ABSTRACT
Recent genome-wide screens reveal that the host cells express an arsenal of proteins that inhibit replication of plus-stranded RNA viruses by functioning as cell-intrinsic restriction factors of viral infections. One group of cell-intrinsic restriction factors against tombusviruses contains tetratricopeptide repeat (TPR) domains that directly interact with the viral replication proteins. In this paper, we find that the TPR domain-containing Hop-like stress-inducible protein 1 (Sti1p) cochaperone selectively inhibits the mitochondrial membrane-based replication of Carnation Italian ringspot tombusvirus (CIRV). In contrast, Sti1/Hop does not inhibit the peroxisome membrane-based replication of the closely related Tomato bushy stunt virus (TBSV) or Cucumber necrosis virus (CNV) in a yeast model or in plants. Deletion of STI1 in yeast leads to up to a 4-fold increase in CIRV replication, and knockdown of the orthologous Hop cochaperone in plants results in a 3-fold increase in CIRV accumulation. Overexpression of Sti1p derivatives in yeast reveals that the inhibitory function depends on the TPR1 domain known to interact with heat shock protein 70 (Hsp70), but not on the TPR2 domain interacting with Hsp90. In vitro CIRV replication studies based on isolated mitochondrial preparations and purified recombinant proteins has confirmed that Sti1p, similar to the TPR-containing Cyp40-like Cpr7p cyclophilin and the Ttc4 oncogene-like Cns1 cochaperone, is a strong inhibitor of CIRV replication. Sti1p interacts and colocalizes with the CIRV replication proteins in yeast. Our findings indicate that the TPR-containing Hop/Sti1 cochaperone could act as a cell-intrinsic virus restriction factor of the mitochondrial CIRV, but not against the peroxisomal tombusviruses in yeast and plants.
IMPORTANCE The host cells express various cell-intrinsic restriction factors that inhibit the replication of plus-stranded RNA viruses. In this paper, the authors find that the Hop-like stress-inducible protein 1 (Sti1p) cochaperone selectively inhibits the mitochondrial membrane-based replication of Carnation Italian ringspot tombusvirus (CIRV) in yeast. Deletion of STI1 in yeast or knockdown of the orthologous Hop cochaperone in plants leads to increased CIRV replication. In addition, overexpression of Sti1p derivatives in yeast reveals that the inhibitory function depends on the TPR1 domain known to interact with heat shock protein 70 (Hsp70), but not on the TPR2 domain interacting with Hsp90. In vitro CIRV replication studies based on isolated mitochondrial preparations and purified recombinant proteins have confirmed that Sti1p is a strong inhibitor of CIRV replication. The authors' findings reveal that the Hop/Sti1 cochaperone could act as a cell-intrinsic restriction factor against the mitochondrial CIRV, but not against the related peroxisomal tombusviruses.
INTRODUCTION
Cells produce a yet-unknown number of cell-intrinsic restriction factors that limit replication of plus-stranded RNA [(+)RNA] viruses. The cellular restriction factors could be virus specific or components of the cell-intrinsic innate systems of the host through targeting diverse pathogens (1–7). Cellular factors are also recruited by (+)RNA viruses to aid viral replication, which takes place in membrane-bound viral replicase complexes (VRCs) in the cytoplasm of infected cells (8–16). The diverse, often opposite, roles of host factors are reflected by the identification of stimulatory as well as inhibitory host proteins in genome-wide screens with various hosts and viruses, such as Tomato bushy stunt virus (TBSV), West Nile virus, Brome mosaic virus (BMV), Hepatitis C virus (HCV), Dengue virus, and Drosophila virus C (17–25). However, the detailed functions of the majority of the identified host proteins in (+)RNA virus replication have not been fully revealed.
TBSV is a plant-infecting (+)RNA virus used extensively to study virus replication, recombination, and virus-host interactions based on a yeast (Saccharomyces cerevisiae) model (26–29). We have performed several genome-wide screens of yeast genes by using different global proteomics approaches that have led to the identification of over 500 host genes/proteins putatively involved in TBSV replication or recombination (17, 19, 30–40). The above-mentioned systematic screens have identified host stimulatory and restriction factors of TBSV replication. For example, the Cyp40-like Cpr7p cyclophilin and the Ttc4 oncogene-like Cns1p cochaperone are strong inhibitors of TBSV replication in yeast and in vitro (41, 42). Additional cellular cyclophilins, such as the CypA, and the related Ess1p parvulin also decrease TBSV RNA accumulation in yeast and plants (36, 41, 43). Moreover, the cellular nucleolin, an RNA-binding protein, inhibits TBSV replication by blocking the recruitment of the viral RNA into replication (44). Another group of cellular restriction factors is the WW motif-containing host proteins, such as Rsp5p Nedd4-like E3 ubiquitin ligase, which regulate the degradation of tombusviral p92pol in yeast cells and inhibit the activity of VRC in vitro (45, 46). Cellular kinases, such as Pkc1p, could also restrict TBSV replication in yeast (32). Taken altogether, studies of cellular restriction factors could help to unravel the full arsenal of the native cell-intrinsic innate immune system in the host cell.
Similar to other (+)RNA viruses, tombusviruses, such as TBSV, use intracellular membranes for replication. Interestingly, TBSV utilizes the peroxisomal membrane, while the closely related Carnation Italian ringspot virus (CIRV) takes advantage of the outer mitochondrial membranes to build VRCs in infected plants and yeast (47–49). The two viral replication proteins (i.e., p33 and p92pol for TBSV and p36 and p95pol in the case of CIRV) is known to coopt 8 to 10 host proteins to assemble the tombusvirus VRC (37–39, 50–52). The highly homologous p33 of TBSV and p36 of CIRV replication proteins are master regulators of replication, playing a multifunctional role in recruitment of the tombusviral (+)RNA to the site of replication, the assembly of the VRC, and viral RNA synthesis by acting as RNA chaperones (50, 53–57). The RdRp protein p92pol of TBSV and p95pol of CIRV are also components of the functional VRCs (28, 55, 57–59). The subverted host proteins have been shown to bind to the viral RNA and the viral replication proteins (8, 39, 60). Detailed studies showed that heat shock protein 70 (Hsp70), eukaryotic elongation factor 1A (eEF1A), and several members of the endosomal sorting complexes required for transport (ESCRT) family of host proteins are required for the assembly of VRCs (52, 61–64). Additional subverted host proteins include the DDX3-like Ded1p/AtRH20 and the human p68-like Dbp2, the eIF4AIII-like Fal1/AtRH2 and DDX5-like Dbp3/AtRH5 DEAD-box RNA helicases, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), eEF1Bγ, and eEF1A, all of which have been shown to affect viral RNA synthesis (51, 60, 61, 65–68).
Previous works with TBSV revealed the unexpected inhibitory function for several tetratricopeptide repeat (TPR) domain-containing proteins, such as the Cyp40-like Cpr7p cyclophilin and Ttc4-like Cns1p cochaperone in yeast and in vitro (41, 42). Mechanistic studies showed that the inhibitory effect of Cpr7p was due to its interaction with the RNA-binding domain of the tombusviral p33 replication protein that leads to inhibition of p33/p92pol-based recruitment of the TBSV (+)RNA for replication and a decrease of the efficiency of the VRC assembly. Importantly, the key element in Cpr7p was not the cyclophilin domain, but its TPR domain consisting of three TPR modules in Cpr7p (41). Similarly, via its TPR domain, Cns1p bound to the tombusviral p33 and p92pol replication proteins and inhibited VRC assembly and reduced TBSV replication in yeast and in vitro based on a yeast cell-free extract (CFE) assay (42). However, in case of Cns1p, the interaction targeted the p33-p33/p92 interaction domain, suggesting that TPR-containing cellular proteins might restrict TBSV replication via different mechanisms.
The TPR domain consists of repeats of a 34-amino-acid sequence adopting a right-handed helical helix-loop-helix structure with an amphipathic channel; such channels are involved in many protein-protein interactions (69, 70). Although the TPR domains are highly variable, which likely affects substrate specificity, the canonical TPR domain contains a pattern of small and large hydrophobic amino acids. The TPR domain proteins are abundant in all kingdoms of life, including 246 proteins in Arabidopsis, 338 in mouse, 63 in Caenorhabditis elegans and 26 in yeast (Interpro database, http://www.ebi.ac.uk/interpro/entry/IPR013026/taxonomy) (71). TPR domain proteins function in protein trafficking, protein import to organelles, transposon silencing, apoptosis, and synaptic vesicle fusion (72, 73). Various TPR domain proteins are involved in numerous human diseases, such as cancer, amyloidosis, cystic fibrosis, prion protein propagation, and bacterial pathogenesis (74–79). Several TPR domain proteins have been shown to affect infections by viruses, such as Chikungunya virus, West Nile virus, Vesicular stomatitis virus, Herpes simplex virus, Poxvirus, and baculoviruses (80–85). TPR domain proteins are also important in interferon-induced antiviral responses, including the IFN-induced protein with tetratricopeptide repeats (IFIT) protein family (5, 85–88).
Our previous discoveries invited our attention to TPR-like sequences, including the well-studied stress-induced protein 1 (Sti1p in yeast, Hop protein in mammals and plants) cochaperone. Sti1p, which is a conserved highly abundant protein lacking chaperone activity on its own, is a cochaperone of Hsp70 and Hsp90 chaperones (89, 90). Sti1p contains three TPR domains, which are involved in binding to Hsp90s and Hsp70s. Sti1p plays a role in client protein transfer from the Hsp70 complex to the Hsp90 complex. Interestingly, Sti1p can simultaneously bind to Hsp70 and Hsp90, and by inhibiting the ATPase activity of Hsp90, Sti1p stabilizes the ternary Hsp70-Hsp90-client protein intermediate complex (91, 92).
In this paper, we show that the yeast Sti1p cochaperone has a strong inhibitory function during the mitochondrial CIRV replication but not in the peroxisomal tombusvirus replication. Detailed analysis of Sti1p revealed that it interacted with the RNA-binding domain of CIRV p36 replication protein and ultimately restricted VRC assembly in vitro and CIRV RNA accumulation in yeast and the orthologous Hop inhibited CIRV accumulation in plants. Thus, TPR-containing cellular cochaperone proteins emerge as new cell-intrinsic restriction factors of a mitochondrial (+)RNA virus.
MATERIALS AND METHODS
Yeast strains and expression plasmids.
Yeast strains BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and sti1Δ (single-gene deletion strain) were obtained from Open Biosystems (Huntsville, AL, USA). For tombusviral replication in yeast, pESC-HisCNVp33-DI72, pYES-CNVp92, pESC-C36/DI72, and pYES-C95 were described previously (57). To generate pESC-C36/DI1, CIRV DI-1 (93) (constructed by D. Barajas and P. D. Nagy, unpublished data) was PCR amplified using primer pair 4124 (CCGGAATTCAGAAATATCTCAGGATTTGACCGTCC)/1069(CCGGTCGAGCTCTACCAGGTAATATACCACAACGTGTGT), digested with EcoRI/SacI, and then inserted into EcoRI/SacI-digested pESC-HisCNVp33-DI72, generating pESC-HisCNVp33-DI1. Cucumber necrosis virus (CNV) p33 sequence was then removed by BamHI/XhoI digestion and replaced with BamHI/XhoI-digested CIRV p36 sequence, which was PCR amplified from pESC-C36/DI72 using primer pair 900 (CGACGGATCCGAGGGTTTGAAGGCTGAGTCTACCA)/3230 (CCGCTCGAGCTATTTGACACCGAGGGATT), generating pESC-C36/DI1.
To generate Twin-Strep-tagged CIRV and CNV replication proteins, the primer pairs 5351 (CATCCACAATTCGAAAAATCTGCTGGTGGAGGTGGATCCATGGATACCATCAAGAGGATG)/952 (CCCGCTCGAGTCATGCTACGGCGGAGTCAAGGA), 5350 (GTGGTTCTGGTGGTGGTTCTGGTGGTTCTGCTTGGTCTCATCCACAATTCGAAAAATCTG)/952, and 5349 (GGAAGATCTAAAAATGTCTGCTTGGTCTCATCCACAATTCGAAAAAGGTGGTGGTTCTGGTGGTGGTTCTGGTGG)/952 were sequentially used for PCR using template pYES-CNVp92 to introduce the Twin-Strep tag on CNV p92, generating a Twin-Strep-tagged CNV p92 sequence. The DNA was then digested with BglII/XhoI and inserted into pESC-DI72, generating pESC-StrepCNVp92/DI72. CIRV p36 sequence was PCR amplified using primer pair 900/3230 and digested with BamHI/XhoI and then inserted into BamHI/XhoI-digested pESC-StrepCNVp92/DI72, generating pESC-StrepC36/DI72. To generate pYES-StrepC95, pYES2-NTA (Invitrogen) was digested with HindIII/KpnI and then treated with T4 DNA polymerase and subsequently self-ligated to remove the His6 tag. The modified pYES2-NTA vector without the His6 tag was digested with BamHI/XhoI and used with the BglII/XhoI-digested PCR product of Twin-Strep-tagged CNVp92 for ligation, generating pYES-StrepC92. CIRV p95 sequence was PCR amplified using primer pair 900/970 (CCCGCTCGAGTCAAGCTACGGCGGAGTCGAGGA) from pYES-C95, digested with BamHI/XhoI, and then inserted into BamHI/XhoI-digested pYES-StrepC92, generating pYES-StrepC95.
