Abstract
NEMO (NF-κB essential modulator) is a bridging adaptor indispensable for viral activation of interferon (IFN) antiviral response. Herein, we show that hepatitis A virus (HAV) 3C protease (3Cpro) cleaves NEMO at the Q304 residue, negating its signaling adaptor function and abrogating viral induction of IFN-β synthesis via the retinoic acid-inducible gene I/melanoma differentiation-associated protein 5 (RIG-I/MDA5) and Toll-like receptor 3 (TLR3) pathways. NEMO cleavage and IFN antagonism, however, were lost upon ablation of the catalytic activity of 3Cpro. These data describe a novel immune evasion mechanism of HAV.
TEXT
Innate immune responses are activated through host pattern recognition receptors (PRRs), which recognize pathogen-associated molecular patterns (1). Picornaviruses are sensed by retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), primarily melanoma differentiation-associated protein 5 (MDA5) (2–4) and sometimes RIG-I (5), and by Toll-like receptor 3 (TLR3) (6, 7). Upon engaging viral double-stranded RNAs (dsRNAs), RIG-I and MDA5 recruit mitochondrial antiviral signaling protein (MAVS [also known as IPS-1/VISA/Cardif]), while TLR3 interacts with the Toll–interleukin-1 (IL-1) receptor domain-containing adaptor, inducing interferon beta (IFN-β) (TRIF). Both pathways converge to activate an essential bridging adaptor, NF-κB essential modulator (NEMO), and subsequently classical IKK and IKK-related kinases, leading to phosphorylation of NF-κB and interferon (IFN) regulatory factor 3 (IRF3). These transcription factors directly activate promoters of type I IFNs, such as IFN-β (1).
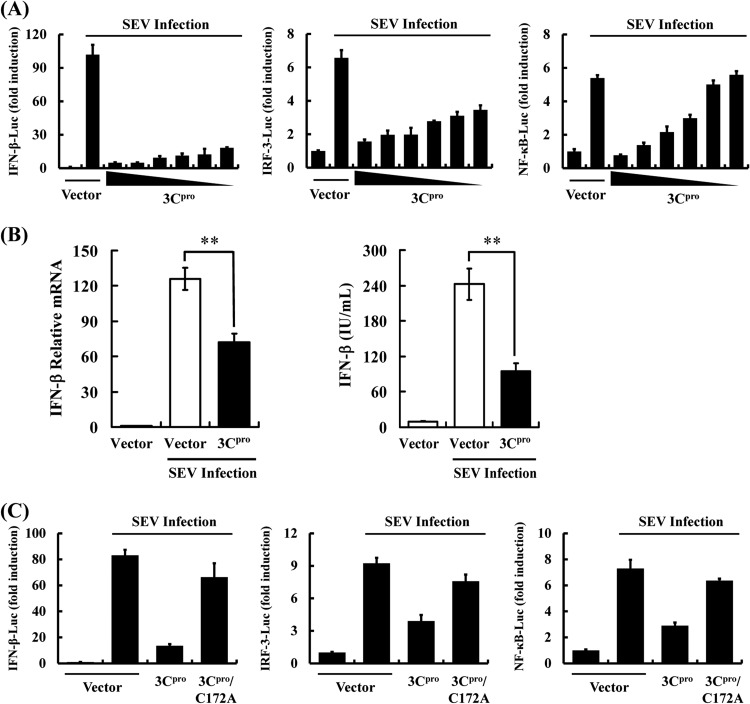
During its coevolution with humans, hepatitis A virus (HAV), a hepatotropic human picornavirus, has acquired mechanisms to subvert host innate immune responses (8–10). Notably, HAV disrupts RLRs signaling by targeting MAVS for proteolysis by a precursor of its 3C protease (3Cpro) cysteine protease, 3ABC (9). HAV also inhibits TLR3 signaling by cleaving TRIF via another precursor, 3CD (10). Herein, we found that mature 3Cpro inhibited Sendai virus (SEV)-induced IFN-β synthesis by using promoter-based luciferase reporter and enzyme-linked immunosorbent assays (ELISAs) (Fig. 1A and B), demonstrating 3Cpro is an additional HAV-encoded IFN antagonist. Furthermore, activation of both IRF3-dependent and NF-κB-dependent promoters was dose-dependently impaired by 3Cpro in human embryonic kidney HEK293T cells, suggesting that 3Cpro likely targets a step in the IFN-inducing pathway prior to the bifurcation of IRFs and NF-κB (Fig. 1A). Similar results were obtained in human hepatoma Huh7 cells (data not shown).
