ABSTRACT
The mechanisms that lead to the tegumentation of herpesviral particles are only poorly defined. The phosphoprotein 65 (pp65) is the most abundant constituent of the virion tegument of human cytomegalovirus (HCMV). It is, however, nonessential for virion formation. This seeming discrepancy has not met with a satisfactory explanation regarding the role of pp65 in HCMV particle morphogenesis. Here, we addressed the question of how the overall tegument composition of the HCMV virion depended on pp65 and how the lack of pp65 influenced the packaging of particular tegument proteins. To investigate this, we analyzed the proteomes of pp65-positive (pp65pos) and pp65-negative (pp65neg) virions by label-free quantitative mass spectrometry and determined the relative abundances of tegument proteins. Surprisingly, only pUL35 was elevated in pp65neg virions. As the abundance of pUL35 in the HCMV tegument is low, it is unlikely that it replaced pp65 as a structural component in pp65neg virions. A subset of proteins, including the third most abundant tegument protein, pUL25, as well as pUL43, pUL45, and pUL71, were reduced in pp65neg or pp65low virions, indicating that the packaging of these proteins was related to pp65. The levels of tegument components, like pp28 and the capsid-associated tegument proteins pp150, pUL48, and pUL47, were unaffected by the lack of pp65. Our analyses demonstrate that deletion of pp65 is not compensated for by other viral proteins in the process of virion tegumentation. The results are concordant with a model of pp65 serving as an optional scaffold protein that facilitates protein upload into the outer tegument of HCMV particles.
IMPORTANCE The assembly of the tegument of herpesviruses is only poorly understood. Particular proteins, like HCMV pp65, are abundant tegument constituents. pp65 is thus considered to play a major role in tegument assembly in the process of virion morphogenesis. We show here that deletion of the pp65 gene leads to reduced packaging of a subset of viral proteins, indicating that pp65 acts as an optional scaffold protein mediating protein upload into the tegument.
INTRODUCTION
The virion of human cytomegalovirus (HCMV), a member of the family Betaherpesvirinae, is composed of a capsid containing the DNA genome, a tegument layer, and an envelope. HCMV virion assembly is an orchestrated process that is initiated with capsid formation and DNA packaging in the nucleus (1, 2). At this stage, a first layer of tegument proteins is attached to the capsid (3). Following translocation through the nuclear membrane by a process of envelopment and de-envelopment, capsids are targeted to specialized sites in the cytosol, termed cytoplasmic viral assembly compartments (4–6). Most of the tegumentation and subsequent envelopment appears to proceed in these structures, which contain components of both the trans-Golgi network and the endoplasmic reticulum (ER)-Golgi intermediate compartments (7). Subsequent to as yet poorly defined processes that lead to HCMV virion envelopment, particles are transported to the cell surface by cellular transport systems. ESCRT (endosomal sorting complexes required for transport) proteins, as well as SNARE (soluble N-ethylmaleimide-sensitive-factor attachment protein receptor complex) proteins, may be involved in HCMV virion transport (1, 8, 9).
Viral proteins contained in the HCMV tegument serve at least three distinct functions. Proteins that are closely attached to the capsid structure may be necessary for the structural integrity of the particle (10, 11). Besides a putative structural function, some of the tegument proteins, mostly located in the outer tegument layers, are also functionally important. These proteins may be essential for the initiation of viral gene expression, for sensing of the cellular environment, or for counteracting innate cellular defense mechanisms (12–16). Finally, tegument interaction with membrane-associated viral glycoproteins is likely to direct final envelopment, as evidenced by the envelopment of subviral dense bodies (DBs) that almost exclusively consist of tegument proteins (17, 18).
The phosphoprotein 65 (pp65) is the major tegument component of HCMV virions (17, 19). It interacts with the cellular protein IFI16, thereby blocking its DNA-sensing function (15, 20). Furthermore, pp65 has been reported to interfere with innate and adaptive immunity, though the discovery of deregulation of pp71 expression in pp65 deletion mutants may lead to a reevaluation of some of these results (21–24). The function of pp65 as a structural component of the viral particle is unknown. Although pp65 may contribute up to 15% of the total virion mass (17), it is dispensable for growth in human fibroblasts (25). Remarkably, the intracellular virion morphology of a mutant lacking pp65 is comparable to that of pp65-competent strains (25, 26). This is surprising, as morphological changes would have been expected with the most abundant constituent of the virus particle lacking. One likely explanation is that a single protein fills the gap in the tegument, thereby complementing a putative structural function of pp65. Alternatively, lack of pp65 could lead to a general increase in the recruitment of other viral proteins into the tegument. We have recently presented preliminary data showing that the overall composition of HCMV virions is not substantially altered in pp65-negative (pp65neg) strains (27). In this communication, we extend these analyses and describe a proteomics approach to address the question about the compensation of the loss of pp65 with regard to individual viral and cellular proteins. The data show that the upload of some tegument proteins is impaired in pp65neg strains, indicating that pp65 serves as an optional tegument scaffold. The loss of pp65 was not compensated for by one or more other viral or cellular proteins, suggesting that the tegument structure in HCMV virions tolerates gross reduction in protein content without abrogating virion infectivity.
MATERIALS AND METHODS
Cells and viruses.
Primary human foreskin fibroblasts (HFF) were cultured as described previously (28). The viruses used in this study were all descendants, as bacterial artificial chromosome (BAC) clones, of the AD169 laboratory strain of HCMV. The RV-HB5 strain was originally cloned by inserting a BAC vector into the US2-US6 gene region of the AD169 strain (29). The deletion of US2 to US6 was repaired to generate RV-HB15 by cre-lox recombination, leaving a single loxp site in the genome (30). The RV-BADwt strain represents a full-length version of the AD169 strain with one loxP site between the US28 and US29 open reading frames (ORFs) (31). RV-Hd65 was constructed by replacing the UL83 (pp65) open reading frame of AD169-BAC with a neomycin resistance cassette (28). AD169-BAC is the BAC clone of RV-HB15. The neomycin resistance gene was excised by using FLP recombinase-mediated excision in Escherichia coli, and the BAC vector was removed by cre-lox recombination at virus reconstitution. The pp65neg strain RV-KB14 was generated by inserting a tetracycline resistance cassette into the UL83 (pp65) ORF of pAD/cre (the BAC clone used for reconstitution of RV-BADwt), thereby deleting the pp65-coding region except for 152 5′ base pairs of the ORF (32). The mutants RV-SB2 and RV-VM1 were generated by inserting the nonapeptide TMYGGISLL from the immediate-early 1 (IE1) protein of HCMV at position S101 or R387, respectively, of pp65, using galK-mediated recombination on the BACmid pHB5 (33, 34). All viruses were characterized previously with respect to their genomic structures and biological properties. Viral stocks were prepared as described previously (35).
Virion preparation.
Virions were purified from late-stage-infected HFF, using glycerol tartrate gradient ultracentrifugation, similar to the method described by Irmiere and Gibson (18). For this, 1.8 × 106 fibroblasts were initially seeded in one 175-cm2 culture flask and grown for 1 day. After that, the cells were infected with 1 ml of a frozen stock in such a way that all the cells showed a cytopathic effect (CPE) at 1 day of infection. At a time when the culture showed the CPE of late HCMV infection (usually at day 7 postinfection [p.i.]), the supernatant of the flask was collected and 1 ml was used to infect each of 10 to 20 175-cm2 flasks of HFF that were seeded 1 day before. After another 7 days of infection, the culture supernatants of these flasks were collected and centrifuged for 10 min at 1,900 × g to remove cells and debris. After that, the supernatant was collected and centrifuged at 95,000 × g (70 min; 10°C) in a SW32Ti rotor in a Beckman Optima L-90K ultracentrifuge. The pellets were resuspended with 2 ml of 1× phosphate-buffered saline (PBS). Na-tartrate gradients were prepared immediately before use. For this, 4 ml of a 35% Na-tartrate solution in 0.04 M Na-phosphate buffer, pH 7.4, was applied to one column, and 5 ml of a 15% Na-tartrate–30% glycerol solution in 0.04 M Na-phosphate buffer, pH 7.4, was applied to the second column of a gradient mixer. The gradients were prepared by slowly dropping the solutions into Beckman Ultra-clear centrifuge tubes (14 by 89 mm), positioned at an angle of 45°. One milliliter of the viral particles was then carefully layered on top of the gradients. Ultracentrifugation was performed without braking in a Beckman SW41 swing-out rotor for 60 min at 90,000 × g and 10°C. The particles were illuminated by light scattering and were collected from the gradient by penetrating the centrifuge tube with a hollow needle below the band. Samples were carefully drawn from the tube with a syringe. The particles for the initial analysis of RV-HB5, RV-SB2, and RV-VM1 virions, using a QTOF Premier mass spectrometer (Waters, Manchester, UK), were washed with 1× PBS and pelleted in an SW41 swing-out rotor for 90 min at 100,000 × g and 10°C. This procedure was repeated once. The particles for the later analyses (RV-HB5, RV-HB15, RV-BADwt, RV-Hd65, and RV-KB14) were washed and pelleted only once. After the last centrifugation step, the pellets were resuspended in 120 to 150 μl 1× PBS. The protein concentration of the purified virions was determined with the Pierce BCA Protein Assay Kit (Thermo Scientific, Bonn, Germany). Again, 20 μg virions were pelleted by ultracentrifugation for 60 min at 100,000 × g at 10°C and stored at −80°C for analysis by mass spectrometry (MS).
Polyacrylamide gel electrophoresis, silver staining, and immunoblot analysis.
SDS-PAGE was performed with 10% polyacrylamide gels containing 0.1% SDS. Proteins in the gel were stained using a silver-staining kit (Roti-Black P-Siberfärbungskit für Proteine; Carl Roth GmbH & Co. KG, Karlsruhe, Germany). Immunoblotting was carried out as described previously (25). Monoclonal antibodies (MAbs) directed against pp65 (65-33), against glycoprotein B (gB; 27-287) (36) (both kindly provided by W. Britt, University of Alabama, Birmingham, AL, USA), and against pUL35 (kindly provided by B. Biegalke, Ohio University, Athens, OH, USA) (37) were used for detection by the Amersham ECL Plus Western Blotting Reagents (GE Healthcare Europe GmbH, Freiburg, Germany).
Determination of the genome-to-infectivity ratio by TaqMan analysis.
Fibroblasts were infected with 100 genomes per cell of the respective viral supernatant. To remove attached viruses, the cells were washed 2 times for 30 s with PBS. The number of genomes in infected cells was measured at 0 h p.i. and 6 h p.i. by TaqMan PCR analysis, as described previously (35). The significance of the values was analyzed with a bidirectional t test.
Negative-staining electron microscopy.
Freshly prepared virions and DBs from glycerol-tartrate gradients were washed once with 1× PBS and centrifuged in an SW41 swing-out rotor for 90 min at 100,000 × g and 10°C. The pellets were resuspended in 100 to 500 μl PBS, and the suspension was used for determination of the protein concentration. Negative staining, using 5% (wt/vol) ammonium-heptamolybdate (pH 7.0), was performed by the single-droplet procedure, as described previously (38, 39). Transmission electron microscopy (TEM) was done with an FEI Tecnai 12 transmission electron microscope at an accelerating voltage of 120 kV and imaged with a TemCam F416.
