ABSTRACT
Influenza A virus (IAV) replication depends on the interaction of virus proteins with host factors. The viral nonstructural protein 1 (NS1) is essential in this process by targeting diverse cellular functions, including mRNA splicing and translation, cell survival, and immune defense, in particular the type I interferon (IFN-I) response. In order to identify host proteins targeted by NS1, we established a replication-competent recombinant IAV that expresses epitope-tagged forms of NS1 and NS2, which are encoded by the same gene segment, allowing purification of NS proteins during natural cell infection and analysis of interacting proteins by quantitative mass spectrometry. We identified known NS1- and NS2-interacting proteins but also uncharacterized proteins, including PACT, an important cofactor for the IFN-I response triggered by the viral RNA-sensor RIG-I. We show here that NS1 binds PACT during virus replication and blocks PACT/RIG-I-mediated activation of IFN-I, which represents a critical event for the host defense. Protein interaction and interference with IFN-I activation depended on the functional integrity of the highly conserved RNA binding domain of NS1. A mutant virus with deletion of NS1 induced high levels of IFN-I in control cells, as expected; in contrast, shRNA-mediated knockdown of PACT compromised IFN-I activation by the mutant virus, but not wild-type virus, a finding consistent with the interpretation that PACT (i) is essential for IAV recognition and (ii) is functionally compromised by NS1. Together, our data describe a novel approach to identify virus-host protein interactions and demonstrate that NS1 interferes with PACT, whose function is critical for robust IFN-I production.
IMPORTANCE Influenza A virus (IAV) is an important human pathogen that is responsible for annual epidemics and occasional devastating pandemics. Viral replication and pathogenicity depends on the interference of viral factors with components of the host defense system, particularly the type I interferon (IFN-I) response. The viral NS1 protein is known to counteract virus recognition and IFN-I production, but the molecular mechanism is only partially defined. We used a novel proteomic approach to identify host proteins that are bound by NS1 during virus replication and identified the protein PACT, which had previously been shown to be involved in virus-mediated IFN-I activation. We find that NS1 prevents PACT from interacting with an essential component of the virus recognition pathway, RIG-I, thereby disabling efficient IFN-I production. These observations provide an important piece of information on how IAV efficiently counteracts the host immune defense.
INTRODUCTION
Influenza virus replication depends on interactions between virus-encoded proteins and host factors, controlling key aspects of host and virus biology, including cell survival, virus production, and the inhibition of virus recognition and host defense (1). The latter aspect of virus host interaction is highlighted by partially characterized functions of the IAV nonstructural protein 1 (NS1), which is essential for virus pathogenicity (2–4). During the last several decades, various cellular NS1 binding partners have been identified, illustrating protein-protein and protein-RNA interactions as modes of NS1 function (1). While an important inhibitory role of NS1 in IFN-I activation has been established for more than 10 years (4), more recent work has started to unravel the molecular mechanisms involved. Initially, the RNA binding property of NS1 has been described as a mechanism to prevent viral RNA from recognition by components of the cellular host defense system, including the interferon-induced, double-stranded RNA (dsRNA)-activated protein kinase (PKR) and the 2′-5′ oligo(A) synthetase (OAS)/RNase L pathway (5, 6). NS1 has also been shown to interfere with virus recognition via the pattern recognition receptor RIG-I, which activates several immune signaling pathways upon viral RNA recognition, including the type I interferon (IFN-I) pathway that mediates antiviral resistance (7). Specifically, NS1 has been described to interact and interfere directly with the ubiquitin ligases TRIM25 and RIPLET, which catalyze the formation of nondegrading polyubiquitin chains that are required for RIG-I activation and recruitment of the downstream effector protein MAVS (8–10). Still, it is not entirely clear whether these functions of NS1 are solely responsible for its inhibitory role in the IFN-I pathway. One protein that has recently been shown to increase RIG-I activity independent of viral RNA is the protein activator of the interferon-induced protein kinase (PACT), which was originally cloned as a binding partner and activator of PKR, an IFN-inducible kinase with established antiviral function (11–13). PACT has also been shown to be required for optimal activity of Dicer, a nuclease required for RNA interference (14, 15). Although the precise mechanism of PACT–RIG-I interaction and function requires further investigation, gain- and loss-of-function experiments established a critical function for PACT in RIG-I activation (15). Very recently, two viral proteins, the herpes simplex virus (HSV) protein Us11 and the Ebola virus protein VP35 were shown to interact with PACT and counteract PACT-mediated IFN activation (16, 17), supporting the idea that PACT-mediated IFN-I activation is a critical part of the antiviral host defense.
In order to identify novel factors involved in IAV biology, we devised a proteomic strategy to analyze protein interactions of NS1 and NS2 during physiological virus replication. While such a strategy has been used for other viruses, IAV have been difficult to approach due to their specific genome packaging mechanism and space constraints. Individual RNA segments of IAV are packed into RNA/protein complexes, where the first approximately 100 bp serve at least two purposes, i.e., as packaging signal and as coding sequence (18). Thus, insertion of epitope tags for protein purification at the 5′ or 3′ end interferes with genome packaging. In order to maintain both functions and allow fusion of an epitope tag for efficient protein purification, we took advantage of the 2A peptide of the porcine teschovirus-1 (P2A), which mediates ribosomal skipping (19), allowing duplication of the first 90 bp of the NS gene segment, thereby functionally separating genome packaging and protein translation (for details, see Fig. 1A) (18). Using a recombinant virus based on this technology, followed by cell infection and quantitative proteomic analysis of NS1-/NS2-interacting proteins, we identified many known NS1-/NS2-interacting proteins, as expected, but also novel factors, including PACT. Here, we describe in detail our strategy to investigate virus-host protein interactions during physiological virus replication and also characterize PACT function during IAV infection, demonstrating that PACT is a critical factor for IAV recognition that is specifically targeted by NS1 to counteract RIG-I-mediated IFN-I activation.
FIG 1.
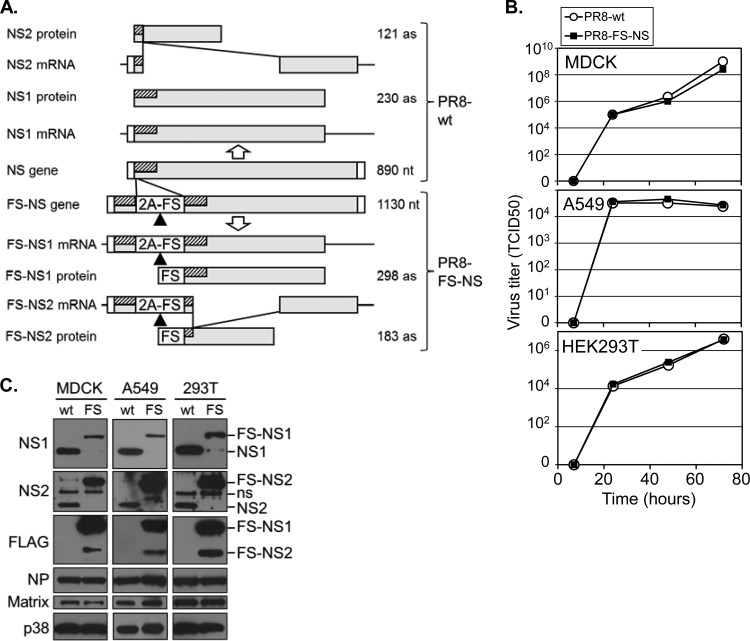
Generation of a recombinant PR8 virus with epitope-tagged NS1- and NS2 (PR8-NS-FS). (A) IAV NS gene arrangement and structure of recombinant PR8-NS-FS virus through the duplication of 90 bp of the 5′ gene segment (hatched), followed by the 2A peptide of the porcine teschovirus-1 (P2A) and a tandem triple-FLAG-StrepOne epitope tag (FS). Different expected mRNA and protein products for PR8-wt (upper part) and PR8-NS-FS (lower part) are shown. Triangle, 2A-encoded “cleavage” site. (B) Replication of PR8-wt and PR8 NS-FS on MDCK, A549, and HEK293T cells was determined by infection at an MOI of 0.001 (MDCK and A549 cells) or at an MOI of 0.01 (HEK293T cells) and determination of the virus titer from the supernatants at the indicated time points. (C) Immunoblot analysis of MDCK, A549, and HEK293T cells, infected with PR8-wt (wt) and PR8-NS-FS (FS) at an MOI of 1 for 24 h. Blots were analyzed with antibodies against proteins indicated (left), and the protein identity is indicated (right).