FIG 1.
HAV 3Cpro inhibits IFN-β promoter activation, and this ability requires the protease activity of 3Cpro. (A) HEK293T cells cultured in 24-well plates were transfected with indicated reporter plasmid (0.1 μg), along with pRL-TK (0.02 μg, for normalization of transfection efficiency) plasmid and increasing quantities (0, 0.03125, 0.0625, 0.125, 0.25, 0.5, or 1 μg) of plasmid encoding HAV 3Cpro, using Lipofectamine 2000. Twenty-four hours after the initial transfection, the cells were further infected or mock infected with SEV. Luciferase assays were performed 16 h after infection. The results represent the mean and standard deviation from three independent experiments. The firefly luciferase activity was normalized to the Renilla reniformis luciferase, and the untreated empty vector control value was set to 1. IFN-β-Luc (left) expresses firefly luciferase under the control of the human IFN-β promoter. 4× IRF3-Luc (middle) and 4× NF-κB-Luc (right) contain four copies of the IRF3- or NF-κB-binding motif in front of a luciferase reporter gene, respectively. (B) HEK293T cells cultured in 24-well plates were transfected with 3Cpro expression plasmids or an empty vector (1 μg). Twenty-four hours after initial transfection, cells were further infected or mock infected with SEV. The cells and supernatants were collected at 16 h postinfection and analyzed for IFN-β levels by real-time RT-PCR (left) and ELISA (right). (C) HEK293T cells cultured in 24-well plates were cotransfected with the indicated reporter plasmid, along with pRL-TK plasmid and the designated 3Cpro expression plasmids (1 μg). An empty vector was used as a control. Twenty-four hours after initial transfection, cells were further infected or mock infected with SEV. Luciferase assays were performed 16 h after infection.
HAV 3Cpro, as a cysteine proteinase, is responsible for most cleavages within the viral polyprotein (11, 12). To determine whether the protease activity is involved in 3Cpro-mediated IFN-β antagonism, a catalytically deficient 3Cpro mutant, C172A (13), was examined. In contrast to wild-type (WT) 3Cpro, 3Cpro-C172A was incapable of suppressing SEV-induced IFN-β promoter, implying that 3Cpro likely proteolytically cleaves a cellular protein(s) to disrupt IFN-β induction (Fig. 1C). Similar results were obtained when cells were transfected with luciferase-tagged IRF3 (IRF3-Luc) or NF-κB-Luc in lieu of IFN-β-Luc (Fig. 1C).
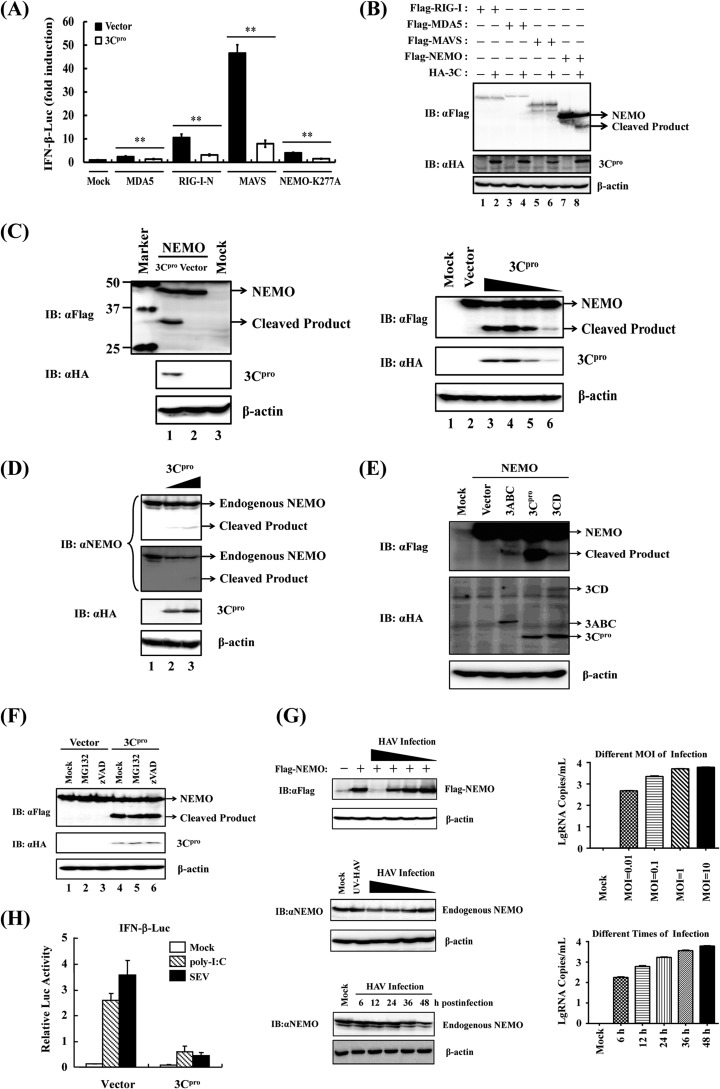
