ABSTRACT
The neutralizing anti-HIV-1 antibody 2G12 is of particular interest due to the sterilizing protection it provides from viral challenge in animal models. 2G12 is a unique, domain-exchanged antibody that binds exclusively to conserved N-linked glycans that form the high-mannose patch on the gp120 outer domain centered on a glycan at position N332. Several glycans in and around the 2G12 epitope have been shown to interact with other potent, broadly neutralizing antibodies; therefore, this region constitutes a supersite of vulnerability on gp120. While crystal structures of 2G12 and 2G12 bound to high-mannose glycans have been solved, no structural information that describes the interaction of 2G12 with gp120 or the Env trimer is available. Here, we present a negative-stain single-particle electron microscopy reconstruction of 2G12 Fab2 in complex with a soluble, trimeric Env at ∼17-Å resolution that reveals the antibody's interaction with its native and fully glycosylated epitope. We also mapped relevant glycans in this epitope by fitting high-resolution crystal structures and by performing neutralization assays of glycan knockouts. In addition, a reconstruction at ∼26 Å of the ternary complex formed by 2G12 Fab2, soluble CD4, and Env indicates that 2G12 may block membrane fusion by induced steric hindrance upon primary receptor binding, thereby abrogating Env's interaction with coreceptor(s). These structures provide a basis for understanding 2G12 binding and neutralization, and our low-resolution model and glycan assignments provide a basis for higher-resolution studies to determine the molecular nature of the 2G12 epitope.
IMPORTANCE HIV-1 is a human virus that results in the deaths of millions of people around the world each year. While there are several effective therapeutics available to prolong life, a vaccine is the best long-term solution for curbing this global epidemic. Here, we present structural data that reveal the viral binding site of one of the first HIV-1-neutralizing antibodies isolated, 2G12, and provide a rationale for its effectiveness. These structures provide a basis for higher-resolution studies to determine the molecular nature of the 2G12 epitope, which will aid in vaccine design and antibody-based therapies.
INTRODUCTION
The envelope glycoprotein (Env) on the HIV-1 viral membrane is responsible for attachment and entry into immune cells. The surface of Env is heavily glycosylated, and the virus also undergoes rapid evolution, allowing HIV-1 to evade adaptive immunity throughout the course of natural infection (1–3). Further contributing to immune evasion, certain conserved epitopes, such as the coreceptor binding site, are only transiently exposed after conformational changes in Env that are triggered by CD4 receptor binding. Nonetheless, a wide variety of broadly neutralizing antibodies (bnAbs) have now been isolated from naturally infected individuals (4–10), and many of these bnAbs have been characterized functionally and structurally, thereby elucidating sites of vulnerability on the Env surface (11–17). Such information is central to structure-based design of immunogens that faithfully mimic the epitopes on the native structure of Env, for use as candidate vaccines that re-elicit such bnAbs by vaccination (18, 19).
Structural studies have been essential to understand the molecular details of Env-bnAb interactions, which often contain significant contributions from N-linked glycans (11–17). Recent cryo-electron microscopy (EM) and X-ray crystallography structures of a cleaved, soluble form of trimeric Env (BG505 SOSIP.664) have begun to describe several of these glycans in a quaternary context and reveal the complex nature of the native glycan shield on Env (20, 21). Although the glycans on the surface of Env are of host origin, their clustered arrangement on the outer domain of gp120 is a unique signature of Env and predominantly contains immature oligomannose glycoforms that form a supersite of vulnerability targeted by very potent bnAbs (13, 20). The bnAb 2G12 is unique, as it was one of the first broadly neutralizing antibodies to be discovered (22) and its epitope is composed entirely of N-linked glycans (23–25). The crystal structure of 2G12 revealed a novel architecture for an antibody in which two Fabs (Fab2) adopted a domain-swapped dimer conformation via exchange of their variable heavy chain domains, creating two primary binding sites and two potential secondary binding sites at the unique VH/VH′ interface (23). This unusual antibody structure maximizes contacts with the tips of the clustered high-mannose glycans on the gp120 outer domain through multivalent interactions with glycans. While many other antibodies have now been found that are more potent and broadly neutralizing than 2G12, the current evidence suggests that 2G12 is an effective prophylactic, even at low serum titers, and does possess therapeutic activity (26–32). Furthermore, additional oligomeric IgG forms of 2G12 have recently been described that can enhance neutralization and protection compared to the more usual monomeric IgG form of the antibody (33–36).
To date, no structures of 2G12 in complex with its full epitope on gp120 or on the Env trimer have been determined. To address this gap in knowledge, we employed hybrid techniques to obtain a structure of 2G12 bound to trimeric Env. Here, we present a negative-stain EM reconstruction of cleaved, soluble, and fully glycosylated trimeric Env in complex with 2G12 Fab2. Docking recent high-resolution structures of trimeric Env into our EM reconstruction, along with previous X-ray crystal structures of 2G12, reveals several key details describing the interaction of 2G12 and Env. Additionally, a negative-stain EM reconstruction of soluble CD4 (sCD4) bound to the 2G12-Env complex suggests that 2G12 may prevent subsequent interaction with the coreceptor via steric hindrance.
MATERIALS AND METHODS
Preparation of the 2G12 Fab2/BG505 SOSIP.664 complex and the 2G12 Fab2/BG505 SOSIP.664/sCD4 complex.
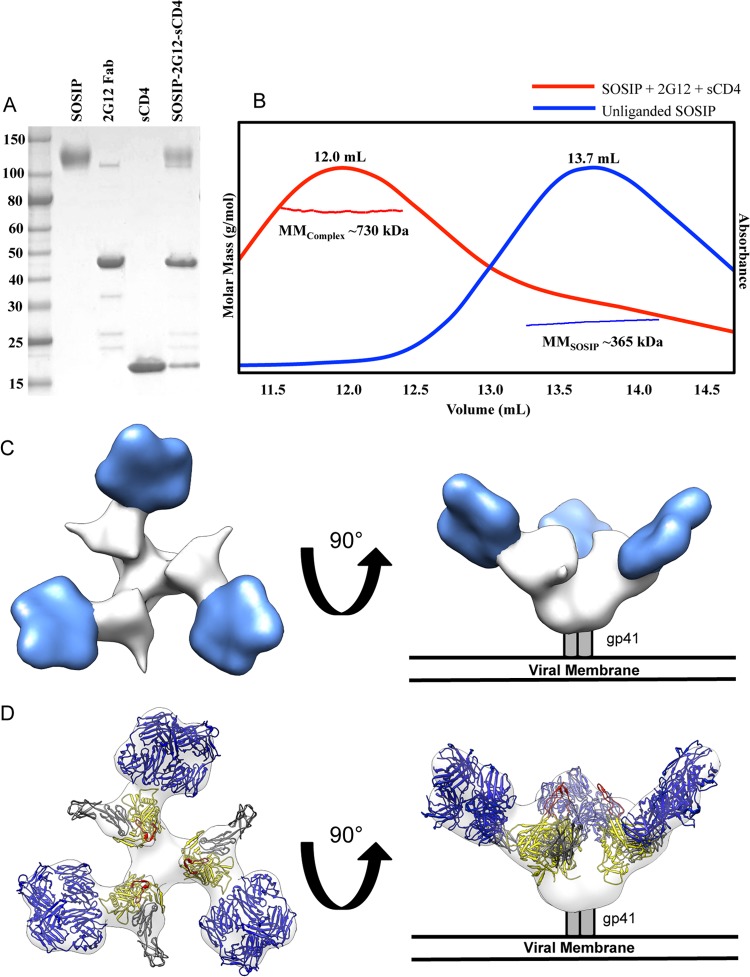
2G12 Fab2 was produced using a protocol similar to that previously described (23). Briefly, human monoclonal antibody 2G12 IgG was produced as a secreted immunoglobulin by coexpression of heavy and light chain genes in suspension-adapted human embryonic kidney 293 cells (HEK293F cells). Following protein A purification, 2G12 IgG was digested with papain. Domain-exchanged 2G12 Fab2 fragments were purified by protein A or protein G affinity, followed by size exclusion chromatography on a Superdex 200 column (GE Healthcare). BG505 SOSIP.664 was expressed and purified from HEK293S N-acetylglucosaminyltransferase I-deficient (GnTI−/−) cells as previously described (47). Briefly, secreted BG505 SOSIP.664 trimers were purified from the supernatant on a 2G12-affinity matrix, eluted with 3 M MgCl2, and separated to homogeneity by gel filtration chromatography using a Superose 6 column (GE Healthcare). Trimers were mixed with 2G12 Fab2 in a 1:3.5 molar ratio, and the trimer/Fab complex was purified by chromatography using a Superose 6 column (GE Healthcare) in a buffer consisting of 20 mM Tris, 150 mM NaCl, pH 8.0 (TBS). To generate the 2G12 Fab2/BG505 SOSIP.664/sCD4 complex, a 10-fold molar excess each of soluble CD4 (sCD4) and 2G12 Fab2 was added to BG505 SOSIP.664 and the mixture incubated at 4°C for 4 h. The complex was then purified by size exclusion chromatography coupled with multiangle light scattering (SEC-MALS) (see below) and imaged by negative-stain EM. sCD4 was prepared as described previously (13).