MATERIALS AND METHODS
Reagents.
Antibodies were used against FLAG (M2 [soluble and bead immobilized]; Sigma-Aldrich), PACT (Cell Signaling), NS1 (mouse monoclonal), clone 18/1 (20), and rabbit polyclonal (GeneTex; used in Fig. 3D only), hemagglutinin (HA; bead immobilized, clone 3F10; Roche), NS2 (GeneTex), matrix protein (mouse monoclonal, clone M2-IC6 [21]), nucleoprotein (GeneTex), and p38 (Santa Cruz).
FIG 3.
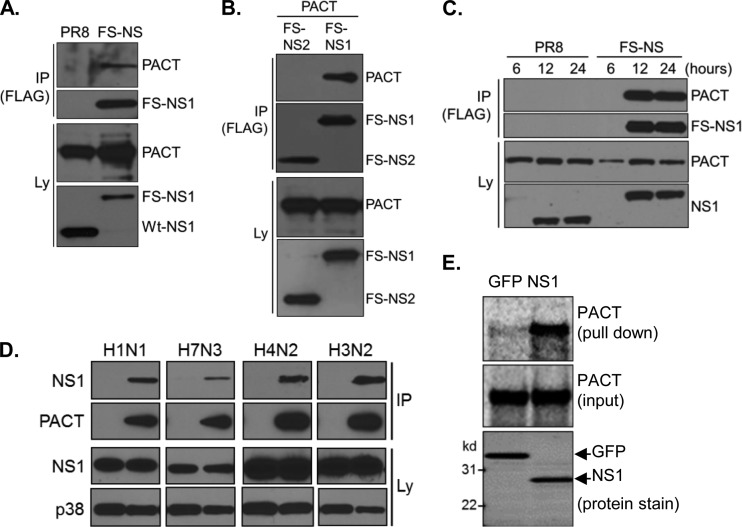
NS1 interacts with PACT during virus replication and in vitro. (A) HEK293T cells were infected with PR8-wt or PR8 NS-FS for 24 h, followed by immunoprecipitation (IP) and immunoblotting (IB) with antibodies against PACT and NS1 (Ly, total lysate). (B) HEK293T cells were transfected with PACT along with FS-tagged NS1 or NS2 for 48 h, followed by IP/IB analysis using antibodies against FLAG (for IP) and antibodies against FLAG (detecting FS-NS1 and FS-NS2) and PACT (for IB). (C) HEK293T cells were infected with PR8-wt or PR8 NS-FS for 6, 12, or 24 h, followed by IP and IB with antibodies against FLAG (for IP) and PACT and NS1 (for IB). (D) HEK293T cells were transfected with control plasmid or HA-tagged PACT, followed by infection with indicated IAV strains for 24 h and IP/IB analysis using antibodies against HA (for IP) and PACT, NS1, and p38 (for IB). H1N1, PR8; H7N3, A/laughing gull/Delaware bay/42/2006; H4N2, A/quail/California/D113023808/2012; H3N2 (A/Perth/16/2009). (E) Pulldown experiment of in vitro-translated PACT using recombinant, bead-immobilized GFP or NS1. A pull-down sample and 10% of input sample were resolved by SDS-PAGE and analyzed by using a phosphorimager or SYPRO Ruby protein staining, respectively.
Plasmids and generation of recombinant virus.
For immunoprecipitation (IP) experiments, FS-tagged NS2 was generated by cloning the full-length coding region of NS2 3-prime of a FS epitope tag as described in Fig. 1, contained in a mammalian expression vector with EF1α-promoter. The same strategy was used for generation of FS-NS1; however, four translationally silent mutations preventing aberrant splicing that target splice donor (G30A, A33T), branch point (A483T), and splice acceptor (A501T) sites were introduced by PCR. A bacterial expression vector for His6-tagged PR8-NS1 was generated by cloning the coding sequence of PR8-NS1 containing described splicing-disabling point mutations C-terminal of a His6 epitope tag into pQE-31 (Qiagen). Sequences of all plasmids were verified by DNA sequencing. A His6-tagged form of TAT-HA-green fluorescent protein (GFP) was kindly provided by Stephen Dowdy (University of California, San Diego) (22). The cDNA of PACT was obtained from Addgene (catalog no. 15667) (23) and cloned 3 prime of a triple HA-epitope tag contained in a pcDNA3-based vector (Invitrogen). The pEF-BOS-based expression vector for F-RIG-I was kindly provided by T. Fujita. The recombinant PR8 viruses used (PR8-wt, PR8-NS-FS, and PR8 ΔNS1) were rescued by reverse genetics as described previously based on plasmids encoding the gene segments of influenza virus A/Puerto Rico/8/34 (PR8) (24). Modifications in the sequence of NS1 (for PR8-NS-FS detailed in Fig. 1A) were introduced by standard molecular biology techniques and verified by DNA sequencing. For PR8 ΔNS1, the sequence encoding AS11-230 was deleted, resulting in one remaining open reading frame encoding NS2, as described previously (4). A high virus titer was produced on Madin-Darby canine kidney (MDCK) cells for proteomic experiments. In experiments where PR8 ΔNS1 was used, viruses were grown on African green monkey kidney (Vero) cells.
Virus titration.
Virus titration of PR8-wt and PR8-NS-FS was done by limiting dilution on MDCK cells, followed by an HA inhibition assay. Where PR8 ΔNS1 was used, the virus titers were determined by intracellular staining in HEK293T cells. To this end, HEK293T cells were seeded on poly-l-lysine-coated 96-well plates (4 × 104 cells/50 μl of infection medium) and infected with 20 μl of virus-containing supernatants (by limiting dilution) for 2 h at 37°C, followed by the addition of 70 μl of medium (DMEM) containing 20% fetal bovine serum (FBS) and incubation at 37°C for another 22 h. Intracellular virus staining was performed on plate by removing the supernatant, washing with phosphate-buffered saline (PBS), fixing the cells with 2% formaldehyde in PBS for precisely 20 min at 25°C, repeated washing with PBS–1% FBS, and staining with biotin-conjugated anti-H1N1 antibody (I7650-05G, US Biological) in PBS containing 1% FBS and 0.5% saponin (PBS-Sap; Sigma-Aldrich) for 20 min at 25°C. After the antiviral antibody was removed, the cells were incubated with streptavidin-phycoerythrin (BD Biosciences) in PBS-Sap for 20 min at 25°C. The cells were washed twice with PBS-Sap and once with PBS, followed by treatment with 30 μl of trypsin-EDTA (Invitrogen) for 3 min at 37°C, resuspension in 170 μl of PBS containing 30% FBS, and flow cytometry.
Cell culture and generation of shRNA knockdown cell lines.
HEK293T, A549, MDCK, and Vero cells were cultured in DMEM high glucose (Invitrogen) supplemented with 10% FBS, 50 mM 2-mercaptoethanol (2-ME), antibiotics (penicillin G at 100 IU/ml and streptomycin sulfate at 100 IU/ml [P/S]), and pyruvate (1 mM). Cell infections were performed in infection medium {DMEM high glucose (Invitrogen) supplemented with 2-ME, P/S, 0.1% bovine serum albumin, and TPCK [l-(tosylamido-2-phenyl) ethyl chloromethyl ketone] trypsin at 1 μg/ml} after the cells were washed twice with PBS.
Retroviral shRNA plasmids for control and human PACT were purchased from Transomix (TRH1000 (RLGH-GN50894 [PACT sh1], RLGH-GN50896 [PACT sh2]), and TRH1103 [sh ctrl]), and nonreplicating retrovirus was produced in HEK293T cells by cotransfection of helper plasmids and VSV-G expression plasmid. Supernatant was directly used for infection of HEK293T cells, followed by puromycin selection of polyclonal populations.