Given the pivotal role of MDA5/RIG-I in sensing of picornaviruses and that RLRs signaling to IRFs and NF-κB diverges at NEMO (14), we investigated whether 3Cpro targets NEMO or its upstream signaling molecules MAVS, RIG-I, and MDA5 for proteolysis. Compared to an empty plasmid control, overexpression of MDA5, a constitutively active RIG-I mutant (RIG-I-N), MAVS, or a constitutively active NEMO mutant (NEMO-K277A) (15) significantly activated the IFN-β promoter. However, such effects were all significantly reduced upon 3Cpro coexpression (Fig. 2A), suggesting that 3Cpro inhibits RLRs signaling at or downstream of NEMO. Interestingly, coexpression of 3Cpro resulted in marked reduction of ectopically expressed Flag-NEMO, concomitant with the appearance of a faster-migrating band with a molecular mass of ∼35 kDa that was detected by an anti-Flag antibody, presumably a 3Cpro cleavage product (Fig. 2B and C). Importantly, the higher the 3Cpro expression level, the more ∼35-kDa product was expressed while less full-length Flag-NEMO was seen (Fig. 2C). In contrast, no cleavage product was observed for RIG-I, MDA5, or MAVS (Fig. 2B). Remarkably, endogenous NEMO protein was similarly cleaved in 3Cpro-expressing cells (Fig. 2D). The processing intermediates encompassing 3Cpro (i.e., 3ABC and 3CD) also processed NEMO with the same cleavage pattern as mature 3Cpro, albeit less efficiently than the latter (Fig. 2E). This implies that although these 3C processor forms have evolved unique ability to target MAVS and TRIF, respectively, they share with mature 3Cpro the capacity to hydrolyze NEMO. Note that ectopic expression of 3CD was less effective in cleaving NEMO than overexpression of 3Cpro, although it produced a comparable amount of mature 3Cpro to the latter. 3D or 3CD could form a complex with 3Cpro matured from 3CD (16–18) impairing the proteolytic activity of 3Cpro toward NEMO. Importantly, 3Cpro-induced cleavage of NEMO was independent of cellular caspases or the proteasome, as not reversed by treatment with ZVAD or MG132 (Fig. 2F). Confirming that 3Cpro produced in the context of virus infection is capable of NEMO cleavage, we found that the protein abundance of both ectopically expressed Flag-NEMO and endogenous NEMO was substantially reduced in HAV-infected HEK293T cells (Fig. 2G). In these experiments, the degree of NEMO reduction correlated with the HAV infection dose, and substantial NEMO degradation coincided with a high level of HAV RNA replication at 36 h postinfection and beyond (Fig. 2G). We also observed that the level of endogenous NEMO protein is higher than that of ectopically expressed Flag-NEMO (data not shown). This could explain why HAV infection appeared to degrade Flag-NEMO more efficiently that it did endogenous NEMO. Consistent with the fact that NEMO is required for activation of the IFN response downstream of both RLRs and TLR3 pathways, extracellular dsRNA [poly(I·C)]-induced activation of the IFN-β promoter in 293-TLR3 cells was blunted by 3Cpro (Fig. 2H).
FIG 2.