To generate yeast vector expressing the STI1 gene, pTEF1 promoter and tCYC1 terminator were PCR amplified from yeast genomic DNA and pESC-C36/DI72 (57), respectively, using primer pair 2764 (CCGCGAGCTCATAGCTTCAAAATGTTTCTAC)/3726 (CCGCGCGGCCGCGTAATTAAAACTTAGATTAGATTGC) or 3728 (CCGCGTCGACGAGGGCCGCATCATGTAA)/3730 (CCGCGGGCCCAGCTTGCAAATTAAAGCCTTC) and digested with SacI/NotI or ApaI/SalI. Digested pTEF1 and tCYC1 were sequentially inserted into SacI/NotI or ApaI/SalI-digested pRS315 vector, generating pRS315-pTEF1 (Zhenghe Li and P. D. Nagy, unpublished data). pRS315-pTEF1 was digested with NotI/SalI and ligated with annealed primer pair 5157 (GGCCGAAAATGAGATCTGGCACTAGTGACTACAAGGACGACGATGACAAGGGTGGCGGTC)/5158 (CTAGGACCGCCACCCTTGTCATCGTCGTCCTTGTAGTCACTAGTGCCAGATCTCATTTTC) to introduce a Flag tag, generating pRS315-NFlag.
Plasmids expressing wild-type (wt) Sti1p or derivatives (C49Y, G325D, ΔTPR2, or ΔTPR1) were generated by using available STI1 mutants (94, 95) in PCR amplifications using primer pairs 2863 (CGCGGGATCCATGTCATTGACAGCCGATG)/2864 (CGCGCTCGAGTTAGCGGCCAGTCCGGATG) or 5156 (CGCGGGATCCAACCCAAAAACTAGCGAAATGATG)/2864, and the obtained PCR products were treated with BamHI/SalI and then were inserted into BamHI/SalI-digested pRS315-NFlag. To visualize Sti1p in yeast, mRFP1 was PCR amplified from pGAD-PEX13-RFP (64) using primer pair 2630 (CGCGGGATCCATGGCCTCCTCCGAGGACGTC)/5159 (GGACTAGTGGCGCC-GGTGGAGTGG), and the PCR product was digested with BamHI/PstI and inserted into BglII/PstI-digested pRS315-Sti1 to generate pRS315-mRFP1-Sti1.
Escherichia coli-based expression plasmids, pGEX-2T-Sti1 and mutated derivatives (C49Y, G325D, ΔTPR1, ΔTPR2) were generated by PCR using primers 2863 (CGCGGGATCCATGTCATTGACAGCCGATG) and 4860 (CGCCGAATTCTTAGCGGCCAGTCCGGATGAT), followed by digestion with BamHI/EcoRI and then ligation into BamHI/EcoRI-digested pGEX-2T.
Arabidopsis thaliana STI1 ortholog AtHOP-1 (96) (AT4G12400, renamed AtHOP3 in reference 131) and the TPR1 deletion version (AtHOP1ΔTPR1) were PCR-amplified from Arabidopsis cDNA using primer pairs 5659 (CGCTGATCAATGGCGGAAGAAGCAAAATCCAAAGG)/5661 (CCGCTCGAGTTACCGGACCTGAACAATTCCGGCACTAACC) and 5660 (CGCTGATCAATGGATCCGGGGACTAGGGTTTATTTGGAG)/5661, respectively, followed by digestion with BclI/XhoI, and the PCR products were inserted into BamHI/SalI-digested pRS315-NFlag, generating pRS315-AtHOP1 and pRS315-AtHOP1ΔTPR1, respectively.
Analysis of CIRV repRNA replication in yeast.
For measuring CIRV replicon RNA (repRNA) accumulation, yeast strains BY4741 and sti1Δ were transformed with plasmids pESC-C36/DI72 and pYES-C95. For complementation and overexpression studies, we transformed yeast strains BY4741 and sti1Δ with pRS315-Sti1 (FLAG-Sti1 plasmid). Tombusvirus repRNA replication was induced by culturing in synthetic complete medium lacking Ura, Leu, and His (sc-ULH−) with 2% galactose medium after overnight culture and then yeast was cultivated for 2 days at 23°C. Total RNA was isolated from yeast and used for detection of repRNA levels by Northern blotting as described previously (97). Replication was calculated by measuring the accumulation of CIRV DI1 repRNA or the TBSV DI-72(+) repRNA relative to the accumulation of 18S rRNA. The tombusvirus replication protein analysis was performed as described previously using an anti-His6 antibody as the primary antibody for the detection of His6-p36 and His6-p95. Detection of Flag-Sti1p and Sti1p was carried out using primary anti-Flag and anti-Sti1 antibody, respectively. The secondary antibody for both primary antibodies was alkaline phosphatase-conjugated anti-mouse immunoglobulin G (Sigma) (41).
Analysis of protein-protein interaction by split-ubiquitin assay.
The bait constructs, pGAD-BT2-N-His36 and pGAD-BT2-N-His33, expressing CIRV replication protein p36 and p33 tombusvirus replication protein, have been published before (36, 38). The PCR products of the STI1 gene and its various truncation versions were digested with BamHI/XhoI and ligated into the pPRN-N-RE vector digested with BamHI/SalI enzymes. The PCR products of AtHop-1 (96) was digested with BclI/XhoI and ligated into the pPRN-N-RE vector digested with BclI/XhoI enzymes. Yeast strain sti1Δ/NMY51 was cotransformed with pGAD-BT2-N-His36 or pGAD-BT2-N-His33 and pPR-N-RE (NubG) or one of the prey constructs carrying the STI1 gene and plated onto Trp−/Leu− (TL−) synthetic minimal medium plates for plasmid selection (36, 38). Yeast colonies were resuspended in 50 μl water and spotted onto Trp−/Leu−/His−/Ade− (TLHA−) plates for 2 to 4 days to detect bait-prey interactions. Plasmid containing the yeast SSA1 Hsp70 gene served as the positive control and an empty vector (pPR-N-RE) as the negative control in this assay (36, 38).
Protein purification from E. coli.
pMAL-p33 (TBSV p33), pMAL-p92 (TBSV p92), pMAL-p36 (CIRV p36), and pMAL-p95 (CIRV p95) (40) were transformed separately into E. coli strain BL21(DE3)CodonPlus. Protein expression was induced using isopropyl β-d-thiogalactopyranoside (IPTG) for 8 h at 16°C, and the cells were harvested by centrifugation at 5,000 rpm at 4°C for 5 min to remove the medium prior to −80°C storage. Affinity columns containing amylose resin (NEB) were used to purify maltose-binding protein (MBP)-tagged recombinant proteins. The frozen pellets were suspended and sonicated in MBP column buffer containing 20 mM Tris-Cl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride (PMSF). The sonicated extract was centrifuged at 15,000 rpm for 5 min, and the supernatant was added to the preequilibrated amylose resin for a 1-h rotating incubation at 4°C. After the resin was washed three times with column buffer and once with a low-salt column buffer (25 mM NaCl), the proteins were eluted with a low-salt column buffer containing 0.18% (vol/wt) maltose and stored at −80°C in 6% (vol/vol) glycerol. Protein fractions used for the replication assays were 95% pure, as determined by 12% SDS-PAGE and staining with Coomassie blue.
Expression of glutathione S-transferase (GST)-tagged proteins Cpr7p, Cpr7-TPRp, Cns1p, Sti1p and their mutated versions (C49Y, G325D, ΔTPR1, and ΔTPR2) were induced using IPTG for 6 h at 23°C, and the cells were harvested by centrifugation at 5,000 rpm at 4°C for 5 min to remove the medium and stored at −80°C. Purification of GST-tagged proteins was carried out using glutathione resin and eluted with 10 mM glutathione–10 mM β-mercaptoethanol in the column buffer following the same protocol as that for MBPs.
In vitro tombusvirus replication assay using yeast mitochondrial preparations.
Yeast intact mitochondria were purified as described previously (40). The purified mitochondrial fraction (1 μl) and different dilutions of GST, Sti1p, Cpr7p, or Cns1p proteins (8, 16, and 32 μM each) were incubated at 25°C for 1 h in 8 μl buffer A (containing 30 mM HEPES-KOH [pH 7.4], 150 mM potassium acetate, 5 mM magnesium acetate, and 0.6 M sorbitol) with 15 mM creatine phosphate, 1 mM ATP, and GTP, 0.1 mg/ml creatine kinase, 0.1 μl of RNase inhibitor, 10 mM dithiothreitol, 0.5 μg DI-72 RNA transcript, and affinity-purified 0.5 μg MBP-36 (CIRV p36) and MBP-p95 (CIRV p95). The volume of the reaction mixture was then adjusted by adding 16 μl buffer B (containing 30 mM HEPES-KOH [pH 7.4], 150 mM potassium acetate, and 5 mM magnesium acetate) with 15 mM creatine phosphate, 1 mM ATP, CTP, and GTP, 0.025 mM UTP, 0.2 μl of [32P]UTP, 0.1 mg/ml creatine kinase, 0.2 μl of RNase inhibitor, 10 mM dithiothreitol, and 0.05 mg/ml actinomycin D. The reaction mixture was incubated at 25°C for 3 h and terminated by adding 100 μl stop buffer (1% sodium dodecyl sulfate and 0.05 M EDTA [pH 8.0]) followed by 100 μl phenol-chloroform extraction and isopropanol-ammonium acetate precipitation overnight at −20°C and washing by 70% ethanol. The newly synthesized 32P-labeled RNA products were incubated at 85°C for 5 min and separated by electrophoresis in a 5% polyacrylamide gel containing 0.5× Tris-borate-EDTA buffer with 8 M urea. Signals were detected using a Typhoon 9400 imaging scanner (GE/Amersham) and quantified by ImageQuant software.
Copurification of host proteins with Twin-Strep-tagged CIRV replication proteins from yeast.
To purify the protein of interest, 200 mg of BY4741 yeast cells were transformed with plasmids pESC-StrepC36/DI72 pYES-StrepC95 and pRS315-Sti. Cultured yeasts were resuspended and homogenized in buffer B (50 mM Tris-HCl [pH 7.5], 15 mM MgCl2, 10 mM KCl, 10 mM β-mercaptoethanol, 1% [vol/vol] yeast protease inhibitor cocktail) by glass beads (modified from reference 28). Membrane fractions from cell homogenates were collected and solubilized with column buffer (50 mM Tris-HCl [pH 7.5], 15 mM MgCl2, 500 mM KCl, 1% Triton X-100, 5% caprylyl sulfobetaine [SB3-10; Sigma], 10 mM β-mercaptoethanol, 1% [vol/vol] yeast protease inhibitor cocktail), and incubated with 40 μl Strep-Tactin Superflow high-capacity 50% resin (IBA Life Sciences) for 1 h at 4°C in a column. Strep-Tactin resin was then washed two times with column buffer, two times with wash buffer (50m M Tris-HCl [pH 7.5], 15 mM MgCl2, 10 mM KCl, 0.1% Triton X-100, 10 mM β-mercaptoethanol, 1% [vol/vol] yeast protease inhibitor cocktail), and eluted with SDS-PAGE loading buffer and then subjected to SDS-PAGE and Western blotting with Strep-Tactin AP conjugate (IBA Life Sciences) and anti-Flag and anti-Hsp70 antibodies (Abcam).
Confocal laser microscopy.
Wild-type (wt) BY4741 or sti1Δ yeast strains were transformed with the following expression plasmids: pESC-GFP-C36/DI72, pYES-C95, or pESC-GFP-C33/DI72, pYES-C92 (57), as well as pRS315-RFP-Sti1p. The yeast cultures were incubated in galactose medium overnight, sampled, and imaged with an Olympus FV1000 confocal laser scanning microscope (Olympus America Inc., Melville, NY). The microscope settings were the following: excitation and emission for green fluorescent protein (GFP) and red fluorescent protein (RFP) were 488-nm laser/500- to 530-nm filter and 543-nm laser/560- to 660-nm filter, respectively.
Virus induced gene silencing of STI1/HOP ortholog gene.
Virus-induced gene silencing (VIGS) in Nicotiana benthamiana was performed as described in reference 98. The C-terminal fragment of the N. benthamiana HOP gene (yeast STI1 ortholog, based on the Arabidopsis AtHOP-1 gene) was PCR amplified from total N. benthamiana cDNA using primer pair 5786 (CGCGGATCCAGGGCATACAGCAACAGGGC)/5787 (CCGCTCGAGTTATTTGACTTGAATAATTCCTGCACTAACCAAC). The obtained PCR product was digested with BamHI/XhoI and inserted into pTRV2 digested with BamHI/XhoI, generating pTRV2-NbHop. As a control for the VIGS experiments, the C-terminal half of the GFP sequence was PCR amplified by primer pair 5353 (CGCGGATCCGAAGGTGATACCCTTGTTAATAGAATCGAG)/3712 (CGGCCTCGAGTTACGCATAGTCAGGAACATCGTATGGGTAGAGTCCGGACTTGTATAGTT) from pESC-GFP-C36/DI72, digested with BamHI/XhoI, and inserted into pTRV2, generating pTRV2–1/2GFP. VIGS-treated N. benthamiana plants were sap inoculated with CIRV or CNV inocula on the 14th day postsilencing. Samples from the inoculated leaves were harvested and subjected to total RNA extraction and Northern blot analysis for viral RNA (64, 98). Efficiency of NbHOP silencing was evaluated by semiquantitative reverse transcription-PCR using NbHOP and Actin gene-specific primer pairs: 5785 (CGCGGATCCAGAGCAGCAAGAGTATTTCGATCCAC)/5787 and 3993 (GGAAGTAGCATAAGATGGCAGATGGAGAGG)/3994 (CCAGATCTTCTCCATATCATCCCAGTTGCTGAC), respectively.
RESULTS
Yeast-based studies reveal that Sti1p cochaperone selectively inhibits mitochondrial CIRV replication but not the peroxisomal TBSV replication.
