ABSTRACT
Kaposi's sarcoma-associated herpesvirus (KSHV, also called human herpesvirus 8) is linked to the development of Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD). KSHV expresses several proteins that modulate host cell signaling pathways. One of these proteins is viral interleukin-6 (vIL-6), which is a homolog of human IL-6 (hIL-6). vIL-6 is able to prevent apoptosis and promote proinflammatory signaling, angiogenesis, and cell proliferation. Although it can be secreted, vIL-6 is mainly an intracellular protein that is retained in the endoplasmic reticulum (ER). We performed affinity purification and mass spectrometry to identify novel vIL-6 binding partners and found that a cellular ER chaperone, hypoxia-upregulated protein 1 (HYOU1), interacts with vIL-6. Immunohistochemical staining reveals that both PEL and KS tumor tissues express significant amounts of HYOU1. We also show that HYOU1 increases endogenous vIL-6 protein levels and that HYOU1 facilitates vIL-6-induced JAK/STAT signaling, migration, and survival in endothelial cells. Furthermore, our data suggest that HYOU1 also modulates vIL-6's ability to induce CCL2, a chemokine involved in cell migration. Finally, we investigated the impact of HYOU1 on cellular hIL-6 signaling. Collectively, our data indicate that HYOU1 is important for vIL-6 function and may play a role in the pathogenesis of KSHV-associated cancers.
IMPORTANCE KSHV vIL-6 is detectable in all KSHV-associated malignancies and promotes tumorigenesis and inflammation. We identified a cellular protein, called hypoxia-upregulated protein 1 (HYOU1), that interacts with KSHV vIL-6 and is present in KSHV-infected tumors. Our data suggest that HYOU1 facilitates the vIL-6-induced signaling, migration, and survival of endothelial cells.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV; human herpesvirus 8) is the causative agent of several human malignancies, including Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD) (1–4). These malignancies often occur in the context of immunosuppression, and as a result, KSHV-associated malignancies have increased in incidence since the onset of the AIDS epidemic (5). KSHV is a member of the gammaherpesvirus subfamily and has a double-stranded DNA genome that expresses over 80 open reading frames (ORFs) (6). KSHV usually exists in a latent state in which a small subset of the viral genome is expressed. When the virus undergoes lytic reactivation, all viral genes are expressed and progeny virions are produced.
It is thought that several latent and lytic genes contribute to modulation of host cell signaling to induce tumorigenesis. One of these genes is ORF K2, which encodes a viral homolog of human interleukin-6 (hIL-6) called viral IL-6 (vIL-6) (7–9). vIL-6 shares 25% identity and 63% similarity to hIL-6 at the amino acid level. vIL-6 is expressed at low levels in latently infected PEL cells and is highly upregulated upon lytic reactivation (10–12). All KSHV-associated malignancies have detectable vIL-6 levels (13–15). vIL-6 expression transforms NIH 3T3 cells, and vIL-6-expressing cells injected into mice form larger tumors than control cells (16). Additionally, transgenic mice engineered to express vIL-6 under the major histocompatibility complex (MHC) class I promoter display a phenotype reminiscent of that of KSHV-associated plasmablastic MCD that is also dependent on mouse IL-6 expression (17). vIL-6 drives production of hIL-6 (18) and vascular endothelial growth factor (VEGF) (16) and can promote angiogenesis (19). Importantly, vIL-6 activates signaling pathways similar to those of human cytokines, including the JAK/STAT, mitogen-activated protein kinase (MAPK), and phosphoinositol 3-kinase (PI3K) pathways (20–22).
vIL-6 differs from hIL-6 in several ways: hIL-6 must bind the IL-6 receptor (IL6R, gp80) before activation of the gp130 signal transducer subunit, whereas vIL-6 can directly bind gp130 to induce signaling (23–25); however, involvement of gp80 can enhance vIL-6 signaling (26). Another difference is that hIL-6 is rapidly secreted from cells but that vIL-6 is retained primarily within the endoplasmic reticulum (ER) (12, 27). In this compartment, vIL-6 binds gp130 in a tetrameric complex to induce intracellular signaling (12). The cellular ER protein calnexin has been shown to interact with vIL-6 to stabilize vIL-6 folding and maintain its intracellular distribution (28). The ER transmembrane protein vitamin K epoxide reductase complex subunit 1 variant 2 (VKORC1v2) was recently identified as an additional intracellular binding partner of vIL-6 (29, 30). vIL-6 binds to VKORC1v2's C terminus, which is present in the ER lumen, but data suggest that this binding domain is not responsible for retention of vIL-6 in the ER. Overexpression of VKORC1v2's vIL-6 binding domain or depletion of VKORC1v2 abrogates vIL-6's progrowth phenotype in PEL cells independently of gp130 signaling (29). Furthermore, it was found that vIL-6 promotes PEL cell survival by suppressing the proapoptotic properties of the VKORC1v2 binding partner cathepsin D (31). This suggests that VKORC1v2 uses a mechanism independent of gp130 signaling to promote vIL-6 function and PEL cell survival.
We performed affinity purification and mass spectrometry (MS) to identify novel binding partners of intracellular vIL-6. We found that a protein called hypoxia-upregulated protein 1 (HYOU1; also called the 150-kDa oxygen-regulated protein, or ORP150) is able to bind vIL-6. HYOU1 is an ER-resident chaperone protein that is a member of the heat shock and ER stress protein families (32). HYOU1 is expressed in many different cell types and can be upregulated by various cellular conditions, including hypoxia and ER stress (32, 33). Furthermore, HYOU1 is upregulated in some human cancers, including head and neck and breast cancers (34, 35). The HYOU1 transcript was originally cloned from astrocytes under hypoxic conditions (36), which makes it a protein relevant to KSHV biology, since hypoxia plays a role in the KSHV life cycle (37). Previous work has indicated that HYOU1 can suppress hypoxia-induced cell death (38) and induce angiogenesis by facilitating VEGF processing (39).
We found that HYOU1's interaction with vIL-6 is important for vIL-6-induced intracellular STAT3 signaling and vIL-6 expression in PEL cells. Furthermore, we show that HYOU1 is required for several vIL-6 biological functions, including promotion of endothelial cell survival and migration. We found that vIL-6 increases extracellular levels of chemokine (C-C motif) ligand 2 (CCL2, also called monocyte chemoattractant protein 1, or MCP1) in a HYOU1-dependent manner. CCL2 is implicated in the migration and metastasis of tumor cells and the extravasation of immune cells (40, 41). Finally, we investigated the impact of HYOU1 on cellular hIL-6 signaling. Our results suggest that by modulating vIL-6 function, HYOU1 may contribute to KSHV-associated tumorigenesis, making HYOU1 an attractive target for the treatment of KSHV-associated malignancies.
MATERIALS AND METHODS
Cell culture and generation of stable cell lines.
Human embryonic kidney 293 (HEK293) and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Corning). BCBL1 PEL cells were cultured in RPMI 1640 medium (Corning) containing 0.05 mM β-mercaptoethanol. TREx BCBL1-inducible cells expressing either empty vector or replication and transcription activator (RTA) protein (42) were cultured in RPMI 1640 medium (Corning) containing tetracycline (Tet) system-approved fetal bovine serum (FBS; Clontech) and 20 μg/ml hygromycin B (Roche). Human telomerase reverse transcriptase-immortalized human umbilical vein endothelial cells (hTERT-HUVEC) were cultured in EBM-2 (Lonza) with the EBM-2 bullet supplement (Lonza) as described previously (43). All media were additionally supplemented with 10% heat-inactivated FBS, 1% penicillin-streptomycin (PS), and 1% l-glutamine. Charcoal-filtered FBS was obtained from Life Technologies. Cells were transfected with X-tremeGENE high-performance (HP) transfection reagent (Roche) at a ratio of 2 μl X-tremeGENE to 1 μg plasmid DNA as per the manufacturer's protocol. Cells were transfected with 50 to 100 nM small interfering RNA (siRNA) by utilizing Lipofectamine RNAiMAX (Invitrogen) per the manufacturer's protocol. For lentiviral transductions, adherent cells were grown to 70% confluence and inoculated with lentivirus in the presence of 8 μg/ml Polybrene. Spinoculation was used for PEL cell transductions as previously described (44). All transfection and transduction media were incubated for 48 to 72 h to allow for protein expression or knockdown. hTERT-HUVEC and HEK293 cells stably expressing an empty vector or vIL-6 were generated by lentiviral transduction. HEK293 cells stably expressing a nontargeting short hairpin RNA (shRNA) or a HYOU1-targeting shRNA plasmid (described below) were also generated by lentiviral transduction. For all stable cells, media were changed 24 h postransduction and the puromycin concentration was increased from 0.1 μg/ml to a final concentration of 0.5 μg/ml for hTERT-HUVEC and 1.0 μg/ml for HEK293 cells over 2 weeks.
Plasmids, lentiviral vectors, shRNAs, and siRNAs.
The pcDNA3.1-vIL-6-His clone was a kind gift from Yuan Chang and Patrick Moore. A C-terminal FLAG tag was added to vIL-6 and cloned into the pcDNA3.1 eukaryotic expression vector (Invitrogen). pSG5-based eukaryotic expression vectors for untagged hIL-6 and hIL-6 with an ER-targeting motif containing KDEL and additional sequences (45) were a kind gift from John Nicholas and were previously described (12). The nontargeting control (NTC) siRNA duplex was purchased from Dharmacon (catalog number D001810-01), and the HYOU1-targeting siRNA duplex was designed and purchased from Invitrogen (catalog number NM_001130991_stealth_455) by utilizing the Block-iT RNA interference (RNAi) designer as previously described (46). Plasmids for the pLKO.1 NTC shRNA and a HYOU1-targeting shRNA (reagent number TRCN0000029220) were purchased from Sigma and used to generate lentivirus. FLAG-tagged vIL-6 was cloned into the lentiviral vector pSuper-CMV puro (Invitrogen). All lentiviruses were produced using the ViraPower lentiviral expression system (Invitrogen) as per the manufacturer's instructions.
MS, immunoprecipitations, and Western blotting.
