ABSTRACT
The downregulation of immune synapse components such as major histocompatibility complex class I (MHC-I) and ICAM-1 is a common viral immune evasion strategy that protects infected cells from targeted elimination by cytolytic effector functions of the immune system. Kaposi's sarcoma-associated herpesvirus (KSHV) encodes two membrane-bound ubiquitin E3 ligases, called K3 and K5, which share the ability to induce internalization and degradation of MHC-I molecules. Although individual functions of K3 and K5 outside the viral genome are well characterized, their roles during the KSHV life cycle are still unclear. In this study, we individually introduced the amino acid-coding sequences of K3 or K5 into a ΔK3 ΔK5 recombinant virus, at either original or interchanged genomic positions. Recombinants harboring coding sequences within the K5 locus showed higher K3 and K5 protein expression levels and more rapid surface receptor downregulation than cognate recombinants in which coding sequences were introduced into the K3 locus. To identify infected cells undergoing K3-mediated downregulation of MHC-I, we employed a novel reporter virus, called red-green-blue-BAC16 (RGB-BAC16), which was engineered to harbor three fluorescent protein expression cassettes: EF1α-monomeric red fluorescent protein 1 (mRFP1), polyadenylated nuclear RNA promoter (pPAN)-enhanced green fluorescent protein (EGFP), and pK8.1-monomeric blue fluorescent protein (tagBFP), marking latent, immediate early, and late viral gene expression, respectively. Analysis of RGB-derived K3 and K5 deletion mutants showed that while the K5-mediated downregulation of MHC-I was concomitant with pPAN induction, the reduction of MHC-I surface expression by K3 was evident in cells that were enriched for pPAN-driven EGFPhigh and pK8.1-driven blue fluorescent protein-positive (BFP+) populations. These data support the notion that immunoreceptor downregulation occurs by a sequential process wherein K5 is critical during the immediately early phase and K3 plays a significant role during later stages.
IMPORTANCE Although the roles of K3 and K5 outside the viral genome are well characterized, the function of these proteins in the context of the KSHV life cycle has remained unclear, particularly in the case of K3. This study examined the relative contributions of K3 and K5 to the downregulation of MHC-I during the lytic replication of KSHV. We show that while K5 acts immediately upon entry into the lytic phase, K3-mediated downregulation of MHC-I was evident during later stages of lytic replication. The identification of distinctly timed K3 and K5 activities significantly advances our understanding of KSHV-mediated immune evasion. Crucial to this study was the development of a novel recombinant KSHV, called RGB-BAC16, which facilitated the delineation of stage-specific phenotypes.
INTRODUCTION
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) is a gammaherpesvirus that causes at least three human diseases: KS, primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD) (1). KSHV is able to establish lifelong infections that are usually asymptomatic in immunocompetent individuals, despite the persistent threat of both innate and adaptive immune surveillance. The formation of immunological synapses (ISs) plays a critical role in orchestrating cell-mediated immune responses, including the activation of cytolytic effector functions of CD8+ T cells and NK cells (2). As a countermeasure, many viruses encode immunoevasins that selectively downregulate major histocompatibility complex class I (MHC-I) molecules (3). KSHV has a remarkable capacity to manipulate host cell machinery; approximately 25% of its genome encodes factors devoted to this activity, including several homologues of cellular genes appropriated by the virus during evolution (4, 5). Many of these genes encode factors that negatively regulate the expression of proteins on the cell surface. Examples of such genes include viral interferon requlatory factor 1 (vIRF1) and vIRF3, which repress transcription of MHC-I and MHC-II, respectively (6, 7); LANA, which interferes with CIITA transcription, resulting in reduced MHC-II expression (8); miR-K12-7, which destabilizes MICB mRNA (9); ORF54, which induces relocalization of NKp44L from the cell surface (10); and two membrane-associated RING-CH (MARCH) family E3 ligases, K3 and K5, which direct the ubiquitination and subsequent internalization and endolysosomal degradation of several different plasma membrane substrates (reviewed in reference 11).
K3 and K5 are prototypic members of the MARCH family of membrane-bound E3 ubiquitin ligases, named for the characteristic amino-terminal C4HC3 zinc-binding domain and type III membrane topology shared by most members (12). Members of this family are part of a growing number of E3 ligases that target plasma membrane proteins for ubiquitin-dependent internalization (13, 14). Several poxviruses and gammaherpesviruses encode MARCH ligases, and 11 homologues (termed MARCH 1 to MARCH 11) have been identified in the human genome (15). Like their viral counterparts, many cellular MARCH proteins appear to play a role in tempering immune responses by targeting IS components and other immune cell activators (15–20).
K3 and K5 proteins were originally identified based on their ability to downregulate surface MHC class I molecules (21–24). Subsequent studies revealed an increasing number of K5 substrates, including the NKT cell ligand CD1d (25); the MHC-I-related molecule HFE (26); the adhesion molecules ICAM-1 (27, 28), PECAM (29), VE-cadherin (30), ALCAM (31), DC-SIGN, and DC-SIGNR (32); the costimulatory molecule B7-2 (27, 28); the NK cell-activating ligands, MICA, MICB, and AICL (33); the cellular restriction factor tetherin (34); the cytokine receptor interferon gamma receptor 1 (IFN-γR1) (35); the plasma membrane t-SNARE syntaxin-4 (36); and the transforming growth factor β (TGF-β) family member BMPRII (37). In addition, Timms et al. recently reported several novel K5 substrates, including 8 verified targets, CD32, CD33, CD99, EPHB4, Plexin A1, PMZL2, Kit (CD117), and IL9R (CD129), and 66 potential new targets, all of which were identified via a quantitative mass spectrometry approach (38). In contrast, substrates of K3 are limited to MHC class I molecules (HLA-A, -B, -C, and -E) (22), CD1d (25), PECAM, ALCAM (31), and IFN-γR1 (35), with HLA-C and HLA-E being the only substrates exclusive to K3.
Although much is known about K3 and K5 in terms of their substrate specificities and molecular mechanisms of downregulation, most studies have relied on overexpression systems, and relatively less is known about the role of K3 and K5 in the context of KSHV-infected cells. At the mRNA level, K3 and K5 are detectable as immediate early transcripts (39–41), although they have also been proposed to be early genes (42, 43). This discrepancy may reflect differences in the concentrations of cycloheximide used to differentiate between immediate early and early kinetics. A latent K3 transcript has also been described (41), although its functional significance is not known. In addition, K5 expression occurs as part of the “lytic burst” that follows de novo infection (44); depletion of K5 during this prelatency stage abrogated KSHV-mediated downregulation of MHC-I and ICAM-I (45). The extent of IS component clearance from the plasma membrane was correlated with the intracellular viral load (46). The characterization of KSHV K3 and K5 deletion mutants confirmed the importance of K5 in surface receptor modulation, but surprisingly, no obvious changes in MHC-I surface expression were apparent in cells infected with K3-deficient viruses (47), indicating that K5, but not K3, plays a dominant role in the context of infected cells. Similar observations were made for KSHV-mediated downregulation of DC-SIGN and DC-SIGNR, two substrates that can be downregulated by either K3 or K5 (32).
To better understand the role of K3 and K5 in KSHV-infected cells, specifically the reasons for the apparent inactivity of K3, we generated recombinant viruses harboring a single copy of K3 or K5 in their authentic loci or at interchanged genomic positions. The ability of these viruses to induce surface receptor downregulation was examined in iSLK cells undergoing lytic replication. In addition, a novel reporter virus harboring an EF1α-mRFP1-pPAN-EGFP-pK8.1-tagBFP expression cassette was used to analyze the role of K3 and K5 in relation to viral promoter activity and lytic replication. This enabled the identification of a significant role for K3-mediated surface receptor downregulation during later stages of lytic replication.
MATERIALS AND METHODS
Viruses and cells.
iSLK and iSLK-BAC16 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, 1 μg/ml puromycin, 250 μg/ml G418, and 1 mg/ml hygromycin B (for stable KSHV-infected cells). Purified bacterial artificial chromosome (BAC) DNA was introduced into iSLK cells via Fugene HD (Promega, Fitchburg, WI) transfection as previously described (47). In brief, BAC DNA was isolated from a 5-ml bacterial culture and resuspended in 40 μl of distilled water. iSLK cells were seeded at 2 × 105 cells/well of a 6-well plate or at ∼70% confluence. The following day, the medium was changed to optiMEM (Gibco) 30 min prior to the addition of the transfection complexes (fetal bovine serum [FBS] and penicillin-streptomycin were excluded from the medium). Transfection complexes were prepared by combining approximately 10 μl BAC DNA (25% of the yield of a 5-ml culture grown overnight) and 90 μl of optiMEM, followed by the addition of 5 μl of Fugene HD (Promega). After 10 min of incubation at room temperature, the complexes were added to the cells. Three hours after the transfection complexes were added, FBS was added to optiMEM to a final concentration of 10%. The following day, transfected cells were trypsinized, transferred into 10-cm dishes, and cultured in the presence of DMEM supplemented with 10% FBS and 1% penicillin-streptomycin without other antibiotics. Two days after transfection, iSLK-BAC cell lines were maintained in the presence of 1 μg/ml puromycin, 250 μg/ml G418, and 600 μg/ml hygromycin B. Once green fluorescent protein-positive (GFP+) colonies were established (∼1 to 2 weeks), the concentration of hygromycin B in the medium was increased to 1,000 μg/ml. Lytic replication was induced in the presence of 1 μg/ml doxycycline and 1 mM sodium butyrate (both purchased from Sigma Life Science, St. Louis, MO) and in the absence of hygromycin, puromycin, and G418.
Virus stocks were prepared from stable iSLK cells as previously described (48), except that iSLK-puro cells were used as a producer cell line. In brief, stably infected iSLK-BAC16 cells were induced into the lytic phase as described above. Four days later, the supernatant was collected and cleared of cells and debris by centrifugation (950 × g for 10 min at 4°C) and filtration (0.45-μm-pore-size filter). Virus particles were pelleted by ultracentrifugation (25,000 × g for 3 h at 4°C) by using an SW32Ti rotor.
Plasmid and BACmid constructs.
