ABSTRACT
The UL128 complex of human cytomegalovirus (CMV) is a major determinant of viral entry into epithelial and endothelial cells and a target for vaccine development. The UL/b′ region of rhesus CMV contains several open reading frames, including orthologs of the UL128 complex. We recently showed that the coding content of the rhesus CMV (RhCMV) UL/b′ region predicts acute endothelial tropism and long-term shedding in vivo in the rhesus macaque model of CMV infection. The laboratory-passaged RhCMV 180.92 strain has a truncated UL/b′ region but an intact UL128 complex. To investigate whether the presence of the UL128 complex alone was sufficient to confer endothelial and epithelial tropism in vivo, we investigated tissue dissemination and viral excretion following experimental RhCMV 180.92 inoculation of RhCMV-seronegative rhesus macaques. We show the presence of at least two virus variants in the RhCMV 180.92 infectious virus stock. A rare variant noted for a nontruncated wild-type-virus-like UL/b′ region, rapidly emerged during in vivo replication and showed high-level replication in blood and tissues and excretion in urine and saliva, features similar to those previously reported in naturally occurring wild-type RhCMV infection. In contrast, the predominant truncated version of RhCMV 180.92 showed significantly lower plasma DNAemia and limited tissue dissemination and viral shedding. These data demonstrate that the truncated RhCMV 180.92 variant is attenuated in vivo and suggest that additional UL/b′ genes, besides the UL128 complex, are required for optimal in vivo CMV replication and dissemination.
IMPORTANCE An effective vaccine against human CMV infection will need to target genes that are essential for virus propagation and transmission. The human CMV UL128 complex represents one such candidate antigen since it is essential for endothelial and epithelial cell tropism, and is a target for neutralizing antibodies in CMV-infected individuals. In this study, we used the rhesus macaque animal model of CMV infection to investigate the in vivo function of the UL128 complex. Using experimental infection of rhesus macaques with a rhesus CMV virus variant that contained an intact UL128 complex but was missing several other genes, we show that the presence of the UL128 complex alone is not sufficient for widespread tissue dissemination and virus excretion. These data highlight the importance of in vivo studies in evaluating human CMV gene function and suggest that additional UL/b′ genes are required for optimal CMV dissemination and transmission.
INTRODUCTION
Human cytomegalovirus (HCMV) is a ubiquitous betaherpesvirus that has coevolved with humans over eons (1). The ∼235-kbp genome of HCMV, the largest of all herpesviruses, consists of unique long (UL) and unique short (US) genomic segments, which are each flanked by terminal (TRL and TRS) and internal repeats (IRL and IRS), that together encode ∼165 open reading frames (ORFs) (2). Genes encoding proteins essential for replication in fibroblasts (termed essential genes) comprise ∼40% of the genome, indicating that the importance of the remaining ORF must be best assessed in either other cell types or in vivo. The latter group (termed nonessential genes) of viral proteins include those directing viral tropism to different cell types and those that modulate host cell antigen presentation, activation, signaling, trafficking, and metabolism (3–6). The distinction in HCMV genes encoding proteins essential for infection/replication in fibroblasts and nonfibroblast cell types was emphasized by the initial discovery of differential viral genome stability in different cell types (7).
In vitro propagation of HCMV on cultured fibroblasts can result in the rapid accumulation of point mutations, rearrangements, and/or deletions in the genome that are primarily confined to ORFs within the right end of the UL genome component, termed UL/b′ (2, 7–10). The UL/b′ region contains multiple genes that are dispensable for viral replication in cultured fibroblasts, and laboratory strains of HCMV, extensively passaged on fibroblasts, typically contain one or more nonfunctional UL/b′ genes (3, 4, 11–14). UL/b′ genes are implicated in various encoded functions, including epithelial and endothelial cell tropism (UL128, UL130, and UL131) (15), viral latency (UL133 and UL138) (16, 17), viral envelope structure (UL132) (18), neutrophilic chemoattraction (UL146) (19), and modulation of several host immune responses (UL141, UL142, and UL144) (20–22). The laboratory-adapted Towne and AD169 HCMV strains have deletions of approximately 13- and 15-kbp segments within the UL/b′ region that correspond to a loss of 19 and 22 ORFs, respectively, present in clinical isolates (7). Despite their ability to induce seroconversion, the Towne and AD169 HCMV strains exhibit limited virulence, as evidenced by absence of systemic infection and hematological abnormalities, reduced replication in several cell types, such as endothelial and epithelial cells, and failure to recover virus from the bodily fluids of experimentally inoculated volunteers (15, 23–27).
Rhesus cytomegalovirus (RhCMV) is similar to HCMV in pathogenesis, genomic content and structure, and genetic mutations in fibroblast-adapted strains (28–41). Two laboratory strains of RhCMV, 68-1 and 180.92, have been generated by propagation of clinical isolates on cultured fibroblasts (33, 39, 42). Sequencing of both strains has revealed divergence within the UL/b′ region compared to RhCMV wild-type (WT) isolates (30, 31, 33). RhCMV 68-1 is missing UL128, the second exon of UL130, and three chemokine-like ORF found in wild-type RhCMV. It contains an inversion of UL146, UL147, UL148, and UL132. RhCMV 180.92, on the other hand, has an intact UL128-UL130-UL131 region (UL128 complex) but contains an ∼8.3-kbp deletion within UL/b′ that generated a fusion between UL148 and rh167 with concomitant deletion of ORFs implicated in viral persistence and modulation of host immune response functions (33). As demonstrated with HCMV strains, both RhCMV strains replicate efficiently in fibroblasts in vitro. However, only RhCMV 180.92 carrying the intact UL128 complex and RhCMV 68-1 with repaired UL128 complex can infect rhesus retinal pigment epithelial cells (43).
Several in vitro studies have identified the UL128 complex as a major determinant of HCMV tropism for endothelial and epithelial cells (11, 44, 45). The product of the UL128-UL131 locus forms a pentamer complex with glycoprotein L (gL) and gH to facilitate endocytic entry into epithelial and endothelial cells (14, 46). We have previously shown that variation in the RhCMV UL/b′ region influences cellular tropism, the nature of the inflammatory response, virus dissemination, shedding, and the transmission of RhCMV in vivo, suggesting that an intact UL128 complex and neutrophil chemotaxis-associated ORF are essential for competent RhCMV infection in vivo (32, 47). Since RhCMV 180.92 contains an intact UL128 complex but has other ORFs missing from the UL/b′ region, we aimed here to investigate strain-specific differences in viral parameters of infection in vivo. Intravenous inoculation of RhCMV-seronegative rhesus macaques with RhCMV 180.92 resulted in detectable RhCMV DNA in plasma and fulminant RhCMV disease in an animal coinfected with simian immunodeficiency virus (SIV). However, PCR analysis showed that viral dissemination and tissue distribution was dominated by a WT-like RhCMV variant (RhCMV-WT) originally present at low levels in the virus stock. Dissemination of the truncated RhCMV 180.92 variant was markedly attenuated with limited to no viral shedding in saliva and urine. Even though the wild-type variant accounted for a minor component in the RhCMV 180.92 virus stock, it constituted the majority of virus detected in vivo in plasma, tissues, saliva, and urine, suggesting that an intact UL128 complex in the absence of other genes within the UL/b′ region is not sufficient for optimal virus dissemination in vivo.
MATERIALS AND METHODS
Virus and animal inoculation.
RhCMV 180.92 was originally isolated from the lungs and gastrointestinal tract of SIV-infected macaque (Mm180.92) at the New England Primate Research Center (NEPRC) and passaged in vitro six times on human fibroblasts (MRC-5) and seven times on primary rhesus macaque fibroblasts (33, 39). The virus stock of RhCMV 180.92 used for experimental infection of RhCMV-seronegative rhesus macaques was derived after transfection of virion DNA purified from infected cells into primary rhesus macaque fibroblast lines. Five SIV-negative and one SIV-infected RhCMV-seronegative rhesus macaques were inoculated intravenously with 2 × 106 50% tissue culture infective doses of RhCMV 180.92 virus. All macaques were housed in the specific-pathogen-free colony of the NEPRC and maintained in accordance with federal and institutional guidelines mandated by the Animal Care and Use Committee of Harvard Medical School. The SIV-infected macaque was euthanized 2 months after RhCMV infection due to disseminated RhCMV infection and simian AIDS.
Sample collection and DNA extraction.