Electron microscopy and sample preparation.
A 4-μl aliquot of each complex, diluted to a concentration of ∼0.03 mg/ml with TBS buffer, was placed for 15 s onto carbon-coated 400-mesh Cu grids which had been plasma cleaned for 20 s (Gatan), blotted off on the edge of the grid, and then immediately stained for 30 s with Nano-W (Nanoprobes). Data were collected either manually or with Leginon (37–39) using an FEI Tecnai F20 electron microscope operating at 120 keV with an electron dose of 30 e−/Å2 and a magnification of ×100,000 or ×52,000 that resulted in a pixel size of 1.09 Å or 2.05 Å at the specimen plane (for the 2G12-Env and 2G12-sCD4-Env complex, respectively). For the 2G12-Env complex, images were acquired using a Gatan 4,000- by 4,000-pixel (4k by 4k) charge-coupled device (CCD) camera with a defocus range of 500 to 800 nm in 5° tilt increments from 0 to 50°. Tilt data were collected in order to offset some slight preferred orientations of the particles on the carbon substrate that we noted in untilted data. Tilting gave better angular sampling, as seen in the Euler angle plot (see Fig. S1 in the supplemental material). Each image was acquired on a new area of the grid so that each particle in the reconstruction received the same electron dose of 30 e−/Å2. For the 2G12-sCD4-Env complex, images were acquired using a Teitz CMOS 4k by 4k CCD camera with a constant defocus value of 1 μm at 0°, which did not require stage tilting as the particles had a generally isotropic orientation on grids at zero degrees as seen in the Euler angle plot (see Fig. S2). Additionally, the EMAN2 refinement script used for both reconstructions weights the number of particles that go into each orientation so that there is not overrepresentation as long as there is generally complete angular sampling, which is the case for both reconstructions.
Image processing of protein complexes.
Particles were picked automatically using DoG Picker (40) and placed into a particle stack using Appion software (41). For the 2G12 Fab2/BG505 SOSIP.664 complex, reference-free 2-dimensional (2-D) class averages were calculated using particles binned by 4 to 4.35 Å/pixel with the Xmipp Clustering 2D Alignment software (42) and sorted into initial classes before being further subclassified into classes with ∼50 particles per class in order to generate the final particle stack used for the reconstruction. Using a map of the unliganded BG505 SOSIP.664 gp140 trimer as an initial model (EMDB accession number EMD-5708) for the 2G12-Env reconstruction, refinement was carried out against a stack of 13,143 raw particles for 10 iterations, imposing C3 symmetry using EMAN software (43). The structure of the 2G12 Fab2/BG505 SOSIP.664/sCD4 complex was generated similarly but with the following parameters: the starting model was generated with templates created from reference-free class averages and EMAN2 common lines (44), the particle stack contained 5,372 particles, which were binned by 2 to 4.1 Å/pixel, and the reconstruction was generated with 10 rounds of refinement using EMAN (44). To determine if there were substoichiometric populations of Env bound to Fab present in our structures, we performed a sorting method similar to a method previously described (21). Although our analysis did show that ∼20% of the particles used for the 2G12-Env structure had only two Fabs bound, we did not see any appreciable difference in the maps of the 2-bound-Fab model and the 3-bound-Fab model at the given resolution (not shown).
Interpretation of EM data.
X-ray structures of available components (gp140 trimer, gp120 monomer, and 2G12 with bound glycans) were used to dock into the EM reconstructions using the Fit in Map function in UCSF Chimera as delineated in the main text. No further refinements of the structure, loops, or glycans were performed. Thus, all interpretations are based on static models.
Plasmid constructs and mutagenesis.
Mutations were introduced using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) following the manufacturer's protocol. Mutants were verified by DNA sequence analysis (Eton Bioscience, San Diego, CA).
Pseudovirus production and neutralization assay.
Pseudoviruses were generated by transfection of HEK293T cells with an HIV-1 Env-expressing plasmid and an Env-deficient genomic backbone plasmid (pSG3ΔEnv) in a 1:2 ratio with the transfection reagent Fugene 6 (Promega). We note that it has been shown that the high-mannose patch is conserved regardless of cell line (HEK293T or HEK293S GnTI−/−), most likely due to the dense clustering of glycans that prevent access by glycan-processing enzymes (45). Pseudoviruses were harvested 72 h posttransfection for use in neutralization assays. Neutralizing activity was assessed using a single round of replication pseudovirus assay and TZM-bl target cells, as described previously (46). Briefly, TZM-bl cells were seeded in a 96-well flat-bottom plate at a concentration of 20,000 cells/well. To this plate was added pseudovirus, which was preincubated with serial dilutions of antibody for 1 h at 37°C. Luciferase reporter gene expression was quantified 48 h after infection by cell lysis and the addition of luciferase substrate (Promega). To determine 50% inhibitory concentrations (IC50), dose-response curves were fitted using nonlinear regression.
SEC-MALS.