Twenty million 293T cells were transfected with pcDNA3 or vIL-6 expression vectors for 48 h. Cells were harvested on ice in NP-40 lysis buffer (0.1% NP-40, 150 mM NaCl, 50 mM Tris HCl, pH 8.0, 30 mM β-glycerophosphate, 50 mM NaF, 1 mM Na3VO4, 1 Roche protease inhibitor tablet per 50 ml), followed by one freeze-thaw cycle. Samples were clarified by centrifugation at 16,000 × g for 10 min, and protein content was determined by Bradford assay (Bio-Rad). Equal amounts of protein were loaded on FLAG antibody-conjugated beads (EZview red anti-FLAG M2 affinity gel; Sigma) and rocked at 4°C overnight. Beads were washed twice with lysis buffer, followed by 2 washes with 50 mM NH4HCO3. Samples were eluted with 3×-FLAG peptide (Sigma) diluted in 50 mM NH4HCO3 and 0.1% PPS Silent surfactant (Protein Discovery). Samples were treated with 5 mM dithiothreitol at 60°C for 15 min. Proteins were trypsinized using the filter-aided sample prep (FASP) protein digestion protocol (Protein Discovery), and tryptic peptides were separated by a nanoAquity UPLC system (Waters Corp.) with a 2-cm trapping column and a self-packed 25-cm analytical column (75-μm internal diameter; Michrom Magic C18 beads of 5.0-μm particle size, 100-Å pore size) at room temperature. The flow rate was 350 nl/min over a gradient of 1% buffer B (0.1% formic acid in acetonitrile) to 35% buffer B in 200 min. A full mass spectrum scan (300 to 2,000 mass/charge ratio [m/z]) was acquired in an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific) at a resolution setting of 60,000; data-dependent tandem mass spectrometry (MS2) spectra were acquired by collision-induced dissociation with the 15 most intense ions. All raw data were searched with the Sorcerer-SEQUEST server (Sage-N Research) against the human UniProtKB/Swiss-Prot sequence database. Search parameters used were a precursor mass between 400 and 4,500 atomic mass units (amu), up to 2 missed cleavages, a precursor ion tolerance of 3 amu, semitryptic digestion, a static carbamidomethyl cysteine modification, and variable methionine oxidation. False discovery rates (FDRs) were determined by ProteinProphet, and minimum protein probability cutoffs resulting in a 1% FDR were used.
For HYOU1 immunoprecipitations, cell lysates containing equal amounts of protein were precleared by incubation with protein A beads and normal rabbit IgG. Beads were pelleted, and supernatants were incubated with 8 μl HYOU1 antibody or rabbit IgG for 5 h prior to the addition of 50 μl protein A beads overnight. Beads were pelleted and washed twice with lysis buffer and twice with cold Tris-buffered saline (TBS). Bound proteins were eluted by boiling the beads in Laemmli buffer for 5 min. For FLAG–vIL-6 immunoprecipitation, HEK293 cells stably expressing the empty vector or vIL-6 were transfected with 75 nM NTC or HYOU1-targeting siRNA for 72 h. Equal amounts of cell lysates were precleared by protein A beads, and normal rabbit IgG was then incubated with 50 μl FLAG antibody-conjugated beads overnight. Bound proteins were eluted with 3X FLAG peptide. All immunoprecipitation inputs and eluates were resolved on 10% SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were blocked with 5% nonfat dry milk in 1× TBS-0.1% Tween 20 (TBST) followed by overnight incubation with primary antibody at 4°C. Blots were then incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. Blots were developed with Pico West chemiluminescent reagent (Thermo). Primary antibodies used were FLAG (Bethyl Laboratories; catalog number A190-101P), actin (Santa Cruz; sc-1615), tubulin (Cell Signaling; 9099), phospho-STAT3 Tyr705 (Cell Signaling; 9131), total STAT3 (Cell Signaling; 4904), HYOU1 (Abcam; ab134944), human IL-6 (Abcam; ab32530), gp130 (Santa Cruz; sc-655), latency associated nuclear antigen (LANA) (Advanced Biotechnologies; 13-210-100), RTA (a gift from Ren Sun), K8.1 (Advanced Biotechnologies; 13-212-100), and vIL-6 (Advanced Biotechnologies; 13-214-050). Anti-rabbit, anti-mouse, anti-rat (Cell Signaling), and anti-goat (Santa Cruz) HRP-conjugated secondary antibodies were used. Normal rabbit IgG (Santa Cruz; sc-2027) and protein A beads (Santa Cruz; sc-2003) were used for preclearing immunoprecipitation samples.
Immunohistochemistry.
Immunohistochemical staining was carried out as described previously (47). PEL xenograft tumors were obtained from a previous study (47). Prepared sections of confirmed KSHV-positive lesions from patients with KS (specimens 30035001 and 30035006) were obtained from the AIDS and Cancer Specimen Resource (ACSR). For HYOU1 staining, slides were deparaffinized and rehydrated and then incubated in 3% H2O2 in 10% methanol. Antigen retrieval was accomplished by boiling sections in 1 mM EDTA (pH 8.0) for 10 min. Slides were incubated with blocking buffer (10% normal horse serum, 5% bovine serum albumin [BSA], 0.3% Triton X-100) for 1 h, followed by overnight incubation with HYOU1 antibody (1:100) or blocking buffer as a negative control. Sections were washed and incubated with biotinylated goat anti-rabbit secondary antibody for 1 h, followed by signal amplification with the Vectastain ABC rabbit IgG kit (Vector Laboratories). Sections were stained with 3,3′-diaminobenzidine (DAB) peroxide substrate according to the manufacturer's protocol (Vector Laboratories). For LANA staining, antigen retrieval was accomplished by boiling sections in Retrievagen A, pH 6.0 (BD Pharmingen). Slides were incubated in blocking buffer (1.5% rabbit serum, 1% BSA, 0.1% Triton X-100, 0.05% Tween 20, 0.1% gelatin from cold-water fish skin in 1× phosphate-buffered saline [PBS]) for 1 h, followed by overnight incubation with LANA antibody diluted 1:100 in antibody buffer (1% BSA, 0.1% Triton X-100, 0.1% gelatin from cold-water fish skin in 1× PBS) or plain antibody buffer for the negative control. Sections were washed and incubated with biotinylated rabbit anti-rat secondary antibody for 30 min, followed by signal amplification with the Vectastain ABC rat IgG kit. Slides were stained with NovaRED substrate according to the manufacturer's protocol (Vector Laboratories). All slides were counterstained with hematoxylin. Slides were imaged using a Leica DM LA histology microscope with 20×/0.25-numeric-aperture (NA) and 40×/0.75-NA objectives.
Scratch assay.
Stable empty-vector- or vIL-6-expressing hTERT-HUVEC were plated at 1.5 × 105 cells/well of a 6-well dish. The following day, cells were transduced with lentivirus expressing NTC or HYOU1-targeting shRNA. Media were changed after 24 h, and 48 h postransduction, cells were serum starved overnight with plain EBM-2. The monolayer was scratched with a P10 pipette tip, and a Nikon Eclipse Ti inverted microscope was used to obtain bright-field images of the cells at ×40 or ×100 magnification at the time points indicated below. ImageJ software (NIH) was used to quantify the area of each scratch over time.
CCL2 ELISA.
Stable empty-vector- or vIL-6-expressing hTERT-HUVEC were plated at 1.2 × 105 cells/well of a 6-well dish. The following day, cells were transfected with 100 nM NTC or HYOU1-targeting siRNA. Seventy-two hours posttransfection, media were replaced with serum-free EBM-2 without supplements for 48 h. Following the harvest, the media were cleared of debris by centrifugation and a CCL2 enzyme-linked immunosorbent assay (ELISA) was carried out per the manufacturer's protocol (Life Technologies). Samples were all run in triplicate. Absorbance was read at 450 nm on a VersaMax tunable microplate reader (Molecular Devices), and a standard curve was generated using a best-fit power trendline in Microsoft Excel. Error bars represent the standard deviations, and CCL2 concentrations were compared using a two-tailed Student t test, with a P of <0.05 considered significant. Results are representative of at least 3 experiments.
Survival assay.
Stable empty-vector- or vIL-6-expressing hTERT-HUVEC were plated at 1.2 × 105 cells/well of a 6-well dish. The following day, cells were transfected with 100 nM NTC or HYOU1-targeting siRNA. Seventy-two hours posttransfection, media were replaced with serum-free EBM-2 without supplements and a Nikon Eclipse Ti inverted microscope was used to obtain bright-field images of the cells at a ×40 or ×100 magnification. Media were replaced with fresh serum-free media before each time point to remove dead cell debris.
Cytotoxicity assay.
Three thousand cells/well of stable empty-vector- or vIL-6-expressing hTERT-HUVEC were plated and transfected in two white-walled 96-well plates using RNAiMAX and 100 nM NTC or HYOU1-targeting siRNA according to the manufacturer's protocol for reverse transfection (Invitrogen). Samples were transfected in triplicate. Three days posttransfection, one plate was used to measure baseline cytotoxicity at day 0 using the CytoTox-Glo kit (Promega) per the manufacturer's instructions with a FLUOstar Optima luminometer (BMG Labtech). The medium of the second plate was replaced with serum-free EBM-2 without supplements for 6 days, followed by a second cytotoxicity assay. The percentage of dead cells at each time point was calculated from the raw data. Data are shown as fold changes in cell death, which was calculated by dividing the percentage of dead cells at day 6 by the percentage at day 0. Error bars represent the standard deviations of the means, and a one-way analysis of variance (ANOVA) with Tukey's post hoc test was used to compare all samples, with a P value of <0.05 considered significant.
RESULTS
Identification of HYOU1 as a vIL-6 binding partner.