The KSHV genome was modified by using a two-step “scarless” homologous recombination procedure that was described previously (49, 50). We used Escherichia coli GS1783 (a gift from Greg Smith, Northwestern University) harboring BAC16, a BAC clone of the KSHV genome (47, 49). PCR amplification was used to generate linear DNA fragments that included a kanamycin resistance gene expression cassette, an I-SceI cleavage site, and flanking sequences for homologous recombination. Linear fragments were amplified by using a High Fidelity PCR Master kit (Roche). In order to reintroduce K3 and K5 into their original genomic positions, we used the K3in and K5in primer sets and the pTracer-K3in and pTracer-K5in plasmid templates, respectively, all of which were described previously (47). For the reintroduction of K3 into the K5 locus, the pTracer-K3in template was amplified by using primers K3→K5loc-For (CACTCTGCTCACCTCCCCTTTCCCTTTTTCAGACTTCCACCCCAGCTGCAGAGATGGAAGATGAGGATGTTCCTGTCTG) and K3→K5loc-Rev (GGTGCATAACACCCAGGGCGTCACGTCACATATCTCTGTGCACCCAAGTGGTTGTTCAATGGTGATGGTGATGATGACCGGTACGCGTAG). For the reintroduction of K5 into the K3 locus, the pTracer-K5in template was amplified by using primers K5→K3loc-For (CACTTGTTGCAGGGGTTAATGCCATGTTTTATTGTGGGTTCTCTCTCAGGATATGGCGTCTAAGGACGTAGAAGAGG) and K5→K3loc-Rev (CGAGGGTATAGGTAAACACCACCAACCACACAGTGTGCTCTTATATACTTTCAATGGTGATGGTGATGATGACCGGTAC). For the replacement of enhanced GFP (EGFP) with monomeric red fluorescent protein 1 (mRFP1), the pEPmRFP1-in template (50) was amplified by using primers mRFP1replEGFP-For (AAGCTGGCTAGGTAAGCTTGGTACCGAGCTCGGATCCACTAGTCCGCCACCATGGCCTCCTCCGAGGACGTCATC) and mRFP1replEGFP-Rev (GAGGGAGAGGGGCGGAATTCCTCTAGTGCGGCCGAGTCGCGGCCGCTTTACTTGTACAAGGCGCCGGTGGAGTG). The pK8.1-tagBFP-Kanr-pPAN-EGFP-pA cassette was amplified from plasmid ppK8.1-tagBFP-Kanr-pPAN-EGFP-pA (described below) by using primers GB-For (GTGGGCGATGTGCGCTCTGCCCACTGACGGGCACCGGAGCGCTTCGAATTCCGGCAGCAATA) and GB-Rev (GTTGGGTGCCACATAACTTCGTATAATGTATGCTATACGAAGTTATCCACAACTAGAATGCAGTGA). This expression cassette was inserted next to the EF1α promoter. Primers were purchased as ultramers from Integrated DNA Technologies.
To construct the pK8.1-tagBFP-Kanr-pPAN-EGFP-pA insertion cassette, the PAN promoter (nucleotides [nt] 28470 to 28640; GenBank accession number GQ994935) and a Kanr cassette (including an adjacent I-SceI cleavage site) were positioned adjacently by overlapping PCR and cloned into the multiple-cloning site (MCS) of pEGFP-N1 (Clontech) to generate pKanR-pPAN-EGFP. A primer that included the K8.1 promoter (51) (nt 75673 to 75732; accession number GQ994935) was used to PCR amplify tagBFP (Evrogen) and clone it into the MCS of pKanR-pPAN-EGFP-pA to generate ppK8.1-tagBFP-Kanr-pPAN-EGFP-pA. This plasmid also included a 40-bp duplication of the polyadenylated nuclear RNA promoter (pPAN) promoter positioned in between monomeric blue fluorescent protein (tagBFP) and the Kanr cassette, allowing scarless removal of the Kanr cassette during the subsequent recombination steps in GS1783-BAC16.
BAC DNA isolation and analysis.
BAC DNA was purified from 5-ml cultures of chloramphenicol-resistant colonies by alkaline lysis followed by isopropanol precipitation (52). Purified BAC DNA was digested with KpnI, CpoI, or SbfI and separated on a 1% agarose gel in 0.5× Tris-borate-EDTA (TBE) by using pulsed-field gel electrophoresis (CHEF-DR II; Bio-Rad) under the following conditions: 6 V/cm for 15 h; initial and final switch times of 1 and 5 s, respectively; and 14°C. To further verify recombinant BACmids, modified regions were PCR amplified and sequenced.
Quantification of infectious virus and KSHV DNA levels in cells.
Various amounts of cell-free virus supernatants were diluted in fresh medium (1-ml final volume/well) and used to inoculate 293A cells that were seeded at approximately 2 × 105cells/well into 12-well plates 12 h prior to infection. Following inoculation, plates were immediately centrifuged (2,000 × g for 45 min at 30°C) and then placed back into the CO2 incubator. One hour later, the inoculum was removed and replaced with fresh medium. Cells were collected 24 h later and washed once with cold phosphate-buffered saline (PBS). The percentage of GFP-positive cells was determined by using a FACSCanto II instrument (BD Bioscience, San Jose, CA). Infectious units (IU) are expressed as the number of GFP-positive cells in each well at the time of analysis. Purification of genomic DNA from infected cells and quantification of KSHV DNA levels were described previously (53). KSHV ORF11-specfic primers (ORF11-For [GGCACCCATACAGCTTCTACGA] and ORF11-Rev [CGTTTACTACTGCACACTGCA]) were used to quantify the amount of intracellular KSHV DNA in stable iSLK cell lines in the absence of or following 36 or 72 h of doxycycline and sodium butyrate treatment. The KSHV DNA amount was normalized to the amount of purified host cellular DNA (HS1-For [TTCCTATTTGCCAAGGCAGT] and HS1-Rev [CTCTTCAGCCATCCCAAGAC]).
RNA isolation and qPCR.
Total cellular RNA was extracted with Tri reagent (Sigma) according to the manufacturer's instructions. One microgram of total RNA was treated with DNase I (Sigma) and reverse transcribed by using an iScript cDNA synthesis kit (Bio-Rad). cDNA was quantified by using iQ SYBR green supermix (Bio-Rad) and a CFX96 quantitative PCR (qPCR) machine (Bio-Rad). The relative quantification of gene expression was calculated by using the comparative threshold cycle (CT) method (2−ΔΔCT) (54), and 18S RNA was used as a reference. Quantitative PCR (qPCR) graphs were made based on the averages of data from at least two independent experiments. qPCR primers used were 18S-For (TTCGAACGTCTGCCCTATCAA), 18S-Rev (GATGTGGTAGCCGTTTCTCAGG), K3-For (TCCTGGTAAGTCAGCCGAGGCA), K3-Rev (GGAAATGAGAGATTTAGAGCCT), K5-For (TAAGCACTTGGCTAACAGTGT), K5-Rev (GGCCACAGGTTAAGGCGACT), K2-For (TCACTGCGGGTTAATAGGATTT), K2-Rev (CATGACGTCCACGTTTATCACT), RTA-For (TTGCCAAGTTTGTACAACTGCT), and RTA-Rev (ACCTTGCAAAGACCATTCAGAT).
Immunoblotting.
Cell pellets were lysed in radioimmunoprecipitation assay (RIPA) buffer. Fifteen micrograms of whole-cell lysate was resolved by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Bio-Rad). The membranes were blocked by using 5% nonfat milk and then probed with antibodies diluted in phosphate-buffered saline with 0.1% Tween 20 (pH 7.4). The following primary antibodies were used: rabbit polyclonal anti-K3 (55), monoclonal mouse anti-K5 and anti-RTA (gifts from at Koichi Yamanishi [Osaka University, Japan]), monoclonal mouse anti-K8 (catalogue number ab36617; Abcam), monoclonal rat anti-LANA (catalogue number 13-210-100; ABI), β-actin (catalogue number 47778; Santa Cruz), mouse anti-V5 (Invitrogen), and anti-GFP (catalogue number 9996; Santa Cruz). The following secondary horseradish peroxidase-conjugated antibodies were used: anti-mouse (catalogue number 7076; Cell Signaling), anti-rabbit (catalogue number 7074; Cell Signaling), and anti-rat (catalogue number 2006; Santa Cruz). Chemiluminescence was detected by using a LAS-4000 digital imaging system (GE Healthcare).
Flow cytometry.
iSLK cells were detached following a short (∼3-min) incubation with 0.25% trypsin. A total of 1 × 106 cells per sample were stained and washed in fluorescence-activated cell sorter (FACS) buffer (PBS with 0.5% bovine serum albumin [BSA] and 1 mM EDTA). The following antibodies were used for surface staining: anti-HLA-ABC biotin (W6/32) and anti-CD54 allophycocyanin (APC) (HA58) (both from BD Bioscience, San Jose, CA) and and-HLA-ABC Alexa 647 (W6/32) (Santa Cruz, Santa Cruz, CA). Following surface staining, cells were either fixed in PBS with 2% paraformaldehyde and set aside for analysis or incubated with permeabilization buffer (PBS with 0.5% BSA, 0.1% saponin, and 0.01% sodium azide), fixed, and stained for intracellular K3 and K5 by using anti-V5 APC (Invitrogen). Biotin conjugates were detected by using streptavidin APC-eFluor 780 (eBioscience, San Diego, CA). In some experiments, a violet dead cell stain (Invitrogen) was used to exclude dead cells. Fluorescent protein expression and antigen staining were analyzed on an 8-color FACSCanto II flow cytometer (BD Bioscience). Data were analyzed by using FlowJo v.6.4.7 software (Tree Star, Ashland, OR).
RESULTS
Construction of recombinant KSHV with altered disposition of K3 and K5 ORFs.
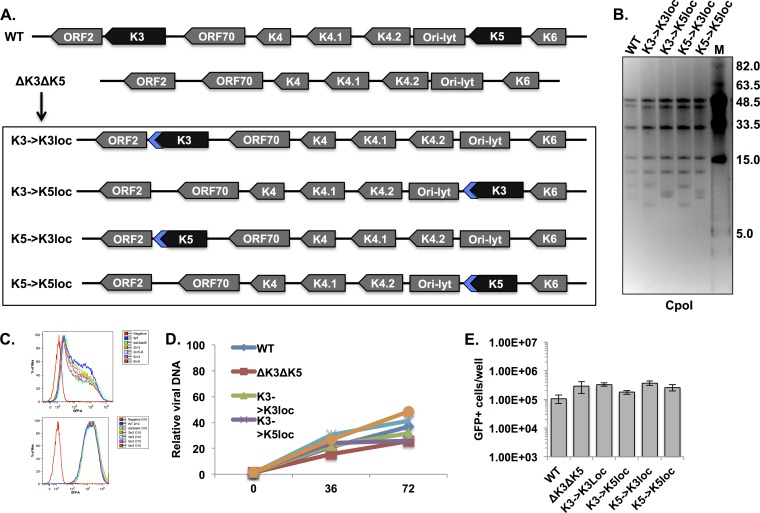
Previously, we found no obvious role for K3 during lytic replication by reverse genetics (47). Based on revertant viruses that express V5-tagged versions of either K3 or K5, we observed that protein levels of K3 were lower than those of K5 (47). Thus, the apparent inactivity of K3 might be due to insufficient protein expression. To address this possibility, a single copy of either the K3 or the K5 protein-encoding sequence was individually reintroduced into ΔK3 ΔK5 KSHV BAC16 (47) either at original genomic locations (K3→K3loc and K5→K5loc) or at interchanged positions (K3→K5loc and K5→K3loc) (Fig. 1A). The reintroduced K3 and K5 open reading frames (ORFs) included a carboxyl-terminal V5 tag to enable comparison of protein levels. Note that only the protein-encoding DNA was removed or reintroduced; 3′ and 5′ regulatory regions were left intact. The recombinant BACmids were verified by pulsed-field gel analysis of CpoI-digested BAC DNA and direct sequencing of the respective modified regions. Pulsed-field gel electrophoresis showed the expected digestion pattern for wild-type (WT) and mutant BACs, including full-length terminal repeat sequences (Fig. 1B). Since the K3 and K5 ORFs reside in CpoI fragments with lengths of 9,272 bp and 7,930 bp, respectively, the sizes of these fragments varied according to the presence or absence of ORFs at each locus (Fig. 1B).
FIG 1.
