were checked in HEK 293T cells and normalized. (D) HEK 293T cells were cotransfected with Myc-VprB, 6 variant samples (Vpr-1, -3, -25, -46, -78, and -100), and His-Ub. After 36 h, MG132 treatment was given for 8 h, and ubiquitinated proteins were enriched by using Ni-NTA beads (as described in Materials and Methods). Immunoblotting was done by using an anti-His antibody to probe whole-cell ubiquitination. GAPDH was used as a loading control.
Redirection of the UPP by Vpr and degradation of antiretroviral restriction factors.
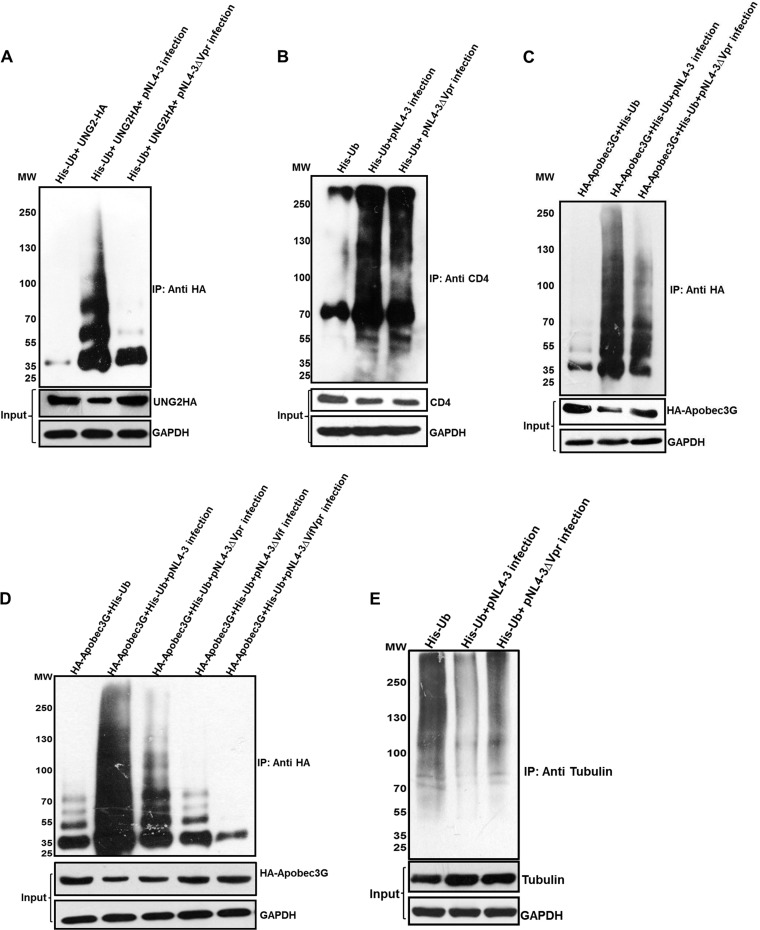
HIV-1 counteracts antiretroviral host factors by degrading them via the UPP. In order to assess the implication of the reduction in whole-cell ubiquitination by Vpr, ubiquitination and degradation of ARVs were checked. Ubiquitination of UNG2 (nuclear uracil-DNA glycosylase), a known target of Vpr degraded through the UPP (48), was checked. HEK 293T cells were cotransfected with His-Ub and UNG2-HA, infected with pNL4-3/pNL4-3Δvpr VSV-G-pseudotyped virus after 12 h, and treated with MG132 after 36 h for 8 h. Immunoblot analysis using anti-HA antibody, after pulldown using Ni-NTA beads, showed that ubiquitination of UNG2-HA was increased in the presence of Vpr in the proviral backbone, whereas its levels were reduced compared to the levels for the control (Fig. 5A). Next, we checked ubiquitination of CD4, which is known to be degraded by multiple viral proteins (Vpu [49], Nef [50], and Env [51]) via the UPP in infected cells. TZM-bl cells were transfected with His-Ub, infected with pNL4-3/pNL4-3Δvpr VSV-G-pseudotyped virus after 12 h, and treated with MG132 after 36 h for 8 h. Despite the reduction in whole-cell ubiquitination, immunoblot analysis using an anti-CD4 antibody confirmed comparable ubiquitination of CD4 in the presence and absence of Vpr (Fig. 5B). Quantitation of the input of CD4 is shown in Fig. S3 in the supplemental material. Also, ubiquitination of APOBEC3G was assessed, for which HEK 293T cells were cotransfected with His-Ub and HA-APOBEC3G plasmids. After 12 h, cells were infected with pNL4-3/pNL4-3Δvpr VSV-G-pseudotyped virus. Immunoblot analysis using an anti-HA antibody, after pulldown using Ni-NTA beads, showed that ubiquitination and degradation of APOBEC3G were increased in the presence of Vpr (pNL4-3) compared to those in the presence of Vpr-null (pNL4-3Δvpr) virus (Fig. 5C). This experiment was repeated in TZM-bl cells by using an APOBEC3G antibody (catalog no. 9963; NIH), and similar results were obtained (see Fig. S4 in the supplemental material). In order to confirm the impact of Vpr on Vif function, we also tested Δvif and Δvif Δvpr pNL4-3 mutants for their effect on APOBEC3G degradation. HEK 293T cells were cotransfected with His-Ub and HA-APOBEC3G plasmids. After 12 h, cells were infected with pNL4-3/pNL4-3Δvpr/pNL4-3Δvif/pNL4-3ΔvifΔvpr VSV-G-pseudotyped virus. When Vif was deleted from the proviral backbone, ubiquitination of APOBEC3G was comparable to that in the uninfected lane (Fig. 5D, lanes 2, 5, and 