ABSTRACT
In polarized epithelial cells, influenza A virus hemagglutinin (HA) and neuraminidase (NA) are intrinsically associated with lipid rafts and target the apical plasma membrane for viral assembly and budding. Previous studies have indicated that the transmembrane domain (TMD) and cytoplasmic tail (CT) of HA and NA are required for association with lipid rafts, but the raft dependencies of their apical targeting are controversial. Here, we show that coexpression of HA with NA accelerated their apical targeting through accumulation in lipid rafts. HA was targeted to the apical plasma membrane even when expressed alone, but the kinetics was much slower than that of HA in infected cells. Coexpression experiments revealed that apical targeting of HA and NA was accelerated by their coexpression. The apical targeting of HA was also accelerated by coexpression with M1 but not M2. The mutations in the outer leaflet of the TMD and the deletion of the CT in HA and NA that reduced their association with lipid rafts abolished the acceleration of their apical transport, indicating that the lipid raft association is essential for efficient apical trafficking of HA and NA. An in situ proximity ligation assay (PLA) revealed that HA and NA were accumulated and clustered in the cytoplasmic compartments only when both were associated with lipid rafts. Analysis with mutant viruses containing nonraft HA/NA confirmed these findings. We further analyzed lipid raft markers by in situ PLA and suggest a possible mechanism of the accelerated apical transport of HA and NA via clustering of lipid rafts.
IMPORTANCE Lipid rafts serve as sites for viral entry, particle assembly, and budding, leading to efficient viral replication. The influenza A virus utilizes lipid rafts for apical plasma membrane targeting and particle budding. The hemagglutinin (HA) and neuraminidase (NA) of influenza virus, key players for particle assembly, contain determinants for apical sorting and lipid raft association. However, it remains to be elucidated how lipid rafts contribute to the apical trafficking and budding. We investigated the relation of lipid raft association of HA and NA to the efficiency of apical trafficking. We show that coexpression of HA and NA induces their accumulation in lipid rafts and accelerates their apical targeting, and we suggest that the accelerated apical transport likely occurs by clustering of lipid rafts at the TGN. This finding provides the first evidence that two different raft-associated viral proteins induce lipid raft clustering, thereby accelerating apical trafficking of the viral proteins.
INTRODUCTION
Influenza virus is an enveloped, negative-stranded, segmented RNA virus belonging to the Orthomyxoviridae family. The virion consists of three integral membrane proteins, hemagglutinin (HA), neuraminidase (NA), and ion channel protein M2. A layer of matrix protein M1 is present underneath the lipid envelope and encases viral ribonucleoprotein (vRNP) complexes. The influenza virus buds from the apical plasma membrane (PM), which is divided by tight junctions in polarized epithelial cells (1). It is considered that all viral components are targeted to the apical PM, where particle budding occurs. HA, NA, and M2 are synthesized at the endoplasmic reticulum (ER) and are transported to the apical PM through the trans-Golgi network (TGN). The apical sorting signals were identified in the transmembrane domains (TMDs) of both HA and NA (2, 3). Many studies indicate that during the apical trafficking, HA and NA are associated with lipid raft microdomains, which are enriched in cholesterol and sphingolipids (3, 4), whereas M2 is excluded from these domains (5, 6). Several studies also indicate that the TMD and the cytoplasmic tail (CT) of HA and NA are important for their association with lipid rafts (3, 5, 7). It has been shown that, in the case of HA, palmitoylation at three conserved cysteines in the TMD-CT region is required for association with lipid rafts (8). A very recent study suggested that M2 was a key player in influenza virus particle budding, which is independent of the endosomal protein sorting complex required for transport (ESCRT) (9).
Lipid rafts are thought to function as platforms for selective concentration of raft-associated proteins to promote protein-protein interactions for their functions (10). Lipid rafts have also been shown to play pivotal roles in apical trafficking in polarized cells (11) and in signal transduction pathways, such as Ras signaling (12) and phosphatidylinositol 4,5-bisphosphate (PIP2) signaling (13). It has been suggested that for influenza virus HA and NA, the association with lipid rafts constitutes a part of the machinery necessary for apical trafficking in polarized cells (14, 15). Previous studies have indicated that disruption of lipid rafts by treatment with methyl-β-cyclodextrin (MβCD) and lovastatin delays the TGN-to-apical PM trafficking of HA and missorts HA to the basolateral membrane, whereas vesicular stomatitis virus G protein, a nonraft-associated basolateral marker, remained unaffected (16), suggesting that raft association is required for apical transport of proteins. However, a number of studies have indicated that some mutations in the HA TMD and CT did not impair apical targeting of HA, irrespective of whether they caused significant reductions in the raft association (5, 7), suggesting that raft association is not essential for apical transport of HA.
Glycosylphosphatidylinositol (GPI)-anchored proteins (GPI-APs) are not integral membrane proteins but are associated with lipid rafts through the GPI moieties. In this group of proteins, the raft association is not a determinant for apical sorting because both apical and basolateral GPI-APs are associated with lipid rafts (17). Interestingly, only apical, but not basolateral, GPI-APs form oligomer complexes when they are associated with lipid rafts. When oligomerization of GPI-APs was impaired by mutations, the GPI-APs were missorted to the basolateral PM domain (17). A recent model has suggested that the oligomerization of GPI-APs promotes their stabilization in lipid rafts, leading to their incorporation into apical transport vesicles (10).
Influenza virus budding and release require the assembly of viral components, which occurs either during their trafficking to the apical PM or at the stage of particle budding (6, 18, 19). During the trafficking, all viral components, HA, NA, M2, M1, and vRNP, either individually or in their complex forms, are targeted to the apical PM and form a higher order of complex (7, 19, 20). Although the apical sorting determinants for individual viral components have been relatively well studied (2, 3), the molecular mechanisms and kinetics involved in their apical targeting have not been elucidated.
In this study, we focused on the kinetics of apical targeting of influenza virus envelope proteins in polarized MDCK cells. We found that HA and NA or M1, but not M2, mutually accelerated their apical PM targeting. Using TMD-CT mutants of HA and NA, we show that the association of HA and NA with lipid rafts is necessary for the acceleration of their apical PM targeting. Our data indicate that HA and NA come into close proximity (clustering) in lipid rafts upon coexpression. Our data also show the clustering of lipid rafts upon coexpression of HA and NA, suggesting a possible mechanism of accelerated apical transport of HA and NA via the clustering of lipid rafts.
MATERIALS AND METHODS
Viruses and plasmids.
The H141Y and E142Q mutations were introduced into the HA gene of the influenza A/Puerto Rico/8/34 (PR8) virus by inverted PCR. This PR8 derivative is referred to as the wild-type (wt) strain in the present study. The authentic PR8 strain was used as the parental virus. The wt and mutant PR8 viruses were generated by a reverse genetics system with PolI plasmids (pHH21) and protein expression plasmids, as described previously (21). Briefly, 293T cells were transfected with PolI plasmids for synthesis of each viral RNA segment and protein expression plasmids for the PB1, PB2, PA, nucleoprotein (NP), HA, and NA. At 6 h posttransfection (hpt), the cell medium was replaced with Opti-MEM I (Gibco) supplemented with 5 μg/ml acetyl trypsin and 0.3% bovine serum albumin (BSA), and the cells were incubated for 2 or 3 days. Recovered viruses were grown in Madin-Darby canine kidney (MDCK) cells stably expressing HA (MDCK-HA) (the kind gift of N. Takizawa from the Institute of Microbial Chemistry, Japan).
The HA and NA constructs with deletion of the CT (ΔCT-HA and ΔCT-NA) for recovery of recombinant viruses have essentially been described elsewhere (22, 23). For ΔCT-HA, three consecutive stop codons were placed at the end of the TMD coding sequence, and the downstream nucleotide sequence was left intact. For ΔCT-NA, the authentic start codon was mutated, and a new start codon was created at the beginning of the TMD coding sequence. The constructs were cloned into pHH21. The HA and NA constructs with alanine substitutions in the CT (residues 557 to 559 of HA [HA557–559], HA560–563, HA564–566, residues 2 to 3 of NA [NA2–3], NA4–6, and NA2–6) and TMD (HA533–535, HA550–552, NA7–10, and NA31–35) and those with cysteine-to-serine substitutions at three palmitoylation sites (HA-SSS) were generated by overlapping PCR and were cloned into pHH21. The viruses with similar alanine substitutions have been described previously (3, 5, 8).
The open reading frames of the wt-HA, NA, M1, and M2 genes were cloned into the eukaryotic expression plasmid pCAGGS. The open reading frames of the HA and NA constructs with deletion of the CT and those with the alanine substitutions were similarly cloned into pCAGGS. The cDNAs encoding CD59, the 75-kDa neurotrophin receptor (p75), and their green fluorescent protein (GFP)-tagged versions (GFP-CD59 and p75-GFP) were also cloned into pCAGGS.
Cell culture, infection, and DNA transfection.
MDCK and 293T cells were maintained in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum. MDCK cells were seeded into 12-well plates (1 × 106 cells/well) and were polarized by a 12-h incubation. The polarized MDCK cells were used throughout this study. The MDCK cells were infected with the wt virus at a multiplicity of infection (MOI) of 0.1 or 1 for 1 h. Transfection of DNA was carried out using Lipofectamine LTX (Invitrogen). To synchronize protein expression from plasmids, DNA-Lipofectamine complexes were sedimented by plate centrifugation at 250 × g for 5 min. In some experiments, MDCK cells were transfected with DNA and subsequently superinfected with the authentic PR8 virus.
Virus growth and plaque assay.
MDCK cells were inoculated with virus at an MOI of 0.1 in Opti-MEM I supplemented with 0.3% BSA for 1 h at 37°C. After being washed, the cells were incubated in Opti-MEM I supplemented with 5 μg/ml acetyl trypsin and 0.3% BSA. The culture medium was harvested at various time points and was subjected to plaque assay on MDCK or MDCK-HA cells.
Cholesterol depletion.
For the inhibition of cholesterol synthesis, 293T and MDCK cells were pretreated with 8 μM lovastatin (Merck) for 12 h. After transfection, the cells were incubated in the presence of 8 μM lovastatin for 12 or 24 h. The cells were further treated with 5 or 10 mM methyl-β-cyclodextrin (MβCD) (Sigma) in the presence of 8 μM lovastatin for 1 h.
Indirect immunofluorescence assay.
Polarized MDCK cells were grown on coverslips in 12-well plates and were either infected with virus or transfected with protein expression plasmids. The cells were fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.5% Triton X-100 (TX-100). Following blocking, the cells were incubated with primary antibodies (Abs) and subsequently with secondary Abs conjugated with Alexa Fluor 488, 568, or 647 (Molecular Probes). The following Abs were used as primary Abs: mouse anti-HA mAb12-1G6 (24), mouse anti-HA Ab (TaKaRa), rabbit anti-NA Ab (Sino Biological), sheep anti-NA Ab (R&D Systems), rabbit anti-M1 polyclonal Ab (25), mouse anti-M2 Ab (Santa Cruz), mouse anti-NP mAb61A5 (26), mouse anti-CD59 Ab (Abcam), rabbit anti-TGN46 Ab (Abcam), rabbit anti-ZO-3 Ab (Invitrogen), and rabbit anti-caveolin-1 Ab (Santa Cruz). For costaining with HA and M2 or vRNP, anti-HA mAb12-1G6 was prelabeled by using a Zenon Alexa Fluor 488 mouse IgG1 labeling kit (Invitrogen), and the mouse anti-HA (TaKaRa), anti-M2, and anti-NP Abs were similarly prelabeled with Alexa Fluor 568, as described previously (24). Nuclear staining was carried out with TO-PRO-3 (Molecular Probes) or 4′,6′-diamidino-2-phenylindole (DAPI; Molecular Probes). The cells were observed with a laser scanning confocal microscope (TCS-SP5II AOBS; Leica). Confocal images were collected at 0.5-μm intervals along the z axis. Reconstitution of an x-z plane was processed using ImageJ software (27). Fifty antigen-positive cells were observed in each experiment, and patterns of antigen distribution were analyzed.
