ABSTRACT
Epstein-Barr virus (EBV) is a human herpesvirus associated with various tumors. Rather than going through the lytic cycle, EBV maintains latency by limiting the expression of viral genes in tumors. Viral microRNAs (miRNAs) of some herpesviruses have been reported to directly target immediate early genes and suppress lytic induction. In this study, we investigated whether BamHI-A rightward transcript (BART) miRNAs targeted two EBV immediate early genes, BZLF1 and BRLF1. Bioinformatic analysis predicted that 12 different BART miRNAs would target BRLF1. Of these, the results of a luciferase reporter assay indicated that only one interacted with the 3′ untranslated region (UTR) of BRLF1: miR-BART20-5p. miR-BART20-5p's effect on gene expression involved two putative seed match sites in the BRLF1 3′ UTR, but a mutant version of the miRNA, miR-BART20-5pm, had no effect on expression. As expected from the fact that the entire 3′ UTR of BZLF1 resides within the 3′ UTR of BRLF1, miR-BART20-5p interacted with the 3′ UTR of BZLF1 as well. BZLF1 and BRLF1 mRNA and protein expression was suppressed in cells of an AGS cell line infected with the recombinant Akata strain of EBV (AGS-EBV) transfected with a miR-BART20-5p mimic. The expression of various EBV early proteins was also suppressed by the miR-BART20-5p mimic. In contrast, BZLF1 and BRLF1 expression in AGS-EBV cells transfected with a miR-BART20-5p inhibitor was enhanced. Furthermore, progeny virus production was suppressed by the miR-BART20-5p mimic and enhanced by the miR-BART20-5p inhibitor in AGS-EBV cells induced for the lytic cycle. Our data suggest that miR-BART20-5p plays a key role in latency maintenance in EBV-associated tumors by directly targeting immediate early genes.
IMPORTANCE Herpesviruses maintain latency using various mechanisms and establish lifelong infection in the host. From time to time, herpesviruses are reactivated and express immediate early genes which trigger a lytic cascade, leading to the production of progeny viruses. Recently, some herpesviruses have been shown to use their own microRNAs (miRNAs) to downregulate immediate early genes to inhibit the lytic cycle. This study presents evidence that EBV also downregulates two immediate early genes by miR-BART20-5p to suppress the lytic cycle and progeny virus production. Overall, this is the first study to report the direct regulation of EBV immediate early genes by an EBV miRNA, implying its likely importance in latency maintenance in EBV-associated tumors.
INTRODUCTION
Epstein-Barr virus (EBV) is a gammaherpesvirus associated with a variety of malignancies, such as Burkitt's lymphoma, Hodgkin's disease, nasal natural killer/T-cell lymphoma (NNL), and gastric carcinoma (1). All herpesviruses, including EBV, have two distinct life cycle phases: latency and lytic replication (2, 3).
Following primary infection, herpesviruses establish persistent latent infection of host cells; by expressing only a few viral genes, they try to avoid host immune surveillance (4, 5). Herpesvirus microRNAs (miRNAs) regulate latency by inhibiting the expression of immediate early genes. For example, herpes simplex virus (HSV) immediate early genes include ICP4, ICP0, ICP27, ICP22, and ICP47 (6). miR-H2 of HSV-1 and HSV-2 downregulates ICP0 expression, while miR-H6 of HSV-1 inhibits ICP4 expression (7). Similarly, human cytomegalovirus (HCMV) miR-UL112-1 targets its immediate early gene IE1, inhibiting viral lytic replication (8). The important transcriptional activators Rta and Zta are encoded by immediate early genes of gammaherpesviruses, including Kaposi's sarcoma-associated herpesvirus (KSHV) and EBV (9). In KSHV, miR-K7, miR-K5, and miR-K9* target ORF50 (Rta) expression to maintain a latent state (10–13).
In EBV, expression of the immediate early genes BZLF1 (which encodes Zta) and BRLF1 (which encodes Rta) can trigger lytic replication (14–18). BZLF1 and BRLF1 are transcribed from two different immediate early promoters, Zp and Rp, respectively. BZLF1 can also be transcribed from Rp as a long bicistronic transcript, as the entire BZLF1 coding sequence as well as its 3′ untranslated region (UTR) resides within the 3′ UTR of BRLF1 (19). However, translation of the BZLF1 gene of the bicistronic mRNA is not very efficient (19). Both BZLF1 and BRLF1 are required for full expression of the early and late EBV proteins (20, 21), leading to the production of progeny virus. Furthermore, these immediate early proteins upregulate each other as well as themselves (14, 22–24).
The expression of BZLF1 and BRLF1 is tightly regulated to maintain latency, and a number of cellular and viral factors play important roles in Zp and Rp regulation (20, 21). Epithelial-to-mesenchymal transition regulators ZEB1 and ZEB2 can bind to the inhibitory elements in Zp, blocking the expression of the gene (25–27). Recently, miR-200b and miR-429 were shown to induce EBV lytic replication in both EBV-infected epithelial and B cells (28). The effects of these cellular miRNAs were exerted by directly targeting ZEB1 and ZEB2 to block their repressing activity on Zp (28). Lin et al. (29) also reported that miR-429 expression in EBV-infected fibroblasts and B cells triggered the lytic cycle through the repression of ZEB1 and the activation of Zp.
EBV generates 44 different mature miRNAs, categorized into BamHI fragment H rightward open reading frame 1 (BHRF1) miRNAs and BamHI-A rightward transcript (BART) miRNAs (30–33). There have been several reports of viral miRNAs directly or indirectly regulating the EBV lytic cycle. For example, in C666-1 and Mutu cells transfected with a miR-BART6-5p antagomir, BZLF1 and BRLF1 mRNA levels increased significantly (34). miR-BART6-5p is thought to suppress expression of EBV immediate early genes indirectly, via silencing effects on Dicer; silencing of Dicer in these cells with a small hairpin RNA (shRNA) substantially downregulated BZLF1 and BRLF1 (34). Others have reported that miR-BHRF1-1 overexpression in SUNE1, a nasopharyngeal carcinoma (NPC) cell line, increases the EBV copy number, following the accumulation of the BZLF1 and BMRF1 lytic proteins (early antigen diffuse component [Ea-D]) (35). In addition, a miR-BHRF1-1 inhibitor reduces BZLF1 and BMRF1 expression in cells in which EBV lytic replication has been induced with 12-O-tetradecanoylphorbol-13-acetate (TPA) (35). The mechanism by which miR-BHRF1-1 regulates the EBV lytic cycle is poorly understood (35). miR-BART2, which is antisense to the 3′ UTR of BALF5, directly targets this EBV DNA polymerase and reduces virus production (36). However, whether individual EBV miRNAs can directly regulate the expression of EBV immediate early genes remains unclear. In this study, we tested whether BART miRNAs which are expressed in all EBV-infected cells can target two EBV immediate early genes, BZLF1 and BRLF1.
MATERIALS AND METHODS
Cell lines and culture conditions.
AGS is an EBV-negative gastric cancer cell line, while SNU-719 and YCCEL1 are gastric carcinoma cell lines naturally infected with EBV (37–39). They were cultured in RPMI 1640 (AGS and SNU-719) or Eagle's minimal essential medium (YCCEL1) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. AGS-EBV is an AGS cell line infected with the recombinant Akata strain of EBV (40, 41). To culture AGS-EBV cells, 400 μg/ml of G418 (Gibco, Carlsbad, CA) was also added to the medium. The human embryonic kidney cell line HEK293T was cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. All cells were incubated at 37°C and supplemented with 5% CO2.
Transfection and TPA treatment.
The miR-BART20-5p mimic and scrambled control were purchased from Genolution Pharmaceuticals (Seoul, South Korea). The locked nucleic acid (LNA)-miR-BART20-5p inhibitor (LNA-miR-BART20-5pi) and negative-control LNA-miRNA inhibitor were purchased from Exiqon (Vedbaek, Denmark). In this experiment, AGS-EBV and HEK293T cells were seeded 24 h prior to transfection in 100-mm-diameter dishes containing 10 ml culture medium. SNU-719 and YCCEL1 cells were seeded 24 h prior to transfection in a 6-well-plate containing 2 ml culture medium. The cells were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA), according to the manufacturer's protocol. After 24 h, cells were treated with 5 nM TPA for 48 or 72 h to induce the lytic cycle.
Plasmid constructs.