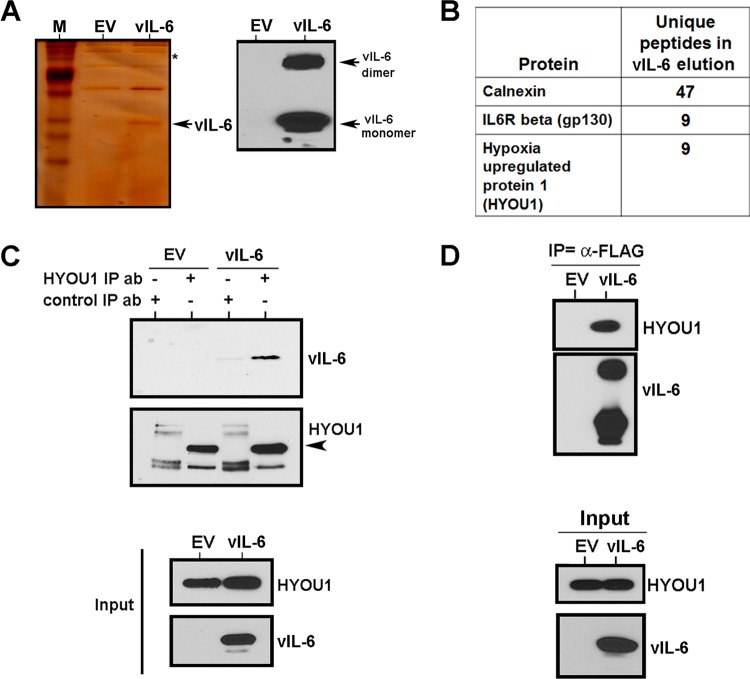
The biological effects of extracellular vIL-6 have been extensively studied (9, 16, 18, 20–22), but less is known about vIL-6's interactions with intracellular proteins. We sought to identify additional cellular proteins that bind to vIL-6 and impact its function. We cloned vIL-6 with an N-terminal FLAG tag into the pcDNA3.1 vector (pcDNA3.1-vIL-6) and transfected the pcDNA3.1 empty vector or pcDNA3.1-vIL-6 into HEK293T cells. Cells were harvested and subjected to an immunoprecipitation with FLAG antibody-conjugated beads. vIL-6-bound proteins were eluted with 3X FLAG peptide. A fraction of the elution was subjected to SDS-PAGE and subsequent silver staining or Western blotting for vIL-6 to ensure successful vIL-6 expression and pulldown (Fig. 1A). The vIL-6 concentration in the lysate was high enough that dimerization of the vIL-6 protein was evident by Western blotting (Fig. 1A, right). The remainder of the eluted samples was digested with trypsin and analyzed by shotgun mass spectrometry (48). Proteins with peptide counts that were higher in the vIL-6–FLAG sample than in the empty-vector sample were identified as potential vIL-6 binding partners (Fig. 1B). Over 40 unique calnexin peptides and 9 unique gp130 peptides were identified in the vIL-6 sample, consistent with previous findings (28). The vIL-6 sample also had 9 unique peptides identified for the protein hypoxia-upregulated protein 1 (HYOU1; ORP150), while no HYOU1 peptides were identified in the empty vector control.
FIG 1.
vIL-6 binds the ER chaperone protein hypoxia-upregulated protein 1 (HYOU1). (A) HEK293T cells were transfected with an empty vector (EV) or FLAG-tagged vIL-6, and lysates were harvested for immunoprecipitation with FLAG antibody. Portions of the eluates were analyzed by SDS-PAGE and subsequent silver staining (left) (lane M contains the molecular size markers) or Western blotting for vIL-6 (right). (B) HYOU1, calnexin, and gp130 were identified as binding partners of vIL-6 by mass spectrometry. (C) HEK293T cells were transfected with the empty vector or vIL-6 constructs, and lysates were harvested and immunoprecipitated (IP) with HYOU1 or rabbit IgG control antibody (ab). Bound proteins were eluted with Laemmli buffer. Samples were analyzed by SDS-PAGE and Western blotting for the indicated proteins. (D) BCBL1 PEL cells were transduced with lentivirus expressing the empty vector or FLAG-tagged vIL-6. Lysates were immunoprecipitated with FLAG antibody, and eluates and inputs were analyzed by SDS-PAGE and Western blotting for the indicated proteins.
To confirm that HYOU1 interacts with vIL-6, we again transfected HE293T cells with the empty vector or FLAG-tagged vIL-6 constructs. Lysates from these cells were immunoprecipitated with HYOU1 antibody or rabbit IgG as a control, and bound proteins were eluted by boiling the plates in Laemmli buffer. Eluates and input samples were resolved on an SDS-PAGE gel and analyzed by Western blotting. As seen in Fig. 1C, vIL-6 coimmunoprecipitated with HYOU1. To determine whether vIL-6 interacts with HYOU1 in a more relevant cell type, BCBL1 PEL cells were transduced with the lentivirus-expressing empty vector or FLAG-tagged vIL-6 and lysates were harvested 48 h later. The FLAG-tagged vIL-6 was immunoprecipitated by FLAG antibody-conjugated beads, and bound proteins were eluted with FLAG peptide. Our data again showed that HYOU1 coimmunoprecipitated with vIL-6, suggesting that there is a bona fide interaction between HYOU1 and vIL-6 in multiple cell types.
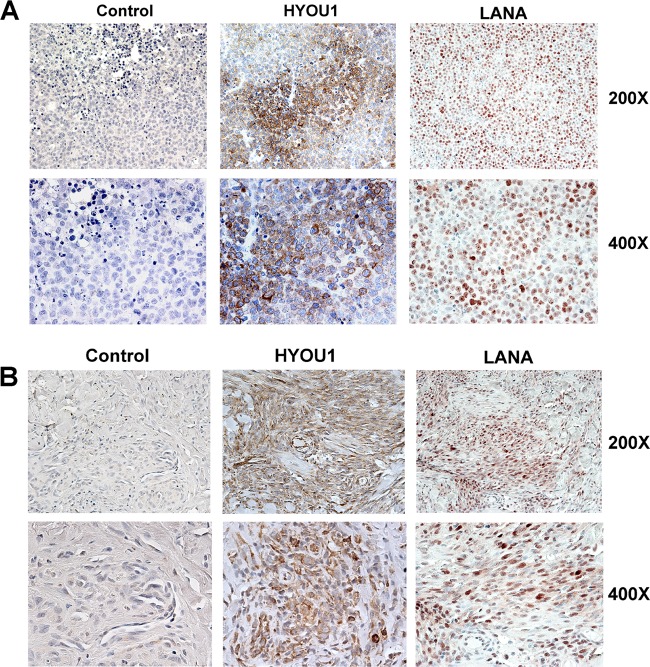
To determine whether HYOU1 is expressed in tumors associated with KSHV infection, sections of BC-1 PEL tumors grown in immunodeficient mice (47) and human KS sections were stained for HYOU1 using the DAB peroxide substrate. Matched tumor sections were also stained for LANA to confirm KSHV infection. HYOU1 and LANA staining were observed in both PEL xenografts and KS lesions (Fig. 2A and B). In the PEL xenografts in particular, HYOU1 displayed distinct perinuclear staining, which is consistent with its ER localization (Fig. 2A).
FIG 2.
HYOU1 is expressed in tissue from KSHV-associated tumors. (A) Immunohistochemical staining for HYOU1 and LANA on BC-1 PEL xenograft sections. The control received no primary antibody. (B) Immunohistochemical staining for HYOU1 and LANA on sections of human KS lesions. The control received no primary antibody. Magnifications are noted at the right.
HYOU1 increases vIL-6 levels.
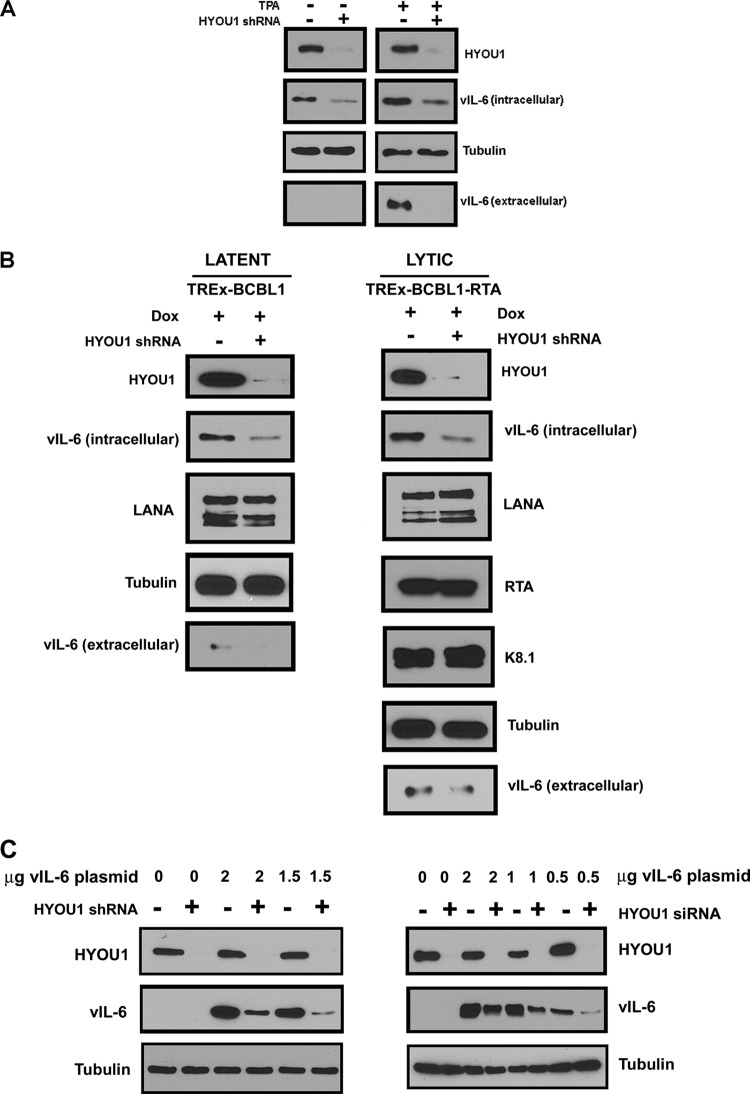
HYOU1 is an ER chaperone protein (32), so we sought to determine whether its interaction with vIL-6 influences endogenous vIL-6 protein levels. BCBL1 PEL cells were transduced with lentivirus expressing a nontargeting control (NTC) shRNA or a HYOU1-targeting shRNA. Three days postransduction, cells were treated with dimethyl sulfoxide (DMSO) to maintain latency or 25 ng/ml 12-O-tetradecanoylphorbol 13-acetate (TPA) to induce lytic reactivation for 24 h. Cell lysates and media were harvested and analyzed by SDS-PAGE and Western blotting. High levels of HYOU1 knockdown were achieved with the HYOU1-targeting shRNA, and as expected, vIL-6 levels were higher overall in lytic samples than in latent samples since vIL-6 is induced during lytic reactivation (10) (Fig. 3A). We found that HYOU1 knockdown decreased endogenous vIL-6 protein levels in both lytic and latent BCBL1 cells (Fig. 3A). Additionally, HYOU1 knockdown decreased levels of secreted vIL-6 during reactivation. While this is likely due to the increased levels of intracellular vIL-6 when HYOU1 is expressed, we cannot rule out the possibility that HYOU1 also promotes the secretion of vIL-6.