For quantitative analysis of HA and NA expression, polarized MDCK cells on coverslips in 12-well plates (1 × 106 cells/well) were singly transfected with an HA expression plasmid or cotransfected with the HA and an NA expression plasmids. After fixation with 4% PFA, the cells were incubated with mouse anti-HA Ab (TaKaRa) (for HA on the apical PM). The cells were refixed with 4% PFA and were permeabilized with 0.5% TX-100. The cells were incubated with rabbit anti-NA Ab (Sino Biological) (for total NA) and subsequently with anti-mouse IgG conjugated with Alexa Fluor 647 and anti-rabbit IgG conjugated with Alexa Fluor 568. For total HA, the cells were refixed with 4% PFA and blocked with nonspecific mouse Ig. The cells were finally incubated with anti-HA mAb12-1G6 prelabeled by using a Zenon Alexa Fluor 488 mouse IgG labeling kit (Invitrogen). Nuclear staining was carried out with DAPI (Molecular Probes). Confocal images were collected at 0.5-μm intervals along the z axis. In each cell, the sum of intensity values of a z stack was calculated as a z-projection image by the “sum slices” command of ImageJ. In each acquired channel, single cell area and mean fluorescence intensities (MFIs; arbitrary units) were measured. The MFIs of the Alexa Fluor 488 and 568 channels were evaluated as total expression levels of HA and NA in each cell, respectively. The total fluorescence intensity (MFI × area) of HA divided by that of NA was evaluated as the expression ratio of HA to NA. The cells (39 to 45 cells) were subjected to analysis of HA distribution patterns (either apical PM alone or PMs plus cytoplasmic compartments). Sparse MDCK cells (1 × 105 cells/well) were also used in the experiment shown in Fig. 3C.
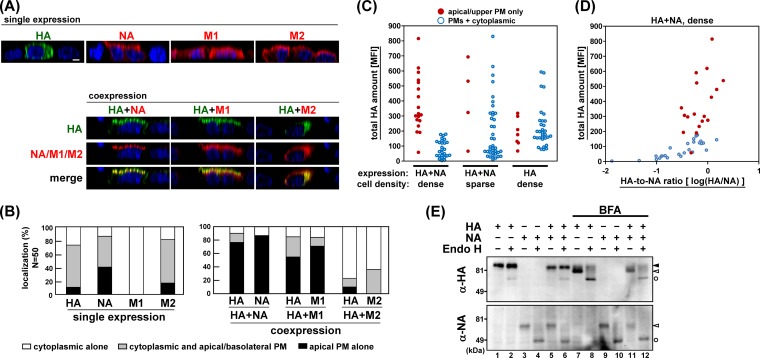
FIG 3.
Acceleration of apical PM targeting of HA by coexpression with NA or M1. (A) Polarized MDCK cells were cotransfected with the combination of an HA expression plasmid and an NA, M1, or M2 expression plasmid. At 12 hpt, the cells were fixed and stained with anti-HA mAb12-1G6 (green) and sheep anti-NA, rabbit anti-M1, or mouse anti-M2 Ab (red). For costaining, anti-HA mAb12-1G6 and mouse anti-M2 Ab were prelabeled with Alexa Fluor dyes. Nuclei were stained with TO-PRO-3 (blue). Images in the x-z planes are shown. All images were taken at the same magnification. Scale bar, 10 μm. (B) In each experiment, 50 antigen-positive cells were subjected to analysis of the viral antigen distribution pattern. In coexpression experiments, 50 antigen double-positive (HA-positive and NA, M1, or M2-positive) cells were chosen and subjected to distribution pattern analysis for each viral protein. (C) Polarized MDCK cells (1 × 106 cells/well) or sparse MDCK cells (1 × 105 cells/well) were singly transfected with an HA or doubly transfected with HA and NA expression plasmids. At 12 hpt, intracellular viral antigens were stained with anti-HA mAb12-1G6 and rabbit anti-NA Ab. Nuclei were stained with DAPI. In each cell, the fluorescence intensities of serial confocal images were accumulated in z direction, and the MFIs were calculated. For HA localization, 39 or 45 HA-positive cells were analyzed and sorted into two categories (either apical PM alone or PMs plus cytoplasmic compartments). (D) The data of coexpression in polarized cells were also sorted by the ratio of the MFI of HA to NA. (E) Polarized MDCK cells were transfected with HA and/or NA expression plasmids. At 12 hpt, cells were harvested, and protein samples were treated with Endo H and analyzed by Western blotting with rabbit anti-HA and sheep anti-NA Abs. ▲, Endo H-resistant (complex glycan) forms; △, Endo H-sensitive (high mannose) forms; ○, Endo H-digested products. Brefeldin A (BFA) was added to the cells as an inhibitor of the transport of the protein from the endoplasmic reticulum to the Golgi compartment.
TX-100 solubilization analysis and coimmunoprecipitation.
293T cells were seeded into 12-well plates (5 × 105 cells/well) or 10-cm-diameter dishes (5 × 106 cells/dish). The cells were singly transfected with HA and NA expression plasmids or cotransfected with a combination of HA and NA, HA and M1, or HA and GFP-CD59 expression plasmids. At 24 hpt, the cells were resuspended with cold TNE buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, and 150 mM NaCl) containing 1 mM dithiothreitol (DTT) and protease inhibitor Complete Mini cocktail (Roche). After brief sonication, the samples were treated with 1% TX-100 at 4°C or 37°C for 30 min and then centrifuged at 17,400 × g for 30 min at 4°C to separate the soluble and insoluble fractions. For TX-100 solubilization analysis, both the supernatants and pellets were adjusted to be the same volume with TNE buffer and analyzed by Western blotting. For coimmunoprecipitation, the supernatants were mixed with 10 μg of anti-HA mAb12-1G6 and were incubated at 4°C for 90 min. The supernatants were subsequently mixed with preblocked protein G-Sepharose beads (GE Healthcare) and were incubated at 4°C for 60 min. After samples were washed with TNE buffer containing 0.1% TX-100 three times, immunoprecipitates were eluted from the beads by boiling with SDS sample buffer and analyzed by Western blotting with sheep anti-NA Ab (R&D Systems) and rabbit anti-M1 polyclonal Ab (25). GFP-CD59 was detected with mouse anti-GFP Ab (Sigma).
In situ PLA.
An in situ proximity ligation assay (PLA) was performed according to the manufacturer's instructions (Olink Biosciences). Briefly, polarized MDCK cells were either transfected with combinations of protein expression plasmids or were infected with virus. At 9 hpt or 3.5 or 5 h postinfection (hpi), the cells were fixed with 4% PFA and permeabilized with 0.5% TX-100. After a blocking step, the cells were incubated with mouse anti-HA mAb12-1G6, rabbit anti-NA Ab, mouse anti-GFP Ab, or rabbit caveolin-1 Ab (Santa Cruz) for 1 h at room temperature. After being washed, the cells were incubated with two species-specific secondary antibodies conjugated with unique oligonucleotides (anti-rabbit Plus and anti-mouse Minus) as PLA probes. Phosphorylated connector oligonucleotides were hybridized to the PLA probes. If the PLA probes are present in close proximity (<40 nm), the phosphorylated oligonucleotides are formed in a circular template by a ligase. This template was subsequently amplified by rolling-circle amplification with DNA polymerase and was detected with fluorescently labeled complementary oligonucleotide probes (Duolink Detection Kit Red). TGN46 was stained with rabbit anti-TGN46 Ab prelabeled by using a Zenon Alexa Fluor 488 rabbit IgG labeling kit (Invitrogen). The PLA signals were observed by confocal microscopy, and the number of signals was automatically counted with BlobFinder software (28). Data are shown as means with standard deviations from two independent experiments (at least 10 cells). Statistical significance was determined by Student's t test. P values of <0.05 were considered statistically significant.
Treatment with Endo H.
Polarized MDCK cells (1 × 106 cells/well) were transfected with HA and/or NA expression plasmids. At 12 hpt, the cells were resuspended with cold phosphate-buffered saline and sonicated. The protein samples were denatured and subsequently digested with endoglycosidase H (Endo H; New England BioLabs) for 1 h at 37°C. The samples were analyzed by Western blotting with rabbit anti-HA Ab (Sino Biological) and sheep anti-NA Ab (R&D Systems). As control, 20 μM brefeldin A was added to the cells at 6 hpt.
RESULTS
Apical PM targeting of HA in singly transfected cells was slower than that of HA in infected cells.
HA is transported to the apical PM via the secretory pathway in polarized cells (1). We have previously generated an anti-HA monoclonal antibody (mAb12-1G6) that specifically binds to the loop (amino acid sequence QGKS at positions 142 to 145) of the head domain in H5 HA (24). We found that mAb12-1G6 failed to detect the authentic PR8 HA but did detect HA when it contained H141Y and E142Q substitutions without any reduction in infectivity (Fig. 1D). When prelabeled mAb12-1G6 and commercial mouse anti-HA Ab (TaKaRa) were used for costaining, their staining patterns were identical (Fig. 1E), suggesting that mAb12-1G6 did not detect a specific HA population. Thus, we used the PR8 derivative containing H141Y and E142Q mutations as the wild-type (wt) strain in the present study and investigated apical PM targeting of HA.
FIG 1.
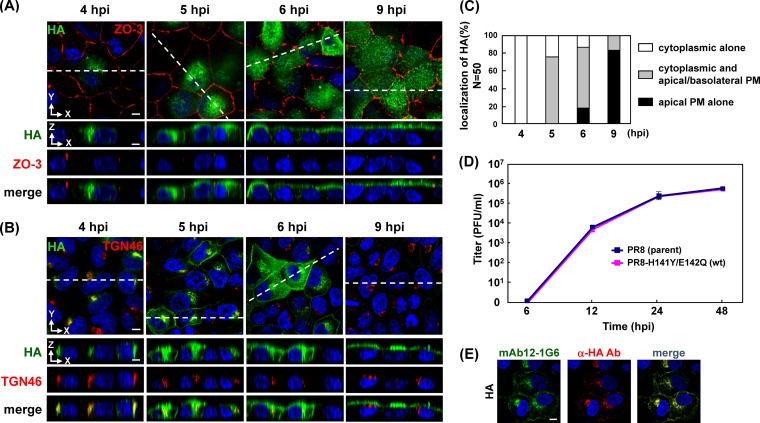
Temporal study of apical PM targeting of HA in infected cells and growth kinetics of virus. Polarized MDCK cells were infected with a PR8 derivative containing H141Y and E142Q substitutions in HA (referred to as wt in this study). At various time points (indicated), cells were stained with anti-HA mAb12-1G6, anti-ZO-3, and anti-TGN46 Abs. Localization of HA (green) and ZO-3 (red) (A) or TGN46 (red) (B) are shown in the x-y and x-z planes. Nuclei were stained with TO-PRO-3 (blue). The dashed lines in x-y images indicate the positions of x-z images. (C) For semiquantification of the HA localization in infected cells, 50 HA-positive cells were observed at each time point, and the number of cells with each distribution pattern of HA was counted. (D) MDCK cells were infected with the authentic PR8 (parent) or the PR8–H141Y/E142Q (wt) virus at an MOI of 0.1. At the indicated time points, an aliquot of the culture medium was harvested, and virus titers were measured by plaque assay on MDCK cells. (E) Polarized MDCK cells were infected with the wt virus. At 5 hpi, the cells were fixed and costained with prelabeled anti-HA mAb12-1G6 (green) and mouse anti-HA Ab (TaKaRa) (red). Nuclei were stained with TO-PRO-3 (blue). All images were taken at the same magnification. Scale bar, 10 μm. α, anti.