Quantitative proteomics analysis. (i) Sample preparation and protein digestion.
Sample preparation and protein digestion of RV-HB15, RV-Hd65, RV-BADwt, RV-KB14, and RV-HB5 virions were performed as described in reference 32. In an independent experiment, purified RV-HB5, RV-SB2, and RV-VM1 virions were digested in the presence of 0.1% RapiGest (Waters) as described in reference 40.
(ii) Ultrahigh-performance LC (UPLC)-MS configuration.
Nanoscale liquid chromatography (LC) separation of tryptic peptides was performed with a nanoAcquity system (Waters) equipped with a BEH C18 1.7-μm, 75-μm by 150-mm analytical reversed-phase column (Waters) in direct-injection mode, as described previously (41). Mobile phases, gradients, and flow rates were chosen as described in reference 32. For RV-HB15, RV-Hd65, RV-BADwt, RV-KB14, and RV-HB5 virions, 0.2 μl of sample (50 ng of total protein) was injected per technical replicate. For RV-HB5, RV-SB2, and RV-VM1 virions, 2 μl of sample (500 ng of total protein) was injected per technical replicate. The running conditions were as described in reference 32.
(iii) Mass spectrometry analysis.
Mass spectrometric analysis of tryptic peptides from RV-HB15, RV-Hd65, RV-BADwt, RV-KB14, and RV-HB5 virions was performed in quintuplicate using a Synapt G2-S mass spectrometer (Waters) with a typical resolution of at least 25,000 full width half maximum (FWHM), as described previously (32). Mass spectrometric analysis of tryptic peptides from two independent preparations of RV-HB5, RV-SB2, and RV-VM1 virions was performed in quadruplicate using a QTOF Premier mass spectrometer (Waters) with a typical resolution of at least 10,000 FWHM, as described previously (41). All analyses were performed in positive-mode electrospray ionization (ESI) using instrument settings and nanoLockspray calibration, as described previously (32).
(iv) Data processing and protein identification.
Continuum LC-MS data were processed and searched using ProteinLynxGlobalSERVER (PLGS) version 2.5.2 (Waters). Protein identifications were obtained by searching a custom-compiled database containing sequences of human and HCMV proteins from the UniProt database. Alternatively, a custom-compiled database containing all translated ORFs derived from a ribosome profiling and transcript analysis of HCMV (42) was searched. Sequence information for enolase 1 (Saccharomyces cerevisiae) and bovine trypsin was added to the databases to normalize the data sets or to conduct absolute quantification, as described previously (43). A database search was performed, allowing a 3-ppm precursor and 10-ppm product ion tolerance (for Synapt G2-S data) and a maximal mass deviation of 15 ppm for precursor ions and 30 ppm for fragment ions (for QTOF Premier data), with one missed cleavage allowed and fixed carbamidomethylcysteine and variable methionine oxidation set as the modifications. For valid protein identification, the following criteria had to be met: at least two peptides detected, together having at least seven fragments. The false-positive rate for protein identification was set to 1%, based on a search of a triple-randomized database. Guideline identification criteria were applied for all searches.
(v) Absolute-quantification approach.
For the absolute quantification of virion proteins, we employed a well-established label-free quantitative-proteomics workflow that we have previously used both for the quantification of nanoparticle protein coronas (41) and for higher-complexity samples, such as myelin (43). Label-free quantification using the TOP3 approach allows both the relative and absolute quantification of proteins. Briefly, the average intensity of the three best ionizing peptides can be regarded as a measure of the absolute molar amount of each of the parent proteins present in the sample (44), using the quantification value of spiked yeast enolase 1 as a reference. Values were normalized across technical replicates and samples using ISOQuant (45). To facilitate the comparison of different virion proteomes, all values obtained were normalized to MCP.
RESULTS
Purification of HCMV virions for proteomic analysis.
The initial purpose of our study was to analyze how the availability of pp65 for packaging influenced the tegument composition of HCMV virions. For this purpose, several BAC-derived variants of the Ad169 strain were selected for proteomic analysis (Fig. 1A). They included three wild-type (wt) controls, RV-HB5 (29), RV-HB15 (30), and RV-BADwt (31); the two pp65neg strains RV-Hd65 and RV-KB14 (28, 32), which fail to produce subviral DBs; and additionally, two viral mutants, RV-SB2 and RV-VM1, in which pp65 was modified by insertion of a nonapeptide sequence at positions S101 and R387, respectively (33, 34). DB formation is also abrogated in HFF infected with RV-SB2 and RV-VM1 (33, 34). RV-VM1-infected HFF display wt pp65 steady-state protein levels (33), whereas RV-SB2 (a pp65low strain)-infected cells show a marked reduction in pp65 levels in relation to other virion proteins (Fig. 1B). Virions of all strains were purified by glycerol tartrate gradient centrifugation and subjected to SDS-PAGE (Fig. 1B). The protein patterns were comparable among the pp65-positive (pp65pos) strains and among the pp65neg strains with respect to the most abundant virion constituents. Subtle differences in the pattern were seen, especially with regard to less abundant proteins. A pronounced difference was found for the mutant RV-SB2. This virus grows to low titers in HFF culture. A larger volume of culture supernatant had to be used for purification to provide sufficient amounts of the material, likely enhancing the contamination with cellular proteins. For this reason, the virus was included only in the proteomic analysis of viral proteins but was excluded from the analyses of the cellular proteins in the virion preparations.
FIG 1.
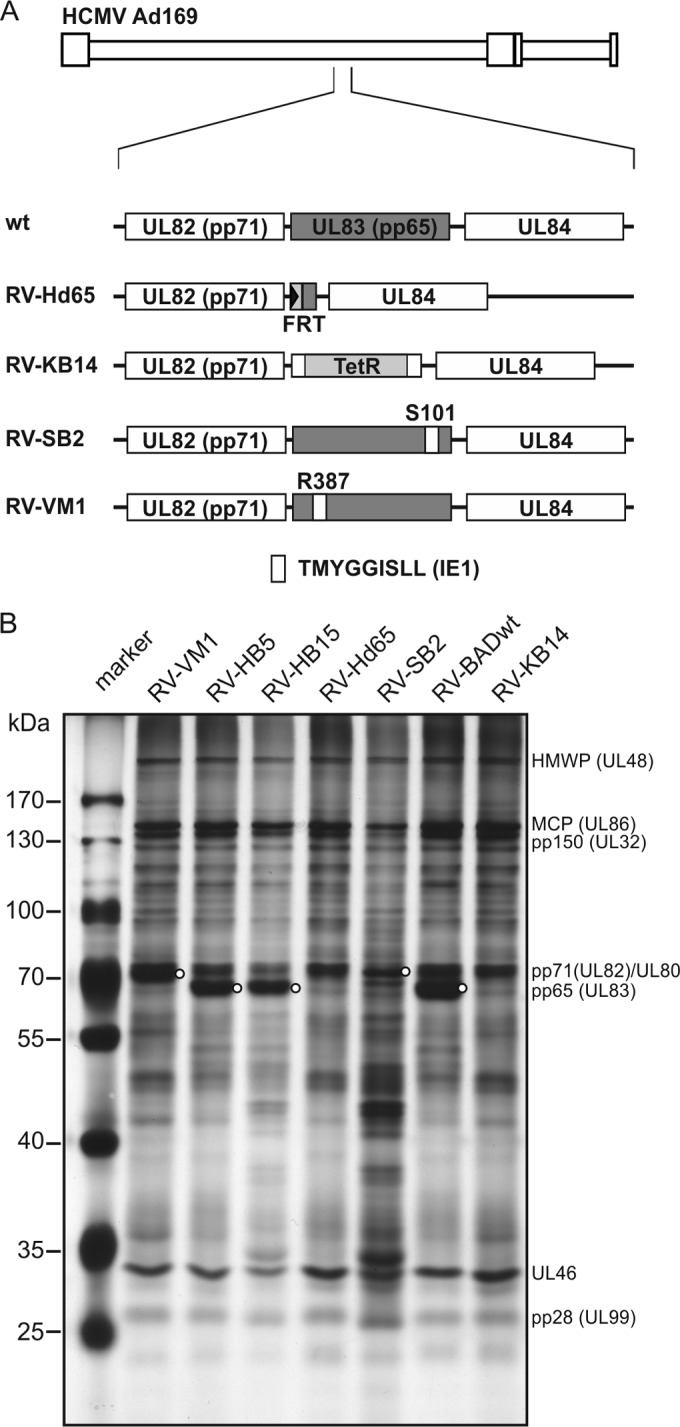
SDS-polyacrylamide gel electrophoresis of purified virions. (A) Schematic representation of the viral mutants used for analysis. The genomic structure of the AD169 strain of HCMV is shown at the top. The location of the genomic region encoding pp65, including the neighboring reading frames UL82 and UL84, is enlarged below. The wt or mutated configuration of pp65 in each strain is shown by the shaded boxes. wt, genomic structure of the wild-type strains RV-HB5, RV-HB15, and BADwt. TetR, tetracycline resistance gene; FRT, recognition site for the Flp recombinase. (B) SDS-polyacrylamide gel electrophoresis of purified virions of the indicated strains. Following electrophoresis, the gel was stained with silver nitrate. The molecular masses of proteins used as a standard are indicated. The locations of abundant viral proteins are tentatively indicated. pp65 in the gel is indicated by circles. The lower electrophoretic mobility of pp65 in lanes RV-VM1 and RV-SB2 is caused by the insertion of a nonapeptide sequence into the primary structure of the tegument protein in these viral mutants. Two micrograms of purified virions were loaded on each lane.
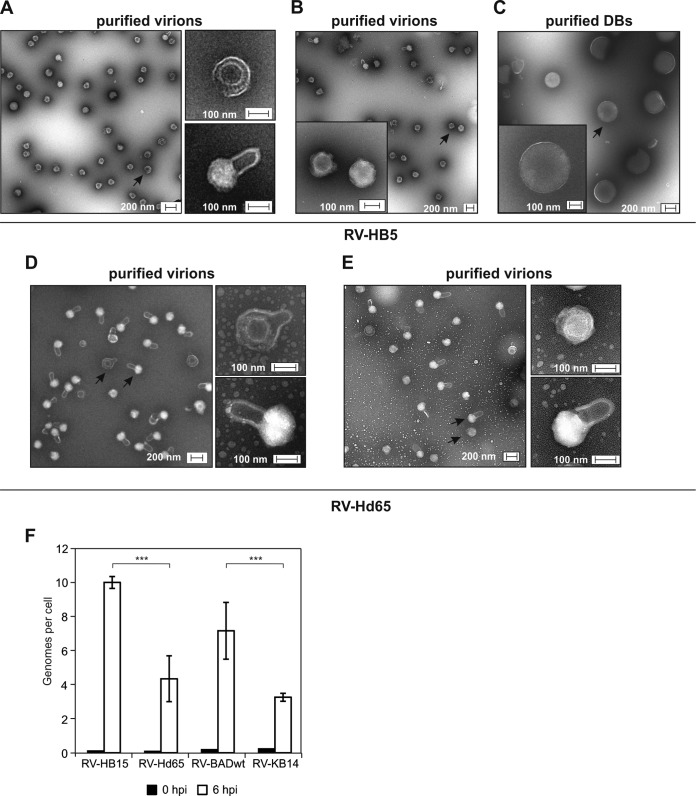
Alteration of virion morphology and genome delivery in the absence of pp65.