Plasma and tissues collected at the time of euthanasia in the SIV-infected macaque, and plasma, saliva, and urine samples collected in the five SIV-negative RhCMV-infected macaques were analyzed for RhCMV DNA by PCR. In addition, DNA was extracted from archived mesenteric lymph node frozen at time of necropsy from animal Mm180.92, from which RhCMV 180.92 was originally isolated. Fixed-volume plasma samples were collected 1 to 6 weeks after RhCMV inoculation, and DNA was extracted from cell-free plasma by using a QIAamp DNA minikit (Qiagen). Saliva and urine samples were collected at 8 or 20 weeks after RhCMV inoculation. Saliva was collected with cotton swabs by using Salivette tubes (Sarstedt) or as mouthwashes using phosphate-buffered saline (PBS). Collected saliva was spun down at 1,500 rpm for 7 min, and the supernatant was concentrated with the Ultracel YM-30 centrifugal filter units (Millipore). DNA was extracted from 200 μl of the extract by using the QIAamp DNA minikit. Free-catch urine samples were centrifuged for 10 min at 1,500 rpm at 4°C to remove debris, and 2 ml of clarified urine was concentrated by using YM-30 centrifugal filter units. DNA was extracted from 140 μl of concentrated urine by using a QIAamp Viral RNA minikit (Qiagen). DNA was extracted from frozen tissues collected at the time of necropsy by using a DNeasy Blood & Tissue kit (Qiagen).
Histopathology.
Five-micron sections of formalin-fixed paraffin-embedded tissues collected at time of necropsy were utilized for microscopic examination. Tissues were fixed in 10% neutral buffered formalin for 7 days prior to paraffin embedding and staining with hematoxylin and eosin (H&E).
Conventional PCR.
Specific primer pairs were designed to differentially amplify either RhCMV 180.92 or RhCMV-WT as shown in Table 1. A forward-primer (5′-AAAGCGGCCTTGGAGTGTGT-3′) and reverse-primer (5′-TCCATCACGATGCCAGTGGT-3′) pair against rhesus β-actin (GenBank accession number NC_007860) amplified a 270-bp product and was used as a positive control for the presence of cellular DNA in the sample. Up to 200 ng of DNA was amplified (5 min at 94°C, followed by 39 cycles of 30 s at 94°C, 1 min at 65°C, 1 min at 72°C [or 5 min at 68°C for larger products], with a final extension for 5 min at 72°C) using GoTaq Green master mix (Promega) according to the manufacturer's instructions.
TABLE 1.
Conventional PCR primers for the detection of RhCMVa
| Primer pair | Primer sequence (5′-3′) and location |
Expected amplicon size |
||||
|---|---|---|---|---|---|---|
| Forward primer | Location | Reverse primer | Location | RhCMV-WT | RhCMV 180.92 | |
| IE1 | TCAGGGTGGCATGGAACTCAT | UL123 | TGCACACTCCCATTACCATGC | UL123 | 171 | 171 |
| UL146 | TCAAACACCGACGGGTTACG | UL146 | AGGGTTCGGAACTACGTTGCAG | UL146 | 172 | Absent |
| ΔCED | GCACAGTAGATGATGATACCCAAGAG | UL132 | GGCTTTCTGCATATCAGTGCTTTGGACGGG | Upstream rh167 | 9,179 | 912 |
| ULb′-1 | GCACAGTAGATGATGATACCCAAGAG | UL132 | GCACCATTCCAAATGGTAACGG | rh161.2 | 3,875 | Absent |
| ULb′-2 | GCACAGTAGATGATGATACCCAAGAG | UL132 | GCAGCGTACTTCGTGTGCTGTACCGAAATC | UL146 | 1,977 | Absent |
| ULb′-3 | GTGCGATGTACACTCGCAGGAAGTCT | Upstream UL146b | CTACATATGGCAAGATGCTCACGGCCTACATGTAC | UL144 | 1,868 | Absent |
| ULb′-4 | GGCTTTCTGCATATCAGTGCTTTGGACGGG | Upstream UL145 | CACCATCGTAATGGTGAGCCTGAC | UL141 | 1,859 | Absent |
Quantitative real-time PCR and plasmid construction.
Primer-probe sets to distinguish RhCMV 180.92 from RhCMV-WT by real-time PCR were designed. A 912-bp PCR product amplified from RhCMV 180.92 and spanning the region of genomic truncation within the UL/b′ region was cloned into pGEM-T Easy Vector (Promega) to generate a plasmid standard for absolute quantification (Fig. 1). Product insertion and sequence homogeneity to parental virus were confirmed by sequencing the inserted amplicon using T7 and SP6 primers (data not shown). The primer-probe set specifically amplifying RhCMV 180.92 (accession number DQ120516), referred to as RhCMV 180.92 primer-probe set, consisted of a forward primer corresponding to residues 166276 to 166299 located within UL148 (5′-TTCAGGTGAATGGAGTGGTTTCGG-3′), a reverse primer corresponding to residues 166383 to 166358 within rh167 (5′-GCTTGACGAGGATGTCTTCGAAGTGT-3′), and a probe spanning a region of truncation corresponding to residues 166329 to 166353 (5′-FAM-TTTGCCCAG-ZEN-GATGGGTGCCGCATCT-IABkFQ-3′) and an amplicon size of 108 bp. A second primer-probe set, referred to as B-region primer-probe set, targets a conserved region within the B segment of UL/b′ (Fig. 1) present in all known strains of RhCMV (RhCMV strains 180.92, 68-1, UCD52, and UCD59) and RhCMV endemic in macaque breeding colonies (30, 33). The forward primer for the B-region primer-probe set corresponded to residues 165948 to 165971 (5′-TTGTCGCAGTAAGAACGGTGGTGA-3′) within UL132, the reverse primer corresponded to residues 166140 to 166117 (5′-TTAATACCGACGCGGGTTCACAGT-3′) within the intron spanning UL132 and UL148, and the probe corresponded to residues 166027 to 166052 (5′-FAM-AGTGCTGTT-ZEN-GGCAGTGGTGGTATTGT-IABkFQ-3′) and an amplicon size of 193 bp. Viral DNA was quantitated in triplicate using Platinum Quantitative PCR SuperMix-UDG (Invitrogen), according to the manufacturer's instructions, with an ABI 7900HT sequence detection system (Applied Biosystems). The PCR run conditions were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The copy numbers of the test samples were determined by extrapolating from a standard curve generated with serial 10-fold dilutions of the plasmid standard (107 copies/reaction to 100 copy/reaction). The sensitivity of detection of the two PCRs was ≤10 DNA copies per run. The total RhCMV copy numbers were calculated based on the B-region primer-probe set amplification, whereas the RhCMV 180.92 copy numbers were based on the 180.92 primer-probe set amplification.
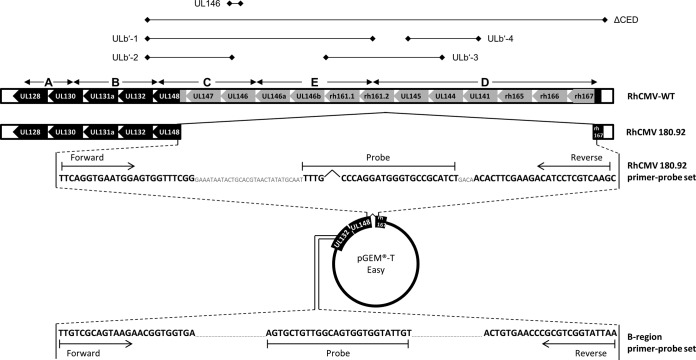
FIG 1.
Molecular characterization of RhCMV 180.92 by variant-specific conventional and quantitative real-time PCR. Schematic representation of the UL/b′ region ORF and alignment of fibroblast-adapted RhCMV 180.92 (accession number DQ120516) to wild-type (WT) RhCMV (EF990255). Conventional PCR primer pair names (Table 1), along with positions of their respective amplicons, are shown above the UL/b′ region alignments. Control pGEM-T Easy plasmid for quantitative real-time PCR and location of the RhCMV 180.92 and B-region primer-probe sets are shown in the bottom. Forward and reverse primers for the RhCMV 180.92 amplicon are located within UL148 and rh167, respectively. Forward and reverse primers for the B-region amplicon are located within UL132 and the intron spanning UL132 and UL148, respectively.
Immunohistochemistry.