Tandem affinity purification.
A549 cells (five 15-cm cell culture dishes) were infected with PR8-wt or PR8-NS-FS at a multiplicity of infection (MOI) of 0.5 in infection medium. After 1 h of incubation at 37°C, medium was replaced by regular cell growth medium, followed by incubation for 24 h at 37°C. Medium was replaced by ice-cold PBS and collected by cell scraping and centrifugation. Cell pellets were incubated with lysis buffer (LB; 20 mM HEPES/KOH [pH 7.5], 150 mM KCl, 1.5 mM MgCl, 1 mM EDTA, 10% glycerol, 1 mM orthovanadate, 10 mM β-glycerophosphate, 5 mM 4-nitrophenyl-phosphate, 10 mM sodium fluoride, Complete protease inhibitors [Roche]) supplemented with 1% NP-40 for 20 min. Samples were sonicated with ten 3-s pulses (level 2) on a Virsonic 60 cell disruptor (Virtis) to disrupt nuclei and then cleared by centrifugation. Cleared lysates were loaded five times over anti-FLAG-bead-containing columns. Unbound proteins were removed by washing column with LB plus 0.1% NP-40, and proteins were eluted in 1 ml of LB plus 0.1% NP-40 supplemented with 3×FLAG peptide (0.2 mg/ml; Sigma-Aldrich). Eluted proteins were loaded five times over a StrepTactin column (IBA), unbound proteins were removed by washing the column with LB plus 0.1% NP-40, and proteins were eluted in 0.9 ml of LB plus desthiobiotin (5 mM; Sigma-Aldrich), followed by concentration using 5K NMWL spin columns (Millipore), separation on a 4 to 12% Bis-Tris gradient gel (Bio-Rad), and staining with SYPRO Ruby protein stain (Invitrogen). Individual bands were cut, in-gel digested using trypsin, and analyzed by MALDI-TOF (matrix-assisted laser desorption ionization–time of flight) mass spectrometry (MS) or liquid chromatography-coupled mass spectrometry (LC-MS) using an electrospray ionization (ESI) coupled to a linear ion trap mass spectrometer (LTQ-XL; Thermo Fisher), as indicated in the figure legends.
One-step protein purification using SILAC-labeled cells.
For stable isotope labeling by amino acids in cell culture (SILAC), HEK293T cells were cultured in arginine- and lysine-free DMEM (Invitrogen) supplemented with 10% dialyzed FBS (Invitrogen), penicillin-streptomycin, and either l-arginine and l-lysine (light) or l-arginine-HCl (13C6; CLM-2265 [R6]) and l-lysine-2HCl (4,4,5,5 D4; DLM-2640 [K4]) (medium) (Cambridge Isotope Labs). For complete incorporation of labeled amino acids, cells were passaged three times in SILAC medium over a period of 5 days. The labeled HEK293T cells were infected with PR8-wt (light) or PR8-NS-FS (medium) at an MOI of 0.6 in infection medium. After 1 h of incubation at 37°C, the medium was replaced by SILAC medium, followed by incubation at 37°C for 24 h. The medium was replaced by ice-cold PBS and collected by cell scraping and centrifugation. Cell pellets were incubated with LB plus 1% NP-40 for 20 min. Samples were sonicated with ten 3-s pulses (level 2) on a Virsonic 60 cell disruptor (Virtis) to disrupt nuclei and cleared by centrifugation. Cleared lysates were loaded five times over anti-FLAG-bead-containing columns. Unbound proteins were removed by washing column with LB plus 0.1% NP-40, and proteins were eluted at pH 3.5 in water supplemented with 100 mM glycine, 50 mM KCl, 0.1% NP-40, and Roche complete protease inhibitors. The proteins were concentrated by trichloroacetic acid precipitation, followed by separation on a 4 to 12% Bis-Tris gradient gel (Bio-Rad) and staining with SYPRO Ruby protein stain (Invitrogen). The entire lane was cut into individual bands (see Fig. 2B) and analyzed by LC-tandem MS (LC-MS/MS) using ESI coupled to an LTQ Orbitrap XL mass spectrometer (Thermo Fisher).
FIG 2.
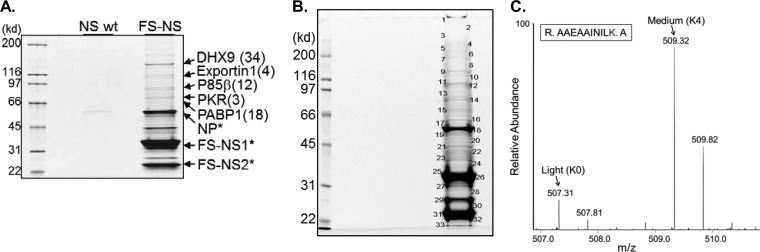
Purification of NS1/2 from IAV-infected cells and identification of associated proteins by MS. (A) SYPRO Ruby-stained SDS-PAGE of samples derived from A549 cells that were infected with PR8-wt and PR8-FS-NS for 24 h at an MOI of 0.5, followed by TAP. (B) SYPRO Ruby-stained SDS-PAGE of purified protein samples from SILAC-labeled HEK293T cells. HEK293T cells were labeled with Lys0/Arg0 (light) and Lys4/Arg6 (medium) and infected with PR8-wt (light) or PR8-NS-FS (medium), followed by protein purification and SDS-PAGE. (C) Bands subjected to LC-MS/MS analysis are indicated. Mass spectrum of a peptide representing PACT, whose differentially labeled forms (Light from PR8-wt-infected cells and medium from PR8-NS-FS-infected cells) can be identified by their specific mass difference of 4 Da (= 2 m/z of double-charged peptide, for lysine). Arrows indicate the C12 monoisotopic peaks of each peptide.
MALDI-TOF MS analysis.
Proteins were reduced and alkylated with iodoacetamide, and a tryptic digest was prepared. MS analysis was performed using a model 4700 proteomics analyzer from Applied Biosystems. The digest was introduced into the instrument in a crystalline matrix of α-cyano-4-hydroxycinnamic acid also containing 2 mM ammonium citrate to suppress ionization of matrix clusters. Database searches were performed using GPS explorer software (Applied Biosystems), which uses the Mascot search engine. Protein assignments were made on the basis of both MS and MS/MS spectra. The Swiss-Pro database (number 090207) was used for protein identification.
Sample preparation and ESI LC.
The protein gel bands were reduced and alkylated with iodoacetamide and digested with trypsin enzyme. The digest was introduced into the instrument via on-line chromatography using reversed-phase (C18) ultra-high-pressure LC on a nanoACQUITY apparatus (Waters). The column used was a Waters BEHC18 with an inner diameter of 75 μm and a bed length of 10 cm. The particle size was 1.7 μm. Tryptic peptides were gradient eluted over a gradient (0 to 70% B for 60 min and 70 to 100% B for 10 min, where B = 70% [vol/vol] acetonitrile, 0.2% formic acid) at a flow rate of 250 nl/min into the linear ion trap (LTQ-XL or LTQ Orbitrap XL) through a noncoated spray needle with voltage applied to the liquid junction.
MS/MS analysis with LTQ XL (Thermo Fisher) and database analysis.
Data-dependent scanning was incorporated to select the 10 most abundant ions (one microscan per spectrum; precursor isolation width, 2.0 Da; 35% collision energy; 30-ms ion activation; 30-s exclusion duration; 15-s repeat duration; and a repeat count of 2) from a full-scan mass spectrum for fragmentation by collision-activated dissociation. Database searches were performed using RAW files in combination with the Mascot search engine. Protein/peptide assignments were made on the basis MS/MS spectra. The NCBInr 20070908 database was used for protein identification. Product ions (b/y-type ions) were queried in an automated database search against a protein database (NCBInr 20070908 [5,454,477 sequences; 1,888,849,002 residues, all species]) by the Mascot search algorithm. The following residue modifications were allowed in the search: carbamidomethylation on cysteine (fixed modification) and oxidation on methionine (variable modification). Mascot was searched with a precursor ion tolerance of ±1 Da and a fragment ion tolerance of ±0.7 Da and using the automatic decoy database-searching tool in Mascot. The identifications from the automated search were verified by manual inspection of the raw data.