To control for the efficiency of the virion purification procedure, TEM was performed. Virion preparations of the wt strain RV-HB5 contained numerous particles corresponding in shape and size to HCMV virions (Fig. 2A). These particles were characterized by an electron-dense core, corresponding to the capsid; by a brighter halo, representing the tegument; and by an envelope (Fig. 2A, higher magnifications). There were also particles detectable in the preparations of RV-HB5 virions that showed a homogeneous central structure with an outer envelope attached (Fig. 2B). These particles were the size of virions, approximately 180 nm. Depending on the micrograph inspected, 0 to 10% of the particles showed such a bright internal structure. They were clearly distinct from the RV-HB5 DBs that were separated from the virions by the centrifugation protocol (Fig. 2C). These DBs were evenly spherical with a diameter of approximately 400 to 450 nm and a sharply defined outer edge. Preparations of RV-Hd65 also contained particles displaying the typical morphology of virions (Fig. 2D). However, in two independent biological replicates, the virion preparations of this pp65neg virus also contained particles that presented with single handle-like blebs (Fig. 2D and E). Although the nature of these structures is unknown, they likely represent disrupted viral envelopes. Similar structures were also found in virions of the parental strain, but at an overall frequency of roughly 5% (Fig. 2A). The samples shown in Fig. 2A and E were prepared in parallel, thus rendering artificial alterations due to different handling conditions of the specimens unlikely. Strikingly, the cores of these particles showed a bright appearance upon TEM (Fig. 2A, D, and E, higher magnifications). This indicates that the appearance of the blebs corresponded to changes in the staining of the particles by molybdate.
FIG 2.
TEM and infectivity of purified particles. (A and B) TEM micrographs of purified virions of RV-HB5. (C) TEM micrographs of purified DBs of RV-HB5 used for comparison. Higher magnifications of selected forms of particles, indicated by arrows in the lower-magnification micrographs, are presented on the right (A) or as insets (B and C). (D and E) TEM micrographs of two independent preparations of virions of RV-Hd65. Higher magnifications of selected forms of particles, indicated by arrows in the lower-magnification micrographs, are shown on the right. (F) Genome delivery into HFF by different viruses. Viral genomes in cells infected with the indicated viruses at 100 genomes per cell were calculated by TaqMan PCR at zero h p.i. and 6 h p.i. Two independent DNA preparations for each sample were performed and analyzed in triplicate. Significance was analyzed with a bidirectional t test, ***, highly significant results (two-tailed P value, <0.0001 for RV-HB15 and RV-Hd65 and <0.0002 for RV-BADwt and RV-KB14). The error bars indicate standard deviations.
To further address whether the differences in the morphologies of pp65pos and pp65neg virion preparations had an impact on the efficiency of infection, the capacities of the different strains for genome delivery were tested. Note that the quantification of cells expressing viral IE proteins could not be applied here as a measure for infection, as the strains differed in the virion content of the transactivator pp71 (32), which increases IE gene expression levels. Cells were infected with doses of the different viruses that were normalized to DNA copy numbers. Intracellular viral DNA was measured at 0 and 6 h p.i., using quantitative PCR. In accordance with the morphological changes seen in electron microscopy, both pp65neg strains showed a significant reduction in the capacity to translocate their genomes into fibroblasts (Fig. 2F). These results indicate that the lack of pp65 reduced the infectivity of the virions.
Mass spectrometry provides a reliable measure of individual HCMV virion proteins.
Using label-free quantitative mass spectrometry, a data set representing the proteomes of the purified virions of the different strains was generated. All proteins were quantified using the TOP3 quantification approach (44). The major capsid protein (MCP) was used for reference. This protein should be present in equimolar amounts in each virion particle. We therefore normalized all values to MCP in each sample (i.e., referring to the MCP values as 1) and used these data for subsequent comparative proteomic analysis.
To initially verify the accuracy of the method, we analyzed the data set for the relative amounts of the capsid constituents. At this point, it should be mentioned that the relative TOP3 signal intensity obtained by mass spectrometry of a given polypeptide depends on a number of parameters, such as the efficiency of tryptic digestion or the ionization efficiency of the peptides that can be measured. These parameters determine the ratio of the measured TOP3 intensity and the protein amount present in the sample (i.e., the relative instrument response, which is typically constant and specific for each individual protein). This allows direct comparison of the amounts measured for each individual protein in different samples (e.g., the minor capsid protein [mCP] in sample A against mCP in sample B). Accordingly, the mCP (pUL85), the minor capsid protein binding protein (mC-BP; pUL46), and the smallest capsid protein (SCP; pUL48A), as well as UL80 and the portal protein pUL104, were all found in amounts that were comparable between the different strains (Fig. 3). The detectability of the portal protein, pUL104, which is included in only 12 copies in the capsid, demonstrated the high level of sensitivity of our approach. The low variation in the relative frequencies of capsid proteins in individual samples and the high degree of technical reproducibility between individual LC-MS results underscored the accuracy of the method in comparing the relative amounts of individual proteins between strains (see Fig. S1 in the supplemental material). Overall, TOP3 quantification is considered to be directly proportional to protein abundance over the full quantification range and therefore is the preferred method in the absence of reference protein measurements (46).
FIG 3.
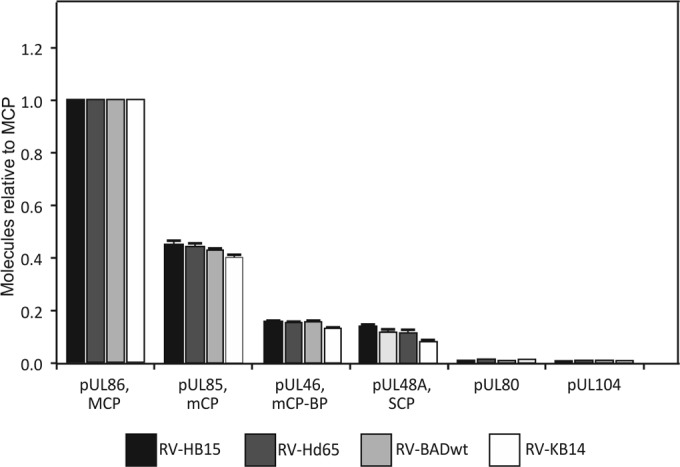
Mass spectrometry of capsid proteins contained in purified virions. The strains used for analyses are indicated. Each sample was analyzed in quintuplicate. Proteins were calculated in molar ratio relative to MCP (set as 1). The error bars represent deviations of the mean.
In contrast to calculations based on cryoelectron microscopy (11), a comparison of the content of one protein to that of another (e.g., the MCP to the mCP) in a given preparation proved to be less reliable in the mass spectrometry analysis performed in this study. This was likely due to protein-specific efficiencies of the tryptic digest, i.e., the number of peptides generated from an individual protein, and to different ionization efficiencies of individual peptides. Consequently, the theoretical relative values of the proteins in a capsid (i.e., 955 copies of the MCP, 640 copies of the mCP, 320 copies of the mC-BP, and 900 copies of the SCP [47, 48]) and the number of molecules calculated from the data set showed striking discrepancies (Fig. 3). This was seen, for instance, in the number of MCP and the SCP molecules, which should roughly match in one capsid (955 versus 900 copies). In contrast, the number of SCP molecules was measured to be less than 20% of the number of molecules found for MCP (Fig. 3). A closer inspection of the data set revealed that there was only one SCP peptide (of 4 peptides in total) that could reliably be detected in mass spectrometry, leading to a clear underestimation of the SCP content in the sample relative to MCP, which was identified by 86 peptides (Table 1). Taken together, our label-free quantitative-MS approach allowed an accurate comparison of the relative representation of a single protein in different virion preparations but was less reliable in comparing absolute amounts of different proteins within a single preparation.
TABLE 1.
Capsid and tegument proteins detected by nanoscale UPLC-MS in virions from pp65pos and pp65neg strainsa
| ORF | Synonym | Molecular wt | Maximum scoreb | No. of reported peptidesc | Copy no.d |
Relative copy no. ratioe |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| BADwt | KB14 | HB15 | Hd65 | KB14/BADwt | Hd65/HB15 | |||||
| Capsid proteins | ||||||||||
| UL86 | MCP | 155,298 | 73,485 | 84 | 1.000 | 1.000 | 1.000 | 1.000 | 1.00 | 1.00 |
| UL85 | mCP | 34,937 | 3,632 | 21 | 0.430 | 0.402 | 0.452 | 0.444 | 0.94 | 0.98 |
| UL46 | mCP-BP | 33,426 | 70,542 | 19 | 0.157 | 0.132 | 0.159 | 0.159 | 0.84 | 1.00 |
| UL48A | SCP | 8,538 | 69,236 | 4 | 0.119 | 0.085 | 0.143 | 0.122 | 0.71 | 0.85 |
| UL93 | CVC2 | 68,919 | 3,521 | 25 | 0.056 | 0.039 | 0.069 | 0.061 | 0.70 | 0.88 |
| UL77 | CVC1 | 71,643 | 12,522 | 27 | 0.019 | 0.013 | 0.024 | 0.019 | 0.68 | 0.79 |
| UL80 | PR | 74,365 | 114,690 | 13 | 0.011 | 0.013 | 0.011 | 0.015 | 1.18 | 1.36 |
| UL104 | PORT | 79,148 | 3,688 | 33 | 0.008 | 0.007 | 0.009 | 0.008 | 0.86 | 0.88 |
| UL52 | 75,661 | 738 | 8 | 0.004 | 0.005 | 0.005 | 0.006 | 1.25 | 1.20 | |
| Tegument proteins | ||||||||||
| UL83 | pp65 | 63,468 | 116,877 | 34 | 5.373 | 0.036 | 4.353 | 0.024 | 0.01 | 0.01 |
| UL82 | pp71 | 62,233 | 96,626 | 26 | 1.596 | 1.456 | 0.906 | 1.759 | 0.91 | 1.94 |
| UL32 | pp150 | 113,087 | 95,639 | 63 | 0.877 | 0.793 | 0.866 | 0.815 | 0.90 | 0.94 |
| UL94 | 39,864 | 42,351 | 13 | 0.629 | 0.445 | 0.557 | 0.478 | 0.51 | 0.86 | |
| UL25 | 74,281 | 46,163 | 37 | 0.499 | 0.026 | 0.304 | 0.031 | 0.05 | 0.10 | |
| UL43 | 48,445 | 16,921 | 29 | 0.310 | 0.003 | 0.313 | 0.004 | 0.01 | 0.01 | |
| UL45 | RR1 | 103,665 | 45,242 | 59 | 0.242 | 0.056 | 0.120 | 0.024 | 0.23 | 0.20 |
| UL48 | HMW-P | 254,877 | 30,652 | 106 | 0.187 | 0.147 | 0.229 | 0.151 | 0.79 | 0.66 |
| UL99 | pp28 | 21,208 | 39,446 | 8 | 0.156 | 0.095 | 0.188 | 0.123 | 0.61 | 0.65 |
| UL47 | HMW-BP | 110,662 | 43,659 | 53 | 0.144 | 0.115 | 0.159 | 0.111 | 0.80 | 0.70 |
| UL88 | 48,260 | 35,928 | 26 | 0.134 | 0.109 | 0.163 | 0.107 | 0.81 | 0.66 | |
| UL26 | 25,038 | 55,753 | 16 | 0.083 | 0.008 | 0.053 | 0.011 | 0.10 | 0.21 | |
| UL71 | 40,399 | 10,927 | 18 | 0.066 | 0.015 | 0.059 | 0.016 | 0.23 | 0.27 | |
| UL35 | 73,043 | 29,556 | 34 | 0.047 | 0.080 | 0.032 | 0.072 | 1.70 | 2.25 | |
| TRS1 | 84,723 | 11,451 | 29 | 0.045 | 0.040 | 0.012 | 0.013 | 0.88 | 1.08 | |
| UL97 | 79,944 | 12,626 | 31 | 0.037 | 0.016 | 0.029 | 0.015 | 0.43 | 0.52 | |
| IRS1 | 91,847 | 10,492 | 31 | 0.034 | 0.023 | 0.106 | 0.007 | 0.68 | 0.07 | |
| UL24 | 34,816 | 6,300 | 15 | 0.013 | 0.004 | 0.030 | 0.005 | 0.31 | 0.17 | |
| UL103 | 29,092 | 6,979 | 9 | 0.011 | 0.004 | 0.011 | 0.006 | 0.36 | 0.55 | |
| US22 | 656,057 | 3,265 | 20 | 0.010 | 0.005 | 0.009 | 0.004 | 0.50 | 0.44 | |
| UL69 | 83,477 | 1,247 | 11 | 0.006 | 0.001 | 0.004 | 0.001 | 0.16 | 0.25 | |
| UL50 | 43,757 | 380 | 5 | 0.002 | 0.001 | 0.002 | 0.001 | 0.50 | 0.50 | |
Classification according to reference 52.