Double immunohistochemistry was performed to identify the cell types supporting RhCMV infection, using a polyclonal antiserum for the RhCMV immediate-early protein (IE1) and cell-specific markers for endothelial cells (CD31), epithelial cells (cytokeratin), mesenchymal (fibroblasts) cells (vimentin), and macrophages (CD68). Slides were deparaffinized and rehydrated automatically using the Leica ST5020 Multistainer. They were then incubated in 3% hydrogen peroxide–PBS for 5 min, followed by incubation in Vector antigen unmasking solution (citrate based; catalog no. H-3300) and heating in a microwave for 20 min. Slides were cooled to 25°C for 20 min and then washed in Tris-buffered saline–Tween 20 (TBS-T; TBS–0.05% Tween 20). To reduce nonspecific binding, tissue sections were treated for 10 min with Protein Block Serum-Free (Dako), and then the slides were incubated with primary antibody (1:1,600 in Dako antibody diluent with background reducing component) for 30 min at 25°C. A rabbit polyclonal antiserum to RhCMV IE1 (exon 4) was used for the detection of RhCMV (48). After washing in TBS-T, slides were incubated with secondary antibody (biotinylated goat anti-rabbit, 1:200; Vector) for 30 min at 25°C. The slides were washed again in TBS-T, and avidin-biotin complex–alkaline phosphatase (Vectastain ABC-AP; Vector) was added, followed by vector red substrate (Vector) as a substrate until optimal color developed. Color development was terminated by incubating the slides in distilled water. Tissues were then processed by a similar staining procedure with markers specific for endothelial cells (CD31; 1:50 dilution with overnight incubation at 4°C; Dako), epithelial cells (cytokeratin AE1/AE3; 1:140 dilution with overnight incubation at 4°C; Dako), macrophages (CD68; 1:100 with overnight incubation at 4°C; Dako), or fibroblasts (vimentin; 1:200 for 30 min at room temperature; Dako). After washing in TBS-T, the slides were incubated with biotinylated secondary antibody (biotinylated horse anti-mouse, 1:200; Vector) for 30 h at 25°C and then incubated with ABC-peroxidase (Vectastain Elite ABC; Vector), followed by diaminobenzidine (Dako). After optimal color development, the slides were washed in distilled water to stop additional color development and then counterstained with three short dips in 10% aqueous Mayer's hematoxylin solution (Dako). Finally, tissues were dried overnight and then covered with coverslips. Because microwave-based antigen retrieval is incompatible with CD31 antigen, a pressure cooker antigen retrieval in Trilogy solution (Cell Marque) was performed instead for tissues double labeled for CD31 and RhCMV-IE1.
Statistical analysis.
Mean values for RhCMV 180.92 variant and total RhCMV titers in plasma, urine, and saliva were compared by using a paired Student t test and Prism data analysis software (GraphPad Software).
Nucleotide sequence accession number.
The nucleotide sequence of PCR amplicons from UL147, UL146, UL146a, UL146b, rh161.1, rh161.2 and UL145 of RhCMV 180.92 WT-like variant has been deposited in the GenBank database and assigned accession number KJ882284.
RESULTS
Genomic heterogeneity of the UL/b′ region in RhCMV 180.92 virus stock.
RhCMV strain 180.92 was originally isolated from a naturally CMV-seropositive rhesus macaque at NEPRC that developed AIDS following experimental SIV infection. The genomic sequence of RhCMV 180.92 was obtained from virus stock derived after serial passage in human and primary macaque fibroblast lines as previously described (33). The genome was cloned as a set of overlapping cosmid clones from which the annotated sequence was derived (33). Subsequent to this, purified RhCMV 180.92 virion DNA transfected into primary rhesus macaque fibroblast lines was used to generate infectious virus stock for in vivo infection experiments in rhesus macaques. When a series of PCR primer pairs that amplify different portions of the UL/b′ region were tested against this RhCMV 180.92 virus stock, we unexpectedly detected amplicons corresponding to the region of UL/b′ that was apparently deleted in the RhCMV 180.92 annotation (data not shown). These data suggested that the stock of RhCMV 180.92 was not composed of a single variant. Rather, the data suggested that there was some proportion of the stock containing wild-type RhCMV-like full-length version of UL/b′ (Fig. 1).
To better characterize the UL/b′ region within the stock of 180.92, primer pairs were designed to evaluate the presence or absence of various ORF of the UL/b′ region by conventional PCR (Table 1; Fig. 1, top panel) and by quantitative real-time PCR (Fig. 1, bottom panel). The RhCMV 180.92 primer-probe set designed to specifically amplify the 180.92 viral genome spanning the annotated 8,267-bp deletion in the C-E-D segments of the UL/b′ region enabled quantitation of the genome copy number of the truncated variant, whereas the B-region primer-probe set targeting a conserved region present in all known RhCMV strains enabled the quantitation of total RhCMV genome copies (Fig. 1).
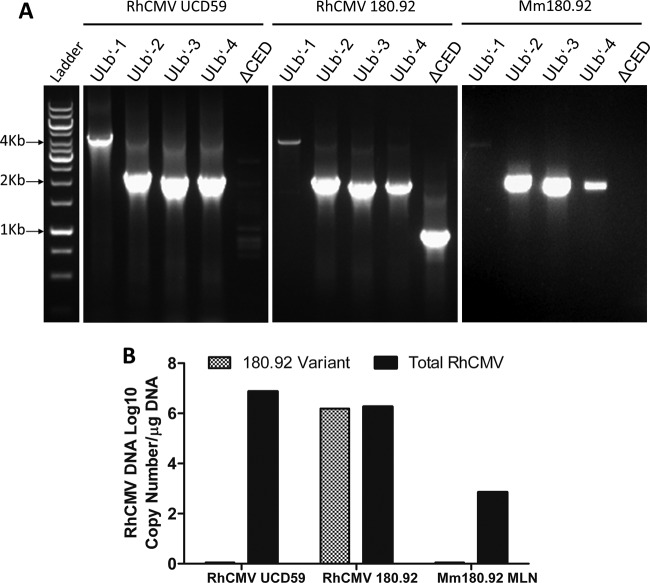
Since the RhCMV 180.92 virus stock was not plaque purified and preliminary analyses suggested a nonhomogeneous viral population, the UL/b′ genetic content in the virus stock used for experimental infections was characterized in parallel to RhCMV strain UCD59, an epitheliotropic strain of RhCMV carrying the complete UL/b′ region (32, 49). Conventional PCR on RhCMV UCD59 (accession number EU130540) using primer pairs ULb′-1 to ULb′-4 (Table 1 and Fig. 1) amplified large segments within the UL/b′ region, a finding consistent with the expected product sizes of RhCMV-WT (Fig. 2A, left panel). The ΔCED primer pair, which should only optimally amplify a 180.92-specific amplicon and not the expected 9,179-bp amplicon of RhCMV-WT (Table 1) under the PCR run conditions, yielded only barely detectable bands of multiple sizes (Fig. 2A, left panel, lane 5). In contrast to RhCMV UCD59, PCR amplification of RhCMV 180.92 with the ΔCED primer pair amplified a 912-bp band size consistent with the expected product of RhCMV 180.92 genome (Fig. 2A, middle panel). In addition, the primer pairs ULb′-1 to ULb′-4 amplified bands in RhCMV 180.92 identical in size to those amplified with RhCMV UCD59 (Fig. 2A, middle panel). Direct sequencing of PCR amplicons from RhCMV 180.92 in the region spanned by the ULb′-1 through ULb′-4 primer pairs revealed a 94 to 100% identity in UL147, UL146, UL146a, UL146b, rh161.1, rh161.2, and UL145 with RhCMV-WT (data not shown). These data confirmed the presence of at least two viral variants in the RhCMV 180.92 virus stock that were distinguished by the presence or absence of the ΔCED region of UL/b′.
FIG 2.
Conventional and real-time PCR amplification of RhCMV-specific segments within the UL/b′ region. DNA extracted from RhCMV UCD59 virus stock, RhCMV 180.92 virus stock, and frozen mesenteric lymph node (MLN) tissue collected at necropsy from the SIV-infected rhesus macaque Mm180.92, the monkey from whom RhCMV 180.92 was originally recovered, were used. (A) Conventional PCR runs with UL/b′ region primers on RhCMV UCD59 (left panel), RhCMV 180.92 (middle panel), and MLN from Mm180.92 (right panel). Segments within the UL/b′ region corresponding to the low-passage-number WT-like isolate RhCMV UCD59 (ULb′-1 to -4) are present in RhCMV strain 180.92 (middle panel). Amplicon specific to the truncated sequence of RhCMV 180.92 (ΔCED) is minimal to completely absent from DNA isolated from RhCMV UCD59 (left panel) and animal Mm180.92 (right panel), respectively. (B) Quantitative real-time PCR showing total and 180.92 truncated variant-specific RhCMV copy numbers per μg of DNA extracted from RhCMV UCD59, RhCMV 180.92, and MLN of Mm180.92. Median values of three replicate wells shown.