The full-length 3′ UTRs of BZLF1 and BRLF1 were amplified from the cDNA of AGS-EBV cells. The 3′ UTRs of BZLF1 and BRLF1 were then cloned into XhoI/NotI sites between the Renilla luciferase-coding sequence and the poly(A) site of the psiCHECK-2 plasmid (Promega, Madison, WI) to produce psiC-BZLF1 and psiC-BRLF1, respectively. Primers used for amplification were as follows: for BZLF1, 5′-TCGACTCGAGCGAGGATCTCTTAAATTTCTAACTCC-3′ and 5′-GGCCGCGGCCGCCAAAGAGAGCCGACAGGAAG-3′; for BRLF1, 5′-TCGACTCGAGGAGCCACAGGCATTGCTAA-3′ and 5′-GGCCGCGGCCGCCAAAGAGAGCCGACAGGAAG-3′. Mutations were introduced into the seed sequences of psiC-BZLF1 and psiC-BRLF1 using an EZchange site-directed mutagenesis kit (Enzynomics, Daejeon, South Korea). The primers used for this purpose were as follows: for BZLF1m1 and BRLF1m1, 5′-CACGCCTCGTTTACTAATGGAATATTAATAAATAT-3′ and 5′-ATCGAGCCGTGGTTTCAATAACG-3′; for BRLF1m2, 5′-CTGAGAATGCTTATCAAGCTTATGCAGCAC-3′ and 5′-GTCGAGCCTGAGGGGCAGGAAACCACG-3′. In order to exclude the effect of BZLF1, which is encoded within the 3′ UTR of BRLF1, in the luciferase assay for BRLF1, a −1 frameshift mutation was introduced. This was carried out by deleting 1 nucleotide from the start codon of BZLF1 within psiC-BRLF1 using EZchange site-directed mutagenesis. The primers used for this purpose were as follows: for psiC-BRLF1(ΔBZLF1), 5′-GGACCCAAACTCGACTTCTGAAGAT-3′ and 5′-TCATCTTCAGCAAAGATAGCAAAGGTC-3′.
Luciferase reporter assay.
To investigate the effect of miR-BART20-5p upon the expression of immediate early genes, HEK293T cells or AGS cells were seeded in a 96-well plate (5 × 103 cells/well). After 24 h, the cells were cotransfected with a psiCHECK reporter vector containing a BZLF1 or BRLF1 3′ UTR fragment and 10 nM miR-BART20-5p or a miR-BART20-5p mutant: miR-BART20-5pm. Luciferase activity was measured at 48 h posttransfection using a Dual-Glo luciferase reporter assay system (Promega). For each sample, Renilla luciferase activity was normalized using firefly luciferase activity.
Quantitative reverse transcription-PCR (qRT-PCR).
AGS-EBV cells were harvested, and total RNA was extracted using the RNAzol B reagent (Tel-Test, Friendswood, TX), according to the manufacturer's instructions. cDNA was synthesized using 3 μg total RNA, oligo(dT) primers (Macrogen, Seoul, South Korea), and Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen). Real-time PCR for the indicated genes was carried out using a SYBR green quantitative PCR (qPCR) kit (TaKaRa, Tokyo, Japan) with an Mx3000p real-time PCR system (Stratagene, La Jolla, CA). The sequences of the primers were as follows: for BZLF1, 5′-GGCTAACCAAGGACAACAGC-3′ and 5′-GAAGCCACCCGATTCTTGTA-3′; for BRLF1, 5′-GTGTTCCACAGCCTGCAC-3′ and 5′-GAAGCCACCCGATTCTTGTA-3′; and for GAPDH (the glyceraldehyde-3-phosphate dehydrogenase gene), 5′-ATGGGGAAGGTGAAGGTCG-3′ and 5′-GGGGTCATTGATGGCAACAATA-3′. PCR conditions were 95°C for 10 min, followed by 40 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. To confirm specific amplification of the PCR product, dissociation curves were checked routinely. For this, reaction mixtures were incubated at 95°C for 60 s and ramped from 60°C to 95°C at a heating rate of 0.1°C/s, with fluorescence measured continuously. Relative gene expression was calculated using the comparative threshold cycle (CT) method, using GAPDH as an internal standard.
Quantitative real-time PCR for miRNA analysis.
miRNA cDNA was synthesized using a Mir-X miRNA First-Strand synthesis kit (Clontech, Mountain View, CA) according to the manufacturer's instructions. Real-time quantitative PCRs were performed using a SYBR green qPCR kit (TaKaRa, Tokyo, Japan). Specific miRNA sequences in the cDNA were quantified using miRNA-specific sequences as 5′ primers. The forward primer used for miR-BART20-5p was 5′-TAGCAGGCATGTCTTCATTCC-3′. All amplifications were performed in triplicate, and values were normalized to the value for an endogenous control, U6, which was supplied in the kit.
Western blot analysis.
Cell lysate in radioimmunoprecipitation assay (RIPA) buffer (5 μg) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, and 10 μg/ml aprotinin) was mixed with loading buffer (5×) (Fermentas, Waltham, MA) and heated at 95°C for 5 min. Samples were separated electrophoretically on 8% sodium dodecyl sulfate (SDS)-polyacrylamide gels, and the separated proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA). The effect of miR-BART20-5p upon the expression of EBV-lytic proteins was assessed using anti-BZLF1 (1:500; Dako, Denmark), anti-BRLF1 (1:500; Argene, France), anti-BMRF1 (1:500; Novocastra, United Kingdom), anti-BHRF1 (1:250; monoclonal antibody 3E8), and anti-BALF5 (1:300) antibodies (42, 43). After washing, the blots were incubated with horseradish peroxidase-conjugated antimouse secondary antibodies or horseradish peroxidase-conjugated antirabbit secondary antibodies (Amersham Biosciences, Piscataway, NJ) at a dilution of 1:5,000 for 1 h at room temperature. Protein bands were visualized using an enhanced chemiluminescence detection system (Amersham Bioscience), and the membrane was exposed to X-ray film (Agfa, Mortsel, Belgium). α-Tubulin antibody (Cell Signaling Technology, MA) was used to confirm that loading was comparable between gel lanes. The density of each protein band was read and quantified using Fujifilm Multi Gauge software (version 3.0) (44).
siRNA knockdown of BZLF1 and BRLF1 expression.
Small interfering RNAs (siRNAs) specific for BZLF1 (siBZLF1) and BRLF1 (siBRLF1), as well as a negative-control siRNA lacking any known target gene product, were synthesized by Genolution Pharmaceuticals. The sequence of the negative-control siRNA was 5′-ACGUGACACGUUCGGAGAAUU-3′. The sequences of the siRNAs were as follows: for siBZLF1, 5′-CGACATAACCCAGAATCAACA-3′, and for siBRLF, 5′-ATCTTGGATACATTTCTAAATGATT-3′. AGS-EBV cells (1 × 106 cells/dish) were transfected with 10 nM siRNA using Lipofectamine 2000 (Invitrogen) in 100-mm-diameter dishes. Following transfection, lytic induction proceeded for 48 h. Then, cells were harvested to analyze BZLF1 and BRLF1 expression.
Quantitative PCR to assess EBV genome copy numbers.
AGS-EBV cells (1 × 106) were seeded in a 100-mm-diameter dish. After 24 h, the cells were transfected with the miR-BART20-5p mimic (10 nM) or LNA-miR-BART20-5p inhibitor (30 nM). After 24 h the cultures were refreshed with new RPMI 1640 medium that included 5 nM TPA. The cells were incubated at 37°C in a 5% CO2 incubator for 3 days, allowing the production of viral particles. Cells were harvested by centrifuging them for 5 min at 800 × g at room temperature, and the supernatant was then passed through a 0.45-μm-pore-size filter (Nalgene, Rochester, NY). Subsequently, the supernatant was ultracentrifuged using an SW41 rotor (Beckman Instruments, Fullerton, CA) at 75,000 × g for 2 h at 4°C, and the pellet was resuspended in 200 μl 0.2× phosphate-buffered saline and then heated to 95°C for 15 min. Next, 20 μl of 20 mg/ml proteinase K was added to the solution and the mixture was incubated at 56°C for 1 h, followed by heat inactivation at 95°C for 30 min. Real-time PCR amplification of EBNA-1 was carried out using a SYBR green qPCR kit (TaKaRa, Tokyo, Japan) with an Mx3000P real-time PCR system (Stratagene). The sequences of the EBNA-1 primers were 5′-AGTCGTCTCCCCTTTGGAAT-3′ and 5′-TCCTCACCCTCATCTCCATC-3′. The PCR conditions were 95°C for 30 s, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. For the dissociation curve, the reaction mixtures were incubated at 95°C for 60 s and then ramped from 55°C to 95°C at a heating rate of 0.1°C/s, with fluorescence being measured continuously. The relative viral copy number was calculated according to the comparative CT method.