MS/MS analysis with LTQ XL Orbitrap (Thermo Fisher) (for SILAC) and MaxQuant analysis.
Data-dependent scanning was incorporated to select the 20 most abundant ions (one microscan per spectrum; precursor isolation width, 2.0 Da; 35% collision energy; 30-ms ion activation; 30-s exclusion duration; 15-s repeat duration; and a repeat count of 2) from a full-scan mass spectrum collected in the Orbitrap at a 60,000 resolution for fragmentation by collision-activated dissociation in the ion trap. Database searches are performed using the Andromeda search engine that is part of the MaxQuant software (version 1.2.0.34) developed at the Max Planck Institute (25). MaxQuant was also used to quantitate peptides and proteins and to provide ratios and generate a report in Excel format. Protein assignments were made on the basis of both MS and MS/MS spectra, whereas peptide quantitation was based solely on MS data.
Production of recombinant proteins.
His6-tagged recombinant proteins were expressed in Escherichia coli strain BL21(DE3). Protein expression was induced by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) in Turbo Prime media (Athena Enzyme Systems) at 16°C for 16 h. Cells were lysed in MCAC-0 buffer (50 mM Tris-Cl [pH 7.2], 500 mM NaCl, 10% [wt/vol] glycerol) and lysed by passage through a microfluidizer. The lysate was cleared by centrifugation, and proteins were captured using nickel beads (Gold Biotechnology), followed by elution with imidazole and dialysis.
In vitro translation and pulldown experiments.
In vitro translation and radioactive [35S]methionine labeling of PACT was performed using the TNT SP6 high-yield wheat germ protein expression system (Promega) according to the manufacturer's instructions. Pulldown assays were performed by incubating 5 μg of His-GFP or His-NS1 fusion proteins immobilized on Ni-NTA beads (Qiagen) with in vitro-translated protein for 4 h at 4°C. Bead-immobilized proteins were washed five times with lysis buffer supplemented with 0.1% NP-40, and proteins were eluted with sample buffer (Bio-Rad). Proteins were separated by SDS-PAGE and incubated with Amplify Reagent (GE Healthcare) for signal amplification. Gels were dried, scanned by using a phosphorimager (Typhoon FLA 9500; GE Healthcare), and analyzed by using ImageQuant TL software (GE Healthcare).
Immunoblotting (IB) and IP.
Cells were lysed in lysis buffer supplemented with 0.5% NP-40 for 20 min. Lysates were precleared with Sepharose beads (Sigma-Aldrich) for 1 h, followed by IP overnight using anti-Flag M2 resin (Sigma-Aldrich) or anti-HA affinity matrix (3F10; Roche). The immunoprecipitated samples or cell lysates were resolved by SDS-PAGE (Bio-Rad) and transferred onto nitrocellulose membranes. Membranes were probed with the indicated antibodies and visualized using enhanced chemiluminescence (Pierce) for detection.
Dual luciferase reporter assays.
HEK293T cells were transfected with an IFN-β promoter (−125) reporter (firefly luciferase) vector, Renilla luciferase vector (pRL-TK; Promega), and the indicated expression plasmids using Lipofectamine 2000 (Invitrogen). At 24 h after transfection, the cells were lysed in passive lysis buffer (Promega). The luciferase activity was determined using a dual-luciferase kit (Promega), and the firefly luciferase activity values were normalized to the Renilla luciferase activity.
IFN bioassay.
To inactivate virus, 120 μl of virus-containing cell culture supernatant was treated in 96-well plates by UV light (15 W, 302 nm, XX-15M UV bench lamp; Upland) for 10 min at a 6-cm distance, resulting in the reduction of IAV infectivity below the detection limit. UV-treated cell culture supernatant and recombinant human IFN-β (hIFN-β; PBL) in limiting dilution as a standard was added to Caki cells (kindly provided by L. Pfeffer) that were stably cotransduced with lentiviral vectors containing ISRE-firefly luciferase and EF1α-Renilla luciferase cassettes. The luciferase activity was determined after 24 h using a dual-luciferase kit (Promega), and the firefly luciferase activity values were normalized to the Renilla luciferase activity. IFN values were determined based on the hIFN-β standard. The detection limit of this bioassay was 1.5 U/ml.
Statistical analysis.
Data presented in reporter assays and IFN bioassays are expressed as means ± the standard deviations and were compared using the Student t test. A P value of <0.05 was considered significant.
RESULTS
Generation of recombinant IAV with epitope-tagged NS1 and NS2.
In order to establish epitope-tagged forms of NS1 and NS2 that could be purified by different affinity purification strategies, including tandem affinity purification (TAP), we established a fusion TAP tag, consisting of a triple FLAG tag and a StrepOne tag, here referred to as the FS tag. The coding sequence of this tag was introduced into the influenza virus A/PR/8/34 (PR8) NS gene as illustrated in Fig. 1A. The upper part of Fig. 1A depicts the NS gene and the two mRNA splice variants encoding NS1 and NS2. To introduce the FS tag at the N terminus of NS1/2, we duplicated 90 bp of the 5′ coding region of the NS1 gene in order to avoid interference with efficient vRNA packaging (18). To trigger removal of these additional 30 amino acids from the mature NS1/2 proteins, we introduced downstream thereof the 2A peptide of the porcine teschovirus-1 (P2A), which confers near-complete ribosomal “skipping” (19) that is functionally equivalent to cleavage immediately upstream of the FS tag (Fig. 1A, lower part). This NS gene (and in parallel the PR8 wild-type [PR8-wt] gene as control) was used, along with the other PR8 gene segments to establish recombinant virus by reverse genetics (26). The recombinant virus, referred to as PR8-NS-FS, displayed growth characteristics on MDCK, A549, and HEK293T cells similar to those of the PR8-wt virus (Fig. 1B), suggesting the functional activity of the recombinant FS proteins. We infected the different cell lines with these viruses and determined the expression of virus proteins by IB. NS1 and NS2 expression of both viruses was detected, with the FS-tagged forms displaying the expected increase in molecular size (6 kDa) due to the FS tag (Fig. 1C). Although the expression of matrix protein and nucleoprotein was comparable for both viruses on all cell types investigated, the expression of the FS-tagged form of NS1 was reduced, whereas expression of FS-NS2 was increased compared to their wild-type forms (Fig. 1C). FS-NS1 and FS-NS2 expression from PR8 NS-FS was also detected when using FLAG-specific antibodies (Fig. 1C, right side), and the identity of both proteins was confirmed by MS analysis (see below). As such, the strategy of duplicating part of the genomic sequence, followed by a P2A peptide, can be used to introduce an epitope tag into full-length viral proteins maintaining virus replication, at least under in vitro conditions, albeit the relative expression levels of wild-type and epitope-tagged forms of proteins may be affected.
Proteomic analysis of NS1 and NS2 during virus replication.