To understand the overall kinetics of intracellular trafficking of HA, we initially carried out a time course study. Polarized MDCK cells were infected with the wt PR8 virus and were subjected to immunofluorescent assays at various time points (4, 5, 6, and 9 hpi) with mAb12-1G6 and anti-ZO-3 (for tight junctions) or anti-TGN46 (for TGN) Ab (Fig. 1A and B). Anti-ZO-3 Ab was used to differentiate the apical from the basolateral PM. The TGN is the major sorting organelle in the cytoplasmic trafficking pathways (29). Serial confocal z sections were collected at 0.5-μm intervals from the top to the bottom of cells. As shown in the x-z plane (Fig. 1A and B), three patterns of HA distribution (cytoplasmic compartments; cytoplasmic compartments and both apical and basolateral PMs; only at the apical PM) were observed. We observed 50 HA-positive cells at each time point and sorted them into the three categories (Fig. 1C). At 4 hpi, all HA antigens were observed in the cytoplasmic compartments (100% of HA-positive cells), most likely at the TGN. At 5 hpi, HA was observed both in the cytoplasmic compartments and at the apical/basolateral PMs (76% of HA-positive cells). At 6 hpi, the majority of HA was still localized to the cytoplasmic compartments and at the apical/basolateral PMs (70% of HA-positive cells), but a population of HA had accumulated at the apical PM in some cells (17% of HA-positive cells). HA accumulation at the apical PM became evident at 9 hpi (83% of HA-positive cells) (Fig. 1B and C). HA has been suggested to be transported initially to the basolateral PM and then to the apical PM via transcytosis (30). Overall, these data indicated that the HA in infected cells was accumulated at the apical PM within another 4 or 5 h after the appearance of HA in the cytoplasmic compartments, such as the TGN.
To examine whether singly expressed HA followed similar transport kinetics, polarized MDCK cells were transfected with a plasmid expressing wt-HA and were subjected to immunofluorescence assays. The majority of HA was localized at the TGN at 9 hpt (84% of HA-positive cells), similar to the HA distribution pattern in infected cells at 4 hpi. Surprisingly, HA was broadly distributed in the cytoplasmic compartments even at 12 hpt. HA accumulation at the apical PM was observed in some cells at 24 hpt (40% of HA-positive cells) and became prominent at 48 hpt (Fig. 2A and C), suggesting that the apical targeting of HA in singly transfected cells was very slow compared with that of HA in infected cells.
FIG 2.
Temporal study of apical PM targeting of HA in transfected cells and acceleration of the apical targeting upon superinfection. (A) Polarized MDCK cells were transfected with a plasmid expressing wt HA. At various time points (indicated), the cells were stained with anti-HA mAb12-1G6 and anti-TGN46 Ab. Localizations of HA (green) and TGN46 (red) are shown in the x-z plane. (B) Polarized MDCK cells were similarly transfected with a plasmid expressing wt-HA. At 6 hpt, the cells were infected with the authentic PR8 strain (parent) of influenza virus. At 12 hpt, the cells were stained with prelabeled mAb12-1G6 (green; only reactive with wt-HA) and anti-NP Ab (red; for detection of superinfection). Nuclei were stained with TO-PRO-3 (blue). Localizations of HA (green) and vRNP (red) are shown in the x-z plane. All images were taken at the same magnification. Scale bar, 10 μm. (C) For semiquantification of the HA localization, 50 mAb12-1G6-reactive (transfected) HA-positive cells were observed at each time point, and the number of cells with each distribution pattern of HA was counted.
Apical targeting of transfected HA was accelerated by superinfection.
To investigate whether efficient apical targeting of HA required coexpression of viral components other than HA, polarized MDCK cells were transfected with the wt-HA expression plasmid, and, at 6 hpt, the cells were superinfected with the authentic PR8 virus, the HA of which was not reactive with mAb12-1G6. Immunostaining revealed that upon superinfection, the transfected HA became accumulated at the apical PM at 12 hpt (50% of HA-positive cells) (Fig. 2B and C). In contrast, transfected HA without superinfection was still localized in the cytoplasmic compartments (Fig. 2A and C). These data suggested that the apical PM targeting of HA was accelerated by the presence of other viral components.
Apical targeting of HA was accelerated by coexpression with NA or M1.
Integral membrane proteins of influenza virus (HA, NA, and M2) are transported to the apical PM via the secretory pathway (31, 32, 33). Previous studies identified the apical sorting signal in the TMD of HA and NA (2, 3). M1 lacks an intrinsic apical sorting signal but has been suggested to be transported to the apical PM through association with HA, NA, and M2 CTs (7, 20). To identify the viral components responsible for the acceleration of apical targeting of HA, polarized MDCK cells were cotransfected with a combination of the HA expression plasmid and an NA, M1, or M2 expression plasmid. The cells were fixed at 12 hpt and were subjected to immunostaining with the anti-HA mAb12-1G6 and sheep anti-NA, anti-M1, or anti-M2 Ab (Fig. 3A). In singly transfected cells, a relatively large fraction of NA was accumulated at the apical PM at 12 hpt (40% of NA-positive cells). M1 was diffusely distributed in the cytoplasm and the nucleus, consistent with previous studies (34). M2 was observed both in the cytoplasmic compartments and at the apical/basolateral PMs. In doubly transfected cells, we found that HA was more accumulated at the apical PM when it was coexpressed with NA or M1 (79% in HA and NA double-positive cells; 52% in HA and M1 double-positive cells), suggesting that apical targeting of HA was accelerated by coexpression with NA or M1. Interestingly, apical targeting of NA and M1 was also accelerated by coexpression with HA, compared with the level in the singly transfected cells (85% versus 40% in the case of NA; 73% versus 0% in the case of M1) (Fig. 3A and B), suggesting that coexpression of HA with NA and M1 mutually accelerated their apical PM targeting.
In contrast, when HA was coexpressed with M2, apical targeting of HA and M2 was rather inhibited (Fig. 3A and B). The inhibition was also observed at a later time point (48 hpt) (data not shown). In cells expressing HA alone, HA was accumulated at the apical PM. In contrast, HA was distributed in the cytoplasmic compartments when M2 was coexpressed, suggesting that HA and M2 mutually interfered with their apical transport. This observation is in accordance with a previous study in which M2 inhibited the TGN release of apical secreted proteins (35). However, such inhibition was not apparent in infected cells. At 9 hpi, both HA and M2 accumulated at the apical PM (data not shown), suggesting that such inhibition may have been cancelled in the infected cells.
It is possible that high-level expression of HA and NA facilitates their apical targeting. To explore this possibility, intracellular expression levels of HA and NA were measured. In each cell, serial confocal z sections were collected at 0.5-μm intervals, and the MFI was calculated by summing the z stack. The MFIs were evaluated as total expression levels of HA and NA, and the cells were subjected to analysis of HA distribution patterns (either apical PM alone or PMs plus cytoplasmic compartments) (Fig. 3C). In singly transfected cells, apical accumulation of HA was not linked with the expression level of HA. In some cells, apical accumulation of HA was not seen even when the expression level of HA was high. In coexpressed cells, apical accumulation of HA was observed at various levels of HA expression (Fig. 3C) although the apical targeting was rarely seen at a very low level of HA expression (below the threshold of visible fluorescence intensity). These data suggested that the expression level of HA was not fully responsible for the apical targeting of HA. When the expression ratio of HA-to-NA was calculated from the total fluorescence intensities of HA and NA, no apparent relationship between the apical targeting of HA and the expression ratio of HA to NA was observed (Fig. 3D).
The endoplasmic reticulum-to-Golgi compartment transport is responsible for the apical protein trafficking. We employed Endo H digestion assays to explore whether HA coexpressed with NA was transported to the Golgi compartment faster than HA expressed alone. Results indicated that the sensitivity of HA to Endo H in the coexpressed cells was very similar to that of HA in singly expressed cells (Fig. 3E), suggesting that the acceleration of apical targeting of HA occurred at or after the Golgi compartment. The sensitivity of NA to Endo H in the coexpressed cells was also similar to that of NA in singly expressed cells (Fig. 3E) although the fact that a large fraction of NA seemed to be Endo H sensitive in Western blotting suggested that antibody-based detection in two different assays may not have detected the identical population of NA.
Mutations of HA/NA involved in the association with lipid rafts abrogated acceleration of their apical targeting.
HA and NA are intrinsically associated with lipid raft microdomains, whereas M2 is excluded from these domains (5, 6). HA is a type I transmembrane (TM) protein and contains the regions responsible for lipid raft association in the CT and TMD (5, 7). Previous studies have shown that hydrophobic amino acids in the outer leaflet of the TMD (4, 5) and palmitoylation at three cysteine residues in the TMD-CT (8) are required for the association of HA with lipid rafts. One study has shown that deletion of the CT in HA reduces the lipid raft association without affecting the apical transport (7). NA is a type II TM protein, and the residues located at the TMD and CT are also involved in the association of NA with lipid rafts (3, 7).
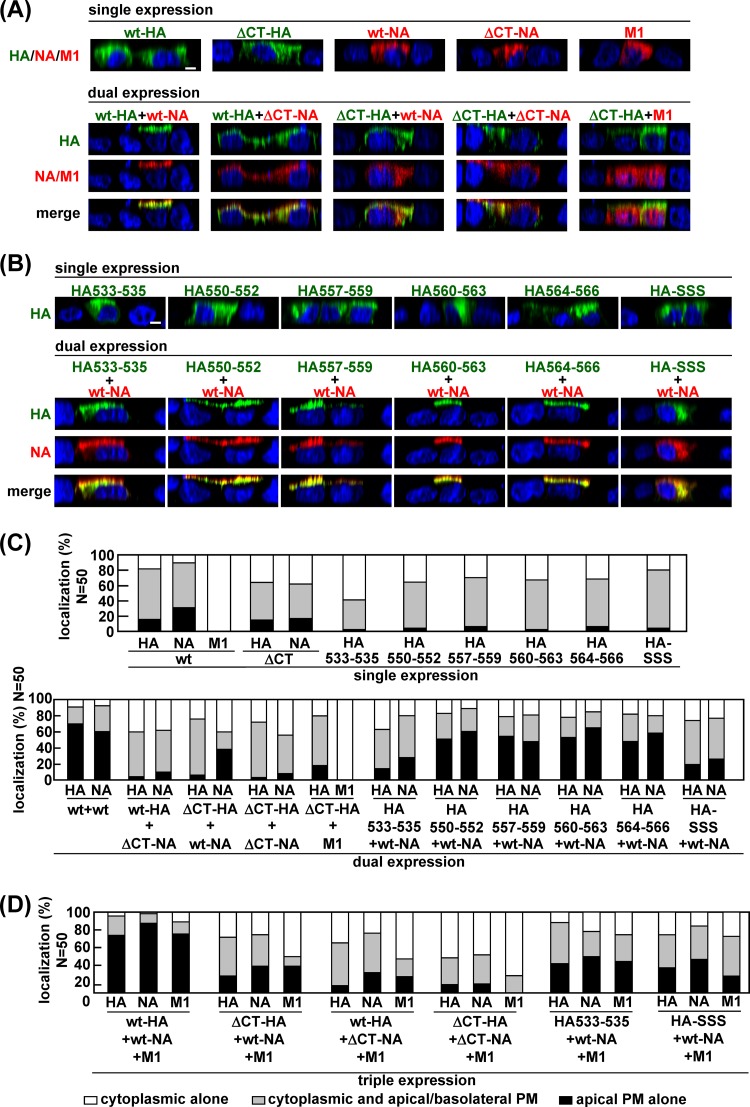
To examine whether the lipid raft association of HA and NA was linked to the acceleration of their apical transport, we constructed HA and NA mutants with CT deletions (ΔCT-HA and ΔCT-NA) (Fig. 4). Polarized MDCK cells were cotransfected with a combination of these constructs and subjected to confocal microscopy at 12 hpt (Fig. 5A). In the singly transfected cells, ΔCT-HA was predominantly observed in the cytoplasmic compartments, and only 15% of HA-positive cells showed HA accumulation at the apical PM. This distribution pattern was similar to the localization pattern of wt-HA. ΔCT-NA was also localized mostly to the cytoplasmic compartments, suggesting less efficient apical transport than for the wt-NA (17% versus 30% of NA-positive cells) (Fig. 5A and C). When polarized MDCK cells were cotransfected with a combination of the wt-HA plus wt-NA, wt-HA plus ΔCT-NA, ΔCT-HA plus wt-NA, or ΔCT-HA plus ΔCT-NA constructs, we found that apical targeting of ΔCT-HA was not accelerated despite the coexpression with wt-NA (apical accumulation in 6% of ΔCT-HA and wt-NA double-positive cells), and, similarly, that apical targeting of ΔCT-NA was not accelerated despite the coexpression of wt-HA (apical accumulation in 10% of wt-HA and ΔCT-NA double-positive cells) (Fig. 5A and C). These results suggested that both CT domains were necessary for the mutual acceleration of apical targeting of HA and NA. Several studies have shown that M1 is intrinsically not associated with the membrane but is targeted to the apical PM through the interaction with the CT domains of HA and NA (7, 20). Consistent with these reports, when M1 was coexpressed with ΔCT-HA, M1 failed to target the apical PM (compare Fig. 3 and 5).
