ABSTRACT
Members of the family Partitiviridae have bisegmented double-stranded RNA (dsRNA) genomes and are not generally known to cause obvious symptoms in their natural hosts. An unusual partitivirus, Sclerotinia sclerotiorum partitivirus 1 (SsPV1/WF-1), conferred hypovirulence on its natural plant-pathogenic fungal host, Sclerotinia sclerotiorum strain WF-1. Cellular organelles, including mitochondria, were severely damaged. Hypovirulence and associated traits of strain WF-1 and SsPV1/WF-1 were readily cotransmitted horizontally via hyphal contact to different vegetative compatibility groups of S. sclerotiorum and interspecifically to Sclerotinia nivalis and Sclerotinia minor. S. sclerotiorum strain 1980 transfected with purified SsPV1/WF-1 virions also exhibited hypovirulence and associated traits similar to those of strain WF-1. Moreover, introduction of purified SsPV1/WF-1 virions into strain KY-1 of Botrytis cinerea also resulted in reductions in virulence and mycelial growth and, unexpectedly, enhanced conidial production. However, virus infection suppressed hyphal growth of most germinating conidia of B. cinerea and was eventually lethal to infected hyphae, since very few new colonies could develop following germ tube formation. Taken together, our results support the conclusion that SsPV1/WF-1 causes hypovirulence in Sclerotinia spp. and B. cinerea. Cryo-EM (cryo-electron microscopy) reconstruction of the SsPV1 particle shows that it has a distinct structure with similarity to the closely related partitiviruses Fusarium poae virus 1 and Penicillium stoloniferum virus F. These findings provide new insights into partitivirus biological activities and clues about molecular interactions between partitiviruses and their hosts.
IMPORTANCE Members of the Partitiviridae are believed to occur commonly in their phytopathogenic fungal and plant hosts. However, most partitiviruses examined so far appear to be associated with latent infections. Here we report a partitivirus, SsPV1/WF-1, that was isolated from a hypovirulent strain of Sclerotinia sclerotiorum and describe its biological and molecular features. We have demonstrated that SsPV1 confers hypovirulence. Furthermore, SsPV1 can infect and cause hypovirulence in Botrytis cinerea. Our study also suggests that SsPV1 has a vigorous ability to proliferate and spread via hyphal contact. SsPV1 can overcome vegetative incompatibility barriers and can be transmitted horizontally among different vegetative compatibility groups of S. sclerotiorum, even interspecifically. Cryo-EM reconstruction of SsPV1 shows that it has a distinct structure with similarity to closely related partitiviruses. Our studies exploit a novel system, SsPV1 and its hosts, which can provide the means to explore the mechanisms by which partitiviruses interact with their hosts.
INTRODUCTION
Plant pathogenic fungi cause many destructive crop diseases worldwide, and the control of these diseases poses one of the greatest challenges to sustainable agriculture (1, 2). Since it is often difficult to obtain disease-resistant cultivars for many crops, farmers rely heavily on fungicides to control fungal diseases. However, fungicide overuse is likely to enhance the emergence of fungicide-resistant pathogens in the field. Furthermore, excessive use of fungicides has the potential to cause serious problems for the environment and for human health. Consequently, there is an urgent need to develop novel methods to reduce the dosage of chemicals used to control plant diseases. In nature, some mycoviruses are known to be responsible for debilitation/hypovirulence of plant pathogens, and hence, they represent potential agents that can be exploited in combating fungal disease (3–5). Moreover, a classical example of mycovirus-based biocontrol was implemented successfully in Europe to help manage chestnut blight caused by Cryphonectria parasitica (6). Recently, a bipartite, double-stranded RNA (dsRNA) mycovirus, Rosellinia necatrix megabirnavirus 1 (RnMBV1), and a DNA mycovirus, Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1 (SsHADV-1), have been shown to be very promising in controlling fungal diseases (7–9). Therefore, mycoviruses have attracted the attention of many phytopathologists, and a growing number of mycoviruses that reduce virulence and suppress growth of their hosts have been identified in plant pathogenic fungi (3–5, 8, 10, 11). Moreover, mycovirus-associated hypovirulence also provides opportunities for elucidating fundamental aspects of mycovirus-host interactions and the potential of fungal virus spread in nature (12–14).
Members of family Partitiviridae are characterized by having genomes comprised of two linear, monocistronic dsRNA segments (1.4 to 2.4 kbp in length) (15, 16). Most partitiviruses examined to date are typically associated with latent infections of their fungal, plant, and protozoan hosts (16). Partitiviruses that infect fungi are transmitted vertically via spores or horizontally via hyphal anastomosis (16, 17). As more partitiviruses were isolated and their genomes were sequenced, it became clear that some fungal partitiviruses are phylogenetically more closely related to plant partitiviruses than to some other fungal partitiviruses (18, 19); therefore, the host-based taxonomic classification of partitiviruses is no longer suitable. To address this inconsistency, the classification of genera in this family has recently been reevaluated (20–22). A proposal to have the family Partitiviridae be reorganized to include four new genera, Alphapartitivirus, Betapartitivirus, Gammapartitivirus, and Deltapartitivirus, was recently approved by the International Committee for Taxonomy of Viruses (http://talk.ictvonline.org/files/proposals/taxonomy_proposals_fungal1/m/fung04/4772.aspx). In the new reorganization, the genera Alphapartitivirus and Betapartitivirus contain a mixture of fungal and plant viruses, whereas the genera Gammapartitivirus and Deltapartitivirus contain only fungal or plant viruses, respectively (22). Recently, phylogenetic analysis of the coat protein sequences of some partitiviruses revealed that incidences of horizontal gene transfer had occurred between viruses and eukaryotic organisms (18, 19). A system in which partitivirus particles (i.e., Rosellinia necatrix partitivirus 1-W8 [RnPV1-W8]) can be transfected into fungal protoplasts has been successfully established and was utilized to confirm that RnPV1-W8 has a broad host range in Sordariomycetes (23, 24). Particle structures determined by cryo-electron microscopy (cryo-EM) and three-dimensional (3D) image reconstruction, as well as one determined by X-ray crystallography, have shown three different partitivirus capsids to have a so-called “T=2” organization comprising 60 capsid protein (CP) dimers arranged on a T=1 icosahedral lattice (25–27). These developments and technological advances have facilitated research on different aspects of the biology of fungal partitiviruses, such as virus-host interactions, virion structure, virus evolution, and host range.
Sclerotinia sclerotiorum is a typical necrotrophic, destructive, soilborne, plant pathogenic fungus that infects a large number of economically important crops, such as rapeseed (Brassica napus), soybean (Glycine max), and the model plant Arabidopsis thaliana (28). The diseases caused by S. sclerotiorum result in substantial losses of yield in crop production each year. Because there is a lack of resistant cultivars, Sclerotinia diseases are controlled at low efficiency via fungicides and cultural approaches, e.g., crop rotation (29). Since hypovirulence and dsRNA elements (possibly mycoviruses) in S. sclerotiorum were first reported in the early 1990s (30), many novel mycoviruses, including members of the families Hypoviridae, Alphaflexiviridae, Partitiviridae, and Narnaviridae and an unassigned, single-stranded DNA (ssDNA) mycovirus, were isolated recently from S. sclerotiorum and are well characterized (5, 8, 9, 31). Furthermore, most of these viruses were shown to be responsible for reducing the virulence and growth of S. sclerotiorum (5, 9, 31, 32). These hypovirulence-associated mycoviruses of S. sclerotiorum provide an opportunity for developing innovative approaches to control annual crop diseases caused by S. sclerotiorum. Here we report the discovery and characterization of a novel partitivirus, Sclerotinia sclerotiorum partitivirus 1 from hypovirulent strain WF-1 (SsPV1/WF-1), which exhibits unique characteristics that distinguish it from all previously reported partitiviruses. Our experiments demonstrate that SsPV1/WF-1 does cause hypovirulence in S. sclerotiorum, Sclerotinia nivalis, Sclerotinia minor, and Botrytis cinerea. Furthermore, this virus can be transmitted and spread via hyphal contact among different vegetative compatibility groups of S. sclerotiorum and can also be transmitted interspecifically to S. nivalis and S. minor, two sister species of S. sclerotiorum. The cryo-EM structure of SsPV1/WF-1 was also analyzed and is discussed.
MATERIALS AND METHODS
Fungal strains and growth conditions.
Sclerotinia sclerotiorum strain WF-1 was originally isolated from a sclerotium collected from a diseased Japanese thistle (Cirsium japonicum) in Suizhou County, Hubei Province, People's Republic of China. Ep-1PNA367hyg is a normal, virus-free strain obtained from single-ascospore-isolation progeny of strain Ep-1PN, and virus-free strain Sunf-MAhyg is a single-ascospore-isolation progeny of strain Sunf-M that was originally isolated from sclerotia on a diseased sunflower plant (Helianthus annuus). These two S. sclerotiorum strains were labeled with a hygromycin resistance gene by using Agrobacterium tumefaciens-mediated transformation, and the hygromycin-resistant isolates showed no significant differences in colony morphology from their parental isolates. Virus-free strain 1980 was kindly supplied by Jeffrey A. Rollins (Department of Plant Pathology, University of Florida). S. nivalis strain Let-19 was isolated from diseased lettuce at Shennongjia, Hubei Province, People's Republic of China (33). S. minor strain SM-1 was infected with a new dsRNA virus, Sclerotinia minor RNA virus 1 (SmRV1/SM-1) (data not shown). B. cinerea strain KY-1, a virus-free strain, was originally isolated from diseased blueberry and was identified via sequencing of the contiguous internal transcribed spacer region. All strains were grown on potato dextrose agar (Difco) medium amended with 0.5% yeast extract (PDAY medium) at 20°C to 22°C and stored on PDAY slants at 4°C to 6°C. Sclerotia produced on PDAY plates were collected, air dried, and stored at −20°C.
Phenotypic characteristics and virulence assay.
To assess the growth characteristics of the different fungal strains used in this study, agar plugs of fresh mycelium were transferred from colony margins of old cultures onto fresh PDAY medium. The colony radius of each strain was measured daily for 3 days, and the growth rate of each strain was calculated. To evaluate the virulence of the fungal strains on their hosts, freshly grown mycelial agar plugs were inoculated onto the stem or detached leaves of rapeseed, A. thaliana, soybean, or tomato plants and incubated at 20°C with 90% humidity. There were more than three replicates for each treatment, and all biological characterization experiments were conducted at least twice.
