ABSTRACT
Enterovirus 71 (EV71) causes hand, foot, and mouth disease in young children and infants. Severe infection with EV71 can lead to various neurological complications or fatal diseases. However, the mechanism of EV71 pathogenesis is poorly understood. Emerging evidence suggests that EV71 modulates type I interferon (IFN) and cytokine responses. Here, we show that EV71 disables components of the TAB2 complex through the 3C protein. When expressed in mammalian cells, EV71 3C interacts with TAB2 and TAK1, which inhibits NF-κB activation. Furthermore, 3C mediates cleavage of TAB2 and its partners, which requires the protease activity. H40D or C147S substitution in the 3C active sites abolishes its activity, whereas R84Q or V154S substitution in the RNA binding domain has no effect. The 3C protein targets TAB2 at Q113-S114, TAK1 at Q360-S361, TAB1 both at Q414-G415 and Q451-S452, and TAB3 at Q173-G174 and Q343-G344. Importantly, overexpression of TAB2 inhibits EV71 replication, whereas addition of cleaved fragments has no effect. Thus, an equilibrium between the TAB2 complex and EV71 3C represents a control point of viral infection. These results suggest that TAK1/TAB1/TAB2/TAB3 cleavage mediated by EV71 may be a mechanism to interfere with inflammatory responses.
IMPORTANCE The TAK1 complex plays a critical role in the activation of NF-κB and cytokine production. However, little is known about its connection to enterovirus 71 (EV71). We demonstrate that EV71 3C suppresses cytokine expression via cleavage of the TAK1 complex proteins. EV71 3C interacts with TAB2 and TAK1. Furthermore, overexpression of TAB2 inhibits EV71 replication, whereas addition of cleaved fragment has no effect. These results suggest that the interplay of EV71 and the TAK1 complex influences the outcome of viral infection.
INTRODUCTION
Enterovirus 71 (EV71) is a causative agent of hand, foot, and mouth disease (HFMD) in young children and infants. Severe infection with EV71 can lead to various neurological complications or even fatal diseases (1). To date, there is no effective treatment for EV71 infection. EV71 is a member of the Picornaviridae family with a single, positive-stranded RNA genome which encodes a single polyprotein precursor. This precursor is proteolytically cleaved to four structural and seven nonstructural proteins during virus infection (1). The nonstructural protein 3C is essential for the precursor cleavage and viral replication (2–4). It also possesses RNA-binding activity (3). Recent reports show that EV71 3C inhibits type I interferon (IFN) responses by targeting innate immune factors, such as RIG-I, TRIF, interferon regulatory factor 7 (IRF7), and IRF9 (5–8).
Transforming growth factor-β-activated kinase 1 (TAK1) is a member of the mitogen-activated protein kinase kinase kinase (MAP3K) family, which is activated by various stimuli, including Toll-like receptor (TLR) ligands, tumor necrosis factor alpha (TNF-α), and interleukin-1β (IL-1β) (9, 10). In mammalian cells, TAK1 constitutively binds to the TAK1 binding protein 1 (TAB1), which is necessary for its activation (11). In this process, TAB2 and TAB3 bind to TAK1-TAB1, forming a TAK1/TAB1/TAB2/TAB3 complex which then activates IKKαβγ, p38, and c-Jun N-terminal kinase (JNK) (12–18). TAK1, TAB1, TAB2, and TAB3 are all essential for downstream NF-κB activation. In the TNF-α pathway, TAB2 and TAB3 facilitate recruitment of TAK1 to adaptor proteins, including TRADD, TRAF2/5, and RIP (19, 20). In the IL-1β pathway, TAB2 and TAB3 facilitate recruitment of TAK1 to adaptor proteins, including TRAF6 and IRAK4 (21). This leads to the activation of TAK1 and ultimately the downstream IKK complex and NF-κB. Although the TAK1 complex activation has been studied extensively, its connection to EV71 remains elusive.
Here, we report that EV71 inhibits NF-κB activation by targeting the TAK1/TAB1/TAB2/TAB3 complex. This involves virally induced cleavage of components associated with this complex. We provide evidence that the 3C protease mediates cleavage of the four proteins of the TAK1/TAB1/TAB2/TAB3 complex, which inhibits NF-κB activation. Furthermore, we show that overexpression of TAB2 inhibits EV71 replication, but the cleaved fragments are incapable of inhibiting virus replication. These results suggest that the interaction of EV71 3C and the TAK1 complex may alter the outcome of EV71 infection.
MATERIALS AND METHODS
Cell lines and viruses.
RD cells, 293T cells, and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% heated-inactivated fetal bovine serum (FBS) (HyClone, Logan, UT), 100 U/ml penicillin, and 100 μg/ml streptomycin. All the cells were cultured at 37°C in a 5% CO2 humidified atmosphere. Enterovirus 71 infection was carried out as described previously (8).
Plasmids.
The plasmids pEGFP (where EGFP is enhanced green fluorescent protein), pEGFP-3C, pEGFP-3C variants, and pCDNA3.1-3C have been described elsewhere (8). The plasmids expressing TAK1, TAB2, and TAB3 were purchased from Origene (Rockville MD). The TAK1, TAB1, TAB2, or TAB3 variants were constructed by site-directed mutagenesis using Pfu DNA polymerase (Strategene, La Jolla, CA). These variants include amino acid substitution mutants Q337, Q356, Q359, and Q360 of TAK1, Q48, Q83, Q113, and Q116 of TAB2, Q328, Q414, Q437, Q444, and Q451 of TAB1, and Q116, Q173, Q237, Q246, Q251, Q255, Q335, and Q343 of TAB3. All variants were verified by nucleotide sequence analysis.
Antibodies and reagents.
Antibodies against Flag, GFP, and β-actin were purchased from Sigma (St. Louis, MO). Anti-TAK1, anti-TAB1, and anti-TAB2 antibodies were purchased from NOVUS (Charles, MO). Anti-TAB3 antibody was purchased from Abcam (Cambridge, United Kingdom). Mouse anti-enterovirus 71 was purchased from Chemicon (Billerica, MA). Goat-anti mouse or rabbit secondary antibodies were purchased from Li-Cor (Li-Cor, Inc., Lincoln, NE). The general caspase inhibitor Z-VAD-FMK (carboxybenzyl-Val-Ala-Asp-fluoromethylketone) was purchased from Sigma (St. Louis, MO). Rupintrivir was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Luciferase reporter assays.
This assay was performed as described previously (22). Briefly, 293T cells were seeded on 24-well plates at a cell density of 3 × 105 cells per well. The following day, cells were transfected with a control plasmid or plasmid expressing TAK1, TAB1, TAB2, and TAB3, along with 200 ng of PGL3-NF-κB-Luc (where Luc is luciferase) and 5 ng of pRL-SV40 (where SV40 is simian virus 40) using Lipofectamine 2000 (Invitrogen), as indicated in the figure legends. The total amount of DNA was kept constant by adding empty control plasmid. At 24 h after transfection, cells were lysed, and luciferase activities were analyzed using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to the manufacturer's instructions.
Reverse transcription-PCR.
Total RNA was extracted from cells by using TRIzol reagent (Invitrogen, Carlsbad, CA). RNA samples were treated with DNase I (Pierce, Rockford, IL), and reverse transcription was carried out using a Superscript cDNA synthesis kit (Invitrogen) according to the manufacturer's instructions. cDNA samples were subjected to real-time PCR by using SYBR green. The primers used were the following: human IL-6, GCCCTGAGAAAGGAGACAT and CTGTTCTGGAGGTAC; IL-8, TTTGAAGAGGGCTGAGAA and TGTTCTGGATATTTCATGG; IL-12, TGCTCCAGAAGGCCAGAC and TTCATAAATACTACTAAGGCACAGG; IL-1β ACAGATGAAGTGCTCCTTCCA and GTCGGAGATTCGTAGCTGGAT; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), CGGAGTCAACGGATTTGGTCGTA and AGCCTTCTCCATGGTGGTGAAGAC. Levels of gene mRNAs were normalized to GAPDH mRNA. Results are reported as fold change using the ΔΔCT (where CT is threshold cycle) method.
Western blot analysis.