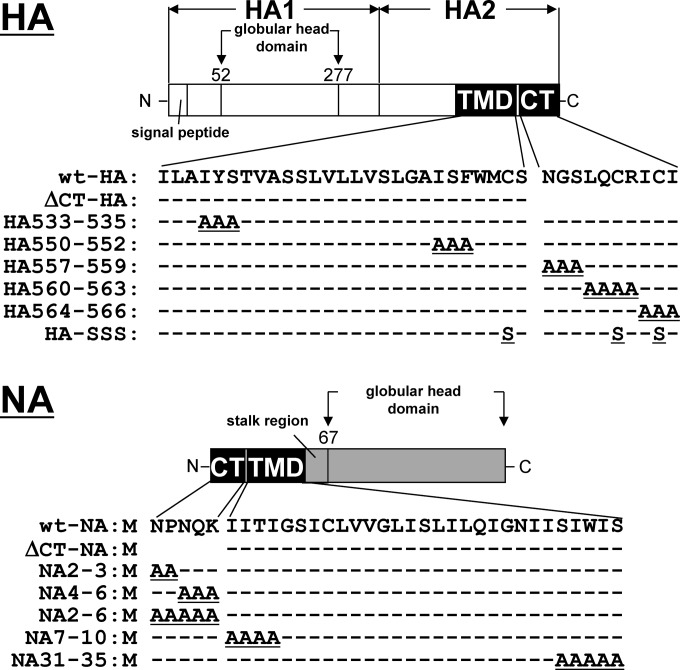
FIG 4.
Schematic representation of HA and NA mutants. HA consists of the N-terminal HA1 and C-terminal HA2 subunits, the latter of which contains the TMD (27 amino acids) and CT (10 amino acids). NA consists of the N-terminal CT (5 amino acids) and TMD (29 amino acids), the stalk region, and the C-terminal globular head domain. Deletions are shown as blanks, and alanine or serine substitutions are underlined. Dashed lines indicate an absence of amino acid substitutions.
FIG 5.
Apical PM targeting of HA and NA mutants with CT deletions and HA TMD mutants. Polarized MDCK cells were cotransfected with double (A and B) and triple (D) combinations of wt and HA and NA mutant expression plasmids, as indicated above the panels. At 12 hpt, the cells were stained with anti-HA mAb12-1G6 (green) and sheep anti-NA or anti-M1 Ab (red). Nuclei were stained with TO-PRO-3 (blue). All images were taken at the same magnification, and images in the x-z planes are shown. Scale bar, 10 μm. For semiquantification, 50 HA and NA or M1 double-positive cells (C) and 50 HA, NA, and M1 triple-positive cells (D) were subjected to antigen distribution pattern analysis, and the distribution patterns of each of HA, NA, and M1 were analyzed separately.
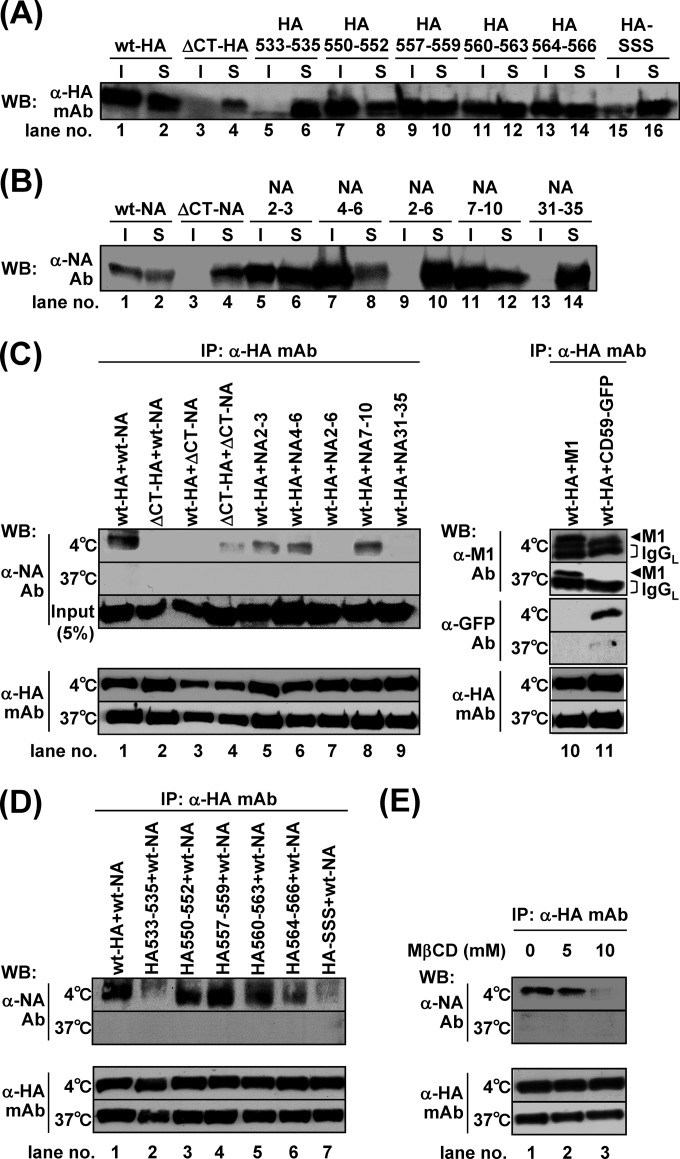
Previous studies have shown that the amino acid sequences of the HA and NA TMDs spanning the outer leaflet of the lipid bilayer were critical for association with lipid rafts, whereas the sequences spanning the inner leaflet were less important (3, 5). We carried out alanine or serine substitution in the TMD-CT of HA (HA533–535, HA550–552, HA557–559, HA560–563, HA564–566, and HA-SSS) and NA (NA2–3, NA4–6, NA2–6, NA7–10, and NA31–35) (Fig. 4). The lipid rafts are defined as the membranes insoluble to nonionic detergent at 4°C (15, 36). We expressed these mutants in 293T cells and solubilized the membrane in the presence of 1% TX-100 at 4°C. TX-100 solubilization analysis showed that the ΔCT-HA, HA533–535, HA-SSS (with mutations at the three palmitoylation sites) (Fig. 6A, lanes 4, 6, and 16), ΔCT-NA, NA2–6, and NA31–35 (Fig. 6B, lanes 4, 10, and 14) were mainly distributed to the detergent-soluble fractions, suggesting the lack of lipid raft association. Next, they were expressed in polarized MDCK cells, and at 12 hpt the antigen localizations were analyzed by confocal microscopy (Fig. 5). In singly transfected cells, none of the HA mutants was accumulated at the apical PM, which was similar to the distribution pattern for the wt-HA (percentages of the respective cell populations are 2% for HA533–535, 4% for HA550–552, 6% for HA557–559, 2% for HA560–563, 6% for HA564–566, and 4% for HA-SSS) (Fig. 5A and C). When the HA mutants were coexpressed with wt-NA, apical targeting of HA550–552, HA557–559, 560–563, and HA564–566 was accelerated (51%, 54%, 53%, and 48% in HA and NA double-positive cells, respectively) (Fig. 5B and C). It should be noted that in these combinations, apical targeting of wt-NA also appeared to be accelerated. In contrast, apical targeting of HA533–535 (with mutations of the amino acids spanning the outer leaflet of the lipid bilayer) and HA-SSS was not accelerated, despite the coexpression of wt-NA (Fig. 5B and C).
FIG 6.
TX-100 solubility of HA and NA and interaction of HA and NA through lipid rafts. For the TX-100 solubilization test, 293T cells were singly transfected with mutant HA (A) or NA (B) expression plasmids (indicated). The cells were lysed with TNE buffer containing 1% TX-100 at 4°C for 30 min. Insoluble (I) and soluble (S) fractions were separated by centrifugation. For the interaction of HA and NA, cells were cotransfected with wt-HA and mutant NA expression plasmids (indicated) (C) or with mutant HA and wt-NA expression plasmids (indicated) (D). M1 was used as a positive control for direct interaction with HA, and GFP-CD59 was used as a marker for lipid rafts. The cells were lysed with TNE buffer containing 1% TX-100 at 4°C or 37°C for 30 min and were subjected to coimmunoprecipitation with anti-HA mAb12-1G6. Precipitates were analyzed by Western blotting using sheep anti-NA, anti-M1, and anti-GFP Abs and anti-HA mAb12-1G6. (E) For the cholesterol depletion experiment, 293T cells were pretreated with lovastatin for 12 h, cotransfected with wt-HA and wt-NA expression plasmids, and incubated at 37°C in the presence of lovastatin. At 24 hpt, the cells were further treated with lovastatin plus 5 mM or 10 mM MβCD for 1 h and were subjected to coimmunoprecipitation experiments. WB, Western blotting; mAb, mAb12-1G6; IP, immunoprecipitation.
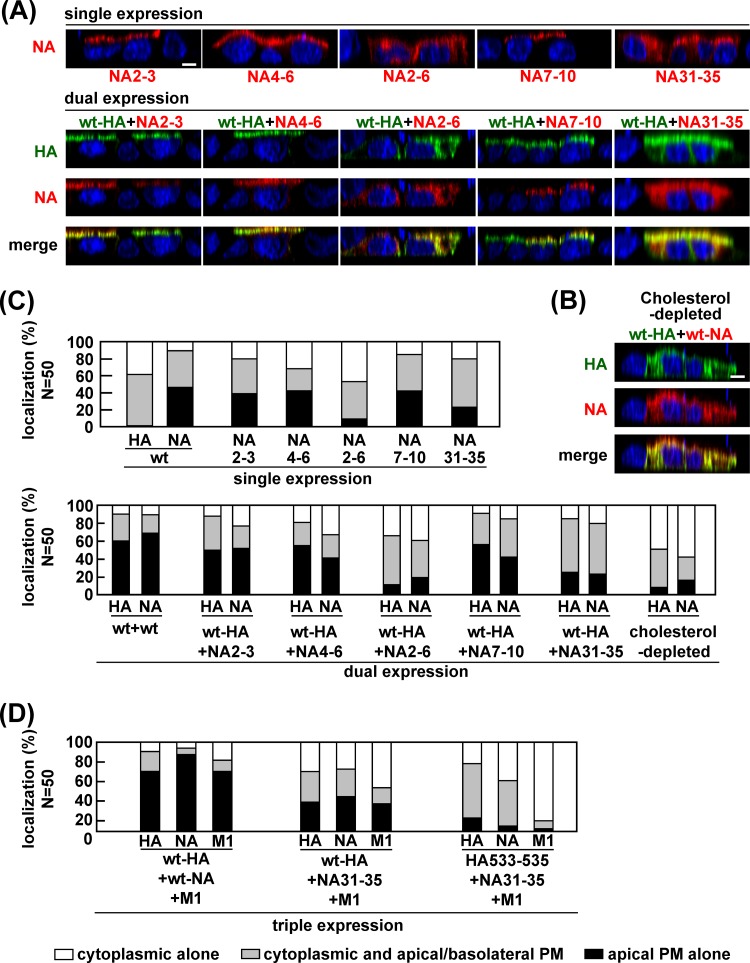
We similarly investigated apical transport of the NA mutants. In singly transfected cells, apical targeting of NA2–6 and NA31–35 was slightly impaired (9% and 23% of NA-positive cells, respectively), compared with wt-NA, NA2–3, NA4–6, and NA7–10 (46%, 39%, 44%, and 42% of NA-positive cells, respectively) (Fig. 7A and C). When wt-HA was coexpressed with these NA mutants, the apical transport of wt-HA was accelerated by coexpression of NA2–3, NA4–6, and NA7–10 (50%, 55%, and 56% in HA and NA double-positive cells, respectively) but not by coexpression of NA2–6 or NA31–35 (11% and 25%, respectively) (Fig. 7A and C), suggesting that the entire CT and the region spanning the outer leaflet of the lipid bilayer in NA were important for acceleration of the apical transport of HA as well as NA. Since some studies have indicated that the association of HA with lipid rafts is dispensable for its apical targeting (5, 7), we examined the intracellular localization of the HA/NA mutants that failed to target to the apical PM (e.g., ΔCT-HA, HA533–535, HA-SSS, ΔCT-NA, NA2–6, and NA31–35) at later time points. As expected, even in the singly transfected cells, these HA/NA mutants were accumulated at the apical PM in more than 70% of HA- or NA-positive cells at 48 hpt, indicating that their apical transport was not blocked but was slowed (data not shown).