FIG 3.
HYOU1 increases endogenous vIL-6 levels. (A) BCBL1 PEL cells were transduced with lentivirus expressing a nontargeting control (NTC) or HYOU1-targeting shRNA. Three days later, cells were treated with DMSO or 25 ng/ml TPA to induce reactivation for 24 h. Lysates and media were collected and analyzed by SDS-PAGE and Western blotting for the indicated proteins. (B) TREx BCBL1 or TREx BCBL1-RTA PEL cells expressing the doxycycline (Dox)-inducible empty vector or RTA, respectively, were transduced with lentivirus expressing an NTC or HYOU1-targeting shRNA. Three days postransduction, 1 μg/ml doxycycline was added for 24 h. Lysates and media were harvested and analyzed by SDS-PAGE and Western blotting for the indicated proteins. (C) HEK293 cells stably knocked down for HYOU1 (left) or HEK293 cells first transfected with NTC or HYOU1-targeting siRNA (right) were then transfected with a titration of FLAG-tagged vIL-6 plasmid for 48 h. Lysates were harvested and analyzed by SDS-PAGE and Western blotting for the indicated proteins.
We also performed this experiment using TREx BCBL1 and TREx BCBL1-RTA PEL lines that express a doxycycline-inducible empty vector or RTA, respectively (42). The two TREx BCBL1 cell lines were transduced with lentivirus expressing an NTC or a HYOU1-targeting shRNA. After 72 h, cells received 1 μg/ml doxycycline for 24 h to induce lytic replication in the TREx BCBL1-RTA cells while the TREx BCBL1 cells remained latent. Cell lysate and media were harvested and analyzed by SDS-PAGE and Western blotting for the proteins indicated in Fig. 3B. Knockdown of HYOU1 in both latent TREx BCBL1 and lytic TREx BCBL1-RTA cells reduced levels of intracellular and extracellular vIL-6, similar to the results seen in BCBL1 PEL cells. Conversely, levels of LANA did not significantly change with HYOU1 knockdown in both the latent and the lytic cells. We also examined the levels of the lytic proteins RTA and K8.1 in the reactivated cells and found that HYOU1 knockdown did not impact the levels of these lytic proteins.
To further investigate whether HYOU1 knockdown impacts levels of vIL-6, we used HEK293 cells stably expressing an NTC or a HYOU1-targeting shRNA. These cells were then transfected with a titration of FLAG-tagged vIL-6 plasmid for 48 h. We also performed a similar experiment by transiently transfecting HEK293 cells with NTC or HYOU1-targeting siRNA for 24 h, followed by transfection with a titration of FLAG-tagged vIL-6 plasmid for 48 h. Lysates from both of these experiments were harvested and analyzed by Western blotting (Fig. 3C). In both of these experiments, knockdown of HYOU1 prior to expression of FLAG-tagged vIL-6 resulted in a reduction in vIL-6 expression compared to that of cells that expressed endogenous levels of HYOU1. These results closely match the phenotype that we see in PEL cells endogenously expressing vIL-6 (Fig. 3A and B). Interestingly, we find that when FLAG-tagged vIL-6 is expressed prior to knockdown of HYOU1, vIL-6 levels do not appear to be significantly changed by knockdown of HYOU1 (Fig. 4, 5C, 6B, and 7B). For subsequent experiments, we chose to overexpress FLAG-tagged vIL-6 before knocking down HYOU1 to keep the amount of vIL-6 the same and eliminate differences in vIL-6 levels as a variable contributing to the observed results. This approach allowed us to determine whether HYOU1 actually affects vIL-6 function as opposed to simply affecting vIL-6 protein levels as a mechanism of action.
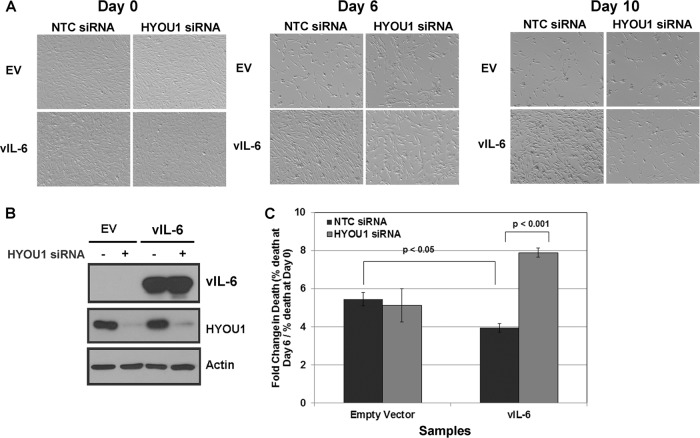
FIG 4.
HYOU1 enhances vIL-6-induced STAT3 signaling. (A) HEK293 cells were transfected with an empty vector (EV) or FLAG-tagged vIL-6 plasmids followed by a transfection with nontargeting control (NTC) or HYOU1-targeting siRNA 24 h later. Twenty-four hours after siRNA transfection, cells were serum starved for another 24 h. Lysates were harvested and analyzed by SDS-PAGE and Western blotting for the indicated proteins. (B) hTERT-HUVEC stably expressing the empty vector or FLAG-tagged vIL-6 were transfected with NTC or HYOU1-targeting siRNA for 3 days. Media were replaced with plain EBM-2 with 2% charcoal-filtered FBS for 24 h. Lysates were harvested and analyzed by SDS-PAGE and Western blotting. (C) HEK293 stably expressing the empty vector or FLAG-tagged vIL-6 were transfected with 75 nM NTC or HYOU1 siRNA for 3 days. Lysates were harvested and subjected to a FLAG immunoprecipitation overnight. Bound proteins were eluted with 3X FLAG peptide, and eluates and input samples were analyzed by SDS-PAGE and Western blotting for the indicated proteins.
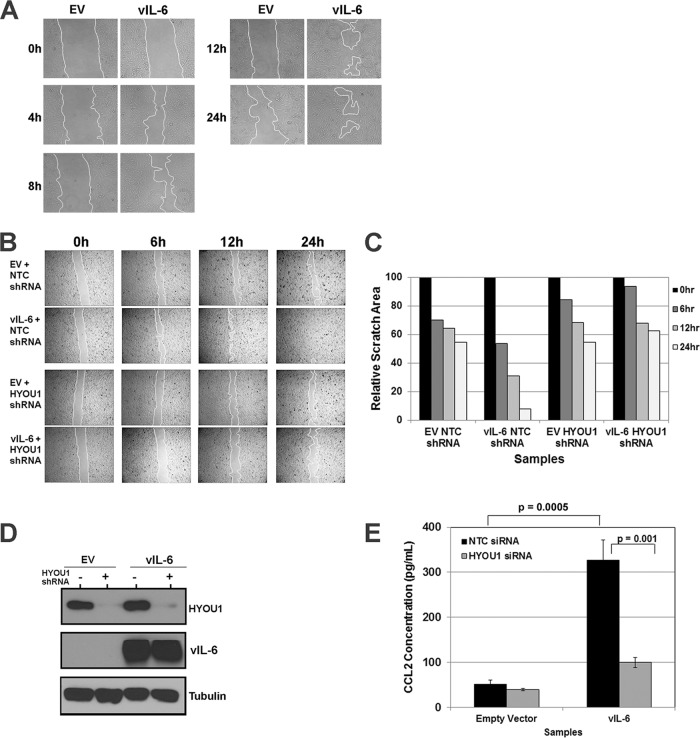
FIG 5.