In order to analyze NS-interacting proteins during virus replication, we infected A549 lung epithelial cells with the recombinant PR8-NS-FS and PR8-wt in parallel, followed by TAP, SDS-PAGE, and SYPRO Ruby staining. Several distinct bands were visible following TAP of PR8 NS-FS-infected cells, which were largely missing in the TAP sample of PR8-wt-infected cells, demonstrating the purification power of the FS-tag (Fig. 2A). Several bands were analyzed by MS, identifying unequivocally NS1 and NS2, but also several other proteins with a significant number of unique peptides (Fig. 2A). These included known NS1- and NS2-interacting proteins, such as the regulatory beta-subunit of phosphatidylinositol 3-kinase (p85β) and Exportin-1 (XPO1), respectively (1, 27, 28), indicating that the recombinant PR8-FS-NS virus can be used to identify NS1/2-interacting proteins during the natural course of infection. In this TAP-based experiment, the assessment of true NS1/2-interacting proteins relied largely on quantification by protein staining. Unequivocal identification of less-abundant NS1/2-interacting proteins will require more precise methods of quantification. In the last decade, substantial progress in quantitative forms of proteomics has been made, allowing the separation of true interacting proteins from contaminating proteins. One of these techniques is based on SILAC (29). In this procedure, cells are labeled with different, nonradioactive heavy isotopes of certain amino acids. For example, control cells are “labeled” with regular, “light” amino acids, and cells to be infected are “labeled” with “medium” or “heavy” arginine and lysine. After infection, the lysates of both cell populations are combined, and all following steps, including protein purification and MS analysis, are performed with one, mixed sample. The source of proteins will then unequivocally be identified by the specific mass differences of individual peptides (29, 30). One prerequisite for this approach is cell culture in the presence of dialyzed FBS, which allows complete replacement of unlabeled amino acids with exogenous, labeled amino acids. When culturing A549 cells in the presence of different isotopic amino acids followed by IAV infection, we noted a profound reduction in infection efficiency by more than 10-fold, preventing complete cell infection (data not shown). We therefore tested HEK293T cells and found these cells much less affected by SILAC conditions. We therefore used this cell type for the following experiment. HEK293T cells were labeled with different isotopes, followed by infection with PR8-wt and PR8-FS-NS. At 18 h postinfection, lysates were prepared, combined, and subjected to one-step purification based on anti-FLAG antibodies, which was followed by acidic elution and LC-MS. The purified samples were separated by SDS-PAGE, and 33 bands were analyzed individually by LC-MS, followed by MaxQuant-based analysis (Fig. 2B). Some of the proteins identified are depicted in Table 1 (for complete analysis see Table S1 in the supplemental material). As expected, several known NS1- and NS2-interacting proteins were identified specifically as NS1/NS2-interacting proteins, as expressed by the ratio of medium- and light-labeled proteins, representing proteins derived from PR8-FS-NS- and PR8-wt-infected cells, respectively. For example, p85β was identified at 5.9-fold-higher levels from PR8-FS-NS- than PR8-wt-infected cells. As expected, the PIK3R2-associated alpha-subunit, PIK3R1, was also enriched, possibly copurifying with the regulatory β-subunit. Other known NS1/NS2-interacting proteins, such as PABP-1 and the very recently identified NS1-interacting protein dsRNA-specific adenosine deaminase (ADAR), as well as the NS2-interacting Exportin-1 (XPO1), were identified as specifically associated with the NS-FS-expressing virus (Table 1) (28, 31). Notably, a number of proteins not characterized as NS1- or NS2-interacting proteins were identified, including the protein PACT, which was identified unequivocally with four unique peptides at a ratio of 2.98 over the control (Table 1 and see Table S1 in the supplemental material). The MS spectrum of one PACT-derived peptide is shown in Fig. 2C, illustrating the relative abundance of PACT from PR8-NS-FS (medium) and PR8-wt (light). Taken together, the data demonstrate that the recombinant virus can be used to specifically identify virus-interacting host proteins in a quantitative proteomic approach. Certainly, in the case of various splice products, as observed for NS proteins, differentiation between NS1- or NS2-interacting proteins needs to be established in independent experiments (see below).
TABLE 1.
MS-/MaxQuant-based identification of NS1- and NS2-interacting proteins using SILAC
| Protein | Gene | No. of unique peptides | M/L ratio (normalized)a | Sequence coverage (%) |
|---|---|---|---|---|
| IFN-inducible dsRNA-dependent protein kinase activator A (PACT) | PRKRA | 4 | 2.98 | 14.1 |
| Phosphatidylinositol 3-kinase regulatory subunit beta | PIK3R2 | 23 | 5.90 | 40.7 |
| Phosphatidylinositol 3-kinase regulatory subunit alpha | PIK3R1 | 4 | 2.87 | 7 |
| Double-stranded RNA-specific adenosine deaminase | ADAR | 34 | 3.08 | 30.6 |
| Polyadenylate-binding protein 1 (PABP-1) | PABPC1 | 7 | 1.90 | 39.8 |
| Exportin-1 | XPO1 | 7 | 2.05 | 7.3 |
| ATP-dependent RNA helicase A | DHX9 | 41 | 0.99 | 37.1 |
M, medium (proteins derived from PR8-NS-FS-infected cells); L, light (proteins derived from PR8-wt-infected cells).
NS1 interacts with PACT.
We confirmed interaction of NS1/NS2 with endogenous PACT during virus infection in small scale experiments using PACT-specific antibodies. After 24 h of infection, PACT copurified specifically with FS-tagged proteins but was not identified in samples from PR8-wt-infected cells (Fig. 3A). To determine whether this interaction could be recapitulated in the absence of virus infection and also to determine whether NS1 or NS2 interacted with PACT, we performed transient-transfection experiments with epitope-tagged proteins. As shown in Fig. 3B, NS1, but not NS2, interacted robustly with PACT. Analyzing the kinetics of PACT-NS1 interaction by infecting cells for different periods of time, we found protein interaction at the earliest time point of NS1 expression, indicating immediate and efficient interaction of the two proteins (Fig. 3C). To determine whether binding of PACT is a more general property of IAV NS1 proteins, we infected cells with different IAV strains and analyzed PACT-NS1 interaction by IP/IB analysis. Similar to PR8 NS1, NS1 of the three other investigated IAV strains interacted robustly with PACT, suggesting that PACT binding is a conserved function of NS1 (Fig. 3D). To determine whether NS1 could interact directly with PACT, we established pulldown assays based on bacterially expressed recombinant NS1 and in vitro-translated PACT. NS1 precipitated PACT robustly, whereas GFP, used as a negative control, did not (Fig. 3E). As such, NS1 interacts with PACT during IAV infection.
NS1 inhibits PACT binding to RIG-I and blocks RIG-I/PACT-mediated IFN-β promoter activity.
To study the impact of NS1 on PACT biology, we coexpressed NS1 together with PACT and RIG-I and measured the activity of a cotransfected IFN-β-promoter luciferase reporter. RIG-I alone activated the IFN-β-promoter, whose activity was further increased by coexpression of PACT, as expected (Fig. 4A) (15). However, IFN-β promoter activity induced by either RIG-I alone or PACT/RIG-I was strongly reduced by NS1, demonstrating that NS1 interfered with the PACT/RIG-I pathway (Fig. 4A).
FIG 4.