FIG 7.
Apical PM targeting of NA TMD mutants and the effect of cholesterol depletion. Polarized MDCK cells were cotransfected with double (A) and triple (D) combinations of wt HA and NA and mutant expression plasmids, as indicated above the panels. (B) For the cholesterol depletion experiment, polarized MDCK cells were pretreated with lovastatin for 12 h and then cotransfected with wt-HA and wt-NA expression plasmids. After 11 h of incubation at 37°C with lovastatin, the cells were further treated with MβCD for 1 h at 37°C. At 12 hpt, the cells were stained with anti-HA mAb12-1G6 (green) and sheep anti-NA Ab (red). Nuclei were stained with TO-PRO-3 (blue). All images were taken at the same magnification, and images in the x-z planes are shown. Scale bar, 10 μm. For semiquantification, 50 HA and NA double-positive cells (C) and 50 HA, NA, and M1 triple-positive cells (D) were subjected to distribution pattern analysis as described in the legend of Fig. 5.
Our data suggested that the acceleration of apical targeting of HA and NA was linked with their lipid raft associations. To confirm this, MDCK cells coexpressing wt-HA and wt-NA were treated with MβCD and lovastatin for depletion of cholesterol and were similarly analyzed by confocal microscopy. As expected, HA and NA were poorly accumulated at the PM (8% and 16%, respectively) (Fig. 7B and C).
We also tested whether these defective mutants were targeted to the apical PM when they were triply expressed with their wt-HA/NA counterparts plus additional M1 (Fig. 5D and 7D). In the cells triply transfected with wt-HA, wt-NA, and M1, apical targeting of wt-HA and wt-NA was accelerated at a level similar to that in cells with dual expression of wt-HA and wt-NA, indicating that M1 had no additive effect on the apical targeting of wt-HA and wt-NA (Fig. 5D and 7D). Similarly, when either HA or NA contained CT deletions, their apical targeting was hardly accelerated by coexpression with additional M1. This was most likely due to the lack of M1 binding sites within the CTs of HA and NA. Only slight acceleration of apical targeting was observed when HA533–535, NA31–35, or HA-SSS was coexpressed with its wt-HA/NA counterpart plus M1 (Fig. 5D and 7D). These results indicated that M1 did not sufficiently rescue the apical targeting defects of the HA and NA mutants.
Interaction of HA with NA via lipid rafts was required for acceleration of their apical targeting.
To analyze the association of HA and NA via lipid rafts, the HA and NA mutants were coexpressed in 293T cells and were subjected to coimmunoprecipitation using anti-HA mAb12-1G6 (Fig. 6C and D). We first coexpressed the wt-HA and GFP-CD59 (a lipid raft marker) and solubilized the membrane with 1% TX-100. GFP-CD59 was coimmunoprecipitated with wt-HA at 4°C but not at 37°C, indicating that both CD59 and wt-HA were associated with lipid rafts (Fig. 6C, lane 11). We used M1 as a positive control in the coimmunoprecipitation assays because M1 has been shown to interact directly with HA (7). We found that M1 was coimmunoprecipitated with wt-HA even when the lipid rafts were disrupted by TX-100 at 37°C (Fig. 6C, lane 10). When wt-HA was coexpressed with wt-NA or mutant NAs, some NA mutants (e.g., NA2–3, NA4–6, and NA7–10) were coimmunoprecipitated with wt-HA at 4°C although the efficiencies varied (Fig. 6C, lanes 1, 5, 6, and 8). In contrast, wt-HA failed to coprecipitate ΔCT-NA, NA2–6, and NA31–35 at 4°C, all of which were incapable of accelerating the apical targeting of wt-HA (Fig. 6C, lanes 3, 7, and 9). Conversely, when wt-NA was coexpressed with HA mutants and was similarly subjected to immunoprecipitation analysis, we found that HA550–552, HA557–559, HA560–563, and HA564–566 coprecipitated wt-NA at 4°C (Fig. 6D, lanes 3, 4, 5, and 6). In contrast, ΔCT-HA, HA533–535, and HA-SSS, all of which failed to accelerate the apical transport of wt-NA, coprecipitated wt-NA inefficiently (Fig. 6C, lane 2, and D, lanes 2 and 7). In all combinations, NA was not coimmunoprecipitated with HA when the lipid rafts were solubilized by treatment with TX-100 at 37°C. Thus, we depleted cholesterol from the cells by treatment with MβCD and lovastatin to disrupt the lipid rafts. wt-NA was not coimmunoprecipitated with wt-HA when the cells were treated with 10 mM MβCD (Fig. 6E, lane 3). Taken together, these data indicated that the interaction of HA with NA via lipid rafts was linked to the acceleration of their apical transport.
Proximal accumulation of HA and NA in lipid rafts.
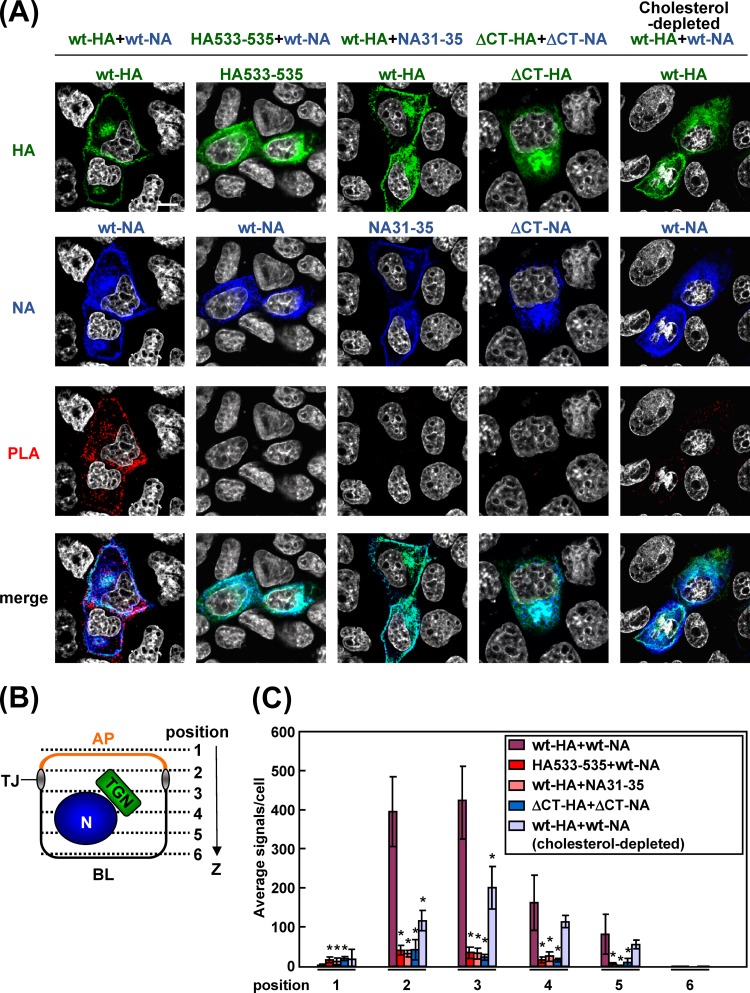
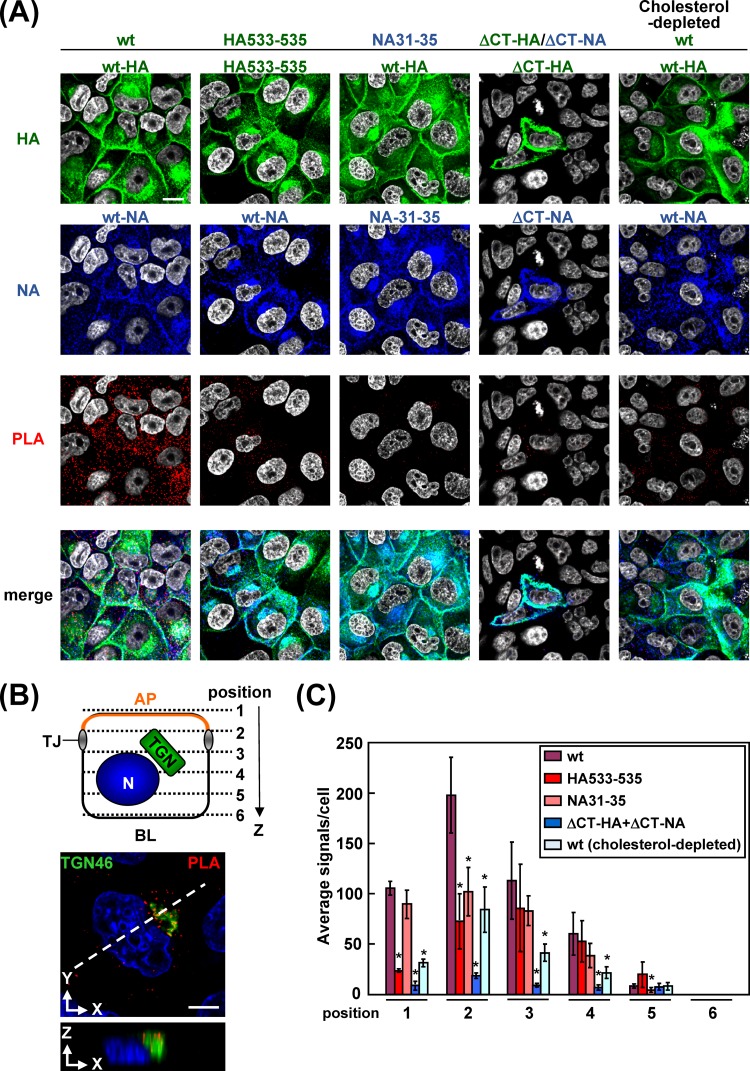
It has been considered that the TGN and apical recycling endosome are sorting platforms to route proteins to their destinations (37, 38). Recent studies have suggested that protein oligomerization or clustering in lipid rafts plays a pivotal role for segregation of apical cargos from basolateral cargos at the TGN and generation of intracellular transport vesicles (17, 39, 40). To examine whether HA and NA were accumulated in close proximity in lipid rafts, we prepared transfected cells at 9 hpt, when the majority of antigens were localized at the TGN, and performed in situ PLA. This method uses two primary antibodies specific for two target proteins and two oligonucleotide-conjugated and species-specific secondary antibodies (PLA probes) and detects the two targets in close proximity (<40 nm). After the PLA reaction, the cells were costained with Alexa Fluor-conjugated secondary Abs. Many PLA signals were observed in the cells coexpressing wt-HA and wt-NA (Fig. 8A). Confocal images were taken at 6 z slices (position 1 to 6) from the apical to basolateral aspects (Fig. 8B). The PLA signals in each x-y plane were automatically counted with BlobFinder software (Fig. 8C). The majority of PLA signals produced by the combination of the wt-HA and wt-NA were present at z positions 2 and 3, corresponding to the location of the TGN (see Fig. 10B), and some signals were in the cytoplasm (Fig. 8B). In contrast, PLA signals were rarely seen when either HA or NA was defective in lipid raft association (e.g., ΔCT-HA, HA533–535, ΔCT-NA, or NA31–35) (Fig. 8A). The numbers of their PLA signals were reduced more than 10-fold, compared with that obtained by the combination of the wt-HA and wt-NA (Fig. 8C). No significant differences in the expression levels of HA and NA were observed between the wt and mutants (Fig. 8A). These data indicated that HA and NA came into very close proximity, most likely clustering in lipid rafts, when their TMDs and CTs were intact. To confirm this, cholesterol was depleted from the cells by treatment with MβCD and lovastatin. The PLA signals in cells coexpressed with wt-HA and wt-NA were dramatically decreased by the disruption of lipid rafts (Fig. 8A and C). These data suggested that the lipid rafts were platforms for cytoplasmic accumulation of HA and NA.
FIG 8.