These findings were confirmed by real-time PCR (Fig. 2B). Amplification with the conserved B-region primer-probe set amplified ∼107 genome copies/μg of DNA purified from the UCD59 virus stock (mean ± standard deviation [SD] = 7.3 × 106 ± 3.6 × 105 copies/μg of DNA), whereas the RhCMV 180.92 primer-probe set amplified <1 copy/μg of UCD59 DNA (mean ± SD 0.9 ± 0.3 copies/μg of DNA in three positive replicate wells). In contrast, the RhCMV 180.92 virus stock showed strong amplification with both primer-probe sets (Fig. 2B). The copy number with the conserved B-region primer-probe set was comparable to the RhCMV 180.92 primer-probe set (mean ± SD = 1.9 × 106 ± 0.6 × 105 versus 1.5 × 106 ± 0.9 × 104 copies/μg of DNA, respectively), indicating that the truncated 180.92 variant was predominant in the RhCMV 180.92 virus stock (Fig. 2B).
The availability of archived frozen tissues from rhesus macaque Mm180.92, from which RhCMV 180.92 virus was originally recovered, enabled evaluation of any UL/b′-related size polymorphisms within virus present in infected tissue. When a similar PCR was performed on DNA extracted from mesenteric lymph node using amplification conditions identical to those used for amplification on DNA prepared from viral stocks, primer pairs ULb′-2 to ULb′-4 resulted in amplifications consistent with RhCMV UCD59 (Fig. 2A, right panel). A faint 3.8-kb band of predicted size was seen with the primer pair ULb′-1, but there was no detectable amplification with the ΔCED primers (Fig. 2A, right panel). Real-time PCR did not reveal any amplification with the RhCMV 180.92 primer-probe set, thus confirming the absence of RhCMV 180.92 variant in the mesenteric lymph node (Fig. 2B) and suggesting that the in vivo RhCMV virus did not have a truncated UL/b′ region. In all, these data confirmed that the RhCMV 180.92 truncated variant was generated during the in vitro propagation on fibroblasts, as has been demonstrated for HCMV. As a result, the RhCMV 180.92 virus stock was heterogeneous and contained a mixed population of the annotated and truncated 180.92 variant and RhCMV-WT variants, with RhCMV 180.92 being the dominant variant in terms of copy number.
Attenuation and limited dissemination of RhCMV 180.92 in vivo.
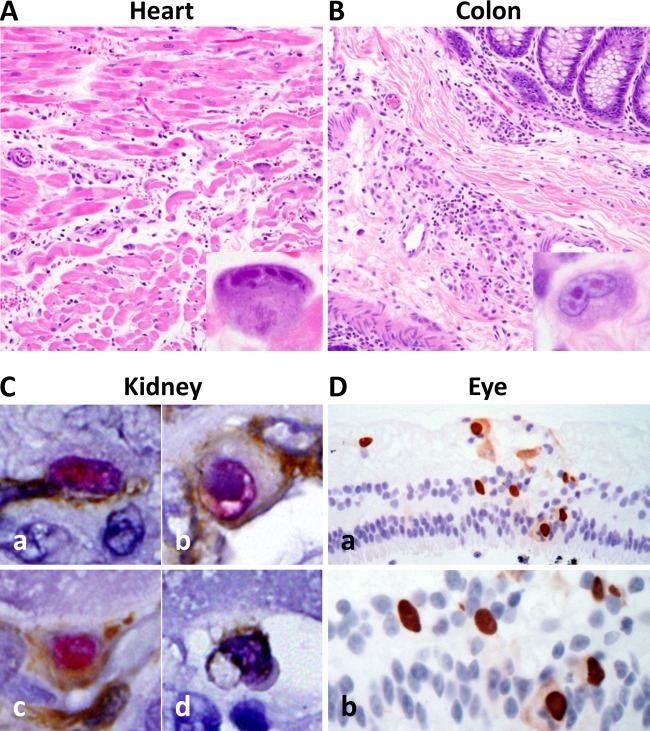
In HCMV, genetic polymorphisms have been implicated in strain-specific outcomes of infection (50–52). To investigate the clinical significance of genetic divergence within RhCMV strain 180.92, the kinetics and pathogenicity of viral dissemination were analyzed in one RhCMV-seronegative SIV-infected macaque that was inoculated with the mixed RhCMV 180.92 stock 17 weeks following SIV infection. At 8 weeks after RhCMV inoculation, the animal was euthanized due to clinical signs of AIDS. At necropsy, histopathological examination revealed cytomegalic cells containing pathognomonic owl-eye CMV intranuclear inclusion bodies in multiple tissues, including heart, lung, colon, liver, tongue, spleen, and kidney tissues. These inclusion bodies were occasionally associated with mild to moderate inflammation predominantly consisting of mononuclear cells, specifically within heart and colon tissues (Fig. 3A and B).
FIG 3.
Histopathologic analysis of CMV localization in tissues of a SIV-infected CMV-seronegative rhesus macaque experimentally inoculated with RhCMV 180.92. Photomicrographs of H&E-stained and immunohistochemically stained tissue sections from a rhesus macaque with simian AIDS and disseminated CMV after SIV and RhCMV 180.92 coinfection. (A and B) RhCMV-induced inflammation in the heart (A) and colon (B) characterized by mild edema and tissue infiltration by lymphocytes and histiocytes admixed with cytomegalic and karyomegalic cells containing multiple large round to oval magenta-colored CMV intranuclear inclusion bodies surrounded by clear halo and chromatin margination (magnification, ×200; inset magnification, ×600). (C) Immunohistochemical analysis of kidney tissue showing distribution of RhCMV 180.92 infection in a wide range of cell types identified by double immunostaining of CMV-IE1 (magenta) with CD31 for endothelial cell identification (brown [a]), cytokeratin for epithelial cell identification (brown [b]), vimentin for fibroblast identification (brown [c]), and CD68 for macrophage identification (brown [d]). Magnification, ×600. (D) RhCMV-induced retinitis characterized by the presence of numerous immunohistochemically positive cells for CMV-IE1 (brown) associated with focal retinal degeneration and disruption of retinal internal and external nuclear layers shown at low (×200 [a]) and high (×400 [b]) magnifications.
Viral localization using immunohistochemical staining of RhCMV IE1 protein expanded the list of infected organs to include the sacral spinal cord, the kidneys, the right and left eyes, the axillary and mesenteric lymph nodes, and the jejunum (Fig. 3C-D and data not shown). RhCMV-induced retinitis was characterized by focal retinal degeneration, disruption of the retinal nuclear layers and numerous CMV IE1-positive cells detected by immunohistochemistry (Fig. 3D). The detection of RhCMV in the eyes of an SIV-infected animal is especially noteworthy because, to our knowledge, this is the first demonstration of RhCMV associated with retinitis in the context of SIV-induced AIDS. Viral colocalization studies in kidney tissue identified RhCMV IE1 protein expression within multiple cell types, including endothelial cells (CD31), epithelial cells (cytokeratin), fibroblasts (vimentin), and macrophages (CD68) (Fig. 3Ca to d, respectively). These findings indicated that experimental inoculation of the RhCMV 180.92 virus stock induced disseminated CMV disease that recapitulated the cellular tropism of wild-type RhCMV (38, 47).
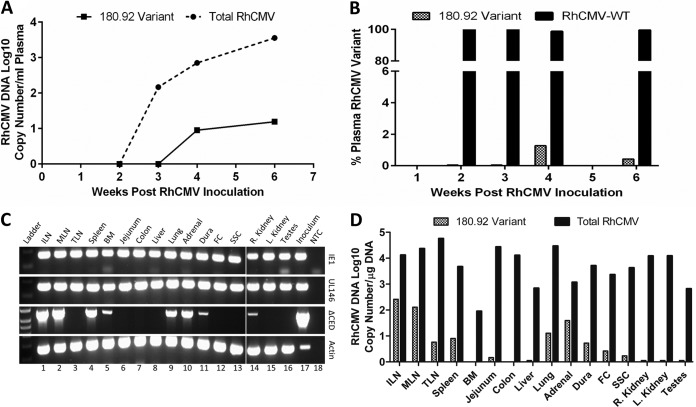
We next used variant-specific PCR analysis of plasma and tissues to determine the relative contribution of RhCMV 180.92 and RhCMV-WT virus toward in vivo pathogenicity. RhCMV 180.92 inoculation induced viremia as indicated by the exponential increase in plasma RhCMV DNA copy numbers starting at 3 weeks postinfection (Fig. 4A). At all time points, the total RhCMV copy number measured with the B-region primer-probe set exceeded that obtained with the 180.92 primer-probe set by more than one to two logs (Fig. 4A). Based on quantitation by real-time PCR, the truncated variant accounted for <5% of the total circulating RhCMV strains in the first 6 weeks postinfection (Fig. 4B), suggesting that its in vivo replication capacity was markedly impaired compared to WT-like RhCMV.