Maximum PLGS identification score; Tandem MS Search Algorithm (Waters).
Number of peptides detected for each protein.
Calculated copy numbers of molecules per MCP molecule.
Ratios of the relative copy numbers of individual proteins (pp65neg versus pp65pos strains). The proteins are listed according to decreasing frequencies in virions of RV-BADwt. Ratios of more than 2.0 and less than 0.5 are indicated by boldface.
Deletion of pp65 has limited impact on the packaging of the tegument proteins pp150, pUL48, pUL47, and pp28.
To address the question of whether one or more of the HCMV tegument proteins were complementing a putative structural function of pp65 in the virion, the amounts of all tegument proteins were pairwise evaluated and calculated relative to that of MCP. The amounts of individual tegument proteins were found to be either unaffected, increased, or decreased, depending on pp65 (Table 2).
TABLE 2.
Glycoproteins and proteins without virion assignment detected by nanoscale UPLC-MS in virions from pp65pos and pp65neg strainsa
| ORF | Synonym | Molecular wt | Maximum scoreb | No. of reported peptidesc | Copy no.d |
Relative copy no. ratioe |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| BADwt | KB14 | HB15 | Hd65 | HB15/Hd65 | BADwt/KB14 | |||||
| Glycoproteins | ||||||||||
| UL55 | gB | 102,004 | 66,727 | 55 | 0.450 | 0.311 | 0.429 | 0.436 | 1.02 | 0.69 |
| UL100 | gM | 42,861 | 13,786 | 8 | 0.301 | 0.197 | 0.291 | 0.216 | 0.74 | 0.66 |
| UL73 | gN | 15,096 | 6,470 | 2 | 0.170 | 0.085 | 0.170 | 0.110 | 1.55 | 2.00 |
| RL10 | 19,033 | 26,157 | 9 | 0.166 | 0.138 | 0.143 | 0.095 | 0.66 | 0.83 | |
| UL75 | gH | 84,452 | 24,272 | 25 | 0.131 | 0.086 | 0.098 | 0.095 | 0.97 | 0.66 |
| US27 | 41,994 | 18,769 | 10 | 0.051 | 0.037 | 0.060 | 0.032 | 0.53 | 0.73 | |
| UL132 | 29,972 | 16,423 | 12 | 0.046 | 0.018 | 0.062 | 0.023 | 0.37 | 0.39 | |
| UL115 | gL | 30,831 | 17,991 | 11 | 0.039 | 0.025 | 0.029 | 0.029 | 1.00 | 0.64 |
| UL119/118 | 38,709 | 2,862 | 6 | 0.025 | 0.017 | 0.013 | 0.014 | 1.08 | 0.68 | |
| UL74 | gO | 54,577 | 4,629 | 14 | 0.023 | 0.013 | 0.014 | 0.016 | 0.88 | 1.77 |
| UL41A | 8,953 | 8,978 | 2 | 0.021 | 0.009 | 0.024 | 0.010 | 0.42 | 0.43 | |
| UL33 | 46,295 | 7,163 | 8 | 0.020 | 0.014 | 0.044 | 0.030 | 0.68 | 0.70 | |
| UL116 | 34,139 | 3,132 | 4 | 0.015 | 0.009 | 0.017 | 0.013 | 0.77 | 0.60 | |
| IR11 | 26,660 | 448 | 2 | 0.001 | 0.001 | 0.002 | 0.001 | 0.50 | 1.00 | |
| Proteins without virion assignment | ||||||||||
| UL44 | 46,233 | 9,752 | 19 | 0.012 | 0.020 | 0.031 | 0.017 | 0.55 | 1.66 | |
| UL122 | 62,852 | 878 | 11 | 0.003 | 0.003 | 0.006 | 0.003 | 0.50 | 1.00 | |
| UL84 | 65,428 | 753 | 16 | 0.003 | 0.004 | 0.003 | 0.003 | 1.00 | 1.33 | |
| UL98 | 65,204 | 1,298 | 11 | 0.002 | 0.002 | 0.003 | 0.001 | 0.33 | 1.00 | |
| UL112 | 70,112 | 398 | 4 | 0.002 | 0.002 | 0.002 | 0.001 | 0.50 | 1.00 | |
| UL114 | 28,369 | 567 | 2 | 0.000 | 0.000 | 0.001 | 0.001 | 1.00 | ||
Classification according to reference 52.
Maximum PLGS identification score; Tandem MS Search Algorithm (Waters).
Number of peptides detected for each protein.
Calculated copy numbers of molecules per MCP molecule.
Ratios of the relative copy numbers of individual proteins (pp65neg versus pp65pos strains). The proteins are listed according to decreasing frequencies in virions of RV-BADwt. Ratios of more than 2.0 and less than 0.5 are indicated by boldface.
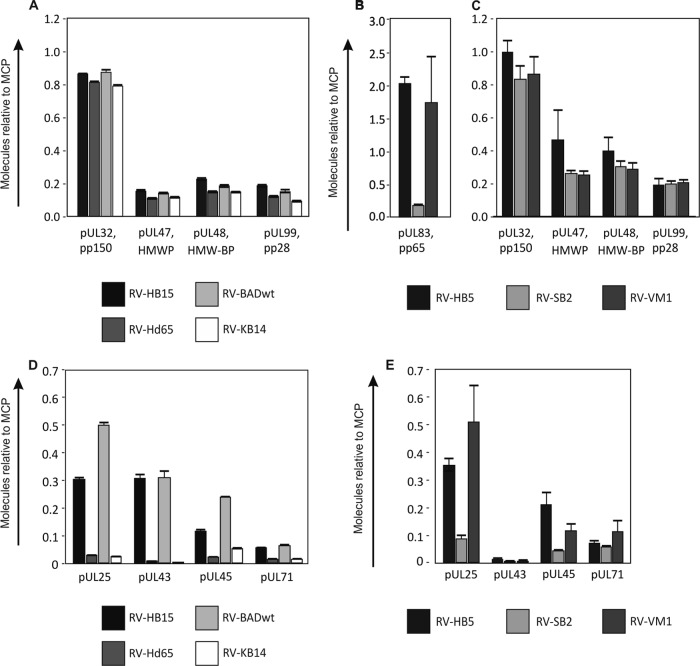
When the data obtained from the wt strains RV-HB15 and RV-BADwt were compared with those of their respective pp65 deletion mutants, the amounts of pp28 (pUL99) and the inner tegument proteins pp150, pUL47, and pUL48 were comparable (Fig. 4A). This suggested that packaging of these proteins was independent of pp65. There were, however, slightly smaller relative amounts of each tegument protein found in the strains with pp65 deleted versus the parental strains. To determine if this was truly an effect of the pp65 loss or due to contamination with DBs, HCMV strains RV-SB2 and RV-VM1 were analyzed. Both strains fail to produce DBs (33, 34). RV-SB2 is also deficient in pp65 packaging into virions, whereas RV-VM1 virions contain normal amounts of the tegument protein (Fig. 4B). The levels of pp150, pUL47, pUL48, and pp28 were comparable between RV-SB2 and RV-VM1 virions (Fig. 4C). These results indicated that the four proteins were neither complementing the pp65 loss nor were they dependent on pp65 for their inclusion during assembly.
FIG 4.
Mass spectrometry of tegument proteins contained in purified virions. The strains used for analyses are indicated. The virus strains RV-HB15, RV-Hd65, RV-BADwt, and RV-KB14 were analyzed in quintuplicate on a Synapt G2-S mass spectrometer. Each of the virus strains RV-HB5, RV-SB2, and RV-VM1 was independently prepared twice and analyzed in quadruplicate on a QTOF Premier mass spectrometer. Proteins were calculated in molar ratio relative to MCP (set as 1). The error bars represent deviations of the mean. (A) Inner tegument proteins contained in pp65neg and pp65pos strains. (B) pp65 contained in virions of a pp65pos strain and pp65low strains. (C) Inner tegument proteins contained in pp65low and pp65pos strains. (D) Outer tegument proteins contained in pp65pos and pp65neg strains. (E) Outer tegument proteins contained in pp65pos and pp65low strains.
Several proteins, including pUL43, pUL45, and pUL71, are significantly reduced in the tegument of pp65-negative viruses.
Next, the data were evaluated to identify proteins with loss of packaging in pp65neg or pp65low virions. The packaging of pUL25 was found to be dependent on pp65 (Fig. 4D), as had been reported by others using immunoblot analyses (26). This confirmed the accuracy of our analysis. Striking reductions were also found for pUL43, pUL45, and pUL71, suggesting that inclusion of these proteins was impaired in the absence of pp65 (Fig. 4D and 5). The results were confirmed by analyzing the data sets from RV-SB2 and RV-VM1 (Fig. 4E), although pUL43 was found at much lower levels in this experiment.
FIG 5.