Biophysical analysis by size exclusion chromatography coupled with multiangle light scattering (SEC-MALS) was completed as described previously (11). Briefly, the sCD4/2G12 Fab2/BG505 SOSIP.664 complex was loaded on a Superose 6 10/30 SEC column (GE Healthcare), which was coupled in-line on an AKTA Avant fast protein liquid chromatography (FPLC) system (GE Healthcare) with the following calibrated detection systems: (i) HP1 1050 Hewlett-Packard UV detector; (ii) MiniDawn Treos MALS detector (Wyatt); (iii) quasielastic light scattering (QELS) detector (Wyatt); and (iv) Optilab T-reX refractive index (RI) detector (Wyatt). Analysis of the light scattering data coupled to 280-nm UV and refractive index protein concentration measurements allowed determination of the molar mass of the eluting protein by using the protein conjugate template in Astra 6, as reported previously (47).
EM reconstructions have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-5982 and EMD-5983.
RESULTS AND DISCUSSION
Structure of 2G12 Fab2 bound to HIV-1 Env revealed by negative-stain single-particle EM.
To determine where 2G12 binds in the context of trimeric Env, negative-stain single-particle EM was used to calculate a reconstruction of 2G12 Fab2 bound to the BG505 SOSIP.664 trimer at ∼17-Å resolution (Fig. 1; see also Fig. S1 in the supplemental material). BG505 SOSIP.664 is a soluble, trimeric construct of Env stabilized by a disulfide bond introduced between gp120 and gp41 (SOS) and an isoleucine-to-proline mutation (IP) in the heptad repeat 1 (HR1) region of gp41 (48). The native BG505 virus does not contain a glycan at Env position N332; therefore, wild-type (WT) Env does not bind 2G12 and is not susceptible to neutralization. Restoration of this glycan by the mutation T332N restores the ability of 2G12 to bind and neutralize the virus, consistent with previous data suggesting that N332 is critical for 2G12 binding (24, 25). Thus, for the experiments described here, the T332N form of BG505 SOSIP.664 will be referred to as WT.
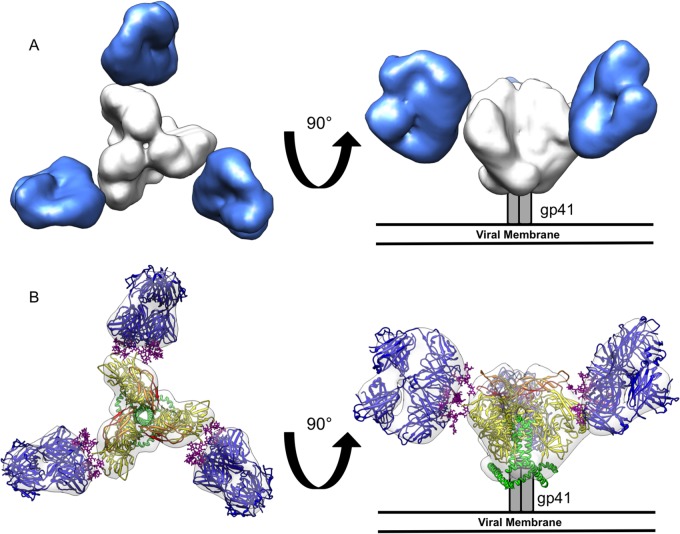
FIG 1.
Negative-stain single-particle EM reconstruction of BG505 SOSIP.664 Env bound to 2G12 Fab2. (A) Top (left) and side (right) views of the reconstruction are rendered with the segmented densities corresponding to 2G12 Fab2 in blue and BG505 SOSIP.664 Env in white. (B) Crystal structures of 2G12 Fab2 (PDB ID 1OP5) (23) and Env (PDB ID 4NCO) (20) were fit into the EM density map. All glycans in the Env crystal structure were removed for clarity. Top (left) and side (right) views of the fitted maps are shown with gp120 rendered in yellow, V1/V2 loops in orange, V3 loops in red, gp41 in green, and 2G12 Fab2 in dark blue, with bound glycans in purple.
The EM reconstruction clearly shows that three domain-exchanged 2G12 Fab2 bind Env, one per protomer (Fig. 1A). Binding occurs on the outer surface of Env, which corresponds to the high-mannose cluster on the gp120 outer domain. Crystal structures of both 2G12 Fab2 and HIV-1 Env were docked into the EM reconstruction using UCSF Chimera (49) and exhibited excellent fits (Fig. 1B). The epitope is composed of glycans; indeed, the closest protein-protein distance between 2G12 and Env, based on our docking and distance measurements with Chimera, is the ∼7-Å distance between one of the VH domains (HC1) of 2G12 Fab2 and residues in the V1 loop of Env (Fig. 2A). We cannot, however, rule out some interaction, even transient, with the V4 loop. In the existing structures of gp120 core monomers and HIV Env SOSIP, the V4 loop is flexible, resulting in a lack of clear density for this region (20, 21, 50). Therefore, at the resolution of the current EM analysis, it cannot be determined whether the V4 loop affects 2G12 binding, either directly or indirectly. The angle at which 2G12 Fab2 interacts with Env indicates that the upper 2G12 Fab of the Fab2 is closer to the apex of the trimer and the heavily glycosylated gp120 outer domain (∼8 to 12 Å) than the lower Fab (∼12 to 25 Å) (Fig. 2A). This suggests that although domain exchange is crucial for binding (51), interaction with Env is biased toward one half of the Fab2 in terms of contact surface.
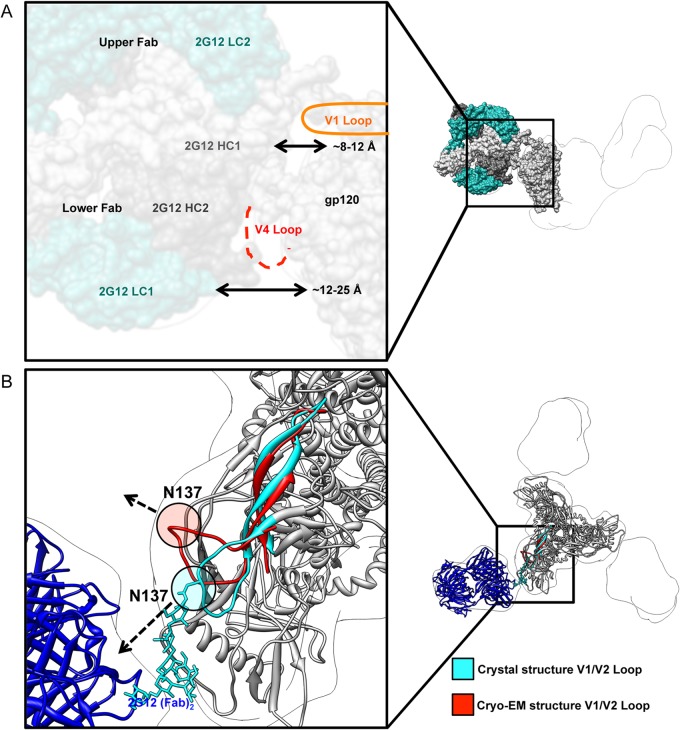
FIG 2.