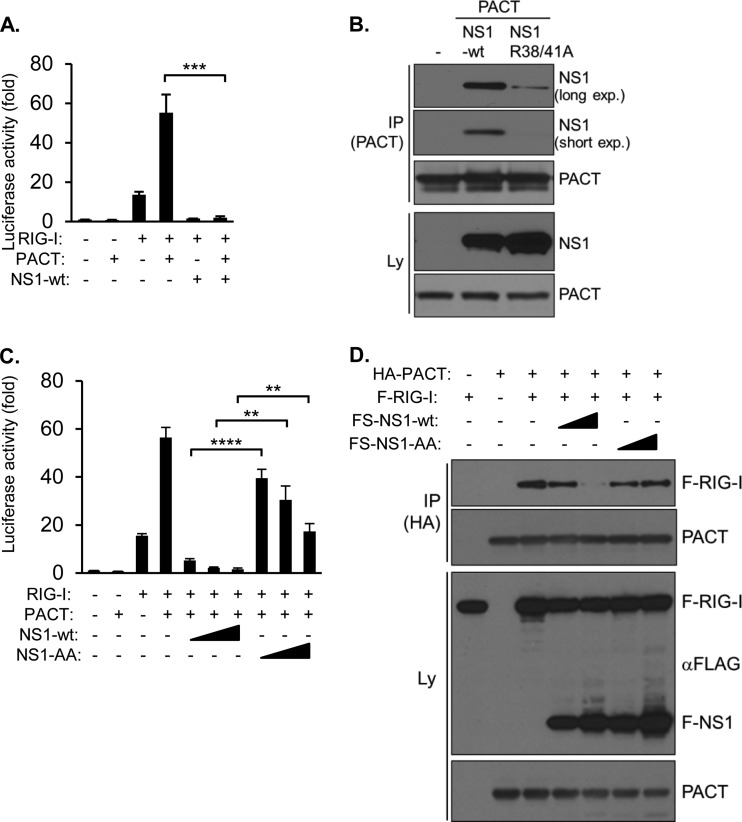
NS1 prevents interaction between PACT and RIG-I and blocks PACT/RIG-I-mediated IFN-β activation. (A) IFN-β reporter assay. HEK293T cells were transfected with an IFN-β promoter luciferase vector, either alone (control) or along with expression vectors for RIG-I, PACT, and NS1, as indicated, followed by analysis of luciferase activity after 48 h. Values are presented as the fold changes over the control. Error bars represent the standard deviations of three transfection replicates. ***, P < 0.001. (B) HEK293T cells transfected with PACT, along with FS-NS1 or FS-NS1 (R38A/K41A) (NS1-AA), for 48 h, followed by IP/IB analysis with antibodies against PACT (for IP) and NS1 and PACT (for IB). Long and short exposures of films are shown, as indicated. (C) IFN-β reporter assay. HEK293T cells were transfected with an IFN-β promoter luciferase vector, either alone (control) or along with expression vectors for RIG-I, PACT, NS1, and NS1-AA as indicated, followed by analysis of luciferase activity after 48 h. Values are presented as the fold changes over the control. Error bars represent the standard deviations of three transfection replicates. **, P < 0.01; ****, P < 0.0001. (D) HEK293T cells were transfected with expression vectors for the indicated proteins for 48 h and analyzed by IP/IB with antibodies against HA (for IP) and antibodies against RIG-I, PACT, and FLAG (recognizing FLAG-RIG-I and FS-NS1) (for IB).
Previously, the RNA binding domain of NS1 has been demonstrated to be involved in NS1-mediated inhibition of virus-mediated IFN-I activation, where two highly conserved amino acids, which confer binding of dsRNA, i.e., R38 and K41 of PR8-NS1, were found to be essential for inhibition of IFN-I activation (32, 33). To determine whether these amino acids might be involved in PACT binding, we coexpressed NS1-wt and a R38A/K41A mutant protein (NS1-AA), along with PACT in HEK293T cells, followed by IP. Although the expression levels of both NS1 proteins were comparable, the interaction between PACT and the mutant protein was strongly reduced, suggesting that the integrity of the RNA binding domain is important for protein interaction (Fig. 4B). To substantiate these findings in functional assays, we coexpressed RIG-I, PACT, and NS1 proteins in IFN-β reporter assays. Although NS1 blocked RIG-I/PACT-induced IFN-β-promoter activity at all of the concentrations used, NS1-AA exhibited strongly reduced, albeit not completely abolished, inhibitory activity (Fig. 4C). Given that direct interaction of PACT with RIG-I is required for RIG-I activation (15), we sought to determine whether NS1 could directly interfere with this protein interaction. Indeed, whereas PACT precipitated RIG-I efficiently in transient-transfection experiments, coexpression of NS1-wt, but not NS1-AA, reduced RIG-I–PACT interaction significantly (Fig. 4D). We conclude that the functional integrity of the RNA binding domain of NS1 is important for PACT binding and interference with RIG-I–PACT interaction and, as a consequence, the inhibition of PACT/RIG-I-mediated IFN activation.
PACT is required for virus recognition and IFN activation in the absence of NS1.
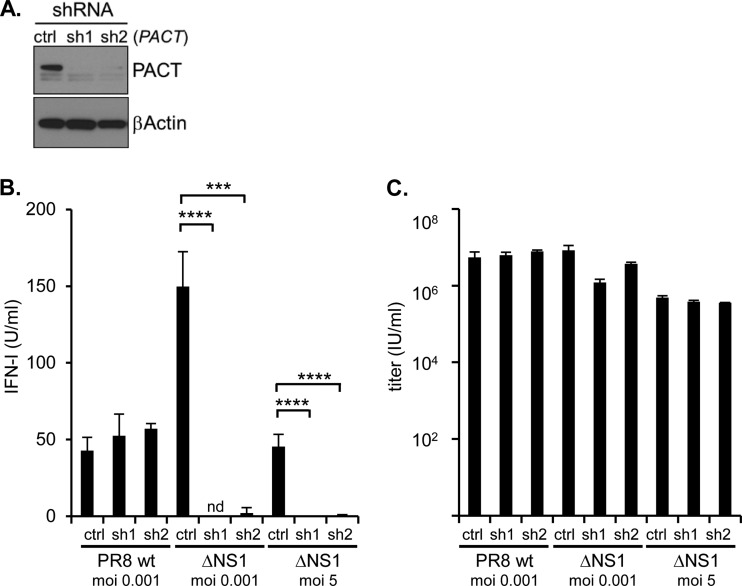
PACT has previously been demonstrated to contribute to Sendai virus-mediated IFN-I activation (15). To study the impact of PACT for IAV-induced IFN activation, we established HEK293T cell lines wherein PACT expression was stably suppressed based on retrovirally delivered shRNA against endogenous PACT mRNA. Two independent, polyclonal PACT shRNA cell lines (and shRNA control cells) were generated, both of which exhibited strongly reduced PACT protein levels compared to control cells (Fig. 5A). These cells were used for infection with PR8-wt and a recombinant PR8 mutant, where the NS1-specific coding region of the NS gene segment was deleted (PR8 ΔNS1), followed by analysis of IFN-I secretion in a quantitative bioassay. Both viruses induced IFN-I in control cells, although the PR8 ΔNS1 induced more IFN-I, as expected (4) (Fig. 5B). However, PR8 ΔNS1-induced IFN-I secretion in cell lines with compromised PACT expression was strongly reduced (Fig. 5B). In contrast, IFN-I secretion upon PR8-wt infection was minimally affected by the PACT expression levels, which is consistent with the idea that NS1 efficiently counteracted PACT-mediated IFN activation, which could not further be reduced by additional suppression of PACT (Fig. 5B). It is currently unclear why PR8-wt induced more IFN-I than PR8 ΔNS1 on PACT shRNA cells (Fig. 5B). Apparently, this phenomenon was not related to virus replication since both viruses multiplied to a similar extent during the time period analyzed, and a slightly reduced proliferative capacity of PR8 ΔNS1 on PACT shRNA cells at a lower MOI was not apparent at a high MOI, where IFN-I release was still fully ablated (Fig. 5B and C). Taken together, the data show that PACT is critical for IAV recognition in the absence of NS1. The data are consistent with the interpretation that PR8-wt-mediated NS1 expression interferes efficiently with PACT biology.
FIG 5.
PACT is required for efficient IFN-I production upon IVA infection in the absence of NS1. (A) Stable knockdown of PACT in HEK293T cells by two different, retrovirally delivered shRNAs against PACT mRNA (sh1, sh2) and control shRNA (ctrl), analyzed by IB with antibodies against PACT and β-actin. (B and C) The HEK293T cells described in panel A were infected with PR8-wt and PR8 ΔNS1 at an MOI of 0.001 for 72 h or with PR8 ΔNS1 at an MOI of 5 for 24 h as indicated. The IFN-I activity (B) and virus titer (C) were determined by IFN bioassay and limiting dilution, after 72 h (MOI of 0.001) and 24 h (MOI of 5), respectively. nd, not detectable; ***, P < 0.001; ****, P < 0.0001.