Cells were washed and lysed in buffer containing 150 mM NaCl, 25 mM Tris (pH 7.4), 1% NP-40, 0.25% sodium deoxycholate, and 1 mM EDTA with protease inhibitor cocktail (Roche, Indianapolis, IN). The lysates were centrifuged at 16,000 × g for 10 min at 4°C. Aliquots of cell lysates were electrophoresed on 12% SDS-PAGE gels and were transferred to a nitrocellulose membrane (Pall, Port Washington, NY). The membranes were blocked with 5% nonfat dry milk, and then proteins on the membrane were incubated with primary antibodies at 4°C overnight as indicated in the figure legends. This was followed by incubation with corresponding IRD Fluor 800-labeled IgG or IRD Fluor 680-labeled IgG secondary antibody (Li-Cor, Inc., Lincoln, NE) for 1 h at room temperature. After the membranes were washed, they were scanned with the Odyssey Infrared Imaging System (Li-Cor, Lincoln, NE) at a wavelength of 700 to 800 nm, and the molecular sizes of the developed proteins were determined by comparison with prestained protein markers (Ferments, MD).
Immunoprecipitation.
Transfected cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl buffer [pH 7.4] containing 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate) containing protease inhibitor cocktail (Roche, Indianapolis, IN). Lysates of cells were incubated with anti-Flag antibody (Sigma, St. Louis, MO) in 500 μl of RIPA buffer at 4°C overnight on a rotator in the presence of protein A/G-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA). Immunocomplexes captured on the protein A/G-agarose were fractionated by 10 to 12% SDS-PAGE and transferred to nitrocellulose membranes for analysis.
Transfection and immunofluorescence.
Plasmids expressing TAK1, TAB1, TAB2, TAB3, and IRF3 were added to 96-well plates. Then these plasmids were mixed with Lipofectamine 2000. After 30 min, RD cells were plated in every well. Empty plasmid was used as a control. After 48 h of transfection, cells were mock infected or infected with EV71 at a multiplicity of infection (MOI) of 0.2 PFU per cell. After 24 h, the medium was removed, and the cells were then fixed with 4% formalin. After cells were washed with phosphate-buffered saline (PBS), they were blocked and stained with anti-EV71 antibody (MAB979; Millipore), followed by an Alexa Fluor 488 goat anti-mouse secondary antibody at 1:500. The cells were imaged on an Operetta instrument (PerkinElmer), and the fluorescence intensity or images were analyzed using Harmony software (PerkinElmer).
Statistics.
A Student t test was used for two-group comparisons. P values of <0.05, <0.01, and <0.001 were considered significant, as indicated in the figure legends.
RESULTS
The expression levels of TAB2, TAK1, TAB1, and TAB3 are reduced in EV71-infected cells.
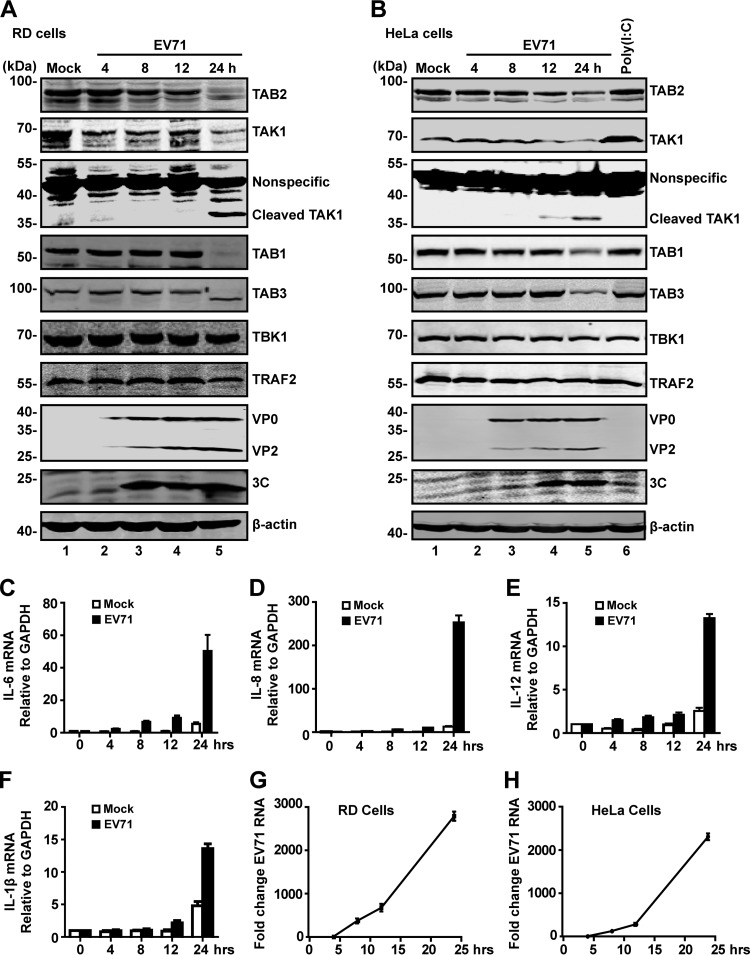
The TAK1 complex mediates cytokine expression in response to danger signals. To investigate the impact of EV71, we analyzed the expression of TAB2, TAK1, TAB1, and TAB3. RD and HeLa cells were mock infected or infected with EV71. At different time points postinfection, cell lysates were processed for Western blot analysis. As shown in Fig. 1A, TAB2, TAK1, TAB1, and TAB3 were expressed in mock-infected RD cells (lane 1). EV71 infection induced degradation or cleavage of these proteins with a different kinetics (Fig. 1A, lanes 2 to 5). The cleaved TAK1 fragment appeared as a 35-kDa band in both EV71-infected RD and HeLa cells (Fig. 1A and B). Western blot analysis verified the expression of 3C, VP0, and VP2 (lanes 2 to 5). A similar pattern was also observed in HeLa cells (Fig. 1B). TAK1 complex reduction paralleled late expression of IL-6, IL-8, IL-12, and IL-1β in EV71-infected cells (Fig. 1C, D, E, and F) as viral RNA replication progressed (Fig. 1G and H). These results show that EV71 reduces the expression of TAB2, TAK1, TAB1, and TAB3 in infected cells, but poly(I·C) has no effects on the expression of these proteins.
FIG 1.
EV71 infection induces cleavage of TAB2, TAK1, TAB1, and TAB3. RD (A) or HeLa (B) cells were mock infected or infected with EV71 (MOI of 1 PFU per cell for RD; MOI of 2 PFU per cell for HeLa). HeLa cells were treated with poly(I·C) for 4 h, which was added to the medium. At 4, 8, 12, and 24 h postinfection, cell lysates were prepared and analyzed by Western blotting with antibodies for TAB2, TAK1, TAB1, TAB3, TBK1, TRAF2, EV71, EV71-3C, and β-actin. Data are representative of three independent experiments. (C to F) HeLa cells were mock infected or infected with EV71. At 24 h after infection, total RNA extracted for cells was analyzed for the expression of IL-6 (C), IL-8 (D), IL-12 (E), and IL-1β (F) by quantitative real-time PCR using SYBR green. (G and H) RD cells or HeLa cells were infected with EV71. At different time points after infection, total RNA was extracted, and the viral RNA levels of EV71 were evaluated by quantitative real-time PCR using SYBR green. Data are expressed as fold change of the EV71 RNA level relative to that at the 4-h time point using the ΔΔCT method as described in Materials and Methods.
EV71 3C cleaves TAB2, TAK1, TAB1, and TAB3 in mammalian cells and inhibits their function.