Interaction of 2G12 with Env BG505 SOSIP.664. (A) Distance measurements between pairs of atoms were made between the closest protein residues in the docked 2G12 Fab2 (PDB ID 1OP5) and the crystal structure of Env BG505 SOSIP.664 trimer (PDB ID 4NCO), which were then averaged for each Fab. The closest protein interaction with 2G12 occurs within the V4 loop, which is not completely modeled in the docked crystal structure because of disorder (dashed red line). All glycans were removed for clarity. (B) Although the Env structures in complex with antibodies against different sites of vulnerability as determined by X-ray crystallography and cryo-EM are overall very similar, their V1 loop positions are different. The V1/V2 motif from the cryo-EM structure (red) (PDB ID 3J5M) was overlaid on the X-ray crystallography structure (cyan and gray) (PDB ID 4NCO). Residue N137 is highlighted in each structure, and the dashed arrows indicate the direction that the residue is pointing relative to 2G12. No glycan is modeled in the cryo-EM structure, but there is one in the X-ray crystallography structure (which is bound by PGT122 Fab). The V1 loop in the X-ray crystal structure points toward 2G12 and into the density of the negative-stain EM structure. The V1 loop in the cryo-EM structure, however, is pointed away from 2G12. These differences demonstrate that antibody interaction with Env variable loops, especially with glycans, may alter the position of these loops and influence binding/neutralization.
Glycan epitope of 2G12 on trimeric HIV-1 Env.
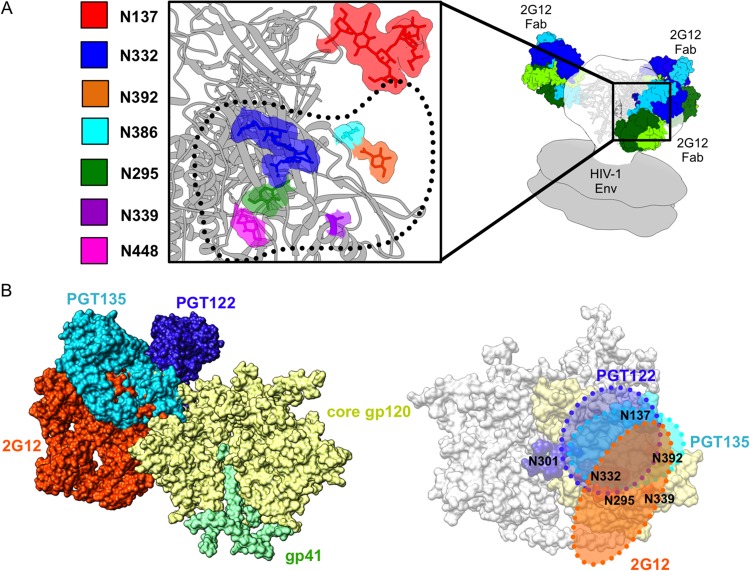
As predicted (13), the 2G12 epitope overlaps with several other bnAbs that target the N332 supersite of vulnerability on Env (Fig. 3). Glycans previously implicated in 2G12 recognition (24, 25) fit well into the density map between 2G12 and Env, namely, N295, N332, N392, N386, N339, and N448 (Fig. 3A). Glycans N295, N332, N392, and N339 are centrally located within the footprint of the antibody, while N448 and N386 are on the periphery, consistent with previous models of the gp120 epitope (24, 25).
FIG 3.
Details of the glycan epitope of 2G12. (A) A closeup view of the interface between BG505 SOSIP.664 Env (PDB ID 4NCO) and 2G12 Fab2 (PDB ID 1OP5) (contact interface highlighted with dotted black line) showing the 2G12-relevant glycans. Glycans from the Env crystal structure that fall within the 2G12 epitope are rendered in space-filling mode and highlighted by different colors as indicated in the key. gp120 is rendered in gray ribbon. Our 2G12-Env EM reconstruction was fit into a model of unliganded membrane-anchored HIV-1 trimer (gray surface) (EMDB accession numbers EMD-5019 and EMD-5021) (60) in order to fit the Fab2 crystal structures. The BG505 SOSIP.664 crystal structure was subsequently fit into our EM density map in order to determine which glycans fell within the 2G12 epitope (20). The crystal structures used in the docking were solved after partial deglycosylation, such that some of the N-linked glycans only harbor an N-acetylglucosamine (NAG), while others contain high-mannose glycans (20). (B) On the left, we show a superimposition of glycan-dependent monoclonal antibody (MAb) cocrystal structures of PGT135 (PDB ID 4JM2) (13) and PGT122 (PDB ID 4NCO) fit onto the Env trimer crystal structure along with our own EM fitting of 2G12 to expand on the mapping of this glycan supersite of vulnerability on Env, showing the overlap of these three antibodies. The extents of these epitopes are detailed on the right, further emphasizing the large size of the 2G12 epitope on the trimeric surface of Env and where it overlaps with other glycan-dependent MAbs.
In the crystal structure of 2G12, two Man9GlcNAc2 glycans are bound at the primary combining sites of the 2G12 Fab2 and two additional symmetry-related Man9GlcNAc2 glycans are bound at the unique VH/VH′ interface created by the domain swap (PDB ID 1OP5) (23). When this structure is modeled into the EM density along with the trimeric structure of Env (13, 20), the assignment of these glycans becomes possible (Fig. 4). The primary combining sites of 2G12 are most likely occupied by the Man9GlcNAc2 glycans in the docked 2G12 crystal structure at N392 and N295 on Env (Fig. 5). The glycan at N392 has previously been predicted to bind at this site (23) and has been shown to be critical for binding (24, 25). In our docked structure, the Fab2 is closest to the N392 glycan at the top of Env and further away than the incompletely modeled glycan at N295, which resides toward the bottom of the gp120 outer domain. Superposition of a gp120 structure that has a complete N392 glycan modeled (not shown) (13), along with distance measurements (Fig. 4), shows that N392 overlays well with the uppermost glycan bound to 2G12. The positioning of N295 is consistent with our distance measurements (Fig. 4) and with previous data, suggesting that N295 plays an indirect role in binding (23).
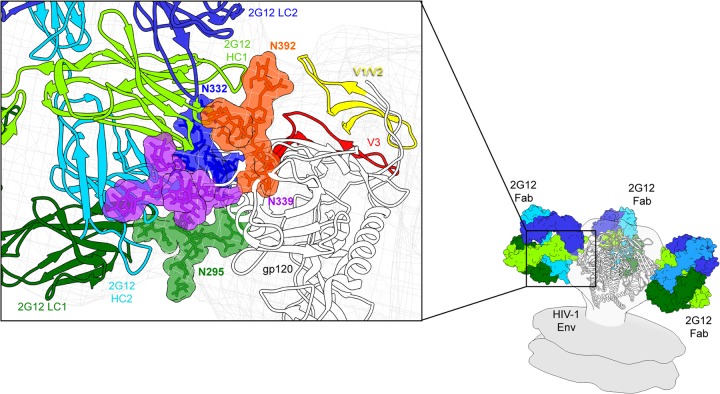
FIG 4.