Construction and replication of recombinants with altered dispositions of the K3 and K5 ORFs. (A) Schematic depiction of the KSHV genomic region harboring the K3 and K5 ORFs (WT) and the BAC16-derived recombinants (ΔK3 ΔK5, K3→K3loc, K3→K5loc, K5→K3loc, and K5→K5loc) generated for this study (not drawn to scale). Targeted deletions in the ΔK3 ΔK5 recombinant span the entire coding sequences of K3 and K5, including start and stop codons. Blue arrows indicate carboxyl-terminal V5-tagged coding sequences included with the reintroduced K3 and K5 ORFs. (B) Pulsed-field gel electrophoresis of CpoI-digested BAC DNAs. CpoI fragment sizes of WT BAC16 were 51,393, 45,802, 34,729, 18,240, 12,497, 9,272, 7,930, and 243 bp. Lane M, molecular weight markers. (C) Flow cytometry analysis of GFP fluorescence at 24 h (top) and 10 days (bottom) after infection with the indicated recombinant viruses. (D) Relative amounts of viral DNA were quantified in latently infected cells (0 h) and lytically replicating cells (36 and 72 h after treatment with 1 μg/ml of doxycycline and 1 mM sodium butyrate). ORF11-specific primers were used to detect viral DNA, and values were normalized to cellular DNA levels. The graph shows relative viral DNA copy numbers, with the level of WT viral DNA during latency set to 1. (E) Cell-free supernatants were collected from iSLK cells after 72 h of lytic induction, and infectious virus particles were quantified by infecting 293A cells and counting GFP+ cells by flow cytometry. The experiment was performed in triplicate; error bars represent the standard deviations between replicates.
Recombinant viruses were produced by introducing BAC DNA into iSLK cells as previously described (47). We have observed significant differences in viral replication from iSLK-BAC16 cell lines that were independently established by transfection of the same WT BAC DNA, even when each cell line was transfected and selected at the same time and under the same conditions (data not shown). It is possible that a low BAC DNA transfection efficiency leads to the selection of distinct subpopulations of iSLK cells, and this may ultimately lead to extraneous differences in viral replication. In any case, to exclude this potential source of variation, the recombinant viruses were purified from cell-free supernatants, and infections were initiated at the same time by applying virus to naive iSLK cells at a multiplicity of infection (MOI) of ∼1. Cells were subsequently maintained under hygromycin selection for ∼2 weeks in order to establish latently infected cell lines and allow sufficient time for transient lytic gene expression to subside. To verify that an equivalent dose of each virus was delivered, infected iSLK cells were analyzed for GFP expression at 24 h postinfection and again at 10 days postinfection (Fig. 1C). At 24 h after infection, the percentages of GFP-positive cells were comparable (62 to 73%) among the different recombinants (Fig. 1C, top). Following 10 days of selection, all cells were GFP positive and showed similar distributions of fluorescent intensities (Fig. 1C, bottom).
In order to characterize the replication of these recombinant viruses, iSLK cell lines were harvested following induction with a combination of doxycycline and sodium butyrate for 0, 36, and 72 h. Comparable amounts of viral DNA were detected in cells carrying WT and mutant viruses during both latent infection and lytic reactivation (Fig. 1D). Furthermore, infectious virus production was quantified by seeding equivalent cell numbers of each cell line, inducing lytic replication for 72 h, and applying serial dilutions of cell-free supernatants to naive iSLK cells (Fig. 1E). Infectious virus production was not significantly altered by the presence, absence, or altered location of K3 or K5, verifying that these genes are not required for KSHV lytic replication in iSLK cells (Fig. 1E).
The K5 locus is more conducive to high protein expression levels.
K3 and K5 are expressed during lytic replication and can be transcribed with immediate early kinetics (39, 40). We previously observed that the level of K3 was markedly lower than that of K5 (47). To further examine the influence of genomic context on the expression of K3 and K5, iSLK stable cell lines harboring the K3 and K5 positional mutant viruses were harvested at 0, 36, and 72 h post-lytic induction. Protein levels of the latent gene LANA, the constitutively expressed GFP marker, and the immediate early genes K8 and RTA were similar among the different recombinants (Fig. 2A). Recombinant RTA is produced from a doxycycline-inducible expression cassette stably integrated within the iSLK cell genome and therefore expressed in the absence of KSHV infection (48). Recombinants harboring K3 or K5 in the K5 locus produced higher levels of protein than did those harboring K3 or K5 in the K3 locus (Fig. 2A). Based on densitometry analysis, the average difference in expression levels between the two loci was >30-fold, suggesting that the genomic context of these genes is an important determinant of their expression levels. An internal initiation codon within the K3 gene was recently identified (56). It is in frame with the primary ORF and produces a smaller protein, termed K3A, predicted to be 18.8 kDa (22.0 kDa in the carboxyl-terminally tagged recombinants). However, the expression of this gene product was not detected, suggesting that levels of K3A may be too low to be detected (Fig. 2A).
FIG 2.
The K5 locus is more conducive to high protein expression levels than the K3 locus. (A) iSLK cells or iSLK cells carrying the different recombinant KSHVs were induced with doxycycline and sodium butyrate for 36 or 72 h, and protein levels from whole-cell lysates were analyzed by immunoblotting by using the indicated antibodies. (B) mRNA was analyzed by RT-qPCR using the indicated primers. Values were normalized to 18S levels and are the averages of values from two independent experiments. The graphs shows relative expression levels, with WT levels set to 1.
K3-encoding transcripts were generated by the K3→K3loc and K3→K5loc recombinants, and K5-encoding transcripts were generated by the K5→K3loc and K5→K5loc viruses. Since both sets of viruses harbored identical sequences in two different loci, the amounts of mRNA produced at each locus were compared by qPCR using K3-specific as well as K5-specific primers (Fig. 2B). Analysis of total RNA isolated from cells after 72 h of lytic replication showed slightly (∼2- to 3-fold) higher levels of the K5 locus transcripts than of the K3 locus transcripts, a result that was consistent using either K3- or K5-specific primers (Fig. 2B). On the other hand, K3 protein levels in K3→K5loc-infected cells were higher than those infected with the WT, as shown by K3-specific Western blot analysis (Fig. 2A, compare lanes 5 and 6 to lanes 14 and 15), even though the K3→K5loc recombinant showed lower K3 mRNA levels than the WT (Fig. 2B). Thus, while a modest difference in mRNA expression levels between the K3 and K5 loci was evident, it does not fully explain the discrepancy in protein levels, suggesting that low K3 protein levels may be largely due to additional regulation besides transcriptional activity.
At low K3 and K5 expression levels, K5 is more effective than K3 at MHC-I downregulation.
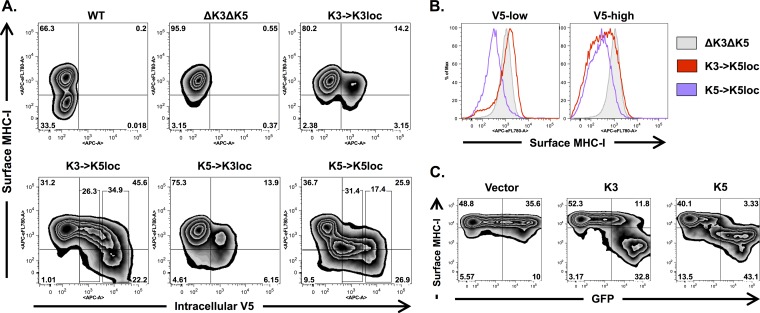
MHC class I molecules can be downregulated upon overexpression of either K3 or K5 (21–23). However, we previously reported that K3 is not required for MHC class I downregulation in the context of infected cells (47). Given the low levels of protein expression from the K3 locus, we examined whether the genomic position of K3 and K5 was also important for KSHV-mediated downregulation of MHC-I. Of particular interest was whether the higher K3 expression levels detected in the K3→K5loc recombinant were sufficient to restore WT levels of MHC-I downregulation activity during lytic replication. iSLK cells harboring different K3 and K5 positional mutants were collected after 3 days of doxycycline and sodium butyrate treatment, and MHC-I surface expression was measured by using an HLA-ABC-specific antibody (W6/32). Cells were subsequently permeabilized and stained with an APC-conjugated V5-specific antibody to monitor intracellular levels of recombinant K3 and K5. FACS analysis showed impaired downregulation of MHC-I by the ΔK3 ΔK5 recombinant compared to the WT (Fig. 3A). This defect was significantly reversed when either K3 or K5 was expressed from the K5 locus (Fig. 3A). In contrast, iSLK cells harboring the K3→K3loc and K5→K3loc recombinants showed minimal MHC-I downregulation (Fig. 3A). Under these conditions, the MHC-I level was reduced in ∼8% of K5→K3loc virus-infected cells, while only 3% of K3→K3loc virus-infected cells showed reduced MHC-I expression levels, suggesting stronger downregulation activity of K5 than of K3, even at low expression levels (Fig. 3A). Likewise, although the K3→K5loc recombinant could significantly restore downregulation activity (∼22% of cells showed reduced MHC-I levels), the K5→K5loc virus induced more extensive MHC-I downregulation (∼34%) (Fig. 3A).
FIG 3.
MHC-I is more susceptible to K5-mediated downregulation than to K3-mediated downregulation. (A) iSLK cells harboring different K3 and K5 positional mutant viruses were induced with doxycycline and sodium butyrate for 3 days and subsequently analyzed by flow cytometry. MHC-I surface expression was detected by using a biotin-conjugated HLA-ABC-specific antibody (W6/32) and APC-e780-conjugated streptavidin. V5-tagged recombinant K3 and K5 were detected by using an APC-conjugated V5-specific antibody. Boxed areas of the K3→K5loc and K5→K5loc flow plots represent the V5-low and V5-high gated cell populations used for panel B. (B) MHC-I surface expression among V5-low and V5-high cell populations of iSLK cells carrying the indicated mutant viruses. (C) BJAB cells were analyzed by flow cytometry at 24 h postelectroporation with an empty vector or K3 or K5 expression plasmids and analyzed by FACS 24 h later. An Alexa 647-conjugated HLA-ABC-specific antibody (W6/32) was used for surface staining.
Interestingly, intracellular staining of K3 in these infected cells revealed a distinctive, nonlinear relationship between MHC-I surface levels and K3 expression levels; low levels of K3 expression were completely insufficient to induce MHC-I downregulation, a phenomenon that was abruptly reversed once K3 reached higher expression levels (Fig. 3A and B). In striking contrast, low levels of K5 expression were just as effective at MHC-I downregulation as high levels of K5 expression (Fig. 3A and B). Notably, MHC-I downregulation was also evident in cells expressing K5 at levels that were below the detection limit (Fig. 3A). These results show an intrinsic difference between K3- and K5-mediated downregulation of MHC-I and suggest that K3 is less effective than K5 for MHC-I downregulation unless it reaches a certain expression level threshold. To confirm this, BJAB cells were electroporated with K3 or K5 expression plasmids. A cotransfected GFP-expressing plasmid was used as a surrogate indication of K3 and K5 expression levels. MHC-I surface expression in these transfected BJAB cells showed a similar pattern: K3 affected MHC-I surface expression only in cells expressing high levels of GFP, while K5 induced MHC-I downregulation in both low- and high-level GFP-expressing cells (Fig. 3C). This suggests that at low expression levels, K3 and K5 show differing abilities to affect MHC-I surface expression (Fig. 3C).
Positioning of K5 within the K3 locus causes delayed ICAM-1 downregulation.