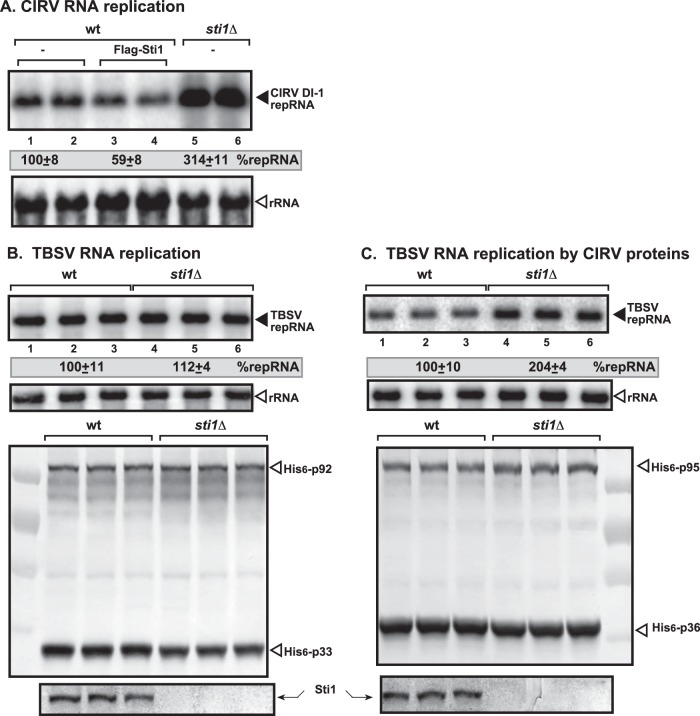
Based on our previous findings that two abundant cytosolic TPR-containing cellular proteins, namely, Cpr7p cyclophilin and Cns1p cochaperone, showed robust restriction activity against TBSV (41, 42), we also tested the abundant TPR-containing protein Sti1p cochaperone for possible effects on the accumulation of TBSV and CIRV replicon RNAs (repRNAs) in sti1Δ yeast versus wt yeast cells. Interestingly, sti1Δ yeast supported CIRV repRNA accumulation at an ∼3-fold higher level than that for wt yeast (Fig. 1A, lanes 5 and 6 versus lanes 1 and 2). However, replication of the TBSV repRNA was comparable in sti1Δ and wt yeast (Fig. 1B, lanes 4 to 6 versus lanes 1 to 3), suggesting that Sti1p has a CIRV-specific inhibitory effect. To test if Sti1p-based inhibition targets the CIRV RNA, we also tested TBSV repRNA accumulation in the presence of CIRV p36 and p95pol replication proteins, which are capable of supporting the replication of the heterologous TBSV repRNA (57), in sti1Δ yeast versus wt yeast cells. The obtained data showed an ∼2-fold increased level of TBSV repRNA accumulation (Fig. 1C, lanes 4 to 6 versus lanes 1 to 3), demonstrating that the inhibitory effect of Sti1p is targeted against CIRV p36 and p95pol and not the viral RNA. Comparison of the accumulation of CIRV p36 and p95pol in sti1Δ yeast versus wt yeast cells revealed similar replication protein levels (Fig. 1C), arguing that Sti1p is unlikely to affect translation or stability of CIRV p36 and p95pol in yeast cells.
FIG 1.
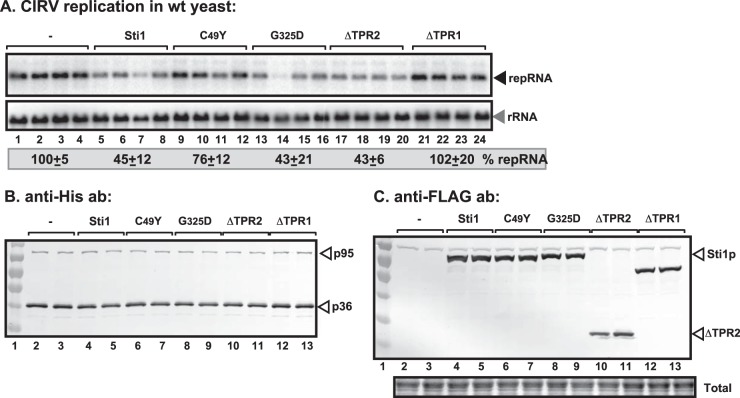
Increased CIRV replication in sti1Δ yeast. (A) Northern blot analysis of accumulation of CIRV DI-1 repRNA in sti1Δ or wt yeast strains at 23°C. We launched CIRV repRNA replication by expressing CIRV His6-p36 and His6-p95 from the galactose-inducible GAL1 promoter and DI-1 (+)repRNA from the galactose-inducible GAL10 promoter in sti1Δ and the parental (wt; BY4741) yeast strains. We also overproduced the FLAG-tagged Sti1p in wt yeast to test its inhibitory function. Note that the data were normalized based on 18S rRNA. Each experiment was repeated three times. (B) Northern blot analysis of accumulation of TBSV DI-72 repRNA in sti1Δ or wt yeast strains. TBSV repRNA replication was launched by expressing CNV His6-p33 and CNV His6-p92 from the GAL1 promoter and DI-72 (+)repRNA from the GAL10 promoter in sti1Δ and the parental (wt; BY4741) yeast strains. See further details in panel A. Bottom images, Western blot analysis of CNV His6-p33, CNV His6-p92 accumulation by anti-His antibody, and Sti1p accumulation by anti-Sti1 antibody. (C) Top images, Northern blot analysis of the CIRV p36/p95-driven TBSV DI-72 RNA accumulation in sti1Δ or wt yeast strains. Same as panel A except DI-72 was used as a repRNA with CIRV His6-p36 and His6-p95, which support viral RNA replication on mitochondrial membrane surfaces. Bottom images, Western blot analysis of CIRV His6-p36, CIRV His6-p95 accumulation by anti-His antibody, and Sti1p accumulation by anti-Sti1 antibody. Each experiment was repeated three times.
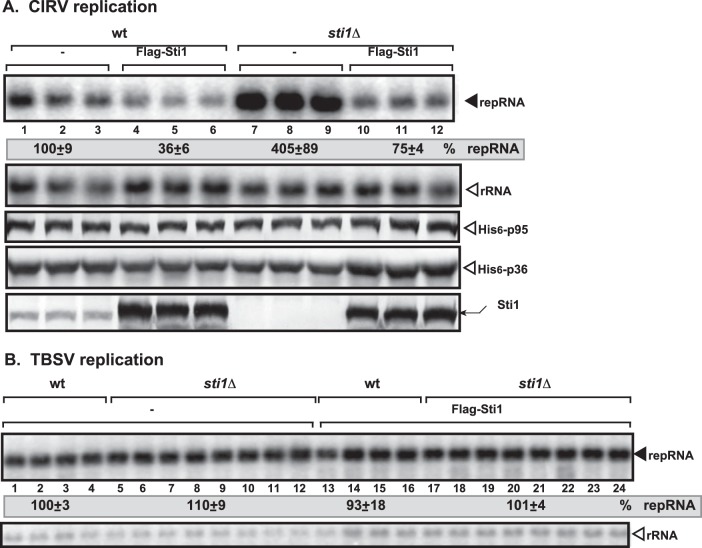
To further test if Sti1p can inhibit CIRV replication in vivo, we overexpressed N-terminally FLAG-tagged Sti1p in yeast supporting CIRV or TBSV accumulation. We found that overexpression of Sti1p reduced CIRV accumulation up to ∼3-fold in wt yeast (Fig. 2A, lanes 4 to 6 versus lanes 1 to 3) and ∼5-fold in sti1Δ yeast (Fig. 2A, lanes 10 to 12 versus lanes 7 to 9). The level of CIRV p36 and p95pol replication proteins was comparable in sti1Δ and wt yeasts overexpressing FLAG-Sti1p (Fig. 2A), suggesting that Sti1p is unlikely to affect the stability of these viral proteins in yeast. In contrast, replication of the TBSV repRNA was not affected by the overexpression of Sti1p in wt (Fig. 2B, lanes 13 to 16) or sti1Δ (lanes 17 to 24) yeasts. Altogether, these data support the idea that Sti1p is a strong inhibitor of the CIRV p36 and p95pol replication proteins, while Sti1p seems to be ineffective against the tombusviral p33 and p92pol replication proteins in yeast.
FIG 2.
Overexpression of Sti1p inhibits CIRV accumulation in yeast. (A) Top panel, Northern blot analysis of CIRV RNA accumulation in wt or sti1Δ yeasts overproducing the FLAG-tagged Sti1p. Second panel, Northern blot analysis to demonstrate the comparable level of rRNA loading in the yeast samples. Bottom panels, Western blot analysis of CIRV His6-p95 and CIRV His6-p36 accumulation by anti-His antibody and Sti1p accumulation by anti-Sti1 antibody. (B) Northern blot analysis of TBSV DI-72 repRNA accumulation in wt or sti1Δ yeasts overproducing the FLAG-tagged Sti1p in the presence of peroxisomal CNV p33/p92 replication proteins. Each experiment was repeated three times. See further details in legend to panel A.
The binding of Sti1p involves different regions in CIRV p36 and the TBSV p33 replication proteins.
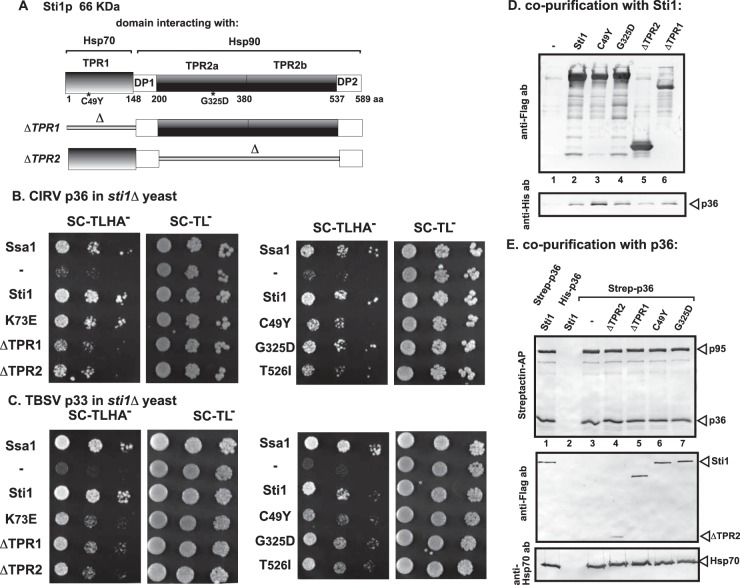
Sti1p contains three TPR domains (Fig. 3A) that are predicted to interact with the tombusviral replication proteins. To test if Sti1p can interact with the CIRV p36 versus the TBSV p33 replication proteins, we first used the split-ubiquitin-based two-hybrid assay with sti1Δ yeast (99, 100). We observed a strong interaction between Sti1p and p36 (Fig. 3B) and Sti1p and p33 (Fig. 3C). We confirmed the interaction between Sti1p and p36 replication protein in sti1Δ yeast (Fig. 3D, lane 2) using a copurification assay with recombinant Sti1p. The reciprocal copurification assay with Strep-tagged p36 also resulted in copurification of Flag-Sti1p from sti1Δ yeast (Fig. 3E, lane 1).
FIG 3.
Interaction between Sti1p and CIRV p36 replication protein in yeast and in vitro. (A) Domain structure of the yeast Sti1p. Tetratricopeptide repeat 1 (TPR1) sequence interacts with Hsp70, while dipeptide repeat of aspartic acid and proline 1 (DP1) might stabilize the bound client protein. TPR2A and TPR2B bind to Hsp90 and together inhibit the ATPase activity of Hsp90. TPR2B also binds to Hsp70, but only in concert with Hsp90 binding to TPR2A. The debilitating mutations are marked with an asterisk, and deletion constructs are shown schematically at the bottom of the panel. (B) A split-ubiquitin MYTH assay was used to test intracellular interaction between CIRV p36 and the wt or mutated yeast Sti1p. The bait p36 was coexpressed with the prey Sti1p protein in sti1Δ yeast. The SSA1 gene (Hsp70 chaperone) and the empty prey vector (NubG) were used as positive and negative controls, respectively. (C) Same split-ubiquitin MYTH assay as that shown in panel B, except that TBSV p33 was used as a bait protein. (D) Copurification of CIRV p36 replication protein with the yeast Sti1p from yeast cells. The membrane fraction of yeast coexpressing the wt or mutated FLAG-Sti1p and His6-p36 was solubilized, and the Sti1p variants were purified using a FLAG column. The eluted proteins were tested using Western blotting with anti-FLAG antibody (top image) and anti-His6 antibody (bottom image). (E) Reciprocal copurification of the yeast Sti1p with CIRV p36 and p95 replication proteins from yeast cells. Details are as described for panel D, except yeast coexpressed the Twin-Strep-tagged CIRV p36 and p95 and Flag-Sti1p. The purification was based on Strep-Tactin columns. The eluted proteins were tested using Western blotting with anti-Strep-Tactin-AP conjugate (top image), anti-Flag antibody (middle image), and anti-Hsp70 antibody (bottom image). Note that the coopted Hsp70 is a permanent member of the tombusvirus replicase complex. Each experiment was repeated three times.
To test what region(s) of Sti1p interacts with p36, we used well-characterized Sti1p mutants lacking particular functional domains (94, 95) as shown in Fig. 3A. The split-ubiquitin assay showed that the interaction with p36 was not eliminated by deletion (ΔTPR1) or mutation (C49Y and K73E) in the TPR1 region (Fig. 3B), which binds to Hsp70 (89, 101). Similarly, deletion (ΔTPR2) or mutation (G325D or T526I) in the TPR2 region (Fig. 3B), which binds to Hsp90, did not debilitate interaction with p36 replication protein. These findings were confirmed in the reciprocal copurification experiments (Fig. 3D and E), demonstrating that Sti1p could use both the TPR1 and TPR2 sequences to bind to the p36 replication protein. Interestingly, binding of Sti1p to the TBSV p33 replication protein showed features similar to those of p36 binding (Fig. 3C versus Fig. 3B). Thus, the binding characteristics of Sti1p to CIRV p36 versus TBSV p33 do not explain why Stip1 can selectively inhibit CIRV replication, but not TBSV replication, in yeast cells.