6). When Vif was present, ubiquitination of APOBEC3G increased, but it was more pronounced when Vpr was also present along with Vif (Fig. 5D, lanes 3 and 4). Finally, in order to verify the specificity of Vpr-mediated inhibition of ubiquitination for housekeeping host proteins, ubiquitination of tubulin was assessed upon infection. HEK 293T cells were transfected with His-Ub, infected with pNL4-3/pNL4-3Δvpr VSV-G-pseudotyped virus after 12 h, and treated with MG132 after 36 h for 8 h. Immunoblot analysis using anti-tubulin antibody, after pulldown using Ni-NTA beads, showed that ubiquitination of tubulin was decreased in the presence of Vpr (pNL4-3) compared to the control and in the absence of Vpr (pNL4-3Δvpr) (Fig. 5D). Quantitation of the input blot is shown in Fig. S5 in the supplemental material. This led us to conclude that although whole-cell ubiquitination is significantly reduced, ubiquitination of antiretroviral restriction factors still occurs and in some instances is increased (APOBEC3G), indicating redirection of ubiquitination for HIV-1-specific outcomes during infection.
FIG 5.
Ubiquitination and degradation of different proteins. (A) HEK 293T cells were cotransfected with HA-UNG2 and His-Ub. After 12 h, the cells were infected with equal multiplicities of infection of pNL4-3/pNL4-3Δvpr VSV-G-pseudotyped virus. After 36 h, MG132 treatment was given for 8 h, and ubiquitinated proteins were enriched by using Ni-NTA beads. Immunoblot analysis was done by using an anti-HA antibody. (B) TZM-bl cells were transfected with His-Ub. After 12 h, the cells were infected with equal multiplicities of infection of pNL4-3/pNL4-3Δvpr VSV-G-pseudotyped virus. After 36 h, MG132 treatment was given for 8 h, and ubiquitinated proteins were enriched by using Ni-NTA beads. Immunoblot analysis was done by using anti-CD4 antibody. (C) HEK 293T cells were cotransfected with HA-APOBEC3G and His-Ub. After 12 h, the cells were infected with equal multiplicities of infection of pNL4-3/pNL4-3Δvpr VSV-G-pseudotyped virus. After 36 h, MG132 treatment was given for 8 h, and ubiquitinated proteins were enriched by using Ni-NTA beads. Immunoblot analysis was done by using an anti-HA antibody. (D) HEK 293T cells were cotransfected with HA-APOBEC3G and His-Ub. After 12 h, the cells were infected with equal multiplicities of infection of pNL4-3/pNL4-3Δvpr/pNL4-3Δvif/pNL4-3ΔvifΔvpr VSV-G-pseudotyped virus. After 36 h, MG132 treatment was given for 8 h, and ubiquitinated proteins were enriched by using Ni-NTA beads. Immunoblot analysis was done by using an anti-HA antibody. (E) HEK 293T cells were transfected with His-Ub. After 12 h, the cells were infected with equal multiplicities of infection of pNL4-3/pNL4-3Δvpr VSV-G-pseudotyped virus. After 36 h, MG132 treatment was given for 8 h, and ubiquitinated proteins were enriched by using Ni-NTA beads. Immunoblot analysis was done by using an anti-tubulin antibody. Levels of proteins are shown as the input (without MG132 treatment). GAPDH was used as a loading control.