HAV 3Cpro disrupts RLR signaling by cleaving NEMO. (A) HEK293T cells cultured in 24-well plates were cotransfected with IFN-β-Luc, pRL-TK plasmid, and plasmid encoding 3Cpro (0.3 μg), together with the MDA5, RIG-I-N, MAVS, or NEMO-K277A expression vector (0.7 μg). Luciferase assays were performed 36 h after transfection. **, P < 0.01 (considered highly significant). (B) HEK293T cells cultured in 60-mm-diameter dishes were transfected with Flag-tagged RIG-I, MDA5, MAVS, or NEMO expression plasmid (4 μg) along with empty vector or plasmid encoding hemagglutinin (HA)-tagged 3Cpro (0.5 μg). Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting (immunoblotting [IB]). (C) (Left) HEK293T cells cultured in 60-mm-diameter dishes were transfected with Flag-tagged NEMO expression plasmid (2 μg) along with empty vector or plasmid encoding HA-tagged 3Cpro (1 μg). Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting. The lane with protein markers (Bio-Rad, catalog no. 161-0376) includes 50-, 37-, and 25-kDa molecular mass bands. (Right) HEK293T cells cultured in 60-mm dishes were transfected with Flag-tagged NEMO expression plasmid (2 μg), along with increasing quantities (0, 0.125, 0.25, 0.5, or 1 μg) of plasmid encoding HA-tagged 3Cpro. Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting. (D) HEK293T cells cultured in 60-mm dishes were transfected with increasing quantities (0, 2, or 4 μg) of plasmid encoding HA-tagged 3Cpro. Cells were lysed 30 h posttransfection and analyzed by Western blotting. (Two different exposures of the blot are shown.) (E) HEK293T cells cultured in 60-mm dishes were transfected with Flag-tagged wild-type NEMO as indicated (2 μg), along with HA-3Cpro or 3Cpro-containing precursors (1 μg). Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting. (F) HEK293T cells cultured in 60-mm dishes were cotransfected with Flag-tagged NEMO expression plasmid (2 μg) and plasmid encoding 3Cpro or empty vector (1 μg). Twenty-four hours after transfection, MG132 or zVAD-FMK was added to a final concentration of 20 μM. Cell lysates were prepared 8 h after treatment and analyzed by Western blotting. (G) (Top) HEK293T cells cultured in 60-mm dishes were transfected with Flag-tagged NEMO (Flag-NEMO) and infected with different doses (multiplicities of infection [MOI] of 0.01, 0.1, 1, and 10) of HAV (L-A-1 attenuated vaccine strain) 6 h posttransfection. The cells were lysed 36 h postinfection and analyzed by Western blotting. (Middle) HEK293T cells were infected with different doses of HAV or, as a control, UV light-inactivated HAV (UV-HAV). The cells were lysed 36 h postinfection and analyzed by Western blotting. (Bottom) HEK293T cells were infected with HAV (MOI of 1), lysed at different times of HAV postinfection, and analyzed by Western blotting. Expression of Flag-NEMO conjugated protein and endogenous NEMO protein was verified by mouse anti-Flag antibody (Macgene, China) or rabbit anti-NEMO (Abclonal, China), respectively. Mouse anti-HA (Abclonal, China) was used to confirm the expression of 3Cpro, and mouse anti-β-actin antibody (Beyotime, China) was used to detect β-actin, which serves as a protein loading control. (H) Activation of the IFN-β promoter by extracellular poly(I·C) (20 μg/ml) or SEV (100 hemagglutinating units [HAU]/ml) in HEK293-TLR3 cells transiently expressing 3Cpro or a control vector.
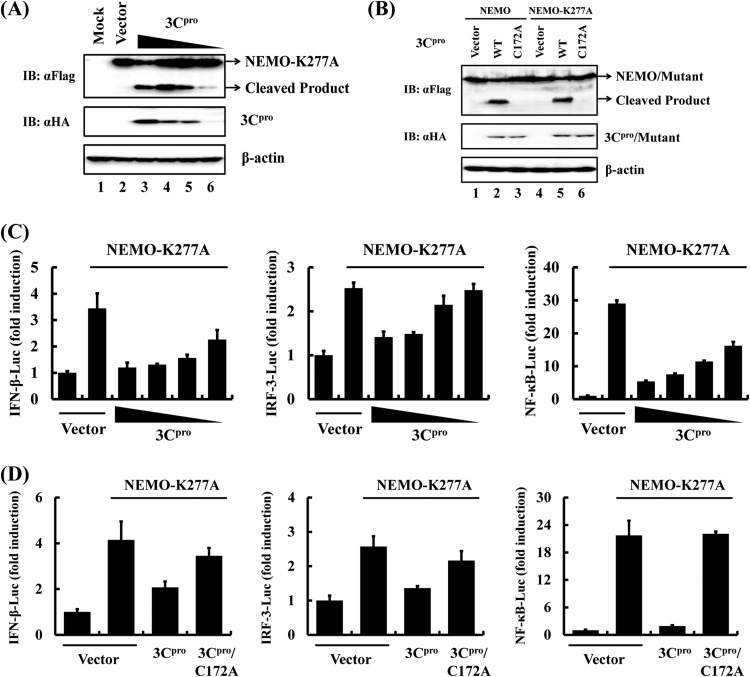
Since WT NEMO did not significantly activate IRFs and NF-κB to induce IFN-β (19), we used a constitutively active NEMO mutant, NEMO-K277A, to examine the effect of 3Cpro-mediated NEMO cleavage on IFN-β induction. NEMO-K277A remained susceptible to WT 3Cpro cleavage but not to the C172A mutant (Fig. 3A and B). As a result, overexpression of 3Cpro dose-dependently inhibited NEMO-K277A-mediated IFN-β promoter activation by disrupting activation of IRF3 and NF-κB (Fig. 3C). In contrast, the catalytically inactive 3Cpro-C172A mutant was without effect (Fig. 3D), consistent with the inability of 3Cpro-C172A to cleave NEMO (or NEMO-K277A) (Fig. 3B).