Statistical analysis.
Data were analyzed using the Student t test. Curve fit and analysis were performed using GraphPad Prism software (GraphPad Software, San Diego, CA). P values of <0.05 were considered statistically significant. All results were expressed as means ± standard deviations (SDs).
RESULTS
Screening of BART miRNAs targeting EBV immediate early genes.
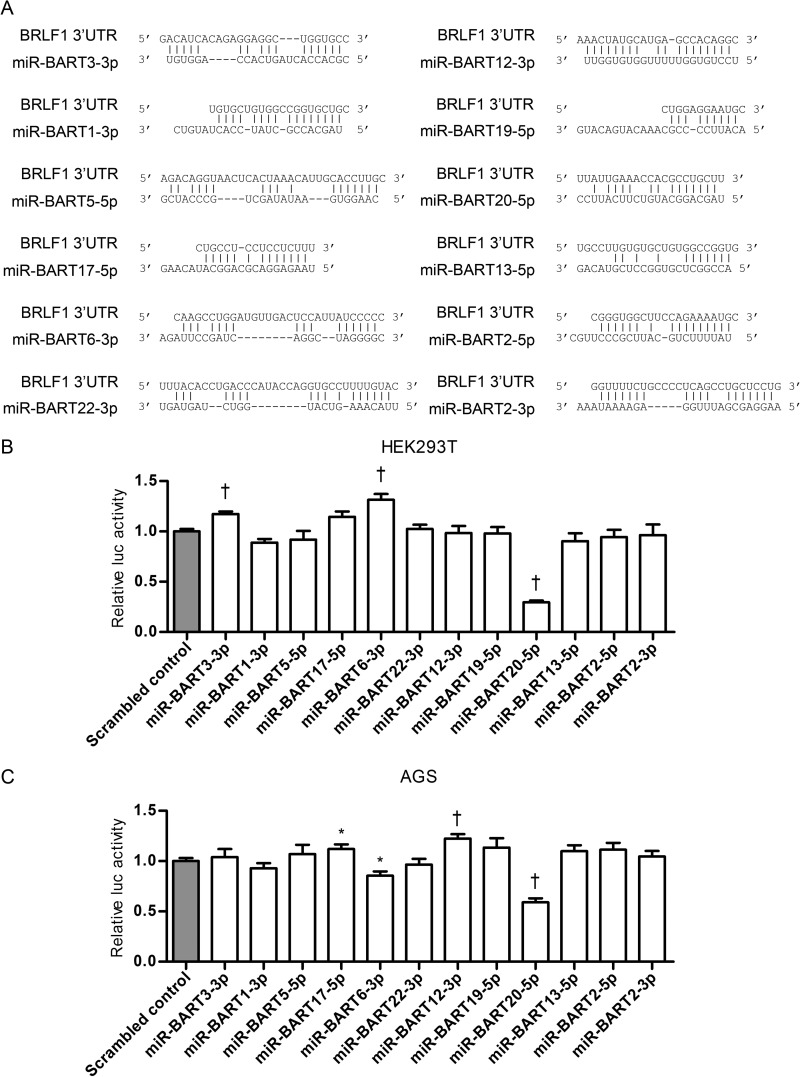
To examine whether BART miRNAs contained seed matches for the 3′ UTR of the EBV immediate early genes BZLF1 and BRLF1, we used the publicly available RNA hybrid program (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). Twelve BART miRNAs (miR-BART3-3p, miR-BART1-3p, miR-BART5-5p, miR-BART17-5p, miR-BART6-3p, miR-BART22-3p, miR-BART12-3p, miR-BART19-5p, miR-BART20-5p, miR-BART13-5p, miR-BART2-5p, and miR-BART2-3p) seed matched the BRLF1 3′ UTR (Fig. 1A).
FIG 1.
Screening of BART miRNAs targeting EBV immediate early genes. (A) Schematic overview of seed matches between 12 BART miRNAs and the 3′ UTR of BRLF1. (B, C) Luciferase (luc) activity in cells cotransfected with psiC-BRLF1 and BART miRNA mimics (n = 3 per experiment). HEK293T (B) or AGS (C) cells were cotransfected with each of the 12 BART miRNA mimics and psiC-BRLF1. Luciferase activity was normalized using internal firefly luciferase activity and expressed as a ratio of the luciferase activity to the activity obtained from the scrambled control-transfected cells. Error bars indicate SDs. *, P < 0.05; †, P < 0.01.
Next, we carried out a luciferase reporter assay to assess whether the 12 identified BART miRNAs targeted the 3′ UTR of BRLF1. First, HEK293T cells were cotransfected with each of the 12 BART miRNA mimics and a luciferase reporter vector, psiC-BRLF1. Only the miR-BART20-5p mimic was able to significantly reduce the luciferase activity of the reporter vector compared to that of the scrambled control (Fig. 1B). miR-BART20-5p suppressed luciferase activity when the experiment was carried out using AGS cells as well (Fig. 1C). Unexpectedly, the luciferase activity in HEK293T cells transfected with the miR-BART3-3p and miR-BART6-3p mimics increased by roughly 20 to 30% (Fig. 1B). However, these results were not repeated when AGS cells (Fig. 1C) instead of HEK293T were used. Thus, miR-BART20-5p was chosen for further investigation.
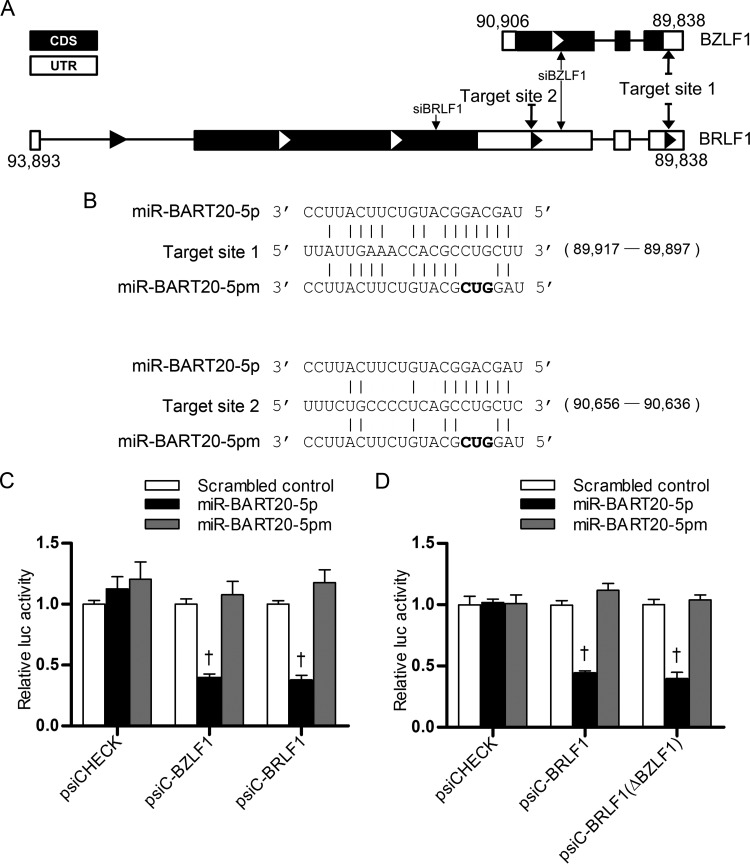
miR-BART20-5p directly targets both the BZLF1 and BRLF1 3′ UTRs.
BZLF1 is located between nucleotides 89,838 and 90,906 of the EBV genome, while BRLF1 is located between nucleotides 89,838 and 93,893 (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/). Thus, the whole BZLF1 sequence, including its 3′ UTR, resides within the 3′ UTR of BRLF1 (Fig. 2A). This makes miR-BART20-5p also seed match the 3′ UTR of BZLF1 (Fig. 2A). Luciferase activity was inhibited in HEK293T cells cotransfected with a miR-BART20-5p mimic and either psiC-BZLF1 or psiC-BRLF1 relative to that in cells cotransfected with the scrambled control and either reporter vector (Fig. 2C). In contrast, no alteration in luciferase activity was observed in HEK293T cells cotransfected with a reporter vector and miR-BART20-5pm, which contains mutations at nucleotides 4 to 6 of the miR-BART20-5p sequence (Fig. 2B and C).
FIG 2.