Transmission electron microscopy of hyphal cells.
Ultrastructural characterization of hyphal cells of Ep-1PNA367hyg and WF-1 was done by using transmission electron microscopy (TEM) and a protocol described previously by Zhang et al. (34).
Virion purification and dsRNA isolation.
Fungal strains were grown in stationary cultures containing potato dextrose broth. Mycelium (∼100 g [wet weight]) of each fungal strain was harvested after an 8- to 10-day culture period by straining through Miracloth in a Buchner funnel. The protocol for extracting viral particles was performed according to a method described previously by Tang et al. (27). After two cycles of high-speed centrifugation, virus particles were purified by centrifugation through a layer of 36% (wt/wt) CsCl in buffer A (0.05 M Tris-HCl [pH 7.6] containing 150 mM NaCl and 5 mM EDTA) for 17 h at 45,000 rpm in a Beckman SW55 rotor. Purified virus particles were collected by puncturing the centrifuge tube with a needle and a syringe. The harvested sample was diluted 1:1 with buffer A and centrifuged at 40,000 rpm for 4 h in a Beckman 50Ti rotor. The final pellet was resuspended in buffer A and then used for dsRNA extraction, structural analysis of virus particles, and infectivity assays.
The dsRNA was isolated from purified virus particles by using phenol-chloroform extraction, whereas dsRNA was isolated from fungal mycelia by CF-11 cellulose (Sigma-Aldrich, Dorset, England) chromatography, as described previously (35, 36). The dsRNA nature of the samples was further confirmed based on resistance to DNase I and S1 nuclease (TaKaRa, Dalian, People's Republic of China). The isolated dsRNA was analyzed by electrophoresis on 1% agarose or 15% nondenaturing polyacrylamide gels. The dsRNA samples were further purified with a gel extraction kit (Axygen Biosciences) and stored at −80°C before use.
Viral genome cloning, sequencing, and bioinformatics analyses.
The purified viral genomic dsRNA was used as a template for the construction of cDNA libraries according to a protocol described previously by Xie et al. (37), using a cDNA synthesis kit (Fermentas) with tagged random primers-dN6. To obtain the complete viral genome sequence, a series of reverse transcription-PCRs (RT-PCRs) and viral terminal rapid amplification of cDNA ends (RACE) PCRs were conducted according to the manufacturer's protocols to amplify overlapping fragments that covered the entire viral genome. All expected PCR fragments were ligated into the pMD18-T vector (TaKaRa) and sequenced. M13 universal primers or sequence-specific primers (see Table S1 in the supplemental material) were used for sequencing or RT-PCR, and each base was identified by sequencing of at least three independent, overlapping clones in both orientations.
The sequences of the dsRNA genomes of previously reported partitiviruses referenced in this paper were retrieved from the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/genomes) and used for comparative analyses. Multiple-sequence alignments were computed by using the DNAMAN, CLUSTAL_W, and BLAST software packages (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Based on these alignments, we constructed phylogenetic trees using the neighbor-joining method, as computed with MEGA version 4.0 programs (38). Phylogenetic trees were plotted with TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) and manually edited.
RNA extraction and RT-PCR.
Total RNA was prepared by using TRIzol reagent (Invitrogen) according to the manufacturer's instructions and treated with DNase I. To verify the presence of dsRNA in virus-infected colonies, cDNAs were synthesized by using Moloney murine leukemia virus (M-MLV) transcriptase (Promega) and an oligo(dT) primer. Four primers (CP1049F, CP1348R, RAPD1260F, and RAPD1579R) were used to detect coat protein and RNA-dependent RNA polymerase (RdRp) (see Table S1 in the supplemental material).
Cryo-EM and 3D image reconstruction of SsPV1.
Small aliquots (2 to 3 μl) of purified SsPV1 virions were vitrified, and cryo-EM image data were recorded essentially as described previously for the partitivirus Penicillium stoloniferum virus S (PsV-S) (25). A total of 23 micrographs were recorded at 200 keV under low-dose conditions (∼15 e/Å2) at underfocus settings ranging between 3.8 and 11 μm on a Gatan Ultrascan 4K2 charge-coupled-device (CCD) camera with an FEI Polara Tecnai microscope at a nominal magnification of ×76,900 (yielding an effective pixel size of 1.8 Å).
The RobEM program (http://cryoem.ucsd.edu/programDocs/runRobem.txt) was used to extract individual particle images from the micrographs and to estimate the defocus level of each micrograph. A total of 1,266 particle images were boxed, and all of them were used to compute the final 3D reconstruction. Image processing was carried out essentially as described previously for structural studies of PsV-F and PsV-S (27). Briefly, the current version of Auto3DEM was used to compute an ab initio 3D reconstruction from 150 particle images by using the Random Model Computation procedure (39, 40). This initial reconstruction was used as a reference model to refine the origin and orientation parameters for all 1,266 particles and led to a 3D reconstruction at an ∼18-Å resolution. The subprograms P3DR and PO2R were next used as standalone applications to refine and improve the map by manual processing to a final resolution of 12 Å, as estimated from a Fourier shell correlation plot and a 0.5-threshold criterion. These manual processing steps involved the iterative use of different combinations of inverse temperature factors, as described previously (27). An inverse temperature factor of 1/(200 Å2) was applied to the final, reconstructed density map, and the absolute hand of the SsPV1 structure was set to be the same as those found for PsV-S, PsV-F, and Fusarium poae virus 1 (FpV1).
Transmission test of SsPV1/WF-1.
To verify that the mycovirus SsPV1/WF-1 causes hypovirulence in S. sclerotiorum strain WF-1 and to determine if SsPV1/WF-1 can be transmitted among different vegetative groups of S. sclerotiorum, horizontal transmission of SsPV1/WF-1 from strain WF-1 to S. sclerotiorum strains Ep-1PNA367hyg, Sunf-MAhyg, and R6V was carried out by dual cultures on PDAY medium, as described previously by Zhang et al. (34).
To test if SsPV1/WF-1 can be transmitted horizontally to S. nivalis or S. minor, two sister species of S. sclerotiorum, strain WF-1 of S. sclerotiorum and strain Let-19 of S. nivalis (or strain SM-1 of S. minor) were dually cultured on a PDAY plate at 20°C to 22°C. Ten mycelial agar discs were cut from the Let-19 (or SM-1) colony and transferred individually into fresh PDAY plates. Based on the colony morphology of Let-19 (or SM-1), which differs significantly from that of S. sclerotiorum, new cultures of Let-19 (or SM-1) were subcultured and stored on PDAY slants at 4°C to 6°C for further study. Subcultures of Let-19 (or SM-1) were subjected to dsRNA analysis to verify infection with SsPV1/WF-1.
Transfection of protoplasts with viral particles.
Protoplast preparations of virus-free S. sclerotiorum strain 1980 and B. cinerea strain KY-1 were made according to the method described previously by Zhang et al. (34). For transfection, approximately 5 × 106 protoplasts in 100 μl STC buffer (1.2 M sorbitol, 10 mM Tris-HCl [pH 7.5], 10 mM CaCl2) were mixed gently with 10 to 20 μg of purified SsPV1/WF-1 particles and incubated at 20°C. After 20 to 30 min, 200-, 200-, and 800-μl aliquots of polyethylene glycol (PEG) solutions (60% PEG 400 in 10 mM Tris [pH 7.5] and 50 mM CaCl2) were gradually added to the protoplast suspension containing SsPV1/WF-1 particles in three steps and incubated at 20°C for 20 min. The protoplast suspension containing SsPV1/WF-1 particles was diluted with 5 to 10 ml STC buffer and centrifuged at 3,000 rpm at 4°C for 10 min. The pellet was resuspended in 500 μl STC buffer, and the protoplast suspension was spread onto regeneration medium and incubated at 20°C for 7 to 8 days. More than 10 mycelial plugs were cut from the regenerated colonies with a cork borer at random and transferred onto fresh PDAY plates. To verify that the transferred cultures were transfected by SsPV1/WF-1, the colony morphologies of newly transfected isolates of S. sclerotiorum strain 1980 and B. cinerea strain KY-1 were compared with those of the original, virus-free strains 1980 and KY-1, respectively. These transfections were further confirmed by dsRNA extraction and/or PCR amplification of viral genomic fragments using gene-specific primers (see Table S1 in the supplemental material), which were designed based on the sequence of the coat protein or RdRp.
Data analyses.
Experimental data were subjected to analysis of variance by using the SAS8.0 program. Treatment means were compared by using the least-significant-difference test at a significance level of a P value of 0.05.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of SsPV1 reported here have been deposited with the EMBL/GenBank/DDBJ databases under accession numbers JX297510 and JX297511.
RESULTS
Hypovirulence traits associated with dsRNA in Sclerotinia sclerotiorum.
Strain WF-1 exhibited abnormal colony morphology compared to virus-free strains Ep-1PNA367hyg (Fig. 1A) and Sunf-MAhyg (data not shown). Strain WF-1 grew slowly on PDAY medium and showed excessive hyphal branching and cytoplasmic extrusion at most hyphal tips (Fig. 1B). Furthermore, strain WF-1 was less virulent in its hosts, including rapeseed, soybean, tomato, and A. thaliana (Fig. 1C). In addition, the sclerotia of strain WF-1 failed to produce apothecium in a rapeseed field or under laboratory conditions (data not shown). Thus, these biological characteristics suggest that strain WF-1 is a hypovirulent strain.