FIG 3.
HAV 3Cpro disrupts NEMO-mediated IFN-β induction. (A) HEK293T cells cultured in 60-mm dishes were transfected with Flag-tagged NEMO-K277A expression plasmid (2 μg) along with plasmid encoding HA-tagged 3Cpro (1 μg). Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting. (B) HEK293T cells cultured in 60-mm dishes were transfected with Flag-tagged NEMO expression plasmid (2 μg), along with the indicated 3Cpro expression plasmids (1 μg). Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting. (C) HEK293T cells cultured in 24-well plates were transfected with the indicated reporter plasmid (0.1 μg), pRL-TK (0.02 μg), and Flag-tagged NEMO-K277A expression plasmid (0.5 μg), along with increasing quantities (0, 0.0625, 0.125, 0.25, or 0.5 μg) of plasmid encoding 3Cpro. Luciferase assays were performed at 36 h after the transfection. (D) HEK293T cells cultured in 24-well plates were cotransfected with the indicated reporter plasmid, pRL-TK plasmid, and Flag-tagged NEMO-K277A expression plasmid (0.5 μg) along with the designated 3Cpro expression plasmids (0.5 μg). An empty vector (pCMV-HA) was used as a control. Cell extracts were collected 36 h after transfection and analyzed for firefly and Renilla luciferase expression.
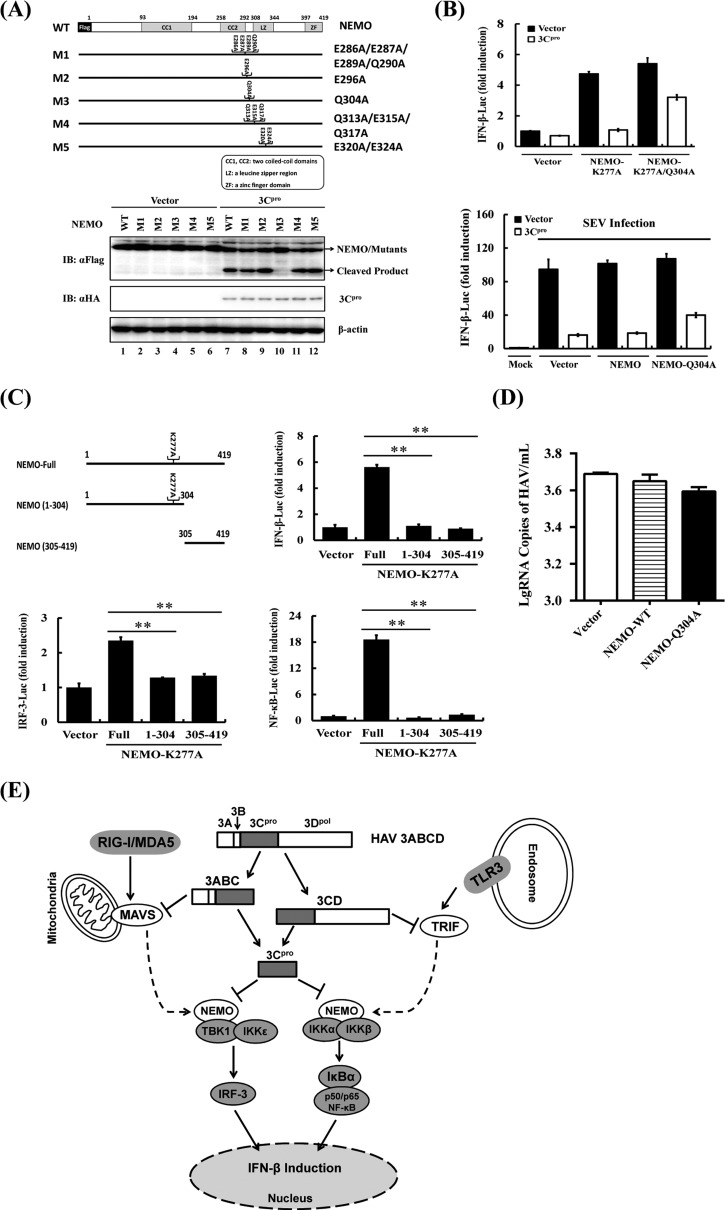
Because NEMO cleavage produced an ∼35-kDa product and the 3Cpro substrate specificity documented a preference for glutamine (Gln [Q]) or glutamic acid (Glu [E]) at the P1 position (20), we examined the sequence of NEMO for potential 3Cpro cleavage sites that could potentially generate fragments of the appropriate size following 3Cpro scission and constructed a cluster of NEMO mutants in which the invariant Gln or Glu at each potential P1 position was replaced with alanine (Ala [A]) (Fig. 4A, upper panel). 3Cpro-mediated NEMO cleavage was not affected by E286A E287A E289A Q290A, E296A, Q313A E315A Q317A, or E320A E324A mutations. In contrast, the Q304A substitution blocked the appearance of the cleavage product as well as the reduction in abundance of full-length NEMO (Fig. 4A, lower panel, lane 10), suggesting 3Cpro cleaved NEMO at Gln304. As further evidence supporting this notion, the Q304A mutation also rendered NEMO-K277A resistant to 3Cpro cleavage (data not shown). We found that the constitutively active NEMO-K277A bearing the Q304A mutation significantly overcame the 3Cpro blockade on activation of the IFN-β promoter compared with its 3Cpro-sensitive counterpart (Fig. 4B). Furthermore, ectopic expression