Luciferase activity assays testing if miR-BART20-5p targets the EBV immediate early genes BZLF1 and BRLF1. (A) Schematic drawing of the 3′ UTRs of BZFL1 and BRLF1. (B) Seed matches between miR-BART20-5p and the 3′ UTRs of BZLF1 and BRLF1. Nucleotides 4 to 6 of the miR-BART20-5p were mutated to create miR-BART20-5pm. Mutated sequences are written in bold. Coordinates of target site 1 and target site 2 are shown in parentheses (target site 1 was nucleotides 89,897 to 89,917 and target site 2 was nucleotides 90,636 to 90,656 in the sequence with GenBank accession number NC_007605.1). (C) Direct targeting of the BZLF1 and BRLF1 3′ UTRs by miR-BART20-5p. Luciferase activity was measured in HEK293T cells cotransfected with psiC-BZLF1 or psiC-BRLF1 and the miR-BART20-5p mimic. (D) Effect of BZLF1 on the luciferase activity of psiC-BRLF1. To avoid the possible expression of BZLF1 from the psiC-BRLF1 vector, a frameshift mutation was introduced at the start codon of BZLF1 to produce psiC-BRLF1(ΔBZLF1). Luciferase activity was measured in HEK293T cells cotransfected with psiC-BRLF1 or psiC-BRLF1(ΔBZLF1) and the miR-BART20-5p mimic. The scrambled control and miR-BART20-5pm were used to confirm sequence-specific binding between miR-BART20-5p and the 3′ UTRs. Luciferase activity was normalized using firefly luciferase activity and expressed as a ratio of the luciferase activity to the activity obtained from the scrambled control-transfected cells. Error bars indicate SDs (n = 3 per experiment). †, P < 0.01.
Even though translation of BZLF1 from the bicistronic transcript is not efficient, BZLF1 might be expressed from psiC-BRLF1 and affect the luciferase activity as part of Zp (nucleotides 90,906 to 91,106) as well as the whole BZLF1 sequence included in psiC-BRLF1. To avoid this complication, we introduced a −1 frameshift mutation at the start codon of BZLF1 in psiC-BRLF1 to produce psiC-BRLF1(ΔBZLF1). When the luciferase assay was carried out in HEK293T cells cotransfected with psiC-BRLF1(ΔBZLF1) and miR-BART20-5p, results comparable to those shown in Fig. 2C were obtained (Fig. 2D).
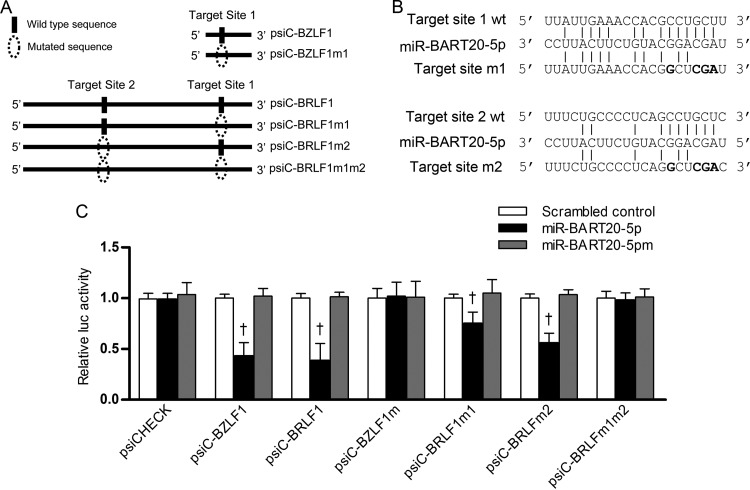
Target sites for miR-BART20-5p in the BZLF1 and BRLF1 3′ UTRs.
There is one additional seed match sequence (target site 2; Fig. 2A) in the 3′ UTR of BRLF1, in addition to the seed match sequence (target site 1) shown in Fig. 1A. Meanwhile, miR-BART20-5p possesses only one putative seed match sequence to the 3′ UTR of BZLF1 (target site 1). Site-directed mutagenesis was performed to produce mutant versions of 3′ UTR reporter vectors (Fig. 3A and B). Each of these vectors was cotransfected with the miR-BART20-5p mimic into HEK293T cells, and the luciferase assay was carried out. No alteration in luciferase activity was observed in cells transfected with the miR-BART20-5p mimic together with either psiC-BZLF1m1 or psiC-BRLF1m1m2 (Fig. 3C). However, luciferase activity was partially reduced in cells transfected with the miR-BART20-5p mimic together with either psiC-BRLF1m1 or psiC-BRLF1m2. The reduction in activity observed was less than that seen in cells cotransfected with the miR-BART20-5p mimic and wild-type psiC-BRLF1 (Fig. 3C). As expected, luciferase activity was not affected in cells cotransfected with wild-type or mutant BZLF1 and BRLF1 3′ UTR reporter vectors and miR-BART20-5pm or the scrambled control (Fig. 3C).
FIG 3.
Target site search for miR-BART20-5p in the BZLF1 and BRLF1 3′ UTRs. (A) Illustration showing (i) the location of possible seed match sites between miR-BART20-5p and the 3′ UTR regions and (ii) the sites altered to produce mutant forms of psiC-BZLF1 and psiC-BRLF1. Site-directed mutagenesis was performed to produce mutant versions of each individual 3′ UTR BRLF1 seed match sequence (psiC-BRLF1m1, psiC-BRLF1m2) and both sequences simultaneously (psiC-BRLF1m1m2). A mutant version of the seed match sequence in the BZLF1 3′ UTR was also produced (psic-BZLF1m1). (B) Seed matches between miR-BART20-5p and the mutated 3′ UTRs of BZLF1 and BRLF1. wt, wild type. (C) Target sites for miR-BART20-5p in the 3′ UTRs of BZLF1 and BRLF1. Luciferase activity was measured in HEK293T cells cotransfected with the miR-BART20-5p mimic and a luciferase reporter vector containing the wild-type or mutated 3′ UTRs of BZLF1 and BRLF1. A scrambled control and miR-BART20-5pm were used to confirm sequence-specific binding between miR-BART20-5p and the 3′ UTRs. Luciferase activity was normalized using firefly luciferase activity and expressed as a ratio of the luciferase activity to the activity obtained from the scrambled control-transfected cells. Error bars indicate SDs (n = 3 per experiment). †, P < 0.01.
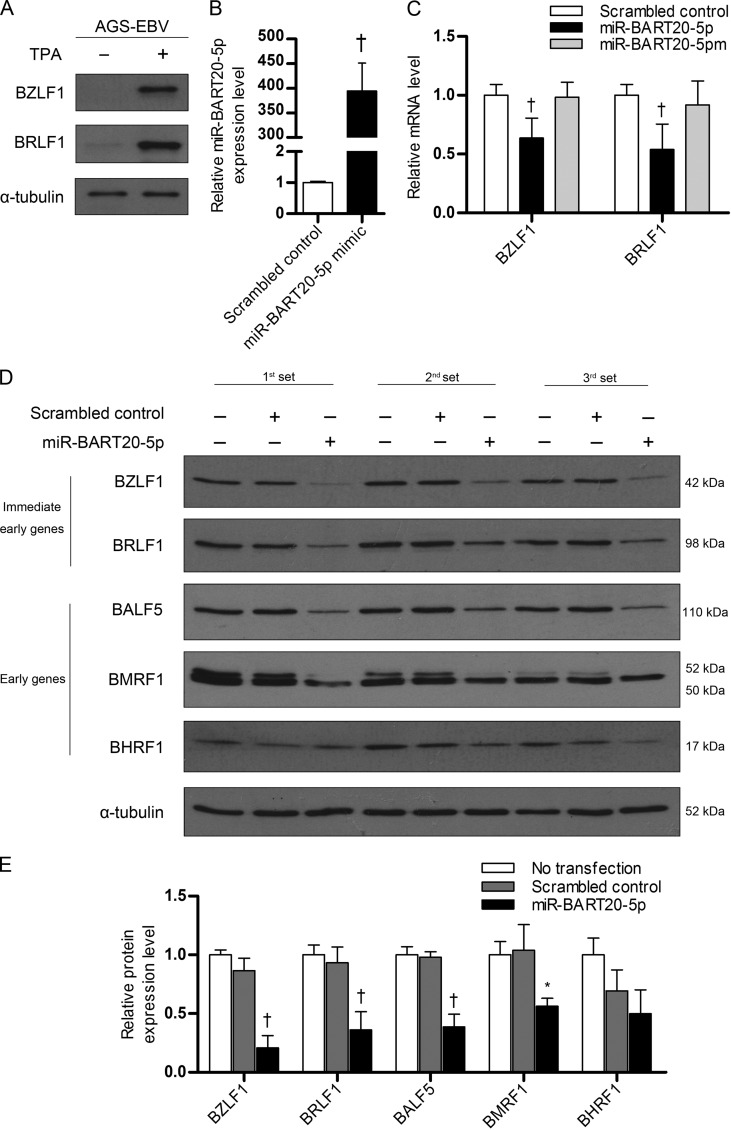
miR-BART20-5p inhibits BZLF1 and BRLF1 mRNA and protein expression.