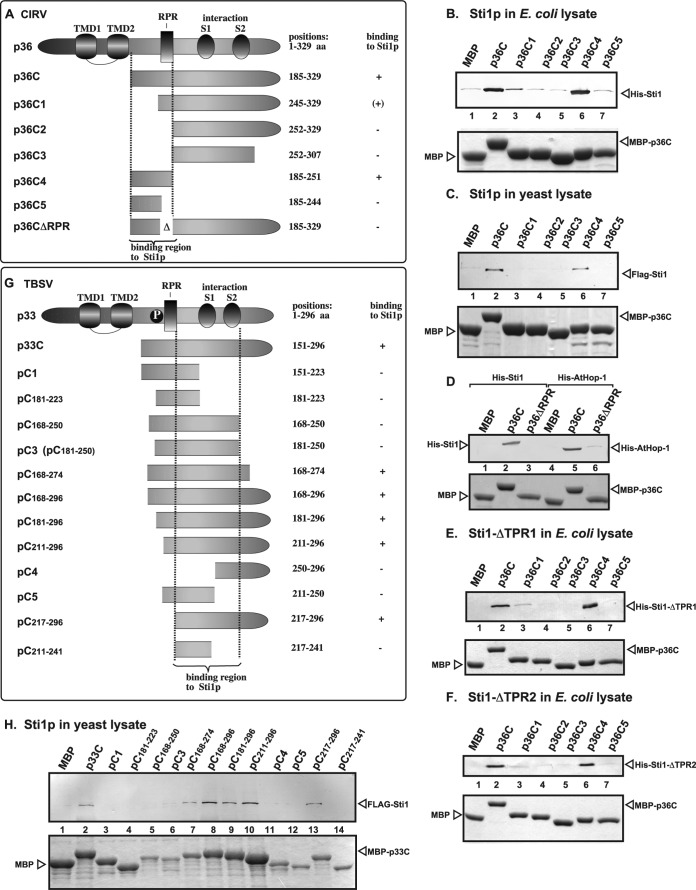
To map the Sti1p-binding site in the CIRV p36 replication protein, we have used pulldown experiments with immobilized MBP-p36 truncation derivatives (Fig. 4A) and Sti1p present in either E. coli lysate (Fig. 4B) or yeast extract containing Flag-Sti1p (Fig. 4C). These experiments revealed that Sti1p binds to a region that includes the arginine- and proline-rich (RPR) motif of p36, which is involved in RNA binding (Fig. 4B and C, lanes 6; Fig. 4A, construct p36C4; and Fig. 4D, construct p36ΔRPR). The RPR motif in replication proteins is required for specific viral (+)RNA recruitment and replicase assembly, and it also binds to Cpr7p Cyp40-like cyclophilin (41, 102–104). Interestingly, the same RPR-containing region in p36 replication protein could bind to both the TPR1 and TPR2 sequences in Sti1p (Fig. 4E and F, lanes 6).
FIG 4.
Defining the sequence within the tombusvirus replication proteins needed for binding to Sti1p in vitro. (A) Schematic representation of the CIRV p36 and its truncated derivatives used in the binding assay. The various domains include the transmembrane domain (TMD), the arginine- and proline-rich (RPR) RNA-binding domain, the phosphorylated serine and threonine (P) domain, and the S1 and S2 subdomains involved in p36-p36/p95 interaction. (B to F) Affinity binding (pulldown) assay to detect interaction between Flag- or His6-Sti1p, ΔTPR1, ΔTPR2, AtHop1, and the MBP-tagged CIRV p36 protein derivatives. The MBP-tagged viral proteins produced in E. coli were immobilized on amylose affinity columns. The recombinant Sti1p and derivatives expressed in E. coli (panels B, D, E, and F) or in yeast (panel C) were then passed through the amylose affinity columns with immobilized MBP-tagged p36 protein (its truncated versions). The affinity-bound proteins were eluted with maltose from the columns (shown in the bottom image). Top images in each panel, eluted proteins were analyzed by Western blotting with anti-Flag or anti-His antibodies to detect the amount of Flag- or His6-Sti1p specifically bound to MBP-tagged viral proteins. Bottom images, SDS-PAGE analysis of the viral protein and its truncated derivatives after elution from the amylose affinity columns. Note that the MBP has a C-terminal extra tail sequence (not present in the fusion protein constructs) due to the sequence in the original cloning vector. (G) Schematic representation of the TBSV p33 and its truncated derivatives used in the binding assay. (H) Affinity binding (pulldown) assay to detect interaction between FLAG-Sti1p and the MBP-tagged viral p33 protein (the soluble C-terminal portion). The MBP-tagged viral protein or MBP control produced in E. coli was immobilized on amylose affinity columns. See further details in legend to panel B. The eluted proteins were analyzed by Western blotting with anti-FLAG antibody to detect the amount of FLAG-Sti1p specifically bound to MBP-tagged viral protein.
To map the Sti1p-binding site in the TBSV p33 replication protein, we have used similar pulldown experiments with immobilized MBP-p33 truncation derivatives (Fig. 4G) in combination with yeast extract containing Flag-Sti1p or its truncation derivatives (Fig. 4H). We found that Sti1p binds to the C-terminal region of TBSV p33 containing the p33-p33/p92 interaction sequences (Fig. 4G). Deletion of the RPR motif, which is involved in RNA binding, did not inhibit p33 binding to Sti1p (Fig. 4H, lane 13). Thus, there is a major difference in Sti1p binding to CIRV (the RPR-containing sequence) and TBSV replication proteins (the C-terminal region in TBSV p33), suggesting that the mechanism of inhibition of CIRV replication by Sti1p could be based on blocking the RNA-binding function of CIRV p36 replication protein.
Sti1p is colocalized with CIRV p36 in yeast cells.
To study if the mostly cytosolic Sti1p is recruited to the mitochondrial membranes, where CIRV replication takes place (49, 57), by the CIRV p36, we coexpressed GFP-p36 with RFP-Sti1p in wt or sti1Δ yeast cells. Confocal laser microscopy revealed the robust recruitment of RFP-Sti1p by CIRV p36 to punctate structures, representing the mitochondrial membranes as shown before (48, 49, 57) in both wt and sti1Δ yeast cells (Fig. 5A). In contrast, the GFP-tagged p33 did not efficiently recruit RFP-Sti1p to p33-containing punctate structures (Fig. 5B), which represent peroxisomal membranes (55, 105). RFP-Sti1p showed a diffused, mostly cytosolic distribution in yeast expressing p33 replication protein or in the absence of viral proteins (Fig. 5C). Based on these data, we suggest that unlike p33, the CIRV p36 replication protein efficiently recruits Sti1p to the site of replication, leading to robust inhibition of CIRV replication.
FIG 5.

Relocalization of yeast Sti1p cochaperone when coexpressed with CIRV p36 replication protein in yeast. (A) Confocal laser microscopy images show the partial colocalization of RFP-Sti1 with CIRV GFP-p36 in wt (BY4741; top two panels), or in sti1Δ (bottom panels) yeast strains. The merged images show the colocalization within punctate structures, representing mitochondria, which are the sites of CIRV replication. Differential interference contrast (DIC) images are shown on the right. (B) Absence of colocalization of RFP-Sti1 and GFP-p33 in yeast. Note the cytosolic distribution of RFP-Sti1, while GFP-p33 is present in punctate structures representing the peroxisomes. (C) Mostly cytosolic distribution of RFP-Sti1p in the absence of viral proteins in BY4741 yeast. Each experiment was repeated.
The TPR1 domain in Sti1p is required to inhibit CIRV replication in isolated mitochondrion-based assay and in yeast cells.
To test what domain of Sti1p is required to block CIRV replication, we expressed mutated versions of Sti1p in wt yeast. We observed a ∼2-fold inhibition of CIRV repRNA accumulation by ΔTPR2 and G325D mutants, comparable to that obtained with the full-length Sti1p (Fig. 6A), while expression of ΔTPR1 or C49Y mutants had no detectable and lesser inhibitory effects, respectively (lanes 9 to 12 and 21 to 24 in Fig. 6A). All these mutated versions of Sti1p were expressed at comparable levels in wt yeast without substantially affecting CIRV p36 or p95 levels (Fig. 6B and C). Altogether, based on these data, we suggest that the TPR1 domain of Sti1p is required to inhibit CIRV replication in yeast.
FIG 6.
Functional TPR1 domain of Sti1p is required for inhibition of CIRV replication in yeast. (A) Northern blot analysis of CIRV accumulation in wt yeast overproducing the FLAG-tagged Sti1p or derivatives. Bottom panel, Northern blot analysis to demonstrate the comparable level of rRNA loading in the yeast samples. (B) Western blot analysis of CIRV His6-p36, and CIRV His6-p95 accumulation by anti-His antibody from yeast overproducing Sti1p or derivatives. (C) Detection of the overproduced FLAG-Sti1p or its derivatives in yeast by Western blotting using anti-FLAG antibody. Each experiment was repeated three times.
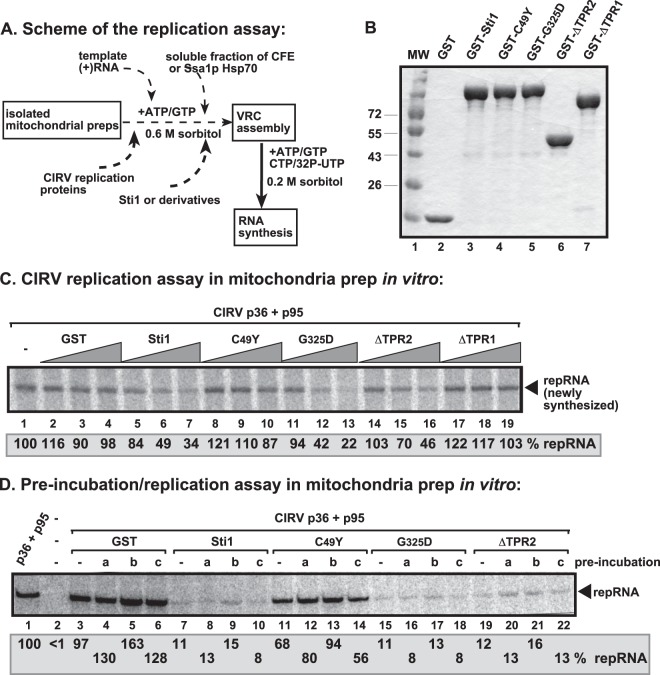
To further test the roles of the TPR sequences of Sti1p in CIRV replication, we applied an isolated mitochondrion-based replication assay, which takes advantage of purified recombinant CIRV p36, p95pol replication proteins, and repRNA transcripts to support full CIRV replication in vitro (Fig. 7A) (57, 58, 104). Addition of the affinity-purified full-length recombinant GST-Sti1p (Fig. 7B) decreased the production of repRNA up to 3-fold (Fig. 7C, lanes 5 to 7 versus lanes 2 to 4), confirming that Sti1p has an inhibitory effect on CIRV replication in vitro. Preincubation of Sti1p either with Ssa1p Hsp70 chaperone or p36 replication protein to facilitate protein complex formation did not alter the inhibitory effect of Sti1p (Fig. 7D), suggesting that Sti1p has a robust effect on CIRV replication in vitro. The presence of recombinant Sti1p lacking a functional TPR2 domain (e.g., ΔTPR2 or mutant G325D) was also inhibitory, reducing CIRV replication up to ∼5-fold in the isolated mitochondrion-based assay (Fig. 7C, lanes 11 to 16, and 7D, lanes 15 to 22). In contrast, inactivation of TPR1 (ΔTPR1 or mutant C49Y) resulted in the loss of the inhibitory function of Sti1p (Fig. 7C, lanes 8 to 10 and 17 to 19, and 7D, lanes 11 to 14) in the isolated mitochondrion-based replication assay, thus emphasizing the critical role of the TPR1 sequence in Sti1p.
FIG 7.
Functional TPR1 domain of Sti1p blocks CIRV replication in vitro. (A) Scheme of in vitro reconstitution of the CIRV replicase in yeast mitochondrial preparation. The purified GST-tagged Sti1p or derivatives, the purified recombinant CIRV MBP-p36 and MBP-p95 proteins and the TBSV-derived (+)repRNA were used in isolated mitochondrial preparations. (B) Coomassie blue-stained SDS-PAGE was used for analysis of affinity-purified GST-tagged Sti1 or derivatives. MW, molecular weight (noted on y axis; in thousands). (C) Denaturing PAGE analysis of the 32P-labeled repRNA products obtained in the replication assays with the isolated yeast mitochondrial preparation and the soluble fraction of yeast CFE that provided soluble host factors. The arrowhead indicates synthesized full-length repRNA. The replication assay with CIRV p36 and p95 (without added Sti1) was chosen as 100% (lane 1). The recombinant proteins were added in 8, 16, and 32 μM amounts. Each experiment was repeated. (D) CIRV mitochondrial replication assays were performed (see panel A) to test the effect of preincubation of various components. Lanes “a” show samples for which the purified Ssa1p Hsp70 (from the sti1Δ yeast strain) was preincubated for 10 min with a comparable amount of GST, GST-Sti1p, or mutants (from E. coli), while in lanes “b,” the MBP-p36/MBP-p95 of CIRV (from E. coli) was preincubated with Ssa1p. In lanes “c,” the MBP-p36/MBP-p95 of CIRV was preincubated with GST, GST-Sti1p, or mutants, while in lanes “-,” no preincubation was performed. In each experiment, we used comparable amounts of each component for preincubation that lasted for 10 min in the reaction buffer. After the preincubation step, we added the missing components and performed the CIRV replication assay (see panel A). Each experiment was repeated at least twice.