DISCUSSION
Owing to their limited genome size, viruses rely heavily on host cellular machinery for survival and propagation. Exploitation of host cell pathways is mediated by viral proteins, which have an enormous ability to interact with host proteins and alter cellular physiology. Ubiquitination is a key process of the host cell that regulates not only protein turnover but also protein localization, protein function, and protein-protein interactions. Hence, ubiquitination is a critical pathway that has been extensively modulated by viruses (52). In addition to modifying ubiquitination of substrates, many viruses also encode components of the multienzyme ubiquitin pathway (11).
The ubiquitination proteasomal pathway (UPP) can both positively and negatively potentiate viral processes based on the cellular state (11). The exact role of the ubiquitination pathway during HIV-1 pathogenesis remains controversial. On the one hand, this host process positively regulates the degradation of antiretroviral restriction factors (ARVs), receptor-mediated endocytosis, and viral maturation and budding (12, 23, 32). On the other hand, the use of ubiquitination machinery for NF-κB signaling can limit virus by modulating the immune response (28). Also, cell cycle progression mediated by the degradation of cell cycle host proteins is not favorable for the virus, as viral replication is maximal in G2/M-arrested cells (53). Several pathogenesis studies have already determined that altered E3 substrate specificity is responsible for the modulation of the UPP during HIV-1 infection, but those studies focused on the perturbation/hijacking of a single E3 ligase by viral proteins. However, we were interested in gaining an understanding of how HIV-1 modulates ubiquitination at the whole-cell level and its functional consequences. We observed that HIV-1 infection (pNL4-3) reduces whole-cell ubiquitination. Furthermore, out of 6 auxiliary genes, Vpr alone was found to be responsible for reducing whole-cell ubiquitination. HIV-1 Vpr is a 96-amino-acid-long multifunctional protein with no known enzymatic activity (37). Vpr facilitates nuclear import of the viral preintegration complex (54) and induces G2 arrest (38), which increases viral replication (55). In addition, Vpr is known to suppress host immune responses by reduction of T cell proliferation and inhibition of NF-κB activation, which results in decreased levels of proinflammatory cytokines (56–58). The Vpr-mediated perturbation in cellular ubiquitination (visible at 12 h post-Vpr transfection) (Fig. 2C) precedes all known Vpr-mediated events, such as LTR transactivation and cell cycle arrest, that are detectable only after 48 h of transfection (59). The transfection-based results were validated in T cells by using VSV-G-pseudotyped HIV-1 (pNL4-3 and its vpr deletion mutant pNL4-3Δvpr). Moreover, this observation was also consistent for the endogenous system (using ubiquitin antibody).
Having established this novel function of Vpr, the next goal was to find the determinants of Vpr responsible for this effect. NMR studies revealed that the Vpr structure is characterized by three alpha helices surrounded by flexible N- and C-terminal domains (60). Each of these alpha helices, when deleted, independently resulted in a reversion of reduction in whole-cell ubiquitination. Functional point mutants were made based on data reported in the literature (37, 41–44) and were verified (Fig. 3B and C). The ability of Vpr to reduce whole-cell ubiquitination was independent of both LTR transactivation and cell cycle arrest (R80A mutant). This is summarized in Table 1 and is consistent with the observation that reduction of whole-cell ubiquitination is evident as soon as intracellular expression of Vpr is detectable. We thus conclude that the determinants of Vpr-mediated host ubiquitination reduction lie in the three helical regions of the protein. Recent studies have also revealed that the structural features of Vpr (the three alpha-helical domains) are crucial for mediating diverse biological functions (60). Consistent with this finding, point mutations made in any of the three leucine-rich helical regions did not inhibit whole-cell ubiquitination, while mutations made outside the helices (R80A) retained this ability.