DISCUSSION
Using a novel strategy to identify IAV NS1/2-interacting host proteins, we identified a number of uncharacterized NS1/2-interacting proteins, including PACT. The specific IAV construct developed retains the full-length structure of the tagged proteins and allows efficient one- and two-step purifications. It is likely that a similar approach is feasible for other IAV gene segments. While two-step purifications yield highly purified samples, the evaluation of identified proteins as true interactors depends on MS-based spectral counting or quantitative assessment of the protein staining, both of which are difficult for low-abundance proteins. In addition, the time and sample dilution required for TAP is likely to result in the loss of low-affinity interactors. In contrast, a one-step approach reduces the purification time significantly but depends for thorough interpretation, particularly of low-abundance proteins, on quantitative MS approaches, e.g., SILAC. Although our data demonstrate the feasibility of the approach, we believe that at least two improvements can be made. First, we noted that some proteins, e.g., DHX9, which was previously identified as NS1-interacting protein in a nonquantitative proteomic approach, a finding confirmed in our TAP approach (Fig. 2A), was identified in the SILAC experiment at similar ratios from the differentially labeled cells and thus would be ranked nonspecific (Table 1) (34). This phenomenon is most likely due to interaction of DHX9 with FS-NS1 during the purification process in the mixed samples, a known problem for dynamic protein interactions (35). As such, our approach has most likely sacrificed the identification of such protein interactions in favor of the reduction of false-positive identifications. A relatively simple remedy to this problem will be the separate purification of individual samples, followed by mixing of purified samples for SDS-PAGE and MS, also referred to as “MAP (mixing after purification)-SILAC” (35). We also noted an unexpected reduction in the infection efficiency of SILAC-labeled A549 cells, which led us to use HEK293T cells. For A549 cells (and possibly other cell types), other forms of quantitative MS seem therefore more suitable, particularly post-tryptic labeling procedures, such as the use of tandem mass tags (TMT) and isobaric tags for relative and absolute quantitation (iTRAQ), which have made major progress during the last few years, overcoming initial quantification problems (36).
One protein unequivocally identified by the proteomic approach and further confirmed as NS1-interacting partner is PACT, a protein whose binding to PKR and RIG-I has been characterized in some detail before (11, 15). We found in luciferase reporter assays based on coexpression of PACT and RIG-I that NS1 interfered efficiently with IFN-β promoter activation. Although PACT upregulated RIG-I-induced IFN-β promoter activity significantly, which was almost completely blocked by NS1, this experimental setting does not differentiate between NS1-mediated interference with PACT-induced RIG-I activation and TRIM25-mediated RIG-I ubiquitination, which was previously shown to increase RIG-I activity (8, 10). Two sets of experiments, however, indicate that NS1-PACT interaction is important. First, knockdown of PACT strongly reduced RIG-I-mediated IFN activation in the absence of NS1, demonstrating that PACT contributes significantly to RIG-I activation in IAV-infected cells and, second, NS1 interfered directly with complex formation between PACT and RIG-I, which mediates RIG-I activation (Fig. 4A and C) (15). As such, it appears that NS1 has acquired the capacity to interfere with distinct mechanisms that control RIG-I activation at different levels of its signaling cascade.
As mentioned, NS1 has also been shown to interact directly with PKR (37) and was found to block PKR activation by dsRNA and by PACT through interaction with the N-terminal region of PKR which contains two dsRNA binding domains (38). In the same publication, the authors noted that NS1 pulled down PACT when overexpressed in HEK293T cells. The interpretation of these findings was complicated by the fact that other proteins, including PKR, in these lysates might have contributed to this interaction. Our results based on recombinant NS1 and in vitro-translated PACT supports the interpretation that the NS1/PACT interaction is direct. An interesting observation is that the two exposed, charged amino acids R38/K41 contained in the RNA binding domain of NS1 support PACT interaction and contribute to NS1-mediated interference with PACT-RIG-I activity (Fig. 4B and C). This observation contrasts to the mentioned NS1/PKR interaction, where a NS1 dsRNA binding mutant with a single amino acid substitution (R38A) retained its inhibitory activity against dsRNA- and PACT-induced PKR activation (38). Although it needs to be formally ruled out that the additional point mutation of K41 used in our report may account for the PKR-inhibitory function by NS1, it appears that the dsRNA binding domain is more specifically involved in interference with the IFN-I pathway than PKR activation and that both functions of NS1 can thus be separated on a molecular level. Despite these molecular differences, it is important to note that PACT represents a clear common denominator of two well-characterized antiviral defense systems, i.e., the RIG-I and PKR pathways, both of which are disabled upon NS1 expression. Interestingly, another mechanism for how these two pathways may act interdependently has been proposed based on the observation that critical components thereof, including PKR and RIG-I, colocalize upon virus infection in a compartment referred to as antiviral stress granules (avSGs) (39). IAV-mediated avSG formation and IFN-I production, most likely RIG-I dependent, was found to depend on (i) the presence of PKR and (ii) the absence of NS1 (39). One possible interpretation of these findings is that NS1-mediated interference with PKR prevents the formation of a structural compartment required for orchestration of the antiviral defense, including RIG-I activation. Although the assembly of the different protein components in avSGs could not be assessed due to solubility issues, it appears that in this model PKR acts upstream of RIG-I (39). As such, it will be interesting to see how PACT-mediated activation of PKR and RIG-I fits into this model, which we would assume act in a parallel rather than serial fashion. Certainly, the two scenarios are not mutually exclusive and may represent independent mechanisms, possibly acting in a cell-type-specific or time-dependent way. In fact, it seems possible that initial virus recognition and IFN-I activation depends on PKR (and RIG-I) in avSGs, while at later time points during infection and upon priming with small amounts of IFN-I, a PACT-RIG-I-dependent (avSG-independent) mechanism prevails. This hypothesis is supported (i) by data from PKR-deficient cells, where a deficiency of IFN-I production upon poly(I·C) treatment could be overcome by pretreatment with IFN-I, suggesting a restriction of PKR function to the initial step of IFN-I activation (40), and (ii) by data from HEK293T cells, where PACT overexpression strongly supported IFN-β-induced IFN-β promoter activity, suggesting that PACT represents a critical part of a secondary, IFN-I-mediated RIG-I activation mechanism, akin to the previously characterized positive feedback regulation of IFN production (15, 41). Certainly, more work will be required to assess the time-dependent, relative contributions of PACT to RIG-I versus PKR activation.
Although our data indicate a critical role of R38/K41 in protein interaction, these amino acids are certainly also critical for NS1-mediated dsRNA binding, a function that has been proposed as mechanism to prevent recognition of viral dsRNA emerging during replication (6, 42). The existence of such immunostimulatory dsRNA, particularly in the cytoplasm, where RIG-I-mediated virus sensing most likely occurs, however, has been questioned for negative-strand RNA viruses such as IAV, at least in the presence of functional IAV nucleoprotein (43, 44). As such, the binding of viral dsRNA by NS1 and its relevance for virus recognition requires further investigation. Although our in vitro binding assay of NS1 and PACT does not completely rule out the possibility that trace amounts of RNA contained in the in vitro translation sample contribute to protein interaction, another emerging possibility is that the RNA binding domain represents a protein interaction motif. In this context it is important to note that the two viral proteins, i.e., HSV Us11 and Ebola VP35, which have very recently been shown to counteract PACT-mediated IFN activation (16, 17), both contain dsRNA binding domains that are critical for PACT interaction. dsRNA seemed dispensable for Us11-PACT and VP35-PACT interaction, supporting the idea that the RNA binding domains may act as protein interaction motifs. For VP35, critical residues involved in RNA binding have indeed been found to mediate intermolecular VP35 interactions, providing evidence for the principle possibility of protein interaction of involved RNA binding domains. More detailed information based on structural experiments will be required to confirm this hypothesis.
Taken together, the data shown in the present study contribute to our understanding of how NS1 interferes with pathogen recognition, highlighting yet another host target, PACT, whose inactivation along with other proteins acting at various stages of virus recognition, including TRIM25, Riplet, and PKR, compromises and delays host immune defense against IAV infection (8, 9).
Supplementary Material
ACKNOWLEDGMENTS
We thank Youming Shao and Richard Heath (St. Jude Protein Production Facility) for the production of the recombinant proteins. We thank Ruiqiong Wu and Liying Chi for excellent technical support.
E.R.H. was supported by the Deutsche Forschungsgemeinschaft. This study was supported by the American Lebanese Syrian Associated Charities.
Footnotes
Published ahead of print 4 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00830-14.