Comparison of the inhibitory effects of host proteins carrying TPR domains on CIRV replication in isolated mitochondrion-based replication assay.
Two other cellular proteins with TPR domains, namely, Cpr7p cyclophilin and Cns1p cochaperone, have been shown to inhibit TBSV replication (41, 42). To test if these host proteins have activities comparable to those of Sti1p in the inhibition of CIRV replication, we used the isolated mitochondrion-based in vitro replication assay and purified recombinant cellular proteins and CIRV replication proteins (Fig. 8A and B). Interestingly, all these TPR-containing proteins inhibited CIRV replication in vitro with Sti1p and the TPR region of Cpr7p, showing up to an ∼10-fold reduction in repRNA production in the isolated mitochondrion-based replication assay (Fig. 8A, lanes 6 to 8 and 12 to 14 versus lanes 3 to 5). Cns1p was the least effective in this assay (Fig. 8A, lanes 15 to 17), but this could be due to the smaller amount of recombinant GST-Cns1p obtained from E. coli (Fig. 8B). However, the purified recombinant GST-Cns1p was the most effective inhibitor of TBSV replication, reducing TBSV replication up to ∼20-fold in a cell-free extract (CFE)-based replication assay (Fig. 8C) (42). Altogether, the TPR-containing Cpr7p seems to have a strong inhibitory effect against both TBSV and CIRV replication, while Sti1p efficiently inhibits CIRV, but its effect on TBSV replication in vitro is only moderate.
FIG 8.
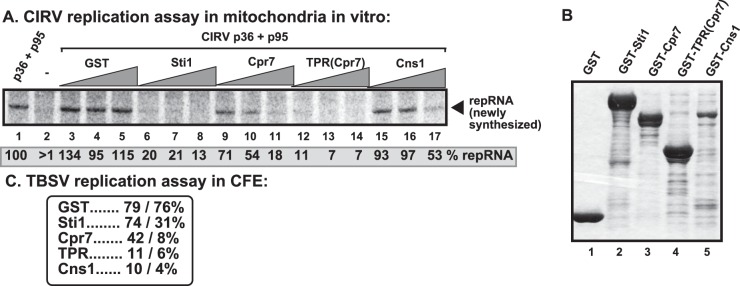
Comparison of the inhibitory effects of TPR-containing cellular proteins on CIRV replication in isolated mitochondria in vitro. (A) Denaturing PAGE analysis of the 32P-labeled repRNA products obtained in the replication assays with the isolated yeast mitochondrial preparations. The purified GST-tagged Sti1p, the yeast Cpr7p Cyp40-like cyclophilin, or the TPR domain of Cpr7p or Cns1p cochaperone (8, 16, and 32 μM) was added in combination with purified recombinant CIRV MBP-p36 and MBP-p95 proteins and the TBSV-derived (+)repRNA to the isolated mitochondrial preparations to perform the in vitro replication assay. The synthesized full-length repRNA is marked by an arrow. See further details in the legend to Fig. 7. (B) Coomassie blue-stained SDS-PAGE was used for analysis of affinity-purified GST-tagged Sti1p, Cpr7p, and Cns1p. (C) The level of in vitro TBSV repRNA replication in total yeast cell extracts in the presence of purified TBSV p33 and p92 replication proteins and purified GST-tagged Sti1p, Cpr7p, the TPR domain of Cpr7p or Cns1p (16 and 32 μM). Each experiment was repeated three times.
The plant Hop ortholog of the yeast Sti1p inhibits CIRV replication in yeast and plants.
Arabidopsis thaliana has three orthologs of Sti1p cochaperone, namely, AtHop1-3, that carry TPR1 and TPR2 domains (96, 106). AtHop-1 has 558 amino acids that show 40% identity and 57% similarity with ScSti1, which is 589 amino acids long. Using the membrane yeast two-hybrid (MYTH) assay, we showed a strong interaction between CIRV p36 replication protein and the AtHop-1 ortholog (Fig. 9A). These data confirmed the pulldown experiments with recombinant AtHop-1 and CIRV p36 that also showed the involvement of the RPR domain in p36 in the interaction with AtHop-1 (Fig. 4D).
FIG 9.
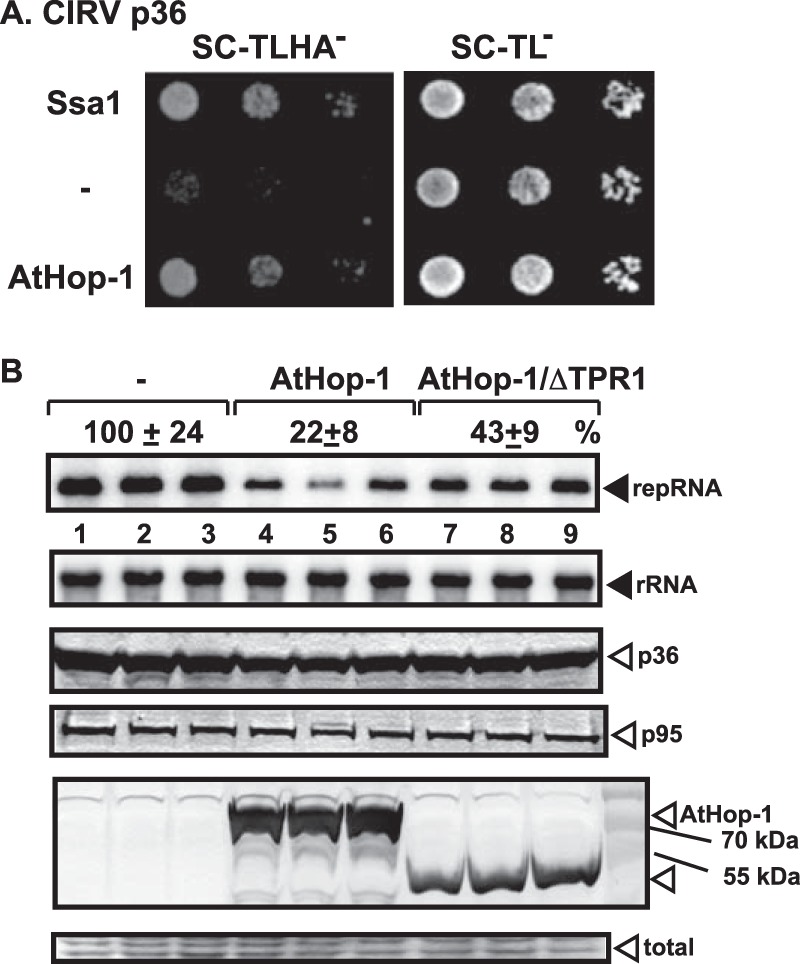
Inhibition of CIRV replication by expression of the orthologous AtHop-1 in yeast. (A) A Split-ubiquitin MYTH assay was used to test intracellular interaction between CIRV p36 and the AtHop-1 ortholog of the yeast Sti1p. The bait p36 was coexpressed with the prey AtHop-1 protein in yeast. The SSA1 gene and the empty prey vector (NubG) were used as positive and negative controls, respectively. (B) Northern blot analysis of CIRV RNA accumulation in wt yeast overproducing the FLAG-tagged AtHop-1 or its TPR1-deletion derivative. Second panel, Northern blot analysis to demonstrate the comparable level of rRNA loading in the yeast samples. Third and fourth panels, Western blot analysis of CIRV His6-p36 and CIRV His6-p95 accumulation by anti-His antibody. Bottom panels, detection of the overproduced FLAG-AtHop-1 in yeast by Western blotting using anti-FLAG antibody and the Coomassie blue-stained SDS-PAGE as a loading control. Each experiment was repeated three times.
To test if AtHop-1 could inhibit CIRV replication, we expressed it in yeast. Similar to results with the yeast Sti1p, AtHop-1 inhibited CIRV accumulation ∼5-fold in yeast (Fig. 9B, lanes 4 to 6). Deletion of the TPR1 sequence made AtHop-1 less effective inhibitor of CIRV replication (Fig. 9B, lanes 7 to 9), suggesting that the TPR1 sequence is important for the inhibitory function of AtHop-1.
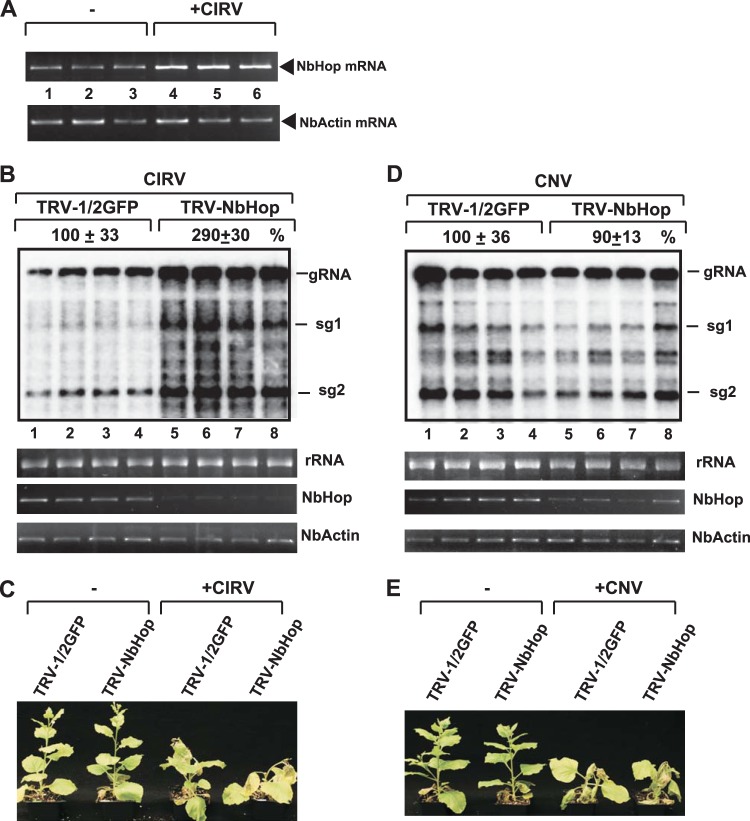
To test the relevance of the plant Hop protein in tombusvirus replication, we tested the accumulation level of Hop mRNA in the Nicotiana benthamiana host. The semiquantitative reverse transcription-PCR (RT-PCR) analysis revealed the induction of NbHop mRNA upon infection with CIRV (Fig. 10A). In addition, knockdown of NbHop level via VIGS in N. benthamiana led to ∼3-fold-increased CIRV genomic RNA accumulation (Fig. 10B). As expected, due to the high level of CIRV accumulation, the Hop-knockdown N. benthamiana plants developed more severe symptoms than control plants when infected with CIRV (Fig. 10C). In contrast, the accumulation of the genomic RNA of the related CNV (a peroxisomal replicating tombusvirus, closely related to TBSV) was not significantly affected by Hop-knockdown N. benthamiana plants (Fig. 10D). Also, the symptom severity of CNV-infected knockdown or control plants was comparable (Fig. 10E). Based on these in planta experiments, we suggest that the plant Hop ortholog plays a potent inhibitory role, similar to that of the yeast Sti1p, in the mitochondrial CIRV replication, but not in the peroxisomal CNV replication.
FIG 10.
Knockdown of Hop gene by VIGS increases CIRV accumulation in whole plants. (A) Semiquantitative RT-PCR analysis of the accumulation of NbHOP mRNAs in CIRV-infected N. benthamiana plants 3 days postinoculation (dpi) and in the control mock-inoculated plants. Semiquantitative RT-PCR analysis of the NbActin mRNA level served as a control. (B) Top image, accumulation of CIRV genomic and subgenomic RNAs in the inoculated leaves of HOP-knockdown N. benthamiana plants at 2 dpi, based on Northern blot analysis. VIGS was performed via agroinfiltration of TRV vectors carrying NbHOP sequence or the TRV vector carrying the C-terminal half of a GFP insert (as a control). Inoculation with CIRV gRNA was done 14 days after VIGS. Second image, rRNA level in the samples used as loading control. Bottom images, Semiquantitative RT-PCR analysis of the accumulation of NbHOP mRNA in the knockdown N. benthamiana plants and in the control plants 2 days after inoculation with CIRV. Semiquantitative RT-PCR analysis of the NbActin mRNA from the same samples served as a control. (C) Development of more severe CIRV-induced symptoms in the NbHOP-knockdown plant (shown on the right) at 7 dpi than those in the control plant infiltrated with the control TRV vector. Note the minor phenotypic effect in the uninfected NbHOP-knockdown N. benthamiana plants for the control plants (left), which were agroinfiltrated with the pTRV1/2GFP vector. (D) Accumulation of CNV gRNA in the inoculated leaves of HOP-knockdown N. benthamiana plants 2 days postinoculation, based on Northern blot analysis. See further details in panel B. Note that the peroxisome-localized CNV and TBSV replication proteins behave similarly in yeast cells (not shown) in regard to lack of effect by the Sti1p on CNV or TBSV replication. (E) Comparable CNV-induced symptom development in the HOP-knockdown and control plants. See further details in panel C. Each experiment was repeated three times.
DISCUSSION
Identification of the Hop-like Sti1p cochaperone as a novel cell-intrinsic restriction factor against CIRV replication in mitochondria.
Cellular protein chaperones are important for virus replication and during other steps of the infectious process (10, 107–113). For example, Hsp70 has been shown to affect the intracellular localization and membrane insertion of TBSV replication proteins and the assembly of the tombusviral VRCs (62, 63, 104). Although Hsp70 interacts directly with the tombusviral replication proteins, it is possible that other cellular factors could affect the subversion of Hsp70s by TBSV. Since cochaperones facilitate selection and delivery of client proteins to the major Hsp70 and Hsp90 chaperones (114–116), some cochaperones might also be involved in viral infections, as demonstrated in this paper and earlier (10, 107–113).