Due to its high genetic variability, HIV-1 is classified into three major phylogenetic groups (groups M, N, and O) that arose independently after cross-species transmission (61, 62). Of these, group M, which is responsible for the majority of infections worldwide, is further subdivided into 9 subtypes or clades (subtypes A to K) (63). Previous reports (46, 64–66) suggested that there are differences between subtypes B and C in the accessory genes, leading to specific differences in their abilities to carry out a particular function. Hence, we were interested to see if there is any such difference with respect to inhibition of whole-cell ubiquitination by Vpr. When the two subtypes of HIV-1 (subtypes B and C) were compared, it was observed that Vpr proteins of both subtypes were equally potent in reducing whole-cell ubiquitination, as the three helical domains were conserved. We further verified conservation of this activity across natural isolates of Vpr derived from HIV-1-infected patients in Northern India. Genetic analysis of natural variants from HIV-1-infected individuals from Northern India revealed that Vpr from these samples was closely related to Vpr of either B, C, or BC recombinants (data not shown). Five out of six natural variants retained the function of reducing whole-cell ubiquitination, indicating the significance of this function in HIV-1 pathogenesis. Sequence alignment showed that the single natural variant (Vpr-1) that was able to revert the effect on whole-cell ubiquitination harbors an L64P mutation, which distorts the third helical region that is critical for mediating this effect, consistent with the mutagenesis data showing that the three helical regions are critical for this function.
Although UPP modulation in the case of HIV-1 remains questionable, numerous independent reports suggested that its positive modulation brings about the degradation of ARVs during the course of infection (12, 22–24, 67). Interestingly, although Vpr seems to perturb the ubiquitin machinery and reduce the whole-cell ubiquitination level, our results show that this phenomenon exempts degradation of UNG2, CD4, and APOBEC3G, which could be possible when there is redirection. The ubiquitination of CD4 is comparable in the presence and absence of Vpr; however, the ubiquitination of UNG2 and APOBEC3G increases, while that of tubulin decreases. Ubiquitination of APOBEC3G, CD4, and UNG2, despite a drastic reduction of the whole-cell ubiquitination profile in the presence of Vpr, rules out the possibility of inhibition at the core ubiquitin machinery (E1/E2/E3/proteasome). Moreover, Vpr is not known to possess any deubiquitinase activity, and there is no change in the level of free ubiquitin when Vpr is overexpressed in cells. It is well established that the functional activity of Vpr is known to depend on its interaction with host cell proteins, many of which are unidentified (60). We thus hypothesize that an unknown cellular partner interacts with the leucine-rich region of Vpr, which, in natural cells, is supposed to perform a cofactor job for the UPP, which is sequestered in the presence of Vpr. The identity of this protein remains unknown, but we propose that it might be involved in regulating the ubiquitination of a large subset of cellular proteins, and hence, further efforts toward the identification of such a molecule may give us some new insights into how ubiquitination itself is regulated in normal cells.
Taken together, our study has generated new insights into the regulation of the host ubiquitin system by HIV-1. Previous reports have shown manipulation of this pathway by the HIV-1 Vif, Vpr, and Vpu proteins (12, 22, 23, 27, 48), but we have extended this understanding at the whole-cell level. We have shown that Vpr has a unique ability to redirect cellular ubiquitination. Reduction of whole-cell ubiquitination by HIV-1 Vpr may also contribute to the modulation of NF-κB function and the double-strand break signaling pathway, both of which are regulated primarily through the UPP. Furthermore, a recent report demonstrated that host protein translation is suppressed during HIV-1 infection (68). Therefore, reduction of whole-cell ubiquitination by HIV-1 Vpr may also contribute to the maintenance of cellular levels of housekeeping proteins and other proteins essential for HIV-1 replication. Hence, through redirection of the UPP, the most versatile pathway of cellular homeostasis, HIV-1 can alter the host cell environment in order to maximize viral replication and pathogenesis. Our discovery opens new dimensions to identify the mechanistic basis for such an evolutionarily conserved selective perturbation of ubiquitination during HIV-1 infection.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge K. Strebel, NIH; Serge Benichou, Institute Cochin, CNRS, France; Dimritis Xirodimas, University of Dundee, United Kingdom; and Kathleen Boris-Lawrie, Ohio State University, for plasmid constructs. We thank Aalia S. Bano, Sanket S. Ponia, Richa Kapoor, Binod Kumar, and Jyotsna Singh for helpful discussions and technical assistance. Several reagents were obtained from the AIDS Reagent Program, NIAID, NIH.
This work was supported by the Department of Biotechnology (grant no. BT/PR8322/Med/14/1245/2006) and the Indian Council of Medical Research (grant no. HIV/50/142/9/2011-ECD-II), Government of India, to A.C.B. and the NII, New Delhi. We also acknowledge the fellowship received from the CSIR, Government of India.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 4 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00619-14.