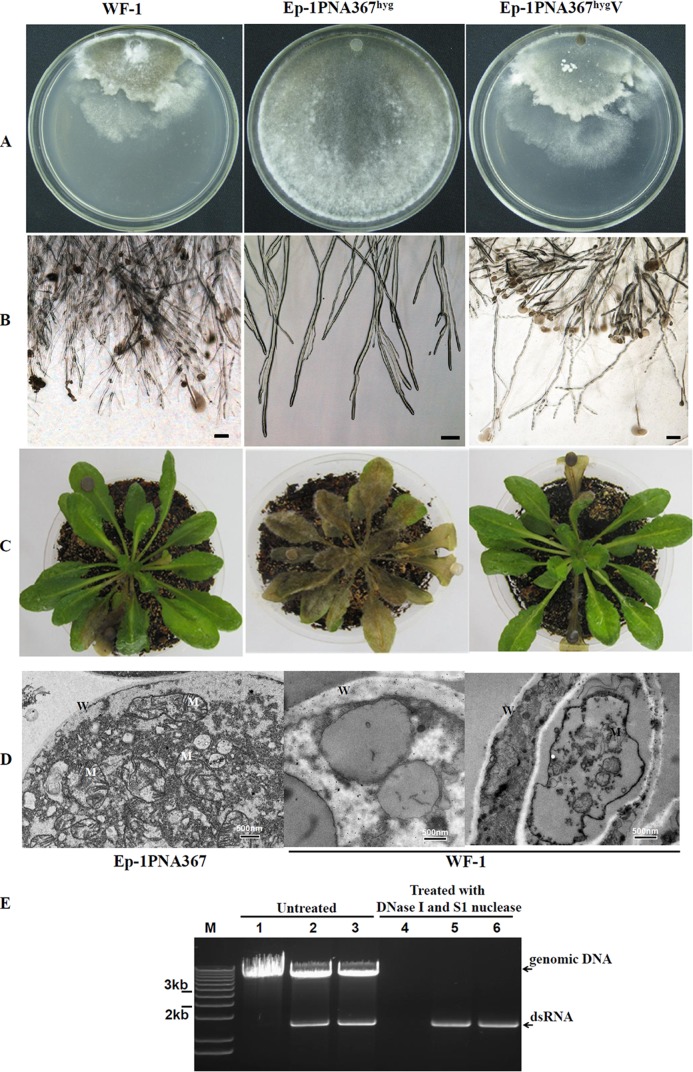
FIG 1.
Phenotypes associated with hypovirulence of different strains of S. sclerotiorum. (A) Colony morphology of strains WF-1, Ep-1PNA367hyg, and Ep-1PNA367hygV. The latter strain represents the strain that arises from contact between Ep-1PNA367hyg and hypovirulent strain WF-1. All strains were cultured on plates with 20 ml PDAY medium at 20°C to 22°C for 3 days prior to photography. Compared to strain Ep-1PNA367hyg, strains WF-1 and Ep-1PNA367hygV exhibit low growth rates. (B) Strains WF-1 and Ep-1PNA367hygV exhibit excessive branching compared to strain Ep-1PNA367hyg. Moreover, cytoplasm was observed to extrude from some mycelial tips in the strains that contain the 2.3-kb dsRNA segments. (C) Assay of A. thaliana leaves that show hypovirulence when infected with strains WF-1 and Ep-1PNA367hygV (20°C; 4 days postinfection). (D) Ultrastructure of cells of S. sclerotiorum strain WF-1 examined by thin-section transmission electron microscopy (TEM). The ultrastructure of the hyphal tip of strain Ep-1PNA367 shows a well-distributed cytoplasm with many mitochondria whose membranes appear normal and contain many intact cristae. The ultrastructure of the hyphal tips in hypovirulent strain WF-1 shows a granulated cytoplasm and damaged mitochondria with abnormal cristae; only a few normal mitochondria remained. W, cell wall; M, mitochondria. (E) Detection of dsRNA in mycelia of strains Ep-1PNA367hyg (lanes 1 and 4), WF-1 (lanes 2 and 5), and Ep-1PNA367hygV (lanes 3 and 6). Lane M, DNA size markers.
The hyphal ultrastructure of strain WF-1 was compared with that of strain Ep-1PNA367hyg. Strain WF-1 exhibited a granulated cytoplasm, and only a few normal mitochondria were observed, with all others containing abnormal cristae (Fig. 1D), suggesting that strain WF-1 was likely infected by at least one kind of mycovirus. Furthermore, dsRNA with an estimated size of 2.3 kb was extracted and fractionated by agarose gel electrophoresis from hypovirulent strain WF-1. The double-stranded nature of WF-1 dsRNA was confirmed based on its resistance to digestion with DNase I and S1 nucleases (Fig. 1E).
Horizontal transmission of hypovirulence and dsRNA.
Following hyphal contact of strain WF-1 with recipient strains Ep-1PNA367hyg and Sunf-MAhyg, mycelial agar discs were obtained from the colony contact zones and placed onto hygromycin-containing PDAY medium. Several derivative cultures that developed from hygromycin-containing PDAY medium were examined for hypovirulence and dsRNA content. The results of these experiments showed that hypovirulence and its associated traits, as well as the 2.3-kb dsRNA element of strain WF-1, were cotransmitted to virulent strains Ep-1PNA367hyg (Fig. 1A) and Sunf-MAhyg (data not shown). These results were further confirmed by dsRNA extraction (Fig. 1E). Moreover, strain WF-1 is vegetatively incompatible with strain Ep-1PNA367hyg (31) and Sunf-MAhyg (data not shown). Therefore, since the 2.3-kb dsRNA segment is associated with hypovirulence and other related traits of strain WF-1, this dsRNA segment also overcomes transfer barriers between vegetatively incompatible strains.
Properties of mycoviral particles and mycoviral genome analysis.
Viral particles isolated from mycelia of strain WF-1 via differential centrifugation were negatively stained and examined by TEM; they were shown to be spherical and ∼40 nm in diameter (Fig. 2A). Nucleic acids extracted from viral particles had the same electrophoretic mobility as those extracted directly from mycelium of strain WF-1 (Fig. 2C and D). SDS-PAGE analysis of purified viral particles showed a single major protein band with a molecular mass of ∼75 kDa (Fig. 2B). Although the 2.3-kb dsRNA segment appeared as a single band on the 1% agarose gel, two clearly separated dsRNA bands (Fig. 2C and D) were detected on nondenaturing 15% polyacrylamide gels. Thus, strain WF-1 contains two similarly sized dsRNA segments, which we named dsRNA-1 and dsRNA-2. These segments were recovered and purified from 1% agarose gels and subjected to cDNA cloning and sequence analysis.
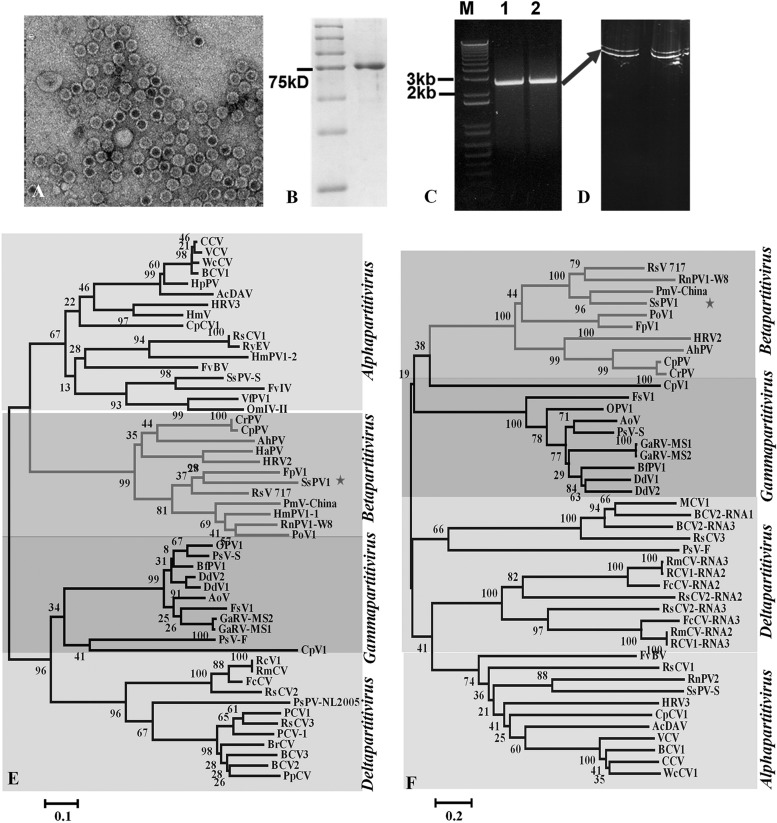
FIG 2.
Characterization of SsPV1/WF-1 isolated from hypovirulent strain WF-1. (A) TEM images of negatively stained particles of SsPV1/WF-1. (B) SDS-PAGE of purified SsPV1/WF-1 virus particles showing one prominent band (right lane) corresponding to the viral CP. (C) Agarose gel electrophoresis of dsRNA extracted from SsPV1/WF-1 (lane 1) and mycelia of strain WF-1 (lane 2). (D) Ethidium bromide-stained, nondenaturing 15% polyacrylamide gel electrophoresis of dsRNA extracted from purified SsPV1/WF-1 (lane 1) and mycelia of strain WF-1 (lane 2). All dsRNA samples were treated with DNase I and S1 nuclease prior to electrophoresis. (E and F) Neighbor-joining phylogenetic trees constructed based on the complete amino acid sequences of the viral RdRp (E) or CP (F) of SsPV1/WF-1 and numerous other members of the family Partitiviridae. Bootstrap values (percent) obtained with 2,000 replicates are indicated on the branches, and branch lengths correspond to genetic distances; the scale bar at the bottom left corresponds to the genetic distance. Abbreviations of virus names and GenBank accession numbers are listed in Table S2 in the supplemental material.
The complete nucleotide sequences of dsRNA-1 and dsRNA-2 were determined by using RT-PCR and terminal RACE amplification clones (data not shown). Excluding an interrupted poly(A) tail, dsRNA-1 contains 2,287 bases and a single, large open reading frame (ORF), which encodes a putative protein of 704 amino acids (aa) with a calculated molecular mass of 82 kDa. The coding region of dsRNA-1 is flanked by a 5′ untranslated region (UTR) (nucleotides [nt] 1 to 92) and a 3′ UTR (nt 2208 to 2287), with the latter being followed by a poly(A) tail. Also excluding an interrupted poly(A) tail, dsRNA-2 contains 2,274 bases, a 5′ UTR (nt 1 to 82), a 3′ UTR (nt 2120 to 2274), and a single ORF (nt 83 to 2119) that encodes a putative 678-aa protein with a calculated molecular mass of 74 kDa. Furthermore, the CAA repeat motif was found at the 5′ UTRs of both dsRNAs (data not shown). These CAA motifs are similar to the translational enhancer elements present at the 5′ UTRs of some plant viruses and some mycoviruses, including Rhizoctonia solani virus 717 and Penicillium chrysogenum virus (36, 41, 42).