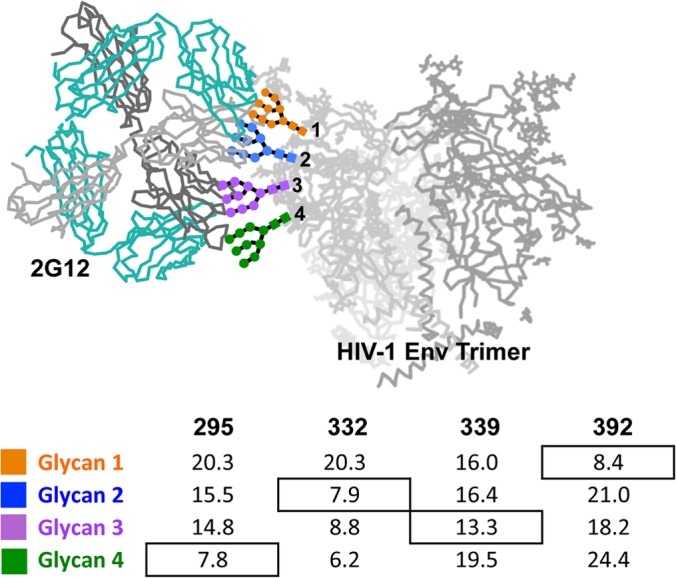
Assignment of glycan positions. Crystal structures of 2G12 bound to four Man9GlcNAc sugars and Env were fit into the 2G12-Env EM density. Using UCSF Chimera, distance measurements were made from the OH of C-1 on the terminal GlcNAc of 2G12-bound sugars and the OH of C-4 from the terminal GlcNAcs of N295, N332, N339, and N392 that were modeled in the crystal structure (PDB ID 4NCO). There was no sugar modeled at position N339 in the crystal structure; therefore, the structure of PGT135 (PDB ID 4JM2), which has a terminal GlcNAc modeled at this position, was overlaid onto Env. Glycan assignments bound to 2G12 were assigned on the surface of Env according to the shortest distance, with the exception of N332. Three independent crystal structures with antibodies bound to N332 were overlaid onto Env, and in each case, N332 aligned nearly exactly with glycan 2 and not glycan 4. The structure of PGT135 also contains glycan N392, which overlays with glycan 1, in agreement with our distance measurements. Distances in angstroms are tabulated below the structure.
FIG 5.
Model of the 2G12 epitope in the context of trimeric Env. Our model was constructed using the hybrid methods of docking separate crystal structures of Env and glycan-bound 2G12 Fab2 into our low-resolution EM reconstruction, distance measurements (Fig. 3), and previous biochemical data (24, 25). We place glycans N392 and N295 within the primary combining sites of 2G12, while N332 and N339 interact with the secondary combining sites, which are formed by the VH-VH interactions arising from the VH/VH′ domain swap. Glycans linked to positions N392 and N332 both bind to the upper Fab in the Fab2 structure. The upper Fab is also closer to the variable loops and the gp120 outer domain than the lower Fab. The glycan at N339 likely stabilizes these interactions, although it is not required for 2G12 to recognize Env (as shown by the N339A glycan knockout below), while the glycan at N295 may be important for glycan processing.
The unique 2G12 secondary binding sites at the VH/VH′ interface are occupied by glycans in the 2G12 crystal structure that likely correspond to positions N332 and N339 (Fig. 4 and 5). The glycan at N332 is fully modeled in three separate structures and consistently points toward this binding site in each case (13, 15, 20). N332 is also critical for 2G12 binding, although it had been predicted to bind in the primary binding site (23). The glycan at N339 is not critical for 2G12 recognition; however, it is thought to interact with the VH/VH′ interface (23), consistent with what we observe in our model. The glycan at N339 is modeled in the structure of PGT135 (PDB ID 4JM2) as a single GlcNAc; therefore, its position is inferred and based on distance measurements (Fig. 4).
The glycans at N448 and N386, which have been implicated in the binding of 2G12 before but are not critical for recognition, do not appear to directly interact with 2G12 but may have some strain-dependent influence over primary glycan recognition. A seventh glycan, N137, within the V1 loop, also resides in close proximity and may interact with a primary binding site of 2G12 at complementarity-determining regions (CDRs) L1, L2, and L4, which are clustered together and do not form primary interactions with glycans in the crystal structure (23). Glycans in V4 that were not modeled in previous crystal structures might also play a role in shaping the interaction of 2G12 and Env and, thus, should be taken into account for immunogen design.
A large and diverse group of broadly neutralizing antibodies have now been discovered that target a conserved epitope region anchored by the N-linked glycan of Asn 332. Structural information has been limited, however, regarding the exact location and extent of the epitopes within this site and the relative position and arrangement of the glycans in the context of the trimeric Env. The recently determined structures of Env, however, have allowed a more accurate description of this epitope which resides on the side of Env in a location that is distinct from the CD4-binding site (20, 21, 52). Our results here expand the definition of this epitope by identifying the location of the 2G12-binding site and its angle of approach in the context of trimeric Env (Fig. 3B).
Glycan knockout analysis.
To measure the relative contribution of each glycan to 2G12 binding in the context of BG505 and to corroborate our structure, we conducted individual Asn-to-Ala knockouts of several important glycans and determined the effect on neutralization (Fig. 6). Because BG505 Env does not natively contain a glycan at position N332, a T332N mutation was introduced in each of our knockout experiments in order to restore the full 2G12 epitope, which we call WT for the context of this study. The T332 virus alone cannot be neutralized by 2G12. Additionally, the elimination of glycans at N295 and N392 completely abrogates the ability of 2G12 to neutralize the WT BG505 virus (Fig. 6). The importance of these glycans is consistent with previous data and our own structure (Fig. 5) (24, 25).
FIG 6.
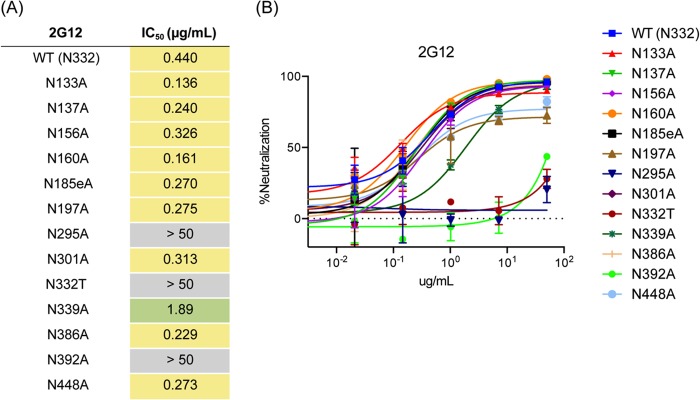
Glycan knockout analysis of BG505 SOSIP.664. (A) Individual glycan sites on BG505 pseudovirus were removed by alanine mutagenesis and tested for neutralization in a TZM-bl assay (46). Each glycan site was then removed and tested for binding to WT BG505 virus. Here, we define wild-type (WT) virus as that with an Asn residue at position 332. Tabulated neutralization IC50 are presented. Glycan knockouts with an IC50 less than 1 were not considered important for 2G12 binding, those with an IC50 greater than 1 and less than 50 were considered important but not critical to binding, and those with an IC50 greater than 50 were considered critical for binding. (B) Glycans at positions N332, N295, and N392 are critical for 2G12-based neutralization of HIV, with N339 also playing an important role.
Although a knockout of N386 and N448 did not affect 2G12 neutralization, mutagenesis of N339 did affect the ability of 2G12 to neutralize the virus (Fig. 6). Residues N386, N448, and N339 have previously been implicated in mutagenesis studies as having some contribution to 2G12 binding, although these studies were conducted with JR-FL (24) or JR-CSF (25) strains of HIV-1 and the relative contribution of these residues differed slightly between each strain. The N339 glycan may make contacts with 2G12 less critical for neutralization but may regulate the 2G12 angle of approach such that the Fab2 makes better contacts with N332 and N392 glycans. Generally, the position of glycans in our model relative to the center of the domain-swapped Fab2 was predictive of their effect on neutralization; i.e., the closer to the center of the docked 2G12 Fab2, the more important that glycan is for 2G12 neutralization (Fig. 3A). However, the extent of 2G12 dependency on the peripheral glycans (N386, N448, and N339) for neutralization appears isolate dependent, as the type and arrangement of these glycans differs slightly across strains.