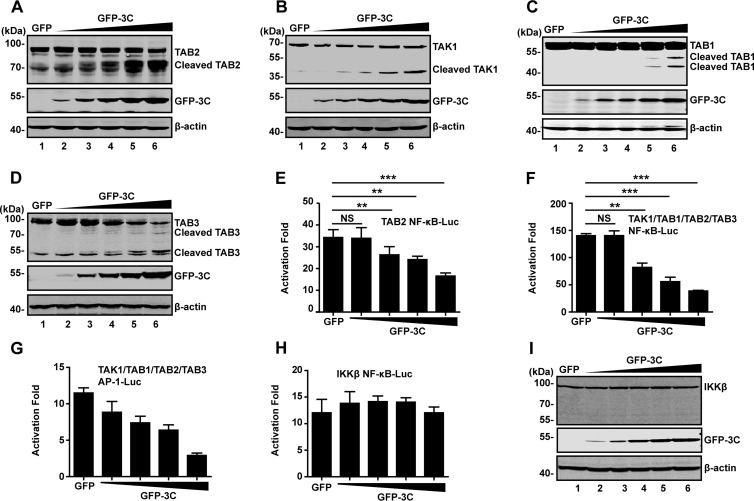
EV71 3C can affect functions of host factors via proteolytic cleavage (6, 7, 23). To explore whether EV71 3C targets the TAK1 components, we carried out cleavage assays. 293T cells were transfected with increasing amounts of GFP-3C plasmid, along with a plasmid expressing TAB2, TAK1, TAB1, or TAB3. At 24 h after transfection, cell lysates were processed to assess 3C-mediated cleavage by Western blotting. As shown in Fig. 2A, GFP-3C reduced TAB2 expression in a dose-dependent manner compared to the GFP control; this paralleled the appearance of a smaller protein band (65 kDa) (Fig. 2A, lanes 3 to 6). Next, we determined the effect of 3C on TAK1, TAB1, and TAB3. As shown in Fig. 2B, C and D, TAK1, TAB1, and TAB3 were cleaved by GFP-3C but not GFP. TAK1 cleavage resulted in a smaller band of 30 kDa. TAB1 cleavage led to cleaved bands of 45 kDa and 50 kDa. Moreover, TAB3 cleavage produced products of 45 kDa and 60 kDa. However, ectopic expression of GFP-3C had no effect on TRAF2 or TRAF6 (data not shown).
FIG 2.
The 3C protease of EV71 mediates cleavage of TAK1 complex components. (A to D) 293T cells were transfected with plasmids encoding TAB2 (A), TAK1 (B), TAB1(C), or TAB3 (D), along with GFP (lane 1) or increasing amounts of GFP-3C (lanes 2 to 6). At 24 h after transfection, lysates of cells were analyzed by Western blotting with antibodies against Flag, GFP, Myc, or β-actin. β-Actin was used as a protein loading control. (E and F) EV71 3C inhibits NF-κB promoter activation. 293T cells were transfected with TAB2 alone or a mixture of TAK1, TAB1, TAB2, and TAB3, along with NF-κB-Luc and increasing amounts of GFP-3C. A plasmid expressing GFP or pRL-SV40 was used as a control. At 24 h after transfection, cell lysates were assayed for luciferase activities. (G) EV71 3C inhibits AP-1 promoter activation. 293T cells were transfected with TAK1, TAB1, TAB2 and TAB3, along with AP-1-Luc and increasing amounts of GFP-3C. Data are a representative of three independent experiments with triplicate samples. (H) 293T cells were transfected with plasmids encoding IKKβ, along with NF-κB-Luc and increasing amounts of GFP-3C plasmid. At 24 h after transfection, cell lysates were assayed for luciferase activities. (I) Cell lysates from panel H were analyzed by Western blotting with antibodies against Flag, GFP, or β-actin, respectively. **, P < 0.01; ***, P < 0.001; NS, nonsignificant.
As the TAK1 complex activates NF-κB, we tested whether 3C-mediated cleavage has a functional role. As illustrated in reporter assays, TAB2 alone activated the NF-κB promoter by approximately 35-fold (Fig. 2E). Addition of EV71 3C reduced the TAB2 activity. Further analysis showed that coexpression of TAK1, TAB1, TAB2, and TAB3 robustly mediated NF-κB promoter activation. However, when ectopically expressed, EV71 3C also suppressed NF-κB activation (Fig. 2F). Similar phenotypes were observed for AP-1 promoter activation (Fig. 2G). Picornavirus 3C is known to contribute to virally induced host transcription and translation shutoff by cleavage of host factors including TATA-binding protein (TBP), CREB, and poly(A)-binding protein (PABP) (24–27). To test whether the observed effects were due to general inhibition of transcription and translation by EV71 3C, we evaluated IKKβ-stimulated NF-κB activation. As shown in Fig. 2H, overexpression of EV71 3C did not inhibit NF-κB activation where the 3C protein failed to cleave the IKKβ (Fig. 2I). Taken in combination, these results suggest that EV71 3C targets the TAK1 complex and negatively modulates NF-κB activation.
The protease activity of 3C is required for cleavage of the TAK1 components.
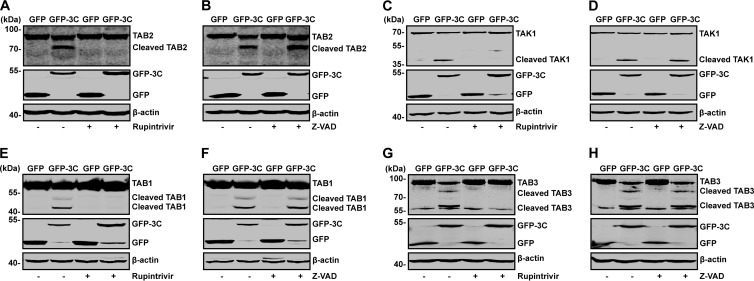
To investigate whether the protease activity of EV71 3C is necessary for the cleavage of TAK1 components, we evaluated the impact of rupintrivir, which is an inhibitor against 3C protease. As indicated by the results shown in Fig. 3A, GFP-3C mediated TAB2 cleavage, resulting in a 65-kDa protein band (lane 2). This was blocked by rupintrivir (lane 4). However, the caspase inhibitor Z-VAD-FMK had no effects (Fig. 3B, lane 4,). Similarly, rupintrivir blocked GFP-3C-mediated cleavage of TAK1 (Fig. 3C), TAB1, and TAB3 (Fig. 3E and G). In contrast, the caspase inhibitor Z-VAD-FMK had virtually no effect (Fig. 3D, F, and H). Neither the lysosome inhibitor NH4Cl nor the proteasome inhibitor MG132 blocked the cleavage of TAK1 components by 3C (data not shown).
FIG 3.
Effects of selected inhibitors on cleavage of TAB2, TAK1, TAB1, and TAB3. 293T cells were transfected with plasmids encoding TAB2 (A and B), TAK1 (C and D), TAB1 (E and F), and TAB3 (G and H), along with plasmids encoding GFP or GFP-3C. At 4 h after transfection, cells were incubated with the protease inhibitor rupintrivir (A, C, E, and G) (2 nM) or caspase inhibitor Z-VAD (B, D, F, and H) (20 μM). At 24 h after transfection, cell lysates were then processed for Western blot analysis with antibodies specific for Flag, GFP, or Myc.