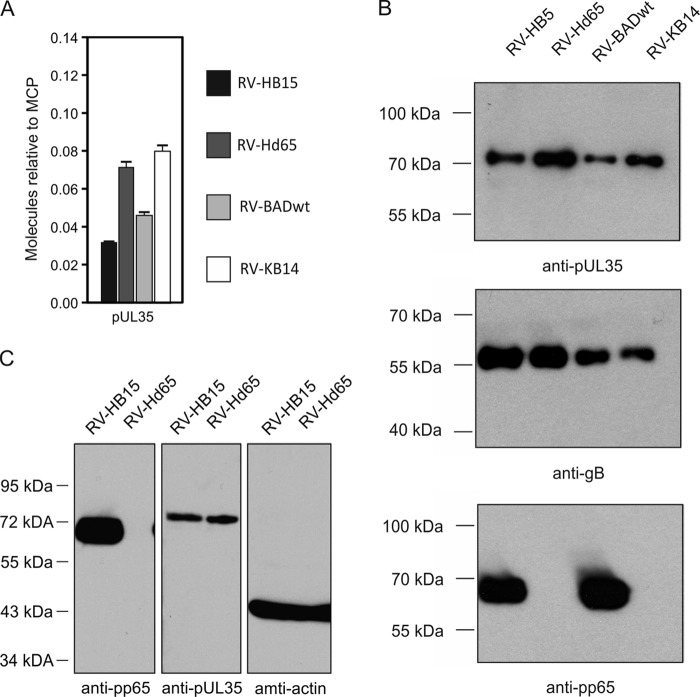
Analysis of pUL35 packaging, (A) Mass spectrometry of purified virions from pp65pos and pp65neg strains. The strains used for analyses are indicated. The virus strains were analyzed in quintuplicate on a Synapt G2-S mass spectrometer. pUL35 was calculated in molar ratio relative to MCP (set as 1). The error bars represent deviations of the mean. (B) Enhanced chemiluminescence (ECL) immunoblot analysis of purified virions from pp65pos and pp65neg HCMV strains. The antibodies used for detection are indicated. The exposure times for X-ray films were 3 s for pp65 and for gB and 1 min for pUL35. (C) ECL immunoblot analyses of fibroblasts at 4 days after infection with RV-HB15 or RV-Hd65 (multiplicity of infection [MOI], 1). A total of 105 infected cells were applied to each lane. The antibodies used for detection are indicated.
Besides these four proteins, other tegument proteins were also found to be reduced. Some of these, like pUL88 or pUL94, were found in considerable amounts in virions, but the rate of reduction was moderate (Table 1). Others were found in only small amounts, like pUL24 or pUL26, yet pp65-dependent reduction of these proteins appeared to be severalfold. Taken together, our data suggest that the loss of pp65 leads to impairment of the packaging of several of the known tegument-associated proteins, indicating that their association with virions was supported by pp65.
Only pUL35 is preferentially packaged into pp65neg and pp65low virions.
The data set was next analyzed for proteins that were increased in pp65neg virions. pUL35 was found at roughly 2-fold-higher frequencies in the pp65neg virions of RV-Hd65 and RV-KB14 than in virions of the parental strains (Fig. 5A). An immunoblot analysis was used to corroborate these findings (Fig. 5B). The increased packaging of pUL35 was independent of the steady-state levels of pUL35 in infected cells, which were comparable between the strains (Fig. 5C). In addition to pUL35, pp71 (pUL82) was found at an elevated level, but only in the virions of RV-Hd65 and not in those from RV-KB14 (Table 1). In a recent publication, we were able to show that enhanced packaging of pp71 into virions was due to pp71 overexpression in RV-Hd65-infected cells (32). Thus, it appears that pUL35 is the only virion protein that is more efficiently packaged into pp65neg virions. However, the increase in pUL35 appeared unsuitable to replace any structural function of pp65 in HCMV virions. Single pp65neg virions contain roughly 40 molecules of pUL35 in excess compared to pp65pos virions, leading to a gain in mass of 0.03 MDa (Table 1). A single virion of a pp65pos strain, however, contains a calculated number of 4,500 to 5,000 pp65 molecules (Table 1). The lack of pp65 consequently results in a loss of protein mass of an individual virion of roughly 286 to 317 MDa. Thus, the gain in pUL35 mass by no means matches the pp65 loss. However, replacement of a putative regulatory function of pp65 by pUL35 cannot be formally excluded. Taken together the data suggest that neither a single viral protein nor a set of viral proteins replaces pp65 as a structural component of HCMV virions in pp65neg strains.
Viral envelope glycoproteins and proteins without assignment to virions.
Although this study did not particularly focus on analyzing viral glycoproteins, some findings nevertheless merit mention. All major viral envelope glycoproteins that had been reported previously as components of the HCMV virion were also detectable in this analysis (49). Furthermore, only limited variations were seen in the different preparations regarding gB, gH, gL, gO, and gM, indicating that pp65 was not essential for the incorporation of any of these glycoproteins into virions. Contrary to a previous report (17), gM was consistently found in smaller amounts than gB in all virion preparations. An analysis optimized for the detection of membrane-associated proteins is warranted to resolve the discrepancy between our results and those of Varnum and colleagues (17). Finally, gO was consistently found in these preparations, confirming that the protein is an integral constituent of extracellular HCMV particles (50, 51). Taken together, these results suggest that pp65 has no dramatic impact on the incorporation of viral glycoproteins into particles.
A few proteins without assignment to virions (52) were also found in the virion preparations (Table 2). Except for UL98 and UL114, these proteins were also found in a previous proteomic analysis by Varnum and colleagues, using different methodologies (17). This argues in favor of a regular association of these proteins with HCMV virions. UL98 and UL114 were found at low frequencies, and further analyses must be performed with respect to the significance of their association with HCMV virions.
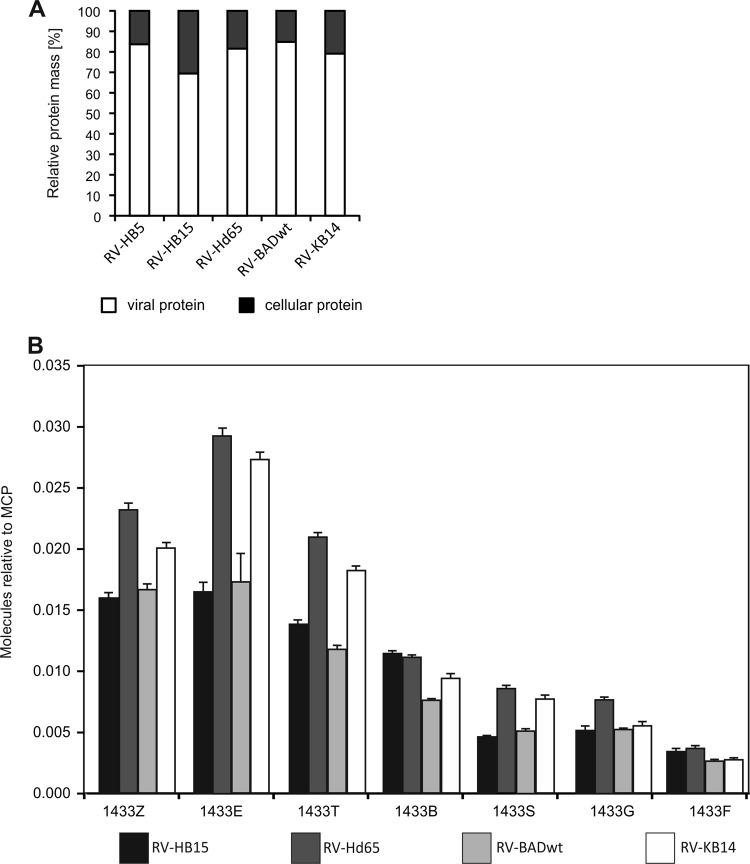
Deletion of pp65 leads to a preferential reduction of cytoskeletal proteins and heat shock proteins in virion preparations.
Cellular proteins have been found to be associated with purified HCMV virions (17, 49). We consequently evaluated how pp65 deletion would alter the pattern of virion-associated cell proteins. The fraction of cellular proteins in the total protein mass of the samples was roughly 20%, except for RV-HB15, where host cell-derived proteins added up to 30% (Fig. 6A). The likely explanation for this discrepancy is a higher degree of contaminating host cell proteins in RV-HB15 virion preparations. This was considered in the subsequent analyses of the data, where RV-HB5, the parental strain of both RV-Hd65 and RV-HB15, was used as a reference for RV-Hd65 instead of RV-HB15 (Table 3). Again, for comparison of the different virion preparations, the number of MCP molecules was normalized to 1 and the relative abundances of cellular proteins were determined, using this capsid protein as reference. The differences between pp65pos and pp65neg strains were calculated and are provided in Table 3.
FIG 6.
Mass spectrometry of cellular proteins in relation to viral proteins and of 14-3-3 proteins. (A) Relative masses of total viral and cellular proteins contained in pp65pos and pp65neg virions. (B) Amounts of 14-3-3 isoforms relative to MCP contained in each virion preparation. The error bars represent deviations of the mean.
TABLE 3.