Although the removal of N137 did not significantly alter neutralization by 2G12, it and other V1 glycans may subtly modulate binding in a strain-dependent manner. V1, in both the X-ray and EM structures (20, 21), lies in close proximity to 2G12, although the V1 loops in these structures are in different positions, likely because of the different antibodies to which Env is bound in the respective structures. In the crystal structure of Env in complex with PGT122 (20), the V1 loop is positioned close to 2G12 in our model, while in the EM structure in complex with PGV04 (21), the loop is further away from 2G12 (Fig. 2B). While the glycan at N137 is not modeled in the EM Env-PGV04 structure, the V1 loop appears to point away from 2G12. These observations suggest that the binding of Fabs significantly affects the position of the V1 loop and that this loop is flexible enough to interact with a range of antibodies, including 2G12, contributing either positively or negatively to the interaction, and this should be taken into account for immunogen design.
Binding of sCD4 to 2G12-bound Env.
The binding of CD4 induces a large conformational change in trimeric Env, exposing the V3 loop, which is primed to bind the coreceptor (53). Additionally, in contrast to PGT121, which allosterically blocks CD4 engagement (12), it has been shown previously that binding of 2G12 to Env does not block access to the CD4 binding site on gp120 (54). To determine if 2G12 might interfere with coreceptor binding, we added excess two-domain soluble CD4 (sCD4) and 2G12 Fab2 to BG505 SOSIP.664 and purified the complex by size exclusion chromatography with inline multiangle light scattering (SEC-MALS), followed by SDS-PAGE analysis, which indicated that 2G12 Fab2, sCD4, and SOSIP all comigrated as a single complex with a molecular mass of ∼730 kDa, consistent with 3 Fab2 (300-kDa), 3 sCD4 (69-kDa), and 1 SOSIP (360-kDa) trimer per complex (Fig. 7A and B). Subsequent negative-stain 2D class averages of the purified material indicated that, upon sCD4 binding, the trimer had undergone a large structural change, opening the trimer in a manner similar to previously published results (see Fig. S2A and B in the supplemental material) (55). A reconstruction at ∼26-Å resolution of 2G12/sCD4-bound SOSIP.664 Env by negative-stain EM indicates a large shift in antibody binding angle in conjunction with the opening of Env (Fig. 7C and D; see also Fig. S2A and B). Fitting crystal structures into this density map of CD4-bound gp120 and comparison of unbound and sCD4-bound structures (both in the presence of 2G12) demonstrated that, upon CD4 binding, the V3 loops are likely exposed and oriented toward the host membrane where the coreceptor resides (although the V3 loops were not explicitly resolved in our EM density map). 2G12 shifts significantly upward during this conformational change (Fig. 7C and D).
FIG 7.
Negative-stain single-particle EM reconstruction of BG505 SOSIP.664 Env bound to 2G12 Fab2 and soluble 2-domain CD4 (sCD4). (A) Coomassie-stained, nonreducing SDS-PAGE gel analysis of the complex components and the complex purified by size exclusion chromatography. BG505 SOSIP.664, 2G12 Fab, and sCD4 were stained similarly with equal amounts of protein. Note that some 2G12 F(ab′)2 remained with 2G12 Fab2 during purification and subsequently bound to SOSIP; because 2G12 is domain exchanged, these two products are virtually identical except on a nonreducing SDS-PAGE gel. (B) SEC-MALS was used to determine that the observed shift in elution volume and molar mass for the SOSIP-2G12 Fab-sCD4 (MMComplex, red) complex in comparison to those of the unliganded trimer (MMSOSIP, blue) correspond to one trimer binding three Fab2 and three sCD4 molecules simultaneously. (C) Structural characterization of BG505 SOSIP.664 Env bound to three 2G12 Fab2 and three sCD4 by negative-stain single-particle EM. Top (left) and side (right) views of the reconstruction are rendered with the segmented densities corresponding to 2G12 Fab2 in blue and BG505 SOSIP.664 Env in white. (D) Crystal structures of 2G12 Fab2 (PDB ID 1OP5) and gp120 bound to sCD4 (PDB ID 2B4C) (53) were fit into the EM density map. All glycans were removed for clarity. Top (left) and side (right) views of the fitted maps are shown with gp120 rendered in yellow, V3 loops in red, sCD4 in gray, and 2G12 Fab2 in blue.
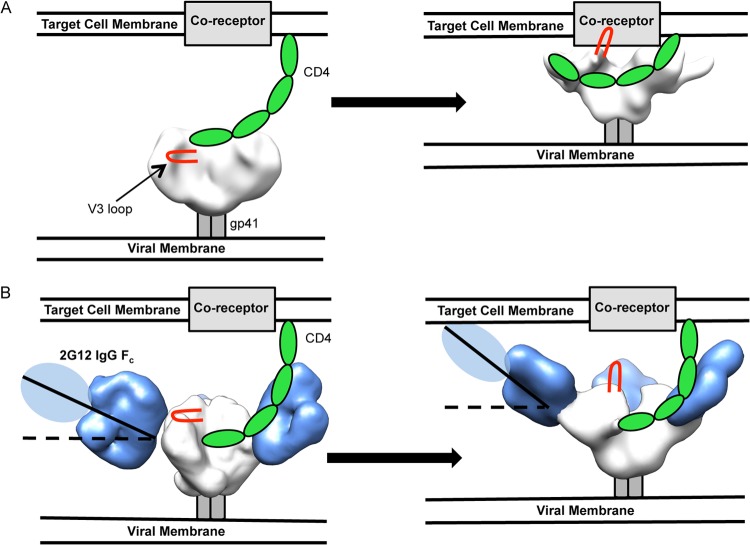
Our EM analysis indicates that, prior to CD4 engagement, the initial angle of 2G12 binding to Env is small relative to the plane of the top of Env. Upon CD4 binding to Env, which leads to a rotation in gp120, the angle of binding increases significantly. In such an orientation, 2G12 would sterically block access to the V3 loop for binding its coreceptors CXCR4 or CCR5, both integral membrane proteins which are closely associated with the target cell membrane (56–58) (Fig. 8). According to our measurements using gp120 bound to CD4 docked into our structure (PDB ID 2B4C), the 2G12 Fab2 potentially creates an additional ∼20-Å space between the tip of the V3 loop and the host-cell surface. The V3 loop, in a model with CD4 bound (PDB ID 2QAD), can extend up to 30 Å from the surface of gp120 (56).
FIG 8.
Proposed model of 2G12 steric blockade of gp120-coreceptor interaction. (A) Unliganded HIV-1 Env contacts CD4 from a target cell (left), which causes conformational changes in Env, exposes the V3 loops (red), and makes Env competent to bind its coreceptor and initiate viral fusion (right). (B) Env is bound by 2G12 at an initially shallow angle relative to the plane of the membrane and is still capable of binding CD4 (left); however, conformational changes and gp120 rotation brought about by CD4 binding cause the angle of 2G12 in relation to the viral membrane to increase significantly. This increase in angle may lead to steric clashes with the target cell membrane, such that the V3 loop is prevented from contacting the coreceptor. Env is in white, the V3 loop in red, CD4 in green, the membrane proximal external region in gray, and 2G12 in blue. Unliganded Env was solved previously by negative-stain EM (12). The CD4-bound Env reconstruction is deposited in EMDB under accession number EMD-5708.