A BLASTp search of the NCBI protein database and multiple-sequence alignments were performed with the putative proteins encoded by dsRNA-1 and dsRNA-2. The protein encoded by dsRNA-1 is closely related to the RdRps encoded by other mycoviruses of the genus Betapartitivirus, such as Fusarium poae virus 1 and Rhizoctonia solani virus 717, with 47% and 45% identities, respectively. The protein encoded by dsRNA-2 has high sequence similarity to the capsid proteins (CPs) of mycoviruses belonging to the genus Betapartitivirus, such as Rhizoctonia solani virus 717 and Rosellinia necatrix partitivirus 1-W8, with 33% and 31% identities, respectively. Hence, these dsRNA-1 and dsRNA-2 sequences represent the genome of a novel mycovirus species belonging to family Partitiviridae, and it was given the name Sclerotinia sclerotiorum partitivirus 1 (SsPV1/WF-1). Furthermore, phylogenetic analysis based on the full-length amino acid sequences of the putative RdRp and CP viral proteins of SsPV1/WF1 compared to other recognized partitiviruses further confirmed that SsPV1 is a new member of the genus Betapartitivirus (Fig. 2E and F).
Cryo-EM and 3D image reconstruction of SsPV1/WF-1.
Twenty-three transmission electron micrographs of unstained, vitrified samples of purified SsPV1/WF-1 virions were recorded (see Materials and Methods). These micrographs showed fields of regularly sized, intact, spherical particles with diameters of nearly 400 Å, consistent with our assessment of negatively stained samples (Fig. 3A). A total of 1,266 images of individual SsPV1/WF-1 virions was extracted from these micrographs and used to compute a 3D reconstruction at an estimated resolution of ∼12 Å (Fig. 3A and D).
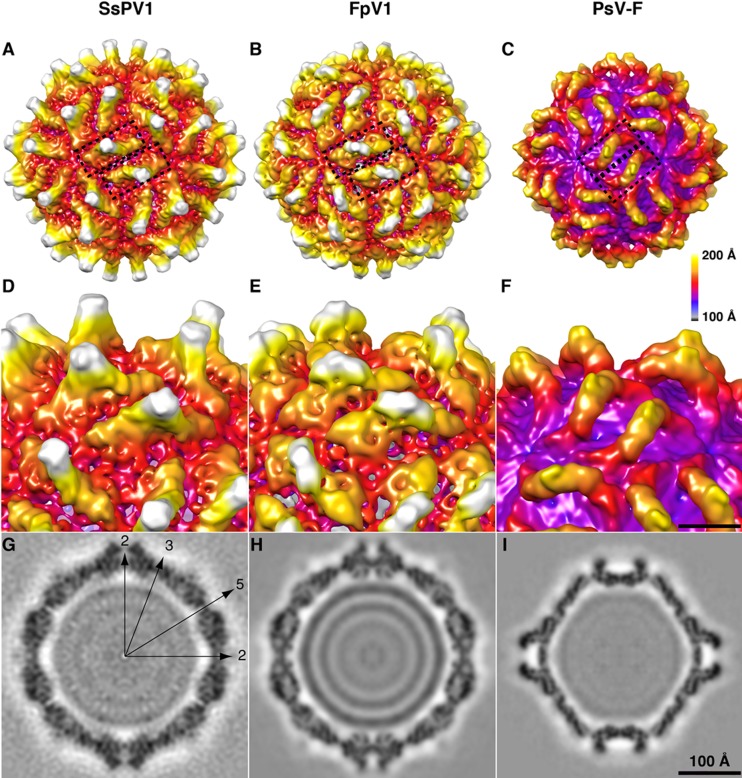
FIG 3.
Three-dimensional structures of SsPV1/WF-1 capsids. (A to C) Surface view of three related partitiviruses, SsPV1 (A), FpV1 (B), and PsV-F (C). Two dimers in SsPV1 are highlighted in blue-shaded regions in the center. (D to F) Close-up view of the surface protrusions in SsPV1 (D), FpV1 (E), and PsV-F (F). (G to I) Central section of SsPV1 (G), FpV1 (H), and PsV-F (I) showing RNA genomes inside the particles. The capsid is radially depth cued, with the radial color bar shown on the right. The surface view of the reconstruction of SsPV1 was rendered with UCSF Chimera together with FpV1 and PsV-F.
The outer surface topography of the SsPV1/WF-1 capsid is quite convoluted and resembles that seen for other partitiviruses such as FpV1 (Fig. 3B and E) and PsV-F (Fig. 3C and F). All three partitivirus capsids are characterized by deep depressions at each of the icosahedral symmetry axes (I2, I3, and I5), with the broadest pits encompassing the 5-fold (I5) and 3-fold (I3) axes. Each icosahedral 2-fold (I2) axis is crossed by a long and narrow linear canyon, with capsid structures rising steeply on both sides. A second long and narrow linear canyon traverses the distal side of each of these elevated regions and runs nearly parallel to the I2 canyon. SsPV1/WF-1 is about the same size (∼420-Å maximum outer diameter) as FpV1 but is noticeably larger than PsV-F (∼376-Å diameter).
Just as was found for the capsid structures of FpV1, PsV-S, and PsV-F (20), SsPV1/WF-1 has 60 identical, elevated regions (2 of which are outlined by boxes in Fig. 3A to C), each of which exhibits a strong, 2-fold-symmetric appearance and a centrally located protrusion that extends to a large radius. In SsPV1/WF-1, the elevated regions closely resemble those of FpV1 in morphology and orientation (Fig. 3A, B, D, and E), but the protrusions in SsPV1/WF-1 rise a bit more steeply than those in FpV1. In contrast, PsV-F has narrow, arch-like structures that adopt a distinctly different orientation (i.e., more vertical than horizontal, as seen in the views of SsPV1/WF-1 and FpV1 depicted in Fig. 3A and B). The presence of 60 dimeric structural features in SsPV1 is completely consistent with the conclusion that the SsPV1 capsid contains 120 CP monomers, organized as 60 CP dimers in a pseudo-T=2 icosahedral lattice, which appears to be a universal signature of dsRNA viruses (20).
Equatorial cross-sectional views of the SsPV1/WF-1 (Fig. 3G), FpV1 (Fig. 3H), and PsV-F (Fig. 3I) virions demonstrate that all three capsids have inner surfaces with topographies that are much smoother than the outer surfaces. The innermost diameters of these capsids are about 270, 270, and 240 Å, respectively. In addition, the equatorial view of each density map shows interior density features that represent the packaged, genomic dsRNA. In FpV1 (Fig. 3H), this density appears as four well-defined rings (progressively decreasing in density from high to low radii), which indicates that the dsRNA is tightly packed into concentric shells. However, the packing of dsRNA in the cores of SsPV1 (Fig. 3G) and PsV-F (Fig. 3I) is less defined (i.e., more diffuse) and therefore more disorganized than that in FpV1. Consistent with the reorganized taxonomy of partitiviruses, structural differences were shown to be greater between the genera Betapartitivirus and Gammapartitivirus (FpV1 and SsPV1 versus PsV-F) than within the genus Betapartitivirus (FpV1 versus SsPV1). SsPV1 and FpV1 have similar-sized particles with the same central dimer orientation, while PsV-F is much smaller, with a tilted central dimer.
Confirmation of SsPV1/WF-1-associated hypovirulence of Sclerotinia sclerotiorum by completion of Koch's postulates using a transfection assay.
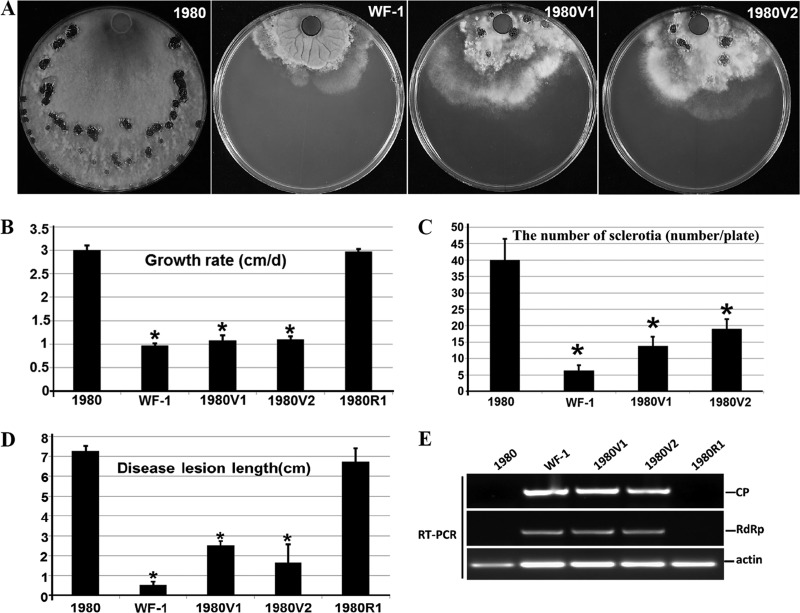
S. sclerotiorum strain 1980 is a virus-free strain isolated from Phaseolus vulgaris in Nebraska, and its genome was recently completely sequenced (43). Strain 1980 develops normal colonies at an average growth rate of 3.0 cm/day on PDAY medium and causes typical necrotic lesions on soybean stems, with an average lesion length of 7.2 cm at 96 h postinoculation (hpi) (Fig. 4). To determine whether SsPV1/WF-1 causes hypovirulence and its associated traits in S. sclerotiorum, protoplasts of strain 1980 were transfected with SsPV1/WF-1 particles by using PEG-mediated protoplast transfection. More than 10 regenerated colonies selected at random from five plates were assayed to verify mycovirus infection. RT-PCR analysis and dsRNA extraction indicated that all regenerated colonies were infected with SsPV1/WF-1, but no mycovirus was detected in noninoculated control strain 1980 (Fig. 4E). The regenerated colonies infected by SsPV1/WF-1 were designated 1980V1 and 1980V2. SsPV1/WF-1 in strain 1980 remained unchanged during subculturing of 1980V1 and 1980V2. The colony morphologies of isolates 1980V1 and 1980V2 differed significantly from that of parental strain 1980 in showing irregular hyphal extensions at the colony margin (Fig. 4A). Hyphal extension of isolates 1980V1 and 1980V2 occurred at a significantly lower rate than for strain 1980 (Fig. 4B). Isolates 1980V1 and 1980V2 produced fewer sclerotia in the mature colony, with average numbers reaching only 20 sclerotia/plate, compared to the >30 sclerotia/plate for strain 1980 (Fig. 4C). Moreover, similarly to SsPV1-infected strain Ep-1PNA367hygV, cytoplasm leaked out from the hyphal tips of isolates 1980V1 and 1980V2, but this phenomenon did not occur in wild-type strain 1980 (data not shown). The virulence of strains 1980V1 and 1980V2 was significantly reduced, and lesions induced by strains 1980V1 and 1980V2 on stems and detached leaves of soybean were significantly smaller than those induced by wild-type strain 1980 (Fig. 4D). The results of the viral transfection assay provided strong, direct evidence that SsPV1/WF-1 infection was responsible for hypovirulence and its associated traits in S. sclerotiorum.