FIG 4.
Viral load and tissue distribution of RhCMV in a SIV-infected CMV-seronegative rhesus macaque experimentally inoculated with RhCMV 180.92. Real-time and conventional PCR data shown. (A) Median DNA copy numbers of total RhCMV plasma virus burden (solid line) and RhCMV 180.92 variant (dashed line). (B) Percentage of RhCMV 180.92 variant copy number (hashed column) compared to percentage of WT-like RhCMV variant (RhCMV-WT) copy numbers (solid column), as measured by variant-specific real-time PCR at 2, 3, 4, and 6 weeks after RhCMV 180.92 experimental inoculation. (C) Genomic DNA extracted from various tissues was assayed to differentiate total RhCMV (IE1), RhCMV-WT variant (UL146), and RhCMV 180.92 variant (ΔCED) using a variant-specific conventional PCR assay. Lane 18 is nontemplate control (water [NTC]) serving as a negative control, and DNA extracted from RhCMV 180.92 virus stock (Inoculum, lane 17) and β-actin amplifications (Actin) served as positive controls. (D) Quantitative analysis of RhCMV 180.92 variant DNA copy number/μg of DNA as measured by real-time PCR of 100 ng of tissue genomic DNA. Median values determined for three replicate wells are shown. ILN, inguinal lymph node; MLN, mesenteric lymph node; TLN, tracheobronchial lymph node; BM, bone marrow; FC, frontal cortex; SSC, sacral spinal cord.
An essential step in the pathogenesis of CMV is the ability of progeny virions to disseminate to organs essential for viral shedding, such as salivary glands and the genitourinary system. To further dissect the spreading potential of each RhCMV variant to tissues, variant-specific PCR analysis was also performed on DNA extracted from tissues frozen at the time of necropsy (Fig. 4C and D). RhCMV was detected in every major organ system tested, with viral loads ranging between 90 and 5.7 × 104 copies per μg of tissue genomic DNA (Fig. 4D). Furthermore, comparative analysis showed the same attenuated pattern of truncated 180.92 distribution in tissues as was observed in plasma (Fig. 4C and D). RhCMV DNA was detected in 8 of 16 tissues examined (Fig. 4C, third row). However, quantitation by real-time PCR showed that the RhCMV-WT variant was present at more than 1-log-higher levels compared to the RhCMV 180.92 truncated variant in all tissues (Fig. 4D). The RhCMV 180.92 truncated variant was primarily detected in secondary lymphoid organs (lymph nodes and spleen), lung, and adrenal gland. In contrast, it was present at levels below the limit of detection of the real-time PCR assay in tissues such as liver, colon, kidneys, and testes (Fig. 4D). The detection of the truncated variant in the secondary lymphoid organs may reflect some type of viral sequestration in which one strain outcompetes another strain in some tissues during a mixed infection. In the case of 180.92, it appears that the full-length UL/b′ variant rapidly outcompeted the truncated 180.92 variant, albeit to a lesser extent in some tissues. The mechanism of this is unknown, but we have observed this phenomenon previously with RhCMV 68.1, another strain with an incomplete UL/b′ coding content. Inoculation of naive macaques with RhCMV 68.1 resulted in virus dissemination to multiple tissues throughout the body when just this single strain was used (38). However, when animals were inoculated with a mixture of three strains (68-1 and two epitheliotropic strains [UCD52 and UCD59]), detection of 68-1 in tissues was restricted to inguinal lymph nodes, whereas UCD52 and UCD59 were widely disseminated (32).
Limited shedding of RhCMV 180.92 in urine and saliva.
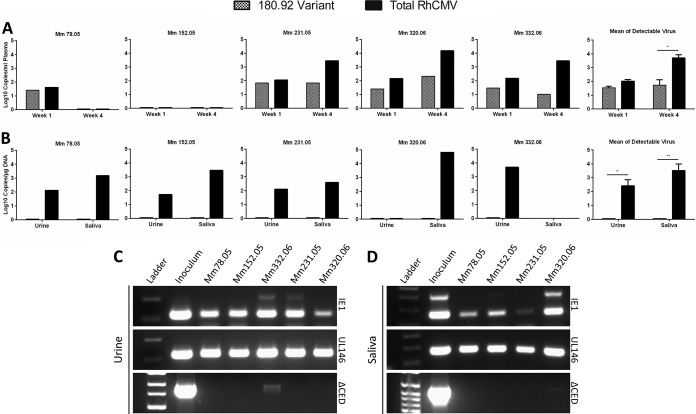
RhCMV 180.92 dissemination and shedding was also examined in the plasma, saliva, and urine of five SIV naive RhCMV-seronegative rhesus macaques inoculated with RhCMV 180.92 via the intravenous route. Four of five animals showed detectable plasma DNAemia of both RhCMV 180.92 truncated and RhCMV-WT variants in the first 4 weeks postinfection (Fig. 5A). However, the RhCMV 180.92 truncated variant was detected at 0.2- to 2.4-log-lower levels compared to total circulating RhCMV, and this difference was statistically significant at 4 weeks postinfection (Fig. 5A). The ability of cytomegalovirus to shed in body fluids is a crucial step in its natural history, and genes in the UL/b′ region are implicated in virus shedding. Hence, the shedding patterns of the RhCMV 180.92 truncated and WT-like variants were measured in urine and saliva at 8 to 20 weeks post RhCMV inoculation at available time points of maximal excretion. All animals shed high titers of WT-like RhCMV in one or both bodily secretions (Fig. 5B). In marked contrast to plasma viremia, minimal to no RhCMV 180.92 variant was detectable in the saliva and urine of all animals (Fig. 5B to D).
FIG 5.
Kinetics of plasma CMV DNA load and patterns of shedding of RhCMV 180.92 by variant-specific conventional and real-time PCR in five SIV-negative CMV-seronegative rhesus macaques experimentally inoculated with RhCMV 180.92. (A) DNA copy number of total RhCMV plasma virus burden (solid column) and RhCMV 180.92 variant (hashed column) measured by variant-specific real-time PCR at 1 and 4 weeks after RhCMV inoculation. Individual monkey data represent the median values obtained for three replicates. The graph at the far right shows a comparison of the mean total and variant RhCMV plasma copy number + the standard errors of the mean for animals with detectable CMV loads at 1 and 4 weeks after RhCMV inoculation. (B) Shedding of RhCMV 180.92 variant (hashed column) compared to total RhCMV (solid column) in urine and saliva samples collected at 8 weeks (Mm231.05, Mm320.06, and Mm332.06) or 20 weeks (Mm78.05 and Mm152.05) after RhCMV inoculation and measured by variant-specific real-time PCR. Individual monkey data represent the median values obtained for three replicates. The graph at the far right compares the mean total and variant copy numbers + the standard errors of the mean in urine and saliva of animals with detectable CMV load. A significant difference in CMV load was determined by using a paired Student t test. *, P < 0.05; **, P < 0.01. (C and D) RhCMV shedding patterns in urine (C) and saliva (D) samples were assayed to differentiate total RhCMV (IE1), RhCMV-WT variant (UL146), and RhCMV 180.92 variant (ΔCED) contributions by using a variant-specific conventional PCR assay. DNA extracted from RhCMV 180.92 virus stock (Inoculum) served as a positive control.
DISCUSSION
Although viral proteins encoded by genes located in the UL/b′ region of HCMV are considered important for viral persistence and dissemination, the preponderance of these observations is based on in vitro studies that do not necessarily reflect the complexity of HCMV infection in its human host. The availability of the rhesus macaque model of HCMV infection and two fully sequenced, laboratory-passaged strains of RhCMV with defined genetic deletions in RhCMV UL/b′ (31, 33) has led to studies addressing how coding content differences between these strains and a prototypical wild-type strain of RhCMV encoding a full-length UL/b′ alter viral replication and dissemination in vivo. Using experimental infection with WT-like RhCMV and the laboratory-passaged RhCMV strain 68-1 in rhesus macaques, we recently showed the importance of the UL128 complex and the chemokine-like ORF located within the UL/b′ region on acute and persistent parameters of infection, particularly in regard to cell types acutely supporting RhCMV infection and the long-term shedding of RhCMV in bodily fluids (32, 47). Another laboratory-passaged strain, RhCMV 180.92, is distinguished from RhCMV 68-1 in retaining an intact UL128 complex, while other UL/b′-encoded ORF implicated in viral persistence, immune evasion, and neutrophilic chemotaxis were apparently deleted during serial passage in fibroblasts (33). In the present study, we investigated virus dissemination and shedding in vivo following RhCMV 180.92 infection to determine how an intact UL128 complex within an otherwise truncated UL/b′ region affects RhCMV growth and replication in a primate host.