of NEMO-Q304A, but not WT NEMO, substantially restored Sendai virus-induced IFN-β promoter activity (Fig. 4B). The incomplete restoration of IFN activation by the Q304A mutant NEMO could be attributed to the ability of HAV 3Cpro to associate with NEMO and inhibit assembly of NEMO-containing signaling complexes, in addition to NEMO cleavage, as has been observed with foot-and-mouth-disease virus 3Cpro (19). This possibility needs to be investigated in future studies.
FIG 4.
HAV 3Cpro-mediated NEMO cleavage is involved in the inhibition of IFN-β induction. (A) Schematic representation of wild-type NEMO and its derivatives (top). HEK293T cells cultured in 60-mm-diameter dishes were transfected with Flag-tagged wild-type NEMO or NEMO mutants as indicated, along with HA-3Cpro or empty vector. Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting. (B) HEK293T cells cultured in 24-well plates were cotransfected with IFN-β-Luc, pRL-TK plasmid, and plasmid encoding 3Cpro (0.3 μg) together with the empty vector, NEMO-K277A, or the NEMO-K277A/Q304A expression vector (0.7 μg). Luciferase assays were performed 36 h after transfection (top). HEK293T cells cultured in 24-well plates were cotransfected with IFN-β-Luc, pRL-TK plasmid, and plasmid encoding 3Cpro (0.3 μg) together with the empty vector, NEMO, or the NEMO-Q304A expression vector (0.7 μg). Twenty-four hours after the initial transfection, the cells were further infected or mock infected with SEV. Luciferase assays were performed at 16 h after infection (down). (C) HEK293T cells cultured in 24-well plates were cotransfected with IFN-β-Luc, pRL-TK plasmid and either 1 μg of plasmid encoding Flag-fused NEMO-K277A (Full), putative 3Cpro-induced cleavage fragments of NEMO-K277A, or empty vector. Cell extracts were collected 36 h after transfection and analyzed for firefly and Renilla luciferase expression. **, P < 0.01 (considered highly significant). (D) HEK293T cells cultured in 24-well plates were transfected with the indicated NEMO-expressing plasmids and infected with HAV (L-A-1 attenuated vaccine strain; MOI of 1) 12 h posttransfection. The cells were lysed 36 h postinfection and analyzed by RT-qPCR according to the reference methods (21). (E) Type I interferon signaling pathways disrupted by HAV 3Cpro and its precursors. HAV 3Cpro and its processing intermediates disrupt IFN-β induction by 3ABC-mediated proteolysis of MAVS (9), 3CD-mediated proteolysis of TRIF (10), and 3Cpro-mediated proteolysis of NEMO in this study.