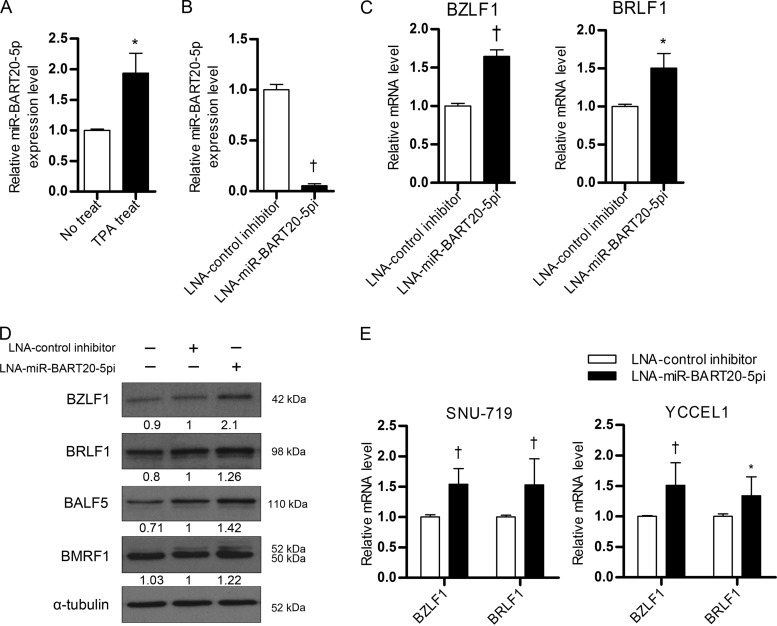
We examined whether miR-BART20-5p modulates BZLF1 and BRLF1 expression in AGS-EBV cells. TPA treatment was used to induce the EBV lytic cycle, as AGS-EBV cells did not express lytic genes in the basal state. Lytic induction by TPA treatment was confirmed via Western blotting, which demonstrated the presence of the BZLF1 and BRLF1 proteins (Fig. 4A). AGS-EBV cells were transfected with the miR-BART20-5p mimic and treated with TPA. After lytic induction, we compared the levels of miR-BART20-5p in the mimic-transfected and the scrambled control-transfected AGS-EBV cells. The level of miR-BART20-5p was increased over 400-fold by transfection of the mimic compared with the level achieved by transfection of the scrambled control (Fig. 4B).
FIG 4.
Effect of miR-BART20-5p on BZLF1 and BRLF1 mRNA and protein levels. (A) Induction of lytic gene expression by TPA treatment. To induce the EBV lytic cycle, AGS-EBV cells were treated with 5 nM TPA for 48 h, and EBV immediate early gene products were detected by Western blotting. Anti-α-tubulin antibody (1:500) was used to demonstrate comparable loading between lanes. (B) Relative level of miR-BART20-5p in mimic-transfected cells. AGS-EBV cells were transfected with 10 nM the miR-BART20-5p mimic or the scramble control. At 24 h after transfection, the cells were treated with 5 nM TPA. Real-time RT-PCR was carried out 48 h later to assess the level of miR-BART20-5p. (C) Reduction in BZLF1 and BRLF1 mRNA levels by miR-BART20-5p. AGS-EBV cells were transfected with the miR-BART20-5p mimic, miR-BART20-5pm, or the scrambled control. The cells were then treated with TPA for 48 h and harvested for real-time RT-PCR. (D) Effect of mR-BART20-5p upon the expression of various EBV proteins. AGS-EBV cells were transfected with the miR-BART20-5p mimic or the scrambled control, treated with TPA for 48 h, and then harvested for analysis. Western blotting was carried out using three sets of independently transfected AGS-EBV cells. Untransfected AGS-EBV cells were also used for comparison. (E) The Western blotting results shown in panel D were normalized to those for α-tubulin and are expressed as ratios of the protein expression level to the level obtained from untransfected controls. Error bars indicate the SDs. *, P < 0.05; †, P < 0.01.
Cells were harvested after 48 h to detect the expression of BZLF1 and BRLF1. Real-time reverse transcription-PCR (RT-PCR) revealed that BZLF1 and BRLF1 mRNA levels were reduced by 35 to 45% following transfection with the miR-BART20-5p mimic (Fig. 4C). Similarly, Western blotting showed that the level of BZLF1 and BRLF1 expression was reduced by transfection of the miR-BART20-5p mimic relative to that achieved by transfection of a scrambled control (Fig. 4D and E). As early and late gene expression follows the expression of immediate early genes upon lytic reactivation, we examined whether the protein products of EBV early genes were also reduced by miR-BART20-5p. We found that BMRF1, BALF5, and BHRF1 levels were decreased in cells transfected with the miR-BART20-5p mimic relative to those in cells transfected with the scrambled control (Fig. 4C and D).
The EBV miR-BART20-5p inhibitor increases BZLF1 and BRLF1 mRNA and protein expression.
qRT-PCR results showed that the expression of miR-BART20-5p in AGS-EBV cells which were treated with TPA was about 2-fold higher than that in untreated AGS-EBV cells (Fig. 5A). When AGS-EBV cells were transfected with LNA-miR-BART20-5pi to deplete endogenous miR-BART20-5p, the level of miR-BART20-5p was reduced by over 90% compared to that in the control inhibitor-transfected cells (Fig. 5B).
FIG 5.
Effect of LNA-miR-BART20-5pi on BZLF1 and BRLF1 expression. (A) miR-BART20-5p expression before and after lytic induction. To induce the lytic cycle, AGS-EBV cells were treated with 5 nM TPA for 48 h. (B, C) Effect of LNA-miR-BART20-5pi on the level of miR-BART20-5p (B) and the BZLF1 and BRLF1 mRNAs (C). AGS-EBV cells were transfected with 30 nM LNA-miR-BART20-5pi. At 24 h after transfection, the cells were treated with 5 nM TPA. Real-time RT-PCR was carried out 48 h later to assess the level of miR-BART20-5p (B) or the BZLF1 and BRLF1 mRNAs (C). (D) Effect of LNA-miR-BART20-5pi on expression of various EBV proteins. The same sets of cells used for the assays whose results are presented in panels B and C were used for Western blotting. The numbers under each band represent the ratio of the level of expression to that for cells transfected with the control inhibitor. (E) Experiments similar to those described for panel C were also performed using naturally EBV-infected gastric carcinoma cell lines SNU-719 and YCCEL1. For SNU-719 and YCCEL1 cells, 50 nM LNA-miR-BART20-5pi was transfected. Error bars indicate SDs. *, P < 0.05; †, P < 0.01.
BZLF1 and BRLF1 mRNA levels and the corresponding protein levels were then measured after lytic induction in cells transfected with the LNA inhibitor. BZLF1 and BRLF1 mRNA levels increased by 50% after treatment with LNA-miR-BART20-5pi (Fig. 5C). Western blotting revealed that expression of the two immediate early proteins, as well as BALF5 and BMRF1, increased following transfection with LNA-miR-BART20-5pi (Fig. 5D). The mRNA levels of BZLF1 and BRLF1 were also increased by LNA-miR-BART20-5pi transfection compared to the levels achieved by control LNA transfection in naturally EBV-infected gastric carcinoma cell lines, SNU-719 and YCCEL1 (Fig. 5E).
miR-BART20-5p reduced EBV particle production.