The current models of CXCR4/CCR5 binding to CD4-bound Env predict that sulfated tyrosine residues on the N terminus of membrane-bound coreceptors interact with the base of the V3 loop, near the core of gp120 (56–58). For CCR5, this is an ∼19-residue portion with a predicted alpha-helical structure approximately at residues 10 to 14, where interaction with the base of V3 occurs (56, 58). Taking into account the potential extent of V3 reach, 30 Å, and the additional space that 2G12 Fab2 alone creates, 20 Å, the 14-residue N terminus, which is partially structured over this length, must span at least 50 Å in order for a productive interaction to occur between V3 and CCR5. Therefore, binding of the bulky 2G12 Fab2, in addition to its increased binding angle upon CD4-induced conformational changes in gp120, likely abrogates coreceptor interactions. The Fc portion of the full 2G12 IgG likely results in further steric hindrance of V3/coreceptor interaction (Fig. 8). The Fc portion of the IgG could not be visualized experimentally due to the flexible nature of the hinge regions of 2G12 IgG (see Fig. S3 in the supplemental material). We cannot rule out the possibility that CD4-bound trimers that are not saturated with 2G12 could still interact with coreceptors or that cell membrane ruffling may place the coreceptor in proximity to the exposed V3 loop even with 2G12 bound. Our model, however, is well supported by previous work that suggests that 2G12 competitively inhibits the interaction of gp120's V3 loop and the N terminus of CCR5 (54) and now provides a structural basis for this observation. It is also possible that 2G12 alters the conformation of the V3 loop, abrogating coreceptor engagement. While it has been previously shown that some antibodies can block primary receptor engagement (12), 2G12 uses a different mechanism by blocking coreceptor engagement.
A dimeric form of 2G12 IgG is more potent at neutralizing and protecting from HIV-1 in rodent models (26, 34, 36). Low-resolution crystal structures (∼8 Å) indicated that avidity is likely responsible for the enhanced neutralization and protection relative to those of the monomeric form of 2G12 IgG (33). In the electron density of unliganded 2G12 IgG dimers, Wu et al. were able to detect different forms of the IgG dimers, indicating dynamic states of the molecule (33). EM data further supported a flexible structure (33). Docking these forms of dimeric 2G12 IgG into the density maps reported here (not shown) indicates that the additional bulk of dimeric 2G12 would enhance steric hindrance for coreceptor binding, in accordance with our proposed model.
It was previously suggested that 2G12 IgG dimers may bind bivalently within a single Env trimer, making weak interactions with glycans on an adjacent protomer and thereby increasing avidity to the HIV-1 viral surface, which lacks the density of viral spikes that other viruses, such as influenza virus or Ebola virus, contain (33, 59). 2G12 IgG dimers have a nanomolar affinity for gp120 that is comparable to that of 2G12 monomers (23, 34), although they do exhibit higher apparent affinity when gp120s are clustered in a surface plasmon resonance assay, due to increased avidity resulting from cross-linking adjacent gp120 monomers (33). Our docking suggests, however, that bivalent binding within a single HIV Env trimer may be very limited due to the angle of primary contact with Env. Instead, the increased potency of the IgG dimers may result from cross-linking adjacent Env trimers that have clustered at the virion-host cell junction.
Our structural studies of 2G12 help to further describe the extent of the N332 site of vulnerability on the surface of Env (Fig. 5) and provide a model for 2G12-mediated neutralization of HIV-1 (Fig. 8). Our structures are consistent with a wide body of work on 2G12 and reveal how the Fab2 architecture of 2G12 is uniquely suited for recognition of the high-mannose patch on the outer domain of gp120. The high quality of the negative-stain EM reconstruction also makes this complex a good candidate for higher-resolution cryo-EM studies that will be valuable for describing the structures of several glycans on and around the high-mannose patch on the surface of Env.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeong Hyun Lee, Dmitry Lyumkis, and Natalia de Val Alda for helpful discussions and for describing and sharing data processing scripts.
This research was supported by HIVRAD P01 AI082362 (I.A.W., J.P.M., and A.B.W.), Scripps CHAVI-ID UM1 AI100663, a National Science Foundation Graduate Student Research Fellowship (C.D.M.), and the International AIDS Vaccine Initiative (IAVI), who obtain part of their funding through the generous support of the American people through the United States Agency for International Development (USAID).
EM data were collected at the National Resource for Automated Molecular Microscopy at the Scripps Research Institute, which is supported by the Biomedical Technology Research Center program (GM103310) of the National Institute of General Medical Sciences.
Footnotes
Published ahead of print 25 June 2014
This article is manuscript 27050 from The Scripps Research Institute.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01229-14.
REFERENCES
- 1.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705–711. 10.1038/31514 [DOI] [PubMed] [Google Scholar]
- 2.McCaffrey RA, Saunders C, Hensel M, Stamatatos L. 2004. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J. Virol. 78:3279–3295. 10.1128/JVI.78.7.3279-3295.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson WE, Desrosiers RC. 2002. Viral persistence: HIV's strategies of immune system evasion. Annu. Rev. Med. 53:499–518. 10.1146/annurev.med.53.082901.104053 [DOI] [PubMed] [Google Scholar]
- 4.Mouquet H, Klein F, Scheid JF, Warncke M, Pietzsch J, Oliveira TY, Velinzon K, Seaman MS, Nussenzweig MC. 2011. Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PLoS One 6:e24078. 10.1371/journal.pone.0024078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640. 10.1038/nature07930 [DOI] [PubMed] [Google Scholar]
- 6.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637. 10.1126/science.1207227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol G Principal Investigators. Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. 10.1038/nature10373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR, Protocol G Principal Investigators 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. 10.1126/science.1178746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. 10.1126/science.1187659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Program NCS, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476. 10.1038/nature12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. 2013. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl. Acad. Sci. U. S. A. 110:4351–4356. 10.1073/pnas.1217537110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, Katpally U, Depetris RS, Stanfield RL, McBride R, Marozsan AJ, Paulson JC, Sanders RW, Moore JP, Burton DR, Poignard P, Ward AB, Wilson IA. 2013. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 9:e1003342. 10.1371/journal.ppat.1003342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. 2013. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat. Struct. Mol. Biol. 20:796–803. 10.1038/nsmb.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. 10.1038/nature10696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103. 10.1126/science.1213256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, Burton DR, Wilson IA. 2010. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. U. S. A. 107:11483–11488. 10.1073/pnas.1004600107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. 10.1126/science.1192819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, Connell MJ, Stevens E, Schroeter A, Chen M, Macpherson S, Serra AM, Adachi Y, Holmes MA, Li Y, Klevit RE, Graham BS, Wyatt RT, Baker D, Strong RK, Crowe JE, Johnson PR, Schief WR. 2014. Proof of principle for epitope-focused vaccine design. Nature 507:201–206. 10.1038/nature12966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. 2013. Rational HIV immunogen design to target specific germline B cell receptors. Science 340:711–716. 10.1126/science.1234150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. 10.1126/science.1245625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. 10.1126/science.1245627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065–2071. 10.1126/science.1083182 [DOI] [PubMed] [Google Scholar]