REFERENCES
- 1.Clapham PR, McKnight A. 2001. HIV-1 receptors and cell tropism. Br. Med. Bull. 58:43–59. 10.1093/bmb/58.1.43 [DOI] [PubMed] [Google Scholar]
- 2.Cummins NW, Badley AD. 2010. Mechanisms of HIV-associated lymphocyte apoptosis: 2010. Cell Death Dis. 1:e99. 10.1038/cddis.2010.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cullen BR, Greene WC. 1990. Functions of the auxiliary gene products of the human immunodeficiency virus type 1. Virology 178:1–5. 10.1016/0042-6822(90)90373-Y [DOI] [PubMed] [Google Scholar]
- 4.Malim MH, Emerman M. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398. 10.1016/j.chom.2008.04.008 [DOI] [PubMed] [Google Scholar]
- 5.Kirchhoff F. 2010. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8:55–67. 10.1016/j.chom.2010.06.004 [DOI] [PubMed] [Google Scholar]
- 6.Pickart CM. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503–533. 10.1146/annurev.biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- 7.Clague MJ, Coulson JM, Urbe S. 2012. Cellular functions of the DUBs. J. Cell Sci. 125:277–286. 10.1242/jcs.090985 [DOI] [PubMed] [Google Scholar]
- 8.Kerscher O, Felberbaum R, Hochstrasser M. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22:159–180. 10.1146/annurev.cellbio.22.010605.093503 [DOI] [PubMed] [Google Scholar]
- 9.Hershko A, Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479. 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 10.Messick TE, Greenberg RA. 2009. The ubiquitin landscape at DNA double-strand breaks. J. Cell Biol. 187:319–326. 10.1083/jcb.200908074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Randow F, Lehner PJ. 2009. Viral avoidance and exploitation of the ubiquitin system. Nat. Cell Biol. 11:527–534. 10.1038/ncb0509-527 [DOI] [PubMed] [Google Scholar]
- 12.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. 10.1126/science.1089591 [DOI] [PubMed] [Google Scholar]
- 13.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792–7798. 10.1074/jbc.M313093200 [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi M, Takaori-Kondo A, Miyauchi Y, Iwai K, Uchiyama T. 2005. Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-Elongin B-Elongin C complex is essential for Vif function. J. Biol. Chem. 280:18573–18578. 10.1074/jbc.C500082200 [DOI] [PubMed] [Google Scholar]
- 15.Iwatani Y, Chan DS, Liu L, Yoshii H, Shibata J, Yamamoto N, Levin JG, Gronenborn AM, Sugiura W. 2009. HIV-1 Vif-mediated ubiquitination/degradation of APOBEC3G involves four critical lysine residues in its C-terminal domain. Proc. Natl. Acad. Sci. U. S. A. 106:19539–19544. 10.1073/pnas.0906652106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehle A, Goncalves J, Santa-Marta M, McPike M, Gabuzda D. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 18:2861–2866. 10.1101/gad.1249904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stopak K, de Noronha C, Yonemoto W, Greene WC. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591–601. 10.1016/S1097-2765(03)00353-8 [DOI] [PubMed] [Google Scholar]
- 18.Belzile JP, Duisit G, Rougeau N, Mercier J, Finzi A, Cohen EA. 2007. HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase. PLoS Pathog. 3:e85. 10.1371/journal.ppat.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeHart JL, Zimmerman ES, Ardon O, Monteiro-Filho CM, Arganaraz ER, Planelles V. 2007. HIV-1 Vpr activates the G2 checkpoint through manipulation of the ubiquitin proteasome system. Virol. J. 4:57. 10.1186/1743-422X-4-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belzile JP, Richard J, Rougeau N, Xiao Y, Cohen EA. 2010. HIV-1 Vpr induces the K48-linked polyubiquitination and proteasomal degradation of target cellular proteins to activate ATR and promote G2 arrest. J. Virol. 84:3320–3330. 10.1128/JVI.02590-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. 1998. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1:565–574. 10.1016/S1097-2765(00)80056-8 [DOI] [PubMed] [Google Scholar]