REFERENCES
- 1.Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. The J. Gen. Virol. 89:2359–2376. 10.1099/vir.0.2008/004606-0 [DOI] [PubMed] [Google Scholar]
- 2.Egorov A, Brandt S, Sereinig S, Romanova J, Ferko B, Katinger D, Grassauer A, Alexandrova G, Katinger H, Muster T. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 72:6437–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kochs G, Koerner I, Thiel L, Kothlow S, Kaspers B, Ruggli N, Summerfield A, Pavlovic J, Stech J, Staeheli P. 2007. Properties of H7N7 influenza A virus strain SC35M lacking interferon antagonist NS1 in mice and chickens. J. Gen. Virol. 88:1403–1409. 10.1099/vir.0.82764-0 [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330. 10.1006/viro.1998.9508 [DOI] [PubMed] [Google Scholar]
- 5.Lu Y, Wambach M, Katze MG, Krug RM. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214:222–228. 10.1006/viro.1995.9937 [DOI] [PubMed] [Google Scholar]
- 6.Min JY, Krug RM. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo(A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. U. S. A. 103:7100–7105. 10.1073/pnas.0602184103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- 8.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449. 10.1016/j.chom.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villan E, Garcia-Sastre A, Gack MU. 2012. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS pathogens 8:e1003059. 10.1371/journal.ppat.1003059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920. 10.1038/nature05732 [DOI] [PubMed] [Google Scholar]
- 11.Patel RC, Sen GC. 1998. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 17:4379–4390. 10.1093/emboj/17.15.4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meurs E, Chong K, Galabru J, Thomas NS, Kerr IM, Williams BR, Hovanessian AG. 1990. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62:379–390. 10.1016/0092-8674(90)90374-N [DOI] [PubMed] [Google Scholar]
- 13.Gale M, Jr, Katze MG. 1998. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol. Ther. 78:29–46 [DOI] [PubMed] [Google Scholar]
- 14.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. 2006. The role of PACT in the RNA silencing pathway. EMBO J. 25:522–532. 10.1038/sj.emboj.7600942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kok KH, Lui PY, Ng MH, Siu KL, Au SW, Jin DY. 2011. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe 9:299–309. 10.1016/j.chom.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 16.Kew C, Lui PY, Chan CP, Liu X, Au SW, Mohr I, Jin DY, Kok KH. 2013. Suppression of PACT-induced type I interferon production by herpes simplex virus 1 Us11 protein. J. Virol. 87:13141–13149. 10.1128/JVI.02564-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luthra P, Ramanan P, Mire CE, Weisend C, Tsuda Y, Yen B, Liu G, Leung DW, Geisbert TW, Ebihara H, Amarasinghe GK, Basler CF. 2013. Mutual antagonism between the Ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell Host Microbe 14:74–84. 10.1016/j.chom.2013.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dos Santos Afonso E, Escriou N, Leclercq I, van der Werf S, Naffakh N. 2005. The generation of recombinant influenza A viruses expressing a PB2 fusion protein requires the conservation of a packaging signal overlapping the coding and noncoding regions at the 5′ end of the PB2 segment. Virology 341:34–46. 10.1016/j.virol.2005.06.040 [DOI] [PubMed] [Google Scholar]
- 19.Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. 2001. Analysis of the aphthovirus 2A/2B polyprotein “cleavage” mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal “skip.” J. Gen. Virol. 82:1013–1025 [DOI] [PubMed] [Google Scholar]
- 20.Brown LE, Hinshaw VS, Webster RG. 1983. Antigenic variation in the influenza A virus nonstructural protein, NS1. Virology 130:134–143. 10.1016/0042-6822(83)90123-X [DOI] [PubMed] [Google Scholar]
- 21.van Wyke KL, Yewdell JW, Reck LJ, Murphy BR. 1984. Antigenic characterization of influenza A virus matrix protein with monoclonal antibodies. J. Virol. 49:248–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker-Hapak M, Dowdy SF. 2003. Protein transduction: generation of full-length transducible proteins using the TAT system. Curr. Protoc. Cell Biol. Chapter 20:Unit 20 22. 10.1002/0471143030.cb2002s18 [DOI] [PubMed] [Google Scholar]
- 23.Kok KH, Ng MH, Ching YP, Jin DY. 2007. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J. Biol. Chem. 282:17649–17657. 10.1074/jbc.M611768200 [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108–6113. 10.1073/pnas.100133697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275–2289. 10.1007/s007050170002 [DOI] [PubMed] [Google Scholar]
- 27.Ehrhardt C, Wolff T, Pleschka S, Planz O, Beermann W, Bode JG, Schmolke M, Ludwig S. 2007. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 81:3058–3067. 10.1128/JVI.02082-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann G, Hughes MT, Kawaoka Y. 2000. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 19:6751–6758. 10.1093/emboj/19.24.6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ong SE, Mann M. 2005. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1:252–262. 10.1038/nchembio736 [DOI] [PubMed] [Google Scholar]
- 30.Harsha HC, Molina H, Pandey A. 2008. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nature protocols 3:505–516. 10.1038/nprot.2008.2 [DOI] [PubMed] [Google Scholar]
- 31.de Chassey B, Aublin-Gex A, Ruggieri A, Meyniel-Schicklin L, Pradezynski F, Davoust N, Chantier T, Tafforeau L, Mangeot PE, Ciancia C, Perrin-Cocon L, Bartenschlager R, Andre P, Lotteau V. 2013. The interactomes of influenza virus NS1 and NS2 proteins identify new host factors and provide insights for ADAR1 playing a supportive role in virus replication. PLoS Pathog. 9:e1003440. 10.1371/journal.ppat.1003440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El Hefnawi M, Alaidi O, Mohamed N, Kamar M, El-Azab I, Zada S, Siam R. 2011. Identification of novel conserved functional motifs across most Influenza A viral strains. Virol. J. 8:44. 10.1186/1743-422X-8-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donelan NR, Basler CF, Garcia-Sastre A. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 77:13257–13266. 10.1128/JVI.77.24.13257-13266.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin L, Li Y, Pyo HM, Lu X, Raman SN, Liu Q, Brown EG, Zhou Y. 2012. Identification of RNA helicase A as a cellular factor that interacts with influenza A virus NS1 protein and its role in the virus life cycle. J. Virol. 86:1942–1954. 10.1128/JVI.06362-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Huang L. 2008. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol. Cell. Proteomics 7:46–57. 10.1074/mcp.M700261-MCP200 [DOI] [PubMed] [Google Scholar]
- 36.Wuhr M, Haas W, McAlister GC, Peshkin L, Rad R, Kirschner MW, Gygi SP. 2012. Accurate multiplexed proteomics at the MS2 level using the complement reporter ion cluster. Anal. Chem. 84:9214–9221. 10.1021/ac301962s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan SL, Katze MG. 1998. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J. Interferon Cytokine Res. 18:757–766. 10.1089/jir.1998.18.757 [DOI] [PubMed] [Google Scholar]
- 38.Li S, Min JY, Krug RM, Sen GC. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13–21. 10.1016/j.virol.2006.01.005 [DOI] [PubMed] [Google Scholar]
- 39.Onomoto K, Jogi M, Yoo JS, Narita R, Morimoto S, Takemura A, Sambhara S, Kawaguchi A, Osari S, Nagata K, Matsumiya T, Namiki H, Yoneyama M, Fujita T. 2012. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One 7:e43031. 10.1371/journal.pone.0043031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams BR, Aguet M, Weissmann C. 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 14:6095–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marie I, Durbin JE, Levy DE. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660–6669. 10.1093/emboj/17.22.6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, Garcia-Sastre A. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989–7996. 10.1128/JVI.74.17.7989-7996.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059–5064. 10.1128/JVI.80.10.5059-5064.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wisskirchen C, Ludersdorfer TH, Muller DA, Moritz E, Pavlovic J. 2011. The cellular RNA helicase UAP56 is required for prevention of double-stranded RNA formation during influenza A virus infection. J. Virol. 85:8646–8655. 10.1128/JVI.02559-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.