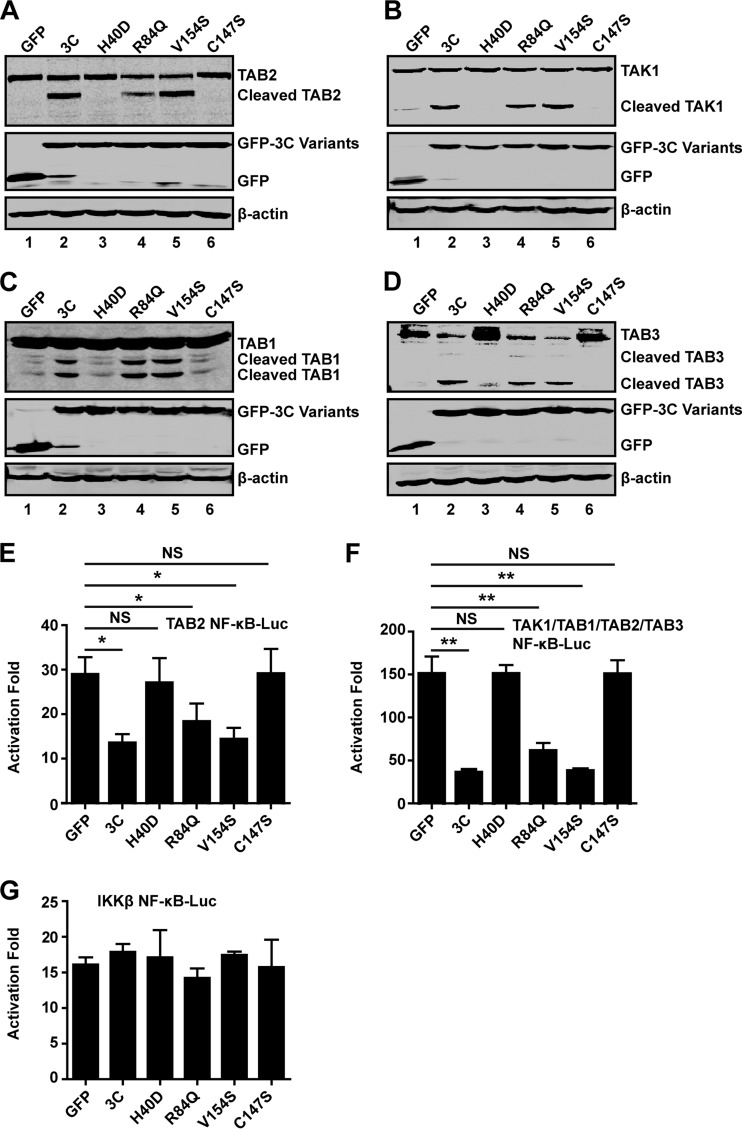
To further address this issue, we carried out a mutational analysis. H40, C147, and E71 are essential components of the catalytic triad of EV71 3C (28, 29). H40D or C147S substitution in the active site of EV71 3C disrupts the protease activity. As illustrated in Fig. 4A, when expressed in 293T cells, wild-type 3C, but not the GFP control, induced TAB2 cleavage (lanes 1 and 2). In contrast, H40D or C147S failed to mediate TAB2 cleavage (lanes 3 and 6). R84Q or V154S, which abolishes the RNA binding activity of 3C, still induced cleavage of TAB2 (lanes 4 and 5). These mutants were expressed at comparable levels as measured by Western blotting (Fig. 4A, middle panel, lanes 2 to 6). Similar results were observed on cleavage of TAK1, TAB1, or TAB3 (Fig. 4B, C, and D). Thus, the protease activity of EV71 3C is essential for the TAK1 complex cleavage.
FIG 4.
Mutational effects on EV71 3C activities. 293T cells were transfected with plasmids encoding TAB2(A), TAK1 (B), TAB1 (C), or TAB3 (D), along with GFP (lane 1) or GFP-3C variants (lanes 2 to 6), as indicated. At 24 h after transfection, cell lysates were analyzed by Western blotting using a Li-Cor Odyssey Dual-Color System (Li-Cor, Lincoln, NE). (E, F, and G) 293T cells were transfected with the TAB2 (E), TAK1 complex (F), or IKKβ (G), along with NF-κB-Luc and GFP-3C variants. A plasmid expressing GFP or pRL-SV40 was used as a control. At 24 h after transfection, cell lysates were assayed for luciferase activities. Data are representative of three independent experiments with triplicate samples. *, P < 0.05; **, P < 0.01; NS, nonsignificant.
We next tested the effect of 3C variants on NF-κB activation by TAB2 or the TAK1 complex in reporter assays. As shown in Fig. 4E and F, wild-type 3C, R84Q, or V154S inhibited the NF-κB promoter activation mediated by TAB2 alone or TAK1 complex. When the TAB2 or TAK1 complex was coexpressed with 3C, NF-κB activation was decreased. Nevertheless, mutant H40D or C147S did not inhibit the NF-κB promoter activation induced by the TAB2 or TAK1 complex. Additionally, none of the 3C variants inhibited IKKβ-induced NF-κB promoter activation (Fig. 4G). These results suggest that the protease activity of 3C is essential to inhibit TAK1 complex-induced NF-κB activation.
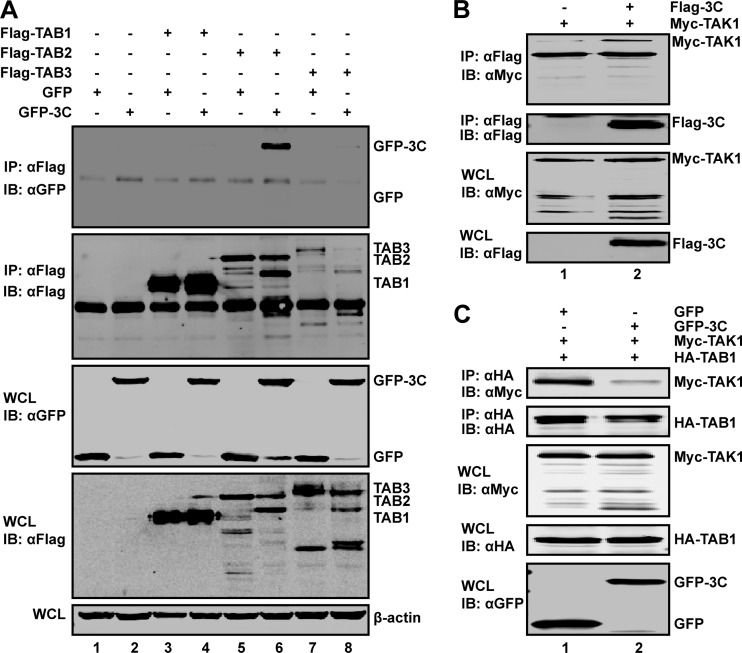
The 3C protease associates with TAB2 or TAK1.
To explore whether the 3C protease interacts with the TAK1 components, we performed immunoprecipitation analysis. Specifically, 293T cells were transiently transfected with GFP-tagged 3C along with Flag-TAB1, -TAB2, or -TAB3. Cell extracts were immunoprecipitated with anti-Flag antibody, followed by Western blotting with anti-GFP antibody. As shown in Fig. 5A, TAB2 associated with GFP-3C (Fig. 5A, lane 6) but not with GFP (Fig. 5A, lane 5). However, TAB1 or TAB3 did not interact with GFP-3C (Fig. 5A, lane 4 or lane 8). Further analysis revealed that TAK1 associated with 3C (Fig. 5B, lane 2). Importantly, ectopic expression of 3C disrupted the interaction between TAK1 and TAB1 (Fig. 5C), suggesting that EV71 3C alters formation of the TAK1 complex. Collectively, these data indicate that the 3C protein associates with TAB2 and TAK1.
FIG 5.
(A) EV71 3C associates with TAB2. 293T cells were transfected with plasmids encoding TAB1 (lanes 3 and 4), TAB2 (lanes 5 and 6), TAB3 (lanes 7 and 8), GFP (lanes 1, 3, 5, and 7), and GFP-3C (lanes 2, 4, 6, and 8). Empty pCMV6 (lanes 1 and 2) vector was used as a control. At 24 h after transfection, cell lysates were immunoprecipitated with antibody against Flag. Samples were then subjected to Western blot analysis. (B) EV71 3C interacts with TAK1. 293T cells were transfected with plasmids encoding Myc-TAK1 (lanes 1 and 2) and Flag-3C (lane 2). The total amount of DNA was kept constant using empty pCDNA3.1 (lane 1). At 24 h after transfection, cell lysates were immunoprecipitated with Flag antibody. Immunoprecipitates and cell lysates were then subjected to Western blot analysis. (C) EV71 3C inhibits the interaction between TAK1 and TAB1. 293T cells were transfected with plasmids encoding Myc-TAK1 (lanes 1 and 2), hemagglutinin (HA)-TAB1 (lanes 1 and 2), and GFP-3C (lane 2). At 24 h after transfection, cell lysates were immunoprecipitated with HA antibody. Immunoprecipitates and cell lysates were then subjected to Western blot analysis. IB, immunoblotting; IP, immunoprecipitation; WCL, whole-cell lysate; α, anti.