- 24.Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293–7305. 10.1128/JVI.76.14.7293-7305.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306–7321. 10.1128/JVI.76.14.7306-7321.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, Koff WC, Watkins DI, Burton DR. 2009. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5:e1000433. 10.1371/journal.ppat.1000433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baba TW, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, Cavacini LA, Posner MR, Katinger H, Stiegler G, Bernacky BJ, Rizvi TA, Schmidt R, Hill LR, Keeling ME, Lu Y, Wright JE, Chou TC, Ruprecht RM. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200–206. 10.1038/72309 [DOI] [PubMed] [Google Scholar]
- 28.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, Lewis MG. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207–210. 10.1038/72318 [DOI] [PubMed] [Google Scholar]
- 29.Armbruster C, Stiegler GM, Vcelar BA, Jager W, Michael NL, Vetter N, Katinger HW. 2002. A phase I trial with two human monoclonal antibodies (hMAb 2F5, 2G12) against HIV-1. AIDS 16:227–233. 10.1097/00002030-200201250-00012 [DOI] [PubMed] [Google Scholar]
- 30.Stiegler G, Armbruster C, Vcelar B, Stoiber H, Kunert R, Michael NL, Jagodzinski LL, Ammann C, Jager W, Jacobson J, Vetter N, Katinger H. 2002. Antiviral activity of the neutralizing antibodies 2F5 and 2G12 in asymptomatic HIV-1-infected humans: a phase I evaluation. AIDS 16:2019–2025. 10.1097/00002030-200210180-00006 [DOI] [PubMed] [Google Scholar]
- 31.Trkola A, Kuster H, Rusert P, Joos B, Fischer M, Leemann C, Manrique A, Huber M, Rehr M, Oxenius A, Weber R, Stiegler G, Vcelar B, Katinger H, Aceto L, Gunthard HF. 2005. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat. Med. 11:615–622. 10.1038/nm1244 [DOI] [PubMed] [Google Scholar]
- 32.Mehandru S, Vcelar B, Wrin T, Stiegler G, Joos B, Mohri H, Boden D, Galovich J, Tenner-Racz K, Racz P, Carrington M, Petropoulos C, Katinger H, Markowitz M. 2007. Adjunctive passive immunotherapy in human immunodeficiency virus type 1-infected individuals treated with antiviral therapy during acute and early infection. J. Virol. 81:11016–11031. 10.1128/JVI.01340-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y, West AP, Jr, Kim HJ, Thornton ME, Ward AB, Bjorkman PJ. 2013. Structural basis for enhanced HIV-1 neutralization by a dimeric immunoglobulin G form of the glycan-recognizing antibody 2G12. Cell Rep. 5:1443–1455. 10.1016/j.celrep.2013.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West AP, Jr, Galimidi RP, Foglesong CP, Gnanapragasam PN, Huey-Tubman KE, Klein JS, Suzuki MD, Tiangco NE, Vielmetter J, Bjorkman PJ. 2009. Design and expression of a dimeric form of human immunodeficiency virus type 1 antibody 2G12 with increased neutralization potency. J. Virol. 83:98–104. 10.1128/JVI.01564-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein JS, Webster A, Gnanapragasam PN, Galimidi RP, Bjorkman PJ. 2010. A dimeric form of the HIV-1 antibody 2G12 elicits potent antibody-dependent cellular cytotoxicity. AIDS 24:1633–1640. 10.1097/QAD.0b013e32833ad8c8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo XM, Lei MYY, Feidi RA, West AP, Balazs AB, Bjorkman PJ, Yang LL, Baltimore D. 2010. Dimeric 2G12 as a potent protection against HIV-1. PLoS Pathog. 6:e1001225. 10.1371/journal.ppat.1001225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carragher B, Kisseberth N, Kriegman D, Milligan RA, Potter CS, Pulokas J, Reilein A. 2000. Leginon: an automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol. 132:33–45. 10.1006/jsbi.2000.4314 [DOI] [PubMed] [Google Scholar]
- 38.Potter CS, Chu H, Frey B, Green C, Kisseberth N, Madden TJ, Miller KL, Nahrstedt K, Pulokas J, Reilein A, Tcheng D, Weber D, Carragher B. 1999. Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy 77:153–161. 10.1016/S0304-3991(99)00043-1 [DOI] [PubMed] [Google Scholar]
- 39.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. 2005. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151:41–60. 10.1016/j.jsb.2005.03.010 [DOI] [PubMed] [Google Scholar]
- 40.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. 2009. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166:205–213. 10.1016/j.jsb.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B. 2009. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166:95–102. 10.1016/j.jsb.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17–24. 10.1006/jsbi.1996.0004 [DOI] [PubMed] [Google Scholar]
- 43.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97. 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- 44.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157:38–46. 10.1016/j.jsb.2006.05.009 [DOI] [PubMed] [Google Scholar]
- 45.Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR, Crispin M, Scanlan CN. 2010. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc. Natl. Acad. Sci. U. S. A. 107:13800–13805. 10.1073/pnas.1006498107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125. 10.1128/JVI.79.16.10108-10125.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Depetris RS, Julien JP, Khayat R, Lee JH, Pejchal R, Katpally U, Cocco N, Kachare M, Massi E, David KB, Cupo A, Marozsan AJ, Olson WC, Ward AB, Wilson IA, Sanders RW, Moore JP. 2012. Partial enzymatic deglycosylation preserves the structure of cleaved recombinant HIV-1 envelope glycoprotein trimers. J. Biol. Chem. 287:24239–24254. 10.1074/jbc.M112.371898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9:e1003618. 10.1371/journal.ppat.1003618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 50.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. 10.1038/31405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doores KJ, Fulton Z, Huber M, Wilson IA, Burton DR. 2010. Antibody 2G12 recognizes di-mannose equivalently in domain- and nondomain-exchanged forms but only binds the HIV-1 glycan shield if domain exchanged. J. Virol. 84:10690–10699. 10.1128/JVI.01110-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.West AP, Jr, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. 2014. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 156:633–648. 10.1016/j.cell.2014.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028. 10.1126/science.1118398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Platt EJ, Gomes MM, Kabat D. 2012. Kinetic mechanism for HIV-1 neutralization by antibody 2G12 entails reversible glycan binding that slows cell entry. Proc. Natl. Acad. Sci. U. S. A. 109:7829–7834. 10.1073/pnas.1109728109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khayat R, Lee JH, Julien JP, Cupo A, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. 2013. Structural characterization of cleaved, soluble HIV-1 envelope glycoprotein trimers. J. Virol. 87:9865–9872. 10.1128/JVI.01222-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934. 10.1126/science.1145373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. 2010. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330:1066–1071. 10.1126/science.1194396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, Li T, Ma L, Fenalti G, Li J, Zhang W, Xie X, Yang H, Jiang H, Cherezov V, Liu H, Stevens RC, Zhao Q, Wu B. 2013. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science 341:1387–1390. 10.1126/science.1241475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klein JS, Bjorkman PJ. 2010. Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 6:e1000908. 10.1371/journal.ppat.1000908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. 10.1038/nature07159 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.