Our finding that the conserved cellular Hop-like Sti1p cochaperone is a restriction factor for CIRV replication in the mitochondria contributes to the emerging complex roles of cellular chaperones in virus replication (10). While deletion of Sti1p led to a 2- to 4-fold increase in CIRV replication in the yeast model host, and knockdown of the orthologous Hop in N. benthamiana increased CIRV accumulation ∼3-fold, overexpression of Sti1p or AtHop-1 in yeast was inhibitory. In vitro CIRV replication experiments based on isolated mitochondria also confirmed the robust inhibitory effect of Sti1p on CIRV. Moreover, the expression of the Sti1 ortholog Hop is increased during CIRV replication in plant leaves. Thus, Sti1p is a new member of the growing family of cell-intrinsic restriction factors.
However, Sti1p did not have a robust effect on replication of the closely related TBSV in yeast or on the replication of CNV in plants, both of which utilize the peroxisomal membranes for replication (55, 105). This contrasting finding with different tombusviruses exploiting different subcellular locations could be due to the difference in accessibility of Sti1/Hop to replication proteins of tombusviruses in their respective cellular environments. For example, it has been shown in plants that Hop/Sti1 is involved in transportation of freshly synthesized mitochondrial and chloroplast proteins from the cytosol into these organelles (117). Moreover, the delivery/import of mitochondrial preproteins from the cytosol to the mitochondria often depends on Hsp70/Hsp90 chaperones and includes Hop/Sti1 and the TPR domain in Tom70 mitochondrial receptor (118, 119). Also, the CIRV p36 replication protein was shown to interact with various Tom receptor proteins, which might have roles in mitochondrial membrane insertion of p36 (47). Based on these studies, we propose that Sti1/Hop might be easily accessible and bind efficiently to the mitochondrion-targeted CIRV replication proteins in cells, while the cellular Sti1/Hop cochaperone has a lesser chance to bind to the peroxisome-targeted TBSV and CNV replication proteins. Accordingly, live-cell imaging showed the relocalization of Sti1p to the mitochondria in the presence of CIRV p36, while Sti1p showed mostly cytosolic localization in yeast cells expressing the CNV p33 replication protein (Fig. 5). Thus, the difference in accessibility of Sti1/Hop could be the major mechanism restricting CIRV but not TBSV or CNV replication.
Mechanisms of Sti1p cochaperone-driven restriction of CIRV replication.
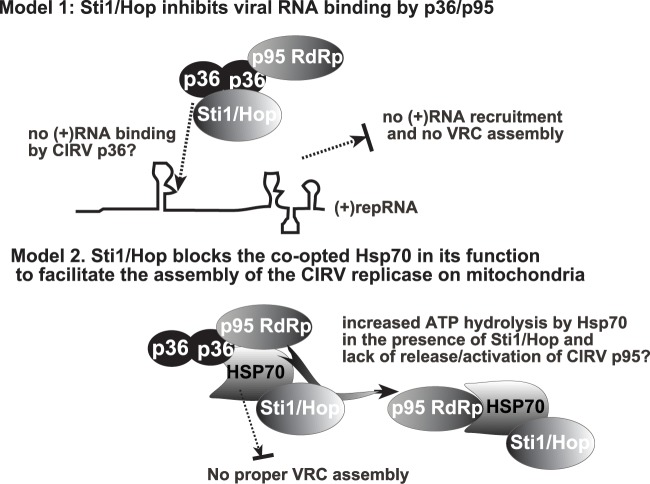
Recruitment of the tombusvirus (+)RNA into replication requires selective binding by the tombusvirus replication proteins via recognition of a RNA recruitment element (named p33RE) within the polymerase gene sequence (54, 120). The same p33RE element is also required for the VRC assembly and activation of the polymerase function of the replication protein (102, 121). The specific recognition of p33RE is performed by arginines within the RPR motif in p33/p92pol (122, 123). Therefore, binding of cellular factors to the RPR motif-containing region could block the ability of tombusvirus replication proteins to bind the viral (+)RNA, thus inhibiting the essential viral processes of (+)RNA recruitment, VRC assembly, and replicase activation (121, 124). Indeed, the TPR domain of Cpr7p binds to the RPR region in the tombusvirus p33/p92pol replication proteins and blocks the above viral processes, thus acting as a restriction factor (41). We find that Sti1p also binds to the RPR region in the CIRV p36/p95pol replication proteins (Fig. 4), and this could explain the strong in vitro inhibitory effect of recombinant Sti1p on CIRV replication based on mitochondrial preparations (Fig. 7). The CIRV p36 interaction with Sti1p also leads to the recruitment of Sti1p to punctate structures (mitochondrial membranes) in yeast cells, suggesting robust p36-Sti1p interaction in cells. Thus, direct interaction between the Sti1p and CIRV p36 might block viral RNA recruitment (Fig. 11, Model 1).
FIG 11.
Models on the inhibitory role of Sti1p cochaperone in CIRV replication. Model 1 predicts that direct interaction between Sti1p and the RPR region of CIRV p36 replication protein blocks the viral (+)RNA recruitment function of p36, and thus blocks replication of CIRV. Model 2 emphasizes the additional role of Sti1p through the coopted Hsp70 chaperone. Binding of the TPR1 domain of Sti1p to Hsp70 within the CIRV replicase might inhibit the function of the subverted Hsp70 in VRC assembly, by possibly stabilizing the Hsp70-p95pol complex. Note that both mechanisms might operate inside the cell.
However, the picture on the mechanism of CIRV inhibition is likely more complex. This is because both the TPR1 and TPR2 regions of Sti1p bind to the RPR domain of CIRV p36/p95pol, yet the expression of TPR1 is inhibitory, while TPR2 is less effective in reducing CIRV accumulation in yeast or CIRV replication in vitro with mitochondrial preparations (Fig. 6 and 7). Thus, the binding to the RPR domain in p36/p95pol is unlikely enough for Sti1p to effectively inhibit p36 or p95pol functions.
Interestingly, Sti1p does not bind to the RPR region of the TBSV p33 replication protein. The binding between p33 and Sti1p involves the C-terminal region of p33 containing the p33-p33/p92 interaction sequence (Fig. 4G to H) and likely the corresponding sequence in p92pol. It is possible that this interaction is not robust or stable enough to interfere with p33/p92pol functions in cells. It is likely that p33 could readily interact with additional p33 molecules, while binding to Sti1p molecules by p33 might be less favored in cells. Indeed, Sti1p is not efficiently relocalized to punctate structures containing the p33 molecules in yeast cells (Fig. 5B). A similar situation was observed with Cpr6p Cyp40-like cyclophilin, which also binds to p33 within the C-terminal domain and does not inhibit p33 functions (41). Yet this rule is not general, since Cns1p cochaperone binds to the p33-p33/p92 interaction sequence in TBSV p33 replication protein and effectively inhibits TBSV replication in yeast (42) and in vitro (Fig. 8C). It seems that the intracellular distribution/accessibility of these TPR-containing host proteins might be a major factor in their ability to inhibit replication of different tombusviruses.
Although direct interaction between the RPR region of CIRV p36 and the TPR1 sequence in Sti1p might explain the inhibitory effect on CIRV replication (Fig. 11, Model 1), it is also possible that Sti1p limits the functions of subverted cellular factors, such as Hsp70, for its antiviral activity. Cytosolic Hsp70s are coopted by tombusviruses, and they are permanent residents in the tombusviral VRCs (39). This model is supported by the observation that, in spite of the binding of both TPR1 and TPR2 sequences to the RPR region of CIRV p36 (Fig. 3), only the expression of the Hsp70-interacting TPR1 region (89, 91) was able to robustly inhibit CIRV replication in yeast and in vitro (Fig. 6 and 7). Moreover, mutation within the TPR1 sequence (i.e., mutant C49Y) that debilitates the interaction with Hsp70, but not with p36 (Fig. 3) had lesser inhibitory effects on CIRV replication when expressed in yeast (Fig. 6). In contrast, a mutation (i.e., G325D) that affects interaction with Hsp90 did not interfere with the inhibitory function of Sti1p in vivo or in vitro. Based on these findings, we propose that the recruited Sti1p cochaperone inhibits the proviral function of the coopted cellular Hsp70 molecules during CIRV replication. For example, the predicted Sti1p-Hsp70 interaction during the formation of VRC or within the assembled VRC might inhibit the Hsp70-driven activation of the polymerase function of p95pol or other steps/functions (Fig. 11, Model 2).
The major role of the Sti1p cochaperone in eukaryotic cells is to bring the Hsp70-client protein complex together with the Hsp90 chaperone to facilitate robust refolding/activation of client proteins by the powerful Hsp90 system (91, 125, 126). This is facilitated by the ability of Sti1p to bind simultaneously to Hsp70 (via the TPR1 sequence) and Hsp90 (via the TPR2A region). However, based on our data, it is unlikely that this function of Sti1p is critical to inhibit CIRV replication. This is because deletion of the entire TPR2 domain from Sti1p did not eliminate the inhibitory function of Sti1p in vitro or in yeast (Fig. 6 and 7). Also, blocking the function of Hsp90 by applying a geldanamycin inhibitor in yeast had no effect on tombusvirus replication (data not shown), arguing against the functional role of Hsp90 in tombusvirus replication. Therefore, the direct effect of Sti1p on CIRV p36/p95pol and the coopted Hsp70 is the best suited to explain the current experimental data (Fig. 11).
Sti1/Hop is the first cellular restriction factor specifically affecting one tombusvirus (i.e., CIRV, which replicates in the mitochondria) but not other tombusviruses (TBSV and CNV, which both replicate in peroxisomal membranes). The previously identified TPR domain-containing cellular proteins, namely, Cpr7p and Cns1p, could inhibit the replication of all these tombusviruses (this work and references 41 and 42). Interestingly, all three cellular factors are part of the Hsp70/Hsp90 chaperone system, suggesting that they, at least in part, inhibit tombusvirus replication via regulating chaperone functions. Because the Hsp70/Hsp90 chaperone system is known to affect many viruses (reviewed in references 10 and 127), it is possible that the identified restriction factor activities of these TPR-containing cellular proteins might be functional against other viruses and pathogens.
Another use of Hop/Sti1 in host innate defense against pathogens is its role in the maturation and transport of rice chitin receptor OsCERK1, which is a pattern recognition receptor (PRR), against rice blast fungus (128). This function of Hop/Sti1 might link the functions of PRRs, small Rho-type GTPases, and resistance against pathogens. Sti1p is also known to affect prion propagation in yeast (129), and its expression is increased in simian virus 40 (SV40)-transformed MRC-5 fibroblasts and some tumor tissues (79, 130). Thus, Hop/Sti1 is emerging as a possibly key component in propagations of several infectious agents and innate defense responses of host cells.
Summary.
The current and recent works (41, 42) with tombusviruses indicate that some members of the large family of TPR-containing proteins might act as cell-intrinsic restriction factors of tombusviruses. The list includes the Hop-like Sti1p and Ttc4 oncogene-like Cns1p cochaperones and Cyp40-like Cpr7p cyclophilin. Yet, based on the yeast Cyp40-like Cpr6p cyclophilin (41), we already know that not all TPR-containing proteins are viral restriction factors in spite of their abilities to interact with tombusvirus replication proteins. Since many TPR-containing proteins are expressed in all eukaryotes, it will be important to identify all the members of this cellular protein family that act as restriction factors during the replication of tombusvirus and other (+)RNA viruses.
ACKNOWLEDGMENTS
Yeast Sti1 mutants and ySti1p polyclonal antibody were provided by Daniel C. Masison (National Institutes of Health, Bethesda, Maryland). We thank Judit Pogany and Chingkai Chuang for critical readings of the manuscript and for very helpful suggestions.
This work was supported by NIAID (5R21AI096323).