RhCMV 180.92 inoculation of RhCMV-seronegative macaques led to persistent infection and virus shedding in SIV naive macaques, and disseminated RhCMV disease in one SIV-infected macaque. Using a real-time PCR strategy that allowed specific quantitation of the truncated 180.92 virus, we show that the RhCMV 180.92 virus inoculum was heterogeneous in genomic content. The majority of RhCMV genomes in the viral inoculum consisted of the annotated version of the 180.92 genome, while the remaining genomes consisted of a WT-like variant(s) with an intact UL/b′. A previous study noted that RhCMV 180.92 replicates efficiently in cultured fibroblasts but is impaired for attachment and entry in a rhesus retinal pigment epithelial cell line, compared to a variant of RhCMV 68-1 engineered to express a functional UL128-UL131 complex (43). Because the heterogeneity of the 180.92 stock was not recognized at the time this particular study was done, no genetic analysis of the progeny virions was performed to directly compare the relative in vitro replication parameters of the different variants. The presence of both the WT-like and truncated 180.92 variants in different tissues indicates that both variants are replication competent in vivo. However, quantitative PCR analysis demonstrated that the RhCMV 180.92 truncated variant is significantly attenuated in vivo for tissue dissemination within the host and for virus shedding. This was evidenced by (i) low plasma RhCMV DNA copy numbers, (ii) limited dissemination and infection of multiple organs, (iii) low RhCMV DNA copy numbers in infected organs and, most remarkably, (iv) minimal to no shedding of virus progeny in saliva and urine of the truncated 180.92 variant. In contrast, the WT-like variant(s), which constituted a minor fraction of the RhCMV 180.92 virus stock, emerged to dominate the RhCMV population detected in plasma, tissues, and salivary and urinary excretions. Thus, despite the presence of an intact UL128-UL130-UL131 coding region, the truncated 180.92 is profoundly attenuated in vivo, suggesting that loss of other ORFs in UL/b′ results in a loss of viral fitness. Of interest, the patterns of dissemination of the WT-like and the truncated 180.92 variants reflect the distinct patterns of viral shedding following inoculation of seronegative macaques with UCD52, UCD59, and 68-1 (32). In particular, only RhCMV variants containing a full complement of UL/b′-encoded genes are capable of disseminating from the sites of inoculation to tissues where progeny virions can be persistently shed in bodily fluids. In this regard, the results with RhCMV reflect the results from humans inoculated with different HCMV strains.
Comparison of clinical patterns of infection in experimentally inoculated volunteers reveals marked phenotypic distinctions between HCMV strains that differ primarily in UL/b′ coding content, including different viral patterns of shedding in bodily fluids. While shedding is profoundly restricted in individuals inoculated with the tissue culture-adapted AD169 and Towne strains, people inoculated with the low-passage-number, clinical Toledo strain exhibit shedding profiles that are more similar to those naturally infected with wild-type HCMV (23–25, 53–55). In this regard, it is noteworthy that the UL/b′ region of the Toledo strain encodes the tumor necrosis factor receptor homolog UL144 and the alpha chemokine genes UL146 and UL147 but has an inverted segment which renders the UL128 gene nonfunctional (10, 56). This suggests that a functional UL128 complex may not be an absolute requirement for viral shedding. Taken together, the results in humans and rhesus monkeys demonstrate that shedding in bodily fluids, a central element in primate CMV natural history, requires the functional integrity of UL/b′ coding capacity. A clinical ramification of this conclusion is that vaccine studies directed against UL/b′-encoded proteins should be especially effective at limiting the potential for HCMV to disseminate from primary sites of mucosal infection to distal sites throughout the body, such as the maternal-fetal interface in the case of vaccinations designed to prevent congenital HCMV infection.
Given the extent of the deletion within the UL/b′ region of the truncated 180.92 variant, there are multiple ORFs that, when absent, could individually or collectively attenuate dissemination. Based on the known functions of the HCMV orthologs of the deleted ORF, potential ORFs that affect dissemination include those involved in modulation of host cell activation (UL141 and UL142) (21, 22, 57, 58), signaling (UL144) (59–61), and trafficking (UL146) (19). In addition, the UL132 ORF, which is involved in virion assembly (UL132) (18), could be another dissemination determinant. Although the coding content of UL132 is extant in the truncated 180.92 variant, there is a functional absence of pUL132 due to deletion of its cognate promoter upstream of UL147 (Y. Yue and P. A. Barry, unpublished data).
Similar to published studies of RhCMV-WT reactivation in SIV-infected rhesus macaques (41, 47, 62), RhCMV 180.92 induced histopathological lesions in multiple organs. However, unique to our study is the identification of RhCMV-infected retinal cells detected by IHC in both eyes of an SIV-infected monkey (Fig. 3D), an observation that has not been documented to date. This lesion was associated with bilateral iridal edema, indicating a concurrent ocular vascular damage. HCMV-retinitis was one of the most common lesions of AIDS patients before the use of highly active antiretroviral therapy (63). HCMV-retinitis is considered a late-onset lesion developing over 12 months postimmunosuppression (64). In contrast, the ocular lesion in our study developed 2 months after RhCMV infection in the immunosuppressed monkey. Although the reason for accelerated ocular lesion in our case is unclear, it is possible that progressive SIV infection combined with RhCMV genetic variation in unknown virulence factors might have enhanced access of RhCMV 180.92 to ocular tissues and accelerated the onset of retinitis (65).
Substantive progress has been achieved in two clinical trials involving vaccination of either pregnant women or transplant recipients with a recombinant glycoprotein B (gB) protein (66, 67). However, protection against natural infection was limited, suggesting that immune response targeting a single HCMV protein might be insufficient for long-term prevention of primary infection. The inclusion of additional antigens in future vaccine candidates, such as those within the UL/b′ region involved in cellular tropism and modulation of the host immune responses, could augment gB-mediated protective immunity. With the preservation of a functionally intact UL128 complex and loss of immune modulation genes, RhCMV 180.92 could be a potentially valuable vaccine candidate to evaluate host immune responses against the UL128 complex. However, due to the mixed nature of the virus stock, in vivo evaluation of a plaque-purified single-strain truncated RhCMV 180.92 variant is required to test its immunogenicity and confirm disease attenuation.
ACKNOWLEDGMENTS
We thank the histopathology core and veterinary staff at the NEPRC for expert technical assistance.
This study was supported by funding from National Institutes of Health grants P51 OD0111103 and T32 OD011064 to NEPRC, R01 AI43890 to A.K., R01 AI49342 to P.A.B., P51 OD011107 to the California National Primate Research Center, and the Margaret Deterding Infectious Disease Research Support Fund to P.A.B.