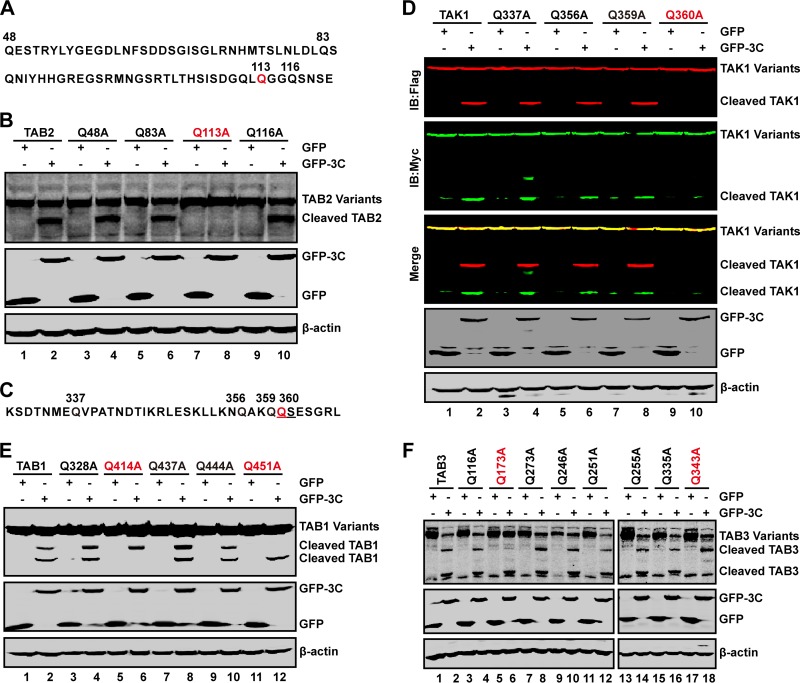
The cleavage site(s) of 3C is localized in TAB2 and its partners.
In order to identify the potential 3C cleavage site(s) within TAB2 and its partners, we carried out mutational analysis. As TAB2 cleavage produced 65-kDa protein species, we inferred that a cleavage site may exist between amino acids 48 and 120 (Fig. 6A). This region has several glutamines which resemble the Q-G/Q-S sequence of proteolytic sites for enterovirus 3C. To test this, we analyzed a series of TAB2 mutants. These mutants were expressed alone with a control GFP or GFP-3C in 293T cells. Cell lysates were subjected to Western blot analysis. As shown in Fig. 6B, wild-type TAB2 was cleaved by GFP-3C, resulting in a 65-kDa band (lane 2). Similarly, Q48A, Q83A, and Q116A substitution mutants were cleaved by GFP-3C (lanes 4, 6, and 10). However, Q113A was resistant to cleavage (lane 8), suggesting that the Q113-G114 pair is a cleavage site of 3C within TAB2.
FIG 6.
The cleavage sites of 3C in TAK1 complex. (A) Primary sequences of amino acids 48 to 120 within TAB2. In this region, glutamines of 48, 83, 113, and 116 sites were replaced with alanines. (B) The 3C cleavage site in TAB2. 293T cells were transfected with plasmids encoding wild-type TAB2 or TAB2 variants as indicated along with GFP (lanes 1, 3, 5, 7, and 9) or GFP-3C (lanes 2, 4, 6, 8, and 10). At 24 h after transfection, cell lysates were subjected to Western blot analysis using antibodies against Flag and GFP. β-Actin was used as a loading control. (C) Primary sequence of amino acids 330 to 366 within TAK1. In this region, glutamines were replaced with alanines. (D) The 3C cleavage site in TAK1. 293T cells were transfected with wild-type TAK1 or TAK1 mutants as indicated along with GFP (lanes 1, 3, 5, 7, and 9) or GFP-3C (lanes 2, 4, 6, 8, and 10). At 24 h after transfection, cell lysates were subjected to Western blot analysis with antibodies using a Li-Cor Odyssey Dual-Color System. Two antibodies that recognize TAK1 (Myc, C terminus of TAK1, 800 nm, green; Flag, N terminus of TAK1, 700 nm, red) were used. β-Actin was used as a loading control. (E and F) The 3C cleavage sites in TAB1 and TAB3. 293T were transfected with wild-type TAB1 or its variants (E) or TAB3 or its variants (F) as indicated along with GFP or GFP-3C. Cell lysates were subjected to Western blot analysis.
To localize the cleavage sites within TAK1, TAB1, and TAB3, we introduced alanine substitutions. When ectopically expressed, 3C induced cleavage of wild-type TAK1, Q337A, Q356A, and Q359A (Fig. 6C and D, lanes 1 to 8). This was not detectable in Q360A (lane 10), suggesting the Q360-S361 pair as a cleavage site. When coexpressed with TAB1, Q328A, Q437A, and Q444A, 3C induced full cleavage (Fig. 6E), resulting in two cleavage bands (lanes 2, 4, 8, and 10). This pattern was not seen in Q414A or Q451A, where a single cleaved band appeared (lanes 6 and 12). These data suggest that Q414-G415 and Q451-S452 within TAB1 are cleavage sites for EV71 3C. Further analysis of TAB3 revealed that 3C induced cleavage of Q116A, Q273A, Q246A, Q251A, Q255A, and Q335A. However, 3C was unable to mediate Q173A and Q343A cleavage (Fig. 6F). These results suggest that the Q173-G174 and Q343-G344 pairs within TAB3 are cleavage sites for EV71 3C.
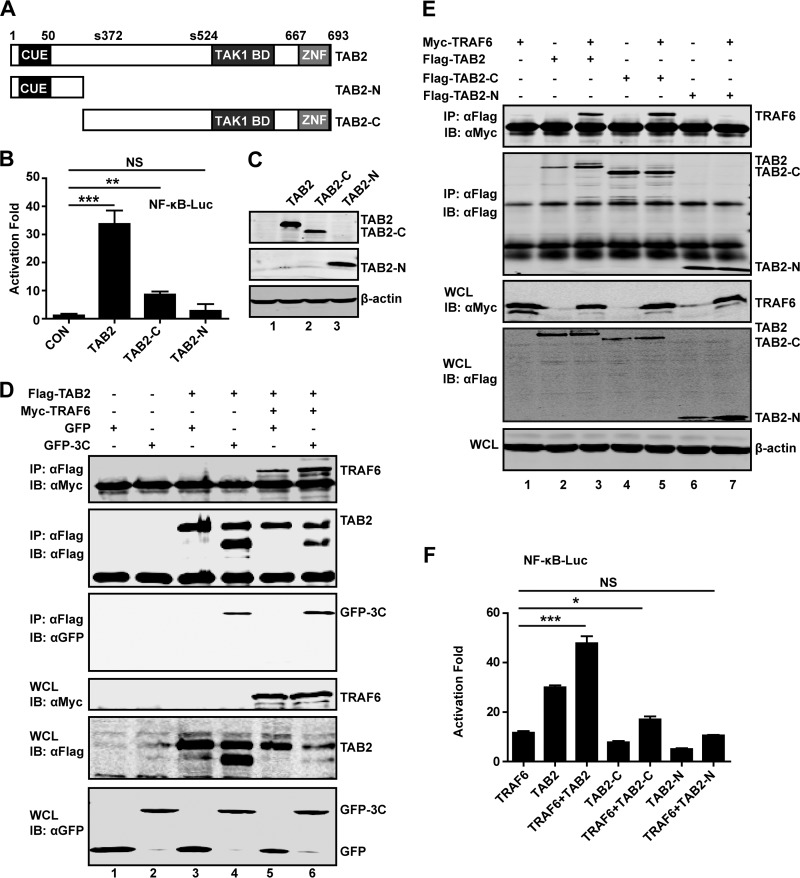
Cleavage of TAB2 by EV71 3C reduces its functional activity.
To assess the impact of TAB2 cleavage, we generated two TAB2 mutants that resemble cleaved products. As illustrated in Fig. 7A, TAB2-N represents the amino-terminal fragment, whereas TAB2-C represents the carboxyl-terminal fragment. In reporter assays, wild-type TAB2 activated the NF-κB reporter. However, neither TAB2-N nor TAB2-C was active (Fig. 7B). Western blot analysis revealed that TAB2 variants were expressed at comparable levels (Fig. 7C). Together, these results suggest that EV71 3C mediates TAB2 cleavage at the Q113-G114 site, resulting in two fragments which are inactivated.
FIG 7.