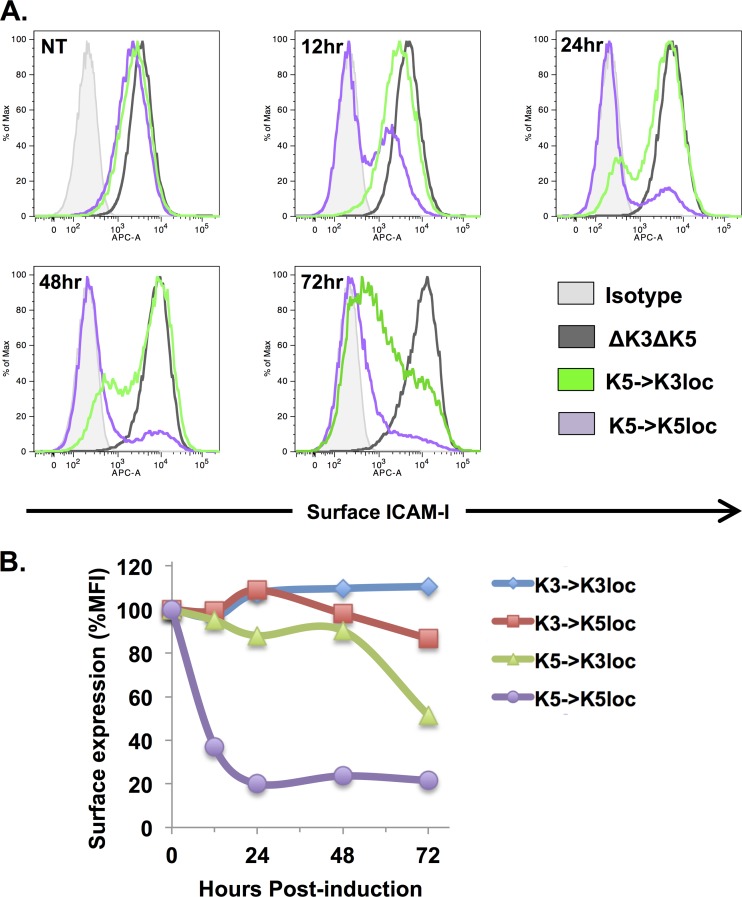
ICAM-1 is robustly cleared from the cell surface by K5 (but not by K3) (27, 28), and its downregulation is evident even in cells that express normal levels of MHC-I (46, 47). Thus, we used ICAM-1 surface expression as a sensitive readout to assess whether positioning of the K5 gene within the K3 locus affects immunoreceptor downregulation kinetics. iSLK cells harboring different K3 and K5 positional mutant viruses were treated with doxycycline and sodium butyrate to induce lytic replication, and ICAM-1 surface expression was measured at 0, 12, 24, 48, and 72 h posttreatment (Fig. 4A and B). While the K5→K5loc recombinant showed prompt ICAM-1 downregulation as early as 12 h postinduction, the ability of the K5→K3loc recombinant to downregulate ICAM-1 surface levels was significantly delayed, with extensive downregulation appearing only after 48 or 72 h of lytic replication (Fig. 4A and B). As expected, the K3→K3loc and K3→K5loc viruses did not affect ICAM-1 surface expression (Fig. 4B). Together, these data suggest that while the K5 locus is equipped for prompt downregulation of surface receptors, the K3 locus imposes inherent limitations on protein expression that result in delayed downregulation kinetics.
FIG 4.
ICAM-1 downregulation is delayed when K5 is expressed from the K3 locus. (A) iSLK cells harboring different K3 and K5 positional mutant viruses were induced with doxycycline and sodium butyrate for 12, 24, 48, and 72 h. ICAM-1 surface expression was detected by flow cytometry analysis using an APC-conjugated ICAM-1 antibody. Dead cells were excluded by using a fixable Live/Dead stain kit. NT, no treatment. (B) Geometric mean fluorescence intensities (MFI) of ICAM-1 surface staining were compared among iSLK cells harboring the indicated recombinants. Mean fluorescence intensity values for ΔK3 ΔK5-infected cells were set to 100%.
Construction and characterization of RGB-BAC16.
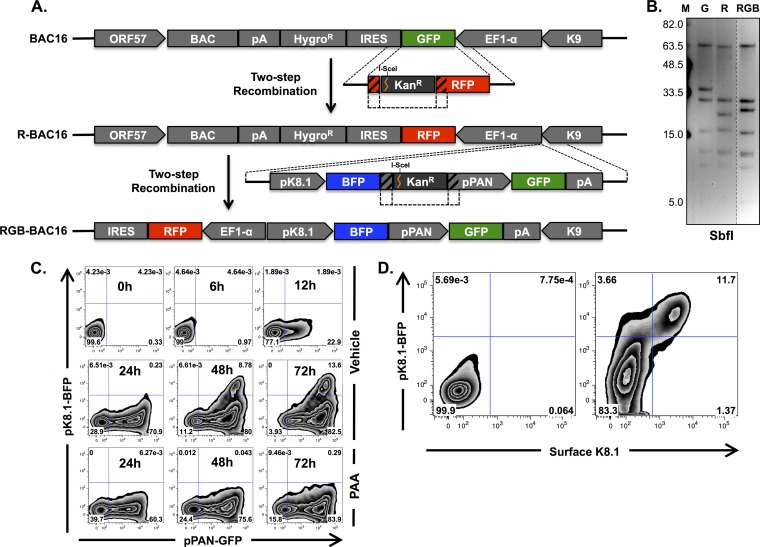
The analysis of positional mutants of K3 and K5 described above indicates that a K3-specific phenotype would likely be limited to an early or late stage of lytic replication. However, lytic replication of KSHV is asynchronous in cultured cells, and furthermore, only between 5 and 20% of cells can be found in a given stage of lytic replication (e.g., immediate early, early, or late) following chemical induction of lytic replication (57, 58). Thus, it would be difficult to pinpoint a stage-specific effect on MHC-I surface expression unless the expression of lytic genes within individual cells was monitored in parallel. To facilitate such an approach, we constructed a novel reporter virus, called red-green-blue-BAC16 (RGB-BAC16) wherein expressions of mRFP1, EGFP, and tagBFP were placed under the control of the constitutively active EF1α, the immediate early PAN, and the late K8.1 promoters, respectively (Fig. 5A). This was accomplished in two steps. First, the coding sequence of EGFP was replaced with that of mRFP1 by using a two-step recombination procedure (49, 50). Next, a subsequent round of two-step recombination was applied to introduce a pPAN-GFP-pK8.1-tagBFP-pA expression cassette (Fig. 5A). Genomic integrity of R-BAC16 and RGB-BAC16 was verified by pulsed-field gel electrophoresis and direct sequencing of the reporter gene regions (Fig. 5B). The presence of an SbfI site within the mRFP1 sequence and the introduction of the lytic gene reporter cassette each resulted in alterations of the SbfI digestion pattern of these BACmids (the 37,839-bp fragment of BAC16 becomes 26,800 bp and 11,001 bp in R-BAC16, and the 26,800-bp fragment of R-BAC16 becomes 28,687 bp in RGB-BAC16) (Fig. 5B). It should be noted that because the genetic origins of tagBFP, EGFP, and mRFP1 are different, there was negligible sequence identity, leaving little or no chance of homologous recombination between these fluorescent protein genes.
FIG 5.
Construction and characterization of RGB-BAC16. (A) Schematic depiction of the cloning strategy used for the construction of R-BAC16 and its descendant, RGB-BAC16 (not drawn to scale). (B) Pulsed-field gel electrophoresis of SbfI-digested BAC DNAs. SbfI digestion of WT BAC16 generates the following fragment sizes: 62,983, 37,839, 32,720, 18,531, 11,989, 9,750, and 6,294 bp. (C) Flow cytometry analysis of EGFP and tagBFP fluorescence of iSLK cells harboring RGB-BAC16 following 0, 6, 12, 24, 48, and 72 h of doxycycline and sodium butyrate treatment in the absence (top two rows) or presence (bottom row) of PAA. (D) Flow cytometry analysis of iSLK-RGB-BAC16 cells without treatment (left) and following 72 h of doxycycline and sodium butyrate treatment (right). K8.1 surface levels were detected by using a K8.1-specific antibody and an APC-e780-conjugated secondary antibody.
The kinetics of reporter gene expression of WT RGB-BAC16 was analyzed by flow cytometry following induction of lytic replication in the presence or absence of the viral DNA replication inhibitor phosphonoacetic acid (PAA) (Fig. 5C). As expected, pPAN-driven EGFP expression was promptly induced, while pK8.1-driven tagBFP expression was detected only after the onset of viral DNA replication, which occurs at between 24 and 48 h postinduction (56; data not shown). Moreover, tagBFP but not EGFP expression was sensitive to PAA treatment, consistent with authentic viral gene expression kinetics, where viral DNA replication is necessary only for the expression of late genes. To further verify that tagBFP accurately marks late-gene-expressing cells, K8.1 surface expression and tagBFP expression were analyzed at 72 h postinduction (Fig. 5D). Indeed, tagBFP expression was coincident with K8.1 expression (Fig. 5D). These results demonstrate that RGB-BAC16 can be used to accurately identify cells at specific stages of lytic replication without necessitating the use of fixatives or antibody-mediated detection.
Construction and characterization of RGB-derived K3 and K5 deletion mutants.
Our previous analysis of K3 and K5 deletion mutants showed that in the absence of K5, KSHV-mediated downregulation of MHC-I is abolished, even in the presence of an intact K3 gene (47). However, those results were limited to relatively early time points of lytic replication (24 and 48 h postinduction) and did not incorporate concurrent analysis of lytic gene expression. Thus, we decided to revisit the role of K3 in KSHV-mediated immunoreceptor downregulation using RGB-BAC16. To this end, the K3 and K5 coding sequences were removed from RGB-BAC16, either individually or in succession, to generate single- and double-deletion mutants (Fig. 6A). The successful removal of K3 and K5 and the overall integrity of the mutant genomes were verified by pulsed-field gel electrophoresis and direct sequencing, as described above (Fig. 6B). The BACmids were introduced into iSLK cells as detailed above. Following 3 days of doxycycline and sodium butyrate treatment, iSLK cells harboring the different recombinants were collected, and reporter gene expression was analyzed by flow cytometry (Fig. 6C). Comparable levels of pPAN- and K8.1-driven gene expression were detected in the recombinant BACmids, as evidenced by similar percentages of cells within the EGFPhigh and GFP+ BFP+ gates, consistent with our previous findings demonstrating that K3 and K5 are dispensable for lytic gene expression and viral replication (47) (Fig. 6C).
FIG 6.
Construction and characterization of K3 and K5 deletion mutants of RGB-BAC16. (A) Schematic depiction of RGB-BAC16 recombinants generated. Complete and scarless deletions (from the start codon to the stop codon) were engineered within the K3 or K5 ORFs, and a ΔK3 ΔK5 BAC was subsequently derived from the ΔK3 recombinant (not drawn to scale). (B) Pulsed-field gel electrophoresis of CpoI-digested BAC DNA. CpoI fragment sizes for WT RGB-BAC16 were 51,390, 45,802, 36,578, 18,240, 12,497, 9,272, 7,930, and 243 bp. (C) EGFP and tagBFP fluorescence of iSLK cells harboring WT or mutant RGB-BACmids following 72 h of doxycycline and sodium butyrate treatment analyzed by FACS. The EGFPhigh and GFP+ BFP+ gates and the corresponding percentages are shown. The EGFPhigh gate was set according to an arbitrary EGFP intensity, and the GFP+ BFP+ gates was set based on a PAA-treated control.