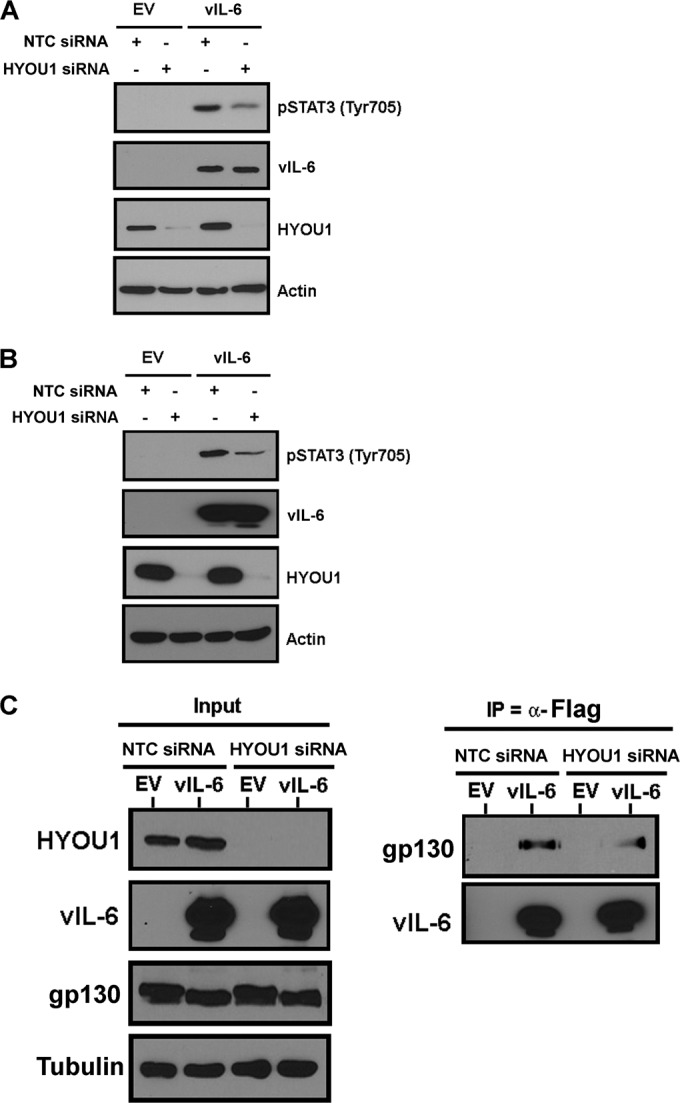
HYOU1 facilitates vIL-6-induced migration of endothelial cells. (A) hTERT-HUVEC stably expressing an empty vector (EV) or FLAG-tagged vIL-6 were serum starved overnight and scratched with a P10 pipette. Scratch closure was monitored at 0, 4, 8, 12, and 24 h by bright-field microscopy at ×100 magnification using a Nikon Eclipse Ti inverted microscope. (B) hTERT-HUVEC stably expressing the empty vector or FLAG-tagged vIL-6 were transduced with lentivirus expressing a nontargeting control (NTC) or HYOU1-targeting shRNA. Cells were serum starved overnight, scratched with a P10 pipette tip, and monitored for scratch closure at 0, 6, 12, and 24 h at ×40 magnification. (C) Quantification of the area of each scratch pictured in panel B normalized to the 0-h time point. (D) Lysates from the cells described in panel B were harvested at the 24-h time point and analyzed by SDS-PAGE and Western blotting to confirm efficient HYOU1 knockdown and vIL-6 expression. (E) hTERT-HUVEC stably expressing the empty vector or FLAG-tagged vIL-6 were transfected with 50 nM NTC or HYOU1-targeting siRNA for 3 days, followed by serum starvation for 48 h. Supernatants were collected, and a CCL2 ELISA was performed, with each sample tested in triplicate. Absorbance was read at 450 nm, and a standard curve was generated using a best-fit power trendline in Microsoft Excel. The concentrations of the samples were calculated, and error bars represent standard deviations of the results from triplicates. A two-tailed Student t test was used to compare the sample concentrations, with a P value of <0.05 considered significant. Results are representative of at least 3 experiments.
FIG 6.
HYOU1 is required for vIL-6-induced endothelial cell survival under serum-starved conditions. (A) hTERT-HUVEC stably expressing an empty vector (EV) or FLAG-tagged vIL-6 were plated as a confluent monolayer and transfected with 100 nM nontargeting control (NTC) or HYOU1-targeting siRNA. Cells were serum starved 72 h posttransfection, and bright-field images at a ×100 magnification were taken at 0, 6, and 10 days poststarvation using a Nikon Eclipse Ti inverted microscope. Replicate samples were harvested 24 h after starvation for analysis by SDS-PAGE and Western blotting to confirm efficient vIL-6 expression and HYOU1 knockdown (B). (C) hTERT-HUVEC stably expressing the empty vector or FLAG-tagged vIL-6 were reverse transfected with 100 nM NTC or HYOU1-targeting siRNA in triplicate in two white-walled 96-well plates. Three days posttransfection, a CytoTox-Glo (Promega) cytotoxicity assay was performed on the first plate and the percentage of dead cells at day 0 was calculated. The second plate was serum starved for 6 days, followed by completion of a cytotoxicity assay used to calculate the percentage of dead cells at day 6. The graph represents a fold change in dead cells calculated by dividing the percentage of dead cells at day 6 by the percentage of dead cells at day 0. Error bars represent standard deviations of the means, and P values were calculated using a one-way ANOVA with Tukey's post hoc test to compare all values to each other, with a P value of <0.05 considered significant. These data are representative of 3 independent experiments.
FIG 7.
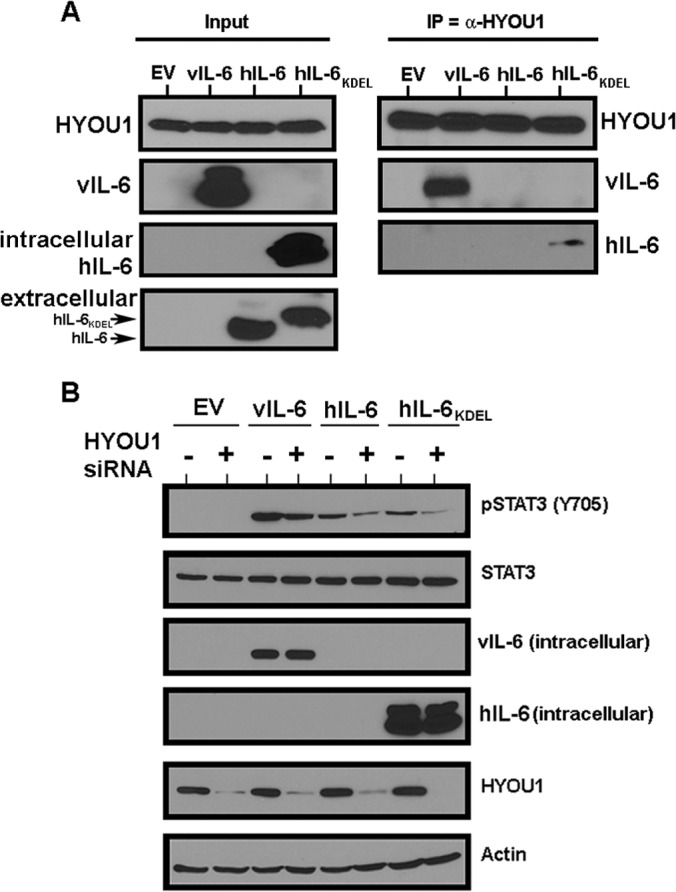
HYOU1 interacts with and influences the signaling of hIL-6. (A) HEK293 cells were transfected with an empty vector (EV), vIL-6, wild-type hIL-6, or hIL-6 with a KDEL motif (hIL-6–KDEL). Lysates were harvested and immunoprecipitated with a HYOU1 antibody. Eluates were subjected to SDS-PAGE and Western blotting for the indicated proteins. Input lysates and media were analyzed for intracellular expression of HYOU1, vIL-6, and hIL-6. (B) HEK293 cells were transfected with the empty vector, vIL-6, wild-type hIL-6, or hIL-6–KDEL for 24 h, followed by transfection with 100 nM nontargeting or HYOU1-targeting siRNA for an additional 24 h. Cells were serum starved overnight, and lysates and media were harvested for analysis by SDS-PAGE and Western blotting for the indicated proteins.
HYOU1 facilitates vIL-6-dependent STAT signaling.
Following activation of gp130, STAT3 becomes phosphorylated at Y705. This causes STAT3 to dimerize and relocate to the nucleus, where it upregulates IL-6-responsive proinflammatory genes. Others have shown that vIL-6 induces STAT3 Y705 phosphorylation through activation of gp130 (21, 28). To determine whether HYOU1's interaction with vIL-6 is involved in the induction of this signaling cascade, HEK293 cells were transfected with the empty vector or the FLAG-tagged vIL-6 plasmid and then transfected with an NTC or HYOU1-targeting siRNA 24 h later. Twenty-four hours after the siRNA transfection, media were replaced with serum-free media for another 24 h. Lysates were harvested and analyzed by SDS-PAGE and Western blotting. vIL-6 increased STAT3 Y705 phosphorylation in the presence of HYOU1, compared to that of cells expressing the empty vector; however, knockdown of HYOU1 diminished this phosphorylation event (Fig. 4A). We generated hTERT-HUVEC (43) stably expressing the empty vector or FLAG-tagged vIL-6 and transfected these cells with NTC or HYOU1-targeting siRNA for 3 days. These cells were treated with plain EBM-2 medium containing 2% charcoal-filtered FBS for 24 h prior to harvest to eliminate background levels of STAT3 phosphorylation. Lysates from the hTERT-HUVEC showed a trend similar to that of HEK293 cells for STAT3 phosphorylation (Fig. 4B). This suggests that HYOU1 is involved in enhancing vIL-6-induced activation of the JAK/STAT signaling pathway in multiple cell types.
Since vIL-6 can bind ER-embedded gp130 to initiate JAK/STAT signaling (12), we hypothesized that HYOU1 may facilitate vIL-6-induced STAT3 phosphorylation by promoting vIL-6's interaction with gp130. To test this, we used HEK293 cells stably expressing an empty vector or FLAG-tagged vIL-6. Cells were transfected with 75 nM NTC or HYOU1-targeting siRNA for 72 h. Lysates were harvested and subjected to an immunoprecipitation with FLAG beads to pull down vIL-6. Bound proteins were eluted with FLAG peptide, and the eluates and input samples were analyzed by SDS-PAGE and Western blotting. We found that gp130 coimmunoprecipitated with vIL-6 as expected but that knockdown of HYOU1 consistently reduced this interaction (Fig. 4C). This suggests that HYOU1 promotes the vIL-6-gp130 interaction, so this may be one mechanism by which HYOU1 facilitates vIL-6-mediated STAT signaling.
HYOU1 is essential for vIL-6-induced endothelial cell migration.
To further understand the impact of HYOU1 on vIL-6 activity, we developed biological assays for vIL-6 function in endothelial cells. KS is a cancer of endothelial cell origin, and vIL-6 can be detected in patients with this malignancy (13). Therefore, endothelial cells are a relevant model for studying vIL-6 function. We first investigated whether vIL-6 can influence endothelial cell migration in a scratch assay. Equivalent numbers of hTERT-HUVEC stably expressing an empty vector or FLAG-tagged vIL-6 were plated as a confluent monolayer and serum starved overnight before the monolayer was scratched with a P10 pipette tip. The scratch was monitored at various time points by utilizing bright-field microscopy on a Nikon Eclipse Ti inverted microscope. Cells expressing vIL-6 were able to close the scratch faster than cells expressing the empty vector (Fig. 5A). The influence of vIL-6 expression on endothelial cell migration has not been previously studied, so these data represent a novel biological function for vIL-6.