To further determine whether either of the 3Cpro-induced cleavage fragments of NEMO would remain active, we ectopically expressed the constitutively active NEMO-K277A, the N-terminal fragment of NEMO-K277A (residues 1 to 304), or the C-terminal fragment of NEMO (residues 305 to 419) that would result from 3Cpro cleavage and examined their abilities to activate IFN-β and IRF3/NF-κB-dependent promoters. In contrast to full-length NEMO-K277A which was competent in inducing all 3 promoters, neither the N-terminal nor C-terminal fragment retained such abilities (Fig. 4C), suggesting that 3Cpro-mediated cleavage of NEMO efficiently inactivates NEMO-mediated downstream signaling. In addition, reconstitution of NEMO signaling by the Q304A mutant NEMO led to a modest reduction (∼20%) in HAV replication (Fig. 4D) as revealed by quantitative reverse transcription-PCR (RT-qPCR) (21).
In summary, our data identify 3Cpro as a novel IFN antagonist encoded by HAV and show that the 3Cpro-mediated proteolytic cleavage of NEMO is directly involved in inhibition of IFN-β transcription. By virtue of this, our study describes a novel mechanism by which HAV counteracts the host's innate antiviral responses. The multipronged targeting of MAVS, TRIF, and NEMO by HAV through 3ABC, 3CD, and 3Cpro illustrates a remarkable example of picornaviral evolution, disarming host innate immunity to the greatest extent possible and offering the virus a maximal survival advantage (Fig. 4E). This also could explain the striking observation that acute HAV infection is associated with a very limited type I IFN response, despite the persistence of intrahepatic viral RNA (8). Previously, we reported that NEMO is a substrate for 3Cpro of another picornavirus, foot-and-mouth disease virus (19). Recently, 3Cpro of enterovirus 71 was reported to target interferon regulatory factor 7 (IRF7) for proteolysis (22). We did not find this viral protease to cleave NEMO (data not shown), indicating that targeting NEMO for immune evasion is adopted by some but not all picornaviruses. Exactly how the differential targeting of NEMO affects innate immune responses and/or picornaviral pathogenesis in vivo will require further study.
ACKNOWLEDGMENTS
This research was supported by the National Natural Sciences Foundation of China (31225027 and 31121004), the National Basic Research Program (973) of China (2014CB522700), the Fundamental Research Funds for the Central Universities (2013PY043), and the U.S. National Institutes of Health (AI069285).
Footnotes
Published ahead of print 11 June 2014
REFERENCES
- 1.Kawai T, Akira S. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131–137. 10.1038/ni1303 [DOI] [PubMed] [Google Scholar]
- 2.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 103:8459–8464. 10.1073/pnas.0603082103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Husser L, Alves MP, Ruggli N, Summerfield A. 2011. Identification of the role of RIG-I, MDA-5 and TLR3 in sensing RNA viruses in porcine epithelial cells using lentivirus-driven RNA interference. Virus Res. 159:9–16. 10.1016/j.virusres.2011.04.005 [DOI] [PubMed] [Google Scholar]
- 4.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- 5.Papon L, Oteiza A, Imaizumi T, Kato H, Brocchi E, Lawson TG, Akira S, Mechti N. 2009. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology 393:311–318. 10.1016/j.virol.2009.08.009 [DOI] [PubMed] [Google Scholar]
- 6.Oshiumi H, Okamoto M, Fujii K, Kawanishi T, Matsumoto M, Koike S, Seya T. 2011. The TLR3/TICAM-1 pathway is mandatory for innate immune responses to poliovirus infection. J. Immunol. 187:5320–5327. 10.4049/jimmunol.1101503 [DOI] [PubMed] [Google Scholar]