FIG 4.
Transfection of protoplasts of S. sclerotiorum virulent strain 1980 with purified SsPV1/WF-1 particles. (A) Comparison of colony morphologies of parental strain 1980, strain WF-1, and the SsPV1-transfected isolates 1980V1 and 1980V2. (B) Comparison of hyphal growth rates of strains 1980, WF-1, 1980V1, 1980V2, and 1980R1 (the latter is an isolate from regenerated nontransfected control protoplasts of strain 1980) on PDAY medium (20°C). (C) Numbers of sclerotia per plate produced by strains 1980, WF-1, 1980V1, 1980V2, and 1980R1 on PDAY medium (20°C; 20 days postinfection). (D) Lesion length caused by strains 1980, WF-1, 1980V1, 1980V2, and 1980R1 (20°C; 4 days postinfection) on the stems of soybean (Glycine max). (E) RT-PCR detection of SsPV1/WF-1 in individual isolates. The actin gene is used as an internal control. The predicted lengths of the RT-PCR products for CP and RdRp are 300 and 320 nt, respectively. Results in each of the histograms shown in panels B to D are expressed as arithmetic means ± standard errors of the means. Asterisks indicate a significant difference (P < 0.05) among strains of S. sclerotiorum according to the Student t test.
Horizontal transmission of SsPV1/WF-1 to Sclerotinia nivalis and Sclerotinia minor.
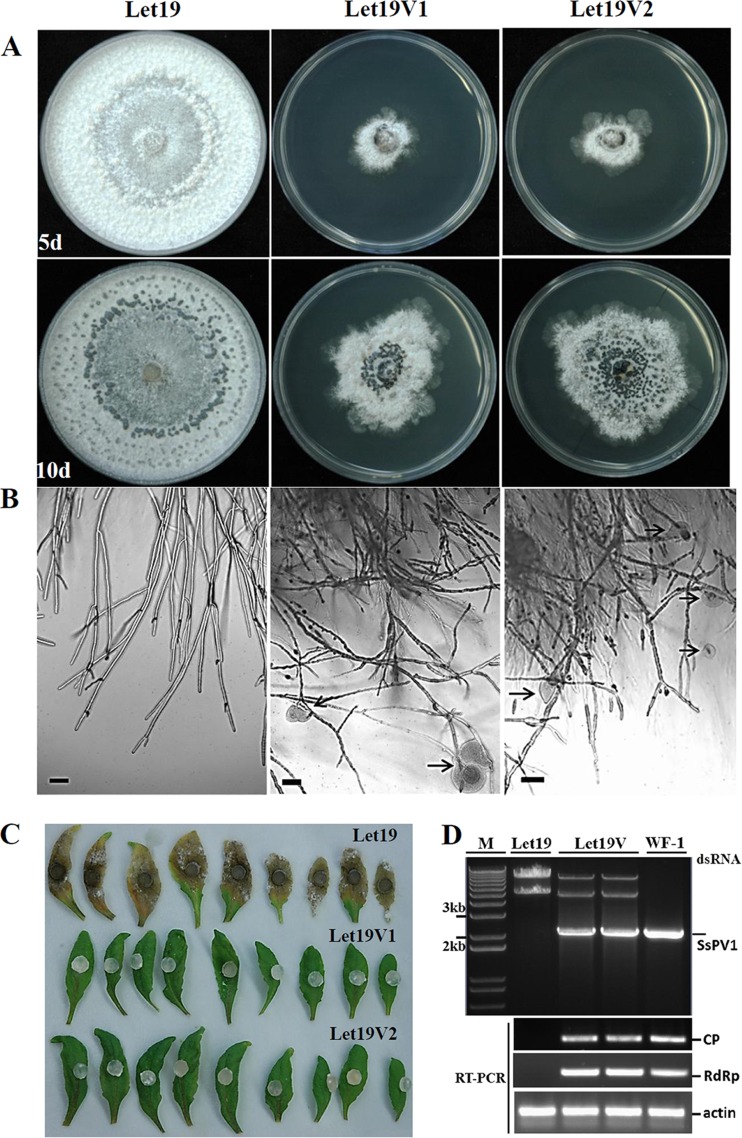
To determine whether SsPV1/WF-1 has the ability to be transmitted between different Sclerotinia species via hyphal contact, S. nivalis strain Let-19 was dually cultured with strain WF-1 on the same PDAY plate. After dual culturing on a PDAY plate, subcultures of S. nivalis were tested for infection with SsPV1/WF-1. All 10 subcultures of strain Let-19 showed an abnormal phenotype of S. nivalis (Fig. 5A), and two of them (Let-19V1 and Let-19V2) were further examined by dsRNA extraction and RT-PCR amplification to confirm infection by SsPV1/WF-1. The dsRNA extraction experiment showed that strain Let-19 carried two dsRNA segments, whereas Let-19V1 and Let-19V2 both carried three dsRNA bands. Two of these segments were similar to the segments in wild-type strain Let-19, while the smallest dsRNA segment was found have a size similar to that of a segment extracted from strain WF-1 (Fig. 5D). RT-PCR amplification further confirmed that SsPV1/WF-1 was interspecifically transmitted successfully to strain Let-19 of S. nivalis (Fig. 5D).
FIG 5.
Interspecies transmission of SsPV1/WF-1 from S. sclerotiorum to S. nivalis via hyphal contact. (A) Comparison of colony morphologies of S. nivalis strain Let-19 and SsPV1-infected isolates Let-19V1 and Let-19V2, which were obtained from Let-19 after the mycelia of Let-19 contacted those of hypovirulent strain WF-1, at 5 and 10 days postinfection. (B) The hyphal tips of strains Let-19V1 and Let-19V2 showed excessive branching compared to strain Let-19. Moreover, cytoplasm (marked with black arrows) was released from some hyphal tips of strains Let-19V1 and Let-19V2. (C) Virulence assays of S. nivalis. Let-19V1 and Let-19V2 did not infect detached A. thaliana leaves at 4 days postinfection, but Let-19 did. (D) RT-PCR detection of SsPV1/WF-1 dsRNA in extracts from strain Let-19, two representative Let-19V isolates, and strain WF-1. All dsRNA samples were treated with DNase I and S1 nuclease prior to electrophoresis. The actin gene was used as an internal control. The predicted lengths of the RT-PCR products for CP and RdRp of SsPV1/WF-1 are 300 and 320 nt, respectively.
Compared to wild-type strain Let-19, virus-infected strain Let-19 exhibited hypovirulence and its associated traits. Unlike wild-type strain Let-19, virus-infected strains Let-19V1 and Let-19V2 did not induce lesions on leaves of A. thaliana or lettuce (Fig. 5C). These strains also grew slowly on PDAY medium and formed colonies with abnormal morphology, and cytoplasm was seen frequently leaking from the hyphal tips (Fig. 5A and B). These results suggest that SsPV1/WF-1 can be transmitted to S. nivalis interspecifically through colony contact and causes hypovirulence in S. nivalis.
Similar results were obtained for S. sclerotiorum strain WF-1 and S. minor strain SM-1. SsPV1/WF-1 also transmitted horizontally from strain WF-1 to strain SM-1 via hyphal contact. Moreover, the phenotypes of strain SM-1V showed hypovirulence in the presence of SsPV1/WF-1 (data no shown).
SsPV1/WF-1 causes hypovirulence in Botrytis cinerea.
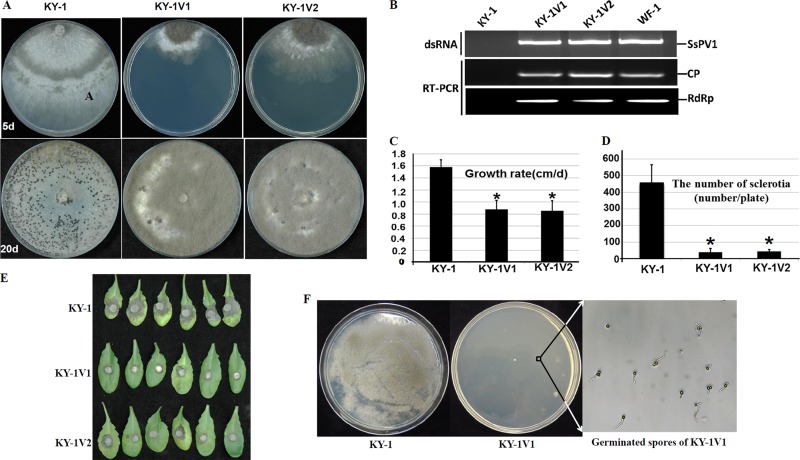
Since our data strongly supported the conclusion that the partitivirus SsPV1/WF-1 is responsible for hypovirulence in S. sclerotiorum, our next goal was to determine whether SsPV1/WF-1 also affects other plant pathogenic fungi such as B. cinerea. Indeed, protoplasts of B. cinerea (strain KY-1) were successfully transfected with SsPV1/WF-1 by using a PEG-mediated method. Mycelial agar discs were selected from a protoplast-regenerated colony and transferred individually into fresh PDAY plates. All new cultures were subsequently found to be infected with SsPV1/WF-1, as verified by dsRNA extraction and RT-PCR amplification analyses (Fig. 6B). Virus-infected strain KY-1 isolates were designated KY-1V1 and KY-1V2, etc.
FIG 6.