We extended this assay to study the effect of HYOU1 on vIL-6-induced endothelial cell migration. hTERT-HUVEC stably expressing the empty vector or FLAG-tagged vIL-6 were transduced with lentivirus expressing an NTC or a HYOU1-targeting shRNA. Two days postransduction, cells were serum starved overnight prior to scratching of the monolayer. Again, when HYOU1 was expressed normally, the vIL-6-expressing cells closed the scratch more rapidly than the empty-vector-expressing cells (Fig. 5B). However, vIL-6-expressing cells knocked down for HYOU1 were unable to close the gap, as with the empty-vector-expressing cells (Fig. 5B). The area of each scratch was quantified with ImageJ software (Fig. 5C). Lysates were harvested from these cells and subjected to SDS-PAGE and Western blotting to confirm HYOU1 knockdown and vIL-6 expression (Fig. 5D). These data suggest that HYOU1 is critical for vIL-6-induced migration of endothelial cells.
To elucidate the mechanism by which vIL-6 induces endothelial cell migration, we investigated whether vIL-6 affects the levels of chemokine (C-C motif) ligand 2 (CCL2, also called monocyte chemoattractant protein 1, or MCP1) since this chemokine is associated with tumor cell migration and metastasis and increased vascular permeability (40, 41). We transfected hTERT-HUVEC stably expressing an empty vector or FLAG-tagged vIL-6 with 100 nM NTC or HYOU1-targeting siRNA for 72 h, followed by serum starvation for 48 h. We performed a CCL2 ELISA on supernatants from these cells and found that vIL-6 expression increases the level of extracellular CCL2 compared to the level with the empty vector (Fig. 5E). Interestingly, knockdown of HYOU1 in vIL-6 cells caused the CCL2 level to decrease substantially, although the level of CCL2 in this sample was still higher than the level seen in hTERT-HUVEC expressing the empty vector (Fig. 5E). This suggests that there may be factors besides CCL2 that are involved in vIL-6-mediated cell migration.
HYOU1 is required for vIL-6-mediated endothelial cell survival.
We next investigated vIL-6's role in endothelial cell survival under serum-starved conditions. hTERT-HUVEC stably expressing the empty vector or FLAG-tagged vIL-6 were transfected with 100 nM NTC or HYOU1-targeting siRNA. Seventy-two hours posttransfection, cells were serum starved and bright-field images were obtained at the time points indicated in Fig. 6. vIL-6 expressing cells expressing HYOU1 maintained normal morphology, remained adherent, and survived significantly longer under serum-free conditions than cells expressing the empty vector (Fig. 6A). HYOU1 knockdown had little impact on survival in cells expressing the empty vector. However, vIL-6-expressing cells with HYOU1 knockdown displayed significantly reduced survival compared to that of vIL-6 cells expressing HYOU1 (Fig. 6A). Lysates from a duplicate set of samples were harvested for analysis by SDS-PAGE and Western blotting to ensure adequate HYOU1 knockdown and vIL-6 expression (Fig. 6B).
To quantify the effect of HYOU1 on vIL-6-induced survival, we performed a luminescence-based cytotoxicity assay. hTERT-HUVEC stably expressing the empty vector or FLAG-tagged vIL-6 were reverse transfected with 100 nM NTC or HYOU1 siRNA in triplicate in two 96-well white-walled plates. Three days posttransfection, the cytotoxicity assay was performed on one plate (day 0 reading). At this time point, the levels of cell death were comparable between all samples (data not shown). The second plate was serum starved for 6 days, and another cytotoxicity assay was performed. The percentages of dead cells at day 0 and day 6 were calculated as per the manufacturer's instructions. A fold change in dead cells was calculated by dividing the percentage of dead cells at day 6 by the percentage at day 0. As with trends seen in the serum-starved survival assay (Fig. 6A), empty vector cells receiving NTC siRNA have a statistically significant increase in cell death after 6 days of serum starvation compared to that of vIL-6-expressing cells receiving the NTC siRNA (Fig. 6C). HYOU1 knockdown had little impact on the survival of cells expressing the empty vector. However, knockdown of HYOU1 in vIL-6-expressing cells caused significantly higher levels of cell death after 6 days of serum starvation than was seen with vIL-6 cells expressing HYOU1 (Fig. 6C). Overall, these data suggest that HYOU1 is critical for vIL-6-mediated survival of serum-starved endothelial cells.
HYOU1 influences the signaling of human IL-6.
After confirming vIL-6's interaction with HYOU1, we wanted to determine whether hIL-6 can also interact with HYOU1. hIL-6 is rapidly secreted (27), so intracellular levels are typically undetectable by Western blotting. hIL-6 constructs that were either wild type or tagged with an ER-targeting KDEL motif (12, 45) were transfected into HEK293 cells. Lysates were harvested and subjected to an immunoprecipitation with HYOU1 antibody, followed by SDS-PAGE and Western blot analysis. We found that hIL-6–KDEL, but not wild-type hIL-6, coimmunoprecipitated with HYOU1 (Fig. 7A). The fact that an immunoprecipitated band for wild-type hIL-6 was not visible is not surprising, since no wild-type hIL-6 was identified in the input lysates, whereas high levels of hIL-6–KDEL were observed in the lysates (Fig. 7A). Therefore, HYOU1 can interact with hIL-6 only when this cytokine is retained in the ER.
Since we detected an interaction between HYOU1 and hIL-6–KDEL, we investigated whether HYOU1 impacts wild-type and KDEL-tagged hIL-6 signaling through the JAK/STAT pathway. HEK293 cells were transfected with an empty vector, FLAG-tagged vIL-6, wild-type hIL-6, or hIL-6–KDEL for 24 h, followed by an additional transfection with either NTC or HYOU1-targeting siRNA. One day later, cells were serum starved for 24 h and lysates were harvested and subjected to SDS-PAGE and Western blotting. Expression of vIL-6 and wild-type hIL-6 increased STAT3 Y705 phosphorylation (Fig. 7B). hIL-6–KDEL also induced STAT3 phosphorylation, which is likely due to leaky expression of hIL-6–KDEL into the media in our experimental system (Fig. 7A). Knockdown of HYOU1 decreased STAT3 phosphorylation mediated by vIL-6, wild-type hIL-6, and hIL-6–KDEL (Fig. 7B). This suggests that HYOU1 not only impacts vIL-6 signaling events but also may influence signaling by hIL-6.
DISCUSSION
KSHV expresses several homologs of human cytokines and chemokines, including vIL-6 and multiple viral CC chemokines (vCCLs, previously called viral macrophage inflammatory proteins [vMIPs]) (9). vIL-6 shares considerable structural and functional homology with hIL-6 and is detectable in all KSHV-associated malignancies (7, 13, 14). Patients with these malignancies also have elevated hIL-6 levels (49), and vIL-6 signaling likely promotes the proinflammatory signaling of hIL-6 to exacerbate disease. Inhibition of hIL-6 signaling to treat KSHV-associated lymphomas has been successful (50), so it is plausible that targeting vIL-6 signaling may also be a viable treatment for KSHV-associated malignancies.
Many previous reports on vIL-6 used exogenously applied vIL-6 (16, 18, 20–22). Although this has given valuable insight into the role of this viral cytokine, recent publications suggest that vIL-6 is retained primarily within the host cell in the ER (12, 27). Evidence that supports this intracellular retention include the fact that vIL-6 is secreted at a much lower rate than hIL-6, has glycosylation patterns distinct from those seen on secreted cytokines, and is capable of binding intracellular gp130 to induce signaling (27, 51).
We utilized affinity purification and mass spectrometry to identify cellular vIL-6 binding partners. HYOU1 stood out as a potential hit because it had the same number of unique peptides as were seen for gp130. HYOU1 is an ER chaperone that facilitates protein processing, is involved in the ER stress response, and protects cells from hypoxia-induced cell death (32, 33, 38). KSHV-associated malignancies often persist in hypoxic environments, such as KS lesions on the lower extremities and PEL cells in oxygen-deprived pleural cavities. Hypoxia is able to induce lytic replication due to hypoxia response elements present in the promoter region of multiple lytic genes (37). Hypoxic conditions in KSHV-infected cells or tumors may create a favorable environment for high HYOU1 expression and function, making HYOU1 a relevant protein to investigate in KSHV pathogenesis.
Based on HYOU1's role as a chaperone protein for other secreted factors (39), we tested whether it modulates the level of vIL-6 in the cell. We found that knockdown of HYOU1 reduces endogenous vIL-6 protein levels in latent and lytic PEL cells as well as in HEK293 cells when HYOU1 is knocked down prior to transfection with a vIL-6 expression construct. Interestingly, in cells that exogenously express vIL-6 before knockdown of HYOU1, the effect of HYOU1 on vIL-6 expression is not significant. Based on HYOU1's role as a known processing factor for VEGF (39), we speculate that HYOU1 may be involved in the processing and/or stability of vIL-6, as well as the biological function of vIL-6.
Our data also show that HYOU1 enhances vIL-6-mediated STAT3 phosphorylation, and it is known that vIL-6 can initiate STAT signaling through ER-associated gp130 (12). We hypothesized that HYOU1's interaction with vIL-6 may place vIL-6 in closer proximity to gp130 as a mechanism to promote signaling. To test this, we performed an immunoprecipitation and found that HYOU1 expression appears to increase the ability of vIL-6 to bind gp130. In addition to increasing intracellular vIL-6 levels, promoting the vIL-6-gp130 interaction may be yet another way that HYOU1 is able to positively influence vIL-6 function.