Footnotes
Published ahead of print 11 June 2014
REFERENCES
- 1.Diamond MS, Gale M., Jr 2012. Cell-intrinsic innate immune control of West Nile virus infection. Trends Immunol. 33:522–530. 10.1016/j.it.2012.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoshi T, Koyama S, Kobiyama K, Akira S, Ishii KJ. 2011. Innate and adaptive immune responses to viral infection and vaccination. Curr. Opin. Virol. 1:226–232. 10.1016/j.coviro.2011.07.002 [DOI] [PubMed] [Google Scholar]
- 3.Jensen S, Thomsen AR. 2012. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 86:2900–2910. 10.1128/JVI.05738-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding SW. 2010. RNA-based antiviral immunity. Nat. Rev. Immunol. 10:632–644. 10.1038/nri2824 [DOI] [PubMed] [Google Scholar]
- 5.Diamond MS, Farzan M. 2013. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 13:46–57. 10.1038/nri3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasunaga A, Hanna SL, Li J, Cho H, Rose PP, Spiridigliozzi A, Gold B, Diamond MS, Cherry S. 2014. Genome-wide RNAi screen identifies broadly-acting host factors that inhibit arbovirus infection. PLoS Pathog. 10:e1003914. 10.1371/journal.ppat.1003914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, Cherry S. 2014. Viruses and antiviral immunity in Drosophila. Dev. Comp. Immunol. 42:67–84. 10.1016/j.dci.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagy PD. 2008. Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 46:217–242. 10.1146/annurev.phyto.121407.093958 [DOI] [PubMed] [Google Scholar]
- 9.Nagy PD, Pogany J. 2008. Multiple roles of viral replication proteins in plant RNA virus replication. Methods Mol. Biol. 451:55–68. 10.1007/978-1-59745-102-4_4 [DOI] [PubMed] [Google Scholar]
- 10.Nagy PD, Wang RY, Pogany J, Hafren A, Makinen K. 2011. Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology 411:374–382. 10.1016/j.virol.2010.12.061 [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Nagy PD. 2011. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol. 8:305–315. 10.4161/rna.8.2.15391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.den Boon JA, Diaz A, Ahlquist P. 2010. Cytoplasmic viral replication complexes. Cell Host Microbe 8:77–85. 10.1016/j.chom.2010.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374. 10.1038/nrmicro1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez-Garcia MD, Mazzon M, Jacobs M, Amara A. 2009. Pathogenesis of flavivirus infections: using and abusing the host cell. Cell Host Microbe 5:318–328. 10.1016/j.chom.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 15.Nagy PD, Pogany J. 2012. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 10:137–149. 10.1038/nrmicro2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischl W, Bartenschlager R. 2011. Exploitation of cellular pathways by Dengue virus. Curr. Opin. Microbiol. 14:470–475. 10.1016/j.mib.2011.07.012 [DOI] [PubMed] [Google Scholar]
- 17.Panavas T, Serviene E, Brasher J, Nagy PD. 2005. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 102:7326–7331. 10.1073/pnas.0502604102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. 2005. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 19:445–452. 10.1101/gad.1267905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y, Serviene E, Gal J, Panavas T, Nagy PD. 2006. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J. Virol. 80:7394–7404. 10.1128/JVI.02686-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kushner DB, Lindenbach BD, Grdzelishvili VZ, Noueiry AO, Paul SM, Ahlquist P. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. U. S. A. 100:15764–15769. 10.1073/pnas.2536857100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242–245. 10.1038/nature07207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. 2009. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc. Natl. Acad. Sci. U. S. A. 106:16410–16415. 10.1073/pnas.0907439106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, Chien M, Weir DB, Russo JJ, Ju J, Brownstein MJ, Sheridan R, Sander C, Zavolan M, Tuschl T, Rice CM. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104:12884–12889. 10.1073/pnas.0704894104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, Garcia-Blanco MA. 2009. Discovery of insect and human dengue virus host factors. Nature 458:1047–1050. 10.1038/nature07967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298–307. 10.1016/j.chom.2009.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy PD, Pogany J. 2006. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology 344:211–220. 10.1016/j.virol.2005.09.017 [DOI] [PubMed] [Google Scholar]
- 27.White KA, Nagy PD. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 78:187–226. 10.1016/S0079-6603(04)78005-8 [DOI] [PubMed] [Google Scholar]
- 28.Panaviene Z, Panavas T, Serva S, Nagy PD. 2004. Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J. Virol. 78:8254–8263. 10.1128/JVI.78.15.8254-8263.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panavas T, Nagy PD. 2003. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology 314:315–325. 10.1016/S0042-6822(03)00436-7 [DOI] [PubMed] [Google Scholar]
- 30.Serviene E, Shapka N, Cheng CP, Panavas T, Phuangrat B, Baker J, Nagy PD. 2005. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc. Natl. Acad. Sci. U. S. A. 102:10545–10550. 10.1073/pnas.0504844102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serviene E, Jiang Y, Cheng CP, Baker J, Nagy PD. 2006. Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J. Virol. 80:1231–1241. 10.1128/JVI.80.3.1231-1241.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah Nawaz-ul-Rehman M, Martinez-Ochoa N, Pascal H, Sasvari Z, Herbst C, Xu K, Baker J, Sharma M, Herbst A, Nagy PD. 2012. Proteome-wide overexpression of host proteins for identification of factors affecting tombusvirus RNA replication: an inhibitory role of protein kinase C. J. Virol. 86:9384–9395. 10.1128/JVI.00019-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nawaz-Ul-Rehman MS, Reddisiva Prasanth K, Baker J, Nagy PD. 2013. Yeast screens for host factors in positive-strand RNA virus replication based on a library of temperature-sensitive mutants. Methods 59:207–216. 10.1016/j.ymeth.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 34.Nagy PD. 2011. The roles of host factors in tombusvirus RNA recombination. Adv. Virus Res. 81:63–84. 10.1016/B978-0-12-385885-6.00008-0 [DOI] [PubMed] [Google Scholar]
- 35.Nagy PD, Pogany J. 2010. Global genomics and proteomics approaches to identify host factors as targets to induce resistance against Tomato bushy stunt virus. Adv. Virus Res. 76:123–177. 10.1016/S0065-3527(10)76004-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendu V, Chiu M, Barajas D, Li Z, Nagy PD. 2010. Cpr1 cyclophilin and Ess1 parvulin prolyl isomerases interact with the tombusvirus replication protein and inhibit viral replication in yeast model host. Virology 406:342–351. 10.1016/j.virol.2010.07.022 [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Pogany J, Panavas T, Xu K, Esposito AM, Kinzy TG, Nagy PD. 2009. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology 385:245–260. 10.1016/j.virol.2008.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z, Barajas D, Panavas T, Herbst DA, Nagy PD. 2008. Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J. Virol. 82:6911–6926. 10.1128/JVI.00702-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serva S, Nagy PD. 2006. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 80:2162–2169. 10.1128/JVI.80.5.2162-2169.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu K, Nagy PD. 2010. Dissecting virus-plant interactions through proteomics approaches. Curr. Proteomics 7:316–327. 10.2174/157016410793611792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin JY, Mendu V, Pogany J, Qin J, Nagy PD. 2012. The TPR domain in the host Cyp40-like cyclophilin binds to the viral replication protein and inhibits the assembly of the tombusviral replicase. PLoS Pathog. 8:e1002491. 10.1371/journal.ppat.1002491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin JY, Nagy PD. 2013. Identification of novel host factors via conserved domain search: Cns1 cochaperone is a novel restriction factor of tombusvirus replication in yeast. J. Virol. 87:12600–12610. 10.1128/JVI.00196-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kovalev N, Nagy PD. 2013. Cyclophilin a binds to the viral RNA and replication proteins, resulting in inhibition of tombusviral replicase assembly. J. Virol. 87:13330–13342. 10.1128/JVI.02101-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang Y, Li Z, Nagy PD. 2010. Nucleolin/Nsr1p binds to the 3′ noncoding region of the tombusvirus RNA and inhibits replication. Virology 396:10–20. 10.1016/j.virol.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barajas D, Li Z, Nagy PD. 2009. The Nedd4-Type Rsp5p ubiquitin ligase inhibits tombusvirus replication by regulating degradation of the p92 replication protein and decreasing the activity of the tombusvirus replicase. J. Virol. 83:11751–11764. 10.1128/JVI.00789-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin J, Barajas D, Nagy PD. 2012. An inhibitory function of WW domain-containing host proteins in RNA virus replication. Virology 426:106–119. 10.1016/j.virol.2012.01.020 [DOI] [PubMed] [Google Scholar]
- 47.Hwang YT, McCartney AW, Gidda SK, Mullen RT. 2008. Localization of the Carnation Italian ringspot virus replication protein p36 to the mitochondrial outer membrane is mediated by an internal targeting signal and the TOM complex. BMC Cell Biol. 9:54. 10.1186/1471-2121-9-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT. 2005. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17:3513–3531. 10.1105/tpc.105.036350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber-Lotfi F, Dietrich A, Russo M, Rubino L. 2002. Mitochondrial targeting and membrane anchoring of a viral replicase in plant and yeast cells. J. Virol. 76:10485–10496. 10.1128/JVI.76.20.10485-10496.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagy PD, Barajas D, Pogany J. 2012. Host factors with regulatory roles in tombusvirus replication. Curr. Opin. Virol. 2:691–698. 10.1016/j.coviro.2012.10.004 [DOI] [PubMed] [Google Scholar]
- 51.Kovalev N, Nagy PD. 2014. The expanding functions of cellular helicases: the tombusvirus RNA replication enhancer co-opts the plant eIF4AIII-like AtRH2 and the DDX5-like AtRH5 DEAD-box RNA helicases to promote viral asymmetric RNA replication. PLoS Pathog. 10:e1004051. 10.1371/journal.ppat.1004051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barajas D, Martin IF, Pogany J, Risco C, Nagy PD. 2014. Noncanonical role for the host Vps4 AAA+ ATPase ESCRT protein in the formation of tomato bushy stunt virus replicase. PLoS Pathog. 10:e1004087. 10.1371/journal.ppat.1004087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonczyk M, Pathak KB, Sharma M, Nagy PD. 2007. Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 362:320–330. 10.1016/j.virol.2007.01.004 [DOI] [PubMed] [Google Scholar]
- 54.Pogany J, White KA, Nagy PD. 2005. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 79:4859–4869. 10.1128/JVI.79.8.4859-4869.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Panavas T, Hawkins CM, Panaviene Z, Nagy PD. 2005. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology 338:81–95. 10.1016/j.virol.2005.04.025 [DOI] [PubMed] [Google Scholar]
- 56.Stork J, Kovalev N, Sasvari Z, Nagy PD. 2011. RNA chaperone activity of the tombusviral p33 replication protein facilitates initiation of RNA synthesis by the viral RdRp in vitro. Virology 409:338–347. 10.1016/j.virol.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu K, Huang TS, Nagy PD. 2012. Authentic in vitro replication of two tombusviruses in isolated mitochondrial and endoplasmic reticulum membranes. J. Virol. 86:12779–12794. 10.1128/JVI.00973-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pogany J, Nagy PD. 2008. Authentic replication and recombination of Tomato bushy stunt virus RNA in a cell-free extract from yeast. J. Virol. 82:5967–5980. 10.1128/JVI.02737-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Panaviene Z, Panavas T, Nagy PD. 2005. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 79:10608–10618. 10.1128/JVI.79.16.10608-10618.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang RY, Nagy PD. 2008. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe 3:178–187. 10.1016/j.chom.2008.02.005 [DOI] [PubMed] [Google Scholar]
- 61.Li Z, Pogany J, Tupman S, Esposito AM, Kinzy TG, Nagy PD. 2010. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 6:e1001175. 10.1371/journal.ppat.1001175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang RY, Stork J, Pogany J, Nagy PD. 2009. A temperature sensitive mutant of heat shock protein 70 reveals an essential role during the early steps of tombusvirus replication. Virology 394:28–38. 10.1016/j.virol.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang RY, Stork J, Nagy PD. 2009. A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J. Virol. 83:3276–3287. 10.1128/JVI.02313-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barajas D, Jiang Y, Nagy PD. 2009. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 5:e1000705. 10.1371/journal.ppat.1000705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang TS, Nagy PD. 2011. Direct inhibition of tombusvirus plus-strand RNA synthesis by a dominant-negative mutant of a host metabolic enzyme, glyceraldehyde-3-phosphate dehydrogenase, in yeast and plants. J. Virol. 85:9090–9102. 10.1128/JVI.00666-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kovalev N, Pogany J, Nagy PD. 2012. A co-opted DEAD-box RNA helicase enhances tombusvirus plus-strand synthesis. PLoS Pathog. 8:e1002537. 10.1371/journal.ppat.1002537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sasvari Z, Izotova L, Kinzy TG, Nagy PD. 2011. Synergistic roles of eukaryotic translation elongation factors 1Bgamma and 1A in stimulation of tombusvirus minus-strand synthesis. PLoS Pathog. 7:e1002438. 10.1371/journal.ppat.1002438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kovalev N, Barajas D, Nagy PD. 2012. Similar roles for yeast Dbp2 and Arabidopsis RH20 DEAD-box RNA helicases to Ded1 helicase in tombusvirus plus-strand synthesis. Virology 432:470–484. 10.1016/j.virol.2012.06.030 [DOI] [PubMed] [Google Scholar]
- 69.D'Andrea LD, Regan L. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28:655–662. 10.1016/j.tibs.2003.10.007 [DOI] [PubMed] [Google Scholar]
- 70.Allan RK, Ratajczak T. 2011. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16:353–367. 10.1007/s12192-010-0248-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haslbeck V, Eckl JM, Kaiser CJ, Papsdorf K, Hessling M, Richter K. 2013. Chaperone-interacting TPR proteins in Caenorhabditis elegans. J. Mol. Biol. 425:2922–2939. 10.1016/j.jmb.2013.05.019 [DOI] [PubMed] [Google Scholar]