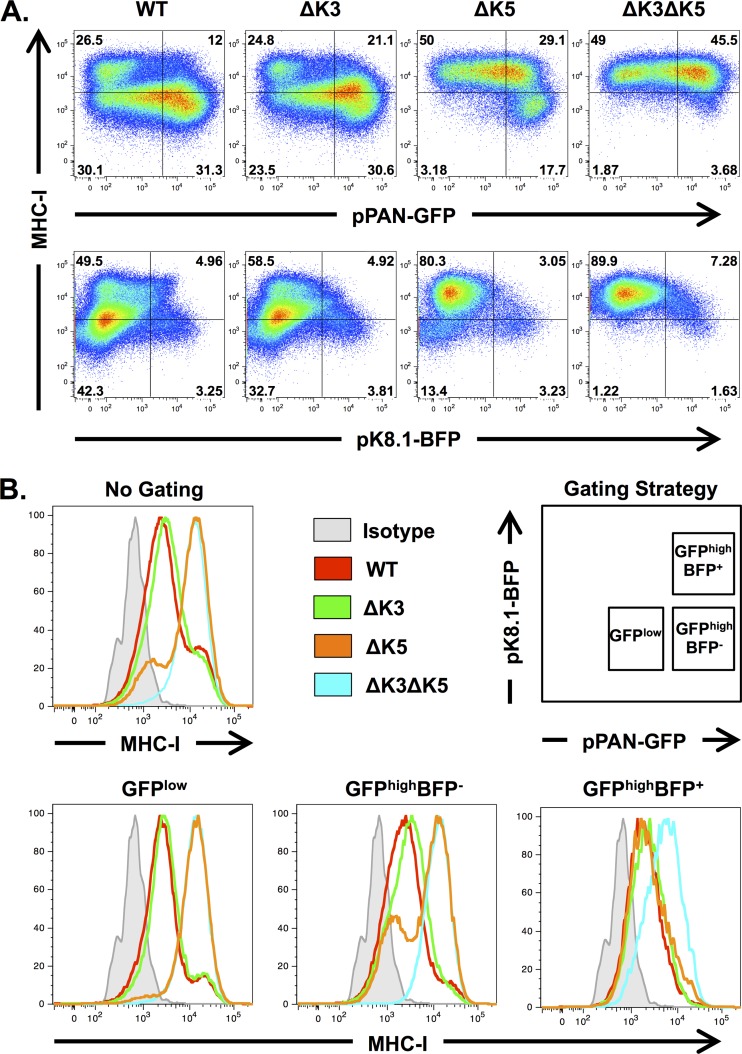
We then examined the immunoreceptor downregulation capabilities of K3 and K5 in the RGB-BAC16 backbone. iSLK cells carrying RGB-derived viruses lacking either or both K3 and K5 were induced with doxycycline and sodium butyrate treatment for 72 h (as described above), and the expressions of MHC-I, EGFP, and tagBFP were analyzed by flow cytometry (Fig. 7A and B). Consistent with our previous findings, K3 was not required for MHC-I downregulation when the K5 gene was intact (Fig. 7A, WT versus ΔK3). In the absence of K5, on the other hand, K3 was required for MHC-I downregulation in a population of cells (∼17.7%) that expressed high levels of pPAN-driven EGFP (EGFPhigh) and were enriched for the late marker tagBFP (∼19%); MHC-I downregulation in this cell population was abolished in ΔK3 ΔK5-infected cells (Fig. 7A, compare ΔK5 and ΔK3 ΔK5 panels). MHC-I surface expression in these cells was also analyzed by gating on specific cell populations that were defined based on EGFP and tagBFP expression levels (Fig. 7B). This analysis demonstrated that the subtle effect of K3 on MHC-I surface expression was more apparent in comparisons of gated cell populations; MHC-I downregulation in ΔK5 virus-infected cells was approximately 38% in EGFPhigh cells and nearly 100% in GFP+ BFP+ cells, both of which were markedly higher than values for nongated populations (Fig. 7B). These results reaffirm the requirement of K5, particularly during the immediate early phase of infection. In addition, the data suggest a novel role for K3 during later stages of lytic replication.
FIG 7.
K3-mediated MHC-I downregulation is enriched in EGFPhigh and tagBFP+ cells. (A) iSLK cells harboring different RGB-BACmids were collected after 3 days of doxycycline and sodium butyrate treatment, and MHC-I, EGFP, and tagBFP were analyzed by flow cytometry. MHC-I surface expression was measured by using a biotin-conjugated HLA-ABC-specific antibody (W6/32) and APC-e780-conjugated streptavidin. (B) Lytic cell populations of iSLK cells infected with the indicated recombinant viruses were gated according to the strategy depicted. Histograms represent MHC-I surface expression levels among the indicated cell populations.
DISCUSSION
In this study, we identified a role for K3 in KSHV-mediated downregulation of MHC class I molecules during the lytic phase of infection. While K5 is essential during the immediate early stage, both K3 and K5 appear to contribute to surface protein downregulation during later stages. Particularly striking was the functional contribution of K3 in late-gene-expressing cells (Fig. 7). Stage-specific activity of immunoevasins has also been described for other herpesviruses: two evolutionarily related glycoproteins of human cytomegalovirus (HCMV), US3 and US11, are sequentially deployed and, in turn, interfere with MHC-I surface expression via distinct mechanisms during the immediate early and early phases of infection, respectively (59). In addition, the BNLF2a protein of Epstein-Barr virus (EBV) was shown to be important for evading recognition by CD8+ T cells specific for immediate early and early epitopes, but not late epitopes, a phenotype that correlated with the transient expression of the BNLF2a protein (60). Thus, stage-specific activity of immunoevasins may be a common feature of many herpesviruses.
The selective advantage of encoding multiple antagonists of antigen presentation is not entirely clear. The sheer diversity of MHC class I molecules among outbred hosts may exert selective pressure on virus species to evolve multiple means of counteraction. In addition, MHC class I molecules, especially HLA-C and HLA-E, are involved in the suppression of NK cell cytotoxicity (61). Many viruses selectively downregulate the expression of HLA-A and HLA-B while sparing HLA-C and HLA-E, and this is thought to be a way of optimally navigating the threats posed by cytotoxic T lymphocytes (CTLs) and NK cells (22, 62–64). In the case of KSHV, K5 selectively removes HLA-A and HLA-B allotypes, while K3 targets HLA-A, HLA-B, HLA-C, and HLA-E (22). In addition, the removal of ligands for activating receptors on NK cells is a common way for viruses to prevent NK cell activation (65), and KSHV encodes several factors, including K5, that reduce the surface expression of such receptors (9, 10, 33). The distinct timing and substrate specificities of K3 and K5 suggest that allotype-specific downregulation of MHC class I molecules occurs during the immediate early stage of lytic replication, while indiscriminant and extensive reduction of surface MHC class I molecules is confined to later stages. This strategy might be particularly relevant after viral DNA replication, when antigenic late proteins are highly expressed (66, 67) and the threat of surveillance from NK cells has been diminished by the activities of K5 and other viral factors (10, 27, 33, 68). The ability of K3 to target HLA-C may be especially significant in light of recent studies showing an important role for HLA-C-restricted CD8+ T cells in the control of viral infections (69, 70), a finding that likely reflects the HLA selectivity of viral immunoevasins such as HIV Nef (71). It is also worth noting that, analogously to K3, the BILF1 protein of EBV was shown to be a broadly specific antagonist of MHC class I molecules: it targets HLA-A, HLA-B, and HLA-E and also shows some ability to downregulate HLA-C albeit weakly (72). Interestingly, BILF1 is expressed late during lytic infection compared to the other major EBV-encoded antagonists of antigen presentation, BNLF2a and BGLF5, and the NK cell antagonist vIL10 (60, 73, 74). Thus, the delayed deployment of broadly specific MHC class I antagonists may represent an evolutionarily conserved immune evasion strategy involving specialized roles for different immunoevasins.
Encoding multiple gene products that interfere with antigen presentation may also be important during infection of different cell types, where conditions could favor the use of one immunoevasin over another, as was shown for the US2 and US11 proteins of HCMV in primary dendritic cells (75). We focused on the role of KSHV in iSLK cells because K3 and K5 are expressed as part of the lytic gene expression program, and this cell line provides a robust means of inducing the lytic phase (47, 48, 56). Although KSHV tends to establish and remain in latency (76), recent advances have demonstrated lytic replication in various cell types, including the B-cell line MC116 (77) and the oral epithelial cell line OEPI (78). In addition, it was recently shown that KSHV-infected lymphatic endothelial cells (LECs) undergo a unique transcription program that includes the expression of several lytic genes (79). Thus, in addition to their stage-specific roles, it may be worthwhile to examine potential cell-type-specific functions of K3 and K5.
Based on current data, K3 and K5 appear to function independently of one another. While the effect of K3 on MHC-I surface expression was apparent only in the absence of K5, it is likely that sustained K5-mediated downregulation overshadows the subtle effects of K3 in WT RGB-BAC16 (Fig. 7). However, we cannot exclude the possibility that K3 and K5 somehow antagonize or cooperate with one another. Collaborative and antagonistic effects on MHC-I surface expression have been demonstrated for the m04, m06, and m152 immunoevasins of murine cytomegalovirus (MCMV) (80, 81). In addition, the US3 protein of HCMV was shown to enhance US2-mediated downregulation (82). In contrast, US3 inhibits US11-mediated downregulation (83). Since the expression of K3 coincides with K5 expression during lytic replication, there is potential for interplay between these proteins. Furthermore, protein purification experiments show that K3 and K5 interact with one another during lytic replication (K. Brulois and J. U. Jung, unpublished data). However, the functional significance of this interaction and the potential interplay between K3 and K5 has yet to be determined.
Although K3 and K5 have redundant expression kinetics at the mRNA level (39, 40), this study shows that inefficient protein production from the K3 locus and the ineffectiveness of K3 at low expression levels contributed to delayed K3-mediated downregulation of MHC-I in the context of KSHV-infected cells. The former mechanism is supported by the finding that lower levels of K3 protein were detected in cells harboring WT BAC16 than in those harboring the K3→K5loc recombinant, even though higher levels of K3-containing mRNA were present in WT-infected cells (Fig. 2). Lower protein levels were observed when either K3 or K5 was expressed from the K3 locus, excluding the possibility that potential differences in codon usage between the K3 and K5 coding sequences are involved in restricting protein expression. K5 is expressed from a single monocistronic mRNA (39, 56). On the other hand, at least five alternatively spliced transcripts have been shown to harbor K3, including both monocistronic and bicistronic transcripts (40, 41). Thus, regulation of alternative splicing may be important for the timing of K3 expression. Alternatively, since K3 is often present as a second cistron, a translation regulatory mechanism may be involved in delayed K3 expression. In fact, the translation of the K3 homologue encoded by mouse herpesvirus 68 (MHV-68) (mK3) is driven by an internal ribosome entry site (IRES)-dependent mechanism (84). It is tempting to speculate that K3 may have evolved to sense increased viral protein synthesis or other alterations in translation that may be coincident with a need for more comprehensive inhibition of antigen presentation. In line with this, K3-mediated downregulation was detectable only in cells with high levels of pPAN-driven gene expression (Fig. 7).