KS lesions are of endothelial cell origin and exist in a highly inflammatory and vascularized environment (52, 53). vIL-6 expression is detectable in the sera of a proportion of KS patients (13), and vIL-6 has been shown to influence important biological functions of endothelial cells, such as differentiation, proliferation, and angiogenesis (19, 54). Our data further show that vIL-6 is able to induce the migration of endothelial cells in a HYOU1-dependent manner. For this assay, we used serum-starved cells to minimize the possibility that proliferating cells, as opposed to migrating cells, filled the gap, since vIL-6 has previously been shown to increase cell proliferation (12, 16, 19, 20). HYOU1 has a known role in facilitating processing of secreted factors, including VEGF (39). We therefore hypothesized that HYOU1 modulates chemokines involved in vIL-6-mediated cell migration. CCL2 is associated with increased migration of tumor cells and immune cells (40, 41). We found that vIL-6 can induce CCL2 expression and that knockdown of HYOU1 substantially reduced this vIL-6-mediated increase in CCL2, but not quite to the level of CCL2 in cells expressing an empty vector. This suggests that HYOU1 may partly influence vIL-6-mediated migration by modulating CCL2 expression but that other chemokines or factors are likely involved. Overall, our data suggest that HYOU1 may have multiple mechanisms by which it promotes vIL-6 function in signaling, endothelial cell migration, and endothelial cell survival under serum-starved conditions.
Since vIL-6 is a homolog of hIL-6, we tested whether HYOU1 can also bind the human cytokine. Because hIL-6 is rapidly secreted, it is present at levels that are undetectable by Western blotting in cell lysates. Therefore, we speculated that it would be difficult to demonstrate an interaction between wild-type hIL-6 and HYOU1, since such an interaction would likely be too transient to detect. We circumvented this issue by using an hIL-6 construct with an ER-targeting KDEL motif that increases the intracellular retention of hIL-6. We detected an interaction between HYOU1 and hIL-6–KDEL, although we could not detect an interaction between wild-type hIL-6 and HYOU1. However, we found that STAT3 signaling mediated by wild-type hIL-6 was diminished when HYOU1 was depleted from cells. Since HYOU1 can impact wild-type hIL-6 signaling, we speculate that HYOU1 might transiently interact with hIL-6 when the cytokine transits through the ER, despite our being unable to see an interaction between HYOU1 and wild-type hIL-6 by immunoprecipitation. Alternatively, HYOU1 might have an effect on other components of the hIL-6 signaling pathway to impact signaling. Overall, our data suggest that HYOU1 may play a role in both vIL-6 and hIL-6 signaling function.
During lytic reactivation of KSHV, vIL-6 is highly expressed, and some is secreted from infected cells, which is likely why patients with MCD and other KSHV-associated malignancies have detectable vIL-6 in their serum (14, 15). Conversely, during latency, vIL-6 is expressed at very low levels that are retained mainly within the cell. Importantly, it has been demonstrated that this low level of latent expression is functional (12). Inhibition of proinflammatory hIL-6 signaling with an anti-IL-6 receptor antibody has been moderately successful for the treatment of MCD (50); however, inhibiting vIL-6 signaling in a similar fashion may be challenging, since a portion of vIL-6 signaling occurs inside the cell beyond reach of an IL6R-targeting antibody. The identification of HYOU1 as a cellular binding partner that is required for facilitating multiple facets of vIL-6 function and hIL-6 activity presents an alternative druggable target for inhibiting IL-6 function in the treatment of KSHV-associated malignancies.
ACKNOWLEDGMENTS
L.G. was supported by training grants T32-CA071341 and T32-AI007419. B.D. is supported by grants CA163217, CA096500, AI107810, AI109965, and DE018281, and M.B.M. is supported by grant DP2OD007149. We declare no competing financial interests. B.D. is a Leukemia and Lymphoma Society Scholar and a Burroughs Wellcome Fund Investigator in Infectious Disease.
We thank John Nicholas for the hIL-6 constructs and for critical reading of the manuscript. We also thank Yuan Chang and Patrick Moore for the pcDNA3.1-vIL-6-His construct and the AIDS and Cancer Specimen Resource (ACSR) for the KS sections. We thank members of the Damania laboratory for discussions.
While this paper was under review, a paper showing similar effects of vIL-6 on migration was published (55).
Footnotes
Published ahead of print 11 June 2014
REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869. 10.1126/science.7997879 [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186–1191. 10.1056/NEJM199505043321802 [DOI] [PubMed] [Google Scholar]
- 3.Gessain A, Sudaka A, Briere J, Fouchard N, Nicola M, Rio B, Arborio M, Troussard X, Audouin J, Diebold J. 1996. Kaposi sarcoma-associated herpes-like virus (human herpesvirus type 8) DNA sequences in multicentric Castleman's disease: is there any relevant association in non-human immunodeficiency virus-infected patients? Blood 87:414–416 [PubMed] [Google Scholar]
- 4.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M, Clauvel J, Raphael M, Degos L, Sigaux F. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276–1280 [PubMed] [Google Scholar]
- 5.Beral V, Peterman TA, Berkelman RL, Jaffe HW. 1990. Kaposi's sarcoma among persons with AIDS: a sexually transmitted infection? Lancet 335:123–128. 10.1016/0140-6736(90)90001-L [DOI] [PubMed] [Google Scholar]
- 6.Renne R, Lagunoff M, Zhong W, Ganem D. 1996. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J. Virol. 70:8151–8154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore PS, Boshoff C, Weiss RA, Chang Y. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739–1744. 10.1126/science.274.5293.1739 [DOI] [PubMed] [Google Scholar]
- 8.Neipel F, Albrecht JC, Ensser A, Huang YQ, Li JJ, Friedman-Kien AE, Fleckenstein B. 1997. Human herpesvirus 8 encodes a homolog of interleukin-6. J. Virol. 71:839–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholas J, Ruvolo VR, Burns WH, Sandford G, Wan X, Ciufo D, Hendrickson SB, Guo H-G, Hayward GS, Reixz MS. 1997. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat. Med. 3:287–292. 10.1038/nm0397-287 [DOI] [PubMed] [Google Scholar]
- 10.Sun R, Lin S-F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandriani S, Ganem D. 2010. Array-based transcript profiling and limiting-dilution reverse transcription-PCR analysis identify additional latent genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 84:5565–5573. 10.1128/JVI.02723-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen D, Sandford G, Nicholas J. 2009. Intracellular signaling mechanisms and activities of human herpesvirus 8 interleukin-6. J. Virol. 83:722–733. 10.1128/JVI.01517-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoki Y, Yarchoan R, Wyvill K, Okamoto S-i, Little RF, Tosato G. 2001. Detection of viral interleukin-6 in Kaposi sarcoma-associated herpesvirus-linked disorders. Blood 97:2173–2176. 10.1182/blood.V97.7.2173 [DOI] [PubMed] [Google Scholar]
- 14.Aoki Y, Tosato G, Fonville TW, Pittaluga S. 2001. Serum viral interleukin-6 in AIDS-related multicentric Castleman disease. Blood 97:2526–2527. 10.1182/blood.V97.8.2526 [DOI] [PubMed] [Google Scholar]
- 15.Parravicini C, Corbellino M, Paulli M, Magrini U, Lazzarino M, Moore P, Chang Y. 1997. Expression of a virus-derived cytokine, KSHV vIL-6, in HIV-seronegative Castleman's disease. Am. J. Pathol. 151:1517–1522 [PMC free article] [PubMed] [Google Scholar]
- 16.Aoki Y, Jaffe E, Chang Y, Jones K, Teruya-Feldstein J, Moore P, Tosato G. 1999. Angiogenesis and hematopoiesis induced by Kaposi's sarcoma-associated herpesvirus-encoded interleukin-6. Blood 93:4034–4043 [PubMed] [Google Scholar]
- 17.Suthaus J, Stuhlmann-Laeisz C, Tompkins VS, Rosean TR, Klapper W, Tosato G, Janz S, Scheller J, Rose-John S. 2012. HHV-8-encoded viral IL-6 collaborates with mouse IL-6 in the development of multicentric Castleman disease in mice. Blood 119:5173–5181. 10.1182/blood-2011-09-377705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori Y, Nishimoto N, Ohno M, Inagi R, Dhepakson P, Amou K, Yoshizaki K, Yamanishi K. 2000. Human herpesvirus 8-encoded interleukin-6 homologue (viral IL-6) induces endogenous human IL-6 secretion. J. Med. Virol. 61:332–335. [DOI] [PubMed] [Google Scholar]
- 19.Zhou F, Xue M, Qin D, Zhu X, Wang C, Zhu J, Hao T, Cheng L, Chen X, Bai Z, Feng N, Gao S-J, Lu C. 2013. HIV-1 Tat promotes Kaposi's sarcoma-associated herpesvirus (KSHV) vIL-6-induced angiogenesis and tumorigenesis by regulating PI3K/PTEN/AKT/GSK-3β signaling pathway. PLoS One 8:e53145. 10.1371/journal.pone.0053145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hideshima T, Chauhan D, Teoh G, Raje N, Treon SP, Tai Y-T, Shima Y, Anderson KC. 2000. Characterization of signaling cascades triggered by human interleukin-6 versus Kaposi's sarcoma-associated herpes virus-encoded viral interleukin 6. Clin. Cancer Res. 6:1180–1189 [PubMed] [Google Scholar]
- 21.Molden J, Chang Y, You Y, Moore PS, Goldsmith MA. 1997. A Kaposi's sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J. Biol. Chem. 272:19625–19631. 10.1074/jbc.272.31.19625 [DOI] [PubMed] [Google Scholar]