Transfection of protoplasts of virulent Botrytis cinerea strain KY-1 with purified SsPV1/WF-1 particles. (A) Comparison of colony morphologies of parental strain KY-1 and SsPV1-transfected isolates KY-1V1 and KY-1V2 at 5 and 20 days postinfection. (B) RT-PCR detection of SsPV1/WF-1 dsRNA in extracts from individual isolates. The predicted lengths of the PCR products for CP and RdRp are 300 and 320 nt, respectively. (C) Comparison of hyphal growth rates for strains KY-1, KY-1V1, and KY-1V2 on PDAY medium (20°C). (D) Comparison of numbers of sclerotia per plate produced by strains KY-1, KY-1V1, and KY-1V2 on PDAY medium (20°C; 20 days postinfection). The results plotted in each histogram are expressed as arithmetic means ± standard errors of the means. Asterisks indicate a significant difference (P < 0.05) among strains of S. sclerotiorum according to the Student t test. (E) Virulence testing on detached leaves of A. thaliana with different isolates of B. cinerea (20°C; 2 days postinfection). Strains KY-1V1 and KY-1V2 show hypovirulence. (F) Infection of B. cinerea conidia with SsPV1 is eventually lethal to infected hyphae. Plates of PDAY medium were seeded with 200-μl conidial suspensions (105 spores) of B. cinerea KY-1 and KY-1V1. Most SsPV1/WF-1-infected conidia of B. cinerea can produce a germination tube after 36 h but are unable to develop into colonies. This phenomenon was further confirmed by light microscopy.
Two virus-infected strains, KY-1V1 and KY-1V2, were further characterized to examine their biological properties. The results showed that compared to wild-type strain KY-1, KY-1V1 and KY-1V2 exhibited abnormal colony morphology (Fig. 6A), low growth rates, production of few sclerotia, excessive hyphal branching (Fig. 6C and D), and leakage of cytoplasm from hyphal tips (data not shown). SsPV1-infected KY-1V1 and KY-1V2 had reduced virulence on detached leaves of tomato (data not shown) or A. thaliana (Fig. 6E). Therefore, the hypovirulence and its traits exhibited by SsPV1-infected strains KY-1V1 and KY-1V2 were very similar to those exhibited by virus-infected S. sclerotiorum strain 1980. Moreover, the hypovirulence and its traits exhibited by KY-1V1 and SsPV1/WF-1 were cotransmitted to virus-free B. cinerea strain BC05.10 via hyphal fusion (data not shown). Therefore, the partitivirus SsPV1/WF-1 also causes hypovirulence in B. cinerea.
Unexpectedly, SsPV1-infected strains KY-1V1 and KY-1V2 produced abundant conidia (Fig. 6A), which germinated on PDAY medium. However, when >50 single germinating conidia were selected individually and transferred onto fresh PDAY plates, no visible colonies appeared on the PDAY plate. A conidial suspension (200 μl) from virus-infected strains KY-1V1 and KY-1V2 was spread onto the PDAY plate at a concentration of 105 spores per PDAY plate; only 13 colonies appeared on five PDAY plates. The phenotypes of these colonies were similar to those of virus-infected strains KY-1V1 and KY-1V2, and all of them were infected with SsPV1/WF-1 (data not shown). These results thus revealed that most of the conidia produced by virus-infected cultures can germinate but fail to develop an intact colony due to the presence of SsPV1/WF-1 (Fig. 6F).
SsPV1/WF-1 is horizontally transmitted to vegetatively incompatible host strains.
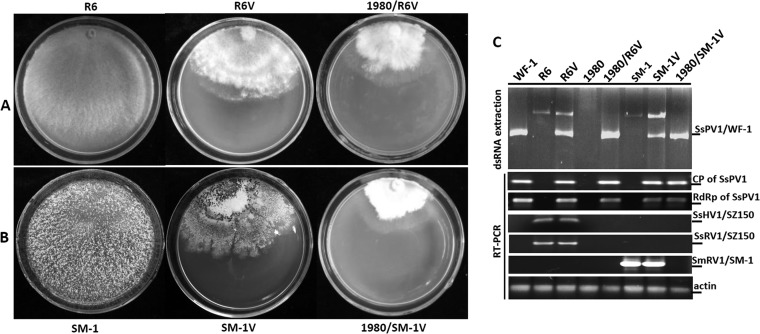
To clarify whether transmission to vegetatively incompatible host strains, i.e., breaking incompatibility transmission barriers, is a function of SsPV1/WF-1, strain WF-1, or a combination of both, SsPV1/WF-1 was horizontally transmitted to S. sclerotiorum strain R6 and S. minor strain SM-1 via hyphal contact. Strain R6 was a protoplast-derived isolate of hypovirulent strain SZ-150, which is coinfected with two viruses, Sclerotinia sclerotiorum RNA virus 1 (SsRV1/SZ150) and Sclerotinia sclerotiorum hypovirus 1 (SsHV1/SZ150) (37). Once strain R6 acquired SsPV1/WF-1 from strain WF-1, SsPV1-infected strain R6 was named strain R6V. When the mycelium of R6V contacted that of virus-free strain 1980 on a PDAY plate, SsPV1/WF-1 was horizontally transferred to strain 1980, but SsRV1/SZ150 and SsHV1/SZ150 were not (Fig. 7A and C). Similar results were observed for the interaction between strain SM-1 and strain 1980. Strain SM-1 carries an uncharacterized dsRNA virus, SmRV/SM-1, and SM-1V is strain SM-1 coinfected with two distant viruses, SsPV1/WF-1 and SsRV/SM-1. When the mycelium of strain SM-1V contacted that of virus-free strain 1980 on a PDAY plate, SsPV1/WF-1 was horizontally transmitted into different Sclerotinia species, but SmRV1 was not (Fig. 7B and C). It is important to note that the hypovirulence-associated traits and SsPV1/WF-1 were cotransmitted between different vegetatively incompatible strains (Fig. 7). Based on the above-described results, we conclude that the transmission properties are a function of SsPV1/WF-1.
FIG 7.
Transmission of SsPV1/WF-1 between vegetatively incompatible host strains. (A) Colony morphology of strains R6, R6V, and 1980/R6V. R6V represents the strain that arises from R6 after contact with hypovirulent strain WF-1, whereas 1980/R6V represents strain 1980 after dual culturing and contact with strain R6V. (B) Colony morphology of strains SM-1, SM-1V, and 1980/SM-1V. SM-1V is a strain of S. minor SM-1 after strain SM-1 was dually cultured with strain WF-1 on a PDAY plate. 1980/SM-1V was a subculture that was cut out from side of strain 1980 after mycelia of strain SM-1V and 1980 contacted on a PDAY plate. (C) dsRNA extraction and RT-PCR detection of SsPV1/WF-1 in individual strains. All dsRNA samples were treated with DNase I and S1 nuclease prior to electrophoresis. The actin gene was used as an internal control. The predicted lengths of the RT-PCR products for CP and RdRp of SsPV1/WF-1 are 300 and 320 nt, whereas the predicted PCR products from SsHV1/SZ150, SsRV/SZ150, and SmRV/SM-1 are 252, 462, and 424 nt, respectively. The primers for virus detection are shown in Table S1 in the supplemental material.
DISCUSSION
In the present study, we isolated and characterized a novel mycovirus, SsPV1/WF-1, from a hypovirulent strain (WF-1) of the plant fungal pathogen S. sclerotiorum. Based on three separate properties (nucleotide sequence, genome organization, and virus structure), SsPV1/WF-1 was unambiguously identified as a new member of the family Partitiviridae. Our experiments convincingly demonstrate that SsPV1/WF-1 causes hypovirulence in S. sclerotiorum. Previously, members of the family Partitiviridae were considered to be associated typically with latent infections, since they seldom have any significant impact on their hosts (16). However, at least three different partitiviruses are likely responsible for inducing abnormal phenotypes or virulence reduction in their hosts. First, Flammulina velutipes browning virus (FvBV) is known to be associated with the cap color of the fruiting bodies of Flammulina velutipes; FvBV was detected in strains that produce browning cups, whereas virus-free strains produce white cups (44). Second, Aspergillus fumigatus partitivirus 1 (AfuPV-1)-infected strain Af-237y-88 of Aspergillus fumigatus showed abnormal colony phenotypes, including slow growth, sectoring, and production of light pigmentation. However, any impact of AfuPV-1 on the virulence of its host is still unknown (45). Third, Rhizoctonia solani partitivirus 2 (RsPV2) was demonstrated to be associated with hypovirulence in Rhizoctonia solani AG-1IA (46). Recently, RnPV2 (Rosellinia necatrix partitivirus 2) without defective interfering dsRNA1 (DI-dsRNA1) was introduced into a Dicer-like 2 knockout mutant (Δdcl-2) and a wild-type strain of the nonnatural host C. parasitica. The RnPV2-infected Δdcl-2 mutant strain displayed disease symptoms, but the RnPV2-infected wild-type strain displayed a normal phenotype (47).
Clearly, SsPV1/WF-1 caused severe weakening of its host. Besides exhibiting reduced virulence, virus-infected strains grew more slowly with frequent sectoring, abnormal colony morphology, reduced sclerotium production, as well as defects in apothecium formation under field conditions and in the laboratory. These altered phenotypes caused by SsPV1/WF-1 are similar to those previously reported for hypovirulent strains of S. sclerotiorum (8, 11, 32, 48) and other phytopathogenic fungi (7, 48–50). Moreover, virus-infected strains showed excessive hyphal branching at the colony margin, and cytoplasmic extrusion at hyphal tips was often observed.
In general, reduced conidium production is a characteristic property of many fungi that are infected by hypovirulence-associated mycoviruses. Depressed production of conidia has been observed for hypovirus-infected strains of C. parasitica (51) and mycovirus-infected strains of Diaporthe ambigua (52). In addition, previous studies indicated that conidium production was reduced significantly in mycovirus-associated, hypovirulent strains of B. cinerea (53, 54). Unexpectedly, we found that although wild-type strain KY-1 produced conidia in a normal fashion, SsPV1-infected strains KY-1V1 and KY-1V2 of B. cinerea produced more conidia than did strain KY-1. Furthermore, although the conidia produced by virus-infected strains could germinate on PDAY medium, most germinating conidia, when transferred into fresh PDAY medium, could no longer develop new colonies. This phenomenon suggested that SsPV1/WF-1 boosts conidium production in an unusual manner but also suppresses mycelial growth of B. cinerea subsequent to germ tube formation.