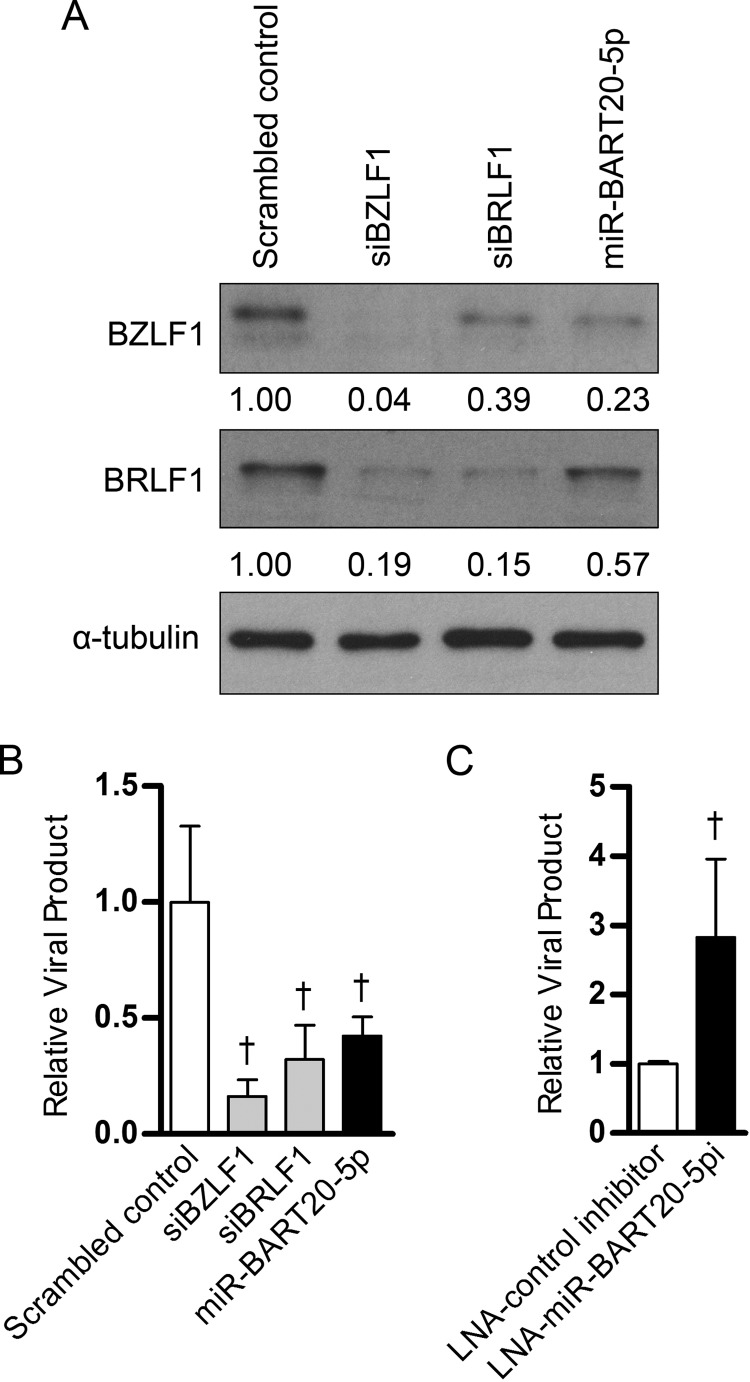
We investigated whether miR-BART20-5p functions in regulating the EBV life cycle by targeting both BZLF1 and BRLF1. First, BZLF1 and BRLF1 protein levels in AGS-EBV cells transfected with siBZLF1 or siBRLF1 were assessed following TPA treatment. Cells transfected with miR-BART20-5p were used for comparison. Western blotting revealed that BRLF1 protein levels were reduced by siBRLF1 transfection (Fig. 6A). Interestingly, siBRLF1 also suppressed BZLF1 expression to some extent. As expected from the fact that the entire BZLF1 sequence is contained within the BRLF1 sequence, both BZLF1 and BRLF1 protein levels were reduced by siBZLF1 transfection (Fig. 6A). As shown in Fig. 4, transfection with the miR-BART20-5p mimic also reduced BZLF1 and BRLF1 protein levels, but the magnitude of the reduction was less than that observed after transfection with the siRNAs (Fig. 6A).
FIG 6.
Effect of miR-BART20-5p and LNA-miR-BART20-5pi on progeny virus production. (A) Effect of siRNAs targeting BZLF1 (siBZLF1) and BRLF1 (siBRLF1) on protein expression. AGS-EBV cells were transfected with the miR-BART20-5p mimic, siBZLF1, siBRLF1, or the scrambled control. After 24 h, lytic replication was induced in AGS-EBV cells with 5 nM TPA. Protein was extracted from the cell pellet 48 h after the lytic induction and assayed by Western blotting. (B) Effect of the miR-BART20-5p mimic, siBZLF1, and siBRLF1 on the production of progeny virus. AGS-EBV cells were transfected with 10 nM the miR-BART20-5p mimic or siRNAs. After 24 h, lytic replication was induced with 5 nM TPA. Viral particles were harvested from the cell culture medium 72 h after the lytic induction and used for the detection of the EBV genome by real-time PCR. The results are shown as ratios of the amount of produced to the amount obtained from the scrambled control-transfected cells. (C) Effects of LNA-miR-BART20-5pi on the production of progeny. AGS-EBV cells were transfected with 30 nM LNA-miR-BART20-5pi and then processed under the same conditions described in the legend to panel B. Error bars indicate SDs. †, P < 0.01.
Next, the effect of miR-BART20-5p on the production of progeny virus production was analyzed in cells transfected with the miR-BART20-5p mimic or the LNA-miR-BART20-5p inhibitor. AGS-EBV cells transfected with siBZLF1, siBRLF1, and the miR-BART20-5p mimic showed a reduction in viral DNA levels of over 50% relative to those in cells transfected with control miRNA (Fig. 6B). In contrast, AGS-EBV cells transfected with LNA-miR-BART20-5pi showed a roughly 2.5-fold increase in the amount of EBV DNA relative to that in cells transfected with the control inhibitor (Fig. 6C).
DISCUSSION
In this study, we found that miR-BART20-5p regulates the expression of the EBV immediate early genes BZLF1 and BRLF1. Results from a luciferase assay using fusions between the luciferase gene and the 3′ UTR of BZLF1 or BRLF1 indicated that miR-BART20-5p directly targets the 3′ UTRs of these genes. miR-BART20-5p reduced the mRNA levels as well as the protein levels of BZLF1 and BRLF1 in TPA-treated AGS-EBV cells. Since BRLF1 and BZLF1 are part of a bicistronic transcript sharing part of the 3′ UTR (45), it is not surprising that miR-BART20-5p targets both mRNAs. miR-BART20-5p seems to have only one binding site (target site 1) in the BZLF1 3′ UTR, as the suppressive effect of the miR-BART20-5p mimic on luciferase activity was completely abolished when this seed match site was mutated. In contrast, miR-BART20-5p appeared to bind to both potential seed match sites in the BRLF1 3′ UTR, as luciferase activity was reduced but not completely abolished when either site was mutated, while it was abolished when both sites were mutated simultaneously. However, the interaction between miR-BART20-5p and target site 1 seemed to be more important than that between miR-BART20-5p and target site 2, as miR-BART20-5p reduced the luciferase activity of psiC-BRLF1m2 more than that of psiC-BRLF1m1 (Fig. 3C).
We found that siBZLF1 inhibited the expression of not only BZLF1 but also BRLF1 (Fig. 6A). This was expected because the whole BZLF1 sequence resides within the 3′ UTR of BRLF1, as depicted in Fig. 2A. However, siBRLF1 suppressed the expression of BZLF1 efficiently as well as strongly inhibited BRLF1 expression, even though siBRLF1 does not directly bind to BZLF1 mRNA. The fact that BZLF1 and BRLF1 can reciprocally induce each other (14, 22–24) may be related to this observation.
In contrast to our findings, which suggest posttranscriptional regulation of BZLF1 and BRLF1 by miR-BART20-5p, Seto et al. (46) have reported that BZLF1 expression does not differ among lymphoblastoid cell lines (LCLs) infected with wild-type EBV, mutant EBV devoid of any EBV miRNAs, or mutant EBV devoid of BHRF1 miRNA. They also found that BZLF1 expression was not altered in LCLs infected with a reconstituted mutant EBV in which an expression cassette encoding all BART miRNAs was introduced into the prototype B95-8 EBV genome by homologous recombination. This discrepancy may be due to differences between our experimental system and their experimental system. First, the cell types used in the experiments were different. We employed an EBV-infected gastric carcinoma cell line, AGS-EBV, while they used LCLs established by infecting B cells with EBV. The effect of miR-BART20-5p upon BZLF1 and BRLF1 expression could differ depending on the cell type. Second, the range of EBV miRNAs investigated differed between the two experiments. Seto et al. (46) examined the combined effect of all EBV miRNAs or all BART miRNAs, while we tested the effect of individual BART miRNAs. In their study, the effect of individual EBV miRNAs, such as miR-BART20-5p, could have been masked by the counterbalancing effects of other miRNAs. Third, the state of the cells used differed between experiments. In our experiment, TPA was used to induce the lytic cycle before the effect of miR-BART20-5p upon immediate early gene expression was investigated; in our hands, AGS-EBV cells showed very low levels of lytic gene expression at the baseline. In contrast, Seto et al. (46) did not artificially induce the EBV lytic cycle during their experiments. Finally, the effect of BART miRNAs upon BZLF1 expression may not have been detectable if BART miRNA expression in the LCLs was not high enough. Seto et al. (46) mentioned that the steady-state levels of miRNAs absent in B95-8-derived EBVs (including miR-BART20-5p) were considerably lower in EBV-infected LCLs reconstituted with BART miRNAs than in control JM LCL cells.