- 22.Osborne J, Moore PS, Chang Y. 1999. KSHV-encoded viral IL-6 activates multiple human IL-6 signaling pathways. Hum. Immunol. 60:921–927. 10.1016/S0198-8859(99)00083-X [DOI] [PubMed] [Google Scholar]
- 23.Aoki Y, Narazaki M, Kishimoto T, Tosato G. 2001. Receptor engagement by viral interleukin-6 encoded by Kaposi sarcoma-associated herpesvirus. Blood 98:3042–3049. 10.1182/blood.V98.10.3042 [DOI] [PubMed] [Google Scholar]
- 24.Chow D-C, He X-L, Snow AL, Rose-John S, Garcia KC. 2001. Structure of an extracellular gp130 cytokine receptor signaling complex. Science 291:2150–2155. 10.1126/science.1058308 [DOI] [PubMed] [Google Scholar]
- 25.Wan X, Wang H, Nicholas J. 1999. Human herpesvirus 8 interleukin-6 (vIL-6) signals through gp130 but has structural and receptor-binding properties distinct from those of human IL-6. J. Virol. 73:8268–8278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu F, Nicholas J. 2006. Signal transduction by human herpesvirus 8 viral interleukin-6 (vIL-6) is modulated by the nonsignaling gp80 subunit of the IL-6 receptor complex and is distinct from signaling induced by human IL-6. J. Virol. 80:10874–10878. 10.1128/JVI.00767-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meads MB, Medveczky PG. 2004. Kaposi's sarcoma-associated herpesvirus-encoded viral interleukin-6 is secreted and modified differently than human interleukin-6: evidence for a unique autocrine signaling mechanism. J. Biol. Chem. 279:51793–51803. 10.1074/jbc.M407382200 [DOI] [PubMed] [Google Scholar]
- 28.Chen D, Choi YB, Sandford G, Nicholas J. 2009. Determinants of secretion and intracellular localization of human herpesvirus 8 interleukin-6. J. Virol. 83:6874–6882. 10.1128/JVI.02625-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen D, Cousins E, Sandford G, Nicholas J. 2012. Human herpesvirus 8 viral interleukin-6 interacts with splice variant 2 of vitamin K epoxide reductase complex subunit 1. J. Virol. 86:1577–1588. 10.1128/JVI.05782-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cousins E, Nicholas J. 2013. Role of human herpesvirus 8 interleukin-6-activated gp130 signal transducer in primary effusion lymphoma cell growth and viability. J. Virol. 87:10816–10827. 10.1128/JVI.02047-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen D, Gao Y, Nicholas J. 2014. Human herpesvirus 8 interleukin-6 contributes to primary effusion lymphoma cell viability via suppression of proapoptotic cathepsin D, a cointeraction partner of vitamin K epoxide reductase complex subunit 1 variant 2. J. Virol. 88:1025–1038. 10.1128/JVI.02830-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozawa K, Tsukamoto Y, Hori O, Kitao Y, Yanagi H, Stern DM, Ogawa S. 2001. Regulation of tumor angiogenesis by oxygen-regulated protein 150, an inducible endoplasmic reticulum chaperone. Cancer Res. 61:4206–4213 [PubMed] [Google Scholar]
- 33.Wang Y, Wu Z, Li D, Wang D, Wang X, Feng X, Xia M. 2011. Involvement of oxygen-regulated protein 150 in AMP-activated protein kinase-mediated alleviation of lipid-induced endoplasmic reticulum stress. J. Biol. Chem. 286:11119–11131. 10.1074/jbc.M110.203323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stojadinovic A, Hooke JA, Shriver CD, Nissan A, Kovatich AJ, Kao TC, Ponniah S, Peoples GE, Moroni M. 2007. HYOU1/Orp150 expression in breast cancer. Med. Sci. Monit. 13:BR231–BR239 [PubMed] [Google Scholar]
- 35.Chiu C-C, Lin C-Y, Lee L-Y, Chen Y-J, Lu Y-C, Wang H-M, Liao C-T, Chang JT-C, Cheng A-J. 2011. Molecular chaperones as a common set of proteins that regulate the invasion phenotype of head and neck cancer. Clin. Cancer Res. 17:4629–4641. 10.1158/1078-0432.CCR-10-2107 [DOI] [PubMed] [Google Scholar]
- 36.Kuwabara K, Matsumoto M, Ikeda J, Hori O, Ogawa S, Maeda Y, Kitagawa K, Imuta N, Kinoshita T, Stern DM, Yanagi H, Kamada T. 1996. Purification and characterization of a novel stress protein, the 150-kDa oxygen-regulated protein (ORP150), from cultured rat astrocytes and its expression in ischemic mouse brain. J. Biol. Chem. 271:5025–5032. 10.1074/jbc.271.9.5025 [DOI] [PubMed] [Google Scholar]
- 37.Haque M, Davis DA, Wang V, Widmer I, Yarchoan R. 2003. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains hypoxia response elements: relevance to lytic induction by hypoxia. J. Virol. 77:6761–6768. 10.1128/JVI.77.12.6761-6768.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozawa K, Kuwabara K, Tamatani M, Takatsuji K, Tsukamoto Y, Kaneda S, Yanagi H, Stern DM, Eguchi Y, Tsujimoto Y, Ogawa S, Tohyama M. 1999. 150-kDa oxygen-regulated protein (ORP150) suppresses hypoxia-induced apoptotic cell death. J. Biol. Chem. 274:6397–6404. 10.1074/jbc.274.10.6397 [DOI] [PubMed] [Google Scholar]
- 39.Ozawa K, Kondo T, Hori O, Kitao Y, Stern DM, Eisenmenger W, Ogawa S, Ohshima T. 2001. Expression of the oxygen-regulated protein ORP150 accelerates wound healing by modulating intracellular VEGF transport. J. Clin. Invest. 108:41–50. 10.1172/JCI11772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolf MJ, Hoos A, Bauer J, Boettcher S, Knust M, Weber A, Simonavicius N, Schneider C, Lang M, Sturzl M, Croner RS, Konrad A, Manz MG, Moch H, Aguzzi A, van Loo G, Pasparakis M, Prinz M, Borsig L, Heikenwalder M. 2012. Endothelial CCR2 signaling induced by colon carcinoma cells enables extravasation via the JAK2-Stat5 and p38MAPK pathway. Cancer Cell 22:91–105. 10.1016/j.ccr.2012.05.023 [DOI] [PubMed] [Google Scholar]
- 41.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, Kaiser EA, Snyder LA, Pollard JW. 2011. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 475:222–225. 10.1038/nature10138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205–4220. 10.1128/JVI.77.7.4205-4220.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Wakisaka N, Tomlinson CC, DeWire SM, Krall S, Pagano JS, Damania B. 2004. The Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) K1 protein induces expression of angiogenic and invasion factors. Cancer Res. 64:2774–2781. 10.1158/0008-5472.CAN-03-3653 [DOI] [PubMed] [Google Scholar]
- 44.West J, Damania B. 2008. Upregulation of the TLR3 pathway by Kaposi's sarcoma-associated herpesvirus during primary infection. J. Virol. 82:5440–5449. 10.1128/JVI.02590-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rose-John S, Schooltink H, Schmitz-Van de Leur H, Mullberg J, Heinrich PC, Graeve L. 1993. Intracellular retention of interleukin-6 abrogates signaling. J. Biol. Chem. 268:22084–22091 [PubMed] [Google Scholar]
- 46.Isono T. 2011. O-GlcNAc-specific antibody CTD110.6 cross-reacts with N-GlcNAc2-modified proteins induced under glucose deprivation. PLoS One 6:e18959. 10.1371/journal.pone.0018959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhatt AP, Bhende PM, Sin S-H, Roy D, Dittmer DP, Damania B. 2010. Dual inhibition of PI3K and mTOR inhibits autocrine and paracrine proliferative loops in PI3K/Akt/mTOR-addicted lymphomas. Blood 115:4455–4463. 10.1182/blood-2009-10-251082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siesser PF, Motolese M, Walker MP, Goldfarb D, Gewain K, Yan F, Kulikauskas RM, Chien AJ, Wordeman L, Major MB. 2012. FAM123A binds to microtubules and inhibits the guanine nucleotide exchange factor ARHGEF2 to decrease actomyosin contractility. Sci. Signal. 5:ra64. 10.1126/scisignal.2002871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oksenhendler E, Carcelain G, Aoki Y, Boulanger E, Maillard A, Clauvel J, Agbalika F. 2000. High levels of human herpesvirus 8 viral load, human interleukin-6, interleukin-10, and C reactive protein correlate with exacerbation of multicentric Castleman disease in HIV-infected patients. Blood 96:2069–2073 [PubMed] [Google Scholar]
- 50.Matsuyama M, Suzuki T, Tsuboi H, Ito S, Mamura M, Goto D, Matsumoto I, Tsutsumi A, Sumida T. 2007. Anti-interleukin-6 receptor antibody (tocilizumab) treatment of multicentric Castleman's disease. Intern. Med. 46:771–774. 10.2169/internalmedicine.46.6262 [DOI] [PubMed] [Google Scholar]
- 51.Dela Cruz CS, Lee Y, Viswanathan SR, El-Guindy AS, Gerlach J, Nikiforow S, Shedd D, Gradoville L, Miller G. 2004. N-linked glycosylation is required for optimal function of Kaposi's sarcoma herpesvirus-encoded, but not cellular, interleukin 6. J. Exp. Med. 199:503–514. 10.1084/jem.20031205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boshoff C, Schulz TF, Kennedy MM, Graham AK, Fisher C, Thomas A, McGee JOD, Weiss RA, O'Leary JJ. 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274–1278. 10.1038/nm1295-1274 [DOI] [PubMed] [Google Scholar]
- 53.Hussein M. 2008. Immunohistological evaluation of immune cell infiltrate in cutaneous Kaposi's sarcoma. Cell Biol. Int. 32:157–162. 10.1016/j.cellbi.2007.08.021 [DOI] [PubMed] [Google Scholar]
- 54.Morris VA, Punjabi AS, Wells RC, Wittkopp CJ, Vart R, Lagunoff M. 2012. The KSHV viral IL-6 homolog is sufficient to induce blood to lymphatic endothelial cell differentiation. Virology 428:112–120. 10.1016/j.virol.2012.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu J, Xu Y, Mo D, Huang P, Sun R, Huang L, Pan S, Xu J. 2014. Kaposi's sarcoma-associated herpesvirus (KSHV) vIL-6 promotes cell proliferation and migration by upregulating DNMT1 via STAT3 activation. PLoS One 9:e93478. 10.1371/journal.pone.0093478 [DOI] [PMC free article] [PubMed] [Google Scholar]