(A) Schematic diagrams of TAB2 and deletion mutants. Domain organization is outlined: the binding ubiquitin-conjugating enzyme domain (CUE), the zinc finger domain (ZNF), and phosphoserine (S372 and S524). TAB2-N and TAB2-C represent cleaved fragments of TAB2 by 3C. (B) The effect of TAB2 and its deletion mutants on NF-κB promoter activation. 293T cells were transfected with plasmids encoding TAB2, TAB2-N, or TAB2-C, along with NF-κB-Luc. pRL-SV40 was used as a control. At 24 h after transfection, cell lysates were assayed for luciferase activities. Data are representative of three independent experiments with triplicate samples. (C) Expression of TAB2 mutants. Cell lysates from panel B were subjected to Western blot analysis with antibodies against Flag and β-actin. (D) 293T cells were transfected with plasmids encoding TAB2 (lanes 3, 4, 5, and 6), TRAF6 (lanes 5 and 6), GFP (lanes 1, 3, and 5), and GFP-3C (lanes 2, 4, and 6). At 24 h after transfection, cell lysates were immunoprecipitated with antibody against Flag. Samples were then subjected to Western blot analysis. (E) 293T cells were transfected with plasmids encoding TRAF6 (lanes 1, 3, 5, and 7), TAB2 (lanes 2 and 3), TAB2-C (lanes 4 and 5), and TAB2-N (lanes 6 and 7). At 24 h after transfection, cell lysates were immunoprecipitated and detected as described in panel D. (F) 293T cells were transfected with plasmids as indicated, along with NF-κB-Luc. pRL-SV40 was used as a control. At 24 h after transfection, cell lysates were assayed for luciferase activities. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, nonsignificant.
As TAB2 activates TAK1 by coupling with TRAF6 (18), we asked whether 3C affected association of TAB2 with TRAF6. As shown in Fig. 7D, while TAB2 interacted with TRAF6, coexpression of 3C had no inhibitory effect, as measured by immunoprecipitation analysis. To reconcile this phenotype, we also analyzed the interaction between TRAF6 with 3C-cleaved fragments of TAB2. As illustrated in Fig. 7E, TAB2-C bound to TRAF6, whereas TAB2-N was unable to so. Indeed, coexpression of wild-type TAB2 and TRAF6 activated NF-κB promoter synergistically in report assays (Fig. 7F). Coexpression of TAB2 mutants with TRAF6 produced little or only marginal stimulation of NF-κB activation. Collectively, these results suggest that EV71 3C-mediated TAB2 cleavage compromises its ability to activate NF-κB.
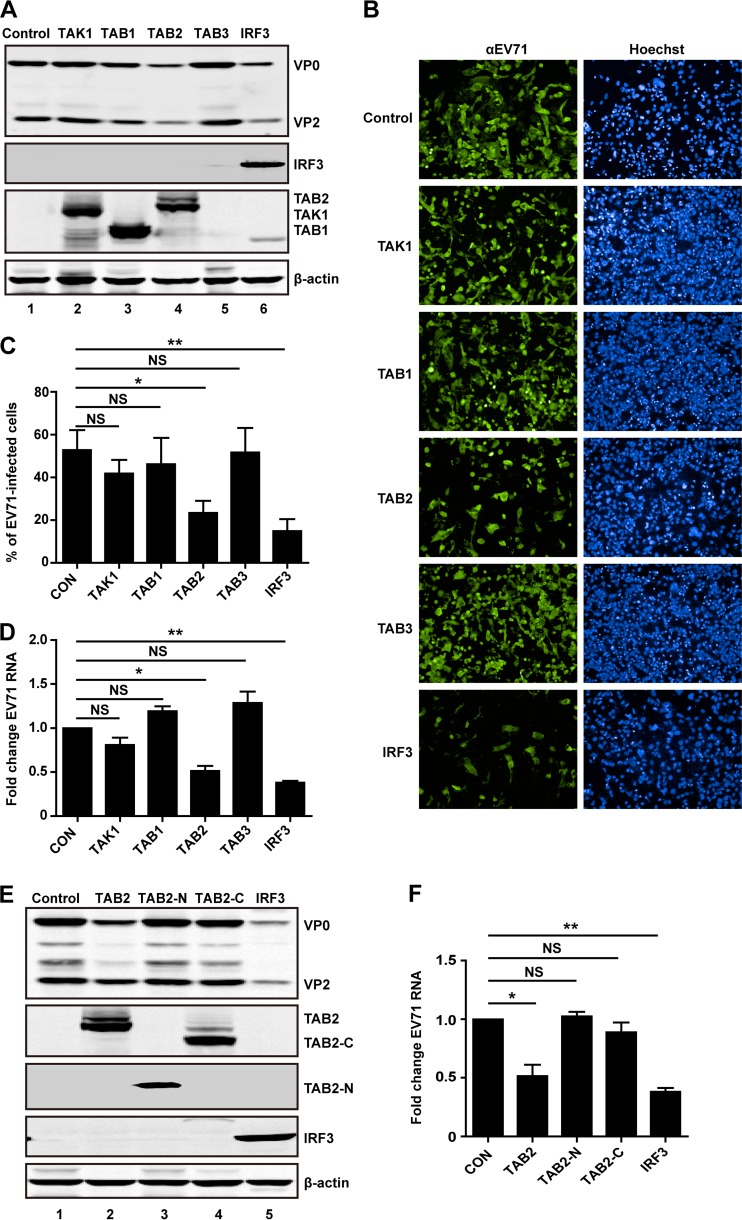
TAB2 but not cleaved TAB2 fragments inhibits EV71 replication.
Finally, we asked whether TAK1 complex is functionally linked to EV71 infection. RD cells were transfected with a vector, TAK1, TAB1, TAB2, TAB3, or IRF3. This was followed by EV71 infection for 24 h and processing of lysates for Western blot analysis. As shown in Fig. 8A, in control cells transfected with a vector, EV71 expressed VP0 and VP2, indicative of viral infection. As a positive control, IRF3 reduced viral protein production. Similarly, TAB2 significantly reduced the expression of VP0 and VP2 (lanes 4 and 6). Nevertheless, TAK1, TAB1, and TAB3 displayed little effect on viral protein production (lanes 2, 3, and 5). Consistently, in immunofluorescence analysis, IRF3 and TAB2 inhibited EV71 replication. In contrast, TAK1, TAB1, and TAB3 virtually displayed no inhibitory effect (Fig. 8B). As illustrated in Fig. 8C, TAB2 reduced viral replication by 50%. Consistently, TAB2 inhibited replication of EV71 RNA in real-time qPCR analysis (Fig. 8D). To further examine the effect of TAB2 on EV71 replication, we assessed two TAB2 mutants that mimic cleaved products. As illustrated in Fig. 8E, unlike wild-type TAB2, neither TAB2-N nor TAB2-C inhibited viral protein production. This correlated well with the levels of EV71 RNA (Fig. 8F). These data demonstrate that overexpression of TAB2 inhibits EV71 replication. However, when it was cleaved by 3C, its inhibitory activity disappeared.
FIG 8.
(A) TAB2 inhibits EV71 replication. RD cells were transfected with plasmids encoding Flalg-TAK1, Flag-TAB1, Flag-TAB2, or Flag-TAB3. Myc-IRF3 used as a control. At 24 h after transfection, cells were treated with EV71 at an MOI of 1 PFU per cell. After 24 h cells were harvested and resolved with 12% SDS-PAGE. Western blot analysis for EV71, Flag, Myc, or β-actin was conducted. (B) RD cells were transfected with plasmids encoding TAK1, TAB1, TAB2, TAB3, or IRF3. At 24 h after transfection, cells were treated with EV71. After 24 h, cells were fixed and stained with antibody against EV71. The images were acquired on an Operetta instrument using a 20× long-working-distance objective. Representative images are shown. (C) The percentage of EV71-infected cells from the experiment shown in panel B was analyzed using Harmony software. (D) RD cells were transfected with plasmids encoding TAK1, TAB1, TAB2, TAB3, or IRF3. Empty vector was used as a control. At 24 h after transfection, cells were infected with EV71. After 24 h, total RNA was extracted, and the viral RNA levels of EV71 were evaluated by quantitative real-time PCR using SYBR green. Data are expressed as fold change of the EV71 RNA level relative to the control using the ΔΔCT method as described in Materials and Methods. (E) RD cells were transfected with TAB2 variants as indicated. IRF3 was used as a control. Cell lysates were detected as described in panel A. (F) RD cells were transfected as described in panel E. At 24 h after transfection, cells were treated with EV71. After 24 h, EV71 RNA levels were detected and expressed as described in panel D. *, P < 0.05; **, P < 0.01; NS, nonsignificant.