Although SsPV1/WF-1 markedly affects its hosts, its productive infection and spread via hyphal contact are evidently maintained in S. sclerotiorum and B. cinerea. We made several attempts to eliminate SsPV1/WF-1 from strain WF-1 using different approaches, as previously described (10), but these attempts proved unsuccessful. In many fungus-mycovirus systems, some conidia do not harbor any virus, and strains developed from these conidia remain virus free (50). Our attempts to cure B. cinerea strains KY-1V1 and KY-1V2 of SsPV1/WF-1 by isolating single conidia also failed. This result was quite unexpected since, as mentioned above, most conidia produced by virus-infected strains did not develop new colonies, although all isolates still carried virus. Furthermore, SsPV1/WF-1 also persists in the mycelia and sclerotia of its new hosts (S. sclerotiorum strain 1980 and B. cinerea strain KY-1) even when transfectants are successively subcultured via hyphal tip isolation or single-sclerotium isolation for more than five rounds. Therefore, SsPV1/WF-1 replicates stably in mycelia and sclerotia of S. sclerotiorum and B. cinerea.
The success in the use of hypoviruses to control chestnut blight disease continues to inspire and stimulate phytopathologists to identify mycoviruses that can be exploited for biological control of other fungal diseases. However, owing to the vegetative incompatibility of fungal pathogens and the subsequent poor transmission of mycoviruses in nature (55), there are major obstacles that must be overcome to achieve effective means of virocontrol of fungal diseases. Previously, we reported that a DNA mycovirus, SsHADV-1, of S. sclerotiorum was easily and efficiently transmitted among different vegetative compatibility groups (8). In the present study, we showed that SsPV1/WF-1 can be transmitted and spread in different vegetative compatibility groups and can also be transmitted to S. nivalis interspecifically. Thus, we believe that other mycoviruses with robust productive infection and spread do exist, regardless of the nature of their genomes (double/single-stranded RNA or ssDNA), and they may be effective agents for controlling fungal diseases, especially those that occur annually.
B. cinerea is closely related phylogenetically to S. sclerotiorum: they are both typical necrotrophic pathogens with wide host ranges and share similar ecological niches (43). Recently, B. cinerea was ranked by fungal pathologists as being the second most important pathogenic fungus based on scientific/economic importance in the journal Molecular Plant Pathology (56). Some hypovirulence-associated mycoviruses from B. cinerea and its related species Botrytis porri have been isolated and characterized (10, 54, 57), but these Botrytis viruses are unlikely to be used for control of diseases caused by these fungi under field conditions because they are easy to be eliminated via asexual propagation (see below). Expansion of the host range of mycoviruses has been accomplished for many fungus-mycovirus systems (7, 8, 24, 58–60). Here we have also shown that SsPV1/WF-1 can transfect B. cinerea protoplasts in the presence of PEG, and we further demonstrated that this virus strongly impacts its new host. Indeed, virus-infected B. cinerea showed hypovirulence and produced many inviable conidia. B. cinerea produces abundant conidia continuously during the infection season, and the prevalence of gray mold in crops is dependent mainly on conidia. Thus, SsPV1/WF-1 can inhibit the disease cycle of gray mold by affecting both plant infection and conidial germination.
The cryoreconstruction of SsPV1/WF-1 clearly shows that SsPV1/WF-1 has a capsid whose structure and arrangement of CP dimers follow the basic principles of all other fungal partitiviruses that have been characterized to date by cryo-EM or X-ray crystallography (25–27). Dimers in partitiviruses, as opposed to those in other T=2 dsRNA viruses, uniquely consist of two CP monomers in a head-to-head arrangement (20). However, in totiviruses and reoviruses, for example, the CPs adopt side-by-side arrangements. Compared to the other partitiviruses, SsPV1/WF-1 has the steepest protrusions, but the functional significance of any of the elevated structural features of partitiviruses remains a mystery, since none of these viruses exist extracellularly, nor do they require binding to a receptor to gain entry into their hosts. It is certainly possible (but unproven) that distinct features of the SsPV1/WF-1 capsid, including the steeper projections, may enable this virus to uniquely transfect fungi of different compatibility groups or interspecifically and cause hypovirulence.
In summary, we have isolated and characterized an unusual partitivirus, SsPV1/WF-1, from S. sclerotiorum and showed that this virus causes hypovirulence in its host. SsPV1/WF-1 is infectious as purified virions and can transmit and spread easily via hyphal contact among different vegetative compatibility groups of its host. It can be transmitted to S. nivalis or S. minor interspecifically through direct contact between strain WF-1 of S. sclerotiorum and strain Let-19 of S. nivalis or strain SM-1 of S. minor. SsPV1/WF-1 virions can also transfect and cause hypovirulence in B. cinerea, which, like S. sclerotiorum, is a member of the family Sclerotiniaceae. The results of this study suggest that partitiviruses significantly impact the viability of their fungal hosts and, hence, that they are potentially useful agents for controlling fungal diseases of plant crops. Finally, these studies highlight an excellent system that can provide the means to explore the mechanisms by which partitiviruses interact with their hosts.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Nature Science Foundation of China (grant 31371982), China National Funds for Distinguished Young Scientists (grant 31125023), the Special Fund for Agro-Scientific Research in the Public Interest (grant 201103016), the Kentucky Science & Engineering Foundation and the Kentucky Soybean Promotion Board (to S.A.G.), and National Institutes of Health grants R37 GM-033050 and 1S10 RR-020016 (to T.S.B.). We also acknowledge support from UCSD and the Agouron Foundation to T.S.B. for establishment and support of the UCSD cryo-TEM facilities.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 25 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01036-14.
REFERENCES
- 1.Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR, Daszak P. 2004. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 19:535–544. 10.1016/j.tree.2004.07.021 [DOI] [PubMed] [Google Scholar]
- 2.Strange RN, Scott PR. 2005. Plant disease: a threat to global food security. Annu. Rev. Phytopathol. 43:83–116. 10.1146/annurev.phyto.43.113004.133839 [DOI] [PubMed] [Google Scholar]
- 3.Ghabrial SA, Suzuki N. 2009. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47:353–384. 10.1146/annurev-phyto-080508-081932 [DOI] [PubMed] [Google Scholar]
- 4.Pearson MN, Beever RE, Boine B, Arthur K. 2009. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 10:115–128. 10.1111/j.1364-3703.2008.00503.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie J, Jiang D. New insights into mycoviruses and exploration for biological control of crop fungal diseases. Annu. Rev. Phytopathol. 10.1146/annurev-phyto-102313-050222 [DOI] [PubMed] [Google Scholar]
- 6.Nuss DL. 2005. Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3:632–642. 10.1038/nrmicro1206 [DOI] [PubMed] [Google Scholar]
- 7.Chiba S, Salaipeth L, Lin Y, Sasaki A, Kanematsu S, Suzuki N. 2009. A novel bipartite double-stranded RNA mycovirus from the white root rot fungus Rosellinia necatrix: molecular and biological characterization, taxonomic considerations, and potential for biological control. J. Virol. 83:12801–12812. 10.1128/JVI.01830-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu X, Li B, Fu Y, Jiang D, Ghabrial SA, Li G, Peng Y, Xie J, Cheng J, Huang J, Yi X. 2010. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. U. S. A. 107:8387–8392. 10.1073/pnas.0913535107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu X, Li B, Fu Y, Xie J, Cheng J, Ghabrial SA, Li G, Yi X, Jiang D. 2013. Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc. Natl. Acad. Sci. U. S. A. 110:1452–1457. 10.1073/pnas.1213755110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu M, Jin F, Zhang J, Yang L, Jiang D, Li G. 2012. Characterization of a novel bipartite double-stranded RNA mycovirus conferring hypovirulence in the phytopathogenic fungus Botrytis porri. J. Virol. 86:6605–6619. 10.1128/JVI.00292-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie J, Ghabrial SA. 2012. Molecular characterization of two mitoviruses co-infecting a hypovirulent isolate of the plant pathogenic fungus Sclerotinia sclerotiorum. Virology 428:77–85. 10.1016/j.virol.2012.03.015 [DOI] [PubMed] [Google Scholar]
- 12.Nuss DL. 2011. Mycoviruses, RNA silencing, and viral RNA recombination. Adv. Virus Res. 80:25–48. 10.1016/B978-0-12-385987-7.00002-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazmierczak P, McCabe P, Turina M, Debora JW, Van Alfen NK. 2012. The mycovirus CHV1 disrupts secretion of a developmentally regulated protein in Cryphonectria parasitica. J. Virol. 86:6067–6074. 10.1128/JVI.05756-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi GH, Dawe AL, Churbanov A, Smith ML, Milgroom MG, Nuss DL. 2012. Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics 190:113–127. 10.1534/genetics.111.133983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nibert ML, Woods KM, Upton SJ, Ghabrial SA. 2009. Cryspovirus: a new genus of protozoan viruses in the family Partitiviridae. Arch. Virol. 154:1959–1965. 10.1007/s00705-009-0513-7 [DOI] [PubMed] [Google Scholar]