Footnotes
Published ahead of print 4 June 2014
REFERENCES
- 1.Cannon MJ, Schmid DS, Hyde TB. 2010. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol. 20:202–213. 10.1002/rmv.655 [DOI] [PubMed] [Google Scholar]
- 2.Fields BN, Knipe DM, Howley PM. 2007. Fields virology, 5th ed. Wolters Kluwer Health/Lippincott/The Williams & Wilkins Co, Philadelphia, PA [Google Scholar]
- 3.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396–12401. 10.1073/pnas.1635160100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223–14228. 10.1073/pnas.2334032100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, Shenk TE. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 100:14976–14981. 10.1073/pnas.2136652100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer D, Emery VC, Griffiths PD, Sinzger C, McSharry BP, Wilkinson GW, Davison AJ. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301–1312. 10.1099/vir.0.79888-0 [DOI] [PubMed] [Google Scholar]
- 7.Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weststrate MW, Geelen JL, Wertheim PM, van der Noordaa J. 1983. Comparison of the physical maps of the DNAs of two cytomegalovirus strains. J. Gen. Virol. 64(Pt 1):47–55 [DOI] [PubMed] [Google Scholar]
- 9.Takekoshi M, Ihara S, Tanaka S, Maeda-Takekoshi F, Watanabe Y. 1987. A new human cytomegalovirus isolate has an invertible subsegment within its L component producing eight genome isomers. J. Gen. Virol. 68(Pt 3):765–776 [DOI] [PubMed] [Google Scholar]
- 10.Prichard MN, Penfold ME, Duke GM, Spaete RR, Kemble GW. 2001. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev. Med. Virol. 11:191–200. 10.1002/rmv.315 [DOI] [PubMed] [Google Scholar]
- 11.Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 78:10023–10033. 10.1128/JVI.78.18.10023-10033.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown JM, Kaneshima H, Mocarski ES. 1995. Dramatic interstrain differences in the replication of human cytomegalovirus in SCID-hu mice. J. Infect. Dis. 171:1599–1603. 10.1093/infdis/171.6.1599 [DOI] [PubMed] [Google Scholar]
- 13.Gerna G, Percivalle E, Lilleri D, Lozza L, Fornara C, Hahn G, Baldanti F, Revello MG. 2005. Dendritic-cell infection by human cytomegalovirus is restricted to strains carrying functional UL131-128 genes and mediates efficient viral antigen presentation to CD8+ T cells. J. Gen. Virol. 86:275–284. 10.1099/vir.0.80474-0 [DOI] [PubMed] [Google Scholar]
- 14.Wang D, Shenk T. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. U. S. A. 102:18153–18158. 10.1073/pnas.0509201102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinzger C, Digel M, Jahn G. 2008. Cytomegalovirus cell tropism. Curr. Top. Microbiol. Immunol. 325:63–83. 10.1007/978-3-540-77349-8_4 [DOI] [PubMed] [Google Scholar]
- 16.Goodrum F, Reeves M, Sinclair J, High K, Shenk T. 2007. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood 110:937–945. 10.1182/blood-2007-01-070078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umashankar M, Petrucelli A, Cicchini L, Caposio P, Kreklywich CN, Rak M, Bughio F, Goldman DC, Hamlin KL, Nelson JA, Fleming WH, Streblow DN, Goodrum F. 2011. A novel human cytomegalovirus locus modulates cell type-specific outcomes of infection. PLoS Pathog. 7:e1002444. 10.1371/journal.ppat.1002444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spaderna S, Kropff B, Kodel Y, Shen S, Coley S, Lu S, Britt W, Mach M. 2005. Deletion of gpUL132, a structural component of human cytomegalovirus, results in impaired virus replication in fibroblasts. J. Virol. 79:11837–11847. 10.1128/JVI.79.18.11837-11847.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penfold ME, Dairaghi DJ, Duke GM, Saederup N, Mocarski ES, Kemble GW, Schall TJ. 1999. Cytomegalovirus encodes a potent alpha chemokine. Proc. Natl. Acad. Sci. U. S. A. 96:9839–9844. 10.1073/pnas.96.17.9839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poole E, King CA, Sinclair JH, Alcami A. 2006. The UL144 gene product of human cytomegalovirus activates NFκB via a TRAF6-dependent mechanism. EMBO J. 25:4390–4399. 10.1038/sj.emboj.7601287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prod'homme V, Sugrue DM, Stanton RJ, Nomoto A, Davies J, Rickards CR, Cochrane D, Moore M, Wilkinson GW, Tomasec P. 2010. Human cytomegalovirus UL141 promotes efficient downregulation of the natural killer cell activating ligand CD112. J. Gen. Virol. 91:2034–2039. 10.1099/vir.0.021931-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wills MR, Ashiru O, Reeves MB, Okecha G, Trowsdale J, Tomasec P, Wilkinson GW, Sinclair J, Sissons JG. 2005. Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J. Immunol. 175:7457–7465. 10.4049/jimmunol.175.11.7457 [DOI] [PubMed] [Google Scholar]
- 23.Plotkin SA, Farquhar J, Horberger E. 1976. Clinical trials of immunization with the Towne 125 strain of human cytomegalovirus. J. Infect. Dis. 134:470–475. 10.1093/infdis/134.5.470 [DOI] [PubMed] [Google Scholar]
- 24.Elek SD, Stern H. 1974. Development of a vaccine against mental retardation caused by cytomegalovirus infection in utero. Lancet i:1–5 [DOI] [PubMed] [Google Scholar]
- 25.Quinnan GV, Jr, Delery M, Rook AH, Frederick WR, Epstein JS, Manischewitz JF, Jackson L, Ramsey KM, Mittal K, Plotkin SA, et al. 1984. Comparative virulence and immunogenicity of the Towne strain and a nonattenuated strain of cytomegalovirus. Ann. Intern. Med. 101:478–483. 10.7326/0003-4819-101-4-478 [DOI] [PubMed] [Google Scholar]
- 26.Plotkin SA, Smiley ML, Friedman HM, Starr SE, Fleisher GR, Wlodaver C, Dafoe DC, Friedman AD, Grossman RA, Barker CF. 1984. Towne-vaccine-induced prevention of cytomegalovirus disease after renal transplants. Lancet i:528–530 [DOI] [PubMed] [Google Scholar]
- 27.Plotkin SA, Huang ES. 1985. Cytomegalovirus vaccine virus (Towne strain) does not induce latency. J. Infect. Dis. 152:395–397. 10.1093/infdis/152.2.395 [DOI] [PubMed] [Google Scholar]
- 28.Yue Y, Kaur A, Zhou SS, Barry PA. 2006. Characterization and immunological analysis of the rhesus cytomegalovirus homologue (Rh112) of the human cytomegalovirus UL83 lower matrix phosphoprotein (pp65). J. Gen. Virol. 87:777–787. 10.1099/vir.0.81516-0 [DOI] [PubMed] [Google Scholar]
- 29.Lilja AE, Chang WL, Barry PA, Becerra SP, Shenk TE. 2008. Functional genetic analysis of rhesus cytomegalovirus: Rh01 is an epithelial cell tropism factor. J. Virol. 82:2170–2181. 10.1128/JVI.02316-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oxford KL, Eberhardt MK, Yang KW, Strelow L, Kelly S, Zhou SS, Barry PA. 2008. Protein coding content of the UL/b′ region of wild-type rhesus cytomegalovirus. Virology 373:181–188. 10.1016/j.virol.2007.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW. 2003. Complete sequence and genomic analysis of rhesus cytomegalovirus. J. Virol. 77:6620–6636. 10.1128/JVI.77.12.6620-6636.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oxford KL, Strelow L, Yue Y, Chang WL, Schmidt KA, Diamond DJ, Barry PA. 2011. Open reading frames carried on UL/b′ are implicated in shedding and horizontal transmission of rhesus cytomegalovirus in rhesus monkeys. J. Virol. 85:5105–5114. 10.1128/JVI.02631-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivailler P, Kaur A, Johnson RP, Wang F. 2006. Genomic sequence of rhesus cytomegalovirus 180.92: insights into the coding potential of rhesus cytomegalovirus. J. Virol. 80:4179–4182. 10.1128/JVI.80.8.4179-4182.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowman JJ, Lacayo JC, Burbelo P, Fischer ER, Cohen JI. 2011. Rhesus and human cytomegalovirus glycoprotein L are required for infection and cell-to-cell spread of virus but cannot complement each other. J. Virol. 85:2089–2099. 10.1128/JVI.01970-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue Y, Barry PA. 2008. Rhesus cytomegalovirus a nonhuman primate model for the study of human cytomegalovirus. Adv. Virus Res. 72:207–226. 10.1016/S0065-3527(08)00405-3 [DOI] [PubMed] [Google Scholar]
- 36.Baroncelli S, Barry PA, Capitanio JP, Lerche NW, Otsyula M, Mendoza SP. 1997. Cytomegalovirus and simian immunodeficiency virus coinfection: longitudinal study of antibody responses and disease progression. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 15:5–15. 10.1097/00042560-199705010-00002 [DOI] [PubMed] [Google Scholar]