- 22.Mangeat B, Gers-Huber G, Lehmann M, Zufferey M, Luban J, Piguet V. 2009. HIV-1 Vpu neutralizes the antiviral factor Tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 5:e1000574. 10.1371/journal.ppat.1000574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douglas JL, Viswanathan K, McCarroll MN, Gustin JK, Fruh K, Moses AV. 2009. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a betaTrCP-dependent mechanism. J. Virol. 83:7931–7947. 10.1128/JVI.00242-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin YJ, Cai CY, Zhang X, Burakoff SJ. 2008. Lysine 144, a ubiquitin attachment site in HIV-1 Nef, is required for Nef-mediated CD4 down-regulation. J. Immunol. 180:7878–7886. 10.4049/jimmunol.180.12.7878 [DOI] [PubMed] [Google Scholar]
- 25.Sette P, Nagashima K, Piper RC, Bouamr F. 2013. Ubiquitin conjugation to Gag is essential for ESCRT-mediated HIV-1 budding. Retrovirology 10:79. 10.1186/1742-4690-10-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akari H, Bour S, Kao S, Adachi A, Strebel K. 2001. The human immunodeficiency virus type 1 accessory protein Vpu induces apoptosis by suppressing the nuclear factor kappaB-dependent expression of antiapoptotic factors. J. Exp. Med. 194:1299–1311. 10.1084/jem.194.9.1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Besnard-Guerin C, Belaidouni N, Lassot I, Segeral E, Jobart A, Marchal C, Benarous R. 2004. HIV-1 Vpu sequesters beta-transducin repeat-containing protein (betaTrCP) in the cytoplasm and provokes the accumulation of beta-catenin and other SCFbetaTrCP substrates. J. Biol. Chem. 279:788–795. 10.1074/jbc.M308068200 [DOI] [PubMed] [Google Scholar]
- 28.Bour S, Perrin C, Akari H, Strebel K. 2001. The human immunodeficiency virus type 1 Vpu protein inhibits NF-kappa B activation by interfering with beta TrCP-mediated degradation of Ikappa B. J. Biol. Chem. 276:15920–15928. 10.1074/jbc.M010533200 [DOI] [PubMed] [Google Scholar]
- 29.Verma S, Ali A, Arora S, Banerjea AC. 2011. Inhibition of beta-TrcP-dependent ubiquitination of p53 by HIV-1 Vpu promotes p53-mediated apoptosis in human T cells. Blood 117:6600–6607. 10.1182/blood-2011-01-333427 [DOI] [PubMed] [Google Scholar]
- 30.Bres V, Kiernan RE, Linares LK, Chable-Bessia C, Plechakova O, Treand C, Emiliani S, Peloponese JM, Jeang KT, Coux O, Scheffner M, Benkirane M. 2003. A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat. Cell Biol. 5:754–761. 10.1038/ncb1023 [DOI] [PubMed] [Google Scholar]
- 31.Iwai K. 2010. Functions of linear ubiquitin chains in the NF-kappaB pathway: linear polyubiquitin in NF-kappaB signaling. Subcell. Biochem. 54:100–106. 10.1007/978-1-4419-6676-6_8 [DOI] [PubMed] [Google Scholar]
- 32.Alroy I, Tuvia S, Greener T, Gordon D, Barr HM, Taglicht D, Mandil-Levin R, Ben-Avraham D, Konforty D, Nir A, Levius O, Bicoviski V, Dori M, Cohen S, Yaar L, Erez O, Propheta-Meiran O, Koskas M, Caspi-Bachar E, Alchanati I, Sela-Brown A, Moskowitz H, Tessmer U, Schubert U, Reiss Y. 2005. The trans-Golgi network-associated human ubiquitin-protein ligase POSH is essential for HIV type 1 production. Proc. Natl. Acad. Sci. U. S. A. 102:1478–1483. 10.1073/pnas.0408717102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayes AM, Qian S, Yu L, Boris-Lawrie K. 2011. Tat RNA silencing suppressor activity contributes to perturbation of lymphocyte miRNA by HIV-1. Retrovirology 8:36. 10.1186/1742-4690-8-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen R, Le Rouzic E, Kearney JA, Mansky LM, Benichou S. 2004. Vpr-mediated incorporation of UNG2 into HIV-1 particles is required to modulate the virus mutation rate and for replication in macrophages. J. Biol. Chem. 279:28419–28425. 10.1074/jbc.M403875200 [DOI] [PubMed] [Google Scholar]
- 35.Xirodimas DP, Stephen CW, Lane DP. 2001. Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp. Cell Res. 270:66–77. 10.1006/excr.2001.5314 [DOI] [PubMed] [Google Scholar]
- 36.Cornall A, Sharma L, Solomon A, Gorry PR, Crowe SM, Cameron PU, Lewin SR. 2010. A novel, rapid method to detect infectious HIV-1 from plasma of persons infected with HIV-1. J. Virol. Methods 165:90–96. 10.1016/j.jviromet.2010.01.010 [DOI] [PubMed] [Google Scholar]