- 72.Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, Reuter M, Yang Z, Berninger P, Palencia A, Benes V, Penninger J, Sachidanandam R, Pillai RS. 2012. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol. Cell 47:970–979. 10.1016/j.molcel.2012.07.019 [DOI] [PubMed] [Google Scholar]
- 73.Stawowczyk M, Van Scoy S, Kumar KP, Reich NC. 2011. The interferon stimulated gene 54 promotes apoptosis. J. Biol. Chem. 286:7257–7266. 10.1074/jbc.M110.207068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cerveny L, Straskova A, Dankova V, Hartlova A, Ceckova M, Staud F, Stulik J. 2013. Tetratricopeptide repeat motifs in the world of bacterial pathogens: role in virulence mechanisms. Infect. Immun. 81:629–635. 10.1128/IAI.01035-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Assimon VA, Gillies AT, Rauch JN, Gestwicki JE. 2013. Hsp70 protein complexes as drug targets. Curr. Pharm. Des. 19:404–417. 10.2174/138161213804143699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nordling E, Abraham-Nordling M. 2012. Colonic amyloidosis, computational analysis of the major amyloidogenic species, serum amyloid A. Comput. Biol. Chem. 39:29–34. 10.1016/j.compbiolchem.2012.06.005 [DOI] [PubMed] [Google Scholar]
- 77.Adams CJ, Pike AC, Maniam S, Sharpe TD, Coutts AS, Knapp S, La Thangue NB, Bullock AN. 2012. The p53 cofactor Strap exhibits an unexpected TPR motif and oligonucleotide-binding (OB)-fold structure. Proc. Natl. Acad. Sci. U. S. A. 109:3778–3783. 10.1073/pnas.1113731109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Banasavadi-Siddegowda YK, Mai J, Fan Y, Bhattacharya S, Giovannucci DR, Sanchez ER, Fischer G, Wang X. 2011. FKBP38 peptidylprolyl isomerase promotes the folding of cystic fibrosis transmembrane conductance regulator in the endoplasmic reticulum. J. Biol. Chem. 286:43071–43080. 10.1074/jbc.M111.269993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kubota H, Yamamoto S, Itoh E, Abe Y, Nakamura A, Izumi Y, Okada H, Iida M, Nanjo H, Itoh H, Yamamoto Y. 2010. Increased expression of co-chaperone HOP with HSP90 and HSC70 and complex formation in human colonic carcinoma. Cell Stress Chaperones 15:1003–1011. 10.1007/s12192-010-0211-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bourai M, Lucas-Hourani M, Gad HH, Drosten C, Jacob Y, Tafforeau L, Cassonnet P, Jones LM, Judith D, Couderc T, Lecuit M, Andre P, Kummerer BM, Lotteau V, Despres P, Tangy F, Vidalain PO. 2012. Mapping of Chikungunya virus interactions with host proteins identified nsP2 as a highly connected viral component. J. Virol. 86:3121–3134. 10.1128/JVI.06390-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fensterl V, Wetzel JL, Ramachandran S, Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW, Sen GC. 2012. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 8:e1002712. 10.1371/journal.ppat.1002712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miettinen JJ, Matikainen S, Nyman TA. 2012. Global secretome characterization of herpes simplex virus 1-infected human primary macrophages. J. Virol. 86:12770–12778. 10.1128/JVI.01545-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Danquah JO, Botchway S, Jeshtadi A, King LA. 2012. Direct interaction of baculovirus capsid proteins VP39 and EXON0 with kinesin-1 in insect cells determined by fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy. J. Virol. 86:844–853. 10.1128/JVI.06109-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jeshtadi A, Burgos P, Stubbs CD, Parker AW, King LA, Skinner MA, Botchway SW. 2010. Interaction of poxvirus intracellular mature virion proteins with the TPR domain of kinesin light chain in live infected cells revealed by two-photon-induced fluorescence resonance energy transfer fluorescence lifetime imaging microscopy. J. Virol. 84:12886–12894. 10.1128/JVI.01395-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Jr, Shi PY, Diamond MS. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. 10.1038/nature09489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iki T, Yoshikawa M, Meshi T, Ishikawa M. 2012. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 31:267–278. 10.1038/emboj.2011.395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu XY, Chen W, Wei B, Shan YF, Wang C. 2011. IFN-induced TPR protein IFIT3 potentiates antiviral signaling by bridging MAVS and TBK1. J. Immunol. 187:2559–2568. 10.4049/jimmunol.1100963 [DOI] [PubMed] [Google Scholar]
- 88.Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, Burckstummer T, Stefanovic A, Krieger S, Bennett KL, Rulicke T, Weber F, Colinge J, Muller M, Superti-Furga G. 2011. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 12:624–630. 10.1038/ni.2048 [DOI] [PubMed] [Google Scholar]
- 89.Flom G, Behal RH, Rosen L, Cole DG, Johnson JL. 2007. Definition of the minimal fragments of Sti1 required for dimerization, interaction with Hsp70 and Hsp90 and in vivo functions. Biochem. J. 404:159–167. 10.1042/BJ20070084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wegele H, Haslbeck M, Reinstein J, Buchner J. 2003. Sti1 is a novel activator of the Ssa proteins. J. Biol. Chem. 278:25970–25976. 10.1074/jbc.M301548200 [DOI] [PubMed] [Google Scholar]
- 91.Schmid AB, Lagleder S, Grawert MA, Rohl A, Hagn F, Wandinger SK, Cox MB, Demmer O, Richter K, Groll M, Kessler H, Buchner J. 2012. The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. EMBO J. 31:1506–1517. 10.1038/emboj.2011.472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Odunuga OO, Hornby JA, Bies C, Zimmermann R, Pugh DJ, Blatch GL. 2003. Tetratricopeptide repeat motif-mediated Hsc70-mSTI1 interaction: molecular characterization of the critical contacts for successful binding and specificity. J. Biol. Chem. 278:6896–6904. 10.1074/jbc.M206867200 [DOI] [PubMed] [Google Scholar]
- 93.Rubino L, Burgyan J, Russo M. 1995. Molecular cloning and complete nucleotide sequence of carnation Italian ringspot tombusvirus genomic and defective interfering RNAs. Arch. Virol. 140:2027–2039. 10.1007/BF01322690 [DOI] [PubMed] [Google Scholar]
- 94.Reidy M, Masison DC. 2010. Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104. Mol. Cell. Biol. 30:3542–3552. 10.1128/MCB.01292-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Song Y, Masison DC. 2005. Independent regulation of Hsp70 and Hsp90 chaperones by Hsp70/Hsp90-organizing protein Sti1 (Hop1). J. Biol. Chem. 280:34178–34185. 10.1074/jbc.M505420200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krishna P, Gloor G. 2001. The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 6:238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pogany J, Panavas T, Serviene E, Nawaz-Ul-Rehman MS, Nagy PD. 2010. A high-throughput approach for studying virus replication in yeast. Curr. Protoc. Microbiol. Chapter 16:Unit 16J.11.11–16J.11.15. 10.1002/9780471729259.mc16j01s19 [DOI] [PubMed] [Google Scholar]
- 98.Jaag HM, Nagy PD. 2009. Silencing of Nicotiana benthamiana Xrn4p exoribonuclease promotes tombusvirus RNA accumulation and recombination. Virology 386:344–352. 10.1016/j.virol.2009.01.015 [DOI] [PubMed] [Google Scholar]
- 99.Iyer K, Burkle L, Auerbach D, Thaminy S, Dinkel M, Engels K, Stagljar I. 2005. Utilizing the split-ubiquitin membrane yeast two-hybrid system to identify protein-protein interactions of integral membrane proteins. Sci. STKE 2005:pl3. 10.1126/stke.2752005pl3 [DOI] [PubMed] [Google Scholar]
- 100.Kittanakom S, Chuk M, Wong V, Snyder J, Edmonds D, Lydakis A, Zhang Z, Auerbach D, Stagljar I. 2009. Analysis of membrane protein complexes using the split-ubiquitin membrane yeast two-hybrid (MYTH) system. Methods Mol. Biol. 548:247–271. 10.1007/978-1-59745-540-4_14 [DOI] [PubMed] [Google Scholar]
- 101.Flom G, Weekes J, Williams JJ, Johnson JL. 2006. Effect of mutation of the tetratricopeptide repeat and asparatate-proline 2 domains of Sti1 on Hsp90 signaling and interaction in Saccharomyces cerevisiae. Genetics 172:41–51. 10.1534/genetics.105.045815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pathak KB, Pogany J, Xu K, White KA, Nagy PD. 2012. Defining the roles of cis-acting RNA elements in tombusvirus replicase assembly in vitro. J. Virol. 86:156–171. 10.1128/JVI.00404-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pathak KB, Pogany J, Nagy PD. 2011. Non-template functions of the viral RNA in plant RNA virus replication. Curr. Opin. Virol. 1:332–338. 10.1016/j.coviro.2011.09.011 [DOI] [PubMed] [Google Scholar]
- 104.Pogany J, Stork J, Li Z, Nagy PD. 2008. In vitro assembly of the Tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc. Natl. Acad. Sci. U. S. A. 105:19956–19961. 10.1073/pnas.0810851105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pathak KB, Sasvari Z, Nagy PD. 2008. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology 379:294–305. 10.1016/j.virol.2008.06.044 [DOI] [PubMed] [Google Scholar]
- 106.Zhang Z, Quick MK, Kanelakis KC, Gijzen M, Krishna P. 2003. Characterization of a plant homolog of hop, a cochaperone of hsp90. Plant Physiol. 131:525–535. 10.1104/pp.011940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dufresne PJ, Thivierge K, Cotton S, Beauchemin C, Ide C, Ubalijoro E, Laliberte JF, Fortin MG. 2008. Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 374:217–227. 10.1016/j.virol.2007.12.014 [DOI] [PubMed] [Google Scholar]
- 108.Kampmueller KM, Miller DJ. 2005. The cellular chaperone heat shock protein 90 facilitates Flock House virus RNA replication in Drosophila cells. J. Virol. 79:6827–6837. 10.1128/JVI.79.11.6827-6837.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mayer MP. 2005. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 153:1–46. 10.1007/s10254-004-0025-5 [DOI] [PubMed] [Google Scholar]
- 110.Okamoto T, Nishimura Y, Ichimura T, Suzuki K, Miyamura T, Suzuki T, Moriishi K, Matsuura Y. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 25:5015–5025. 10.1038/sj.emboj.7601367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tomita Y, Mizuno T, Diez J, Naito S, Ahlquist P, Ishikawa M. 2003. Mutation of host DnaJ homolog inhibits brome mosaic virus negative-strand RNA synthesis. J. Virol. 77:2990–2997. 10.1128/JVI.77.5.2990-2997.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weeks SA, Miller DJ. 2008. The heat shock protein 70 cochaperone YDJ1 is required for efficient membrane-specific flock house virus RNA replication complex assembly and function in Saccharomyces cerevisiae. J. Virol. 82:2004–2012. 10.1128/JVI.02017-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weeks SA, Shield WP, Sahi C, Craig EA, Rospert S, Miller DJ. 2010. A targeted analysis of cellular chaperones reveals contrasting roles for heat shock protein 70 in flock house virus RNA replication. J. Virol. 84:330–339. 10.1128/JVI.01808-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Taipale M, Jarosz DF, Lindquist S. 2010. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11:515–528. 10.1038/nrm2918 [DOI] [PubMed] [Google Scholar]
- 115.Mayer MP. 2010. Gymnastics of molecular chaperones. Mol. Cell 39:321–331. 10.1016/j.molcel.2010.07.012 [DOI] [PubMed] [Google Scholar]
- 116.Kampinga HH, Craig EA. 2010. The HSP70 chaperone machinery: J. proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11:579–592. 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fellerer C, Schweiger R, Schongruber K, Soll J, Schwenkert S. 2011. Cytosolic HSP90 cochaperones HOP and FKBP interact with freshly synthesized chloroplast preproteins of Arabidopsis. Mol. Plant. 4:1133–1145. 10.1093/mp/ssr037 [DOI] [PubMed] [Google Scholar]
- 118.Bhangoo MK, Tzankov S, Fan AC, Dejgaard K, Thomas DY, Young JC. 2007. Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol. Biol. Cell 18:3414–3428. 10.1091/mbc.E07-01-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Young JC, Hoogenraad NJ, Hartl FU. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112:41–50. 10.1016/S0092-8674(02)01250-3 [DOI] [PubMed] [Google Scholar]
- 120.Monkewich S, Lin HX, Fabian MR, Xu W, Na H, Ray D, Chernysheva OA, Nagy PD, White KA. 2005. The p92 polymerase coding region contains an internal RNA element required at an early step in Tombusvirus genome replication. J. Virol. 79:4848–4858. 10.1128/JVI.79.8.4848-4858.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pogany J, Nagy PD. 2012. p33-independent activation of a truncated p92 RNA-dependent RNA polymerase of tomato bushy stunt virus in yeast cell-free extract. J. Virol. 86:12025–12038. 10.1128/JVI.01303-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rajendran KS, Nagy PD. 2003. Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J. Virol. 77:9244–9258. 10.1128/JVI.77.17.9244-9258.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Panaviene Z, Baker JM, Nagy PD. 2003. The overlapping RNA-binding domains of p33 and p92 replicase proteins are essential for tombusvirus replication. Virology 308:191–205. 10.1016/S0042-6822(02)00132-0 [DOI] [PubMed] [Google Scholar]
- 124.Pathak KB, Jiang Z, Ochanine V, Sharma M, Pogany J, Nagy PD. 2013. Characterization of dominant-negative and temperature-sensitive mutants of tombusvirus replication proteins affecting replicase assembly. Virology 437:48–61. 10.1016/j.virol.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 125.Lee CT, Graf C, Mayer FJ, Richter SM, Mayer MP. 2012. Dynamics of the regulation of Hsp90 by the co-chaperone Sti1. EMBO J. 31:1518–1528. 10.1038/emboj.2012.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wegele H, Wandinger SK, Schmid AB, Reinstein J, Buchner J. 2006. Substrate transfer from the chaperone Hsp70 to Hsp90. J. Mol. Biol. 356:802–811. 10.1016/j.jmb.2005.12.008 [DOI] [PubMed] [Google Scholar]
- 127.Mine A, Okuno T. 2012. Composition of plant virus RNA replicase complexes. Curr. Opin. Virol. 2:669–675. 10.1016/j.coviro.2012.09.014 [DOI] [PubMed] [Google Scholar]
- 128.Chen L, Hamada S, Fujiwara M, Zhu T, Thao NP, Wong HL, Krishna P, Ueda T, Kaku H, Shibuya N, Kawasaki T, Shimamoto K. 2010. The Hop/Sti1-Hsp90 chaperone complex facilitates the maturation and transport of a PAMP receptor in rice innate immunity. Cell Host Microbe 7:185–196. 10.1016/j.chom.2010.02.008 [DOI] [PubMed] [Google Scholar]
- 129.Jones G, Song Y, Chung S, Masison DC. 2004. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol. Cell. Biol. 24:3928–3937. 10.1128/MCB.24.9.3928-3937.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Honore B, Leffers H, Madsen P, Rasmussen HH, Vandekerckhove J, Celis JE. 1992. Molecular cloning and expression of a transformation-sensitive human protein containing the TPR motif and sharing identity to the stress-inducible yeast protein STI1. J. Biol. Chem. 267:8485–8491 [PubMed] [Google Scholar]
- 131.Prasad BD, Goel S, Krishna P. 2010. In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in Arabidopsis and rice as putative co-chaperones of Hsp90/Hsp70. PLoS One 5:e12761. 10.1371/journal.pone.0012761 [DOI] [PMC free article] [PubMed] [Google Scholar]