- 16.Ghabrial SA, Nibert ML, Maiss E, Lesker T, Baker TS, Tao YJ. 2012. Partitiviridae, p 523–534 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 17.Ghabrial SA. 1998. Origin, adaptation and evolutionary pathways of mycoviruses. Virus Genes 16:119–131. 10.1023/A:1007966229595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Fu Y, Jiang D, Li G, Xie J, Cheng J, Peng Y, Ghabrial SA, Yi X. 2010. Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J. Virol. 84:11876–11887. 10.1128/JVI.00955-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiba S, Kondo H, Tani A, Saisho D, Sakamoto W, Kanematsu S, Suzuki N. 2011. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS Pathog. 7:e1002146. 10.1371/journal.ppat.1002146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nibert ML, Tang J, Xie J, Collier AM, Ghabrial SA, Baker TS, Tao YJ. 2013. 3D structures of fungal partitiviruses. Adv. Virus Res. 86:59–85. 10.1016/B978-0-12-394315-6.00003-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Fu Y, Xie J, Cheng J, Ghabrial SA, Li G, Peng Y, Yi X, Jiang D. 2012. Evolutionary genomics of mycovirus-related dsRNA viruses reveals cross-family horizontal gene transfer and evolution of diverse viral lineages. BMC Evol. Biol. 12:91. 10.1186/1471-2148-12-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nibert ML, Ghabrial SA, Maiss E, Lesker T, Vainio EJ, Jiang D, Suzuki N. 2014. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res. 188C:128–141. 10.1016/j.virusres.2014.04.007 [DOI] [PubMed] [Google Scholar]
- 23.Sasaki A, Kanematsu S, Onoue M, Oyama Y, Yoshida K. 2006. Infection of Rosellinia necatrix with purified viral particles of a member of Partitiviridae (RnPV1-W8). Arch. Virol. 151:697–707. 10.1007/s00705-005-0662-2 [DOI] [PubMed] [Google Scholar]
- 24.Kanematsu S, Sasaki A, Onoue M, Oikawa Y, Ito T. 2010. Extending the fungal host range of a partitivirus and a mycoreovirus from Rosellinia necatrix by inoculation of protoplasts with virus particles. Phytopathology 100:922–930. 10.1094/PHYTO-100-9-0922 [DOI] [PubMed] [Google Scholar]
- 25.Ochoa WF, Havens WM, Sinkovits RS, Nibert ML, Ghabrial SA, Baker TS. 2008. Partitivirus structure reveals a 120-subunit, helix-rich capsid with distinctive surface arches formed by quasisymmetric coat-protein dimers. Structure 16:776–786. 10.1016/j.str.2008.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan J, Dong L, Lin L, Ochoa WF, Sinkovits RS, Havens WM, Nibert ML, Baker TS, Ghabrial SA, Tao YJ. 2009. Atomic structure reveals the unique capsid organization of a dsRNA virus. Proc. Natl. Acad. Sci. U. S. A. 106:4225–4230. 10.1073/pnas.0812071106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang J, Ochoa WF, Li H, Havens WM, Nibert ML, Ghabrial SA, Baker TS. 2010. Structure of Fusarium poae virus 1 shows conserved and variable elements of partitivirus capsids and evolutionary relationships to picobirnavirus. J. Struct. Biol. 172:363–371. 10.1016/j.jsb.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boland GJ, Hall R. 1994. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 16:93–108. 10.1080/07060669409500766 [DOI] [Google Scholar]
- 29.Bolton MD, Thomma BP, Nelson BD. 2006. Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7:1–16. 10.1111/j.1364-3703.2005.00316.x [DOI] [PubMed] [Google Scholar]
- 30.Boland GJ. 1992. Hypovirulence and double-stranded RNA in Sclerotinia sclerotiorum. Can. J. Plant Pathol. 14:10–17. 10.1080/07060669209500900 [DOI] [Google Scholar]
- 31.Jiang D, Fu Y, Li G, Ghabrial SA. 2013. Viruses of the plant pathogenic fungus Sclerotinia sclerotiorum. Adv. Virus Res. 86:215–248. 10.1016/B978-0-12-394315-6.00008-8 [DOI] [PubMed] [Google Scholar]
- 32.Khalifa ME, Pearson MN. 2013. Molecular characterization of three mitoviruses co-infecting a hypovirulent isolate of Sclerotinia sclerotiorum fungus. Virology 441:22–30. 10.1016/j.virol.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 33.Li G, Wang D, Jiang D, Huang HC, Laroche A. 2000. First report of Sclerotinia nivalis on lettuce in central China. Mycol. Res. 104:232–237 10.1017/S0953756299001045 [DOI] [Google Scholar]
- 34.Zhang L, Fu Y, Xie J, Jiang D, Li G, Yi X. 2009. A novel virus that infecting hypovirulent strain XG36-1 of plant fungal pathogen Sclerotinia sclerotiorum. Virol. J. 6:96. 10.1186/1743-422X-6-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie J, Wei D, Jiang D, Fu Y, Li G, Ghabrial S, Peng Y. 2006. Characterization of debilitation-associated mycovirus infecting the plant-pathogenic fungus Sclerotinia sclerotiorum. J. Gen. Virol. 87:241–249. 10.1099/vir.0.81522-0 [DOI] [PubMed] [Google Scholar]
- 36.Jiang D, Ghabrial SA. 2004. Molecular characterization of Penicillium chrysogenum virus: reconsideration of the taxonomy of the genus Chrysovirus. J. Gen. Virol. 85:2111–2121. 10.1099/vir.0.79842-0 [DOI] [PubMed] [Google Scholar]
- 37.Xie J, Xiao X, Fu Y, Liu H, Cheng J, Ghabrial SA, Li G, Jiang D. 2011. A novel mycovirus closely related to hypoviruses that infects the plant pathogenic fungus Sclerotinia sclerotiorum. Virology 418:49–56. 10.1016/j.virol.2011.07.008 [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299–306. 10.1093/bib/bbn017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan X, Sinkovits RS, Baker TS. 2007. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 157:73–82. 10.1016/j.jsb.2006.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan X, Dryden KA, Tang J, Baker TS. 2007. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J. Struct. Biol. 157:211–225. 10.1016/j.jsb.2006.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallie DR, Walbot V. 1992. Identification of the motifs within the tobacco mosaic virus 5′-leader responsible for enhancing translation. Nucleic Acids Res. 20:4631–4638. 10.1093/nar/20.17.4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strauss EE, Lakshman DK, Tavantzis SM. 2000. Molecular characterization of the genome of a partitivirus from the basidiomycete Rhizoctonia solani. J. Gen. Virol. 81:549–555 [DOI] [PubMed] [Google Scholar]
- 43.Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, Fournier E, Gout L, Hahn M, Kohn L, Lapalu N, Plummer KM, Pradier JM, Quévillon E, Sharon A, Simon A, ten Have A, Tudzynski B, Tudzynski P, Wincker P, Andrew M, Anthouard V, Beever RE, Beffa R, Benoit I, Bouzid O, Brault B, Chen Z, Choquer M, Collémare J, Cotton P, Danchin EG, Da Silva C, Gautier A, Giraud C, Giraud T, Gonzalez C, Grossetete S, Güldener U, Henrissat B, Howlett BJ, Kodira C, Kretschmer M, Lappartient A, Leroch M, Levis C, et al. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230. 10.1371/journal.pgen.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magae Y, Sunagawa M. 2010. Characterization of a mycovirus associated with the brown discoloration of edible mushroom, Flammulina velutipes. Virol. J. 7:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhatti MF, Jamal A, Petrou MA, Cairns TC, Bignell EM, Coutts RH. 2011. The effects of dsRNA mycoviruses on growth and murine virulence of Aspergillus fumigatus. Fungal Genet. Biol. 48:1071–1075. 10.1016/j.fgb.2011.07.008 [DOI] [PubMed] [Google Scholar]
- 46.Zheng L, Zhang M, Chen Q, Zhu M, Zhou E. 2014. A novel mycovirus closely related to viruses in the genus Alphapartitivirus confers hypovirulence in the phytopathogenic fungus Rhizoctonia solani. Virology 456–457:220–226. 10.1016/j.virol.2014.03.029 [DOI] [PubMed] [Google Scholar]
- 47.Chiba S, Lin YH, Kondo H, Kanematsu S, Suzuki N. 2013. Effects of defective interfering RNA on symptom induction by, and replication of, a novel partitivirus from a phytopathogenic fungus, Rosellinia necatrix. J. Virol. 87:2330–2341. 10.1128/JVI.02835-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anagnostakis SL. 1982. Biological control of chestnut blight. Science 215:466–471. 10.1126/science.215.4532.466 [DOI] [PubMed] [Google Scholar]
- 49.Brasier CM. 1983. A cytoplasmically transmitted disease of Ceratocystis ulmi. Nature 305:220–223. 10.1038/305220a0 [DOI] [Google Scholar]
- 50.Chu YM, Jeon JJ, Yea SJ, Kim YH, Yun SH, Lee YW, Kim KH. 2002. Double-stranded RNA mycovirus from Fusarium graminearum. Appl. Environ. Microbiol. 68:2529–2534. 10.1128/AEM.68.5.2529-2534.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi GH, Nuss DL. 1992. A viral gene confers hypovirulence-associated traits to the chestnut blight fungus. EMBO J. 11:473–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smit WA, Wingfield BD, Wingfield MJ. 1996. Reduction of laccase activity and other hypovirulence-associated traits in dsRNA-containing strains of Diaporthe ambigua. Phytopathology 86:1311–1316. 10.1094/Phyto-86-1311 [DOI] [Google Scholar]
- 53.Castro M, Kramer K, Valdivia L, Ortiz S, Castillo A. 2003. A double-stranded RNA mycovirus confers hypovirulence-associated traits to Botrytis cinerea. FEMS Microbiol. Lett. 228:87–91. 10.1016/S0378-1097(03)00755-9 [DOI] [PubMed] [Google Scholar]
- 54.Wu M, Zhang L, Li G, Jiang D, Ghabrial SA. 2010. Genome characterization of a debilitation-associated mitovirus infecting the phytopathogenic fungus Botrytis cinerea. Virology 406:117–126. 10.1016/j.virol.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 55.Milgroom MG, Cortesi P. 2004. Biological control of chestnut blight with hypovirulence: a critical analysis. Annu. Rev. Phytopathol. 42:311–338. 10.1146/annurev.phyto.42.040803.140325 [DOI] [PubMed] [Google Scholar]
- 56.Dean R, van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD. 2012. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13:414–430. 10.1111/j.1364-3703.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearson MN, Bailey AM. 2013. Viruses of Botrytis. Adv. Virus Res. 86:249–272. 10.1016/B978-0-12-394315-6.00009-X [DOI] [PubMed] [Google Scholar]
- 58.Sasaki A, Onoue M, Kanematsu S, Suzaki K, Miyanishi M, Suzuki N, Nuss DL, Yoshida K. 2002. Extending chestnut blight hypovirus host range within Diaporthales by biolistic delivery of viral cDNA. Mol. Plant Microbe Interact. 15:780–789. 10.1094/MPMI.2002.15.8.780 [DOI] [PubMed] [Google Scholar]
- 59.Hillman BI, Supyani S, Kondo H, Suzuki N. 2004. A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the Coltivirus genus of animal pathogens. J. Virol. 78:892–898. 10.1128/JVI.78.2.892-898.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee KM, Yu J, Son M, Lee YW, Kim KH. 2011. Transmission of Fusarium boothii mycovirus via protoplast fusion causes hypovirulence in other phytopathogenic fungi. PLoS One 6:e21629. 10.1371/journal.pone.0021629 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.