We observed differences in the mRNA levels between the K3 and K5 loci (Fig. 2), suggesting that transcriptional regulation is also important for the functional contributions of K3 and K5. Both the K3 and K5 promoters have been shown to be responsive to RTA as well as the Notch-induced transcription factor RBP-Jκ (39, 85, 86). However, unlike the K5 promoter, the K3 promoter is highly sensitive to NF-κB-mediated repression (86). In latently infected B cells, the K3 promoter is free of RNA polymerase (RNAP) II, while the K5 promoter is occupied by stalled RNAP II, showing that the K5 gene is poised for expression and is expressed independently of RTA during latency (87, 88). Recent studies have begun to characterize the 3′ untranslated regions (UTRs) of KSHV protein-encoding genes (89, 90): while the K5 3′ UTR carries a modest stabilizing effect, the K3 3′ UTR exhibits a destabilizing activity (89), miR-K3 has a small inhibitory effect on the a K3 3′ UTR reporter activity (90), and the 3′ UTR of K3 is among the shortest of all the 3′ UTRs in the KSHV genome (89, 90). Finally, while vIRF1 has been shown to bind to a region adjacent to the K3 coding sequence (91), its binding region is 1 kb downstream of the transcription start site (TSS) of K3 (40, 56), raising questions about the mechanism by which vIRF1 might regulate K3 gene expression. It is worth noting that the levels of K3 and K5 transcripts detected in the positional mutants were consistently lower (∼2- to 3-fold) than the corresponding transcripts of the WT, despite comparable levels of RTA and K2 mRNAs (Fig. 2B). It is possible that the extra sequences encoding the carboxyl-terminal epitope tags have a general destabilizing effect on mRNA. Nevertheless, this anomaly is unlikely to affect the main conclusions.
We also showed distinct capabilities of K3 and K5 to downregulate MHC class I molecules, particularly when these viral proteins were present at low levels (Fig. 3). To our knowledge, this is the first study to perform a parallel analysis of both the intracellular level of an endogenously expressed MARCH family member and the surface expression level of its cognate substrate. Although K3 is a more potent downregulator of MHC-I than K5 during overexpression (21, 22), it appears to be the opposite at low expression levels. This subtle observation may have relevance at physiological expression levels during viral infection. The differential effectivenesses of K3 and K5 may have multiple origins; despite significant homology and structural similarities between these proteins (92–94), a number of fundamental differences in their modes of action have been identified. First, while K3 preferentially induces K63-linked ubiquitin chains on MHC class I substrates (95), K5 expression results in K11 and K63 mixed-chain linkages (96). Second, K3 preferentially targets the membrane-distal K340 residue of HLA-A2 molecules (97), while K5 favors the membrane-proximal K335 residue (98). Third, although both K3 and K5 require dynamin and proteolipid protein 2 (PLP2) for their activities (28, 38), they may also use distinct cell machinery to internalize target substrates. K3 requires the E2-conjugating enzymes UbcH5b/c and Ubc13, the ESCRT-1 component Tsg101, and other components, including clathrin, epsin 1, and eps15R (95). Fourth, unlike K3, palmitoylation is required for K5 to properly function; palmitoylation may potentially facilitate its localization to lipid rafts or otherwise modulate its interaction with substrates (99). The use of chimeras of K3 and K5 may help identify the mechanism(s) that contributes to the distinct patterns of MHC-I downregulation observed in our study.
The in vivo significance of viral interference with antigen presentation is still not clear. Deletion of mK3, the only viral MARCH (vMARCH) gene harbored by MHV-68 (100), resulted in abnormally low latent viral loads during primary infection of C57BL/6 mice, a defect which was complemented by depletion of CD8+ T cells (101). Notably, deletion of mK3 did not completely abolish MHV-68-mediated downregulation of MHC-I levels relative to those in uninfected cells, suggesting that (i) other immunoevasins may be encoded by MHV-68 and (ii) only partial recovery of MHC-I surface expression was sufficient to impact latency establishment. The situation in betaherpesviruses is more complicated (102). In immunocompetent hosts, mutants of MCMV and rhesus monkey cytomegalovirus (rhCMV) that completely lack known immunoevasins are able to establish primary infection as efficiently as their WT counterparts (103, 104). In contrast, this rhCMV mutant showed a striking defect in the establishment of secondary infections (103), and thus, it has been suggested that immunoevasins may have evolved for the dissemination of viruses among immune-experienced hosts (102). It was also observed that, during MCMV infection, antigen presentation by uninfected antigen-presenting cells plays an important role in priming CD8+ T cells and that this process is actually enhanced by the presence of immunoevasins (104). Thus, virally mediated immune evasion does not prevent stimulation of CD8+ T cells, but rather, it protects infected cells from already primed CD8+ T cells. More studies are needed to determine how betaherpesvirus immune evasion paradigms are altered in gammaherpesviruses. The role of broadly specific MHC-I antagonists encoded by KSHV and EBV (22, 72) may be particularly important in vivo. However, despite recent advances in the development of in vivo models for KSHV infection (105, 106), their utility is still limited.
In summary, this study uncovered a role for K3 in the removal of MHC class I molecules from the cell surface during lytic replication. In contrast to K5, K3 activity was restricted to the later stages of lytic replication. While a positionally conserved homologue of K3 can be found in several other gammaherpesvirus genomes (100, 107–109), the presence of more than one vMARCH gene is a unique feature of KSHV. As for some other gammaherpesvirus genes (100, 110), it has been suggested that K5 may have originated from a gene duplication event (22). This may have facilitated the evolution of specialized roles for K3 and K5. The rfK3 protein of the closely related Old World primate rhadinovirus retroperitoneal fibromatosis-associated herpesvirus was able to induce endocytosis of MHC-I as well as ICAM-1 (111), suggesting that some of the functions of K3 and K5 were originally combined in one gene and may have been selectively lost as K3 and K5 became more dedicated. Future work on the vMARCH genes of gammaherpesviruses should lead to a better understanding of the evolution of gammaherpesvirus immune evasion.
ACKNOWLEDGMENTS
We thank Yoshihiro Izumiya and Hsing-Jien Kung (UC Davis, Sacramento, CA) for providing the RTA antibody. We thank Isaac R. Rodriguez-Chavez and Samad Amini-Bavil-Olyaee for insightful comments and all members of J.U.J.'s lab for helpful discussions.
The work was partly supported by grants CA082057, CA31363, CA115284, CA180779, DE023926, and AI073099; the GRL Program (K20815000001); the National Research Foundation of Korea; the Hastings Foundation; BaCaTec (A.E. and J.U.J.); DFG SFB796 (A.E.); and the Fletcher Jones Foundation (J.U.J.).
Footnotes
Published ahead of print 4 June 2014
REFERENCES
- 1.Ganem D. 2010. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. J. Clin. Invest. 120:939–949. 10.1172/JCI40567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dustin ML, Long EO. 2010. Cytotoxic immunological synapses. Immunol. Rev. 235:24–34. 10.1111/j.0105-2896.2010.00904.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen TH, Bouvier M. 2009. MHC class I antigen presentation: learning from viral evasion strategies. Nat. Rev. Immunol. 9:503–513. 10.1038/nri2575 [DOI] [PubMed] [Google Scholar]
- 4.Coscoy L. 2007. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Nat. Rev. Immunol. 7:391–401. 10.1038/nri2076 [DOI] [PubMed] [Google Scholar]
- 5.Lee HR, Lee S, Chaudhary PM, Gill P, Jung JU. 2010. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Future Microbiol. 5:1349–1365. 10.2217/fmb.10.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagos D, Trotter MW, Vart RJ, Wang HW, Matthews NC, Hansen A, Flore O, Gotch F, Boshoff C. 2007. Kaposi sarcoma herpesvirus-encoded vFLIP and vIRF1 regulate antigen presentation in lymphatic endothelial cells. Blood 109:1550–1558. 10.1182/blood-2006-05-024034 [DOI] [PubMed] [Google Scholar]
- 7.Schmidt K, Wies E, Neipel F. 2011. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 3 inhibits gamma interferon and major histocompatibility complex class II expression. J. Virol. 85:4530–4537. 10.1128/JVI.02123-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai Q, Banerjee S, Cervini A, Lu J, Hislop AD, Dzeng R, Robertson ES. 2013. IRF-4-mediated CIITA transcription is blocked by KSHV encoded LANA to inhibit MHC II presentation. PLoS Pathog. 9:e1003751. 10.1371/journal.ppat.1003751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. 2009. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5:376–385. 10.1016/j.chom.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 10.Madrid AS, Ganem D. 2012. Kaposi's sarcoma-associated herpesvirus ORF54/dUTPase downregulates a ligand for the NK activating receptor NKp44. J. Virol. 86:8693–8704. 10.1128/JVI.00252-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boname JM, Lehner PJ. 2011. What has the study of the K3 and K5 viral ubiquitin E3 ligases taught us about ubiquitin-mediated receptor regulation? Viruses 3:118–131. 10.3390/v3020118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohmura-Hoshino M, Goto E, Matsuki Y, Aoki M, Mito M, Uematsu M, Hotta H, Ishido S. 2006. A novel family of membrane-bound E3 ubiquitin ligases. J. Biochem. 140:147–154. 10.1093/jb/mvj160 [DOI] [PubMed] [Google Scholar]
- 13.d'Azzo A, Bongiovanni A, Nastasi T. 2005. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 6:429–441. 10.1111/j.1600-0854.2005.00294.x [DOI] [PubMed] [Google Scholar]
- 14.Nathan JA, Lehner PJ. 2009. The trafficking and regulation of membrane receptors by the RING-CH ubiquitin E3 ligases. Exp. Cell Res. 315:1593–1600. 10.1016/j.yexcr.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 15.Bartee E, Mansouri M, Hovey Nerenberg BT, Gouveia K, Fruh K. 2004. Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J. Virol. 78:1109–1120. 10.1128/JVI.78.3.1109-1120.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoer S, Smith L, Lehner PJ. 2007. MARCH-IX mediates ubiquitination and downregulation of ICAM-1. FEBS Lett. 581:45–51. 10.1016/j.febslet.2006.11.075 [DOI] [PubMed] [Google Scholar]