Fifty most abundant cellular proteins detected by nanoscale UPLC-MS in virion preparations from pp65pos and pp65neg strains
| Description | Maximum scorea | No. of reported peptidesb | Copy no.c |
Relative copy no. ratiod |
||||
|---|---|---|---|---|---|---|---|---|
| BADwt | KB14 | HB5 | Hd65 | Hd65/HB5 | KB14/BADwt | |||
| Keratin type II cytoskeletal 2 epidermal (KRT2) | 33,702 | 38 | 0.1717 | 0.0095 | 0.0052 | 0.0033 | 0.64 | 0.06 |
| Myosin 9 (MYH9) | 1,605 | 59 | 0.1505 | 0.1121 | 0.1075 | 0.1485 | 1.38 | 0.75 |
| Keratin type I cytoskeletal 10 (KRT10) | 38,102 | 30 | 0.1501 | 0.0191 | 0.0066 | 0.0057 | 0.86 | 0.13 |
| Actin cytoplasmic 1 (ACTB) | 48,610 | 23 | 0.0933 | 0.2005 | 0.1176 | 0.0719 | 0.61 | 2.15 |
| Heat shock cognate 71-kDa protein (HSPA8) | 31,490 | 35 | 0.0861 | 0.0591 | 0.0574 | 0.0394 | 0.69 | 0.69 |
| Keratin type II cytoskeletal 1 (KRT1) | 23,636 | 35 | 0.0702 | 0.0310 | 0.0247 | 0.0097 | 0.39 | 0.44 |
| Elongation factor 1 alpha 1 (EEF1A1) | 17,000 | 20 | 0.0662 | 0.0975 | 0.0576 | 0.0460 | 0.80 | 1.47 |
| Serum albumin (ALB) | 7,336 | 21 | 0.0532 | 0.0411 | 0.0500 | 0.0288 | 0.58 | 0.77 |
| Glyceraldehyde 3 phosphate dehydrogenase (GAPDH) | 33,212 | 21 | 0.0526 | 0.1042 | 0.0741 | 0.0385 | 0.52 | 1.98 |
| Keratin type I cytoskeletal 17 (KRT17) | 3,539 | 20 | 0.0482 | 0.0298 | 0.0345 | 0.0530 | 1.54 | 1.44 |
| Peptidyl prolyl cis trans isomerase A (PPIA) | 30,655 | 12 | 0.0454 | 0.0338 | 0.0258 | 0.0344 | 1.33 | 0.75 |
| 26S protease regulatory subunit 6A (PSMC30) | 1,023 | 5 | 0.0392 | 0.0249 | 0.0191 | 0.0742 | 3.89 | 0.64 |
| Ras-related protein Rab 43 (RAB43) | 2,637 | 4 | 0.0356 | 0.0076 | 0.0262 | 0.0096 | 0.37 | 0.21 |
| Thioredoxin (TXN) | 30,652 | 6 | 0.0304 | 0.0257 | 0.0195 | 0.0253 | 1.30 | 0.85 |
| Annexin A2 (ANXA2) | 18,059 | 29 | 0.0289 | 0.0270 | 0.0266 | 0.0205 | 0.77 | 0.93 |
| Putative heat shock protein HSP 90 beta 4 (HSP90AB4P) | 2,807 | 8 | 0.0239 | 0.0002 | 0.0094 | 0.0003 | 0.03 | 0.008 |
| Keratin type II cytoskeletal 1b (KRT77) | 3,266 | 14 | 0.0235 | 0.0067 | 0.0132 | 0.0115 | 0.87 | 0.29 |
| Profilin 1 (PFN1) | 24,876 | 8 | 0.0234 | 0.0260 | 0.0212 | 0.0174 | 0.82 | 1.11 |
| Actin gamma enteric smooth muscle (ACTG2) | 22,060 | 16 | 0.0227 | 0.0232 | 0.0138 | 0.0149 | 1.08 | 1.02 |
| Phosphatidylinositol 4 phosphate 3 kinase C2 domain-containing subunit alpha (PIK3) | 796 | 48 | 0.0194 | 0.0173 | 0.0154 | 0.0228 | 1.48 | 0.89 |
| E3 ubiquitin protein ligase TRIM63 (TRIM63) | 889 | 5 | 0.0193 | 0.0174 | 0.0146 | 0.0152 | 1.04 | 0.90 |
| Keratin type I cytoskeletal 19 (KRT19) | 4,111 | 12 | 0.0193 | 0.0055 | 0.0171 | 0.0091 | 0.53 | 0.29 |
| Peripherin (PRPH) | 938 | 7 | 0.0176 | 0.0133 | 0.0120 | 0.0316 | 2.63 | 0.76 |
| E3 SUMO protein ligase RanBP2 (RANBP2) | 969 | 51 | 0.0175 | 0.0031 | 0.0187 | 0.0055 | 0.24 | 0.18 |
| 14-3-3 protein epsilon (YWHAE) | 28,144 | 20 | 0.0173 | 0.0273 | 0.0109 | 0.0293 | 2.69 | 1.58 |
| Heat shock 70-kDa protein 1A 1B (HSPA1A) | 13,281 | 31 | 0.0171 | 0.0116 | 0.0113 | 0.0091 | 0.80 | 0.68 |
| 14-3-3 protein zeta delta (YWHAZ) | 23,199 | 17 | 0.0166 | 0.0201 | 0.0113 | 0.0232 | 2.05 | 1.20 |
| Galectin 1 (LGALS1) | 32,764 | 9 | 0.0160 | 0.0248 | 0.0178 | 0.0131 | 0.74 | 1.55 |
| Alpha enolase (ENO1) | 13,422 | 24 | 0.0155 | 0.0182 | 0.0150 | 0.0086 | 0.57 | 1.17 |
| Cofilin 1 (CFL1) | 19,199 | 13 | 0.0149 | 0.0181 | 0.0155 | 0.0089 | 0.57 | 1.22 |
| Tubulin beta chain (TUBB) | 25,232 | 22 | 0.0145 | 0.0250 | 0.0125 | 0.0098 | 0.78 | 1.74 |
| Keratin type I cuticular Ha6 (KRT36) | 1,115 | 9 | 0.0145 | 0.0009 | 0.0068 | 0.0011 | 0.16 | 0.06 |
| Complement decay-accelerating factor (CD55) | 3,887 | 15 | 0.0143 | 0.0084 | 0.0123 | 0.0103 | 0.84 | 0.59 |
| Keratin type I cytoskeletal 28 (KRT28) | 2,847 | 13 | 0.0140 | 0.0012 | 0.0119 | 0.0028 | 0.24 | 0.09 |
| ADP ribosylation factor 1 (ARF1) | 17,453 | 11 | 0.0140 | 0.0082 | 0.0139 | 0.0098 | 0.71 | 0.59 |
| Pyruvate kinase isozymes M1 M2 (PKM2) | 14,429 | 32 | 0.0132 | 0.0214 | 0.0182 | 0.0099 | 0.54 | 1.60 |
| CD81 antigen (CD81) | 5,301 | 4 | 0.0121 | 0.0088 | 0.0095 | 0.0119 | 1.25 | 0.72 |
| Cell division control protein 42 homolog (CDC42) | 4,853 | 6 | 0.0119 | 0.0035 | 0.0084 | 0.0032 | 0.38 | 0.29 |
| Tubulin beta 8 chain B OS Homo sapiens | 1,482 | 8 | 0.0118 | 0.0003 | 0.0071 | 0.0002 | 0.03 | 0.03 |
| 14 3 3 protein theta (YWHAQ) | 17,722 | 16 | 0.0118 | 0.0183 | 0.0087 | 0.0210 | 2.97 | 1.55 |
| Elongation factor 2 (EEF2) | 7,846 | 38 | 0.0117 | 0.0129 | 0.0126 | 0.0075 | 0.59 | 1.10 |
| Staphylococcal nuclease domain-containing protein 1 (SND1) | 934 | 31 | 0.0116 | 0.0040 | 0.0050 | 0.0029 | 0.58 | 0.35 |
| Tubulin alpha 1A chain (TUBA1A) | 19,169 | 24 | 0.0116 | 0.0213 | 0.0141 | 0.0094 | 0.66 | 1.84 |
| Tubulin alpha 1C chain (TUBA1C) | 19,614 | 23 | 0.0115 | 0.0272 | 0.0110 | 0.0074 | 0.67 | 2.37 |
| Tryptophanyl tRNA synthetase cytoplasmic (WARS) | 453 | 7 | 0.0111 | 0.0108 | 0.0123 | 0.0090 | 0.73 | 0.97 |
| Lactadherin (MFGE8) | 6,952 | 21 | 0.0108 | 0.0064 | 0.0071 | 0.0079 | 1.11 | 0.59 |
| Probable phosphoglycerate mutase 4 (PGAM4) | 1,867 | 4 | 0.0106 | 0.0126 | 0.0031 | 0.0112 | 3.61 | 1.19 |
| Keratin type II cytoskeletal 5 (KRT5) | 8,085 | 24 | 0.0105 | 0.0041 | 0.0044 | 0.0078 | 1.77 | 0.39 |
| Keratin type II cytoskeletal 2 oral (KRT76) | 7,435 | 9 | 0.0103 | 0.0009 | 0.0011 | 0.0003 | 0.27 | 0.09 |
| Protein S100 A6 (S100A6) | 3,706 | 3 | 0.0101 | 0.0101 | 0.0103 | 0.0073 | 0.71 | 1.00 |
Maximum PLGS identification score; Tandem MS Search Algorithm (Waters).
Number of peptides detected for each protein.
Calculated copy numbers of molecules per MCP molecule.
Ratios of the relative copy numbers of individual proteins (pp65neg versus pp65pos strains). The proteins are listed according to decreasing frequencies in virions of RV-BADwt. Ratios of more than 2.0 and less than 0.5 are indicated by boldface.
Looking at the 50 most abundant cellular proteins, a group that appeared to be consistently elevated in preparations of pp65neg viruses consisted of different isoforms of the 14-3-3 protein family (53) (Table 3 and Fig. 6B). The difference between wt virions and pp65neg virions was insufficient to render these proteins candidates that would compensate for a putative structural function of pp65. More than 2-fold-elevated levels were also found for the 26S protease regulatory subunit 6A (PSMC3), for glyceraldehyde 3 phosphate dehydrogenase, for peripherin (PRPH), and for the probable phosphoglycerate mutase 4 (PGAM4). However, differences for these proteins were found for only one of the two virus pairs tested, thus challenging the biological significance of these results with respect to pp65 deletion.
Surprisingly, a number of cellular proteins were found to be reduced in frequency in pp65neg virions (Table 3). Most strikingly, a number of cytoskeleton proteins and heat shock proteins were found at lower levels when pp65 was absent. Other proteins that were consistently found at lower frequencies in both pp65neg strains were the Ras-related protein RAB43, the E3 SUMO protein ligase RanBP2 (RANBP2), and the cell division control protein 42 homolog (CDC42). In contrast, many of the cellular proteins were found at similar frequencies in virion preparations of pp65pos and pp65neg viruses, and most of them had been reported in previous analyses (17, 49, 54–56). A few novel proteins, including phosphatidylinositol 4 phosphate kinase (PIK3), however, were found in both pp65pos and pp65neg strains. In conclusion, these results indicated that only a subset of the HCMV virion-associated cellular proteins were influenced by pp65 loss, and most of those were found at reduced molar ratios in the preparations (for a comprehensive representation of the complete data set, see Tables S1 to S4 in the supplemental material).
ORFL86W is detectable in HCMV virions.
Using ribosome profiling and transcript analysis, Stern-Ginossar and colleagues recently identified hundreds of novel HCMV ORFs (42). To test if proteins encoded by these ORFs were included in HCMV virions, we reanalyzed the data sets from our proteomic analysis by searching against a database that was generated based on the data from their publication. Interestingly, we were able to significantly identify peptides from the translation product of only one of these ORFs, ORFL86W. ORFL86W is positioned in the genomic region between UL30 and UL31 of HCMV and codes for a putative protein of 159 amino acids. In our analysis, we identified three tryptic peptides from ORFL86W, which were present in only relatively small amounts. These peptides were, however, detectable in all wt strains and the two pp65neg strains, arguing in favor of a specific association of the proteins with HCMV virions.
DISCUSSION
Roughly 15% of the total HCMV virion mass is contributed by pp65 (17). Deletion of UL83 in pp65neg strains, however, has little impact on the replication of the mutants in fibroblasts or on the morphology of intracellular virions (25). Here, we provide the first quantitative proteomic analysis to address how the availability of pp65 for virion assembly affects the morphologies and the protein compositions of extracellular HCMV virions.
Impact of pp65 deletion on virion morphology and structural integrity.
Roughly 50% of all RV-Hd65 virions presented with handle-like blebs upon TEM. These structures were not unique to the strain, as they were also apparent, at low frequency, in virion preparations of the p65pos parental strain. Although no formal proof is available, these blebs may represent virion envelopes that partially detached from the tegument layer. The higher magnification of an RV-Hd65 virion in Fig. 2E may support this notion. The virions with blebs showed bright staining of the core. This indicates that alteration of the virion structure, likely with regard to membrane continuity, leads to changes in the staining of particles with ammonium-heptamolybdate in the process of preparing samples for TEM. We cannot exclude the possibility, however, that the bright appearance of the cores was also influenced by the different protein contents in the virions of the two strains.