In our study, a high level of miRNA was achieved by transfecting the miR-BART20-5p mimic. As previously reported and shown here, we detected very low levels of expression of the BZLF1 protein in SNU-719 cells (37) and YCCEL1 cells (39) as well as in AGS-EBV cells (47, 48) in the basal state. These observations suggest that, in addition to miR-BART20-5p, these EBV-infected gastric carcinoma cells may express low levels of miR-200b and miR-429 and/or high levels of ZEB1 and ZEB2. Indeed, downregulation of the miR-200 family, including miR-200b and miR-429, has been observed in EBV-associated gastric carcinomas (49, 50).
Cells transfected with the miR-BART20-5p mimic showed levels of miR-BART20-5p about 400-fold higher than the endogenous level found in AGS-EBV cells. A relatively low basal level of miR-BART20-5p in a variety of EBV-infected cells, including AGS-EBV cells (50, 51), could be the main reason for this big fold increase. In addition, the effects of miR-BART20-5p on BZLF1 and BRLF1 appeared to be undistorted by this high level achieved by mimic transfection. It is because the LNA inhibitor increased the expression of BZLF1 and BRLF1 in AGS-EBV, SNU-719, and YCCEL1 cells, implying that miR-BART20-5p functions at endogenous levels in these cell lines.
When LNA-miR-BART20-5pi was transfected into AGS-EBV cells, the level of miR-BART20-5p was reduced about 90%, as shown in Fig. 5B. However, the mRNA and protein levels of BZLF1 and BRLF1 were increased about 1.25- to 2-fold in these cells. Considering that the endogenous expression level of miR-BART20-5p in AGS-EBV cells is relatively low (50, 51), the effect of the miRNA inhibitor on BZLF1 and BRLF1 expression is not trivial. Furthermore, as miRNAs are known to finely modulate gene expression, blocking an miRNA nearly completely with an inhibitor may not cause a phenomenal increase in target gene expression.
AGS and AGS-EBV cells were reported to be persistently infected with parainfluenza virus type 5 (PIV5; formerly known as simian virus 5 [SV5]) (52). We have not tested whether the cells that we used are also persistently infected with PIV5. Even if the cells were chronically infected with PIV5, it does not seem to affect our conclusion qualitatively, as effects similar to those of LNA-miR-BART20-5pi in AGS-EBV cells were also observed in two other cell lines (SNU-719 and YCCEL1) which were derived independently (38, 39).
Our findings may prove helpful with regard to cancer treatment. Previously, miR-BART20-5p has been shown to target T-bet, the master transcription factor for cytotoxic natural killer (NK) cells, resulting in secondary suppression of p53 in NNL (53). Recently, miR-BART20-5p was shown to target IFNG (gamma interferon [IFN-γ]) as well (54). On the basis of these results, an association between invasive NNL pathogenesis and the inhibition of T-bet and IFNG (IFN-γ) by miR-BART20-5p has been suggested (53, 54). Thus, miR-BART20-5p inhibitors may induce T-bet, IFNG (IFN-γ), and p53 expression as well as EBV lytic antigens in EBV-associated tumors. As BART miRNAs are expressed in all EBV-infected cells regardless of the EBV latency type, targeting of miR-BART20-5p would increase chemosensitivity and susceptibility to immune attack for all varieties of EBV-associated tumors.
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (2012R1A1A2008813), and by the GRRC program of Gyeonggi Province (2013-B06, Development of biomedical lead compounds using RNAi).
Footnotes
Published ahead of print 4 June 2014
REFERENCES
- 1.Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757–768. 10.1038/nrc1452 [DOI] [PubMed] [Google Scholar]
- 2.Greene W, Kuhne K, Ye F, Chen J, Zhou F, Lei X, Gao SJ. 2007. Molecular biology of KSHV in relation to AIDS-associated oncogenesis. Cancer Treat. Res. 133:69–127. 10.1007/978-0-387-46816-7_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller G, El-Guindy A, Countryman J, Ye J, Gradoville L. 2007. Lytic cycle switches of oncogenic human gammaherpesviruses. Adv. Cancer Res. 97:81–109. 10.1016/S0065-230X(06)97004-3 [DOI] [PubMed] [Google Scholar]
- 4.Boss IW, Renne R. 2011. Viral miRNAs and immune evasion. Biochim. Biophys. Acta 1809:708–714. 10.1016/j.bbagrm.2011.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cullen BR. 2009. Viral and cellular messenger RNA targets of viral microRNAs. Nature 457:421–425. 10.1038/nature07757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung P, Panning B, Smiley JR. 1997. Herpes simplex virus immediate-early proteins ICP0 and ICP4 activate the endogenous human alpha-globin gene in nonerythroid cells. J. Virol. 71:1784–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umbach J, Kramer M, Jurak I, Karnowski H, Coen D, Cullen B. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780–783. 10.1038/nature07103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. 2008. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc. Natl. Acad. Sci. U. S. A. 105:5453–5458. 10.1073/pnas.0711910105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller G, El-Guindy A, Countryman J, Ye J, Gradoville L. 2007. Lytic cycle switches of oncogenic human gammaherpesviruses. Adv. Cancer Res. 97:81–109. 10.1016/S0065-230X(06)97004-3 [DOI] [PubMed] [Google Scholar]
- 10.Lin X, Liang D, He Z, Deng Q, Robertson E, Lan K. 2011. miR-K12-7-5p encoded by Kaposi's sarcoma-associated herpesvirus stabilizes the latent state by targeting viral ORF50/RTA. PLoS One 6:e16224. 10.1371/journal.pone.0016224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellare P, Ganem D. 2009. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6:570–575. 10.1016/j.chom.2009.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. 2010. Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J. Virol. 84:2697–2706. 10.1128/JVI.01997-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan Q, Ma X, Shen C, Cao X, Feng N, Qin D, Zeng Y, Zhu J, Gao SJ, Lu C. 2014. Inhibition of Kaposi's sarcoma-associated herpesvirus lytic replication by HIV-1 Nef and cellular microRNA Hsa-miR-1258. J. Virol. 88:4987–5000. 10.1128/JVI.00025-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ragoczy T, Heston L, Miller G. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978–7984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardwick JM, Lieberman PM, Hayward SD. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Countryman J, Miller G. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. U. S. A. 82:4085–4089. 10.1073/pnas.82.12.4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zalani S, Holley-Guthrie E, Kenney S. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. U. S. A. 93:9194–9199. 10.1073/pnas.93.17.9194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu TY, Chang Y, Wang PW, Liu MY, Chen MR, Chen JY, Tsai CH. 2005. Reactivation of Epstein-Barr virus can be triggered by an Rta protein mutated at the nuclear localization signal. J. Gen. Virol. 86:317–322. 10.1099/vir.0.80556-0 [DOI] [PubMed] [Google Scholar]
- 19.Manet E, Gruffat H, Trescol-Biemont MC, Moreno N, Chambard P, Giot JF, Sergeant A. 1989. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 8:1819–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenney SC, Mertz JE. 2014. Regulation of the latent-lytic switch in Epstein-Barr virus. Semin. Cancer Biol. 26C:60–68. 10.1016/j.semcancer.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C, Li D, Guo N. 2009. Regulation of cellular and viral protein expression by the Epstein-Barr virus transcriptional regulator Zta: implications for therapy of EBV associated tumors. Cancer Biol. Ther. 8:987–995. 10.4161/cbt.8.11.8369 [DOI] [PubMed] [Google Scholar]