Accumulation of nonraft HA and NA mutants in transfected cells. (A) Polarized MDCK cells were cotransfected with expression plasmids for a combination of wt-HA plus wt-NA, HA533–535 plus wt-NA, wt-HA plus NA31–35, or ΔCT-HA plus ΔCT-NA. At 9 hpt, the cells were fixed, permeabilized, and incubated with two primary Abs (anti-HA mAb12-1G6 and rabbit anti-NA Ab). For PLA, the cells were incubated with anti-rabbit and anti-mouse proximity ligation secondary Abs conjugated with oligonucleotides (PLA probes). Following ligation of the probes and rolling-circle amplification, the rolling-circle products were detected by hybridization with their complementary oligonucleotides labeled with a fluorophore (excitation wavelength, 594 nm; emission wavelength, 624 nm) (red). The cells were further costained with anti-mouse Alexa Fluor 488 (for HA; green), anti-rabbit Alexa Fluor 647 (for NA; blue), and DAPI (gray). Scale bar, 10 μm. (B) A schematic diagram of six confocal z slices (positions 1 to 6) from the apical (AP) to basolateral (BL) side of the cell is shown. TJ, tight junction; N, nucleus. (C) The PLA signals at positions 1 to 6 were automatically counted using BlobFinder software, and the average number of signals in each x-y plane is shown. In each plane, the numbers in the coexpression samples (indicated) were compared with the number in the sample coexpressed with wt-HA and wt-NA. Asterisks represent statistically significant differences (P < 0.05) by Student's t test. The details of the cholesterol depletion experiment are described in the legend of Fig. 6B.
FIG 10.
Accumulation of HA and NA in cells infected with nonraft mutant viruses. (A) Polarized MDCK cells were infected with the wt virus or HA533–535, NA31–35, or ΔCT-HA/ΔCT-NA mutant virus. At 5 hpi, the cells were fixed, permeabilized, and incubated with two primary Abs (anti-HA mAb12-1G6 and rabbit anti-NA Ab). PLA was carried out as described in the legend of Fig. 8A. Following PLA reaction, the cells were costained with anti-mouse Alexa Fluor 488 (for HA; green) and anti-rabbit Alexa Fluor 647 (for NA; blue). (B) A schematic diagram of six confocal z slices (positions 1 to 6) from the AP to the BL side of the cell is shown. MDCK cells were infected with the wt virus and subjected to PLA with two primary Abs (anti-HA mAb12-1G6 and rabbit anti-NA Ab) at 3.5 hpi. The cells were stained with prelabeled anti-TGN46 Ab. Localizations of the TGN46 (green) and PLA signals (red) are shown in the x-y and x-z planes. The dashed line in the x-y image indicates the positions of the x-z image. Scale bar, 10 μm. (C) The PLA signals at positions 1 to 6 were automatically counted using BlobFinder software. The average number of PLA signals in each x-y plane is shown in the graph. In each x-y plane, the numbers in the mutant virus samples (indicated) were compared with the number in the wt virus sample. Asterisks represent statistically significant differences (P < 0.05) by Student's t test.
Apical transport and proximal accumulation of nonraft-associated HA/NA were also inefficient in infected cells.
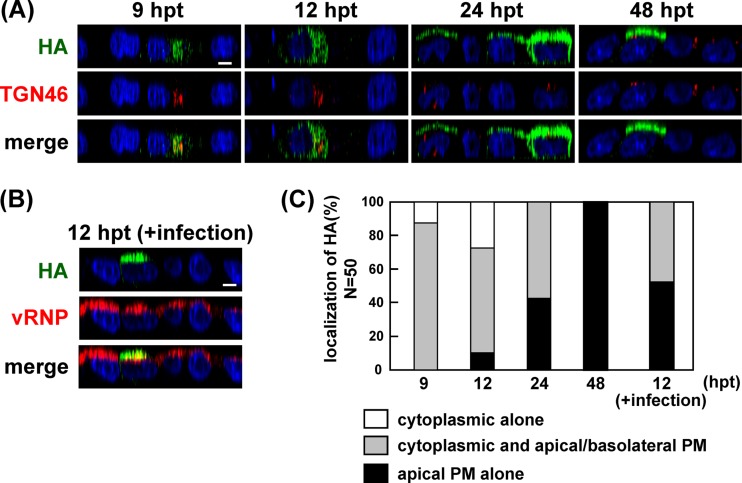
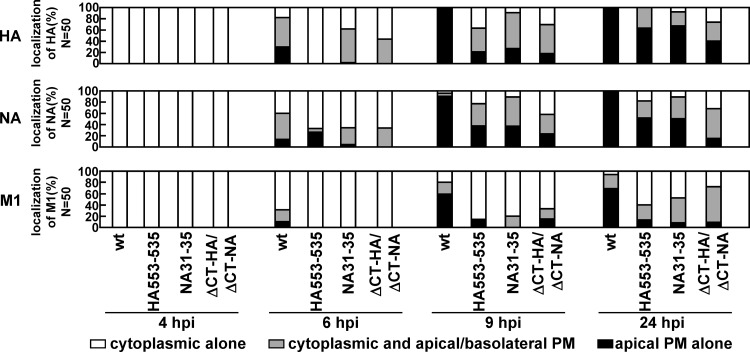
We generated a recombinant virus with a mutation in either the HA or NA TMD (HA533–535 or NA31–35) and a virus with deletion of both CTs (ΔCT-HA/ΔCT-NA) by means of a reverse genetic procedure. Polarized MDCK cells were infected with the wt, HA533–535, NA31–35, or ΔCT-HA/ΔCT-NA virus, and the localization of viral antigens was similarly analyzed by confocal microscopy at 4, 6, 9, and 24 hpi (Fig. 9). In the case of the wt virus, HA and NA were localized to the cytoplasmic compartments at 4 hpi (100% of infected cells). M1 was diffusely distributed throughout the cells. At 9 hpi, HA, NA, and M1 were accumulated at the apical PM (100%, 100%, and 69%). When cells were infected with the nonraft-associated mutant viruses (HA533–535, NA31–35, and ΔCT-HA/ΔCT-NA), apical PM targeting of the viral components was significantly delayed. For example, at 9 hpi, accumulation of HA, NA, and M1 at the apical PM was observed in much smaller numbers of cells (Fig. 9).
FIG 9.
Temporal study of apical PM targeting of HA, NA, and M1 in cells infected with nonraft mutant viruses. Polarized MDCK cells were infected with the wt or nonraft mutant (HA533–535, NA31–35, or ΔCT-HA/ΔCT-NA) virus. At various time points, the cells were costained with anti-HA mAb12-1G6, sheep anti-NA, and anti-M1 Abs. The localizations of HA, NA, and M1 were analyzed in the x-z planes. For semiquantification of HA, NA, and M1 localization, 50 antigen-positive cells were chosen in each experiment, and the distribution patterns of each viral protein were counted separately.
We examined the accumulation/clustering of HA and NA in cells infected with these mutant viruses by in situ PLA (Fig. 10). In the wt-infected cells, PLA signals partially colocalized with TGN46 were observed at 3.5 hpi (Fig. 10B), but no PLA signals were seen at 3 hpi. This result suggested that the lipid rafts functioned as platforms for cytoplasmic accumulation of HA and NA. Many PLA signals were observed broadly in the cytoplasm at 5 hpi (Fig. 10A and C). In contrast, PLA signals were rarely seen when the cells were infected with the HA533–535, NA31–35, or ΔCT-HA/ΔCT-NA virus. The numbers of their PLA signals were reduced more than 2-fold compared with that of the wt virus. Cholesterol depletion by the treatment with MβCD and lovastatin resulted in a similar reduction in the numbers of the PLA signals (Fig. 10A and C). These results were not ascribable to the protein expression levels (Fig. 10A). These data were consistent with the results of the coexpression experiments although the differences in the signal numbers were much more significant in the transfected cells (Fig. 8), suggesting that the lipid raft association of HA and NA in infected cells was still necessary for their efficient apical targeting and clustering.
Lipid raft association of HA and NA was required for efficient virus replication.
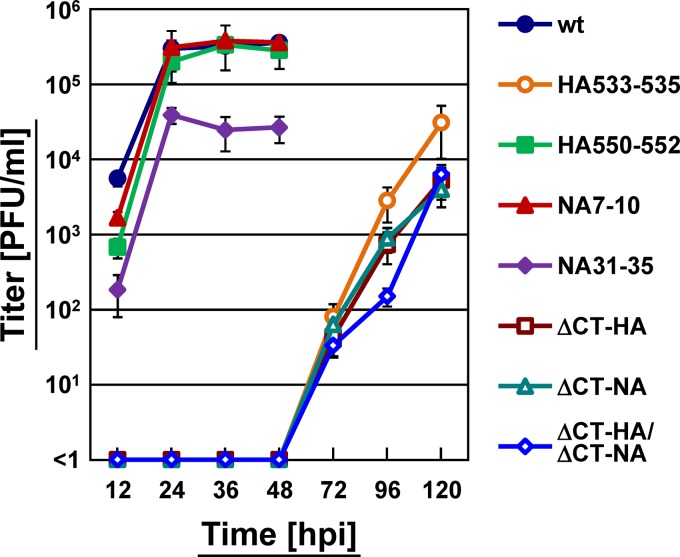
We examined the growth kinetics of the mutant viruses by plaque assay (Fig. 11). Compared with the wt virus, the majority of the nonraft-associated mutant viruses (HA533–535, ΔCT-HA, ΔCT-NA, and ΔCT-HA/ΔCT-NA) grew significantly more slowly. A previous study has shown that nonraft-associated HA viruses do not grow for first 30 h and then grow slowly at low MOIs because of multiple defects: a budding defect and fusion defect (5). The nonraft-associated mutant virus NA31–35 had a titer 10-fold lower than the wt virus. It has been reported that the NA31–35 mutation results in a great reduction in sialidase activity and aggregation of virus particles on the infected cell surface (41). In contrast, raft-associated mutant viruses (HA550–552 and NA7–10) showed growth rates and titers similar to those of the wt virus (Fig. 11). Together, these data are consistent with previous studies in which recombinant viruses containing nonraft-associated HA and/or NA mutants had titers 10- to 1,000-fold lower than their parental virus (5, 42).
FIG 11.
Growth kinetics of HA and NA mutant viruses. MDCK cells were infected with the wt or mutant virus at an MOI of 0.1. At various time points (12, 24, 36, 48, 72, 96, and 120 hpi), an aliquot of the culture medium was harvested, and virus titers were measured by plaque assay on MDCK cells.
Coexpression of HA and NA facilitated apical targeting and clustering of cellular raft-associated proteins.