DISCUSSION
Several lines of evidence suggest that the interplay of EV71 and innate antiviral immunity influences viral infection (30–33). In infected cells, EV71 inhibits type I IFN responses, which involves negative modulation of cytokine expression and signaling (8, 23, 34, 35). In this process, protease 2A targets the mitochondrial antiviral signaling protein (MAVS) and IFN receptor (34, 36). The 2C protein inhibits activation IκB kinase β (35). Moreover, 3C protease suppresses TLR3, cytosolic RNA receptor, and STAT pathways (5–8). In the present study, we demonstrate that EV71 3C protease mediates cleavage of TAB2, TAK1, TAB1, and TAB3. These observations support the notion that EV71 proteins may work coordinately to neutralize innate antiviral immunity upon infection.
As a serine/threonine kinase, TAK1 plays a critical role in cellular responses. TAK1 activation requires TAB1, TAB2, and TAB3 (13, 18, 21). While TAB1 acts as an activator, TAB2 and TAB3 link TAK1 to upstream TRAF6 (18, 21). Once engaged, the TAK1 complex activates p38, c-Jun N-terminal kinase (JNK), and NF-κB, leading to cytokine production (18, 37). Our work suggests that EV71 disables components of the TAK1 complex, thereby reducing cytokine expression. In infected cells, EV71 reduced levels of the TAK1 complex proteins while the levels of TBK1 and TRAF2 remained virtually unchanged. Reduction of TAK1 and TAB2 already took place at 8 and 12 h after EV71 infection, but the cleaved products were only evident at the late stage (24 h). This might be attributable to a low level of cleaved products early in virus infection. Nevertheless, while EV71 perturbs the TAK1 complex, alternative mechanisms may operate to modulate cytokine expression.
It has been reported that EV71 3C functions as an IFN antagonist through cleavage of TRIF, IRF7, and IRF9 (5–7). The 3C proteases of encephalomyocarditis virus (EMCV) and poliovirus cleave G3BP1, a component of stress granules (SGs), to escape SG-mediated antiviral responses (38, 39). In this respect, it is notable that picornavirus 3C also exerts additional activities. For example, enterovirus and poliovirus 3Cs induce translation arrest by cleaving PABP (25, 26). Furthermore, poliovirus 3C inhibits transcription by cleaving TBP and CREB (24, 27). On the other hand, host genes can escape poliovirus-induced transcriptional inhibition (40). In this study, we noted that when ectopically expressed in mammalian cells, EV71 3C mediated cleavage of TAB2, TAK1, TAB1, and TAB3, inhibiting NF-κB promoter activation. Importantly, EV71 3C did not inhibit NF-κB promoter activation induced by IKKβ, a downstream target of the TAK1 complex. As such, we favor the model that EV71 3C acts via cleavage of the TAK1 complex rather than a general inhibition of transcription or translation. However, it remains to be established whether EV71 3C modulates cytokine responses through PABP, TBP, CREB, and G3BP1 in infected cells.
These activities required an active protease, suggesting that 3C protease is functionally linked to the TAK1 complex. It is noteworthy that 3C protease interacted with TAK1 and TAB2. However, no interaction was detectable between 3C protease and TAB1 or TAB3. One possibility is that 3C protease mediates cleavage of TAB1 or TAB3 via an unknown factor. Alternatively, it may act through TAK1 or TAB2 in a protein complex. It should be noted that, without infection data, we cannot exclude the possibility that the cleavage events of 3C overexpression are not physiologically relevant although this is less likely.
Mutational analysis reveals that TAB2 cleavage occurs at the Q113-G114 pair, which results in two cleaved products: the N-terminal fragment with the CUE motif and the C-terminal fragment with TAK1 and zinc binding motifs. Q113A substitution abolished cleavage, suggesting that it is a likely EV71 3C cleavage site. Interestingly, unlike wild-type TAB2, mutants that mimic cleaved TAB2 fragments failed to activate NF-κB-dependent gene expression. As binding of TAB2/3 to TAK1 is necessary for the activation of downstream IKK kinases, it is likely that 3C-mediated cleavage disrupts the integrity of the TAK1 complex. Consistently, we also identified the cleavage sites within TAK1, TAB1, and TAB3. In TAK1, there is a single cleavage site whereas in TAB1 or TAB3 there are two cleavage sites. In this respect, it is notable that EV71 3C also cleaves TRIF to inhibit NF-κB-dependent gene expression (6). The biological basis for this complex layer of 3C activity remains to be established. We speculate that multiple cleavages of components mediated by 3C protease may favor a tight control of cytokine responses in EV71 infection.
A question arises as to why EV71 negatively modulates the components of the TAK1 complex. We noted that when TAB2 was ectopically expressed, it reduced EV71 replication significantly. This suggests that a dynamic equilibrium between EV71 and TAB2 controls EV71 infection. Interestingly, TAK1 as well as TAB1 modestly suppressed EV71 replication. However, TAB3 displayed little inhibitory activity. These different phenotypes might be relevant to the formation of the TAK1 complex where TAB2 plays a key role. Another explanation is that these components may have distinct functions against EV71 infection. Further analysis is needed to elucidate the precise role of this complex in EV71 infection.
ACKNOWLEDGMENTS
This work was supported by grants from the National Basic Research Program of China (973 Project, 2011CB504903), the Chinese National Health and Family Planning Commission Grant (201302006-05), the National Science Foundation for Outstanding Young Scientists (81225014), the National Natural Science Foundation of China (31270200), an intramural grant from the Institute of Pathogen Biology, Chinese Academy of Medical Sciences (2013IPB401), the Program for Changjiang Scholars and Innovative Research Team in University (IRT13007), and the National Institute of Allergy and Infectious Diseases of the United States (AI092230).
Footnotes
Published ahead of print 18 June 2014
REFERENCES
- 1.McMinn PC. 2002. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 26:91–107. 10.1111/j.1574-6976.2002.tb00601.x [DOI] [PubMed] [Google Scholar]
- 2.Li ML, Hsu TA, Chen TC, Chang SC, Lee JC, Chen CC, Stollar V, Shih SR. 2002. The 3C protease activity of enterovirus 71 induces human neural cell apoptosis. Virology 293:386–395. 10.1006/viro.2001.1310 [DOI] [PubMed] [Google Scholar]
- 3.Shih SR, Chiang C, Chen T, Wu CN, Hsu J, Lee J, Hwang MJ, Li M, Chen G, Ho MS. 2004. Mutations at KFRDI and VGK domains of enterovirus 71 3C protease affect its RNA binding and proteolytic activities. J. Biomed. Sci. 11:239–248. 10.1007/BF02256567 [DOI] [PubMed] [Google Scholar]
- 4.Sim AC, Luhur A, Tan TM, Chow VT, Poh CL. 2005. RNA interference against enterovirus 71 infection. Virology 341:72–79. 10.1016/j.virol.2005.06.047 [DOI] [PubMed] [Google Scholar]
- 5.Hung HC, Wang HC, Shih SR, Teng IF, Tseng CP, Hsu JT. 2011. Synergistic inhibition of enterovirus 71 replication by interferon and rupintrivir. J. Infect. Dis. 203:1784–1790. 10.1093/infdis/jir174 [DOI] [PubMed] [Google Scholar]
- 6.Lei X, Sun Z, Liu X, Jin Q, He B, Wang J. 2011. Cleavage of the adaptor protein TRIF by enterovirus 71 3C inhibits antiviral responses mediated by Toll-like receptor 3. J. Virol. 85:8811–8818. 10.1128/JVI.00447-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lei X, Xiao X, Xue Q, Jin Q, He B, Wang J. 2013. Cleavage of interferon regulatory factor 7 by enterovirus 71 3C suppresses cellular responses. J. Virol. 87:1690–1698. 10.1128/JVI.01855-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei X, Liu X, Ma Y, Sun Z, Yang Y, Jin Q, He B, Wang J. 2010. The 3C protein of enterovirus 71 inhibits retinoid acid-inducible gene I-mediated interferon regulatory factor 3 activation. J. Virol. 84:8051–8061. 10.1128/JVI.02491-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai L, Aye Thu C, Liu X, Xi J, Cheung PC. 2012. TAK1, more than just innate immunity. IUBMB Life 64:825–834. 10.1002/iub.1078 [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. 1995. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science 270:2008–2011. 10.1126/science.270.5244.2008 [DOI] [PubMed] [Google Scholar]