- 22.Feederle R, Kost M, Baumann M, Janz A, Drouet E, Hammerschmidt W, Delecluse HJ. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080–3089. 10.1093/emboj/19.12.3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinclair AJ, Brimmell M, Shanahan F, Farrell PJ. 1991. Pathways of activation of the Epstein-Barr virus productive cycle. J. Virol. 65:2237–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu P, Speck SH. 2003. Synergistic autoactivation of the Epstein-Barr virus immediate-early BRLF1 promoter by Rta and Zta. Virology 310:199–206. 10.1016/S0042-6822(03)00145-4 [DOI] [PubMed] [Google Scholar]
- 25.Feng WH, Kraus RJ, Dickerson SJ, Lim HJ, Jones RJ, Yu X, Mertz JE, Kenney SC. 2007. ZEB1 and c-Jun levels contribute to the establishment of highly lytic Epstein-Barr virus infection in gastric AGS cells. J. Virol. 81:10113–10122. 10.1128/JVI.00692-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraus RJ, Perrigoue JG, Mertz JE. 2003. ZEB negatively regulates the lytic-switch BZLF1 gene promoter of Epstein-Barr virus. J. Virol. 77:199–207. 10.1128/JVI.77.1.199-207.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu X, Wang Z, Mertz JE. 2007. ZEB1 regulates the latent-lytic switch in infection by Epstein-Barr virus. PLoS Pathog. 3:e194. 10.1371/journal.ppat.0030194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellis-Connell AL, Iempridee T, Xu I, Mertz JE. 2010. Cellular microRNAs 200b and 429 regulate the Epstein-Barr virus switch between latency and lytic replication. J. Virol. 84:10329–10343. 10.1128/JVI.00923-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Z, Wang X, Fewell C, Cameron J, Yin Q, Flemington EK. 2010. Differential expression of the miR-200 family microRNAs in epithelial and B cells and regulation of Epstein-Barr virus reactivation by the miR-200 family member miR-429. J. Virol. 84:7892–7897. 10.1128/JVI.00379-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen SJ, Chen GH, Chen YH, Liu CY, Chang KP, Chang YS, Chen HC. 2010. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One 5:e12745. 10.1371/journal.pone.0012745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. 10.1371/journal.ppat.0020023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu JY, Pfuhl T, Motsch N, Barth S, Nicholls J, Grasser F, Meister G. 2009. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J. Virol. 83:3333–3341. 10.1128/JVI.01689-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim do N, Chae HS, Oh ST, Kang JH, Park CH, Park WS, Takada K, Lee JM, Lee WK, Lee SK. 2007. Expression of viral microRNAs in Epstein-Barr virus-associated gastric carcinoma. J. Virol. 81:1033–1036. 10.1128/JVI.02271-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iizasa H, Wulff BE, Alla NR, Maragkakis M, Megraw M, Hatzigeorgiou A, Iwakiri D, Takada K, Wiedmer A, Showe L, Lieberman P, Nishikura K. 2010. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J. Biol. Chem. 285:33358–33370. 10.1074/jbc.M110.138362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Chen X, Li L, Liu S, Yang L, Ma X, Tang M, Bode AM, Dong Z, Sun L, Cao Y. 2012. EBV encoded miR-BHRF1-1 potentiates viral lytic replication by downregulating host p53 in nasopharyngeal carcinoma. Int. J. Biochem. Cell Biol. 44:275–279. 10.1016/j.biocel.2011.11.007 [DOI] [PubMed] [Google Scholar]
- 36.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA. 2008. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 36:666–675. 10.1093/nar/gkm1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh ST, Seo JS, Moon UY, Kang KH, Shin DJ, Yoon SK, Kim WH, Park JG, Lee SK. 2004. A naturally derived gastric cancer cell line shows latency I Epstein-Barr virus infection closely resembling EBV-associated gastric cancer. Virology 320:330–336. 10.1016/j.virol.2003.12.005 [DOI] [PubMed] [Google Scholar]
- 38.Park JG, Yang HK, Kim WH, Chung JK, Kang MS, Lee JH, Oh JH, Park HS, Yeo KS, Kang SH, Song SY, Kang YK, Bang YJ, Kim YH, Kim JP. 1997. Establishment and characterization of human gastric carcinoma cell lines. Int. J. Cancer 70:443–449. [DOI] [PubMed] [Google Scholar]
- 39.Kim do N, Seo MK, Choi H, Kim SY, Shin HJ, Yoon AR, Tao Q, Rha SY, Lee SK. 2013. Characterization of naturally Epstein-Barr virus-infected gastric carcinoma cell line YCCEL1. J. Gen. Virol. 94:497–506. 10.1099/vir.0.045237-0 [DOI] [PubMed] [Google Scholar]
- 40.Borza CM, Hutt-Fletcher LM. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594–599. 10.1038/nm0602-594 [DOI] [PubMed] [Google Scholar]
- 41.Huang J, Liao G, Chen H, Wu FY, Hutt-Fletcher L, Hayward GS, Hayward SD. 2006. Contribution of C/EBP proteins to Epstein-Barr virus lytic gene expression and replication in epithelial cells. J. Virol. 80:1098–1109. 10.1128/JVI.80.3.1098-1109.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsurumi T, Kobayashi A, Tamai K, Daikoku T, Kurachi R, Nishiyama Y. 1993. Functional expression and characterization of the Epstein-Barr virus DNA polymerase catalytic subunit. J. Virol. 67:4651–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou SP, Tsai CH, Li LY, Liu MY, Chen JY. 2004. Characterization of monoclonal antibody to the Epstein-Barr virus BHRF1 protein, a homologue of Bcl-2. Hybrid. Hybridomics 23:29–37. 10.1089/153685904322772006 [DOI] [PubMed] [Google Scholar]
- 44.Yang H, Higgins B, Kolinsky K, Packman K, Go Z, Iyer R, Kolis S, Zhao S, Lee R, Grippo JF, Schostack K, Simcox ME, Heimbrook D, Bollag G, Su F. 2010. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res. 70:5518–5527. 10.1158/0008-5472.CAN-10-0646 [DOI] [PubMed] [Google Scholar]
- 45.Chang PJ, Liu ST. 2001. Function of the intercistronic region of BRLF1-BZLF1 bicistronic mRNA in translating the Zta protein of Epstein-Barr virus. J. Virol. 75:1142–1151. 10.1128/JVI.75.3.1142-1151.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W. 2010. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 6:e1001063. 10.1371/journal.ppat.1001063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin HJ, Kim do N, Lee SK. 2011. Association between Epstein-Barr virus infection and chemoresistance to docetaxel in gastric carcinoma. Mol. Cells 32:173–179. 10.1007/s10059-011-0066-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seo JS, Kim TG, Hong YS, Chen JY, Lee SK. 2011. Contribution of Epstein-Barr virus infection to chemoresistance of gastric carcinoma cells to 5-fluorouracil. Arch. Pharm. Res. 34:635–643. 10.1007/s12272-011-0414-7 [DOI] [PubMed] [Google Scholar]
- 49.Shinozaki A, Sakatani T, Ushiku T, Hino R, Isogai M, Ishikawa S, Uozaki H, Takada K, Fukayama M. 2010. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res. 70:4719–4727. 10.1158/0008-5472.CAN-09-4620 [DOI] [PubMed] [Google Scholar]
- 50.Marquitz AR, Mathur A, Chugh PE, Dittmer DP, Raab-Traub N. 2014. Expression profile of microRNAs in Epstein-Barr virus-infected AGS gastric carcinoma cells. J. Virol. 88:1389–1393. 10.1128/JVI.02662-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu J, Cosmopoulos K, Pegtel M, Hopmans E, Murray P, Middeldorp J, Shapiro M, Thorley-Lawson DA. 2011. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog. 7:e1002193. 10.1371/journal.ppat.1002193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young DF, Carlos TS, Hagmaier K, Fan L, Randall RE. 2007. AGS and other tissue culture cells can unknowingly be persistently infected with PIV5; a virus that blocks interferon signalling by degrading STAT1. Virology 365:238–240. 10.1016/j.virol.2007.03.061 [DOI] [PubMed] [Google Scholar]
- 53.Lin TC, Liu TY, Hsu SM, Lin CW. 2013. Epstein-Barr virus-encoded miR-BART20-5p inhibits T-bet translation with secondary suppression of p53 in invasive nasal NK/T-cell lymphoma. Am. J. Pathol. 182:1865–1875. 10.1016/j.ajpath.2013.01.025 [DOI] [PubMed] [Google Scholar]
- 54.Huang WT, Lin CW. 2014. EBV-encoded miR-BART20-5p and miR-BART8 inhibit the IFN-gamma-STAT1 pathway associated with disease progression in nasal NK-cell lymphoma. Am. J. Pathol. 184:1185–1197. 10.1016/j.ajpath.2013.12.024 [DOI] [PubMed] [Google Scholar]