- 17.De Gassart A, Camosseto V, Thibodeau J, Ceppi M, Catalan N, Pierre P, Gatti E. 2008. MHC class II stabilization at the surface of human dendritic cells is the result of maturation-dependent MARCH I down-regulation. Proc. Natl. Acad. Sci. U. S. A. 105:3491–3496. 10.1073/pnas.0708874105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thibodeau J, Bourgeois-Daigneault MC, Huppe G, Tremblay J, Aumont A, Houde M, Bartee E, Brunet A, Gauvreau ME, de Gassart A, Gatti E, Baril M, Cloutier M, Bontron S, Fruh K, Lamarre D, Steimle V. 2008. Interleukin-10-induced MARCH1 mediates intracellular sequestration of MHC class II in monocytes. Eur. J. Immunol. 38:1225–1230. 10.1002/eji.200737902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartee E, Eyster CA, Viswanathan K, Mansouri M, Donaldson JG, Fruh K. 2010. Membrane-associated RING-CH proteins associate with Bap31 and target CD81 and CD44 to lysosomes. PLoS One 5:e15132. 10.1371/journal.pone.0015132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jahnke M, Trowsdale J, Kelly AP. 2012. Ubiquitination of human leukocyte antigen (HLA)-DM by different membrane-associated RING-CH (MARCH) protein family E3 ligases targets different endocytic pathways. J. Biol. Chem. 287:7256–7264. 10.1074/jbc.M111.305961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coscoy L, Ganem D. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. U. S. A. 97:8051–8056. 10.1073/pnas.140129797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishido S, Wang C, Lee BS, Cohen GB, Jung JU. 2000. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 74:5300–5309. 10.1128/JVI.74.11.5300-5309.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haque M, Ueda K, Nakano K, Hirata Y, Parravicini C, Corbellino M, Yamanishi K. 2001. Major histocompatibility complex class I molecules are down-regulated at the cell surface by the K5 protein encoded by Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8. J. Gen. Virol. 82:1175–1180 [DOI] [PubMed] [Google Scholar]
- 24.Stevenson PG, Efstathiou S, Doherty PC, Lehner PJ. 2000. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. U. S. A. 97:8455–8460. 10.1073/pnas.150240097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez DJ, Gumperz JE, Ganem D. 2005. Regulation of CD1d expression and function by a herpesvirus infection. J. Clin. Invest. 115:1369–1378. 10.1172/JCI24041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhodes DA, Boyle LH, Boname JM, Lehner PJ, Trowsdale J. 2010. Ubiquitination of lysine-331 by Kaposi's sarcoma-associated herpesvirus protein K5 targets HFE for lysosomal degradation. Proc. Natl. Acad. Sci. U. S. A. 107:16240–16245. 10.1073/pnas.1003421107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishido S, Choi JK, Lee BS, Wang C, DeMaria M, Johnson RP, Cohen GB, Jung JU. 2000. Inhibition of natural killer cell-mediated cytotoxicity by Kaposi's sarcoma-associated herpesvirus K5 protein. Immunity 13:365–374. 10.1016/S1074-7613(00)00036-4 [DOI] [PubMed] [Google Scholar]
- 28.Coscoy L, Ganem D. 2001. A viral protein that selectively downregulates ICAM-1 and B7-2 and modulates T cell costimulation. J. Clin. Invest. 107:1599–1606. 10.1172/JCI12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansouri M, Douglas J, Rose PP, Gouveia K, Thomas G, Means RE, Moses AV, Fruh K. 2006. Kaposi sarcoma herpesvirus K5 removes CD31/PECAM from endothelial cells. Blood 108:1932–1940. 10.1182/blood-2005-11-4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansouri M, Rose PP, Moses AV, Fruh K. 2008. Remodeling of endothelial adherens junctions by Kaposi's sarcoma-associated herpesvirus. J. Virol. 82:9615–9628. 10.1128/JVI.02633-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manes TD, Hoer S, Muller WA, Lehner PJ, Pober JS. 2010. Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins block distinct steps in transendothelial migration of effector memory CD4+ T cells by targeting different endothelial proteins. J. Immunol. 184:5186–5192. 10.4049/jimmunol.0902938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang SM, Bynoe MO, Karki R, Tartell MA, Means RE. 2013. Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins down regulate both DC-SIGN and DC-SIGNR. PLoS One 8:e58056. 10.1371/journal.pone.0058056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas M, Boname JM, Field S, Nejentsev S, Salio M, Cerundolo V, Wills M, Lehner PJ. 2008. Down-regulation of NKG2D and NKp80 ligands by Kaposi's sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc. Natl. Acad. Sci. U. S. A. 105:1656–1661. 10.1073/pnas.0707883105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansouri M, Viswanathan K, Douglas JL, Hines J, Gustin J, Moses AV, Fruh K. 2009. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi's sarcoma-associated herpesvirus. J. Virol. 83:9672–9681. 10.1128/JVI.00597-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q, Means R, Lang S, Jung JU. 2007. Downregulation of gamma interferon receptor 1 by Kaposi's sarcoma-associated herpesvirus K3 and K5. J. Virol. 81:2117–2127. 10.1128/JVI.01961-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartee E, McCormack A, Fruh K. 2006. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2:e107. 10.1371/journal.ppat.0020107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durrington HJ, Upton PD, Hoer S, Boname J, Dunmore BJ, Yang J, Crilley TK, Butler LM, Blackbourn DJ, Nash GB, Lehner PJ, Morrell NW. 2010. Identification of a lysosomal pathway regulating degradation of the bone morphogenetic protein receptor type II. J. Biol. Chem. 285:37641–37649. 10.1074/jbc.M110.132415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timms RT, Duncan LM, Tchasovnikarova IA, Antrobus R, Smith DL, Dougan G, Weekes MP, Lehner PJ. 2013. Haploid genetic screens identify an essential role for PLP2 in the downregulation of novel plasma membrane targets by viral E3 ubiquitin ligases. PLoS Pathog. 9:e1003772. 10.1371/journal.ppat.1003772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haque M, Chen J, Ueda K, Mori Y, Nakano K, Hirata Y, Kanamori S, Uchiyama Y, Inagi R, Okuno T, Yamanishi K. 2000. Identification and analysis of the K5 gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:2867–2875. 10.1128/JVI.74.6.2867-2875.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rimessi P, Bonaccorsi A, Sturzl M, Fabris M, Brocca-Cofano E, Caputo A, Melucci-Vigo G, Falchi M, Cafaro A, Cassai E, Ensoli B, Monini P. 2001. Transcription pattern of human herpesvirus 8 open reading frame K3 in primary effusion lymphoma and Kaposi's sarcoma. J. Virol. 75:7161–7174. 10.1128/JVI.75.15.7161-7174.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor JL, Bennett HN, Snyder BA, Moore PS, Chang Y. 2005. Transcriptional analysis of latent and inducible Kaposi's sarcoma-associated herpesvirus transcripts in the K4 to K7 region. J. Virol. 79:15099–15106. 10.1128/JVI.79.24.15099-15106.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun R, Lin SF, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu FX, Cusano T, Yuan Y. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishnan HH, Naranatt PP, Smith MS, Zeng L, Bloomer C, Chandran B. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 78:3601–3620. 10.1128/JVI.78.7.3601-3620.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomescu C, Law WK, Kedes DH. 2003. Surface downregulation of major histocompatibility complex class I, PE-CAM, and ICAM-1 following de novo infection of endothelial cells with Kaposi's sarcoma-associated herpesvirus. J. Virol. 77:9669–9684. 10.1128/JVI.77.17.9669-9684.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adang LA, Tomescu C, Law WK, Kedes DH. 2007. Intracellular Kaposi's sarcoma-associated herpesvirus load determines early loss of immune synapse components. J. Virol. 81:5079–5090. 10.1128/JVI.02738-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, Oh TK, Kim JF, Gao SJ, Jung JU. 2012. Construction and manipulation of a new Kaposi's sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol. 86:9708–9720. 10.1128/JVI.01019-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myoung J, Ganem D. 2011. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 174:12–21. 10.1016/j.jviromet.2011.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 634:421–430. 10.1007/978-1-60761-652-8_30 [DOI] [PubMed] [Google Scholar]
- 50.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. 10.2144/000112096 [DOI] [PubMed] [Google Scholar]
- 51.Tang S, Yamanegi K, Zheng ZM. 2004. Requirement of a 12-base-pair TATT-containing sequence and viral lytic DNA replication in activation of the Kaposi's sarcoma-associated herpesvirus K8.1 late promoter. J. Virol. 78:2609–2614. 10.1128/JVI.78.5.2609-2614.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Birnboim HC, Doly J. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513–1523. 10.1093/nar/7.6.1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toth Z, Maglinte DT, Lee SH, Lee HR, Wong LY, Brulois KF, Lee S, Buckley JD, Laird PW, Marquez VE, Jung JU. 2010. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 6:e1001013. 10.1371/journal.ppat.1001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 55.Ciufo DM, Cannon JS, Poole LJ, Wu FY, Murray P, Ambinder RF, Hayward GS. 2001. Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J. Virol. 75:5614–5626. 10.1128/JVI.75.12.5614-5626.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, Ganem D. 2014. KSHV 2.0: a comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 10:e1003847. 10.1371/journal.ppat.1003847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adang LA, Parsons CH, Kedes DH. 2006. Asynchronous progression through the lytic cascade and variations in intracellular viral loads revealed by high-throughput single-cell analysis of Kaposi's sarcoma-associated herpesvirus infection. J. Virol. 80:10073–10082. 10.1128/JVI.01156-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zoeteweij JP, Eyes ST, Orenstein JM, Kawamura T, Wu L, Chandran B, Forghani B, Blauvelt A. 1999. Identification and rapid quantification of early- and late-lytic human herpesvirus 8 infection in single cells by flow cytometric analysis: characterization of antiherpesvirus agents. J. Virol. 73:5894–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahn K, Angulo A, Ghazal P, Peterson PA, Yang Y, Fruh K. 1996. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc. Natl. Acad. Sci. U. S. A. 93:10990–10995. 10.1073/pnas.93.20.10990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Croft NP, Shannon-Lowe C, Bell AI, Horst D, Kremmer E, Ressing ME, Wiertz EJ, Middeldorp JM, Rowe M, Rickinson AB, Hislop AD. 2009. Stage-specific inhibition of MHC class I presentation by the Epstein-Barr virus BNLF2a protein during virus lytic cycle. PLoS Pathog. 5:e1000490. 10.1371/journal.ppat.1000490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lanier LL. 2005. NK cell recognition. Annu. Rev. Immunol. 23:225–274. 10.1146/annurev.immunol.23.021704.115526 [DOI] [PubMed] [Google Scholar]
- 62.Barel MT, Ressing M, Pizzato N, van Leeuwen D, Le Bouteiller P, Lenfant F, Wiertz EJ. 2003. Human cytomegalovirus-encoded US2 differentially affects surface expression of MHC class I locus products and targets membrane-bound, but not soluble HLA-G1 for degradation. J. Immunol. 171:6757–6765. 10.4049/jimmunol.171.12.6757 [DOI] [PubMed] [Google Scholar]
- 63.Llano M, Guma M, Ortega M, Angulo A, Lopez-Botet M. 2003. Differential effects of US2, US6 and US11 human cytomegalovirus proteins on HLA class Ia and HLA-E expression: impact on target susceptibility to NK cell subsets. Eur. J. Immunol. 33:2744–2754. 10.1002/eji.200324182 [DOI] [PubMed] [Google Scholar]
- 64.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661–671. 10.1016/S1074-7613(00)80065-5 [DOI] [PubMed] [Google Scholar]