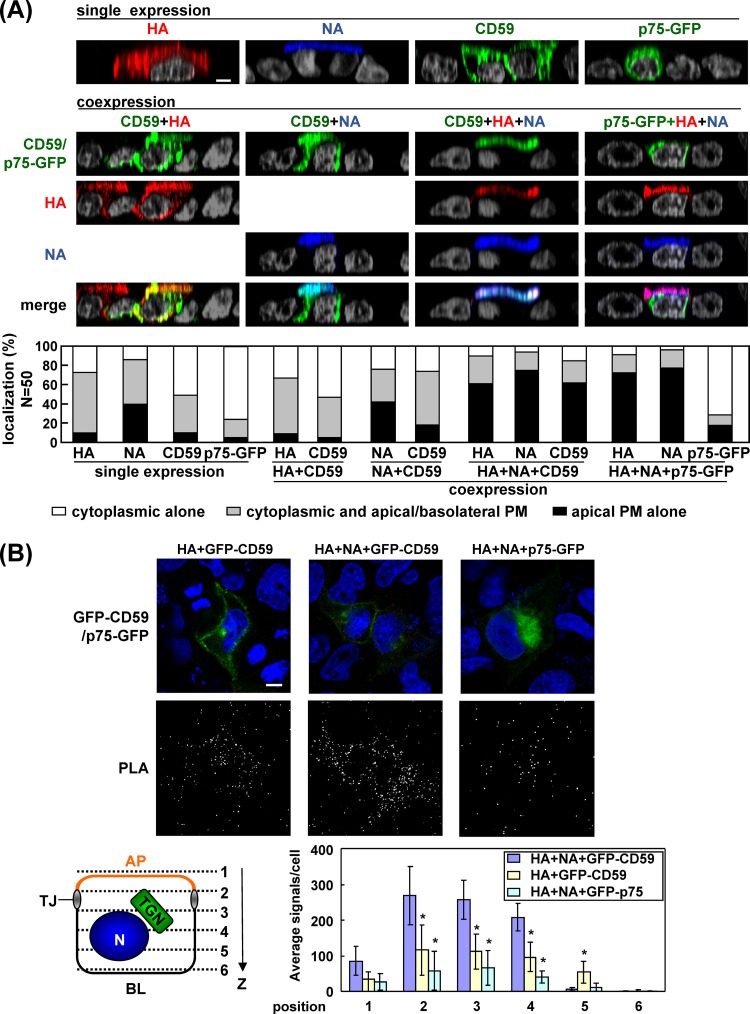
Our data indicated that the coexpression of HA and NA facilitated their apical transport if they were raft associated. To examine whether the apical transport of cellular raft-associated proteins was affected upon coexpression of HA and NA, we carried out dual or triple expression experiments. CD59, a GPI-AP, is a well-known marker for lipid rafts and is transported to the apical PM. In singly transfected cells, the majority of CD59 was localized in the cytoplasmic compartments at 12 hpt (apical PM accumulation in 10% of CD59-positive cells). p75-GFP is also targeted to the apical PM but without lipid raft association (43). p75-GFP was similarly found in the cytoplasmic compartments at 12 hpt (apical accumulation in 5% of p75-GFP-positive cells). When CD59 was coexpressed with HA plus NA, apical targeting of CD59 was significantly accelerated (62% of CD59-positive cells), concomitant with the acceleration of apical targeting of HA/NA, indicating that coexpression of HA and NA similarly accelerated the apical transport of noncognate raft-associated proteins. In sharp contrast, apical targeting of p75-GFP was unaffected (apical targeting in 17% of p75-positive cells) when HA and NA were coexpressed even though apical targeting of HA and NA was accelerated (Fig. 12A). Such acceleration was not seen in coexpression with either HA or NA. These results indicated that coexpression of HA plus NA accelerated the apical targeting of raft-associated CD59 but not that of nonraft p75-GFP.
FIG 12.
Apical targeting and clustering of raft and nonraft proteins by coexpression of HA and NA. (A) Polarized MDCK cells were cotransfected with a combination of wt-HA plus CD59 (apical and raft marker), wt-NA plus CD59, wt-HA and wt-NA plus CD59, or wt-HA and wt-NA plus p75-GFP (apical and nonraft marker) expression plasmids. For the immunofluorescence assay, cells were fixed at 12 hpt and stained with anti-CD59 (green) Ab, anti-HA mAb12-1G6 (red), and sheep anti-NA Ab (blue). Nuclei were stained with DAPI (gray). All images were taken at the same magnification, and images in the x-z plane are shown. (B) For PLA, the cells were fixed at 9 hpt, permeabilized, and incubated with two primary Abs (mouse anti-GFP Ab and rabbit anti-caveolin-1 Ab). PLA was performed as described in the legend of Fig. 8A. Scale bar, 10 μm. A schematic diagram of six confocal z slices (positions 1 to 6) from the AP to the BL side of the cell is shown. The PLA signals in the x-y planes at positions 1 to 6 were automatically counted using BlobFinder software, and the average number of signals in each x-y plane is shown. In each x-y plane, the numbers in the coexpression samples (indicated) were compared with the number in the sample coexpressed with wt-HA and wt-NA plus GFP-CD59. Asterisks represent statistically significant differences (P < 0.05) by Student's t test.
Recent studies have suggested that the clustering of raft-associated proteins may drive the coalescence of lipid rafts, leading to membrane curvature for vesicle budding (17, 39, 44, 45, 46). We investigated this possibility in cells coexpressing HA and NA. Caveolae are a subdomain of lipid raft microdomains (47, 48). We focused on two raft microdomains, caveolin-1-containing rafts and CD59-containing lipid rafts, and examined the coalescence of the two raft fractions by PLA. To this end, we carried out dual or triple expression experiments as before. The cells were fixed at 9 hpt when transfected antigens were present at the cytoplasmic compartments and were analyzed for the PLA signals between CD59 and endogenous caveolin-1. When CD59 was coexpressed with HA, some PLA signals were observed in the cytoplasm. When CD59 was coexpressed with HA plus NA, the PLA signals were significantly increased, indicating that upon coexpression of HA and NA, CD59 and caveolin-1, which were present in distinct raft fractions, came into close proximity. In contrast, when p75-GFP was coexpressed with HA plus NA, the number of PLA signals was lower than that in cells coexpressed with CD59 and HA (Fig. 12B). These findings were confirmed by quantification of the PLA signals in the x-y planes at six z positions (Fig. 12B). These results clearly indicated that coexpression of HA and NA also induced accumulation of lipid raft proteins, caveolin-1 and CD59, but not nonraft p75, suggesting the coalescence of membrane raft microdomains.
DISCUSSION
Coexpression of HA and NA induces lipid raft clustering, leading to efficient apical trafficking.
The influenza virus HA has served as an apical marker in the research field of cell polarity and membrane trafficking (49, 50, 51). NA is also sorted to the apical PM (3, 14). Although the molecular mechanism of the apical transport of these envelope proteins has not been fully understood, previous studies have indicated that their TMD and CT possess determinants for the apical transport and lipid raft association (2, 3, 7). In this study, we found that in polarized cells the apical targeting of HA was accelerated when NA was coexpressed. This acceleration appeared to occur not before but after the Golgi compartment (Fig. 3E), consistent with our findings that the acceleration was linked with the lipid raft association of HA and NA. It is known that lipid rafts are generated at the Golgi compartment in mammalian cells (15). Our quantitative analysis of HA/NA expression revealed that the apical targeting of HA was not linked with the expression level of HA (Fig. 3C). The analysis also indicated that the apical targeting of HA was inefficient without NA but that no certain molar ratio of HA to NA was observed for the acceleration of their apical targeting (Fig. 3D). Interestingly, in sparse, likely nonpolarized MDCK cells, the accumulation of HA to the upper cell surface was very inefficient even with coexpression of NA (upper PM only, 9%; PMs plus cytoplasmic compartments, 91% in HA and NA double-positive cells) (Fig. 3C) although the mechanisms that link the cell density to the apical transport kinetics of cargo proteins remain to be elucidated in future studies.
It has been suggested that the influenza virus utilizes lipid rafts in the apical PM as the sites of virus particle assembly and budding (5, 52). Immunoelectron microscopy has shown that HA is accumulated and clustered in lipid rafts on the cell surface, which provides a sufficient concentration of HA to mediate efficient virus replication (5). Consistent with this finding, our PLA analysis revealed that HA and NA likely accumulated at the TGN before they reached the apical PM (Fig. 10B). This analysis further revealed that the coexpression of HA and NA induced efficient clustering of lipid raft microdomains in the cytoplasmic compartments (Fig. 12). Selective incorporation of certain lipid components (cholesterol and sphingomyelin) into influenza virus particles has been suggested in a previous study based on detergent solubility in viral particle envelopes (52). It is conceivable that the lipid raft clustering induced by coexpression of HA and NA led to an increase in the curvature of the membrane, facilitating the generation of apical sorting/transport vesicles from the TGN, and finally to arrival at the budding site together.
Possible mechanisms of lipid raft clustering by coexpression of HA and NA.
Recent studies have suggested that hydrophobic mismatch, i.e., the difference between the hydrophobic length of the TM proteins and the hydrophobic thickness of the membrane they span, is involved in lipid raft clustering (45). The sphingolipids, such as sphingomyelin, that constitute lipid rafts have saturated fatty acids longer than the unsaturated fatty acids of phospholipids, making the lipid rafts thicker than other membrane domains. In this model, it has been proposed that to avoid unfavorable exposure of the hydrophobic surfaces to hydrophilic environments, the TM proteins are incorporated into lipid microdomains, the hydrophobic thickness of which is equal to the hydrophobic length of the TM proteins (10, 45). The length of the TMD has also been suggested to regulate protein targeting to the PM in Saccharomyces cerevisiae (53, 54). The lengths of the TMDs of PR8 HA and NA are 27 and 29 amino acids, respectively, and are relatively long, considering that TMD helices are usually around 20 amino acids in length. It is conceivable that such longer TMDs in HA and NA contributed to lipid raft clustering, possibly by recruiting and stabilizing sphingolipids containing a bulky head group in the outer leaflet of the lipid bilayer.
A previous in vitro study using two TM peptides, one matching the thickness of raft lipids and the other the thickness of nonraft lipids (23 and 29 amino acids in length), showed that both peptides were primarily localized in nonraft microdomains (55). However, a recent study revealed that cholesterol constrains membranes containing TM peptides, thereby rearranging both lipid acyl chains and TM peptides according to hydrophobic length (56); this suggests that not only hydrophobic mismatch but also cholesterol is involved in the sorting and clustering of TM proteins in the lipid bilayer. We think that this possibility is likely in the case of the influenza virus since previous studies (4, 16) and our study repeatedly found that depletion of cholesterol resulted in a failure of lipid raft association and apical transport of HA.
Cholesterol and sphingolipids are critical components of lipid rafts. Cholesterol is comprised of three structurally distinct regions: a hydroxyl group, a rigid steroid ring, and a flexible alkyl chain. The structure of cholesterol is characterized by a flat and smooth α-face containing only axial hydrogen atoms and a rough β-face containing bulky methyl groups. Lipids, including sphingolipids, generally interact with the α-face of cholesterol, leaving the β-face available for the TMD (57). Cholesterol can also form homodimers through the interaction with their respective smooth α-faces, leaving their opposite β-faces available for protein binding. For example, a cholesterol dimer can recruit two molecules of G protein-coupled receptor, thereby inducing receptor dimerization (58). In this study, we found that HA and NA were accumulated at the cytoplasmic compartments, most likely at the TGN, via their association with lipid rafts, and this accumulation was cholesterol dependent (Fig. 8 and 10).
Several previous studies on the influenza virus used the A/Udorn/72 (H3) strain and indicated that deletions or mutations in the TMD-CT of HA and NA impaired their lipid raft association without affecting apical targeting (5, 7). A kinetic analysis of the apical transport of the HA mutants showed only a 1-h delay to the cell surface compared with wt-HA (59). In contrast, we used the PR8 (H1) strain and showed that our similar nonraft HA mutants exhibited a great delay of transport to the apical PM (Fig. 9A). Another study used the A/Japan/305 (H2) strain and also showed a significant delay of its nonraft HA mutant to the cell surface (2). When we compared the amino acid sequences of the HA TMD-CTs, we found that the PR8 (H1) and A/Japan/305 (H2) shared 75.7% homology, the PR8 (H1) and A/Udorn/72 (H3) shared 37.8% homology, and the A/Japan/305 (H2) and A/Udorn/72 (H3) shared 32.4% homology. Thus, the structure and length of the TMD helices may differ widely among these three HAs. It is possible that the A/Udorn/72 HA may be transported to the cell surface in a less cholesterol-dependent manner.
ACKNOWLEDGMENTS
We thank Yoshihiro Kawaoka (University of Wisconsin—Madison, USA, and University of Tokyo, Japan) for providing us with PolI plasmids (pHH21) used for a reverse genetics system of influenza A virus, Ken Watanabe (Nagasaki University, Japan) for providing us with rabbit anti-M1 polyclonal antibody, and Naoki Takizawa (Institute of Microbial Chemistry, Japan) for MDCK-HA cells. We also thank Hiyori Haraguchi for technical assistance.