- 7.Negishi H, Osawa T, Ogami K, Ouyang X, Sakaguchi S, Koshiba R, Yanai H, Seko Y, Shitara H, Bishop K, Yonekawa H, Tamura T, Kaisho T, Taya C, Taniguchi T, Honda K. 2008. A critical link between Toll-like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. Proc. Natl. Acad. Sci. U. S. A. 105:20446–20451. 10.1073/pnas.0810372105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanford RE, Feng Z, Chavez D, Guerra B, Brasky KM, Zhou Y, Yamane D, Perelson AS, Walker CM, Lemon SM. 2011. Acute hepatitis A virus infection is associated with a limited type I interferon response and persistence of intrahepatic viral RNA. Proc. Natl. Acad. Sci. U. S. A. 108:11223–11228. 10.1073/pnas.1101939108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Liang Y, Qu L, Chen Z, Yi M, Li K, Lemon SM. 2007. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl. Acad. Sci. U. S. A. 104:7253–7258. 10.1073/pnas.0611506104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qu L, Feng Z, Yamane D, Liang Y, Lanford RE, Li K, Lemon SM. 2011. Disruption of TLR3 signaling due to cleavage of TRIF by the hepatitis A virus protease-polymerase processing intermediate, 3CD. PLoS Pathog. 7:e1002169. 10.1371/journal.ppat.1002169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultheiss T, Kusov YY, Gauss-Muller V. 1994. Proteinase 3C of hepatitis A virus (HAV) cleaves the HAV polyprotein P2-P3 at all sites including VP1/2A and 2A/2B. Virology 198:275–281. 10.1006/viro.1994.1030 [DOI] [PubMed] [Google Scholar]
- 12.Schultheiss T, Sommergruber W, Kusov Y, Gauss-Muller V. 1995. Cleavage specificity of purified recombinant hepatitis A virus 3C proteinase on natural substrates. J. Virol. 69:1727–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergmann EM, Mosimann SC, Chernaia MM, Malcolm BA, James MN. 1997. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J. Virol. 71:2436–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, Lin R. 2007. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat. Immunol. 8:592–600. 10.1038/ni1465 [DOI] [PubMed] [Google Scholar]
- 15.Bloor S, Ryzhakov G, Wagner S, Butler PJ, Smith DL, Krumbach R, Dikic I, Randow F. 2008. Signal processing by its coil zipper domain activates IKK gamma. Proc. Natl. Acad. Sci. U. S. A. 105:1279–1284. 10.1073/pnas.0706552105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zell R, Seitz S, Henke A, Munder T, Wutzler P. 2005. Linkage map of protein-protein interactions of porcine teschovirus. J. Gen. Virol. 86:2763–2768. 10.1099/vir.0.81144-0 [DOI] [PubMed] [Google Scholar]
- 17.Shen M, Reitman ZJ, Zhao Y, Moustafa I, Wang Q, Arnold JJ, Pathak HB, Cameron CE. 2008. Picornavirus genome replication. Identification of the surface of the poliovirus (PV) 3C dimer that interacts with PV 3Dpol during VPg uridylylation and construction of a structural model for the PV 3C2-3Dpol complex. J. Biol. Chem. 283:875–888. 10.1074/jbc.M707907200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pathak HB, Arnold JJ, Wiegand PN, Hargittai MR, Cameron CE. 2007. Picornavirus genome replication: assembly and organization of the VPg uridylylation ribonucleoprotein (initiation) complex. J. Biol. Chem. 282:16202–16213. 10.1074/jbc.M610608200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D, Fang L, Li K, Zhong H, Fan J, Ouyang C, Zhang H, Duan E, Luo R, Zhang Z, Liu X, Chen H, Xiao S. 2012. Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J. Virol. 86:9311–9322. 10.1128/JVI.00722-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seipelt J, Guarne A, Bergmann E, James M, Sommergruber W, Fita I, Skern T. 1999. The structures of picornaviral proteinases. Virus Res. 62:159–168. 10.1016/S0168-1702(99)00043-X [DOI] [PubMed] [Google Scholar]
- 21.Qiu F, Zheng H, Yi Y, Jia Z, Cao J, Bi S. 2013. Comparative evaluation of a novel TaqMan real-time reverse transcription-polymerase chain reaction assay for hepatitis A virus detection. J. Int. Med. Res. 41:427–434. 10.1177/0300060513476434 [DOI] [PubMed] [Google Scholar]
- 22.Lei X, Xiao X, Xue Q, Jin Q, He B, Wang J. 2013. Cleavage of interferon regulatory factor 7 by enterovirus 71 3C suppresses cellular responses. J. Virol. 87:1690–1698. 10.1128/JVI.01855-12 [DOI] [PMC free article] [PubMed] [Google Scholar]