- 65.Odom CI, Gaston DC, Markert JM, Cassady KA. 2012. Human herpesviridae methods of natural killer cell evasion. Adv. Virol. 2012:359869. 10.1155/2012/359869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vider-Shalit T, Fishbain V, Raffaeli S, Louzoun Y. 2007. Phase-dependent immune evasion of herpesviruses. J. Virol. 81:9536–9545. 10.1128/JVI.02636-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robey RC, Lagos D, Gratrix F, Henderson S, Matthews NC, Vart RJ, Bower M, Boshoff C, Gotch FM. 2009. The CD8 and CD4 T-cell response against Kaposi's sarcoma-associated herpesvirus is skewed towards early and late lytic antigens. PLoS One 4:e5890. 10.1371/journal.pone.0005890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. 2009. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5:376–385. 10.1016/j.chom.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 69.Apps R, Qi Y, Carlson JM, Chen H, Gao X, Thomas R, Yuki Y, Del Prete GQ, Goulder P, Brumme ZL, Brumme CJ, John M, Mallal S, Nelson G, Bosch R, Heckerman D, Stein JL, Soderberg KA, Moody MA, Denny TN, Zeng X, Fang J, Moffett A, Lifson JD, Goedert JJ, Buchbinder S, Kirk GD, Fellay J, McLaren P, Deeks SG, Pereyra F, Walker B, Michael NL, Weintrob A, Wolinsky S, Liao W, Carrington M. 2013. Influence of HLA-C expression level on HIV control. Science 340:87–91. 10.1126/science.1232685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ameres S, Mautner J, Schlott F, Neuenhahn M, Busch DH, Plachter B, Moosmann A. 2013. Presentation of an immunodominant immediate-early CD8+ T cell epitope resists human cytomegalovirus immunoevasion. PLoS Pathog. 9:e1003383. 10.1371/journal.ppat.1003383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kulpa DA, Collins KL. 2011. The emerging role of HLA-C in HIV-1 infection. Immunology 134:116–122. 10.1111/j.1365-2567.2011.03474.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Griffin BD, Gram AM, Mulder A, Van Leeuwen D, Claas FH, Wang F, Ressing ME, Wiertz E. 2013. EBV BILF1 evolved to downregulate cell surface display of a wide range of HLA class I molecules through their cytoplasmic tail. J. Immunol. 190:1672–1684. 10.4049/jimmunol.1102462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jochum S, Moosmann A, Lang S, Hammerschmidt W, Zeidler R. 2012. The EBV immunoevasins vIL-10 and BNLF2a protect newly infected B cells from immune recognition and elimination. PLoS Pathog. 8:e1002704. 10.1371/journal.ppat.1002704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuo J, Thomas W, van Leeuwen D, Middeldorp JM, Wiertz EJ, Ressing ME, Rowe M. 2008. The DNase of gammaherpesviruses impairs recognition by virus-specific CD8+ T cells through an additional host shutoff function. J. Virol. 82:2385–2393. 10.1128/JVI.01946-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rehm A, Engelsberg A, Tortorella D, Korner IJ, Lehmann I, Ploegh HL, Hopken UE. 2002. Human cytomegalovirus gene products US2 and US11 differ in their ability to attack major histocompatibility class I heavy chains in dendritic cells. J. Virol. 76:5043–5050. 10.1128/JVI.76.10.5043-5050.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bechtel JT, Liang Y, Hvidding J, Ganem D. 2003. Host range of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 77:6474–6481. 10.1128/JVI.77.11.6474-6481.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dollery SJ, Santiago-Crespo RJ, Kardava L, Moir S, Berger EA. 2014. Efficient infection of a human B cell line with cell-free Kaposi's sarcoma-associated herpesvirus. J. Virol. 88:1748–1757. 10.1128/JVI.03063-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Toth Z, Brulois K, Lee HR, Izumiya Y, Tepper C, Kung HJ, Jung JU. 2013. Biphasic euchromatin-to-heterochromatin transition on the KSHV genome following de novo infection. PLoS Pathog. 9:e1003813. 10.1371/journal.ppat.1003813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang HH, Ganem D. 2013. A unique herpesviral transcriptional program in KSHV-infected lymphatic endothelial cells leads to mTORC1 activation and rapamycin sensitivity. Cell Host Microbe 13:429–440. 10.1016/j.chom.2013.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wagner M, Gutermann A, Podlech J, Reddehase MJ, Koszinowski UH. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805–816. 10.1084/jem.20020811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kavanagh DG, Gold MC, Wagner M, Koszinowski UH, Hill AB. 2001. The multiple immune-evasion genes of murine cytomegalovirus are not redundant: m4 and m152 inhibit antigen presentation in a complementary and cooperative fashion. J. Exp. Med. 194:967–978. 10.1084/jem.194.7.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Noriega VM, Tortorella D. 2009. Human cytomegalovirus-encoded immune modulators partner to downregulate major histocompatibility complex class I molecules. J. Virol. 83:1359–1367. 10.1128/JVI.01324-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Noriega VM, Hesse J, Gardner TJ, Besold K, Plachter B, Tortorella D. 2012. Human cytomegalovirus US3 modulates destruction of MHC class I molecules. Mol. Immunol. 51:245–253. 10.1016/j.molimm.2012.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coleman HM, Brierley I, Stevenson PG. 2003. An internal ribosome entry site directs translation of the murine gammaherpesvirus 68 MK3 open reading frame. J. Virol. 77:13093–13105. 10.1128/JVI.77.24.13093-13105.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chang H, Dittmer DP, Shin YC, Hong Y, Jung JU. 2005. Role of Notch signal transduction in Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 79:14371–14382. 10.1128/JVI.79.22.14371-14382.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Izumiya Y, Izumiya C, Hsia DS, Ellison TJ, Luciw PA, Kung HJ. 2009. NF-kappa B serves as a cellular sensor of Kaposi's sarcoma-associated herpesvirus latency and negatively regulates K-Rta by antagonizing the RBP-Jkappa coactivator. J. Virol. 83:4435–4446. 10.1128/JVI.01999-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Okuno T, Jiang YB, Ueda K, Nishimura K, Tamura T, Yamanishi K. 2002. Activation of human herpesvirus 8 open reading frame K5 independent of ORF50 expression. Virus Res. 90:77–89. 10.1016/S0168-1702(02)00142-9 [DOI] [PubMed] [Google Scholar]
- 88.Toth Z, Brulois KF, Wong LY, Lee HR, Chung B, Jung JU. 2012. Negative elongation factor-mediated suppression of RNA polymerase II elongation of Kaposi's sarcoma-associated herpesvirus lytic gene expression. J. Virol. 86:9696–9707. 10.1128/JVI.01012-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McClure LV, Kincaid RP, Burke JM, Grundhoff A, Sullivan CS. 2013. Comprehensive mapping and analysis of Kaposi's sarcoma-associated herpesvirus 3′ UTRs identify differential posttranscriptional control of gene expression in lytic versus latent infection. J. Virol. 87:12838–12849. 10.1128/JVI.02374-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bai Z, Huang Y, Li W, Zhu Y, Jung JU, Lu C, Gao SJ. 2014. Genomewide mapping and screening of Kaposi's sarcoma-associated herpesvirus (KSHV) 3′ untranslated regions identify bicistronic and polycistronic viral transcripts as frequent targets of KSHV microRNAs. J. Virol. 88:377–392. 10.1128/JVI.02689-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Park J, Lee MS, Yoo SM, Jeong KW, Lee D, Choe J, Seo T. 2007. Identification of the DNA sequence interacting with Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 1. J. Virol. 81:12680–12684. 10.1128/JVI.00556-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sanchez DJ, Coscoy L, Ganem D. 2002. Functional organization of MIR2, a novel viral regulator of selective endocytosis. J. Biol. Chem. 277:6124–6130. 10.1074/jbc.M110265200 [DOI] [PubMed] [Google Scholar]
- 93.Coscoy L, Ganem D. 2003. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. 13:7–12. 10.1016/S0962-8924(02)00005-3 [DOI] [PubMed] [Google Scholar]
- 94.Means RE, Lang SM, Jung JU. 2007. The Kaposi's sarcoma-associated herpesvirus K5 E3 ubiquitin ligase modulates targets by multiple molecular mechanisms. J. Virol. 81:6573–6583. 10.1128/JVI.02751-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duncan LM, Piper S, Dodd RB, Saville MK, Sanderson CM, Luzio JP, Lehner PJ. 2006. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 25:1635–1645. 10.1038/sj.emboj.7601056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boname JM, Thomas M, Stagg HR, Xu P, Peng J, Lehner PJ. 2010. Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic 11:210–220. 10.1111/j.1600-0854.2009.01011.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hewitt EW, Duncan L, Mufti D, Baker J, Stevenson PG, Lehner PJ. 2002. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 21:2418–2429. 10.1093/emboj/21.10.2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cadwell K, Coscoy L. 2008. The specificities of Kaposi's sarcoma-associated herpesvirus-encoded E3 ubiquitin ligases are determined by the positions of lysine or cysteine residues within the intracytoplasmic domains of their targets. J. Virol. 82:4184–4189. 10.1128/JVI.02264-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Anania VG, Coscoy L. 2011. Palmitoylation of MIR2 is required for its function. J. Virol. 85:2288–2295. 10.1128/JVI.01961-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Virgin HW, IV, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stevenson PG, May JS, Smith XG, Marques S, Adler H, Koszinowski UH, Simas JP, Efstathiou S. 2002. K3-mediated evasion of CD8(+) T cells aids amplification of a latent gamma-herpesvirus. Nat. Immunol. 3:733–740. 10.1038/ni818 [DOI] [PubMed] [Google Scholar]
- 102.Lemmermann NA, Bohm V, Holtappels R, Reddehase MJ. 2011. In vivo impact of cytomegalovirus evasion of CD8 T-cell immunity: facts and thoughts based on murine models. Virus Res. 157:161–174. 10.1016/j.virusres.2010.09.022 [DOI] [PubMed] [Google Scholar]
- 103.Hansen SG, Powers CJ, Richards R, Ventura AB, Ford JC, Siess D, Axthelm MK, Nelson JA, Jarvis MA, Picker LJ, Fruh K. 2010. Evasion of CD8(+) T cells is critical for superinfection by cytomegalovirus. Science 328:102–106. 10.1126/science.1185350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bohm V, Simon CO, Podlech J, Seckert CK, Gendig D, Deegen P, Gillert-Marien D, Lemmermann NAW, Holtappels R, Reddehase MJ. 2008. The immune evasion paradox: immunoevasins of murine cytomegalovirus enhance priming of CD8 T cells by preventing negative feedback regulation. J. Virol. 82:11637–11650. 10.1128/JVI.01510-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chang H, Wachtman LM, Pearson CB, Lee JS, Lee HR, Lee SH, Vieira J, Mansfield KG, Jung JU. 2009. Non-human primate model of Kaposi's sarcoma-associated herpesvirus infection. PLoS Pathog. 5:e1000606. 10.1371/journal.ppat.1000606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang LX, Kang G, Kumar P, Lu W, Li Y, Zhou Y, Li Q, Wood C. 2014. Humanized-BLT mouse model of Kaposi's sarcoma-associated herpesvirus infection. Proc. Natl. Acad. Sci. U. S. A. 111:3146–3151. 10.1073/pnas.1318175111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bruce AG, Ryan JT, Thomas MJ, Peng X, Grundhoff A, Tsai CC, Rose TM. 2013. Next-generation sequence analysis of the genome of RFHVMn, the macaque homolog of Kaposi's sarcoma (KS)-associated herpesvirus, from a KS-like tumor of a pig-tailed macaque. J. Virol. 87:13676–13693. 10.1128/JVI.02331-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Loh J, Zhao G, Nelson CA, Coder P, Droit L, Handley SA, Johnson LS, Vachharajani P, Guzman H, Tesh RB, Wang D, Fremont DH, Virgin HW. 2011. Identification and sequencing of a novel rodent gammaherpesvirus that establishes acute and latent infection in laboratory mice. J. Virol. 85:2642–2656. 10.1128/JVI.01661-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Herr RA, Wang X, Loh J, Virgin HW, Hansen TH. 2012. Newly discovered viral E3 ligase pK3 induces endoplasmic reticulum-associated degradation of class I major histocompatibility proteins and their membrane-bound chaperones. J. Biol. Chem. 287:14467–14479. 10.1074/jbc.M111.325340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Searles RP, Bergquam EP, Axthelm MK, Wong SW. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Harris S, Lang SM, Means RE. 2010. Characterization of the rhesus fibromatosis herpesvirus MARCH family member rfK3. Virology 398:214–223. 10.1016/j.virol.2009.12.009 [DOI] [PubMed] [Google Scholar]