This work was supported by a Human Science grant from the Ministry of Health, Labor, and Welfare of Japan and by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print 25 June 2014
REFERENCES
- 1.Nayak DP, Balogun RA, Yamada H, Zhou ZH, Barman S. 2009. Influenza virus morphogenesis and budding. Virus Res. 143:147–161. 10.1016/j.virusres.2009.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin S, Naim HY, Rodriguez AC, Roth MG. 1998. Mutations in the middle of the transmembrane domain reverse the polarity of transport of the influenza virus hemagglutinin in MDCK epithelial cells. J. Cell Biol. 142:51–57. 10.1083/jcb.142.1.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barman S, Nayak DP. 2000. Analysis of the transmembrane domain of influenza virus neuraminidase, a type II transmembrane glycoprotein, for apical sorting and raft association. J. Virol. 74:6538–6545. 10.1128/JVI.74.14.6538-6545.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheiffele P, Roth MG, Simons K. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501–5508. 10.1093/emboj/16.18.5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeda M, Leser GP, Russell CJ, Lamb RA. 2003. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. U. S. A. 100:14610–14617. 10.1073/pnas.2235620100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leser GP, Lamb RA. 2005. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 342:215–227. 10.1016/j.virol.2005.09.049 [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Pekosz A, Lamb RA. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634–4644. 10.1128/JVI.74.10.4634-4644.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen BJ, Takeda M, Lamb RA. 2005. Influenza virus hemagglutinin (H3 subtype) requires palmitoylation of its cytoplasmic tail for assembly: M1 proteins of two subtypes differ in their ability to support assembly. J. Virol. 79:13673–13684. 10.1128/JVI.79.21.13673-13684.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossman JS, Jing X, Leser GP, Lamb RA. 2010. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142:902–913. 10.1016/j.cell.2010.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magal LG, Yaffe Y, Shepshelovich J, Aranda JF, de Marco Mdel C, Gaus K, Alonso MA, Hirschberg K. 2009. Clustering and lateral concentration of raft lipids by the MAL protein. Mol. Biol. Cell 20:3751–3762. 10.1091/mbc.E09-02-0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-Boulan E, Kreitzer G, Musch A. 2005. Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell. Biol. 6:233–247. 10.1038/nrm1593 [DOI] [PubMed] [Google Scholar]
- 12.Eisenberg S, Shvartsman DE, Ehrlich M, Henis YI. 2006. Clustering of raft-associated proteins in the external membrane leaflet modulates internal leaflet H-ras diffusion and signaling. Mol. Cell. Biol. 26:7190–7200. 10.1128/MCB.01059-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson CM, Chichili GR, Rodgers W. 2008. Compartmentalization of phosphatidylinositol 4,5-bisphosphate signaling evidenced using targeted phosphatases. J. Biol. Chem. 283:29920–29928. 10.1074/jbc.M805921200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kundu A, Avalos RT, Sanderson CM, Nayak DP. 1996. Transmembrane domain of influenza virus neuraminidase, a type II protein, possesses an apical sorting signal in polarized MDCK cells. J. Virol. 70:6508–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simons K, Ikonen E. 1997. Functional rafts in cell membranes. Nature 387:569–572. 10.1038/42408 [DOI] [PubMed] [Google Scholar]
- 16.Keller P, Simons K. 1998. Cholesterol is required for surface transport of influenza virus hemagglutinin. J. Cell Biol. 140:1357–1367. 10.1083/jcb.140.6.1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paladino S, Sarnataro D, Pillich R, Tivodar S, Nitsch L, Zurzolo C. 2004. Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. J. Cell Biol. 167:699–709. 10.1083/jcb.200407094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali A, Avalos RT, Ponimaskin E, Nayak DP. 2000. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 74:8709–8719. 10.1128/JVI.74.18.8709-8719.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noton SL, Medcalf E, Fisher D, Mullin AE, Elton D, Digard P. 2007. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J. Gen. Virol. 88:2280–2290. 10.1099/vir.0.82809-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen BJ, Leser GP, Jackson D, Lamb RA. 2008. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 82:10059–10070. 10.1128/JVI.01184-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:9345–9350. 10.1073/pnas.96.16.9345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin H, Leser GP, Lamb RA. 1994. The influenza virus hemagglutinin cytoplasmic tail is not essential for virus assembly or infectivity. EMBO J. 13:5504–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Sastre A, Palese P. 1995. The cytoplasmic tail of the neuraminidase protein of influenza A virus does not play an important role in the packaging of this protein into viral envelopes. Virus Res. 37:37–47. 10.1016/0168-1702(95)00017-K [DOI] [PubMed] [Google Scholar]
- 24.Ohkura T, Kikuchi Y, Kono N, Itamura S, Komase K, Momose F, Morikawa Y. 2012. Epitope mapping of neutralizing monoclonal antibody in avian influenza A H5N1 virus hemagglutinin. Biochem. Biophys. Res. Commun. 418:38–43. 10.1016/j.bbrc.2011.12.108 [DOI] [PubMed] [Google Scholar]
- 25.Watanabe K, Takizawa N, Noda S, Tsukahara F, Maru Y, Kobayashi N. 2008. Hsc70 regulates the nuclear export but not the import of influenza viral RNP: a possible target for the development of anti-influenza virus drugs. Drug Discov. Ther. 4:77–84 [PubMed] [Google Scholar]
- 26.Momose F, Kikuchi Y, Komase K, Morikawa Y. 2007. Visualization of microtubule-mediated transport of influenza viral progeny ribonucleoprotein. Microbes Infect. 9:1422–1433. 10.1016/j.micinf.2007.07.007 [DOI] [PubMed] [Google Scholar]
- 27.Abramoff MD, Magelhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 28.Allalou A, Wahlby C. 2009. BlobFinder, a tool for fluorescence microscopy image cytometry. Comput. Methods Programs Biomed. 94:58–65. 10.1016/j.cmpb.2008.08.006 [DOI] [PubMed] [Google Scholar]
- 29.Gu F, Crump CM, Thomas G. 2001. Trans-Golgi network sorting. Cell. Mol. Life Sci. 58:1067–1084. 10.1007/PL00000922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonilha VL, Marmorstein AD, Cohen-Gould L, Rodriguez-Boulan E. 1997. Apical sorting of influenza hemagglutinin by transcytosis in retinal pigment epithelium. J. Cell Sci. 110:1717–1727 [DOI] [PubMed] [Google Scholar]
- 31.Gravotta D, Adesnik M, Sabatini DD. 1990. Transport of influenza HA from the trans-Golgi network to the apical surface of MDCK cells permeabilized in their basolateral plasma membranes: energy dependence and involvement of GTP-binding proteins. J. Cell Biol. 111:2893–2908. 10.1083/jcb.111.6.2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, Li H, Chen Y, Wei H, Gao GF, Liu H, Huang S, Chen JL. 2012. Transport of influenza virus neuraminidase (NA) to host cell surface is regulated by ARHGAP21 and Cdc42 proteins. J. Biol. Chem. 287:9804–9816. 10.1074/jbc.M111.312959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henkel JR, Weisz OA. 1998. Influenza virus M2 protein slows traffic along the secretory pathway. pH perturbation of acidified compartments affects early Golgi transport steps. J. Biol. Chem. 273:6518–6524. 10.1074/jbc.273.11.6518 [DOI] [PubMed] [Google Scholar]
- 34.Bui M, Whittaker G, Helenius A. 1996. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J. Virol. 70:8391–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henkel JR, Gibson GA, Poland PA, Ellis MA, Hughey RP, Weisz OA. 2000. Influenza M2 proton channel activity selectively inhibits trans-Golgi network release of apical membrane and secreted proteins in polarized Madin-Darby canine kidney cells. J. Cell Biol. 148:495–504. 10.1083/jcb.148.3.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown DA, Rose JK. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533–544. 10.1016/0092-8674(92)90189-J [DOI] [PubMed] [Google Scholar]
- 37.Griffiths G, Simons K. 1986. The trans Golgi network: sorting at the exit site of the Golgi complex. Science 234:438–443. 10.1126/science.2945253 [DOI] [PubMed] [Google Scholar]
- 38.Ang AL, Taguchi T, Francis S, Folsch H, Murrells LJ, Pypaert M, Warren G, Mellman I. 2004. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 167:531–543. 10.1083/jcb.200408165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuck S, Simons K. 2004. Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J. Cell Sci. 117:5955–5964. 10.1242/jcs.01596 [DOI] [PubMed] [Google Scholar]
- 40.Paladino S, Pocard T, Catino MA, Zurzolo C. 2006. GPI-anchored proteins are directly targeted to the apical surface in fully polarized MDCK cells. J. Cell Biol. 172:1023–1034. 10.1083/jcb.200507116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barman S, Adhikary L, Chakrabarti AK, Bernas C, Kawaoka Y, Nayak DP. 2004. Role of transmembrane domain and cytoplasmic tail amino acid sequences of influenza a virus neuraminidase in raft association and virus budding. J. Virol. 78:5258–5269. 10.1128/JVI.78.10.5258-5269.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin H, Leser GP, Zhang J, Lamb RA. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16:1236–1247. 10.1093/emboj/16.6.1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeaman C, Le Gall AH, Baldwin AN, Monlauzeur L, Le Bivic A, Rodriguez-Boulan E. 1997. The O-glycosylated stalk domain is required for apical sorting of neurotrophin receptors in polarized MDCK cells. J. Cell Biol. 139:929–940. 10.1083/jcb.139.4.929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Razani B, Woodman SE, Lisanti MP. 2002. Caveolae: from cell biology to animal physiology. Pharmacol. Rev. 54:431–467. 10.1124/pr.54.3.431 [DOI] [PubMed] [Google Scholar]
- 45.Schmidt U, Weiss M. 2010. Hydrophobic mismatch-induced clustering as a primer for protein sorting in the secretory pathway. Biophys. Chem. 151:34–38. 10.1016/j.bpc.2010.04.009 [DOI] [PubMed] [Google Scholar]
- 46.Hogue IB, Grover JR, Soheilian F, Nagashima K, Ono A. 2011. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J. Virol. 85:9749–9766. 10.1128/JVI.00743-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sowa G, Pypaert M, Sessa WC. 2001. Distinction between signaling mechanisms in lipid rafts vs. caveolae. Proc. Natl. Acad. Sci. U. S. A. 98:14072–14077. 10.1073/pnas.241409998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Echarri A, Muriel O, Del Pozo MA. 2007. Intracellular trafficking of raft/caveolae domains: insights from integrin signaling. Semin. Cell Dev. Biol. 18:627–637. 10.1016/j.semcdb.2007.08.004 [DOI] [PubMed] [Google Scholar]
- 49.Puertollano R, Martin-Belmonte F, Millan J, de Marco MC, Albar JP, Kremer L, Alonso MA. 1999. The MAL proteolipid is necessary for normal apical transport and accurate sorting of the influenza virus hemagglutinin in Madin-Darby canine kidney cells. J. Cell Biol. 145:141–151. 10.1083/jcb.145.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tall RD, Alonso MA, Roth MG. 2003. Features of influenza HA required for apical sorting differ from those required for association with DRMs or MAL. Traffic 4:838–849. 10.1046/j.1398-9219.2003.0138.x [DOI] [PubMed] [Google Scholar]
- 51.Cresawn KO, Potter BA, Oztan A, Guerriero CJ, Ihrke G, Goldenring JR, Apodaca G, Weisz OA. 2007. Differential involvement of endocytic compartments in the biosynthetic traffic of apical proteins. EMBO J. 26:3737–3748. 10.1038/sj.emboj.7601813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheiffele P, Rietveld A, Wilk T, Simons K. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038–2044. 10.1074/jbc.274.4.2038 [DOI] [PubMed] [Google Scholar]
- 53.Rayner JC, Pelham HR. 1997. Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 16:1832–1841. 10.1093/emboj/16.8.1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levine TP, Wiggins CA, Munro S. 2000. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell 11:2267–2281. 10.1091/mbc.11.7.2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vidal A, McIntosh TJ. 2005. Transbilayer peptide sorting between raft and nonraft bilayers: comparisons of detergent extraction and confocal microscopy. Biophys. J. 89:1102–1108. 10.1529/biophysj.105.062380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaiser HJ, Orlowski A, Rog T, Nyholm TK, Chai W, Feizi T, Lingwood D, Vattulainen I, Simons K. 2011. Lateral sorting in model membranes by cholesterol-mediated hydrophobic matching. Proc. Natl. Acad. Sci. U. S. A. 108:16628–16633. 10.1073/pnas.1103742108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fantini J, Barrantes FJ. 2013. How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front. Physiol. 4:31. 10.3389/fphys.2013.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC. 2008. A specific cholesterol binding site is established by the 2.8 Å structure of the human beta2-adrenergic receptor. Structure 16:897–905. 10.1016/j.str.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simpson DA, Lamb RA. 1992. Alterations to influenza virus hemagglutinin cytoplasmic tail modulate virus infectivity. J. Virol. 66:790–803 [DOI] [PMC free article] [PubMed] [Google Scholar]