The pp65neg virions were also impaired in genome delivery into cells. This suggests that pp65 deficiency alters virion morphogenesis in such a way that tegument-envelope interaction is disturbed, leading to reduced infectivity. Interestingly, however, the extracellular stability of the particles was not compromised when pp65 was lacking (data not shown). Further analyses are required to investigate if pp65 is involved in the interaction of the tegument layer with the viral envelope.
Malouli and colleagues recently reported on a proteomic analysis of virions from a mutant of the rhesus CMV (RhCMV). The virus had the two genes that share homology with HCMV pp65 (pp65ab) deleted (57). Concordant with our results, little impact of the pp65ab deletion on the overall protein composition of virions was found. A slight decrease in all tegument proteins was detected, confirming the trend that was seen in HCMV virions (Fig. 4 and Table 1). However, in contrast, our analysis revealed that pp65neg HCMV virions display substantial loss of a selected set of tegument proteins. This indicates that pp65 of HCMV and pp65ab of RhCMV differ in their interactions with other tegument proteins. Given a level of conservation between these proteins of 35 to 40%, it may be that only some functions of HCMV pp65 are represented in pp65ab of RhCMV, and other proteins of that virus may selectively interfere with tegument assembly.
Incorporation of cellular proteins.
Roughly 420 different cellular proteins were found in HCMV virions (listed in Table 3 and Tables S1 to S4 in the supplemental material). Those that were consistently elevated in virions of both pp65neg strains were 14-3-3 family members. Individual molecules from that family had been described before as constituents of virion preparations from other herpesviruses (17, 58–61). These proteins are known to be involved in the regulation of signal transduction pathways by modulating the activities of client proteins via interaction with phosphoserine or phosphothreonine. Interestingly, Stegen and colleagues recently showed in a small interfering RNA (siRNA) screen that one of the 14-3-3 family members, 14-3-3 zeta, contained in virions, significantly supported HSV-1 production in cell culture (62). It remains to be determined if 14-3-3 proteins also influence HCMV infection.
A number of cell proteins were consistently underrepresented in pp65neg virions. Cytoskeletal proteins and heat shock proteins were most prominent in this respect. Both have been found in virion preparations of a number of other herpesviruses (58–61, 63–65). The possibility that these proteins represent cellular contaminants of the particle preparations cannot be formally excluded at this point. However, the differences seen between the pp65neg and pp65pos strains argue in favor of pp65-dependent specific inclusion of these proteins in the process of particle morphogenesis.
The majority of the cellular proteins found in the different virion preparations have been described previously as associated with HCMV or murine CMV (MCMV) virions (17, 66) or with the virions of other herpesviruses (reviewed in reference 65). Among those that were not previously identified were the alpha subunit of the phosphatidylinositol 4 phosphate 3-kinase (PI3K) and Rab43. PI3K signaling pathways are induced by HCMV infection and interfere with the host cell stress response (reviewed in reference 67). Rab43 is a member of the family of Rab GTPases that are important effectors in the control of vesicle trafficking and virus assembly (68–70). Interestingly, a number of Rabs, but not Rab43, were found to be associated with virions of other herpesviruses (58, 60, 61, 63). We found Rab43 in the virion preparations of both pp65neg and pp65pos strains, but in reduced amounts when pp65 was absent. It is intriguing to speculate that Rab43 is involved in secondary envelopment by interacting with one of the outer tegument proteins. Concordant with this idea, reduction of Rab43 in pp65neg strains would be in agreement with a phenotype of disturbed membrane-tegument interaction, as suggested from the EM data (Fig. 2). This, however, has to be addressed in future analyses.
Dependence of tegument assembly on pp65.
One obvious explanation for the unaltered morphology of intact RV-Hd65 virions was that one or more viral proteins would replace pp65 during morphogenesis. Among all the viral proteins, only pUL35 was found to be roughly 2-fold elevated in both pp65neg strains. However, that amount of protein was far too little to be regarded as compensation for any structural function of pp65. At this point, it remains unclear if pUL35 could replace a regulatory function of pp65, e.g., its interaction with IFI16 (15). As about 40 molecules of pUL35 would have to replace 5,000 pp65 molecules in single virions, however, it is more likely that pp65 loss in virions is not compensated for at all by any other protein.
The tegument proteins, like pp150 or HMWP, that are considered to be associated with the capsid structure (11) appeared to be unaffected by pp65 loss. Several of the outer tegument proteins, however, were depleted in the absence of pp65. pp65-related reduction had been reported previously for pUL25, pUL69, and pUL97 (26). We could confirm this and extend these findings to show that a number of additional tegument and tegument-associated viral proteins are packaged in a pp65-dependent manner (Table 1 and Fig. 4). Among them were pUL45, pUL24, pUL26, and pUL71. The most striking effects, however, were seen for pUL43 and pUL25, which were found in considerable amounts in pp65pos virions but were virtually absent in pp65neg virions. It has been suggested that pUL25 represents a hub for tegument assembly (71). pUL25, together with pUL43, is an abundant virion constituent and a distinct interaction partner of pp65 (71, 72). Our results are in agreement with a network model of interacting viral proteins consisting of pUL25, pUL43, and pp65 that is assembled in the outer HCMV tegument to form a scaffold structure for further tegument assembly. Through the presence of pp65, this network may gain stability, thereby reinforcing the virion structure. In the absence of pp65, such a scaffold may be impaired, compromising viral and cellular protein upload.
Supplementary Material
ACKNOWLEDGMENTS
The donations of monoclonal antibodies by William Britt, University of Alabama, and Bonita Biegalke, Ohio University, and of BAC clones by Thomas Shenk, Princeton University, Martin Messerle and Eva Borst, Hannover Medical School, and Ulrich Koszinowski and Gaby Hahn, University of Munich, are gratefully acknowledged.
This work was funded by grants from the Else Kröner-Fresenius-Stiftung and from the Deutsche Forschungsgemeinschaft (DFG), Clinical Research Unit 183 (KFO 183), and was supported by the Research Center for Immunology of the University Medical Center of the University of Mainz, Mainz, Germany.
Footnotes
Published ahead of print 11 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01415-14.
REFERENCES
- 1.Tandon R, Mocarski ES. 2012. Viral and host control of cytomegalovirus maturation. Trends Microbiol. 20:392–401. 10.1016/j.tim.2012.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson W, Bogner E. 2013. Morphogenesis of the cytomegalovirus virion and subviral particles, p 230–246 In Reddehase MJ. (ed), Cytomegaloviruses: from molecular pathogenesis to intervention, 2nd ed, vol 1 Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 3.Sampaio KL, Cavignac Y, Stierhof YD, Sinzger C. 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J. Virol. 79:2754–2767. 10.1128/JVI.79.5.2754-2767.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez V, Sztul E, Britt WJ. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-golgi-intermediate compartment. J. Virol. 74:3842–3851. 10.1128/JVI.74.8.3842-3851.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234. 10.1016/j.virusres.2009.03.018 [DOI] [PubMed] [Google Scholar]
- 6.Marschall M, Feichtinger S, Milbradt J. 2011. Regulatory roles of protein kinases in cytomegalovirus replication. Adv. Virus Res. 80:69–101. 10.1016/B978-0-12-385987-7.00004-X [DOI] [PubMed] [Google Scholar]
- 7.Cepeda V, Esteban M, Fraile-Ramos A. 2010. Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell Microbiol. 12:386–404. 10.1111/j.1462-5822.2009.01405.x [DOI] [PubMed] [Google Scholar]
- 8.Tandon R, AuCoin DP, Mocarski ES. 2009. Human cytomegalovirus exploits ESCRT machinery in the process of virion maturation. J. Virol. 83:10797–10807. 10.1128/JVI.01093-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cepeda V, Fraile-Ramos A. 2011. A role for the SNARE protein syntaxin 3 in human cytomegalovirus morphogenesis. Cell Microbiol. 13:846–858. 10.1111/j.1462-5822.2011.01583.x [DOI] [PubMed] [Google Scholar]
- 10.Tandon R, Mocarski ES. 2008. Control of cytoplasmic maturation events by cytomegalovirus tegument protein pp150. J. Virol. 82:9433–9444. 10.1128/JVI.00533-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu X, Shah S, Lee M, Dai W, Lo P, Britt W, Zhu H, Liu F, Zhou ZH. 2011. Biochemical and structural characterization of the capsid-bound tegument proteins of human cytomegalovirus. J. Struct. Biol. 174:451–460. 10.1016/j.jsb.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavalai N, Stamminger T. 2011. Intrinsic cellular defense mechanisms targeting human cytomegalovirus. Virus Res. 157:128–133. 10.1016/j.virusres.2010.10.002 [DOI] [PubMed] [Google Scholar]
- 13.Kalejta RF. 2008. Functions of human cytomegalovirus tegument proteins prior to immediate early gene expression, p 101–115 In Shenk T, Stinski MF. (ed), Human cytomegalovirus, vol 325 Springer, Berlin, Germany: [DOI] [PubMed] [Google Scholar]
- 14.Kalejta RF. 2013. Pre-immediate early tegument protein functions, p 141–151 In Reddehase MJ. (ed), Cytomegaloviruses: from molecular pathogenesis to intervention, 2nd ed. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 15.Li T, Chen J, Cristea IM. 2013. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe 14:591–599. 10.1016/j.chom.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bogdanow B, Weisbach H, von Straschewski S, Voigt S, Winkler M, Hagemeier C, Wiebusch L. 2013. Human cytomegalovirus tegument protein pp150 acts as a cyclin A2-CDK-dependent sensor of the host cell cycle and differentiation state. Proc. Natl. Acad. Sci. U. S. A. 110:17510–17515. 10.1073/pnas.1312235110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, Wang D, Camp DG, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966. 10.1128/JVI.78.20.10960-10966.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irmiere A, Gibson W. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118–133. 10.1016/0042-6822(83)90122-8 [DOI] [PubMed] [Google Scholar]
- 19.Roby C, Gibson W. 1986. Characterization of phosphoproteins and protein kinase activity of virions, noninfectious enveloped particles, and dense bodies of human cytomegalovirus. J. Virol. 59:714–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cristea IM, Moorman NJ, Terhune SS, Cuevas CD, O'Keefe ES, Rout MP, Chait BT, Shenk T. 2010. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J. Virol. 84:7803–7814. 10.1128/JVI.00139-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert MJ, Riddell SR, Plachter B, Greenberg PD. 1996. Cytomegalovirus selectively blocks antigen processing and presentation of its immediate-early gene product. Nature 383:720–722. 10.1038/383720a0 [DOI] [PubMed] [Google Scholar]
- 22.Odeberg J, Plachter B, Branden L, Soderberg-Naucler C. 2003. Human cytomegalovirus protein pp65 mediates accumulation of HLA-DR in lysosomes and destruction of the HLA-DR alpha-chain. Blood 101:4870–4877. 10.1182/blood-2002-05-1504 [DOI] [PubMed] [Google Scholar]