- 11.Ono K, Ohtomo T, Sato S, Sugamata Y, Suzuki M, Hisamoto N, Ninomiya-Tsuji J, Tsuchiya M, Matsumoto K. 2001. An evolutionarily conserved motif in the TAB1 C-terminal region is necessary for interaction with and activation of TAK1 MAPKKK. J. Biol. Chem. 276:24396–24400. 10.1074/jbc.M102631200 [DOI] [PubMed] [Google Scholar]
- 12.Besse A, Lamothe B, Campos AD, Webster WK, Maddineni U, Lin SC, Wu H, Darnay BG. 2007. TAK1-dependent signaling requires functional interaction with TAB2/TAB3. J. Biol. Chem. 282:3918–3928. 10.1074/jbc.M608867200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung PC, Nebreda AR, Cohen P. 2004. TAB3, a new binding partner of the protein kinase TAK1. Biochem. J. 378:27–34. 10.1042/BJ20031794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishitani T, Takaesu G, Ninomiya-Tsuji J, Shibuya H, Gaynor RB, Matsumoto K. 2003. Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J. 22:6277–6288. 10.1093/emboj/cdg605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omori E, Matsumoto K, Sanjo H, Sato S, Akira S, Smart RC, Ninomiya-Tsuji J. 2006. TAK1 is a master regulator of epidermal homeostasis involving skin inflammation and apoptosis. J. Biol. Chem. 281:19610–19617. 10.1074/jbc.M603384200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato S, Sanjo H, Takeda K, Ninomiya-Tsuji J, Yamamoto M, Kawai T, Matsumoto K, Takeuchi O, Akira S. 2005. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 6:1087–1095. 10.1038/ni1255 [DOI] [PubMed] [Google Scholar]
- 17.Shim JH, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, Lee KY, Bussey C, Steckel M, Tanaka N, Yamada G, Akira S, Matsumoto K, Ghosh S. 2005. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 19:2668–2681. 10.1101/gad.1360605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takaesu G, Kishida S, Hiyama A, Yamaguchi K, Shibuya H, Irie K, Ninomiya-Tsuji J, Matsumoto K. 2000. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol. Cell 5:649–658. 10.1016/S1097-2765(00)80244-0 [DOI] [PubMed] [Google Scholar]
- 19.Blonska M, Shambharkar PB, Kobayashi M, Zhang D, Sakurai H, Su B, Lin X. 2005. TAK1 is recruited to the tumor necrosis factor-alpha (TNF-alpha) receptor 1 complex in a receptor-interacting protein (RIP)-dependent manner and cooperates with MEKK3 leading to NF-κB activation. J. Biol. Chem. 280:43056–43063. 10.1074/jbc.M507807200 [DOI] [PubMed] [Google Scholar]
- 20.Jackson-Bernitsas DG, Ichikawa H, Takada Y, Myers JN, Lin XL, Darnay BG, Chaturvedi MM, Aggarwal BB. 2007. Evidence that TNF-TNFR1-TRADD-TRAF2-RIP-TAK1-IKK pathway mediates constitutive NF-kappaB activation and proliferation in human head and neck squamous cell carcinoma. Oncogene 26:1385–1397. 10.1038/sj.onc.1209945 [DOI] [PubMed] [Google Scholar]
- 21.Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ. 2004. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol. Cell 15:535–548. 10.1016/j.molcel.2004.08.008 [DOI] [PubMed] [Google Scholar]
- 22.Feng Z, Cerveny M, Yan Z, Hem B. 2007. The VP35 protein of Ebola virus inhibits the antiviral effect mediated by double-stranded RNA dependent protein kinase PKR. J. Virol. 81:182–192. 10.1128/JVI.01006-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weng KF, Li ML, Huang CT, Shih SR. 2009. Enterovirus 71 3C protease cleaves a novel target CstF-64 and inhibits cellular polyadenylation. PLoS Pathog. 5:e1000593. 10.1371/journal.ppat.1000593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yalamanchili P, Datta U, Dasgupta A. 1997. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J. Virol. 71:1220–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joachims M, Van Breugel PC, Lloyd RE. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol. Cell. Biol. 24:1779–1790. 10.1128/MCB.24.4.1779-1790.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kundu P, Raychaudhuri S, Tsai W, Dasgupta A. 2005. Shutoff of RNA polymerase II transcription by poliovirus involves 3C protease-mediated cleavage of the TATA-binding protein at an alternative site: incomplete shutoff of transcription interferes with efficient viral replication. J. Virol. 79:9702–9713. 10.1128/JVI.79.15.9702-9713.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui S, Wang J, Fan T, Qin B, Guo L, Lei X, Wang M, Jin Q. 2011. Crystal structure of human enterovirus 71 3C protease. J. Mol. Biol. 408:449–461. 10.1016/j.jmb.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Fan T, Yao X, Wu Z, Guo L, Lei X, Wang M, Jin Q, Cui S. 2011. Crystal structures of enterovirus 71 3C protease complexed with rupintrivir reveal the roles of catalytically important residues. J. Virol. 85:10021–10030. 10.1128/JVI.05107-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang L, Huang L, Gau S, Wu Y, Hsia S, Fan T, Lin K, Huang Y, Lu C, Lin T. 2007. Neurodevelopment and cognition in children after enterovirus 71 infection. N. Engl. J. Med. 356:1226–1234. 10.1056/NEJMoa065954 [DOI] [PubMed] [Google Scholar]
- 31.Ho M, Chen ER, Hsu KH, Twu SJ, Chen KT, Tsai SF, Wang JR, Shih SR. 1999. An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N. Engl. J. Med. 341:929–935. 10.1056/NEJM199909233411301 [DOI] [PubMed] [Google Scholar]
- 32.Huang C, Liu C, Chang YC, Chen C, Wang S, Yeh TF. 1999. Neurologic complications in children with enterovirus 71 infection. N. Engl. J. Med. 341:936–942. 10.1056/NEJM199909233411302 [DOI] [PubMed] [Google Scholar]
- 33.Liu ML, Lee YP, Wang YF, Lei HY, Liu CC, Wang SM, Su IJ, Wang JR, Yeh TM, Chen SH, Yu CK. 2005. Type I interferons protect mice against enterovirus 71 infection. J. Gen. Virol. 86:3263–3269. 10.1099/vir.0.81195-0 [DOI] [PubMed] [Google Scholar]
- 34.Wang B, Xi X, Lei X, Zhang X, Cui S, Wang J, Jin Q, Zhao Z. 2013. Enterovirus 71 protease 2Apro targets MAVS to inhibit anti-viral type I interferon responses. PLoS Pathog. 9:e1003231. 10.1371/journal.ppat.1003231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng Z, Li H, Zhang Z, Meng J, Mao D, Bai B, Lu B, Mao P, Hu Q, Wang H. 2011. Enterovirus 71 2C protein inhibits TNF-alpha-mediated activation of NF-kappaB by suppressing IκB kinase beta phosphorylation. J. Immunol. 187:2202–2212. 10.4049/jimmunol.1100285 [DOI] [PubMed] [Google Scholar]
- 36.Lu J, Yi L, Zhao J, Yu J, Chen Y, Lin MC, Kung HF, He ML. 2012. Enterovirus 71 disrupts interferon signaling by reducing the level of interferon receptor 1. J. Virol. 86:3767–3776. 10.1128/JVI.06687-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishitani T, Ninomiya-Tsuji J, Nagai S, Nishita M, Meneghini M, Barker N, Waterman M, Bowerman B, Clevers H, Shibuya H, Matsumoto K. 1999. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature 399:798–802. 10.1038/21674 [DOI] [PubMed] [Google Scholar]
- 38.White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305. 10.1016/j.chom.2007.08.006 [DOI] [PubMed] [Google Scholar]
- 39.Ng CS, Jogi M, Yoo JS, Onomoto K, Koike S, Iwasaki T, Yoneyama M, Kato H, Fujita T. 2013. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J. Virol. 87:9511–9522. 10.1128/JVI.03248-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doukas T, Sarnow P. 2011. Escape from transcriptional shutoff during poliovirus infection: NF-κB-responsive genes IκBa and A20. J. Virol. 85:10101–10108. 10.1128/JVI.00575-11 [DOI] [PMC free article] [PubMed] [Google Scholar]