- 37.Sequar G, Britt WJ, Lakeman FD, Lockridge KM, Tarara RP, Canfield DR, Zhou SS, Gardner MB, Barry PA. 2002. Experimental coinfection of rhesus macaques with rhesus cytomegalovirus and simian immunodeficiency virus: pathogenesis. J. Virol. 76:7661–7671. 10.1128/JVI.76.15.7661-7671.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lockridge KM, Sequar G, Zhou SS, Yue Y, Mandell CP, Barry PA. 1999. Pathogenesis of experimental rhesus cytomegalovirus infection. J. Virol. 73:9576–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaur A, Daniel MD, Hempel D, Lee-Parritz D, Hirsch MS, Johnson RP. 1996. Cytotoxic T-lymphocyte responses to cytomegalovirus in normal and simian immunodeficiency virus-infected rhesus macaques. J. Virol. 70:7725–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaur A, Hale CL, Noren B, Kassis N, Simon MA, Johnson RP. 2002. Decreased frequency of cytomegalovirus (CMV)-specific CD4+ T lymphocytes in simian immunodeficiency virus-infected rhesus macaques: inverse relationship with CMV viremia. J. Virol. 76:3646–3658. 10.1128/JVI.76.8.3646-3658.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaur A, Kassis N, Hale CL, Simon M, Elliott M, Gomez-Yafal A, Lifson JD, Desrosiers RC, Wang F, Barry P, Mach M, Johnson RP. 2003. Direct relationship between suppression of virus-specific immunity and emergence of cytomegalovirus disease in simian AIDS. J. Virol. 77:5749–5758. 10.1128/JVI.77.10.5749-5758.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asher DM, Gibbs CJ, Jr, Lang DJ, Gajdusek DC, Chanock RM. 1974. Persistent shedding of cytomegalovirus in the urine of healthy rhesus monkeys. Proc. Soc. Exp. Biol. Med. 145:794–801. 10.3181/00379727-145-37897 [DOI] [PubMed] [Google Scholar]
- 43.Lilja AE, Shenk T. 2008. Efficient replication of rhesus cytomegalovirus variants in multiple rhesus and human cell types. Proc. Natl. Acad. Sci. U. S. A. 105:19950–19955. 10.1073/pnas.0811063106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patrone M, Secchi M, Fiorina L, Ierardi M, Milanesi G, Gallina A. 2005. Human cytomegalovirus UL130 protein promotes endothelial cell infection through a producer cell modification of the virion. J. Virol. 79:8361–8373. 10.1128/JVI.79.13.8361-8373.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang D, Shenk T. 2005. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J. Virol. 79:10330–10338. 10.1128/JVI.79.16.10330-10338.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adler B, Scrivano L, Ruzcics Z, Rupp B, Sinzger C, Koszinowski U. 2006. Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J. Gen. Virol. 87:2451–2460. 10.1099/vir.0.81921-0 [DOI] [PubMed] [Google Scholar]
- 47.Assaf BT, Mansfield KG, Westmoreland SV, Kaur A, Oxford KL, Diamond DJ, Barry PA. 2012. Patterns of acute rhesus cytomegalovirus (RhCMV) infection predict long-term RhCMV infection. J. Virol. 86:6354–6357. 10.1128/JVI.00607-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barry PA, Alcendor DJ, Power MD, Kerr H, Luciw PA. 1996. Nucleotide sequence and molecular analysis of the rhesus cytomegalovirus immediate-early gene and the UL121-117 open reading frames. Virology 215:61–72. 10.1006/viro.1996.0007 [DOI] [PubMed] [Google Scholar]
- 49.Alcendor DJ, Barry PA, Pratt-Lowe E, Luciw PA. 1993. Analysis of the rhesus cytomegalovirus immediate-early gene promoter. Virology 194:815–821. 10.1006/viro.1993.1323 [DOI] [PubMed] [Google Scholar]
- 50.Coaquette A, Bourgeois A, Dirand C, Varin A, Chen W, Herbein G. 2004. Mixed cytomegalovirus glycoprotein B genotypes in immunocompromised patients. Clin. Infect. Dis. 39:155–161. 10.1086/421496 [DOI] [PubMed] [Google Scholar]
- 51.Puchhammer-Stockl E, Gorzer I, Zoufaly A, Jaksch P, Bauer CC, Klepetko W, Popow-Kraupp T. 2006. Emergence of multiple cytomegalovirus strains in blood and lung of lung transplant recipients. Transplantation 81:187–194. 10.1097/01.tp.0000194858.50812.cb [DOI] [PubMed] [Google Scholar]
- 52.Rasmussen L, Geissler A, Winters M. 2003. Inter- and intragenic variations complicate the molecular epidemiology of human cytomegalovirus. J. Infect. Dis. 187:809–819. 10.1086/367900 [DOI] [PubMed] [Google Scholar]
- 53.Just M, Buergin-Wolff A, Emoedi G, Hernandez R. 1975. Immunization trials with live attenuated cytomegalovirus TOWNE 125. Infection 3:111–114. 10.1007/BF01641052 [DOI] [PubMed] [Google Scholar]
- 54.Plotkin SA, Starr SE, Friedman HM, Gonczol E, Weibel RE. 1989. Protective effects of Towne cytomegalovirus vaccine against low-passage cytomegalovirus administered as a challenge. J. Infect. Dis. 159:860–865. 10.1093/infdis/159.5.860 [DOI] [PubMed] [Google Scholar]
- 55.Fleisher GR, Starr SE, Friedman HM, Plotkin SA. 1982. Vaccination of pediatric nurses with live attenuated cytomegalovirus. Am. J. Dis. Child. 136:294–296 [DOI] [PubMed] [Google Scholar]
- 56.Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, McGeoch DJ, Hayward GS. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17–28. 10.1099/vir.0.18606-0 [DOI] [PubMed] [Google Scholar]
- 57.Smith W, Tomasec P, Aicheler R, Loewendorf A, Nemcovicova I, Wang EC, Stanton RJ, Macauley M, Norris P, Willen L, Ruckova E, Nomoto A, Schneider P, Hahn G, Zajonc DM, Ware CF, Wilkinson GW, Benedict CA. 2013. Human cytomegalovirus glycoprotein UL141 targets the TRAIL death receptors to thwart host innate antiviral defenses. Cell Host Microbe 13:324–335. 10.1016/j.chom.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomasec P, Wang EC, Davison AJ, Vojtesek B, Armstrong M, Griffin C, McSharry BP, Morris RJ, Llewellyn-Lacey S, Rickards C, Nomoto A, Sinzger C, Wilkinson GW. 2005. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat. Immunol. 6:181–188. 10.1038/ni1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benedict CA, Butrovich KD, Lurain NS, Corbeil J, Rooney I, Schneider P, Tschopp J, Ware CF. 1999. Cutting edge: a novel viral TNF receptor superfamily member in virulent strains of human cytomegalovirus. J. Immunol. 162:6967–6970 [PubMed] [Google Scholar]
- 60.Cheung TC, Humphreys IR, Potter KG, Norris PS, Shumway HM, Tran BR, Patterson G, Jean-Jacques R, Yoon M, Spear PG, Murphy KM, Lurain NS, Benedict CA, Ware CF. 2005. Evolutionarily divergent herpesviruses modulate T cell activation by targeting the herpesvirus entry mediator cosignaling pathway. Proc. Natl. Acad. Sci. U. S. A. 102:13218–13223. 10.1073/pnas.0506172102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poole E, Walther A, Raven K, Benedict CA, Mason GM, Sinclair J. 2013. The myeloid transcription factor GATA-2 regulates the viral UL144 gene during human cytomegalovirus latency in an isolate-specific manner. J. Virol. 87:4261–4271. 10.1128/JVI.03497-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baskin GB. 1987. Disseminated cytomegalovirus infection in immunodeficient rhesus monkeys. Am. J. Pathol. 129:345–352 [PMC free article] [PubMed] [Google Scholar]
- 63.Jabs DA. 1995. Ocular manifestations of HIV infection. Trans. Am. Ophthalmol. Soc. 93:623–683 [PMC free article] [PubMed] [Google Scholar]
- 64.Pertel P, Hirschtick R, Phair J, Chmiel J, Poggensee L, Murphy R. 1992. Risk of developing cytomegalovirus retinitis in persons infected with the human immunodeficiency virus. J. Acquir. Immune Defic. Syndr. 5:1069–1074 [PubMed] [Google Scholar]
- 65.Rasmussen L. 1999. Molecular pathogenesis of human cytomegalovirus infection. Transpl. Infect. Dis. 1:127–134. 10.1034/j.1399-3062.1999.010206.x [DOI] [PubMed] [Google Scholar]
- 66.Griffiths PD, Stanton A, McCarrell E, Smith C, Osman M, Harber M, Davenport A, Jones G, Wheeler DC, O'Beirne J, Thorburn D, Patch D, Atkinson CE, Pichon S, Sweny P, Lanzman M, Woodford E, Rothwell E, Old N, Kinyanjui R, Haque T, Atabani S, Luck S, Prideaux S, Milne RS, Emery VC, Burroughs AK. 2011. Cytomegalovirus glycoprotein-B vaccine with MF59 adjuvant in transplant recipients: a phase 2 randomised placebo-controlled trial. Lancet 377:1256–1263. 10.1016/S0140-6736(11)60136-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pass RF, Zhang C, Evans A, Simpson T, Andrews W, Huang ML, Corey L, Hill J, Davis E, Flanigan C, Cloud G. 2009. Vaccine prevention of maternal cytomegalovirus infection. N. Engl. J. Med. 360:1191–1199. 10.1056/NEJMoa0804749 [DOI] [PMC free article] [PubMed] [Google Scholar]