- 37.Thotala D, Schafer EA, Tungaturthi PK, Majumder B, Janket ML, Wagner M, Srinivasan A, Watkins S, Ayyavoo V. 2004. Structure-functional analysis of human immunodeficiency virus type 1 (HIV-1) Vpr: role of leucine residues on Vpr-mediated transactivation and virus replication. Virology 328:89–100. 10.1016/j.virol.2004.07.013 [DOI] [PubMed] [Google Scholar]
- 38.Chang F, Re F, Sebastian S, Sazer S, Luban J. 2004. HIV-1 Vpr induces defects in mitosis, cytokinesis, nuclear structure, and centrosomes. Mol. Biol. Cell 15:1793–1801. 10.1091/mbc.E03-09-0691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawaya BE, Khalili K, Gordon J, Taube R, Amini S. 2000. Cooperative interaction between HIV-1 regulatory proteins Tat and Vpr modulates transcription of the viral genome. J. Biol. Chem. 275:35209–35214. 10.1074/jbc.M005197200 [DOI] [PubMed] [Google Scholar]
- 40.Riccardi C, Nicoletti I. 2006. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat. Protoc. 1:1458–1461. 10.1038/nprot.2006.238 [DOI] [PubMed] [Google Scholar]
- 41.Barnitz RA, Chaigne-Delalande B, Bolton DL, Lenardo MJ. 2011. Exposed hydrophobic residues in human immunodeficiency virus type 1 Vpr helix-1 are important for cell cycle arrest and cell death. PLoS One 6:e24924. 10.1371/journal.pone.0024924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schrofelbauer B, Hakata Y, Landau NR. 2007. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc. Natl. Acad. Sci. U. S. A. 104:4130–4135. 10.1073/pnas.0610167104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belzile JP, Abrahamyan LG, Gerard FC, Rougeau N, Cohen EA. 2010. Formation of mobile chromatin-associated nuclear foci containing HIV-1 Vpr and VPRBP is critical for the induction of G2 cell cycle arrest. PLoS Pathog. 6:e1001080. 10.1371/journal.ppat.1001080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau NR. 1995. Mutational analysis of cell cycle arrest, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J. Virol. 69:7909–7916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hemelaar J, Gouws E, Ghys PD, Osmanov S. 2006. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 20:W13–W23. 10.1097/01.aids.0000247564.73009.bc [DOI] [PubMed] [Google Scholar]
- 46.Douglas JL, Bai Y, Gustin JK, Moses AV. 2013. A comparative mutational analysis of HIV-1 Vpu subtypes B and C for the identification of determinants required to counteract BST-2/Tetherin and enhance viral egress. Virology 441:182–196. 10.1016/j.virol.2013.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verma S, Ronsard L, Kapoor R, Banerjea AC. 2013. Genetic characterization of natural variants of Vpu from HIV-1 infected individuals from Northern India and their impact on virus release and cell death. PLoS One 8:e59283. 10.1371/journal.pone.0059283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahn J, Vu T, Novince Z, Guerrero-Santoro J, Rapic-Otrin V, Gronenborn AM. 2010. HIV-1 Vpr loads uracil DNA glycosylase-2 onto DCAF1, a substrate recognition subunit of a cullin 4A-ring E3 ubiquitin ligase for proteasome-dependent degradation. J. Biol. Chem. 285:37333–37341. 10.1074/jbc.M110.133181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schubert U, Anton LC, Bacik I, Cox JH, Bour S, Bennink JR, Orlowski M, Strebel K, Yewdell JW. 1998. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J. Virol. 72:2280–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.daSilva LL, Sougrat R, Burgos PV, Janvier K, Mattera R, Bonifacino JS. 2009. Human immunodeficiency virus type 1 Nef protein targets CD4 to the multivesicular body pathway. J. Virol. 83:6578–6590. 10.1128/JVI.00548-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujita K, Omura S, Silver J. 1997. Rapid degradation of CD4 in cells expressing human immunodeficiency virus type 1 Env and Vpu is blocked by proteasome inhibitors. J. Gen. Virol. 78(Part 3):619–625 [DOI] [PubMed] [Google Scholar]