- 23.Arnon TI, Achdout H, Levi O, Markel G, Saleh N, Katz G, Gazit R, Gonen-Gross T, Hanna J, Nahari E, Porgador A, Honigman A, Plachter B, Mevorach D, Wolf DG, Mandelboim O. 2005. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat. Immunol. 6:515–523. 10.1038/ni1190 [DOI] [PubMed] [Google Scholar]
- 24.Browne EP, Shenk T. 2003. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc. Natl. Acad. Sci. U. S. A. 100:11439–11444. 10.1073/pnas.1534570100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmolke S, Kern HF, Drescher P, Jahn G, Plachter B. 1995. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J. Virol. 69:5959–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chevillotte M, Landwehr S, Linta L, Frascaroli G, Luske A, Buser C, Mertens T, von Einem J. 2009. Major tegument protein pp65 of human cytomegalovirus is required for the incorporation of pUL69 and pUL97 into the virus particle and for viral growth in macrophages. J. Virol. 83:2480–2490. 10.1128/JVI.01818-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyda S, Buscher N, Tenzer S, Plachter B. 2014. Proteomic analyses of human cytomegalovirus strain AD169 derivatives reveal highly conserved patterns of viral and cellular proteins in infected fibroblasts. Viruses. 6:172–188. 10.3390/v6010172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Besold K, Frankenberg N, Pepperl-Klindworth S, Kuball J, Theobald M, Hahn G, Plachter B. 2007. Processing and MHC class I presentation of human cytomegalovirus pp65-derived peptides persist despite gpUS2-11-mediated immune evasion. J. Gen. Virol. 88:1429–1439. 10.1099/vir.0.82686-0 [DOI] [PubMed] [Google Scholar]
- 29.Borst EM, Hahn G, Koszinowski UH, Messerle M. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320–8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hobom U, Brune W, Messerle M, Hahn G, Koszinowski UH. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720–7729. 10.1128/JVI.74.17.7720-7729.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu D, Smith GA, Enquist LW, Shenk T. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316–2328. 10.1128/jvi.76.5.2316-2328.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hesse J, Reyda S, Tenzer S, Besold K, Reuter N, Krauter S, Büscher N, Stamminger T, Plachter B. 2013. Human cytomegalovirus pp71 stimulates major histocompatibility complex class I presentation of IE1-derived peptides at immediate early times of infection. J. Virol. 87:5229–5238. 10.1128/JVI.03484-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becke S, Fabre-Mersseman V, Aue S, Auerochs S, Sedmak T, Wolfrum U, Strand D, Marschall M, Plachter B, Reyda S. 2010. Modification of the major tegument protein pp65 of human cytomegalovirus inhibits virus growth and leads to the enhancement of a protein complex with pUL69 and pUL97 in infected cells. J. Gen. Virol. 91:2531–2541. 10.1099/vir.0.022293-0 [DOI] [PubMed] [Google Scholar]
- 34.Becke S, Aue S, Thomas D, Schader S, Podlech J, Bopp T, Sedmak T, Wolfrum U, Plachter B, Reyda S. 2010. Optimized recombinant dense bodies of human cytomegalovirus efficiently prime virus specific lymphocytes and neutralizing antibodies without the addition of adjuvant. Vaccine 28:6191–6198. 10.1016/j.vaccine.2010.07.016 [DOI] [PubMed] [Google Scholar]
- 35.Besold K, Wills M, Plachter B. 2009. Immune evasion proteins gpUS2 and gpUS11 of human cytomegalovirus incompletely protect infected cells from CD8 T cell recognition. Virology 391:5–19. 10.1016/j.virol.2009.06.004 [DOI] [PubMed] [Google Scholar]
- 36.Utz U, Britt W, Vugler L, Mach M. 1989. Identification of a neutralizing epitope on glycoprotein gp58 of human cytomegalovirus. J. Virol. 63:1995–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Biegalke BJ. 2002. The human cytomegalovirus UL35 gene encodes two proteins with different functions. J. Virol. 76:2460–2468. 10.1128/jvi.76.5.2460-2468.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pepperl S, Münster J, Mach M, Harris JR, Plachter B. 2000. Dense bodies of human cytomegalovirus induce both humoral and cellular immune responses in the absence of viral gene expression. J. Virol. 74:6132–6146. 10.1128/JVI.74.13.6132-6146.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris JR, Horne RW. 1991. Negative staining, p 203–208 In Harris JR. (ed), Electron microscopy in biology. IRL Press, Oxford, England [Google Scholar]
- 40.Kramer-Albers EM, Bretz N, Tenzer S, Winterstein C, Mobius W, Berger H, Nave KA, Schild H, Trotter J. 2007. Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: trophic support for axons? Proteomics Clin. Appl. 1:1446–1461. 10.1002/prca.200700522 [DOI] [PubMed] [Google Scholar]
- 41.Tenzer S, Docter D, Rosfa S, Wlodarski A, Kuharev J, Rekik A, Knauer SK, Bantz C, Nawroth T, Bier C, Sirirattanapan J, Mann W, Treuel L, Zellner R, Maskos M, Schild H, Stauber RH. 2011. Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. ACS Nano. 5:7155–7167. 10.1021/nn201950e [DOI] [PubMed] [Google Scholar]
- 42.Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, Mann M, Ingolia NT, Weissman JS. 2012. Decoding human cytomegalovirus. Science 338:1088–1093. 10.1126/science.1227919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patzig J, Jahn O, Tenzer S, Wichert SP, de Monasterio-Schrader P, Rosfa S, Kuharev J, Yan K, Bormuth I, Bremer J, Aguzzi A, Orfaniotou F, Hesse D, Schwab MH, Mobius W, Nave KA, Werner HB. 2011. Quantitative and integrative proteome analysis of peripheral nerve myelin identifies novel myelin proteins and candidate neuropathy loci. J. Neurosci. 31:16369–16386. 10.1523/JNEUROSCI.4016-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. 2006. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5:144–156. 10.1074/mcp.M500230-MCP200 [DOI] [PubMed] [Google Scholar]
- 45.Distler U, Kuharev J, Navarro P, Levin Y, Schild H, Tenzer S. 2014. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat. Methods 11:167–170. 10.1038/nmeth.2767 [DOI] [PubMed] [Google Scholar]
- 46.Ahrne E, Molzahn L, Glatter T, Schmidt A. 2013. Critical assessment of proteome-wide label-free absolute abundance estimation strategies. Proteomics 13:2567–2578. 10.1002/pmic.201300135 [DOI] [PubMed] [Google Scholar]
- 47.Chen DH, Jiang H, Lee M, Liu F, Zhou ZH. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10–16. 10.1006/viro.1999.9791 [DOI] [PubMed] [Google Scholar]
- 48.Mocarski ES, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2701–2772 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 49.Caposio P, Streblow DN, Nelson JA. 2013. Cytomegalovirus proteomics, p 86–108 In Reddehase MJ. (ed), Cytomegaloviruses from molecular pathogenesis to intervention, 2nd ed, vol 1 Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 50.Huber MT, Compton T. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J. Virol. 72:8191–8197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou M, Yu Q, Wechsler A, Ryckman BJ. 2013. Comparative analysis of gO isoforms reveals that strains of human cytomegalovirus differ in the ratio of gH/gL/gO and gH/gL/UL128-131 in the virion envelope. J. Virol. 87:9680–9690. 10.1128/JVI.01167-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mocarski ES, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses, p 1960–2014 In Knipe DM, Howley PM. (ed), Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 53.Zhao J, Meyerkord CL, Du Y, Khuri FR, Fu H. 2011. 14-3-3 proteins as potential therapeutic targets. Semin. Cell Dev. Biol. 22:705–712. 10.1016/j.semcdb.2011.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wright JF, Kurosky A, Pryzdial EL, Wasi S. 1995. Host cellular annexin II is associated with cytomegalovirus particles isolated from cultured human fibroblasts. J. Virol. 69:4784–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Michelson S, Turowski P, Picard L, Goris J, Landini MP, Topilko A, Hemmings B, Bessia C, Garcia A, Virelizier JL. 1996. Human cytomegalovirus carries serine/threonine protein phosphatases PP1 and a host-cell derived PP2A. J. Virol. 70:1415–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baldick CJ, Jr, Shenk T. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malouli D, Hansen SG, Nakayasu ES, Marshall EE, Hughes CM, Ventura AB, Gilbride RM, Lewis MS, Xu G, Kreklywich C, Whizin N, Fischer M, Legasse AW, Viswanathan K, Siess D, Camp DG, Axthelm MK, Kahl C, Defilippis VR, Smith RD, Streblow DN, Picker LJ, Fruh K. 2014. Cytomegalovirus pp65 limits dissemination but is dispensable for persistence. J. Clin. Invest. 124:1928–1944. 10.1172/JCI67420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loret S, Guay G, Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605–8618. 10.1128/JVI.00904-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu FX, Chong JM, Wu L, Yuan Y. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:800–811. 10.1128/JVI.79.2.800-811.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kramer T, Greco TM, Enquist LW, Cristea IM. 2011. Proteomic characterization of pseudorabies virus extracellular virions. J. Virol. 85:6427–6441. 10.1128/JVI.02253-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D, Kieff E. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U. S. A. 101:16286–16291. 10.1073/pnas.0407320101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stegen C, Yakova Y, Henaff D, Nadjar J, Duron J, Lippe R. 2013. Analysis of virion-incorporated host proteins required for herpes simplex virus type 1 infection through a RNA interference screen. PLoS One 8:e53276. 10.1371/journal.pone.0053276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bechtel JT, Winant RC, Ganem D. 2005. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:4952–4964. 10.1128/JVI.79.8.4952-4964.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dry I, Haig DM, Inglis NF, Imrie L, Stewart JP, Russell GC. 2008. Proteomic analysis of pathogenic and attenuated alcelaphine herpesvirus 1. J. Virol. 82:5390–5397. 10.1128/JVI.00094-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lippe R. 2012. Deciphering novel host-herpesvirus interactions by virion proteomics. Front. Microbiol. 3:181. 10.3389/fmicb.2012.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kattenhorn LM, Mills R, Wagner M, Lomsadze A, Makeev V, Borodovsky M, Ploegh HL, Kessler BM. 2004. Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 78:11187–11197. 10.1128/JVI.78.20.11187-11197.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alwine JC. 2008. Modulation of host cell stress responses by human cytomegalovirus, p 263–279 In Shenk T, Stinski MF. (ed), Human cytomegalovirus, vol 325 Springer, Berlin, Germany: [DOI] [PubMed] [Google Scholar]
- 68.Zenner HL, Yoshimura S, Barr FA, Crump CM. 2011. Analysis of Rab GTPase-activating proteins indicates that Rab1a/b and Rab43 are important for herpes simplex virus 1 secondary envelopment. J. Virol. 85:8012–8021. 10.1128/JVI.00500-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Indran SV, Britt WJ. 2011. A role for the small GTPase Rab6 in assembly of human cytomegalovirus. J. Virol. 85:5213–5219. 10.1128/JVI.02605-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell. Biol. 10:513–525. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- 71.To A, Bai Y, Shen A, Gong H, Umamoto S, Lu S, Liu F. 2011. Yeast two hybrid analyses reveal novel binary interactions between human cytomegalovirus-encoded virion proteins. PLoS One 6:e17796. 10.1371/journal.pone.0017796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Phillips SL, Bresnahan WA. 2011. Identification of binary interactions between human cytomegalovirus virion proteins. J. Virol. 85:440–447. 10.1128/JVI.01551-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.