- 52.Isaacson MK, Ploegh HL. 2009. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5:559–570. 10.1016/j.chom.2009.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goh WC, Rogel ME, Kinsey CM, Michael SF, Fultz PN, Nowak MA, Hahn BH, Emerman M. 1998. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat. Med. 4:65–71. 10.1038/nm0198-065 [DOI] [PubMed] [Google Scholar]
- 54.Vodicka MA, Koepp DM, Silver PA, Emerman M. 1998. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 12:175–185. 10.1101/gad.12.2.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Groschel B, Bushman F. 2005. Cell cycle arrest in G2/M promotes early steps of infection by human immunodeficiency virus. J. Virol. 79:5695–5704. 10.1128/JVI.79.9.5695-5704.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ayyavoo V, Mahboubi A, Mahalingam S, Ramalingam R, Kudchodkar S, Williams WV, Green DR, Weiner DB. 1997. HIV-1 Vpr suppresses immune activation and apoptosis through regulation of nuclear factor kappa B. Nat. Med. 3:1117–1123. 10.1038/nm1097-1117 [DOI] [PubMed] [Google Scholar]
- 57.Muthumani K, Kudchodkar S, Papasavvas E, Montaner LJ, Weiner DB, Ayyavoo V. 2000. HIV-1 Vpr regulates expression of beta chemokines in human primary lymphocytes and macrophages. J. Leukoc. Biol. 68:366–372 [PubMed] [Google Scholar]
- 58.Muthumani K, Hwang DS, Desai BM, Zhang D, Dayes N, Green DR, Weiner DB. 2002. HIV-1 Vpr induces apoptosis through caspase 9 in T cells and peripheral blood mononuclear cells. J. Biol. Chem. 277:37820–37831. 10.1074/jbc.M205313200 [DOI] [PubMed] [Google Scholar]
- 59.Zhu Y, Gelbard HA, Roshal M, Pursell S, Jamieson BD, Planelles V. 2001. Comparison of cell cycle arrest, transactivation, and apoptosis induced by the simian immunodeficiency virus SIVagm and human immunodeficiency virus type 1 vpr genes. J. Virol. 75:3791–3801. 10.1128/JVI.75.8.3791-3801.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morellet N, Bouaziz S, Petitjean P, Roques BP. 2003. NMR structure of the HIV-1 regulatory protein VPR. J. Mol. Biol. 327:215–227. 10.1016/S0022-2836(03)00060-3 [DOI] [PubMed] [Google Scholar]
- 61.Peeters M, Delaporte E. 1999. Genetic diversity of HIV infection worldwide and its consequences. Med. Trop. (Mars.) 59:449–455 (In French.) [PubMed] [Google Scholar]
- 62.Buonaguro L, Tornesello ML, Buonaguro FM. 2007. Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: pathogenetic and therapeutic implications. J. Virol. 81:10209–10219. 10.1128/JVI.00872-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, Gao F, Hahn BH, Kalish ML, Kuiken C, Learn GH, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp PM, Wolinsky S, Korber B. 2000. HIV-1 nomenclature proposal. Science 288:55–56. 10.1126/science.288.5463.55d [DOI] [PubMed] [Google Scholar]
- 64.Bano AS, Sood V, Neogi U, Goel N, Kuttiat VS, Wanchu A, Banerjea AC. 2009. Genetic and functional characterization of human immunodeficiency virus type 1 VprC variants from north India: presence of unique recombinants with mosaic genomes from B, C and D subtypes within the open reading frame of Vpr. J. Gen. Virol. 90:2768–2776. 10.1099/vir.0.011080-0 [DOI] [PubMed] [Google Scholar]
- 65.Bano AS, Gupta N, Sood V, Banerjea AC. 2007. Vpr from HIV-1 subtypes B and C exhibit significant differences in their ability to transactivate LTR-mediated gene expression and also in their ability to promote apoptotic DNA ladder formation. AIDS 21:1832–1834. 10.1097/QAD.0b013e328277f16b [DOI] [PubMed] [Google Scholar]
- 66.Soares EA, Santos AF, Sousa TM, Sprinz E, Martinez AM, Silveira J, Tanuri A, Soares MA. 2007. Differential drug resistance acquisition in HIV-1 of subtypes B and C. PLoS One 2:e730. 10.1371/journal.pone.0000730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Willey RL, Maldarelli F, Martin MA, Strebel K. 1992. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol. 66:7193–7200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma A, Yilmaz A, Marsh K, Cochrane A, Boris-Lawrie K. 2012. Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 8:e1002612. 10.1371/journal.ppat.1002612 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.